Acquired Antibiotic Resistance Genes: An Overview
- 1Department of Environmental and Occupational Health Sciences, School of Public Health, Seattle, WA, USA
- 2Institute of Farm Animal Genetics, Friedrich-Loeffler-Institut, Neustadt-Mariensee, Germany
- 3National Institute of Health and the Environment (RIVM), Antonie van Leeuwenhoeklaan 9, Bilthoven, Netherlands
A commentary on
Acquired antibiotic resistance genes: an overview
by van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Front. Microbio. 2:203. doi: 10.3389/fmicb.2011.00203
Dr. Marilyn C. Roberts and Dr. Stefan Schwarz have contacted the authors of the original publication with several comments and suggestions to better harmonize the correct nomenclature of the antibiotic resistance genes, as the gene names were not always correctly presented in the various tables given.
Authors often pick their own gene names which in many cases have been approved for use for other genetically distinct genes or give names to determinants which were already given an approved designated name. Therefore, we (Dr. Marilyn C. Roberts and Dr. Stefan Schwarz and Dr. Henk J. M. Aarts on behalf of the authors of the original publication) would like to present here the correct nomenclature and mechanistic features of the antibiotic resistance genes belonging to the following classes: Aminoglycosides (Table 1), Phenicols (Table 3), Macrolides–Lincosamides–Streptogramin B (Table 4), Quinolones (Table 5), Tetracyclines (Table 6), and Trimethoprim (Table 7). In addition some additional information is given on the various classes of antibiotic resistance genes as also a section regarding the antibiotic class Oxazolidinones has been added. Table 2 was correctly displayed by van Hoek et al. (2011) but has been updated.
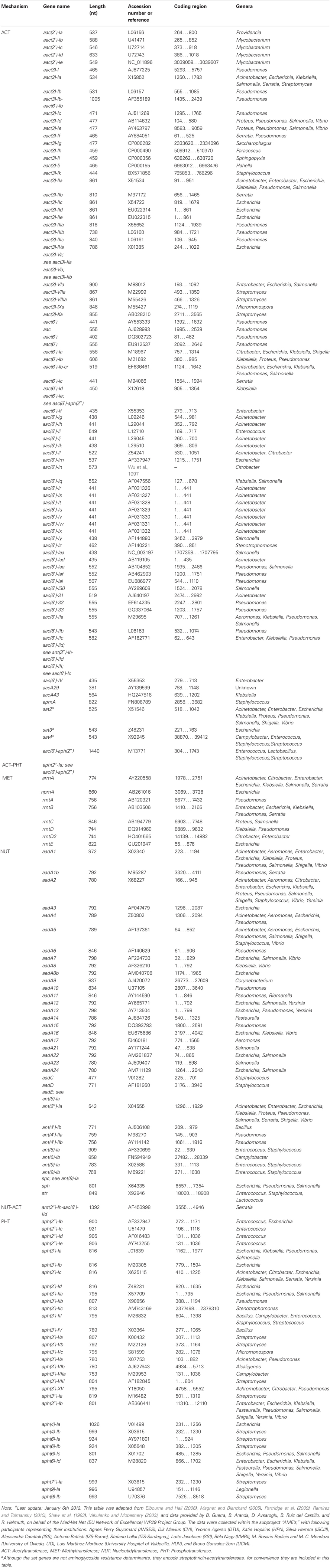
To the subsection dealing with the “Resistance mechanisms” of the AMINOGLYCOSIDES we would like to add that to date six additional methylases have been reported, i.e., npmA, rmtA, rmtB, rmtC, rmtD, and rmtE (Courvalin, 2008; Doi et al., 2008; Davis et al., 2010). Futhermore, that within the three major classes (AAC, ANT, and APH) an additional subdivision can be made based on the enzymes' target sites within the aminoglycoside molecules: i.e., there are four acetyltransferases: AAC(1), AAC(2′), AAC(3), and AAC(6′); five nucleotidyltransferases: ANT(2″), ANT(3″), ANT(4′), ANT(6), and ANT(9); and seven phosphotransferases: APH(2″), APH(3′), APH(3″), APH(4), APH(6), APH(7″), and APH(9).
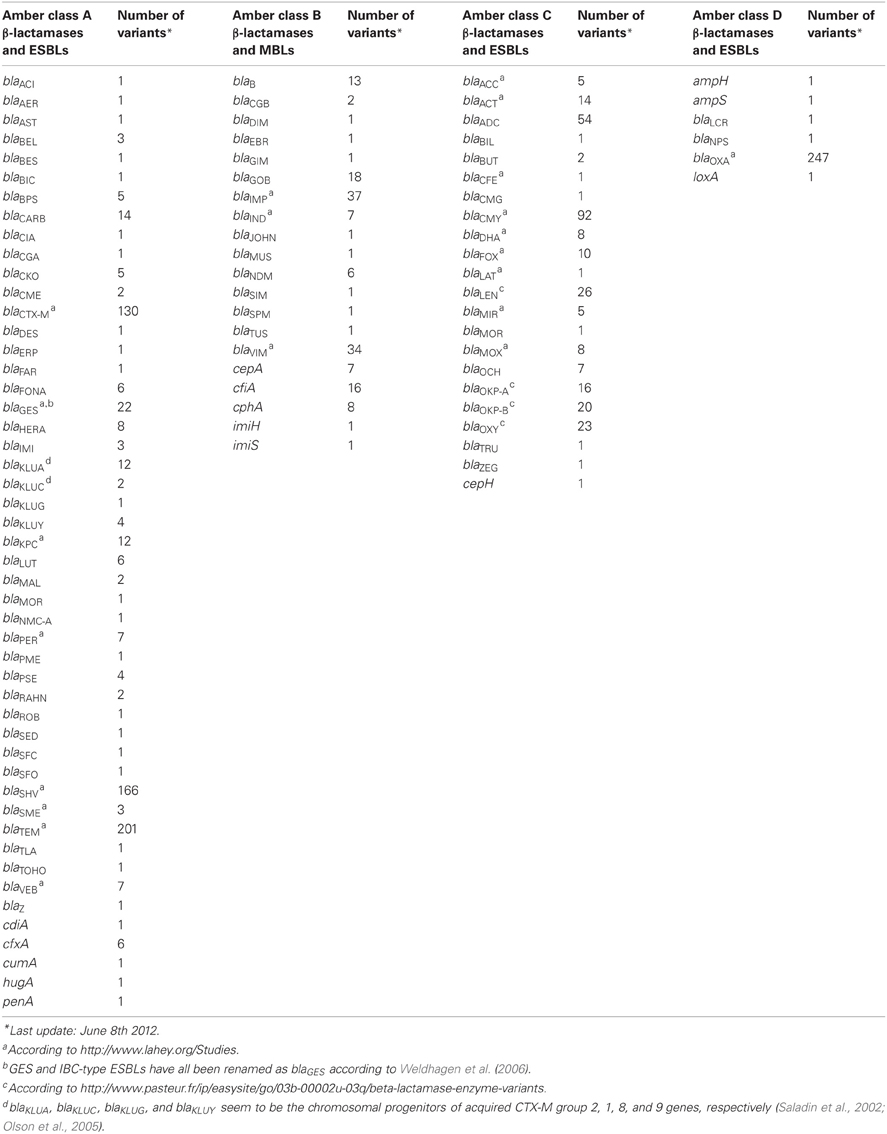
To the subsection β-LACTAM, Resistance, mechanisms we would like to add that in recent years acquired genes encoding ESBLs have become a major concern (Bradford, 2001). Over time, the genes for the parent enzymes blaTEM−1, blaTEM−2, and blaSHV−1 have undergone point mutations which resulted in amino acid substitutions that changed the substrate spectrum to that of ESBLs, starting with blaTEM−3 and blaSHV−2 (Bradford, 2001).
Because chloramphenicol is not an actual antibiotic class the subsection of CHLORAMPHENICOL should be called PHENICOLS. Concerning the history of PHENICOLS, it is worthwhile to know the first antibiotic, chloramphenicol, originally referred to as chloromycetin, was isolated already in 1947 from Streptomyces venezuelae (Ehrlich et al., 1947).
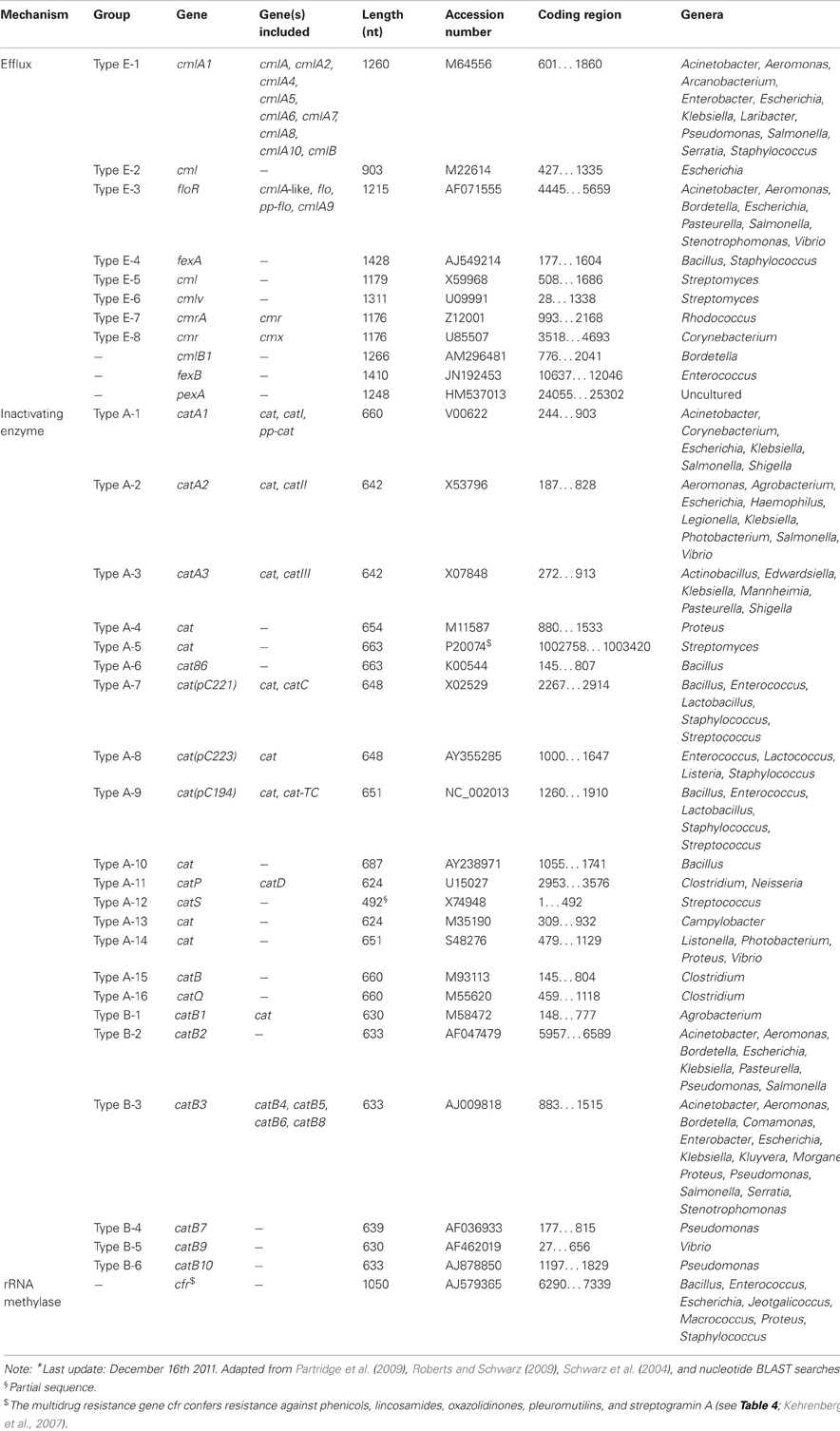
Besides the inactivating enzymes (chloramphenicol acetyltransferases), there are also reports on other phenicol resistance systems, such as the inactivation by phosphotransferases, mutations of the target site, permeability barriers, and efflux systems (Schwarz et al., 2004). Of the latter mechanism, cmlA and floR are the most commonly known genes in Gram-negative bacteria (Bissonnette et al., 1991; Briggs and Fratamico, 1999).
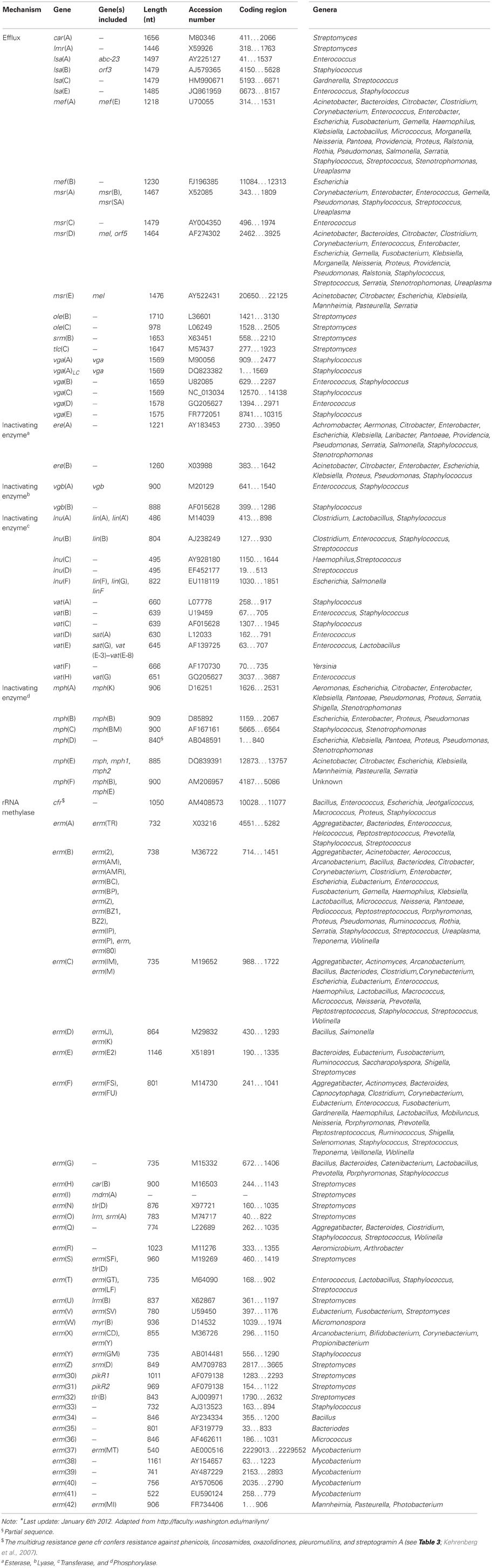
The macrolides (subsection MACROLIDES–LINCOSAMIDES–STREPTOGRAMIN B) have a similar mode of antibacterial action, comparable antibacterial spectra and in part overlapping binding sites at the ribosome as two other antibiotic classes, i.e., lincosamides and streptogramin antibiotics (comprising streptogramin A and B compounds that act synergistically). Consequently, these antibiotics, although chemically distinct, have been clustered together as MLS antibiotics (Roberts, 1996). Macrolides, lincosamides and streptogramins all inhibit protein synthesis by binding to the 50S ribosomal subunit of bacteria (Weisblum, 1995; Roberts, 2002).
To Resistance mechanisms of the subsection MACROLIDES–LINCOSAMIDES–STREPTOGRAMIN B. Shortly after the introduction of erythromycin into clinical setting in the 1950s, bacterial resistance to this antibiotic was reported for the first time in staphylococci (Weisblum, 1995). Since then a large number of bacteria have been identified that are resistant to MLS due to the presence of various different genes. The resistance determinants responsible include rRNA methylases that modify the ribosomal target sites, ABC transporters, and efflux proteins of the Major Facilitator Superfamily, as well as genes for inactivating enzymes (Roberts et al., 1999; Roberts, 2008). The latter group can be further subdivided into esterases, lyases, phosphorylases, and transferases (Table 4).
The most common mechanism of MLSB resistance is due to the presence of rRNA methylases, encoded by the erm genes. These enzymes methylate the adenine residue(s) resulting in MLSB resistance. The methylated adenine(s) prevents the drugs from binding to the 50S ribosomal subunit. The other two mechanisms efflux and enzymatic inactivation result in resistance to only 1 or 2 classes of antibiotics belonging to the MLS group.
There are currently 77 MLS resistance genes recognized. A new MLS gene must have <79% amino acid identity with all previously characterized MLS genes before receiving a unique name (Roberts et al., 1999; Roberts, 2008). For an actual list of the MLS acquired resistance genes we refer to the website of Dr. Marilyn Roberts, http://faculty.washington.edu/marilynr/.
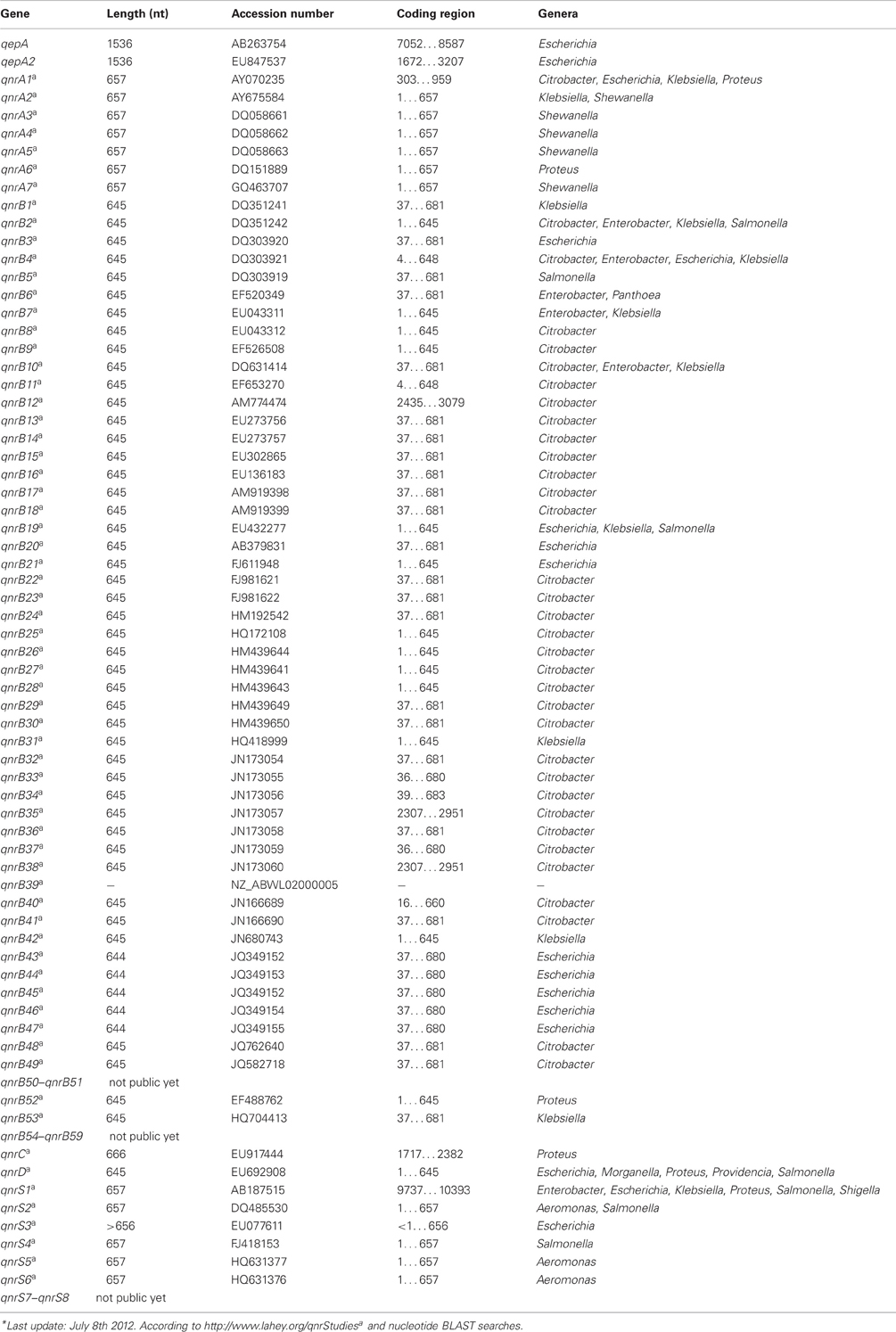
In addition to the subsection of QUINOLONES currently five families of qnr genes have been reported; qnrA (7 subtypes), qnrB (59 subtypes), qnrC (1 subtype), qnrD (1 subtype), and qnrS (8 subtypes) (Jacoby et al., 2008; Cattoir and Nordmann, 2009; Cavaco et al., 2009; Strahilevitz et al., 2009; Torpdahl et al., 2009).
Another mechanism of conferring resistance to quinolones is represented by the plasmid-borne gene qepA, which codes for an efflux pump that can export hydrophilic fluoroquinolones, e.g., ciprofloxacin and enrofloxacin (Périchon et al., 2007; Yamane et al., 2007). A variant of this resistance pump, QepA2, was identified in an E. coli isolate from France (Cattoir et al., 2008).
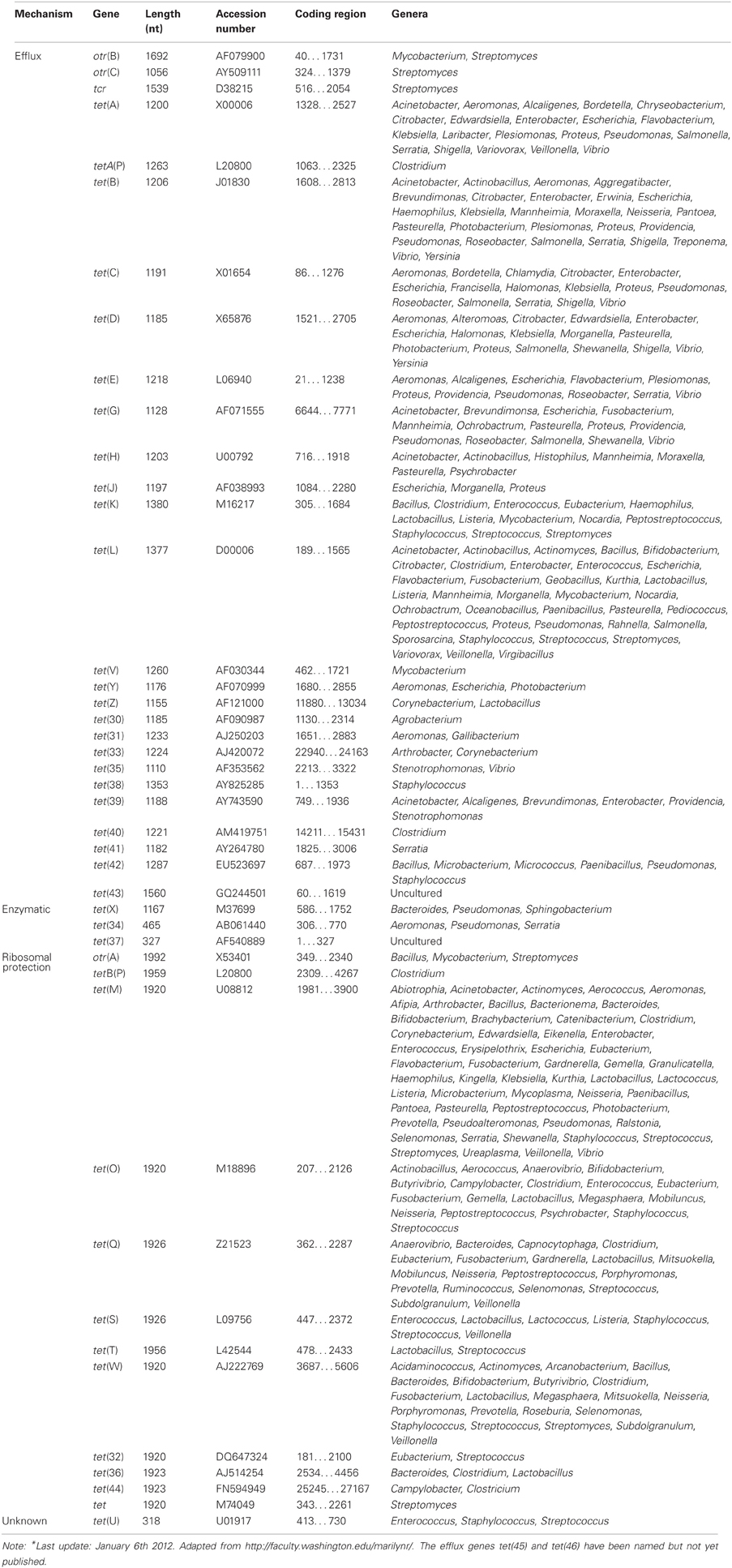
Regarding TETRACYCLINE, Resistance mechanisms, currently there are 45 different acquired tetracycline resistance determinants recognized (Roberts, 1996, 2005; Brown et al., 2008) (Table 6). For an up-to-date list of the acquired tetracycline resistance genes, we refer to the website of Dr. Marilyn Roberts, http://faculty.washington.edu/marilynr/. Among these, 26 of the tet genes, 2 of the otr genes and the only tcr determinant code for efflux pumps, whereas 11 tet genes and 1 otr gene code for ribosomal protection proteins (RPPs). The enzymatic inactivation mechanism can be attributed to 3 tet genes. The tet(U) determinant represents an unknown tetracycline resistance mechanism since its sequence does not appear to be related to either efflux or RPPs, nor to the inactivation enzymes. The efflux and RPP encoding genes are found in members of Gram-positive, Gram-negative, aerobic, as well as anaerobic bacteria. In contrast, the enzymatic tetracycline inactivation mechanism has so far only been identified in Gram-negative bacteria. The tet(M) has the broadest host range of all tetracycline resistance genes, whereas tet(B) gene has the widest range among the Gram-negative bacteria. In recent years published data indicate that there are increasing numbers of Gram-negative bacteria that carry tet genes originally identified in Gram-positive bacteria (Roberts, 2002).
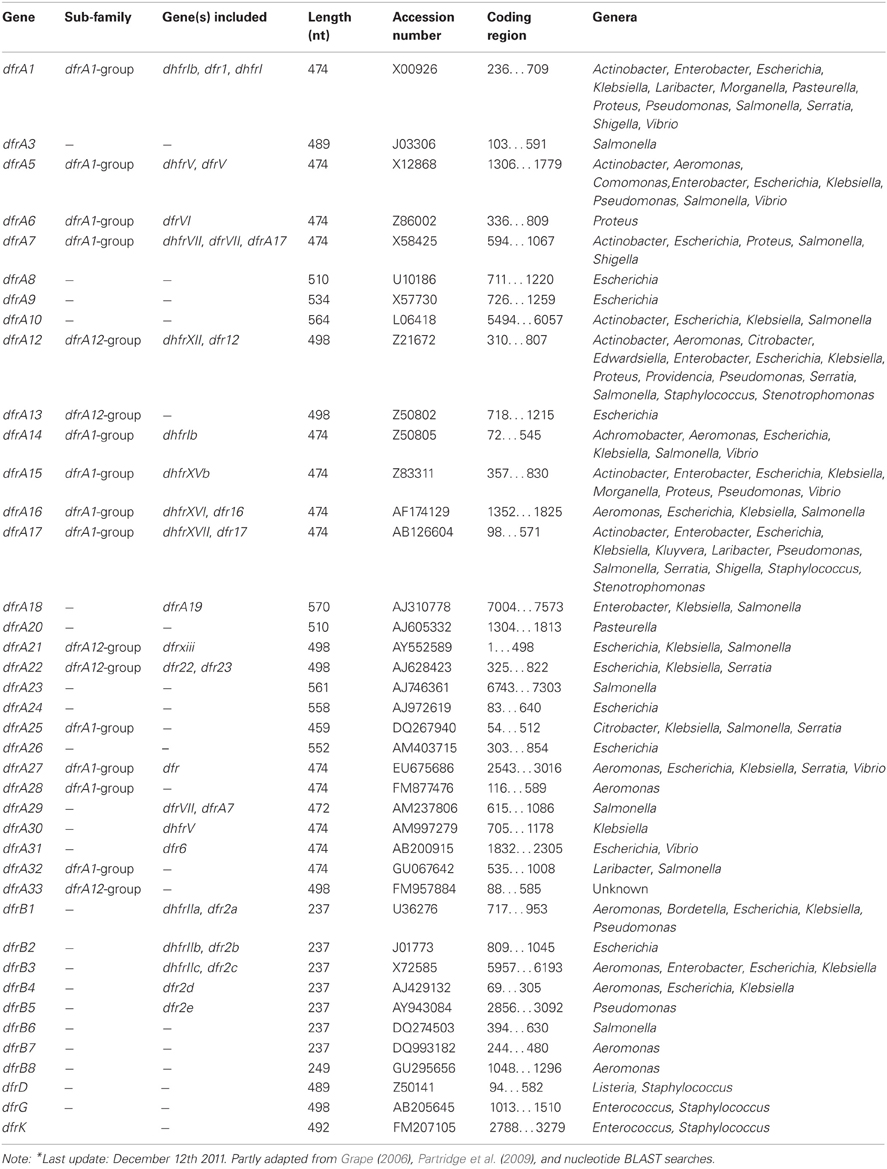
To the subsection TRIMETHOPRIM, Resistance mechanisms. Initially, the acquired DHFRs fell into two distinct families A and B, encoded by the dfrA and dfrB genes (Howell, 2005). Up to now 6 plasmid-mediated families can be distinguished with relatively few dfr determinants originating from Gram-positive bacteria (Table 7). The dfrK and dfrA28 genes are the newest additions to the trimethoprim resistance determinant family (Kadlec and Schwarz, 2009; Kadlec et al., 2011). In contrast to the latest reported DHFRs, the oldest families, dfrA and dfrB, each contain several members (Roberts, 2002; Levings et al., 2006). For example, the dfrA group accomodates over 30 published genes; however, unpublished, dfrA variants are also present in the public DNA libraries and some genes apparently have changed nomenclature (Table 7).
Furthermore, we suggest an additional section concerning oxazolidinones.
Oxazolidinones
History and Action Mechanism
Linezolid is to date the only FDA-approved oxazolidinone (Shaw and Barbachyn, 2011). It was approved in 2000 for the treatment of serious infections caused by Gram-positive bacteria resistant to other antibiotics, such as vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) (Long and Vester, 2012). As such linezolid is considered one of the last resort antimicrobial agents in human medicine. It has not been approved for use in veterinary medicine. Oxazolidinones bind at the P site of the ribosome and inhibit the formation of the initiation complex, which consists of mRNA, f-Met tRNA, and the 50S ribosomal subunit (Shaw and Barbachyn, 2011; Long and Vester, 2012).
Resistance Mechanism
Various mutations located in the peptidyl transferase loop of domain V of 23S rRNA as well as mutations in the genes for the ribosomal proteins L3 and L4, all associated with resistance to oxazolidinones, have been identified (reviewed by Long and Vester, 2012). A single gene, cfr, has been identified to confer transferable resistance to oxazolidinones. This gene codes for a methyltransferase that targets A2503 in 23S rRNA (Kehrenberg et al., 2005). Besides oxazolidinone resistance, it also confers resistance to phenicols, lincosamides, pleuromutilins, and streptogramin A antibiotics. Although initially identified in coagulase-negative staphylococci of animal origin, the gene cfr has now been detected in a wide variety of staphylococci of human and animal origin, including a Panton-Valentin leukocidin-positive MRSA USA300 (Shore et al., 2010) and livestock-associated MRSA ST398 (Kehrenberg et al., 2009). More recently, the cfr gene has also been identified in Bacillus spp. (Dai et al., 2010) and Enterococcus faecalis (Liu et al., 2012), but also in Gram-negative bacteria, such as Proteus vulgaris (Wang et al., 2011) and Escherichia. coli (Wang et al., 2012). Plasmids and insertion sequences seem to play an important role in the spread of this gene across species and genus boundaries.
References
Bissonnette, L., Champetier, S., Buisson, J.-P., and Roy, P. H. (1991). Characterization of the non-enzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J. Bacteriol. 173, 4493–4502.
Bradford, P. A. (2001). Extended-spectrum β-lactamase in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951.
Briggs, C. E., and Fratamico, P. M. (1999). Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43, 846–849.
Brown, M. G., Mitchell, E. H., and Balkwill, D. L. (2008). Tet 42, a novel tetracycline resistance determinant isolated from deep terrestrial subsurface bacteria. Antimicrob. Agents Chemother. 52, 4518–4521.
Cattoir, V., and Nordmann, P. (2009). Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Curr. Med. Chem. 16, 1028–1046.
Cattoir, V., Poirel, L., and Nordmann, P. (2008). Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804.
Cavaco, L. M., Hasman, H., Xia, S., and Aarestrup, F. M. (2009). qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53, 603–608.
Courvalin, P. (2008). New plasmid-mediated resistances to antimicrobial agents. Arch. Microbiol. 189, 289–291.
Dai, L., Wu, C. M., Wang, M. G., Wang, Y., Wang, Y., Huang, S. Y., et al. (2010). First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54, 3953–3955.
Davis, M. A., Baker, K. N. K., Orfe, L. H., Shah, D. H., Besser, T. E., and Call, D. E. (2010). Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob. Agents Chemother. 54, 2666–2669.
Doi, Y., Wachino, J.-I., and Arakawa, Y. (2008). Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob. Agents Chemother. 52, 2287–2288.
Ehrlich, J., Bartz, Q. R., Smith, R. M., Joslyn, D. A., and Burkholder, P. R. (1947). Chloromycetin a new antibiotic from a soil actinomycete. Science 106, 417.
Elbourne, L. D. H., and Hall, R. M. (2006). Gene cassette encoding a 3-N-aminoglycoside acetyltransferase in a chromosomal integron. Antimicrob. Agents Chemother. 50, 2270–2271.
Grape, M. (2006). Molecular Basis for Trimethoprim and Sulphonamide Resistance in Gram Negative Pathogens. Ph.D. Thesis, Stockholm, Sweden: Karolinska Institutet.
Howell, E. E. (2005). Searching sequence space: two different approaches to dihydrofolate reductase catalysis. ChemBioChem 6, 590–600.
Jacoby, G., Cattoir, V., Hooper, D., Martínez-Martínez, L., Nordmann, P., Pascual, A., et al. (2008). qnr gene nomenclature. Antimicrob. Agents Chemother. 52, 2297–2299.
Kadlec, K., and Schwarz, S. (2009). Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53, 776–778.
Kadlec, K., von Czapiewski, E., Kaspar, H., Wallmann, J., Michael, G. B., Steinacker, U., et al. (2011). Molecular basis of sulfonamide and trimethoprim resistance in fish-pathogenic Aeromonas isolates. Appl. Environ. Microbiol. 77, 7147–7150.
Kehrenberg, C., Aarestrup, F. M., and Schwarz, S. (2007). IS21-558 Insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51, 483–487.
Kehrenberg, C., Cuny, C., Strommenger, B., Schwarz, S., and Witte, W. (2009). Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53, 779–781.
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H., and Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073.
Levings, R. S., Lightfoot, D., Elbourne, L. D. H., Djordjevic, S. P., and Hall, R. M. (2006). New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase. Antimicrob. Agents Chemother. 50, 2863–2865.
Long, K. S., and Vester, B. (2012). Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56, 603–612.
Liu, Y., Wang, Y., Wu, C., Shen, Z., Schwarz, S., Du, X. D., et al. (2012). First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56, 1650–1654.
Magnet, S., and Blanchard, J. S. (2005). Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105, 477–497.
Olson, A. B., Silverman, M., Boyd, D. A., McGeer, A., Willey, B. M., Pong-Porter, V., et al. (2005). Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49, 2112–2115.
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons: review article. FEMS Microbiol. Rev. 33, 757–784.
Périchon, B., Courvalin, P., and Galimand, M. (2007). Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51, 2464–2469.
Ramirez, M. S., and Tolmansky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171.
Roberts, M. C. (1996). Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19, 1–24.
Roberts, M. C. (2002). Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechn. 20, 261–284.
Roberts, M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203.
Roberts, M. C. (2008). Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282, 147–159.
Roberts, M. C., and Schwarz, S. (2009). “Tetracycline and chloramphenicol resistance mechanisms,” in Antimicrobial Drug Resistance: Mechanisms of Drug Resistance, ed D. L. Mayers (New York, NY: Humana Press, c/o Springer ScienceCBusiness Media).
Roberts, M. C., Sutcliffe, J., Courvalin, P., Jensen, L. B., Rood, J., and Seppala, H. (1999). Nomenclature for macrolide and macrolide-lincosamide streptogramin B antibiotic resistance determinants. Antimicrob. Agents Chemother. 43, 2823–2830.
Saladin, M., Cao, V. T. B., Lambert, T., Donay, J.-L., Herrmann, J.-L., Ould-Hocine, Z., et al. (2002). Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209, 161–168.
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542.
Shaw, K. J., and Barbachyn, M. R. (2011). The oxazolidinones: past, present, and future. Ann. N.Y. Acad. Sci. 1241, 48–70.
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163.
Shore, A. C., Brennan, O. M., Ehricht, R., Monecke, S., Schwarz, S., Slickers, P., et al. (2010). Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54, 4978–4984.
Strahilevitz, J., Jacoby, G. A., Hooper, D. C., and Robicsek, A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689.
Torpdahl, M., Hammerum, A. M., Zachariasen, C., and Nielsen, E. M. (2009). Detection of qnr genes in Salmonella isolated from humans in Denmark. J. Antimicrob. Chemother. 63, 406–408.
Vakulenko, S. B., and Mobashery, S. (2003). Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16, 430–450.
van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbio. 2:203. doi: 10.3389/fmicb.2011.00203
Wang, Y., He, T., Schwarz, S., Zhou, D., Shen, Z., Wu, C., et al. (2012). Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. doi: 10.1093/jac/dks020. [Epub ahead of print].
Wang, Y., Wang, Y., Wu, C. M., Schwarz, S., Shen, Z., Zhang, W., et al. (2011). Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66, 2521–2526.
Weisblum, B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585.
Weldhagen, G. F., Kim, B., Cho, C.-H., and Lee, S. H. (2006). Definitive nomenclature of GES/IBC-type extended-spectrum β-lactamases. J. Microbiol. Biotechnol. 16, 1837–1840.
Wu, H. Y., Miller, G. H., Guzmán Blanco, M., Hare, R. S., and Shaw, K. J. (1997). Cloning and characterization of an aminoglycoside 6′-N-acetyltransferase gene from Citrobacter freundii which confers an altered resistance profile. Antimicrob. Agents Chemother. 41, 2439–2447.
Citation: Roberts MC, Schwarz S and Aarts HJM (2012) Erratum: Acquired antibiotic resistance genes: an overview. Front. Microbio. 3:384. doi: 10.3389/fmicb.2012.00384
Received: 05 October 2012; Accepted: 15 October 2012;
Published online: 16 November 2012.
Edited by:
Helen Zgurskaya, University of Oklahoma, USAReviewed by:
Helen Zgurskaya, University of Oklahoma, USACopyright © 2012 Roberts, Schwarz and Aarts. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: henk.aarts@rivm.nl