- 1School of Marine Science and Policy, University of Delaware, Newark, DE, USA
- 2Delaware Biotechnology Institute, University of Delaware, Newark, DE, USA
- 3Department of Biological Sciences, Clemson University, Clemson, SC, USA
- 4Department of Biology, University of North Carolina, Charlotte, NC, USA
The process of nitrate reduction via nitrite controls the fate and bioavailability of mineral nitrogen within ecosystems; i.e., whether it is retained as ammonium (ammonification) or lost as nitrous oxide or dinitrogen (denitrification). Here, we present experimental evidence for a novel pathway of microbial nitrate reduction, the reverse hydroxylamine:ubiquinone reductase module (reverse-HURM) pathway. Instead of a classical ammonia-forming nitrite reductase that performs a 6 electron-transfer process, the pathway is thought to employ two catalytic redox modules operating in sequence: the reverse-HURM reducing nitrite to hydroxylamine followed by a hydroxylamine reductase that converts hydroxylamine to ammonium. Experiments were performed on Nautilia profundicola strain AmH, whose genome sequence led to the reverse-HURM pathway proposal. N. profundicola produced ammonium from nitrate, which was assimilated into biomass. Furthermore, genes encoding the catalysts of the reverse-HURM pathway were preferentially expressed during growth of N. profundicola on nitrate as an electron acceptor relative to cultures grown on polysulfide as an electron acceptor. Finally, nitrate-grown cells of N. profundicola were able to rapidly and stoichiometrically convert high concentrations of hydroxylamine to ammonium in resting cell assays. These experiments are consistent with the reverse-HURM pathway and a free hydroxylamine intermediate, but could not definitively exclude direct nitrite reduction to ammonium by the reverse-HURM with hydroxylamine as an off-pathway product. N. profundicola and related organisms are models for a new pathway of nitrate ammonification that may have global impact due to the wide distribution of these organisms in hypoxic environments and symbiotic or pathogenic associations with animal hosts.
Introduction
Nitrate is a major fixed nitrogen pool in the global N-cycle (Falkowski and Godfrey, 2008; Klotz and Stein, 2008). Nitrate can be reduced via denitrification to nitrous oxide or dinitrogen gas, whereby it is lost from terrestrial and aquatic ecosystems, or via ammonification to ammonium that can be retained (Brandes et al., 2007; Klotz and Stein, 2008, 2010). In both pathways, nitrate is reduced to nitrite. In heterotrophic denitrifiers and ammonia oxidizers, nitrite is further reduced by NO-forming nitrite reductases: NirS or NirK. Nitrite reduction to ammonium is catalyzed by assimilatory (NirB/NirA) or respiratory (NrfA) ammonia-forming nitrite reductase (Klotz and Stein, 2008, 2010). Ammonia-oxidizing chemolithotrophs and methanotrophs can also form NO by the oxidation of hydroxylamine, another potential route of nitrogen loss in terrestrial and aquatic ecosystems (Campbell et al., 2011). Several Epsilonproteobacteria that are able to use nitrate for respiration and/or as a sole source of nitrogen for growth do not encode homologs of either nitrite reductase type, suggesting that these organisms possess an unrecognized mechanism for nitrite reduction to ammonium.
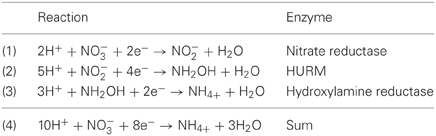
Genome annotation indicates that not only Nautilia profundicola, but several pathogenic Campylobacter spp. among the Epsilonproteobacteria may utilize nitrate as a nitrogen source for growth and/or as a terminal electron acceptor. N. profundicola is a member of the deepest branching lineage of the Epsilonproteobacteria (Campbell et al., 2006; Smith et al., 2008) and its relatives have only been found in deep-sea hydrothermal vent environments. In contrast, Campylobacter spp. are typically associated with animal hosts and their presence in other environments is ill defined. While the genomes of N. profundicola, Campylobacter concisus, C. curvus, and C. fetus contain genes encoding a periplasmic molybdopterin guanine dinucleotide-linked nitrate reductase complex (NAP), there are no genes encoding either ammonium- or NO-forming nitrite reductases. In addition, a napC/cycB gene was identified in the genome of N. profundicola (Campbell et al., 2009), which is unusual as the NAP complex in Epsilonproteobacteria usually lacks a NapC protein (Kern and Simon, 2009; Klotz and Stein, 2010; Simon and Klotz, 2013). Based on genome-informed metabolic reconstruction (Campbell et al., 2009), we recently proposed a novel pathway for nitrate assimilation (Figure 1) whereby nitrite is reduced to hydroxylamine by a hydroxylamine:ubiquinone redox module (HURM, i.e., a quinone-dependent hydroxylamine dehydrogenase), which we term the reverse-HURM pathway. In N. profundicola, the HURM is thought to consist of a NapC/NrfH-related cytochrome cM552 (cycB, NAMH_0559) that mediates electron transfer between the quinone pool and a periplasmic hydroxylamine oxidoreductase (HAO, NAMH_1280). HURM, operating in the forward direction and consisting of hydroxylamine dehydrogenase (EC:1.7.2.6; HaoA3) connected to a cytochrome c protein electron shuttle (c554 and/or cM552 encoded by cycA and cycB, respectively), links the oxidation of hydroxylamine to the quinone pool in aerobic ammonia-oxidizing bacteria (Klotz and Stein, 2008; Simon and Klotz, 2013). In anaerobic ammonia-oxidizing (anammox) bacteria, the redox module consists of hydrazine oxidoreductase and an as yet unidentified cytochrome c (Jetten et al., 2009; Simon and Klotz, 2013). In the reverse-HURM pathway, hydroxylamine generated from nitrite in the periplasm is transported into the cytoplasm and reduced to ammonium via a hydroxylamine reductase, also known as the hybrid cluster protein (Har/Hcp). Thus, the proposed electron accepting reactions of the reverse-HURM pathway are:
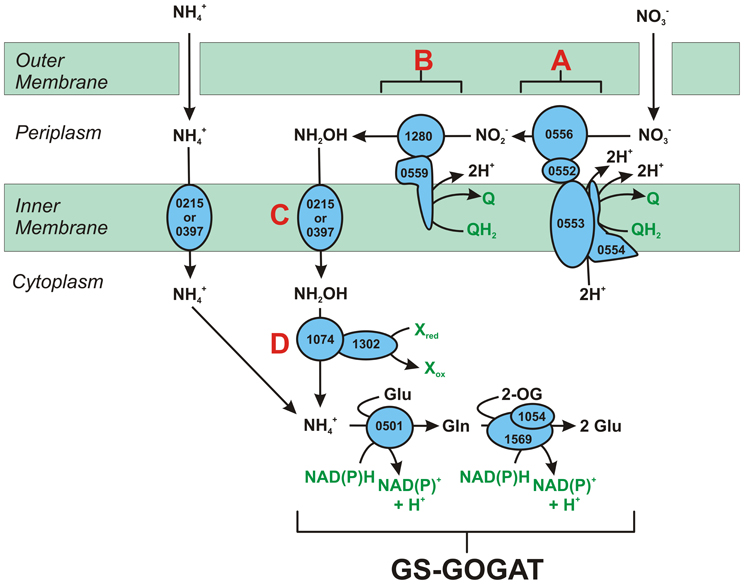
Figure 1. The reverse-HURM pathway in N. profundicola. Individual steps are noted with capital letters in the figure. Numbers in the depicted proteins refer to locus tags from the AmH genome (i.e., 1280 = NAMH_1280 = HaoA). (A) Nitrate reduction by respiratory periplasmic nitrate reductase (NapABGH, 0556-53), (B) HURM (Hydroxylamine:Ubiquinone Redox Module) comprised of a reversely operating hydroxylamine oxidoreductase (HaoA, 1280) functioning as an ocataheme nitrite reductase, that produces hydroxylamine utilizing electrons donated from a tetra-heme cytochrome c in the NapC/NrfH/cM552 family (CycB, 0559). (C) Transport of hydroxylamine via ammonia transporters related to AmtB. (D) Reduction of hydroxylamine to ammonium by a hybrid cluster protein/hydroxylamine reductase (Hcp/Har, 1074) utilizing reducing power from a predicted Fe-S containing protein (1302) whose electron donor (X) is currently unknown. Assimilation of ammonium into biomass occurs via glutamine synthetase and glutamine:2-oxoglutarate aminotransferase (GS-GOGAT).
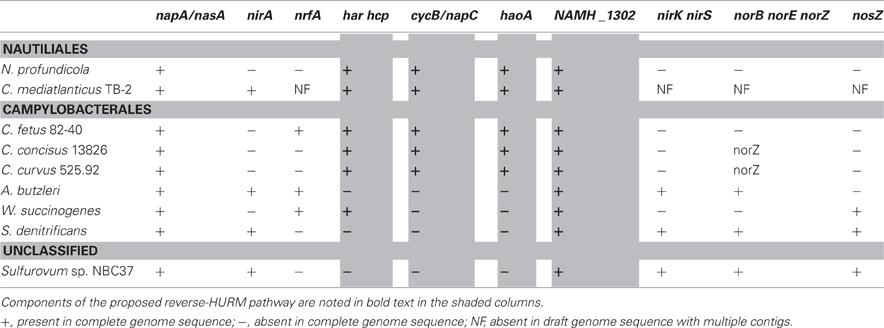
The genomes of N. profundicola, C. concisus, C. curvus, C. fetus, and Caminibacter mediatlanticus each contain genes encoding the enzymes of the reverse-HURM pathway while the genomes of other Epsilonproteobacteria encode homologues of the classical NO-forming NirS/NirK and ammonium-forming assimilatory siroheme NirA, or respiratory pentaheme NrfA nitrite reductases [Table 1, (Kern and Simon, 2009)]. We hypothesize that the reverse-HURM pathway improves the survival and/or dispersal of N. profundicola and Campylobacter spp. in ammonium-deficient host or non-host associated environments. However, for this hypothesis to be true, the reverse-HURM pathway must provide ammonium for biosynthesis and/or facilitate energy conservation in these organisms.
Results
Energetics of Growth in N. profundicola
N. profundicola utilizes both formate and hydrogen as electron donors for energy metabolism and is typically grown on polysulfide as the electron acceptor (Campbell et al., 2001; Smith et al., 2008). Calculations under standard conditions (1 M of all species, 25°C, pH = 7) indicate that when nitrate is reduced by the reactions of the reverse-HURM pathway, the Gibbs free energy (Δ G°) is substantially greater than that available from the reduction of polysulfide (Table 2). For example, the Δ G° per mole of formate oxidized with nitrate is 7.4-fold increased over polysulfide (−237 kJ mol−1 vs. −32 kJ mol−1). From this calculation, one can predict that the growth of N. profundicola should be stimulated in cultures with nitrate as the electron acceptor relative to polysulfide.
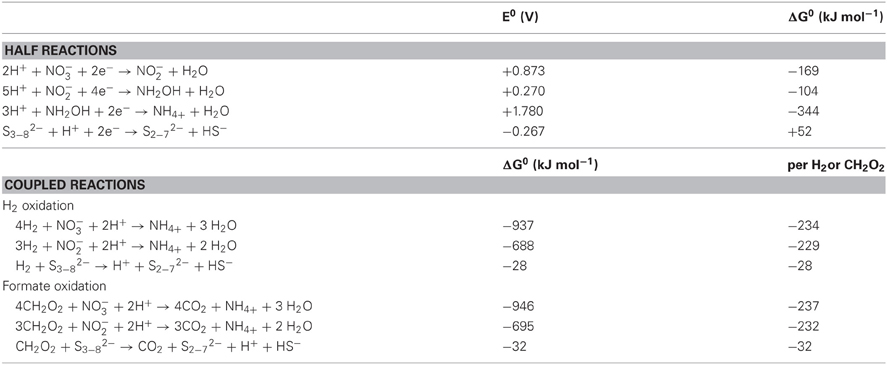
Table 2. Calculated free energies of reaction under standard conditions (1 M all species, 25°C and pH = 7) for electron accepting half reactions of nitrogen compounds and polysulfide and coupled reactions with formate or hydrogen as the electron donor.
Growth of N. profundicola with Nitrate
Prior characterization and growth of N. profundicola utilized media with polysulfide as a combined sulfur source and electron acceptor (Campbell et al., 2001, 2009; Smith et al., 2008). To further clarify the role of nitrate in the energy metabolism of N. profundicola, strain AmH was grown with sulfide as the sole sulfur source so that nitrate was the sole electron acceptor (Figure 2). The electron donor was a mixture of hydrogen and formate in a medium with no other sources of sulfur or nitrogen (Campbell et al., 2001). Growth rates with nitrate as potential electron acceptor and nitrogen source and sulfide (sulfur source) or polysulfide (sulfur source and potential electron acceptor) were more than double the growth rates observed in cultures using ammonium as the sole nitrogen source and polysulfide as an electron acceptor and sulfur source. As expected, negative control (no electron acceptor) cultures provided with ammonium as the nitrogen source and sulfide as the sulfur source were unable to grow.
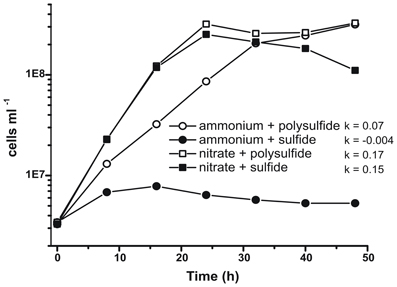
Figure 2. Growth of N. profundicola with indicated electron acceptors, nitrogen compounds, and sulfur sources. Growth rates were determined between 8 and 24 h; k = specific growth rate constant (h−1). Each point is the average of 2 or 3 independent biological replicates.
Nitrogen Balance in N. profundicola Cultures
To confirm that the level of ammonium produced by N. profundicola was in excess of biosynthetic needs, we calculated the biosynthetic nitrogen demand as follows. Assuming a C:N ratio of 32:6.4 for exponentially growing bacterioplankton (Vrede et al., 2002), 288 fg of C μm−3 for exponentially growing Escherichia coli (Loferer-Krossbacher et al., 1998), and a cell volume of 0.021 μm3 calculated from the reported dimensions of N. profundicola (Smith et al., 2008), the production of 1 × 108 cells ml−1 requires ~120 ng ml−1 of N (~9 μM), a figure that would not change regardless of the N-source utilized. N. profundicola did not utilize a significant amount of the ammonium provided during growth with polysulfide as the electron acceptor (Figure 3, open circles), which agrees with the low N requirement calculated to produce the observed cell densities. The ammonium plus sulfide culture did not grow (Figure 2) and also did not detectably consume ammonium (Figure 3, closed circles). In contrast, the amount of ammonium produced by cells grown with 5 mM nitrate was 4.6 mM with polysulfide and 5.2 mM with sulfide (Figure 3, open and closed squares, respectively). If the reverse-HURM pathway in the Epsilonproteobacteria primarily serves to produce ammonium for biosynthesis, we would expect low levels of exogenous ammonium to strongly repress HURM pathway gene expression and nitrate reduction activity. However, the stoichiometric conversion of nitrate to ammonium in excess of the predicted N requirement and observed ammonium consumption in the ammonium + polysulfide cultures is consistent with the hypothesis that nitrate is utilized as a terminal electron acceptor by N. profundicola and that nitrate ammonification is not subject to repression by ammonium.
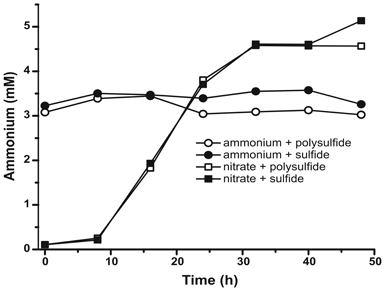
Figure 3. Ammonium concentrations measured in cultures of N. profundicola-containing the indicated electron acceptors, nitrogen compounds, and sulfur sources. Each point is the average of 2 or 3 independent biological replicates.
15N-NO−3 Incorporation into Biomass
The potential for nitrate assimilation was assessed by examining the incorporation of 15N-labeled ammonium or nitrate into N. profundicola biomass during growth with all above combinations of electron donors and potential acceptors. N. profundicola grown on 20 Atom% 15N-labeled ammonium with polysulfide produced biomass enriched to ~10 Atom% 15N after one round of batch culturing (Figure 4), whereas the unlabeled control was not enriched in 15N. Cultures grown with sulfide and 20 Atom% 15N-labeled nitrate produced biomass enriched to 16 Atom% 15N (Figure 4). Nitrate-grown biomass contained approximately 5-fold more N than ammonium-grown biomass (data not shown), which may explain the more extensive enrichment of 15N in the nitrate-grown samples.
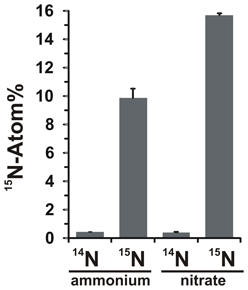
Figure 4. Nitrogen uptake into N. profundicola biomass determined following one round of batch culture with 5 mM of the indicated nitrogen compounds supplied at natural abundance (14N) or at 20 Atom% 15N. Values are averages of 3 independent biological replicates ± the standard deviation.
Transcription of Key Pathway Genes
Transcript abundance for genes that encode enzymes of the proposed reverse-HURM pathway in N. profundicola was determined by quantitative real-time PCR in batch cultures grown with ammonium or nitrate. All components of the proposed pathway displayed a strong increase in transcript abundance in nitrate grown cultures (Figure 5). mRNAs encoding a nitrate reductase subunit (napA) were 4.6-fold (P < 0.05, two-tailed heteroscedastic t-test) more abundant in nitrate + sulfide grown cells relative to ammonium + polysulfide grown cells. Transcripts of both components of the HURM, haoA and cycB/napC, were increased by 8.5- and 7.1-fold (P < 0.05 for both), respectively. Consistent with our prediction that hydroxylamine produced in the periplasm by the HURM would be transported through major facilitator protein channels, mRNA's encoding both homologs of the ammonium/methylammonium transporter amtB were increased by 4.5-fold (amtB-1, NAMH_0397, P < 0.08) and 10.3-fold (amtB-2, NAMH_0215, P > 0.10). Hydroxylamine is a powerful mutagen that must be detoxified as it arrives in the cytoplasm. We propose that this is accomplished by NADH-dependent Har/Hcp based on studies of the hcp gene in the nitrate assimilation gene cluster of Rhodobacter capsulatus E1F1 where it is required for the reduction and assimilation of hydroxylamine produced by a cytoplasmic assimilatory nitrate reductase (Cabello et al., 2004; Pino et al., 2006). Consistent with this proposal, the largest change in transcript abundance was seen for har, which increased by 11.7-fold, but this change was not significant (P > 0.10) due to the high variability in independent measurements of har transcript abundance. Originally, we had proposed that the electron donor to Har was an NADH dehydrogenase encoded by NAMH_0542 (Campbell et al., 2009). However, we now believe a better candidate for this function is NAMH_1302, a predicted 4Fe-4S cluster protein, based on conservation in other Epsilonproteobacteria (Table 1). Consistent with this prediction, transcripts of NAMH_1302 were 4.6-fold more abundant in nitrate grown cells relative to ammonium, but the change was not significant (P > 0.10).
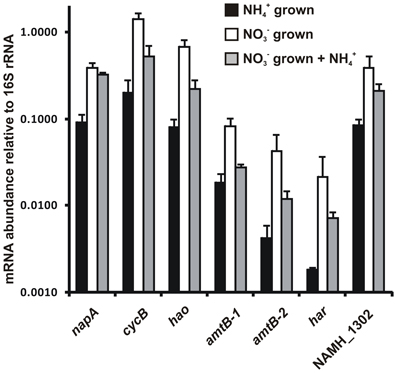
Figure 5. Steady state mRNA levels for key genes encoding enzymes of the reverse-HURM pathway in N. profundicola grown with ammonium + polysulfide (black bars), nitrate + sulfide (white bars), or nitrate + polysulfide after the addition of ammonium. Transcript abundance was determined by fluorescent RT-PCR and normalized to 16S rRNA abundance determined in each sample. Values are averages of 3 independent biological replicates ± the standard deviation.
To test for potential ammonium repression of pathway expression, ammonium was added to nitrate + sulfide grown cultures and the mRNA abundance quantified for the same genes. In general, ammonium addition decreased mRNA abundances of the reverse-HURM pathway encoding genes from 1.2- to 3.6-fold (Figure 5, gray bars), though this effect was only significant (P < 0.10) for haoA and napC/cycB transcripts that encode the HURM subunits. Interestingly, the expression of napA decreased the least (1.2-fold), indicating the need for nitrate respiration in these cells. In no case, did the addition of ammonium reduce transcript levels to those seen in cultures grown in ammonium + polysulfide.
Evidence for a Hydroxylamine Intermediate
Direct measurements of hydroxylamine in filtrates of N. profundicola cultures grown with nitrate found low, but consistently detectable levels of hydroxylamine (5.7 ± 3.5 μM, SD, n = 3). This low level is consistent with an intermediate whose production and consumption rates are closely matched.
To determine if hydroxylamine could be metabolized by N. profundicola, washed suspensions of nitrate-grown cells were incubated in nitrate-free growth medium with hydrogen as the sole electron donor. Hydroxylamine was added as a potential electron acceptor and the concentrations of both hydroxylamine and ammonium were followed over time. The results of this experiment indicate that nitrate-grown cells of N. profundicola rapidly and completely convert hydroxylamine to ammonium (Figure 6). The specific rate of hydroxylamine conversion in this experiment was 28.6 μM min−1 108 cells−1. The ammonium production rate from nitrate calculated for exponential growth in Figure 3 (8–24 h) is ~3 μM min−1 (108 cells)−1. Thus, the measured hydroxylamine conversion rate is far in excess of the ammonium production rate observed during growth of N. profundicola with nitrate. A similarly tight coupling between hydroxylamine production and removal was recently reported for the chemolithotrophic ammonia-oxidizing archaeon, Nitrosopumilus maritimus, where hydroxylamine oxidation is an essential catabolic step (Vajrala et al., 2013).
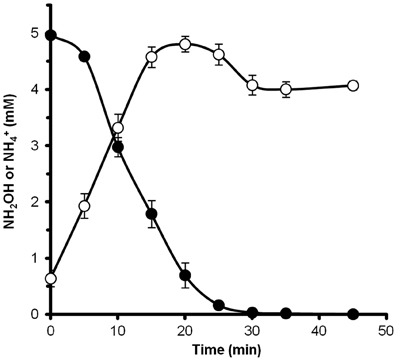
Figure 6. Hydroxylamine is rapidly converted to ammonium by N. profundicola. The concentrations of hydroxylamine (closed circles) and ammonium (open circles) were followed after the addition of 5 mM hydroxylamine to a resting cell suspension. The points are the mean values from three replicates ± the standard deviation.
Discussion
The data presented here are consistent with a new pathway for nitrate reduction in organisms that lack classical NO-forming (NirS or NirK) or ammonium-forming (NirB/NirA) nitrite reductases with N. profundicola strain AmH as a useful model system. A reverse-HURM pathway has been proposed whereby nitrite is reduced to hydroxylamine in the periplasm and subsequently reduced to ammonium in the cytoplasm (Campbell et al., 2009). The original pathway has been slightly modified here to include a more likely candidate for the proximal electron donor to Har/Hcp, a predicted 4Fe-4S containing protein encoded by NAMH_1302 (Figure 1). Homologs of NAMH_1302 are conserved in C. concisus (GenBank accession YP_001466207), C. curvus (YP_001408944), C. fetus (YP_892678), and C. mediatlanticus (ZP_01871213) that possess all other genes for the reverse-HURM pathway. In addition, NAMH_1302 displayed an increase in transcript abundance in N. profundicola cells grown on nitrate relative to ammonium.
While the production of hydroxylamine from nitrate and nitrite both via chemical (Rollefson and Oldershaw, 1932) and biological routes (Lindsey and Rhines, 1932) has long been known and a [HaoA]3 complex was shown to reduce nitrite to hydroxylamine when supplied with reductant (Kostera et al., 2010), the ratio of hydroxylamine vs. ammonium production from nitrite in the reverse-HURM pathway is not yet clear. The uptake of extracellular hydroxylamine by bacteria is known; exogenously supplied hydroxylamine accumulates readily in the anammoxosome of Kuenenia stuttgartiensis (Lindsay et al., 2001; Schmidt et al., 2004), a compartment that can be reached only by crossing both the cell and anammoxosome membranes. The proposed uptake of the highly mutagenic hydroxylamine necessitates protection against DNA lesions, which may partially explain the extensive complement of DNA repair systems identified in N. profundicola (Campbell et al., 2009). Hydroxylamine has also been proposed as an intermediate of ammonium production from nitrite in plants and algae as ferredoxin-dependent nitrite reductase displays high reactivity and specificity for this substrate (Kuznetsova et al., 2004; Hirasawa et al., 2010).
Unlike the quinone reductase function of HURM in ammonia-oxidizers (Klotz and Stein, 2008; Simon and Klotz, 2013), we propose that HURM in N. profundicola acts, similar to NrfAH, as a periplasmic quinol oxidase system that shuttles electrons from the quinol pool to reduce nitrite to hydroxylamine. It is likely that the reverse-HURM-pathway evolved from a common ancestor of the NrfAH and ONR ammonification module in anammox bacteria. We hypothesize that this occurred before HURM—including a covalently bound trimeric HaoA complex capable of hydroxylamine disproportionation—was used as a catabolic module in both anaerobic and aerobic ammonia-oxidizing bacteria (Jetten et al., 2009; Klotz and Stein, 2010; Campbell et al., 2011; Kern et al., 2011; Simon and Klotz, 2013). A similar reversal of function from reducing to oxidizing activity effected by covalent bond-directed complex formation has been discussed for the NO-reducing cytochrome c'-beta (cytS) and the NO-oxidizing cytochrome P460 (cytL) (Elmore et al., 2007). Genes encoding the NrfAH and reverse-HURM modules coexist in only one bacterial genome sequence, C. fetus 82-40 (Table 1). It was shown recently that octaheme cytochrome c hydroxylamine dehydrogenase evolved as a member of a multi-heme cytochrome c protein superfamily with functions in the nitrogen and sulfur cycles and that cytochrome cM552 (CycB), NrfH, and NapC are members of another cytochrome c superfamily (Bergmann et al., 2005; Klotz and Stein, 2008; Kern et al., 2011; Simon and Klotz, 2013). It is also known that NAP complexes in Epsilonproteobacteria generally do not contain NapC subunit proteins (Klotz et al., 2006; Sievert et al., 2008; Kern and Simon, 2009; Klotz and Stein, 2010; Simon and Klotz, 2013). The proposed reverse function of a [HaoA]n—cM552 (CycB/NapC) complex as a nitrite reductase—quinol oxidase complex is feasible given the presence of both transcripts and the absence of the critical tyrosine protein ligand in N. profundicola HaoA (Campbell et al., 2009), which separate the N-oxide reducing NrfA and ONR from the N-oxide oxidizing HAO complexes (Bergmann et al., 2005; Klotz and Stein, 2008; Kern et al., 2011; Simon and Klotz, 2013).
The 15N assimilation data established that N. profundicola incorporates nitrate-nitrogen into biomass during growth when it is provided as the sole nitrogen source. However, the vast excess production of ammonium from nitrate over the calculated demands for biosynthesis leads us to conclude that the primary function of nitrate reduction to ammonium by N. profundicola is that of an efficient respiratory electron acceptor. This conclusion is supported by the increased growth rates and yields in N. profundicola cultures with nitrate as the sole electron acceptor. Furthermore, the gene expression data agree with our proposal (Campbell et al., 2009) that this occurs via the concerted function of Nap, HURM ([HaoA]3 and cM552) and Har/Hcp as a novel N-assimilation and energy conservation pathway: the reverse-HURM pathway. Finally, the data indicate that N. profundicola has the ability to rapidly reduce hydroxylamine to ammonium, consistent with the proposal of free hydroxylamine as an intermediate in the reverse-HURM pathway. The high rate of hydroxylamine uptake observed likely explains why it is not observed consistently during growth of cultures on nitrate. Preliminary measurements of ammonium production from nitrate or nitrite in resting cells are similar to the 3 μM min−1 (108 cells)−1 rate calculated from cultures, ~10-fold lower than the observed hydroxylamine consumption rate. Given these rates, hydroxylamine should be consumed immediately after production in cultures growing on nitrate. As hydroxylamine is mutagenic, we hypothesize that this reflects previous selective pressure to maintain low hydroxylamine levels during growth.
While labeling of biomass has been attempted with 15N-NH2OH in batch N. profundicola cultures, it has not yet been successful (data not shown). Given that hydroxylamine in batch culture must be supplied at low concentrations relative to the unlabeled nitrate to avoid toxicity and that the vast majority of N from hydroxylamine should be converted to ammonium (>90% based on Figure 3), this result is not surprising. Short-term labeling experiments with much higher concentrations of labeled hydroxylamine as the sole N-source may definitively establish this point.
In conclusion, the data presented here are consistent with a model where N. profundicola reduces nitrate via a free hydroxylamine intermediate. These experiments do not yet completely exclude the possibility that the reverse-HURM activity proposed in N. profundicola functions as an ammonium-forming nitrite reductase that transfers 6 electrons without releasing an N-oxide intermediate. Unpublished results indicate that a few point mutations in the nrfA gene can render pentaheme cytochrome c nitrite reductase into a leaky enzyme that releases N oxide intermediates and fails to convert nitrite stoichiometrically into ammonium (Jörg Simon, pers. communi.). Given the evolutionary relationship between nrfA and haoA (Bergmann et al., 2005; Simon and Klotz, 2013), the reverse-HURM module in N. profundicola might naturally function as an ammonium and hydroxylamine-producing enzyme complex whereas extant NrfA operates “leak-free” as an ammonium-producing nitrite reductase. Further experiments using inhibitors of Har/Hcp with nitrate or nitrite as a substrate in whole cells and/or biochemical studies of the reverse-HURM complex to determine the in vitro product should provide additional direct tests of this hypothesis. Irrespective of the specific mechanism, the extensive production of ammonium far in excess of biosynthetic needs by N. profundicola suggests that it and related Epsilonproteobacteria may serve an ecological role as a source of ammonium in nitrate rich, ammonium deprived, hypoxic environments where they are commonly found.
Materials and Methods
Free Energy Calculations
Gibbs free energies (Table 2) were calculated from standard redox potentials of the half reactions by the Nernst equation (Δ G0 = −nFE0) or by the summation of half reaction free energies collected from the literature (Thauer et al., 1977; van der Star et al., 2008). All calculations were performed at standard conditions (1 M all species, 25°C, pH = 7).
Culturing and Nitrogen Compound Analysis
N. profundicola strain AmH (ATCC BAA-1463) was cultured and cells counted as previously described (Smith et al., 2008; Campbell et al., 2009). Ammonium in culture supernatants was quantified after derivatization with dansyl chloride and separation by HPLC with fluorescence detection (Shakila et al., 2001). Nitrate was quantified by HPLC using UV/Vis detection. Hydroxylamine concentrations were determined in culture filtrates (0.2 μm) as described by Frear and Burrell (1955).
15N Labeling Procedures
Stock solutions of 20 Atom% 15N-nitrate and 15N-ammonium were prepared by mixing >99 Atom% and natural abundance salts to prepare medium at a final concentration of 5 mM nitrate or ammonium as needed. Cells were harvested from 15N labeled cultures by centrifugation and the cell pellet washed three times with nanopure water to remove salts. The cell suspension was dried in tin capsules which were sent to the University of California-Davis stable isotope laboratory where 15N Atom % was determined by isotope ratio mass spectrometry. Control samples indicated that no isotopic contamination had occurred during sample processing (data not shown).
Quantitative PCR
Gene-specific primers (Table 3) were used to quantify mRNA abundance in total RNA extracted from N. profundicola as previously described (Campbell et al., 2009).
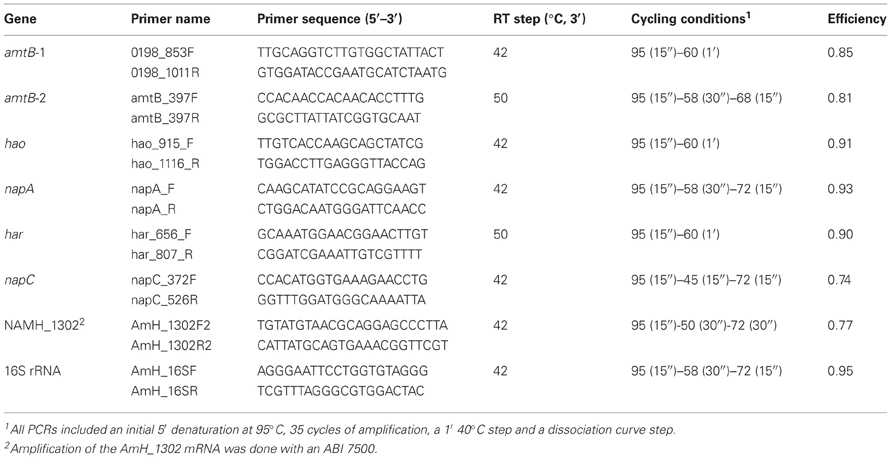
Table 3. Primers and conditions for Q-RT-PCR quantification of specific genes in total RNA samples extracted from N. profundicola.
Hydroxylamine Uptake
N. profundicola was grown on H2 and formate as electron donors and nitrate as the sole electron acceptor to late exponential phase. Cells were harvested by centrifugation. All transfers and cell washings were performed in a Coy Laboratories anaerobic chamber. Cells were resuspended with nitrate-free growth medium to a final concentration of ~109 cells ml−1 and incubated at the 45°C for 2 h under an atmosphere of 80% H2 + 20% CO2 prior to the addition of hydroxylamine (5 mM final concentration) from an anaerobically prepared stock solution. Aliquots of the cell suspension were filtered and assayed for hydroxylamine as described above and for ammonium by HPLC after derivatization with diethyl ethoxymethylenemalonate (Gómez-Alonso et al., 2007).
Author Contributions
Thomas E. Hanson, Barbara J. Campbell, and Martin G. Klotz collaboratively designed experiments and interpreted results, Thomas E. Hanson performed all nitrogen compound analyses and prepared samples for mass spectrometry, Barbara J. Campbell grew all cultures and performed quantitative PCR, Katie M. Kalis performed hydroxylamine uptake experiments, Mark A. Campbell identified NAMH_1302 as a relevant gene. Thomas E. Hanson drafted the paper, which was subsequently revised by Thomas E. Hanson, Barbara J. Campbell, and Martin G. Klotz.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Lisa Y. Stein (Univ. Alberta-Edmonton), Jenn Macalady (Penn State Univ.), David Kirchman and Matt Cottrell (Univ. Delaware) for critical readings of manuscript drafts, Jennifer Biddle for access to an anaerobic chamber and Kathy Coyne for access to an HPLC. This project was supported by grants from the National Science Foundation to Barbara J. Campbell and S. Craig Cary (EF-0333203), to Barbara J. Campbell (DEB-0640414), to Barbara J. Campbell and Thomas E. Hanson (MCB-0950691), and to Martin G. Klotz (MCB-0948202/1202648), by an EPSCoR RII-2 grant to the University of Delaware (EPS-0814251), and by incentive funds provided by the University of North Carolina to Martin G. Klotz.
References
Bergmann, D. J., Hooper, A. B., and Klotz, M. G. (2005). Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria: evidence for their evolutionary history. Appl. Environ. Microbiol. 71, 5371–5382. doi: 10.1128/AEM.71.9.5371-5382.2005
Brandes, J. A., Devol, A. H., and Deutsch, C. (2007). New developments in the marine nitrogen cycle. Chem. Rev. 107, 577–589. doi: 10.1021/cr050377t
Cabello, P., Pino, C., Olmo-Mira, M. F., Castillo, F., Roldán, M. D., and Moreno-Vivián, C. (2004). Hydroxylamine assimilation by Rhodobacter capsulatus E1F1 - Requirement of the hcp gene (hybrid cluster protein) located in the nitrate assimilation nas gene region for hydroxylamine reduction. J. Biol. Chem. 279, 45485–45494. doi: 10.1074/jbc.M404417200
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. doi: 10.1038/nrmicro1414
Campbell, B. J., Jeanthon, C., Kostka, J. E., Luther, G. W., and Cary, S. C. (2001). Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67, 4566–4572. doi: 10.1128/AEM.67.10.4566-4572.2001
Campbell, B. J., Smith, J. L., Hanson, T. E., Klotz, M. G., Stein, L. Y., Lee, C. K., et al. (2009). Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 5:e1000362. doi: 10.1371/journal.pgen.1000362
Campbell, M. A., Nyerges, G., Kozlowski, J., Poret-Peterson, A. T., Stein, L. Y., and Klotz, M. G. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 322, 82–89. doi: 10.1111/j.1574-6968.2011.02340.x
Elmore, B. O., Bergmann, D. J., Klotz, M. G., and Hooper, A. B. (2007). Cytochromes P460 and c'-beta; A new family of high-spin cytochromes c. FEBS Lett. 581, 911–916. doi: 10.1016/j.febslet.2007.01.068
Falkowski, P. G., and Godfrey, L. V. (2008). Electrons, life and the evolution of Earth's oxygen cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2705–2716. doi: 10.1098/rstb.2008.0054
Frear, D. S., and Burrell, R. C. (1955). Spectrophotometric method for determining hydroxylamine reductase activity in higher plants. Anal. Chem. 27, 1664–1665. doi: 10.1021/ac60106a054
Gómez-Alonso, S., Hermosín-Gutiérrez, I., and García-Romero, E. (2007). Simultaneous HPLC analysis of biogenic amines, amino acids, and ammonium ion as aminoenone derivatives in wine and beer samples. J. Agric. Food Chem. 55, 608–613. doi: 10.1021/jf062820m
Hirasawa, M., Tripathy, J. N., Sommer, F., Somasundaram, R., Chung, J. S., Nestander, M., et al. (2010). Enzymatic properties of the ferredoxin-dependent nitrite reductase from Chlamydomonas reinhardtii. Evidence for hydroxylamine as a late intermediate in ammonia production. Photosynth. Res. 103, 67–77. doi: 10.1007/s11120-009-9512-5
Jetten, M. S., Niftrik, L. V., Strous, M., Kartal, B., Keltjens, J. T., and Op den Camp, H. J. (2009). Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44, 1–20. doi: 10.1080/10409230902722783
Kern, M., and Simon, J. (2009). Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim. Biophys. Acta 1787, 646–656. doi: 10.1016/j.bbabio.2008.12.010
Kern, M., Klotz, M. G., and Simon, J. (2011). The Wolinella succinogenes mcc gene cluster encodes an unconventional respiratory sulfite reduction system. Mol. Microbiol. 82, 1515–1530. doi: 10.1111/j.1365-2958.2011.07906.x
Klotz, M. G., Arp, D. J., Chain, P. S., El-Sheikh, A. F., Hauser, L. J., Hommes, N. G., et al. (2006). Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707 Appl. Environ. Microbiol. 72, 6299–6315. doi: 10.1128/AEM.00463-06
Klotz, M. G., and Stein, L. Y. (2008). Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 278, 146–156. doi: 10.1111/j.1574-6968.2007.00970.x
Klotz, M. G., and Stein, L. Y. (2010). “Genomics of ammonia-oxidizing bacteria and insights to their evolution,” in Nitrification, eds B. B. Ward, D. J. Arp, and M. G. Klotz (Washington, DC: ASM Press), 57–93. ISBN: 978-1-55581-481-6.
Kostera, J., McGarry, J., and Pacheco, A. A. (2010). Enzymatic interconversion of ammonia and nitrite: the right tool for the job. Biochemistry 49, 8546–8553. doi: 10.1021/bi1006783
Kuznetsova, S., Knaff, D. B., Hirasawa, M., Sétif, P., and Mattioli, T. A. (2004). Reactions of spinach nitrite reductase with its substrate, nitrite, and a putative intermediate, hydroxylamine. Biochemistry 43, 10765–10774. doi: 10.1021/bi048826r
Lindsay, M. R., Webb, R. I., Strous, M., Jetten, M. S., Butler, M. K., Forde, R. J., et al. (2001). Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175, 413–429. doi: 10.1007/s002030100280
Lindsey, G. A., and Rhines, C. M. (1932). The production of hydroxylamine by the reduction of nitrates and nitrites by various pure cultures of bacteria. J. Bacteriol. 24, 489–492.
Loferer-Krossbacher, M., Klima, J., and Psenner, R. (1998). Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 64, 688–694.
Pino, C., Olmo-Mira, F., Cabello, P., Martínez-Luque, M., Castillo, F., Roldán, M. D., et al. (2006). The assimilatory nitrate reduction system of the phototrophic bacterium Rhodobacter capsulatus E1F1. Biochem. Soc. Trans. 34, 127–129. doi: 10.1042/BST0340127
Rollefson, G. K., and Oldershaw, C. F. (1932). The reduction of nitrites to hydroxylamine by sulfites. J. Am. Chem. Soc. 54, 977–979. doi: 10.1021/ja01342a019
Schmidt, I., Look, C., Bock, E., and Jetten, M. S. M. (2004). Ammonium and hydroxylamine uptake and accumulation in Nitrosomonas. Microbiology 150, 1405–1412. doi: 10.1099/mic.0.26719-0
Shakila, R. J., Vasundhara, T. S., and Kumudavally, K. V. (2001). A comparison of the TLC-densitometry and HPLC method for the determination of biogenic amines in fish and fishery products. Food Chem. 75, 255–259. doi: 10.1016/S0308-8146(01)00173-X
Sievert, S. M., Scott, K. M., Klotz, M. G., Chain, P. S., Hauser, L. J., Hemp, J., et al. (2008). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74, 1145–1156. doi: 10.1128/AEM.01844-07
Simon, J., and Klotz, M. G. (2013). Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta 1827, 114–135. doi: 10.1016/j.bbabio.2012.07.005
Smith, J. L., Campbell, B. J., Hanson, T. E., Zhang, C. L., and Cary, S. C. (2008). Nautilia profundicola sp. nov., a thermophilic, sulfur-reducing epsilonproteobacterium from deep-sea hydrothermal vents. Int. J. Syst. Appl. Microbiol. 58, 1598–1602. doi: 10.1099/ijs.0.65435-0
Thauer, R. K., Jungermann, K., and Decker, K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100–180.
Vajrala, N., Martens-Habbena, W., Sayavedra-Soto, L. A., Schauer, A., Bottomley, P. J., Stahl, D. A., et al. (2013). Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc. Natl. Acad. Sci. U.S.A. 110, 1006–1011. doi: 10.1073/pnas.1214272110
van der Star, W. R., van de Graaf, M. J., Kartal, B., Picioreanu, C., Jetten, M. S., and van Loosdrecht, M. C. (2008). Response of anaerobic ammonium-oxidizing bacteria to hydroxylamine. Appl. Environ. Microbiol. 74, 4417–4426. doi: 10.1128/AEM.00042-08
Keywords: hydroxylamine, hydroxylamine oxidoreductase, nitrite, Epsilonproteobacteria, nitrate ammonification
Citation: Hanson TE, Campbell BJ, Kalis KM, Campbell MA and Klotz MG (2013) Nitrate ammonification by Nautilia profundicola AmH: experimental evidence consistent with a free hydroxylamine intermediate. Front. Microbiol. 4:180. doi: 10.3389/fmicb.2013.00180
Received: 02 May 2013; Accepted: 15 June 2013;
Published online: 04 July 2013.
Edited by:
Boran Kartal, Radboud University, NetherlandsReviewed by:
A. Andrew Pacheco, University of Wisconsin-Milwaukee, USAWouter J. Maalcke, University of Stuttgart, Germany
Copyright © 2013 Hanson, Campbell, Kalis, Campbell and Klotz. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Barbara J. Campbell, Department of Biological Sciences, Clemson University, 132 Long Hall, Clemson, SC 29634, USA e-mail: bcampb7@clemson.edu