Structural characterization of lignin in wild-type versus COMT down-regulated switchgrass
- 1School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA, USA
- 2BioEnergy Science Center, Oak Ridge, TN, USA
- 3Institute of Paper Science and Technology, Georgia Institute of Technology, Atlanta, GA, USA
- 4Forage Improvement Division, The Samuel Roberts Noble Foundation, Ardmore, OK, USA
This study examined the chemical structural characteristics of cellulolytic enzyme lignin isolated from switchgrass focusing on comparisons between wild-type control and caffeic acid 3-O-methyltransferase (COMT) down-regulated transgenic line. Nuclear magnetic resonance techniques including 13C, 31P, and two-dimensional 13C-1H heteronuclear single quantum coherence as well as gel permeation chromatography were employed. Compared to the wild-type, the COMT down-regulated transgenic switchgrass lignin demonstrated a decrease in syringyl (S):guaiacyl (G) ratio and p-coumarate:ferulate ratio, an increase in relative abundance of phenylcoumaran unit, and a comparable content of total free phenolic OH groups along with formation of benzodioxane unit. In addition, COMT down-regulation had no significant effects on the lignin molecular weights during its biosynthesis process.
Introduction
There is a growing focus on innovative technologies that shift bioethanol production to second generation cellulosic biofuels due to the competition between food resources and first generation biofuels derived from agricultural resources such as corn starch (Wyman, 1999; Wang et al., 2007; Rubin, 2008). Although lignocellulosic resources, such as energy crops and agricultural and forest residues, are readily becoming available for bioethanol production, their processing requires a costly pretreatment step to overcome their natural recalcitrance toward biological deconstruction to simple sugars (Sun and Cheng, 2002; Himmel et al., 2007; Pu et al., 2008; Somerville et al., 2010). Lignocellulosic biomass is a complex composite consisting primarily of three biopolymers (i.e., cellulose, hemicelluloses, and lignin) and its recalcitrance has been attributed to several factors such as cellulose accessibility to enzymes, lignin content/structure, lignin-carbohydrate complexes, as well as the presence and structure of hemicelluloses (Wyman et al., 2005; Li et al., 2012; Leu and Zhu, 2013; Pu et al., 2013). The goal of pretreatment is to reduce the recalcitrance of biomass by disrupting/modifying the lignin-polysaccharide matrix. In the past two decades, extensive research efforts have been directed at improving a diverse set of pretreatment technologies including: dilute acid, lime, hot water, steam explosion, ammonia, and organosolv pretreatments. While these pretreatments have achieved various level of success in overcoming the recalcitrance of lignocellulosic biomass, the pretreatment step still remains one of the most expensive steps in the current biomass to bioethanol production platform. Therefore, addressing biomass recalcitrance through alternative approaches is a crucial issue to the widespread, low-cost generation of cellulosic biofuels.
One of the promising approaches for reducing biomass recalcitrance is the development of genetically engineered plants involving down-regulation/overexpression of key enzymes involved in lignin biosynthesis that can achieve an improved sugar release performance with reduced pretreatment severities (Li et al., 2003; Chen and Dixon, 2007; Weng et al., 2008; Pu et al., 2011b; Shen et al., 2012; Tschaplinski et al., 2012). A recent report by Shen et al. (2012) has demonstrated that overexpression of PvMYB4 genes in switchgrass resulted in an approximately threefold increase in sugar release efficiency from transgenic cell wall residues. Chen and Dixon (2007) reported that p-coumarate 3-hydroxylase (C3H) and hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT) down-regulated alfalfa lines showed an improved fermentable sugar yield when compared to wild-type plants. The engineering of these enzyme genes is expected to generate a variation in lignin structures and thus impact the recalcitrance of biomass; therefore, there has been considerable interest in lignin structural characteristics in genetically engineered plants (Marita et al., 1999; Guo et al., 2001; Chen et al., 2004; Pu et al., 2009; Stewart et al., 2009; Lu et al., 2010; Tu et al., 2010; Vanholme et al., 2010; Ziebell et al., 2010). For example, Pu et al. (2009) and Ziebell et al. (2010) isolated ball-milled lignin from C3H and HCT transgenic alfalfa lines and studied their structural features. The results demonstrated that a substantial increase in p-hydroxyphenyl (H) units and a decrease in syringyl (S) and guaiacyl (G) monolignol units were accompanied with a decrease in the presence of β-O-4 aryl ether.
Recently, Fu et al. (2011) generated caffeic acid 3-O-methyltransferase (COMT) down-regulated switchgrass and evaluated the impact of lignin pathway modification in these switchgrass lines on simultaneous saccharification and fermentation (SSF) efficiency and their response to consolidated bioprocessing using Clostridium thermocellum. The fermentation of the control and transgenic lines confirmed that transgenic lines produced more ethanol per gram than the control without pretreatment. Fermentation of select down-regulated switchgrass lines after a moderate dilute acid pretreatment resulted in a 38% increase in ethanol yield over the pretreated control sample under SSF. The reduced recalcitrance in transgenic lines was further demonstrated with a ~300–400% reduction in the required cellulase dosage with respect to the control for equivalent ethanol yield via SSF. In addition, the consolidated bioprocessing of mild acid pretreated COMT transgenic switchgrass was observed to lead to 18% more fermentation products than the control (Fu et al., 2011). The effects of COMT down-regulation on cellulose were investigated and the results showed that the cellulose content in the stems of COMT transgenic lines had small variations at −1 to 3% relatively, whereas the cellulose crystallinity index and degree of polymerization were essentially identical to the control, suggesting that COMT down-regulation had negligible impact on cellulose in switchgrass. Therefore, the lignin structural characteristics in the COMT transgenic switchgrass is of great interest as its alterations due to the COMT down-regulation could be related to the reduced recalcitrance in the transgenic switchgrass. As a continuation of this work, here we investigated and compared the structural features of lignin isolated from wild-type versus COMT down-regulated switchgrass in an attempt to explore the roles that the lignin related structural changes could play in the reduced recalcitrance of transgenic switchgrass.
Materials and Methods
Chemicals
Dimethyl sulfoxide-d6 (DMSO-d6), pyridine, 2-chloro-4,4,5,5-tetramethyl-1,3,2-dioxaphospholane, anhydrous diethyl ether, acetic acid, chloroform, and enzymes were purchased from Sigma-Aldrich and used as received. All other chemicals were obtained from VWR and 1,4-dioxane was distilled over NaBH4 prior to use.
Plant Materials and Sample Collection
A lowland-type switchgrass (Panicum virgatum L.cv. Alamo) was used for genetic transformation and lignin modification (Fu et al., 2011). A COMT RNAi vector was introduced into switchgrass calli by Agrobacterium-mediated transformation. The transgenics were grown in the greenhouse at 26°C with 16 h light (390 μE/m2s). The vegetative development of switchgrass was divided into four elongation stages (E1, E2, E3, and E4) according to the criteria described by Moore et al. (1991) and Shen et al. (2009). Under the greenhouse conditions, switchgrass produced four visible internodes from basal to top before entering into reproductive development.
A transgenic line (T0–2) which had the greatest degree of COMT down-regulation was used to generate T1 progeny by crossing with a wild-type Alamo plant. The T1-generation materials were employed to study the structural differences in the COMT down-regulated transgenic and wild-type control plants. A null segregant (negative T1 plant identified from the progeny of the cross) was used as a control. The first internodes (hereafter referred as I1) from basal were collected for analysis when the plants had reached the E4 stage.
Compositional Analysis
The compositional analysis was performed according to literature procedure (Davis, 1998; Cao et al., 2012). The switchgrass samples were sequentially extracted with water and dichloromethane 24 h each before analysis. The extractive-free switchgrass (~0.175 g) was subjected to a two-step hydrolysis with sulfuric acid (72% w/w, 1.5 mL) for 1 h at 30°C and then at 121°C for additional 1 h after diluting with the addition of water (4% w/w sulfuric acid). The samples were filtered and the filtrates were quantified for neutral monosaccharide contents using high performance anion exchange chromatography with pulsed amperometric detection (Dionex ICS 3000, Dionex Corp., Sunnyvale, CA, USA). The filtrates were diluted 25 times before analysis. Nanopure deionized water was used as the eluent and sodium hydroxide solution (0.4 M NaOH) as post-column rinsing effluent. Fucose was used as an internal standard. Flow rate was 1.0 mL/min. The residues were washed, dried, and weighed to give Klason lignin content. The measurements were carried out in duplicate and the results were reported as the average.
Cellulolytic Enzyme Lignin Isolation
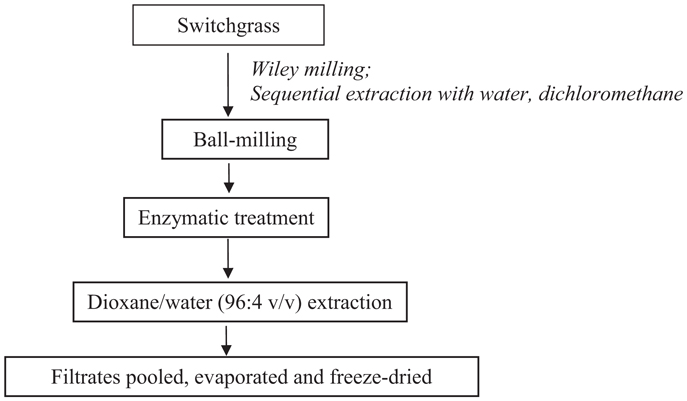
Cellulolytic enzyme lignin was isolated from control and transgenic switchgrass according to a slightly modified literature procedure (Scheme 1) (Chang et al., 1975; Capanema et al., 2004; Hu et al., 2006). In brief, the extractive-free, vacuum dried (40°C overnight) switchgrass samples were ball-milled in a porcelain jar with ceramic balls using a rotatory ball mill running at 96 rpm for 168 h under N2. The milled fine cell wall powder was then subjected to enzyme treatment in acetic acid/ammonium acetate buffer (pH 4.8, 50°C) under continuous agitation at 200 rpm for 48 h. The residue was isolated by centrifugation and was hydrolyzed one more time with freshly added enzymes. The residue obtained was washed with deionized water, centrifuged, and freeze dried. The enzyme-treated residue was extracted with dioxane-water (96% v/v, 10.0 mL/g biomass) for 24 h × 2. The extracted mixture was centrifuged and the supernatant was collected. The extracts were combined, roto-evaporated to reduce the volume under reduced pressure (<45°C), and freeze dried. The obtained lignin samples were dried under vacuum at ~45°C for overnight before nuclear magnetic resonance (NMR) analysis.
NMR Analysis
Nuclear magnetic resonance spectra of isolated lignin samples were acquired in a Bruker Avance/DMX 400 MHz spectrometer operating at a frequency of 100.59 MHz for the 13C nucleus. The 13C NMR acquisition was performed on a QNP probe using a 90° pulse with an inverse-gated decoupling pulse sequence, a 12-s pulse delay, and 12,288 scans at 50°C.
For quantitative 31P NMR, lignin was dissolved in a solvent of pyridine/CDCl3 (1.6/1.0 v/v) and derivatized with 2-chloro-4,4,5,5-tetramethyl-1,3,2-dioxaphospholane. The spectrum was acquired using an inverse-gated decoupling pulse sequence (Waltz-16), 90° pulse, 25-s pulse delay, and 128–256 scans. N-hydroxy-5-norbornene-2,3-dicarboximide was used as an internal standard and the contents of hydroxyl groups were calculated on the basis of the internal standard. A standard Bruker heteronuclear single quantum coherence (HSQC) pulse sequence (hsqcetgp) was used on a BBO probe with the following acquisition parameters: spectra width 10 ppm in F2 (1H) dimension with 2048 time of domain (acquisition time 256.1 ms), 210 ppm in F1(13C) dimension with 256 time of domain (acquisition time 6.1 ms), a 1.5-s delay, a 1JC–H of 145 Hz, and 32 scans. The central DMSO solvent peak was used for chemical shifts calibration (δc 39.5 ppm, δH 2.50 ppm). Relative lignin monomer compositions and interunit linkage abundance were estimated semi-quantitatively using volume integration of contours in HSQC spectra (Ralph et al., 2004; Zhang and Gellerstedt, 2007; Kim et al., 2008; Yelle et al., 2008; Jiang et al., 2010; Kim and Ralph, 2010; Samuel et al., 2011; Mansfield et al., 2012). For monolignol compositions of S, G, H, p-coumarate (pCA), and ferulate (FA) measurements, the S2/6, G2, H2/6, pCA2/6, and FA2 contours were used with G2 and FA2 integrals doubled (Higuchi et al., 1967; Marita et al., 2001; Ralph et al., 2001, 2004; Capanema et al., 2005; Zhang and Gellerstedt, 2007; Kim et al., 2008; Yelle et al., 2008; Jiang et al., 2010; Kim and Ralph, 2010; Mansfield et al., 2012). The Cα signals were used for contour integration for interunit linkages estimation. Bruker’s TopSpin 2.1 software and MestReNova 8.0.0 software were employed for data processing and integrations.
Molecular Weight Distribution Analysis
The lignin molecular weight distribution analysis was performed with gel permeation chromatography (GPC). The lignin samples were acetylated before GPC analysis to dissolve in tetrahydrofuran. Dry lignin sample was dissolved a mixture of acetic anhydride/pyridine (1:1 v/v) and stirred at room temperature for 24 h. After the acetylation, the solvent was removed by rotary evaporation with adding of ethanol (95% v/v). The residue was dissolved in chloroform and precipitated with diethyl ether. The precipitate was centrifuged, washed with diethyl ether (×3), and dried under vacuum prior to GPC analysis. The molecular weight distributions of the acetylated lignin samples were analyzed on a GPC SECurity 1200 system (PSS-Polymer Standards Service, Warwick, RI, USA) operated on Agilent HPLC 1200 with four Waters Styragel columns (HR1, HR2, HR4, and HR6). An UV detector (270 nm) was used for detection. Tetrahydrofuran was used as the mobile phase (flow rate 1.0 mL/min). Polystyrene narrow standards were used for establishing the calibration curve.
Results and Discussion
Compositional Analysis
The compositional analysis results of switchgrass (internode I1) were presented in Figure 1. The COMT down-regulation resulted in a reduction of ca. 14% in Klason lignin content in the transgenic lines when compared to the wild-type control. The observed lignin content changes were consistent with the previous report that transgenic switchgrass had a lower lignin content estimated using acetyl bromide method (Fu et al., 2011). The neutral monosaccharides in both wild-type and transgenic switchgrass were primarily glucose and xylose as well as minor amounts of arabinose and galactose. While the COMT down-regulated transgenic line had slightly higher xylose content with comparison to the wild-type control, both lines appeared to have comparable contents of arabinose, galactose, and glucose which is consistent with an earlier study of COMT down-regulated switchgrass (Fu et al., 2011).
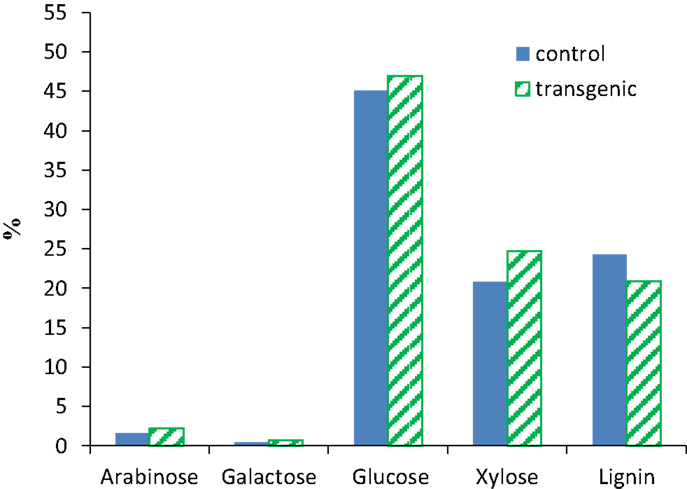
Figure 1. Compositions of wild-type and COMT down-regulated switchgrass (internode I1). Contents expressed on a dry weight basis.
13C NMR Analysis
13C NMR spectra for enzyme lignin samples isolated from the wild-type control and transgenic switchgrass (internode I1) were obtained, as presented in Figure 2. The signal assignments and quantitative analysis of 13C NMR spectra of lignin were performed following literature reports (Higuchi et al., 1967; Marita et al., 2001; Ralph et al., 2001; Capanema et al., 2005; Pu et al., 2005; Hallac et al., 2010). The signals from both guaiacyl and syringyl units were readily observed from the G2, G6, and S2/6 signals centered ~δ 109.5, 119.1, and 104.6 ppm, respectively. Significant amount of p-coumarates in both control and transgenic sample was observed from the presence of its carbonyl peak at δ 166.7 ppm and C4 peak around δ 160.0 ppm. In addition, 13C NMR spectra demonstrated a small peak around δ 122.9 ppm attributing to C6 peak of ferulates, suggesting minor presence of ferulate units in the isolated enzyme lignin from wild-type and transgenic switchgrass. The signals of cinnamyl aldehyde (Cγ at ~194 ppm) and acetyl groups (methyl C around 20.1 ppm and carbonyl C ~170.2 ppm) were observed in both wild-type and COMT transgenic switchgrass lines. Compared to wild-type, the lignin in transgenic lines had a lower S/G ratio as revealed by a signal intensity reduction in S2/6 peaks relative to G2 peak as well as a reduction in S3/5 peak intensity relative to G3/4 peak intensity. The intense peak cluster in the spectral region of δ 79–65 ppm was largely due to the presence of carbohydrates in the isolated enzyme lignin. In addition, the peaks centered around 97.5 and 92.2 ppm arisen from anomeric carbons of xylan were further indicative of the noticeable presence of carbohydrates.
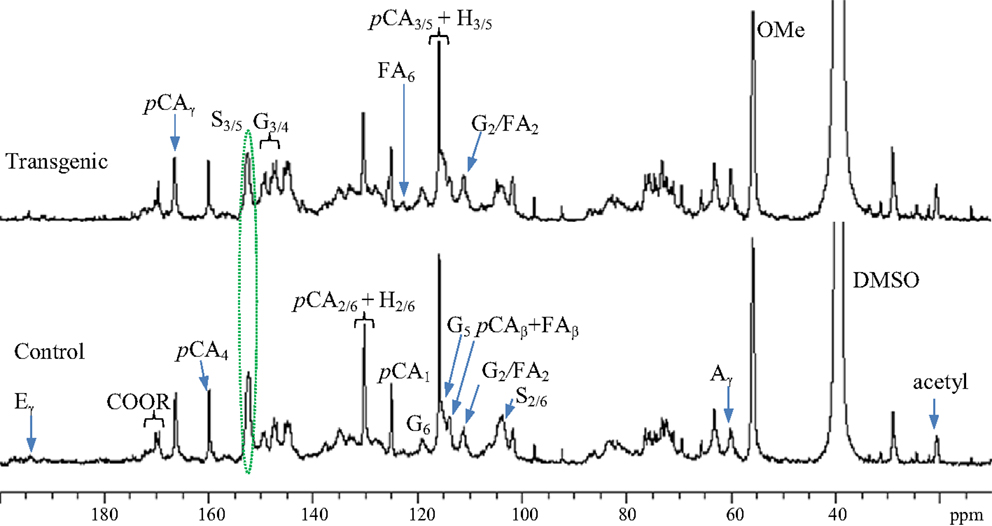
Figure 2. Quantitative 13C NMR spectra of enzyme lignins isolated from switchgrass internode I1 samples (Bottom: control; top: transgenic). A, β-O-4; G, guaiacyl; S, syringyl; H, p-hydroxyphenyl; pCA, p-coumarate; FA, ferulate; E, cinnamyl aldehyde; OMe, methoxyl.
HSQC NMR Analysis
The two-dimensional nature of 13C-1H HSQC NMR can provide valuable information about the chemical structures of lignin by deconvoluting or dispersing the overlapping carbon and proton resonances in the complex spectra over two spectral axes. Figures 3 and 4 presented the aromatic and aliphatic regions of HSQC NMR spectra of lignin samples isolated from control and transgenic switchgrass I1 internodes. The observed lignin sub-units were presented in Figure 5 and signal assignments were summarized in Table 1 (Ralph et al., 2004; Zhang and Gellerstedt, 2007; Kim et al., 2008; Yelle et al., 2008; Jiang et al., 2009; Kim and Ralph, 2010; Mansfield et al., 2012). The correlation signals for various monolignols including syringyl, guaiacyl, p-hydroxyphenyl, p-coumarate, and ferulate units were evidently observed and distributed fairly well in the aromatic region of HSQC spectra. For example, the syringyl, guaiacyl, and p-hydroxyphenyl were readily identified due to the presence of their diagnostic cross peaks around 103.9/6.73 (S2/6), 111.1/6.96 (G2), 118.6/6.79 (G6), and 127.5/7.13 (H2/6) ppm, respectively. The strong cross peak centered at δC/δH 129.4/7.45 ppm was attributed to C2/6/H2/6 of p-coumarate (pCA), confirming the notable presence of p-coumarate with its C3/5/H3/5 correlation peaks overlapping with G5 around 115.3/6.87 ppm. The signals for ferulate units were observed with its indicative cross peaks of FA2 around 111.5/7.49 ppm. In addition, the cross peaks for vinyl carbons in pCA and FA structures were overlapped around Cα/Hα 144.5/7.47 and Cβ/Hβ 113.4/6.32 ppm. HSQC semi-quantitative estimation showed that the transgenic switchgrass lignin had a decrease in S/G ratio than the wild-type (0.45 vs. 0.78), supporting the observations from 13C NMR analysis. Fu et al. (2011) investigated wild-type and transgenic switchgrass stem bulk (without separating the internodes) using GC/MS techniques and reported an S/G ratio decrease by ~32% for transgenic switchgrass. HSQC analysis revealed that p-hydroxyphenyl units were estimated to exhibit no significant changes accounting for 2–3% in wild-type and transgenic lignins. p-Coumarate units were observed to account for ca. 32 and 26% in lignins from wild-type versus transgenic switchgrass. In line with 13C NMR analysis, HSQC spectra also demonstrated a weak signal of ferulate unit, which was estimated to account for ca. 6 and 11% in the isolated lignins of wild-type and transgenic lines, respectively. The pCA:FA ratio in the isolated enzyme lignin samples was observed to decrease (by ca. 50%) for the COMT transgenic switchgrass. Shen et al. (2012) reported a reduced ester-linked pCA:FA ratio in transgenic switchgrass with overexpression of PvMYB4 that demonstrated an increase in sugar release efficiency and suggested that saccharification efficiency was negatively correlated, in part, with cell wall ester-linked pCA:FA ratio.
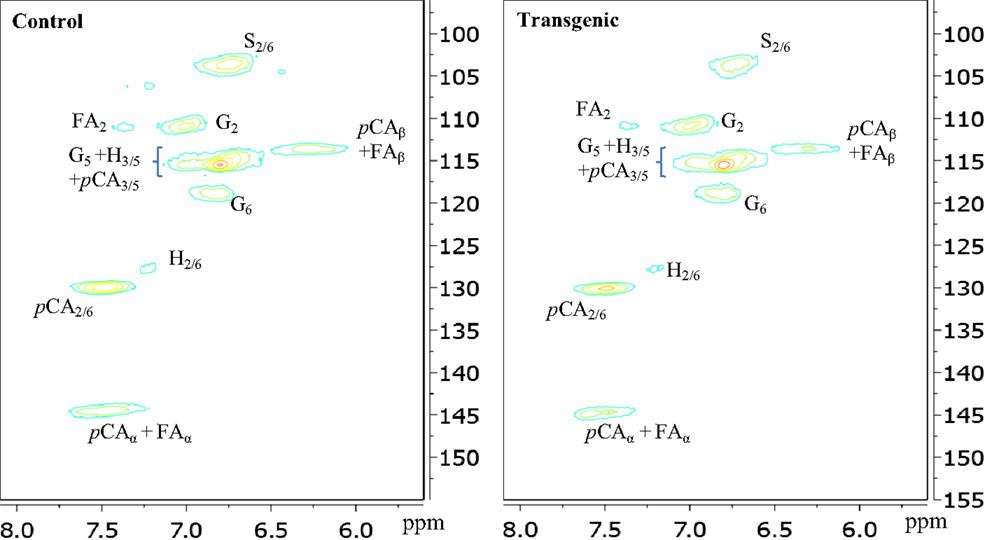
Figure 3. Aromatic regions of 13C-1H HSQC spectra of enzyme lignins isolated from control and transgenic internode I1 switchgrass. G, guaiacyl; S, syringyl; H, p-hydroxyphenyl; pCA, p-coumarate; FA, ferulate.
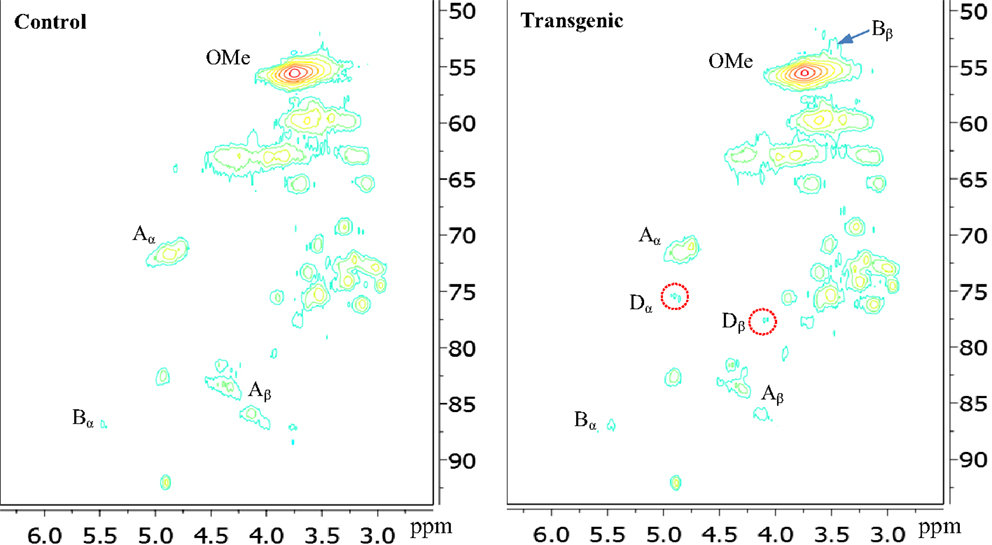
Figure 4. Aliphatic regions of 13C-1H HSQC spectra of enzyme lignins isolated from control and transgenic internode I1 switchgrass. A, β-O-4 ether; B, phenylcoumaran; D, benzodioxane; OMe, methoxyl.
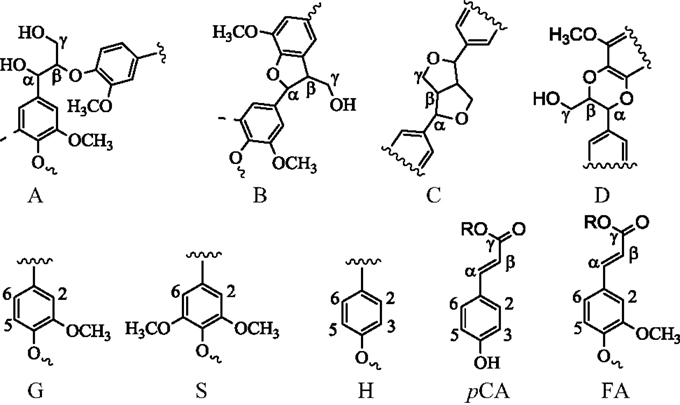
Figure 5. Chemical structures in switchgrass lignin. G, guaiacyl; S, syringyl; H, p-hydroxyphenyl; A, β-O-4 ether; B, phenylcoumaran; C, resinol; D, benzodioxane; pCA, p-coumarate; FA, ferulate.
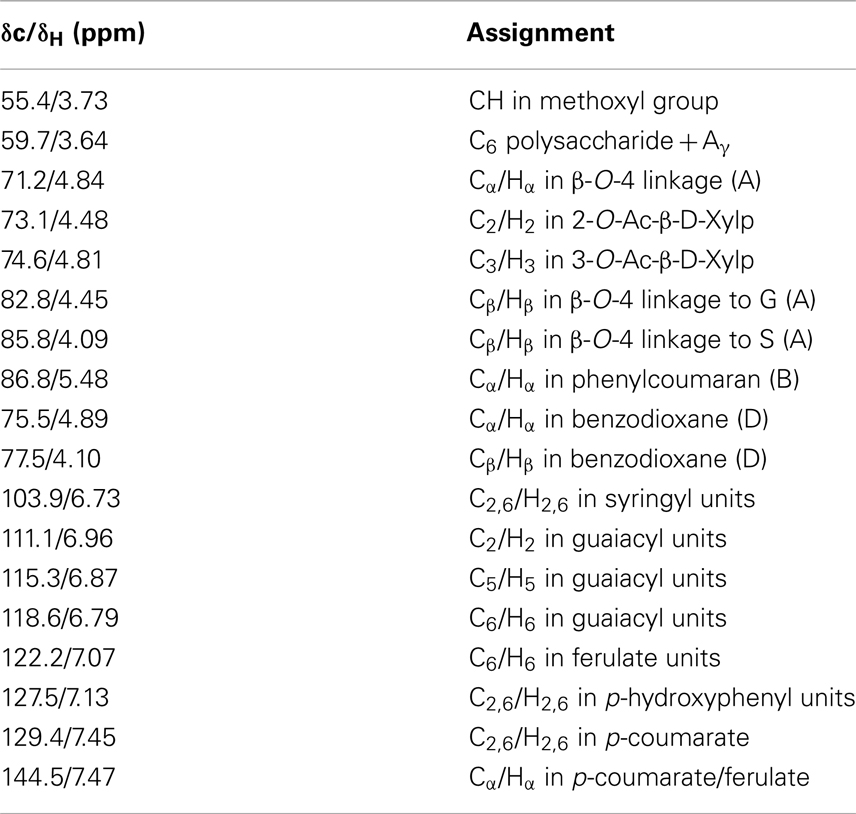
Table 1. Signal assignments of chemical structures in 13C-1H HSQC NMR spectra of control and transgenic switchgrass lignin (Ralph et al., 2004; Zhang and Gellerstedt, 2007; Kim et al., 2008; Yelle et al., 2008; Kim and Ralph, 2010; Mansfield et al., 2012).
In the side chain regions of HSQC spectra, the cross signals for methoxyl and β-O-4 substructures were among the most prominent ones. The phenylcoumaran substructure was evidenced by its Cα/Hα correlations centered at δc/δH 86.8/5.48 ppm. Resinol substructure was detected barely above the noise level in the spectra. In addition, the presence of benzodioxane units (~5% of total interunit linkages) in the transgenic internode I1 lignin was apparently evidenced by the cross peaks at δC/δH 75.5/4.89 ppm (Dα) and 77.5/4.10 ppm (Dβ), which were absent in the wild-type. The observation of benzodioxane substructure in COMT down-regulated switchgrass was consistent with previous reports that down-regulation of COMT during lignin biosynthesis resulted in the incorporation of 5-hydroxy coniferyl alcohol into lignin and generation of novel benzodioxane units (Lu et al., 2010; Vanholme et al., 2010). For example, Lu et al. (2010) reported that down-regulation of COMT in poplar led to the formation of benzodioxane units in lignin. Similarly, Vanholme et al. (2010) observed the newly formed benzodioxane units in COMT down-regulated Arabidopsis. The HSQC analysis showed that the isolated transgenic lignin had a decrease in β-O-4 linkages (by ~10%) and an increase in phenylcoumaran units (by ~57%) when compared to the wild-type.
31P NMR Analysis
The 31P NMR is an effective tool for determining the presence of hydroxyl groups in lignin which can provide quantitative information for various types of major hydroxyl groups including aliphatic, carboxylic, guaiacyl, syringyl, C5-substituted phenolic hydroxyls, and p-hydroxyphenyls (Pu et al., 2011a). The chemical structures of wild-type and COMT transgenic switchgrass lignins were characterized by phosphitylation followed-by 31P NMR spectroscopy and the results were presented in Figures 6 and 7. Figure 6 showed that the aliphatic OH appeared to be the dominant free hydroxyl type in both wild-type and transgenic switchgrass lignin, accounting for ca. 75% of total free hydroxyl groups, part of which can be attributed to sugar hydroxyl groups. The 31P NMR spectra showed that the switchgrass lignin had a sharp and prominent signal of p-hydroxyl phenolic OH, which was mostly attributed from the p-coumarate substructures in switchgrass lignin. Among the free phenolic hydroxyls, the p-hydroxyl was observed to be the most prominent type (0.58 mmol/g in transgenic and 0.70 mmol/g in control), which accounted for 9.1% in transgenic lignin and 11.7% in wild-type control, respectively. Compared to the wild-type, the transgenic switchgrass lignin appeared to have an increase (ca. 8.9%) in aliphatic OH group content. The total amount of free phenolic OH groups in COMT transgenic switchgrass lignin appeared to be comparable with that in the wild-type control, while the amount of p-hydroxyl phenolic OH was observed to decrease in COMT transgenic lignin, in line with the HSQC results that COMT down-regulation resulted in a decrease in lignin p-coumarate substructure. In addition, both lignin samples had a small amount of free carboxylic hydroxyl groups and their contents remained relatively unchanged after COMT down-regulation.
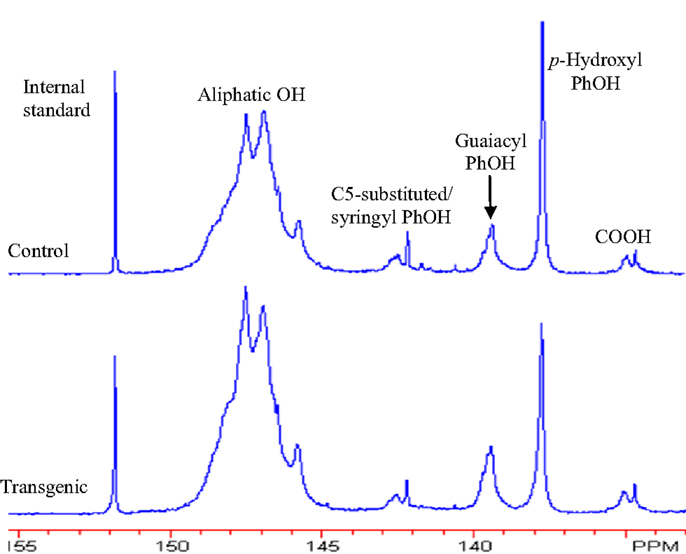
Figure 6. 31P NMR spectra of lignin samples isolated from wild-type and COMT transgenic switchgrass. Top: control; bottom: transgenic.
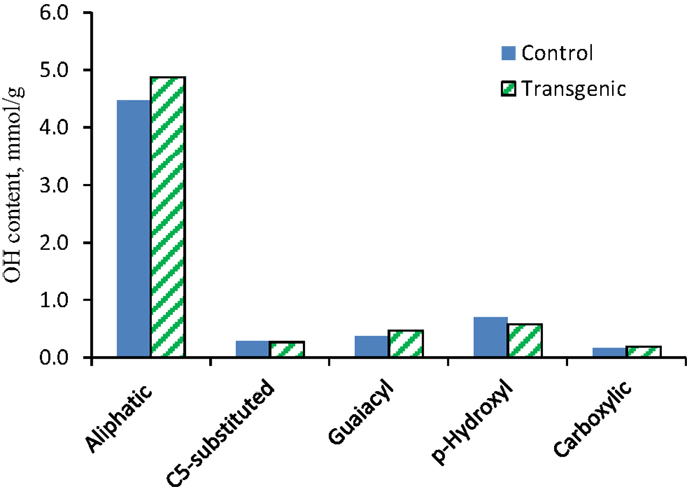
Figure 7. Hydroxyl group contents (millimole per gram) in lignin samples isolated from the wild-type and COMT transgenic switchgrass as determined by 31P NMR analysis.
GPC Characterization
To further understand the effects of COMT down-regulation on the lignin structures in the lignin biosynthesis process, GPC analysis was used to determine the molecule size and molecular weight distribution of lignin. Figure 8 provided the GPC chromatograms of lignin samples isolated from the wild-type and COMT transgenic switchgrass. The number-average molecular weight (Mn) and weight-average molecular weight (Mw) of the lignin fractions were determined by GPC and the results were summarized in Figure 9. Figure 8 showed that the lignin samples demonstrated a similar shape of molecular weight distribution curve, although the COMT transgenic lignin had a slightly higher portion in the low molecular weight range. The GPC analysis showed that the COMT transgenic lignin had comparable molar mass results compared to the wild-type control (Figure 9), indicating that COMT down-regulation under the conditions in this study appeared to have no significant impact on the molecule size and molecular weight distribution of lignin.
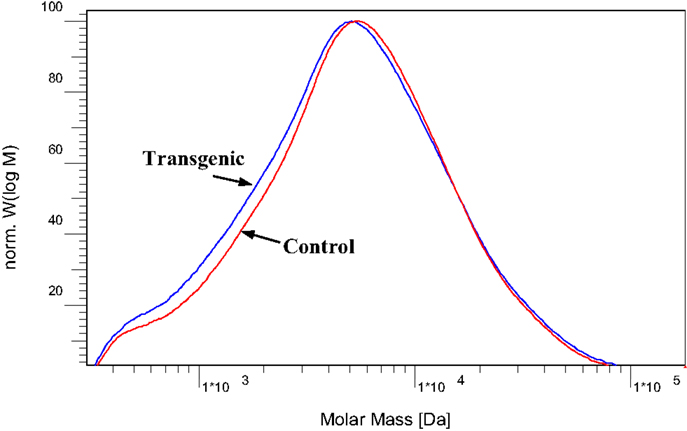
Figure 8. Molecular weight distribution curves of acetylated lignin isolated from the wild-type and COMT transgenic switchgrass.
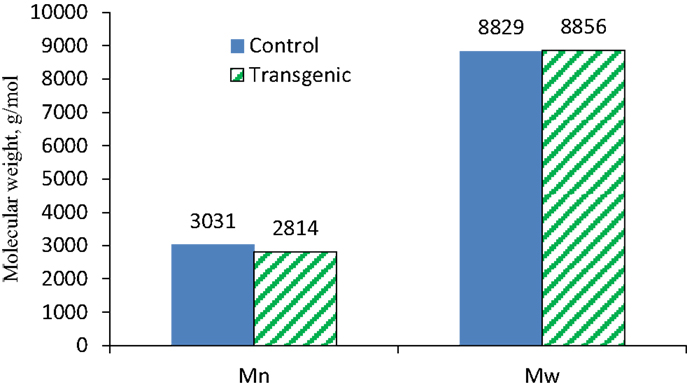
Figure 9. The number-average (Mn) and weight-average (Mw) molecular weights of lignin isolated from the wild-type and COMT transgenic switchgrass as determined by GPC analysis.
Lignin Related Recalcitrance Reduction
To date, several plants have been investigated for the effects of COMT down-regulation on lignin content, structure, and subsequent digestibility of these transgenic lines. For instance, COMT down-regulation of lignin in poplar has resulted in a decrease in lignin content, formation of benzodioxane, and a substantial decrease in syringyl units (Jouanin et al., 2000; Lu et al., 2010). In alfalfa, COMT down-regulation was reported to impact a marginal decrease in lignin content, a slight reduction in guaiacyl units, nearly total loss of syringyl units and the development of minor amount of 5-hydroxy guaiacyl units (Guo et al., 2001). Similarly, Sewalt et al. (1997) reported that COMT down-regulated tobacco had a decrease in S/G ratio and no change in lignin content and exhibited an improved cell wall digestibility. In COMT down-regulated transgenic maize, a strong decrease in lignin content, a decrease in S, H, and p-coumaric acid units, and the occurrence of 5-OH-guaiacyl units were reported to lead to improved digestibility (Piquemal et al., 2002). The lignin related changes observed in this study, such as the decrease in S/G and pCA:FA ratios and a decrease in total lignin content, might collectively contribute, in part, to the reduced recalcitrance in the COMT down-regulated switchgrass.
Conclusion
The COMT down-regulated transgenic switchgrass demonstrated a decrease in lignin content and structural alterations compared to the wild-type. A decreases in both S/G and p-coumarate:ferulate ratios, an increase in the relative abundance of phenylcoumaran units as well as the incorporation of benzodioxane units into lignin were observed in the transgenic switchgrass lignin. In addition, the COMT transgenic switchgrass lignin had an increase (ca. 8.9%) in aliphatic OH group content, while the total amount of free phenolic OH groups appeared to be comparable with that in the wild-type control. No significant effects of COMT down-regulation on the lignin molecular weights were observed. The reduction in recalcitrance of transgenic switchgrass appeared to be related, in part, to a combination of alterations in lignin content and its structures resulting from the COMT down-regulation.
Conflict of Interest Statement
The authors receive funding in this field of research.
Acknowledgments
This work was partially supported and performed as part of the BioEnergy Science Center (BESC). The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
References
Cao, S., Pu, Y., Studer, M., Wyman, C., and Ragauskas, A. J. (2012). Chemical transformations of Populus trichocarpa during dilute acid pretreatment. RSC Adv. 2, 10925–10936. doi: 10.1039/c2ra22045h
Capanema, E. A., Balakshin, M. Y., and Kadla, J. F. (2004). A comprehensive approach for quantitative lignin characterization by NMR spectroscopy. J. Agric. Food Chem. 52, 1850–1860. doi:10.1021/jf035282b
Capanema, E. A., Balakshin, M. Y., and Kadla, J. F. (2005). Quantitative characterization of a hardwood milled wood lignin by nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 53, 9639–9649. doi:10.1021/jf0515330
Chang, H. M., Cowling, E. B., Brown, W., Adler, E., and Miksche, G. (1975). Comparative studies on cellulolytic enzyme lignin and milled wood lignin of sweetgum and spruce. Holzforschung 29, 153–159. doi:10.1515/hfsg.1975.29.5.153
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi:10.1038/nbt1316
Chen, L., Auh, C.-K., Dowling, P., Bell, J., Lehmann, D., and Wang, Z.-Y. (2004). Transgenic down-regulation of caffeic acid O-methyltransferase (COMT) led to improved digestibility in tall fescue (Festuca arundinacea). Funct. Plant Biol. 31, 235–245. doi:10.1071/FP03254
Davis, M. W. (1998). A rapid modified method for compositional carbohydrate analysis of lignocellulosics by high pH anion-exchange chromatography with pulsed amperometric detection (HPAEC/PAD). J. Wood Chem. Technol. 18, 235–252. doi:10.1080/02773819809349579
Fu, C., Mielenz, J. R., Xiao, X., Ge, Y., Hamilton, C. Y., Rodriguez, M., et al. (2011). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 9, 3803–3808. doi:10.1073/pnas.1100310108
Guo, D., Chen, F., Inoue, K., Blount, J. W., and Dixon, R. A. (2001). Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88. doi:10.2307/3871154
Hallac, B. B., Pu, Y., and Ragauskas, A. J. (2010). Chemical transformations of Buddleja davidii lignin during ethanol organosolv pretreatment. Energy Fuels 24, 2723–2732. doi:10.1021/ef901556u
Higuchi, T., Ito, Y., and Kawamura, I. (1967). p-Hydroxyphenylpropane component of grass lignin and role of tyrosine-ammonia lyase in its formation. Phytochemistry 6, 875–881. doi:10.1016/S0031-9422(00)86035-5
Himmel, M. E., Ding, S.-Y., Johnson, D. K., Adney, W. S., Nimlos, M. R., Brady, J. W., et al. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuel production. Science 315, 804–807. doi:10.1126/science.1137016
Hu, Z., Yeh, T., Chang, H. M., Matsumoto, Y., and Kadla, J. F. (2006). Elucidation of the structure of cellulolytic enzyme lignin. Holzforschung 60, 389–397. doi:10.1021/jf2037399
Jiang, N., Pu, Y., Samuel, R., and Ragauskas, A. J. (2009). Perdeuterated pyridinium molten salt (ionic liquid) for direct dissolution and NMR analysis of plant cell walls. Green Chem. 11, 1762–1766. doi:10.1039/b913609f
Jiang, N., Pu, Y., Samuel, R., and Ragauskas, A. J. (2010). Direct dissolution and NMR analysis of the plant cell walls via perdeuterated pyridinium-based ionic liquid. ChemSusChem 3, 1285–1289. doi:10.1002/cssc.201000120
Jouanin, L., Goujon, T., Nadai, V., Martin, M. T., Mila, I., Vallet, C., et al. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123, 1363–1373. doi:10.1104/pp.123.4.1363
Kim, H., and Ralph, J. (2010). Solution state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org. Biomol. Chem. 8, 576–591. doi:10.1039/b916070a
Kim, H., Ralph, J., and Akiyama, T. (2008). Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. Bioenergy Res. 1, 56–66. doi:10.1039/b916070a
Leu, S.-Y., and Zhu, J. Y. (2013). Substrate-related factors affecting enzymatic saccharification of lignocelluloses: our recent understanding. Bioenerg. Res. 6, 405–415. doi:10.1007/s12155-012-9276-1
Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J. M., Ralph, J., et al. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U.S.A. 100, 4939–4944. doi:10.1073/pnas.0831166100
Li, M., Foster, C., Kelkar, S., Pu, Y., Holmes, D., Ragauskas, A. J., et al. (2012). Structural characterization of alkaline hydrogen peroxide pretreated grasses exhibiting diverse lignin phenotypes. Biotechnol. Biofuels 5, 38–52. doi:10.1186/1754-6834-5-38
Lu, F., Marita, J. M., Lapierre, C., Jouanin, L., Morreel, K., Boerjan, W., et al. (2010). Sequencing around 5-hydroxyconiferyl alcohol-derived units in caffeic acid O-methyltransferase-deficient poplar lignins. Plant Physiol. 153, 569–579. doi:10.1104/pp.110.154278
Mansfield, S. D., Kim, H., Lu, F., and Ralph, J. (2012). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 9, 1579–1589. doi:10.1038/nprot.2012.064
Marita, J. M., Ralph, J., Hatfield, R. D., and Chapple, C. (1999). NMR characterization of lignins in Arabidopsis altered in the activity of ferulate 5-hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 96, 12328–12332. doi:10.1073/pnas.96.22.12328
Marita, J. M., Ralph, J., Lapierre, C., Jouanin, L., and Boerjan, W. (2001). NMR characterization of lignins from transgenic poplars with suppressed caffeic acid O-methyltransferase activity. J. Chem. Soc. Perkin Trans. I 1, 2939–2945. doi:10.1039/b107219f
Moore, K. J., Moser, L. E., Vogel, K. P., Waller, S. S., Johnson, B. E., and Pedersen, J. F. (1991). Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83, 1073–1077. doi:10.2134/agronj1991.00021962008300060027x
Piquemal, J., Chamayou, S., Nadaud, I., Beckert, M., Barriere, Y., Mila, I., et al. (2002). Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol. 130, 1675–1685.
Pu, Y., Cao, S., and Ragauskas, A. J. (2011a). Application of quantitative 31P NMR in biomass lignin and biofuel precursors characterization. Energy Environ. Sci. 4, 3154–3166. doi:10.1039/c1ee01201k
Pu, Y., Chen, F., Ziebell, A., Davison, B. H., and Ragauskas, A. J. (2009). NMR characterization of C3H and HCT down-regulated alfalfa lignin. Bioenergy Res. 2, 198–208. doi:10.1007/s12155-009-9056-8
Pu, Y., Hu, F., Huang, F., Davison, B. H., and Ragauskas, A. J. (2013). Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol. Biofuels 6, 15–27. doi:10.1186/1754-6834-6-15
Pu, Y., Kosa, M., Kalluri, U., Tuskan, G., and Ragauskas, A. J. (2011b). Challenges of the utilization of wood polymers: how can they be overcome? Appl. Microbiol. Biotechnol. 91, 1525–1536. doi:10.1007/s00253-011-3350-z
Pu, Y., Samuel, R., and Ragauskas, A. J. (2005). Structural analysis of acetylated hardwood lignins and their photoyellowing properties. Can. J. Chem. 83, 2132–2139. doi:10.1139/v05-231
Pu, Y., Zhang, D., Singh, P. M., and Ragauskas, A. J. (2008). The new forestry biofuels sector. Biofuels Bioprod. Biorefin. 2, 58–73. doi:10.1002/bbb.48
Ralph, J., Lapierre, C., Lu, F., Marita, J. M., Pilate, G., Van Doorsselaere, J., et al. (2001). NMR evidence for benzodioxane structures resulting from incorporation of 5-hydroxyconiferyl alcohol into lignins of O-methyltransferase-deficient poplars. J. Agric. Food Chem. 49, 86–91. doi:10.1021/jf010721x
Ralph, S. A., Ralph, J., and Landucci, L. (2004). NMR database of lignin and cell wall model 497 compounds. Available at: http://ars.usda.gov/Services/docs.htm?docid=10491
Samuel, R., Foston, M., Jiang, N., Cao, S., Allison, L., Studer, M., et al. (2011). HSQC (heteronuclear single quantum coherence) 13C-1H correlation spectra of whole biomass in perdeuterated pyridinium chloride-DMSO system: an effective tool for evaluating pretreatment. Fuel 90, 2836–2842. doi:10.1016/j.fuel.2011.04.021
Sewalt, V. J. H., Ni, W., Jung, H. G., and Dixon, R. A. (1997). Lignin impact on fiber degradation: increased enzymatic digestibility of genetically engineered tobacco (Nicotiana tabacum) stems reduced in lignin content. J. Agric. Food Chem. 45, 1977–1983. doi:10.1021/jf9609690
Shen, H., Fu, C., Xiao, X., Ray, T., Tang, Y., Wang, Z., et al. (2009). Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. Bioenergy Res. 2, 233–245. doi:10.1007/s12155-009-9058-6
Shen, H., He, X., Poovaiah, C. R., Wuddineh, W. A., Ma, J., Mann, D. G., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136. doi:10.1111/j.1469-8137.2011.03922.x
Somerville, C., Youngs, H., Taylor, C., Davis, S. C., and Long, S. P. (2010). Feedstocks for lignocellulosic biofuels. Science 329, 790–792. doi:10.1126/science.1189268
Stewart, J. J., Akiyama, T., Chapple, C., Ralph, J., and Mansfield, S. D. (2009). The effects on lignin structure of over expression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150, 621–635. doi:10.1104/pp.109.137059
Sun, Y., and Cheng, J. (2002). Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour. Technol. 83, 1–11. doi:10.1016/S0960-8524(01)00212-7
Tschaplinski, T. J., Standaert, R. F., Engle, N. L., Martin, M. Z., Sangha, A. K., Parks, J. M., et al. (2012). Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol. Biofuels 5, 71–85. doi:10.1186/1754-6834-5-71
Tu, Y., Rochfort, S., Liu, Z., Ran, Y., Griffith, M., Badenhorst, P., et al. (2010). Functional analysis of a caffeic acid O-methyltransferase from perennial ryegrass. Plant Cell 22, 3357–3373. doi:10.1105/tpc.110.077578
Vanholme, R., Ralph, J., Akiyama, T., Lu, F., Pazo, J. R., Kim, H., et al. (2010). Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down regulation of COMT in Arabidopsis. Plant J. 64, 885–897. doi:10.1111/j.1365-313X.2010.04353.x
Wang, M., Wu, M., and Huo, H. (2007). Life-cycle energy and greenhouse gas emission impacts of different corn ethanol plant types. Environ. Res. Lett. 2, 1–13. doi:10.1088/1748-9326/2/2/024001
Weng, J. K., Li, X., Bonawitz, N. D., and Chapple, C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172. doi:10.1016/j.copbio.2008.02.014
Wyman, C. E. (1999). Biomass ethanol: technical progress, opportunities and commercial challenges. Annu. Rev. Energy Environ. 24, 189–226. doi:10.1146/annurev.energy.24.1.189
Wyman, C. E., Dale, B. E., Elander, R. T., Holtzapple, M., Ladisch, M. R., and Lee, Y. Y. (2005). Coordinated development of leading biomass pretreatment technologies. Bioresour. Technol. 96, 1959–1966. doi:10.1016/j.biortech.2005.01.010
Yelle, D. J., Ralph, J., and Frihart, C. R. (2008). Characterization of nonderivatized plant cell walls using high-resolution solution-state NMR spectroscopy. Magn. Reson. Chem. 46, 508–517. doi:10.1002/mrc.2201
Zhang, L., and Gellerstedt, G. (2007). Quantitative 2D HSQC NMR determination of polymer structures by selecting suitable internal standard references. Magn. Reson. Chem. 45, 37–45. doi:10.1002/mrc.1914
Keywords: switchgrass, COMT transgenic, lignin, NMR, molecular weights
Citation: Samuel R, Pu Y, Jiang N, Fu C, Wang Z-Y and Ragauskas A (2014) Structural characterization of lignin in wild-type versus COMT down-regulated switchgrass. Front. Energy Res. 1:14. doi: 10.3389/fenrg.2013.00014
Received: 10 November 2013; Accepted: 28 December 2013;
Published online: 20 January 2014.
Edited by:
Soo-Jeong Shin, Chungbuk National University, South KoreaReviewed by:
JunYong Zhu, Forest Products Laboratory, USALuis Serrano, University of the Basque Country, Spain
Copyright: © 2014 Samuel, Pu, Jiang, Fu, Wang and Ragauskas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunqiao Pu, Institute of Paper Science and Technology, Georgia Institute of Technology, 500 10th Street Northwest, Atlanta, GA 30332, USA e-mail: yunqiao.pu@ipst.gatech.edu;
Arthur Ragauskas, School of Chemistry and Biochemistry, Georgia Institute of Technology, 500 10th Street Northwest, Atlanta, GA 30332, USA e-mail: arthur.ragauskas@chemistry.gatech.edu
 Reichel Samuel1,2
Reichel Samuel1,2
 Yunqiao Pu
Yunqiao Pu Zeng-Yu Wang
Zeng-Yu Wang Arthur Ragauskas
Arthur Ragauskas