- 1Auckland Cancer Society Research Centre, University of Auckland, Auckland, New Zealand
- 2Department of Molecular Medicine and Pathology, University of Auckland, Auckland, New Zealand
The majority of the human genome is transcribed, even though only 2% of transcripts encode proteins. Non-coding transcripts were originally dismissed as evolutionary junk or transcriptional noise, but with the development of whole genome technologies, these non-coding RNAs (ncRNAs) are emerging as molecules with vital roles in regulating gene expression. While shorter ncRNAs have been extensively studied, the functional roles of long ncRNAs (lncRNAs) are still being elucidated. Studies over the last decade show that lncRNAs are emerging as new players in a number of diseases including cancer. Potential roles in both oncogenic and tumor suppressive pathways in cancer have been elucidated, but the biological functions of the majority of lncRNAs remain to be identified. Accumulated data are identifying the molecular mechanisms by which lncRNA mediates both structural and functional roles. LncRNA can regulate gene expression at both transcriptional and post-transcriptional levels, including splicing and regulating mRNA processing, transport, and translation. Much current research is aimed at elucidating the function of lncRNAs in breast cancer and mammary gland development, and at identifying the cellular processes influenced by lncRNAs. In this paper we review current knowledge of lncRNAs contributing to these processes and present lncRNA as a new paradigm in breast cancer development.
Introduction
Ribonucleic acids were long thought to act mainly as carriers of genetic information between DNA and the protein-synthesizing machinery of the cell. Whole genome technology, including deep sequencing and microarray analyses, has demonstrated that coding regions constitute only 2% of the human genome. On the other hand, 70–90% of the genome is transcribed (Bertone et al., 2004; Carninci et al., 2005; Birney et al., 2007; Kapranov et al., 2010; Djebali et al., 2012). Initially regarded as ‘junk’, these transcripts have been found to possess many regulatory roles and represent a largely unexplored area of cell biology.
Non-coding RNAs (ncRNAs) are defined as those RNAs which are transcribed from the genome but do not encode proteins. They are divided into two broad classes based on their function: housekeeping and regulatory. Housekeeping ncRNAs include ribosomal, transfer, small nuclear, and small nucleolar RNAs (snoRNAs). These ncRNAs are constitutively expressed and crucial to the operation of vital cellular functions. Regulatory ncRNAs are differentially expressed according to tissue and stage of development or disease process. These regulatory ncRNAs are divided into two broad classes based on their size. The first class of regulatory ncRNAs comprises short transcripts <200 nt long and includes microRNA (miRNA), small interfering RNA (siRNA), piwi interacting RNA (piRNA), transcription initiation RNA (tiRNA), and small cajal body-specific RNA (scaRNA). They control a number of developmental and physiological pathways and have been shown to be deregulated in disease processes (Mattick and Makunin, 2006; Carthew and Sontheimer, 2009). The second class of regulatory ncRNAs is long ncRNAs (lncRNA), which is defined as those transcripts >200 nt. Despite making up the majority of the human transcriptome, they were initially dismissed as products of leaky transcription or evolutionary noise due to their lack of open reading frames and poor sequence conservation. However, lncRNAs have now been found to display key characteristics that confirm their roles as functional molecules, and a significant number of papers has been published in recent years focusing on their function in normal development and disease. They are mainly transcribed by RNA polymerase II and the genomic loci of many of them contain chromatin marks, consistent with those of transcribed genes, at their promoters and gene bodies such as H3K9ac, H3K4me3, and H3K36me3. They are also frequently polyadenylated and undergo splicing of exons, demonstrating that they require post-transcriptional processing to form mature transcripts. Furthermore, lncRNA expression is regulated by well characterized transcription factors (reviewed in Guttman and Rinn, 2012).
In general, lncRNAs show poor sequence conservation, due to their rapid evolution, but display conserved tissue specificity and function (Necsulea et al., 2014). Recent work has provided information on the origin and evolution of lncRNAs, suggesting that lncRNA emerge de novo from transposable elements or non-exonic sequences (Derrien et al., 2012; Kapusta et al., 2013). Sequences conserved between different species might be expected to show conserved functions, but a recent study has shown significant differences in stem cell-specific transcription factor binding sites between mouse and human (Odom et al., 2007). This observation suggests that mouse models cannot always be used for studying human lncRNA. Conversely, many lncRNAs display conserved structure between different species while having poorly conserved sequences (Torarinsson et al., 2006). These features of lncRNAs support the hypothesis that they perform biologically robust functions.
Long ncRNAs are further divided into several classes based on their genomic orientation as illustrated in Figure 1 (Ponting et al., 2009). Despite their abundance, the functions of the majority of lncRNAs have yet to be elucidated. Studies of lncRNA mechanism have revealed extensive variety in their function. LncRNAs can act as transcriptional regulators, acting either in cis to regulate nearby genes, or in trans to regulate distal genes (Osato et al., 2007; Wang and Chang, 2011; Bak and Mikkelsen, 2014).
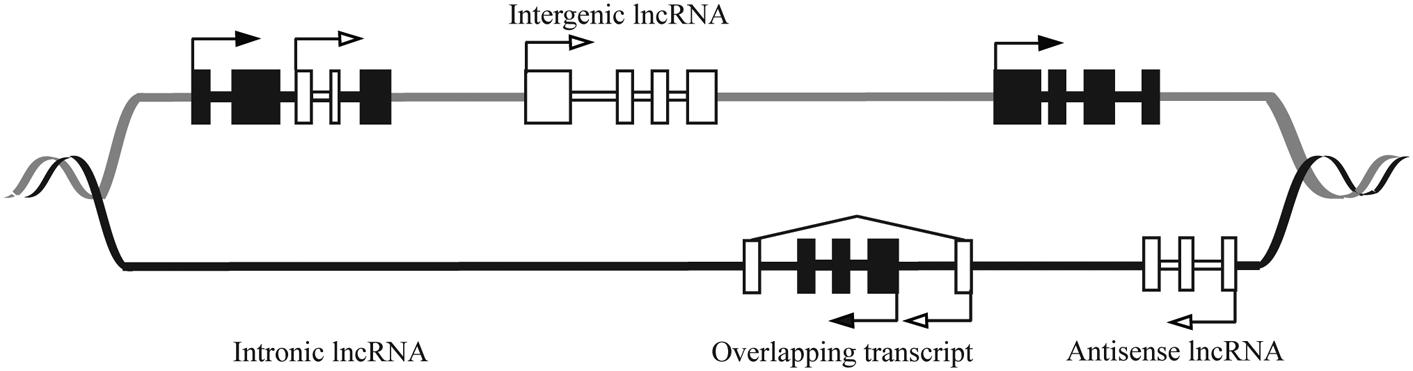
FIGURE 1. Schematic showing genomic organization of different lncRNAs. The DNA strands are shown in black and gray lines. The black and white boxes represent exons of protein coding and lncRNA genes, respectively. LncRNAs are classified as sense (e.g., Intronic or overlapping) or antisense, reflecting the way they overlap with protein coding genes in the same or opposite direction, respectively. The diagram shows that lncRNAs can be antisense to protein coding (antisense lncRNA) or originate from introns of protein coding genes either as by-products of mRNA or as independent transcripts (intronic lncRNA, sense lncRNA). LncRNA also can be transcribed from intergenic regions of the genome (lincRNA). LncRNA genes can overlap the protein-coding genes (overlapping transcript).
The roles of lncRNA in transcription regulation, mRNA processing, and mRNA maturation are depicted in Figures 2A–D. LncRNAs have a broad role in epigenetic control that goes beyond gene regulation at the transcriptional level. Most lncRNA species were previously thought to be restricted to the nucleus, but recent studies have found that a larger proportion than expected is located in the cytoplasm (van Heesch et al., 2014), suggesting post-transcriptional gene regulation, protein shuttling, and localization of proteins and transcripts (Figures 2E–G). A large number of lncRNAs associate with actively translating ribosomes (Guttman et al., 2013), indicating that lncRNA regulation of translation may be a common phenomenon, although the exact mechanisms are yet to be understood.
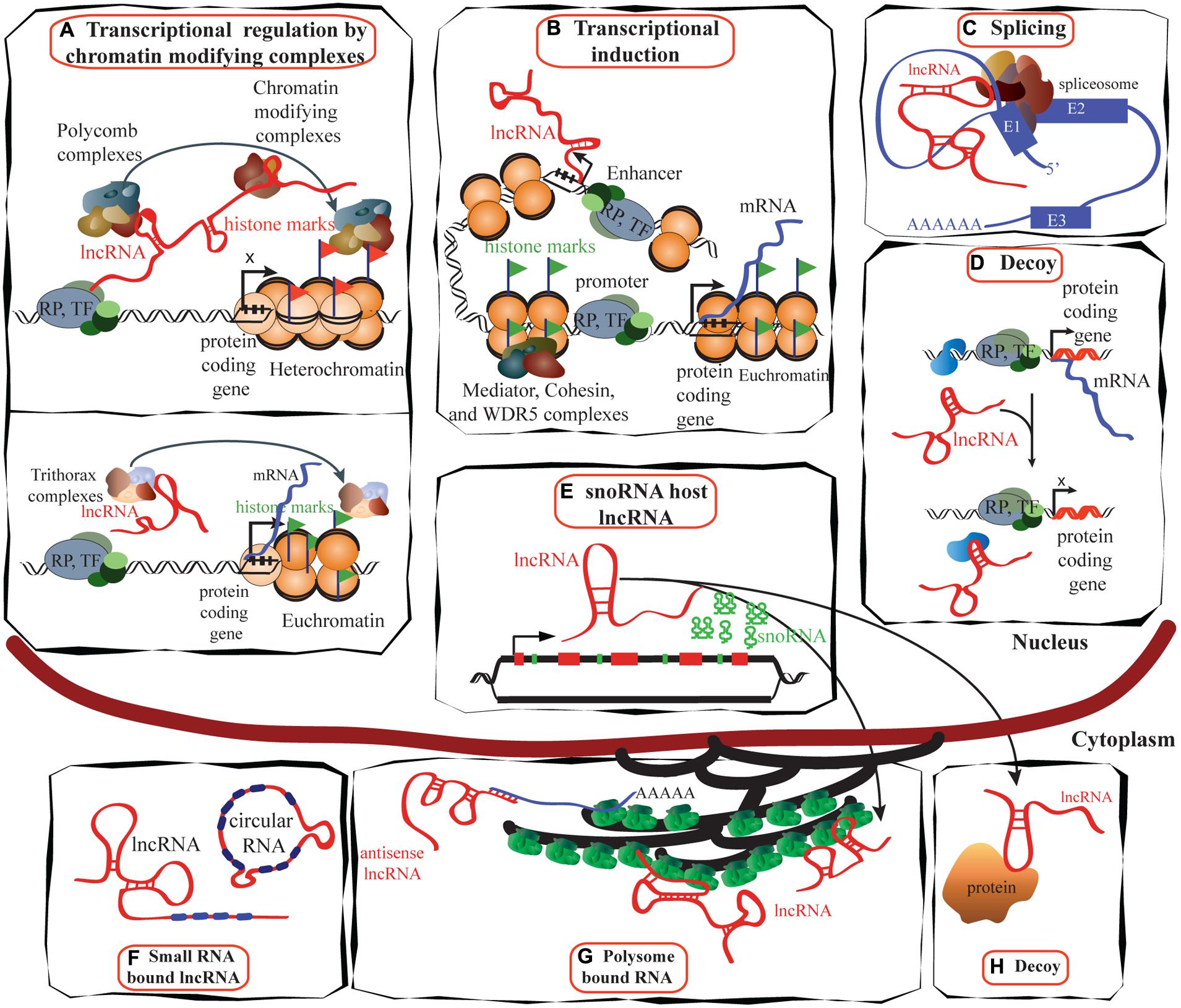
FIGURE 2. Schematic illustrating the functions of lncRNAs. (A) LncRNAs (e.g., HOTAIR) can suppress transcription by interacting with PRC1 and PRC2 complexes as well as with other chromatin modifying proteins, maintaining heterochromatin status and suppressing transcription (reviewed in Gibb et al., 2011; upper panel). Trithorax complexes also can interact with lncRNA (e.g., HOTTIP) and induce transcription (lower panel; reviewed in Jobe et al., 2012). (B) LncRNAs are proposed to be transcribed at enhancer regions and can function in the establishment and maintenance of enhancer–promoter looping and activation of gene expression (Orom and Shiekhattar, 2013). (C) LncRNAs such as MALAT1 can regulate alternative splicing by interacting with the spliceosomal machinery (Tripathi et al., 2013). (D) Specific lncRNAs are transcribed and bind to and titrate away protein factors. Decoy lncRNAs can bind to protein factors such as transcription factors and chromatin modifiers. This leads to broad changes in transcriptomes. (E) Intronic regions of many lncRNAs host snoRNAs. The snoRNAs derived from these lncRNAs remain in the nucleus while the spliced transcript can move to the cytoplasm and bind either to polysomes or to other proteins (Smith and Steitz, 1998). (F) LncRNAs can bind to miRNAs to sequester them and inactivate their repressive functions (reviewed in Gibb et al., 2011). (G) Many lncRNAs are associated with polysomes (van Heesch et al., 2014), while the mechanisms of many are yet to be identified, antisense lncRNAs such as UCHL1AS regulate the translation of associated mRNAs. (H) Decoy lncRNAs such as GAS5 are present in both in the cytoplasm and the nucleus: GAS5 translocates from the cytoplasm into the nucleus with glucocorticoid receptor in response to dexamethasone (Kino et al., 2010). TF, transcription factor; RP, RNA polymerase; E1, exon1.
Although only a few lncRNAs have been studied in detail, their characteristic spatial and temporal expression patterns indicate that they play significant but diverse roles in development (Guttman et al., 2011; Ng et al., 2012; Pauli et al., 2012). Furthermore, numerous studies have shown altered expression of lncRNAs in cancer cells compared to normal tissues (Perez et al., 2007; Du et al., 2013; Qiu et al., 2013), indicating that deregulation of these lncRNAs can facilitate disease progression. This review will explore, in particular, the role of lncRNAs in breast development and cancer.
LncRNAs as Key Regulatory Molecules in Development and Cancer
Development of an organism is a complex process, requiring finely regulated gene expression to determine the precise function and architecture of differentiated tissues. Widespread transcription of the human genome and the emergence of lncRNAs as regulatory molecules suggest that development is regulated not only by proteins but also by an expanded network coordinated by RNAs (reviewed in Mattick, 2007). Microarray analyses of embryonic stem cells have revealed many differentially expressed lncRNAs during induced differentiation, confirming their key role in embryonic stem cell development (Dinger et al., 2008). Several of these lncRNAs are transcribed from regions near developmental genes, indicating a complex relationship between protein coding genes and non-protein coding genes in fine tuning development (Dinger et al., 2008). Systematic knockdown of lncRNAs in mouse embryonic stem cells has been shown to have major effects on gene expression patterns, leading to loss of pluripotency and loss of repression of lineage commitment programs. Additionally, the majority of lncRNAs examined interact with transcription factors associated with pluripotency, and ∼30% interact with chromatin (Guttman et al., 2011). The expression of pseudogenes of embryonic transcription factors could facilitate cancer progression. For example, Oct4 has four related pseudogenes that are found to be expressed in glioma and breast cancer, while Oct4 is not expressed (Zhao et al., 2011). Such interactions suggest that lncRNAs act as components of networks that include established pluripotency factors and DNA to maintain the undifferentiated replicative state of embryonic stem cells. Chromosome architecture and epigenetic memory are regulated by lncRNAs, which recruit histone modifying complexes and DNA methyltransferases to certain loci, maintaining correct gene expression (reviewed in Mattick, 2007; Amaral et al., 2008).
Alterations of lncRNAs involved in cell homeostatic processes such as cell cycle, DNA damage response, survival, proliferation, and migration have been identified in carcinogenesis. Multiple lncRNAs regulate or are regulated by key cancer pathways, such as the p53 pathway, a master regulator of cell cycle and survival. These include lincRNA p21 and PANDA, which are upregulated upon DNA damage and are induced in a p53-dependent manner (Huarte et al., 2010). PANDA interacts with the transcription factor NF-YA to limit expression of proapoptotic genes (Hung et al., 2011). Thus, p53 protein not only induces the activity of lncRNAs to mediate its effects, but is itself regulated by lncRNAs. MEG3 regulates the activity of p53 by facilitating p53 binding and activation of p53 target gene promoters and inhibiting cellular proliferation (Zhou et al., 2007). LncRNAs affect every hallmark of cancer progression (reviewed in Gutschner and Diederichs, 2012) highlighting the shift from a solely proteomic view of tumor biology to one that includes an extensive network of lncRNAs. Here the roles of lncRNAs involved in mammary gland development and the nature of their deregulation in breast cancer development are discussed.
Role of lncRNAs in Breast Development and Cancer
Most organs develop before or around birth, but the mammary gland begins its development in the embryo and continues its growth and differentiation in puberty and adult life (Smith et al., 2012). Mammary gland development in the embryo occurs during mid-gestation and remains quiescent until puberty in which there is further development of the ducts. During pregnancy, the breast tissue undergoes further expansion and branching, and luminal cells rapidly proliferate and differentiate to milk-secreting alveolar cells. These changes underlie the production of milk during lactation. Cessation of lactation is accomplished by involution, in which the breast tissue undergoes extensive apoptosis to return to the quiescent pre-pregnancy state (Watson and Khaled, 2008).
Breast development is orchestrated by a series of genes that regulate these different cellular processes. Alterations to genes encoding growth factors, nuclear transcription factors, cell cycle regulatory proteins and tumor suppressor proteins have been suggested to create favorable conditions to allow the outgrowth of cells. A major focus in breast cancer research is to understand how normal breast epithelium is transformed into malignant epithelium. With the advent of genome-wide sequencing, it became apparent that it is not solely genetic changes that contribute to breast cancer. Epigenetic changes also play a major part in altering gene expression in such a way as to contribute to cancer (Jones and Baylin, 2007). The role of coding genes in the regulation of breast development is well established (Hennighausen and Robinson, 2001; Anderson et al., 2007; Lim et al., 2010), but less researched is the role of lncRNAs. Large scale microarray analyses using mouse mammary gland RNA from different stages of mammary gland development (Askarian-Amiri et al., 2011), tiling arrays (Perez et al., 2008), and transcriptome profiling (Maruyama et al., 2012) of normal tissues compared to breast cancer tissues, have revealed several lncRNAs which are aberrantly expressed in neoplastic tissues. We describe several lncRNAs that have been implicated in breast cancer development, and describe molecular mechanisms by which they may contribute to normal development and cancer progression (Figure 3).
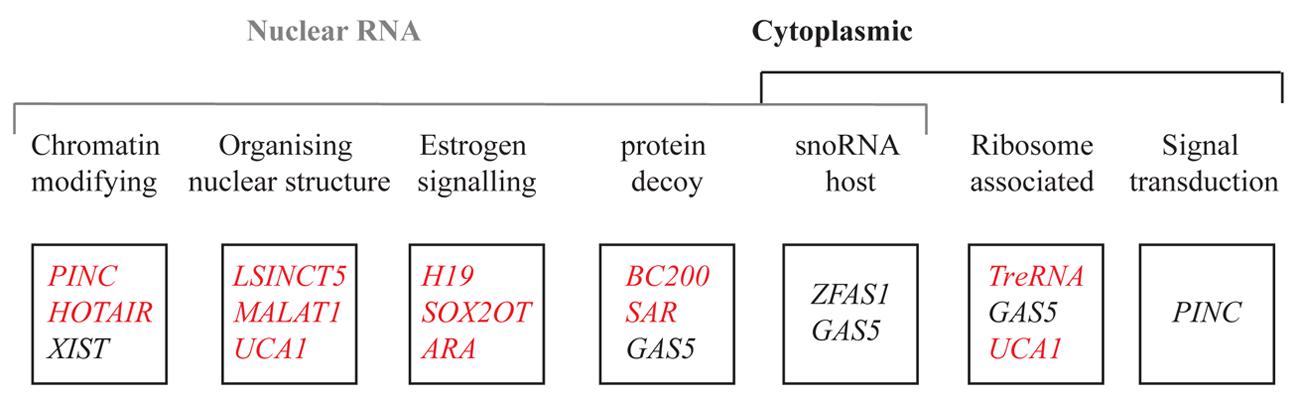
FIGURE 3. List of lncRNAs reported in this review based on their localization, function, and expression pattern. The function of each lncRNA is above each box. The red and black represent the genes upregulated and downregulated respectively in breast cancer.
Chromatin Remodeling Complexes
Of those lncRNAs with identified mechanisms, it is apparent that many function as regulators of transcription through chromatin regulation. These lncRNAs act as scaffolds, interacting with target genes and recruiting factors such as histone modifying complexes for gene silencing (reviewed in Rinn and Chang, 2012). The most commonly identified histone modifying complexes that lncRNAs interact with are polycomb group proteins. PcG proteins form multisubunit complexes; the best characterized are polycomb repressor complex 1 (PRC1) and polycomb repressor complex 2 (PRC2), which modify histones by inducing repressive chromatin marks. PRC1 catalyzes ubiquitination of lysine 119 on histone 2A (H2AUb1) while PRC2 methylates lysine 27 on histone 3 (H3K27me2/3). PRC2 is required for the initial targeting of specific gene silencing, whereas PRC1 maintains the gene silencing initiated by PRC2 (Morey and Helin, 2010). LncRNAs also interact with other chromatin modifying complexes, such as the LSD1–CoREST complex, which mediates the removal of H3K4 mono and dimethylation, a chromatin mark associated with gene activation (Rinn et al., 2007). Other chromatin modifying complexes with which lncRNAs associate include G9a (Nagano et al., 2008; Pandey et al., 2008), a H3K9 methyltransferase, and MLL1 (Bertani et al., 2011; Wang et al., 2011), a histone methyltransferase, which associates with activating trithorax complex 1. Through interacting with chromatin modifying complexes, lncRNAs act as guides to target specific gene regions to modify transcription either in cis or trans (Wang and Chang, 2011). This may be the predominant method in which lncRNAs exert their function, as global analyses have found that a large number of lncRNAs bind to PRC2 and LSD1 (Khalil et al., 2009).
This mechanism of gene regulation by lncRNAs ensures that appropriate genes are switched on or off. Several lncRNAs involved in cancer, including breast cancer, have been shown to promote tumorigenesis by inappropriately silencing genes through deregulated interactions with histone modifying complexes.
Pregnancy Induced Non-Coding RNA (PINC)
Pregnancy induced non-coding RNA (PINC) was identified initially in the rat mammary gland and exhibits conserved synteny in the genomes of other mammals. It is differentially expressed as pregnancy progresses (Ginger et al., 2006), highly expressed in the alveolar cells during pregnancy, but declines during the transition from late pregnancy to early lactation, in which the mammary epithelium terminally differentiates to form milk producing cells, and it is again elevated during involution. PINC inhibits differentiation, and when it is expressed at low levels, mammary cells are able to undergo differentiation (Shore et al., 2012).
Mouse PINC (mPINC) may exert these effects on the mammary gland by regulating cell cycle progression. In HC11 cells, mPINC relocates from the nucleus to the cytoplasm as the cell cycle progresses (Ginger et al., 2006), indicative of a role in nuclear–cytoplasmic signal transduction or shuttling. PINC interacts with retinoblastoma associated protein 46 (RbAp46), a component of PRC2, suggesting that PINC may exert its role in differentiation through modification of chromatin and that it acts to repress gene expression. Several potential target genes, known to be involved in mammary gland development, have been identified, and encode members of the Wnt and Notch signaling pathways (Shore et al., 2012). The role of PINC in human mammary gland development and breast cancer has yet to be ascertained.
HOX Transcript Antisense RNA (HOTAIR)
HOX transcript antisense RNA (HOTAIR) is upregulated in a number of different cancers, including breast (Gupta et al., 2010), colorectal (Kogo et al., 2011), hepatocellular (Geng et al., 2011), and gastrointestinal stromal tumors (Niinuma et al., 2012). In breast cancer, HOTAIR expression is upregulated in both primary and metastatic tumors, and its expression in primary tumors is strongly correlated with later metastases, patient prognosis, and death (Gupta et al., 2010).
HOTAIR is transcribed in an antisense orientation from the HOXC locus. It acts in trans as a repressor of the HOXD locus by recruiting PRC2, leading to trimethylation of H3K27 and subsequent transcriptional silencing. The bordering regions of HOXD are bound by CoREST/REST repressor complexes, which contain the H3K4me2-specific LSD1 demethylase and also maintain appropriate silencing of HOXD. HOTAIR binds to LSD1 at its 3′ domain, whereas the 5′ domain binds to EZH2 or SUZ12, components of the PRC2. It acts bifunctionally as a scaffold that bridges these two repressor complexes. This results in coordinate binding to their target genes for coupled chromatin remodeling (Tsai et al., 2010). Upregulation of HOTAIR leads to genome-wide reprogramming of the PRC2 occupancy pattern from that of breast epithelial cells to that resembling embryonic and neonatal fibroblasts, as well as altered gene expression, that includes the silencing of tumor suppressor genes JAM2 and PCDH (Gupta et al., 2010).
The promoter of HOTAIR contains multiple functional estrogen response elements near the transcription start site, and is transcriptionally induced by estrogen, while repressed by tamoxifen. A number of estrogen receptor coregulators also bind to the HOTAIR promoter region upon treatment with estrogen such as CBP/p300 and histone methyltransferases MLL1 and MLL3. In the basal state, HOTAIR is repressed, but estradiol treatment causes exchange of repressive molecules for transcriptional activators and coregulators, leading to gene activation (Bhan et al., 2013).
An alternative mechanism for HOTAIR’s function in breast cancer is through competitive binding with tumor suppressor genes. BRCA1 is an important breast cancer tumor suppressor gene that functions in DNA damage responses. BRCA1 binds to EZH2, a component of the PRC2 in mouse and human mammary epithelial cells, acting as a competitive inhibitor of HOTAIR. Reduced BRCA1 expression, which is often observed in cancer allows HOTAIR to bind to PRC2, promoting reprogramming of breast epithelial cells to aggressive cancer cells (Wang et al., 2013).
XIST
One of the best studied lncRNAs is XIST, transcribed from the X inactivation center of the X chromosome that is destined to be inactivated in female mammals (Brown et al., 1991), from whence it spreads along the X chromosome to promote heterochromatin formation and ultimately formation of the Barr body. This mechanism ensures dosage compensation with males. Loss of the Barr body is observed consistently in female cancers including breast cancer. Under normal conditions, two active X chromosomes are incompatible with human life, but activation of both X chromosomes may be advantageous to the cancer cell, as the X chromosome contains several oncogenes. XIST expression is lost in female cancers including breast, ovarian, and cervical cancer (reviewed in Froberg et al., 2013). XIST itself is under regulation by several lncRNAs such as Tsix (negatively; Leeb et al., 2009) and Jpx (positively; Tian et al., 2010), highlighting the complexity of the network by which lncRNAs function to induce X chromosome inactivation.
The mechanism by which XIST functions in breast cancer is controversial, with some studies showing interaction of the tumor suppressor gene BRCA1 with XIST, and that loss of BRCA1 is associated with loss of XIST. Upon reactivation of BRCA1, XIST staining by in situ hybridization was restored (Ganesan et al., 2002, 2004). Other studies have shown that XIST and BRCA1 do not physically co-localize and that reintroduction of BRCA1 did not reactivate XIST staining. The same discrepancy of XIST and BRCA1 is observed in human breast tumors, with some studies showing concurrent loss of both BRCA1 and XIST in highly aggressive breast cancers (Richardson et al., 2006) while other studies of breast cancer tissue from patients with germline BRCA1 mutations show expression of XIST in a significant number of samples (Vincent-Salomon et al., 2007). Whether XIST interacts with BRCA1 to regulate cancer progression or if genetic instability due to loss of BRCA1 expression is responsible for reduced XIST expression is yet to be clarified.
Protein Decoys
The ability of lncRNAs to form complex secondary structures gives them the capacity to present a diversity of interfaces for binding to proteins. LncRNAs may recruit or organize proteins as evidenced by their roles as chromatin modifiers, or act as decoys which inhibit protein functions. One such lncRNA, GAS5 which is discussed below, functions as a protein decoy by binding to the glucocorticoid receptor (GR) and preventing it from interacting with the glucocorticoid response element (GRE; Kino et al., 2010; Figure 2H).
BC200
BC200 is normally selectively expressed in brain tissue and germ cells. It exhibits a high degree of tissue specificity in the brain, showing expression in somatodendritic domains in a subset of neurons but not in other somatic cells. BC200 functions to repress translation through its oligo(A) rich region which is able to bind polyA binding protein (PABP), so that it is no longer able to bind translation initiation factors (Kondrashov et al., 2005).
This neuronal specificity is lost in a number of tumors, including breast cancer (Chen et al., 1997). BC200 expression is highly elevated in invasive tumors but not in benign tumors. Furthermore, high BC200 expression is associated with high nuclear grade, an indicator of aggressive cancer cell behavior. Not all DCIS lesions progress to invasive disease, and BC200 could be used as a prognostic indicator to assess the invasive potential in early breast tumors to ensure appropriate treatment is given (Iacoangeli et al., 2004).
Steroid Receptor RNA Activator 1 (SRA1)
Steroid receptor RNA activator 1 (SRA1) acts as a nuclear coactivator for a number of steroid hormone receptors, including the estrogen, progesterone (Lanz et al., 1999), and androgen receptors (Kurisu et al., 2006). It also coregulates the activity of a number of non-steroid receptors and transcription factors. It was discovered as a functional lncRNA which can be alternatively spliced to form a protein coding gene, called steroid receptor RNA activator protein (SRAP) which has unique functions as well as overlapping functions to SRA1 in modulating gene expression. SRA1 forms a stem-loop structure, which enables it to bind to multiple proteins including steroid receptors, transcription factors, RNA helicases, and other molecules involved in adipogenesis, myogenesis, and adrenal gland gene function (reviewed in Colley and Leedman, 2011).
A mouse model was generated to investigate the relationship between SRA1 and mammary gland development. No change was observed during early development but in mature virgin transgenic mice, activation of SRA1 led to an increase in proliferation and differentiation, and to abnormally early development of the ductal epithelium. SRA1 also regulates cell death, as in these transgenic mice, epithelial hyperplasia was accompanied by increased apoptosis (Lanz et al., 2003). These results underline a relationship between the roles of SRA1 and of steroid hormone receptors in mammary gland development, as well as a role in regulating apoptosis to maintain healthy breast function. SRA1 expression is elevated in a number of estrogen responsive tumors such as ovarian (Hussein-Fikret and Fuller, 2005) and breast cancer (Leygue et al., 1999; Murphy et al., 2000). SRA1 is able to activate the estrogen receptor to increase proliferation and its expression could be indicative of tumor grade or type, as higher expression of SRA1 was observed in ER-α-positive/PR-negative tumors compared to ER-α-positive/PR-positive tumors (Leygue et al., 1999), but although SRA1 enhances mammary gland proliferation, it is insufficient for malignant transformation (Lanz et al., 2003). Although the literature suggests that SRA1 promotes cancer progression, a complex interplay with steroid hormone receptor activation and other oncogenic proteins is likely to be required to drive malignant growth.
MicroRNA–lncRNA Interactions
MicroRNAs are small regulatory RNAs of about 22 nt that regulate gene expression by binding to specific mRNAs through sequence complementarity, and targeting them for degradation by the RNA-induced silencing complex (RISC). MiRNAs are regulated transcriptionally and also post-transcriptionally by a class of lncRNAs known as competitive endogenous RNAs (ceRNAs) that act as sponges or decoys to titrate miRNAs away from their target mRNAs and buffer their activity (Bak and Mikkelsen, 2014). Several ceRNAs such HULC in liver cancer cells sequester miR-372 (Wang et al., 2010) and PTENP1 sequesters PTEN-targeting miRNAs through similarities in miRNA binding sites in prostate cancer cells (Poliseno et al., 2010). Non-protein coding regions of the genome contain a high proportion of miRNA targets, and as lncRNAs are often found in intergenic and intronic regions of the genome, could be frequently targeted by miRNAs (Clark et al., 2014). Alignment of several other genes and their corresponding ceRNAs shows that miRNA binding sites are well conserved indicating that miRNA–lncRNA interactions play major roles in biological processes (Poliseno et al., 2010). MiRNAs can target the exons of coding regions and exon–exon junctions, enabling selective targeting of particular splice variants (Tay et al., 2008).
Long ncRNAs, ceRNAs, and miRNAs form regulatory networks across the transcriptome, communicating with each other to regulate gene expression. This includes short motifs known as pyknons, which are a subset of variable length patterns. Pyknons are conserved in the genome, and are found in coding and non-coding regions, but mostly in 3′UTRs. Pyknon sequences contain 40% of known miRNA sequences, suggesting that they regulate miRNA function (Rigoutsos et al., 2006). Different sets of pyknons are associated with disease-associated single nucleotide polymorphisms, suggesting that susceptibility to disease is due to genome wide regulation of miRNAs (Glinsky, 2009).
While some established miRNA–lncRNA interaction sites are conserved to various extents, others lack conservation despite showing confirmed functions (Alaei-Mahabadi and Larsson, 2013). It may potentially be difficult to extrapolate gene effects in animal models to human diseases. To address this challenge, it must first be demonstrated that miRNA binding sites in lncRNAs are conserved before assuming cross-species efficacy (Jackson and Levin, 2012). Paci et al. (2014) have used computational approaches to assess whether specific lncRNAs function as miRNA decoys in normal and invasive breast epithelium. In normal breast tissue, a complex regulatory network of miRNA-mediated interactions exist to bridge target mRNAs and lncRNAs, and this is missing in tumor samples. However, the role of miRNA-decoys in breast cancer remains to be validated with functional studies.
SnoRNA Hosts
Small nucleolar RNAs are a well characterized class of small ncRNAs that predominantly localize to the nucleolus and act there as guides for post transcriptional modification of ribosomal RNAs. These snoRNAs bind modifying proteins, then hybridize with specific rRNA sequences, facilitating rRNA modification. Two major classes of snoRNAs are involved in two different types of post-transcriptional modification: the C/D box snoRNAs which define sites for 2′O ribose methylation, and the H/ACA box snoRNAs which define the sites for pseudo-uridylation (Williams and Farzaneh, 2012).
Genetic searches for cancer genes have revealed that aside from their housekeeping function for rRNA modifications, a number of snoRNAs are also deregulated in cancer. An example is the snoRNA U50, a candidate tumor suppressor gene. It is downregulated and frequently mutated in prostate and breast cancer (Nallar and Kalvakolanu, 2013).
Small nucleolar RNAs are often located within the introns of protein coding and lncRNA genes. The latter may have independent functions, such as involvement in protein synthesis, and may contribute to breast cancer progression.
ZNFX1-Antisense RNA 1 (ZFAS1)
ZNFX1 antisense RNA 1 (ZFAS1) is an antisense lncRNA that overlaps the promoter region of the gene ZNFX1, which encodes a zinc finger protein whose exact role remains to be elucidated. ZFAS1 is host to three snoRNAs in three consecutive introns. ZFAS1 is expressed in a wide variety of tissues, showing highest expression in the mammary gland and lung. It contributes to mammary gland development, showing enrichment in the alveolar structures and ducts and differential expression during different stages of pregnancy in mouse mammary glands. As well as acting as a snoRNA host, Zfas1 has independent regulatory functions. Zfas1 is expressed in both the nucleus and cytoplasm, and knockdown of cytoplasmic Zfas1 by siRNA in mouse mammary epithelial cell lines increases proliferation. This suggests that Zfas1 has dual functions: after splicing in the nucleus to generate snoRNAs, Zfas1 is transported to the cytoplasm where it regulates cellular proliferation. In addition to regulating mammary gland development, ZFAS1 contributes to breast cancer, as its expression is reduced in human ductal carcinoma compared to normal breast epithelium. ZFAS1 plays a role in proliferation as well as differentiation, and its down regulation may promote cancer progression, indicating that it may have tumor suppressor functions (Askarian-Amiri et al., 2011).
Growth Arrest-Specific 5 (GAS5)
Long ncRNA growth arrest-specific 5 (GAS5) containing up to 12 exons was first isolated from growth arrested NIH-3T3 cells, induced upon serum starvation of cells (Schneider et al., 1988; Raho et al., 2000). GAS5 contains a stem-loop structure at its 3′ end that resembles the double stranded GRE and is able to act as a decoy for the GR, binding to it and sequestering it from its DNA response element. GAS5 suppresses the transcription of GR target genes, such as cIAP2, PEPCK, and G6Pase. These genes are crucial for glucose metabolism, and thus GAS5 may function to conserve energy resources in response to starvation (Kino et al., 2010).
Growth arrest-specific 5 acts as a tumor suppressor gene in breast cancer as its expression is significantly reduced compared to normal breast epithelium. It is a regulator of apoptosis as forced overexpression in breast cancer lines induced apoptosis, sensitized cells to apoptotic inducers and repressed proliferation (Mourtada-Maarabouni et al., 2008). GAS5’s function as a tumor suppressor gene is a combination of its two mechanisms of action. Decreased expression of RNU44, a snoRNA derived from GAS5 is associated with more aggressive tumors in breast cancer and head and neck squamous cell carcinoma (Gee et al., 2011). Additionally, many GR target genes are anti-apoptotic (Kino et al., 2010) and through its inhibition, GAS5 promotes apoptosis.
Organizing Nuclear Structures
The nucleus is highly organized into different domains, which provide an essential basis for the complex regulation of nuclear processes and gene expression. In addition to protein factors, some of these subnuclear domains contain lncRNAs. These nuclear lncRNAs are essential for nuclear regulation, with some acting as structural scaffolds, while others directly modulate nuclear compartment function. Nuclear organization is fundamental to modulating the activities of the genome, and this can become disrupted in cancer (Bergmann and Spector, 2014). In breast cancer, lncRNAs involved in organizing nuclear structures become deregulated, and may drive cancer progression, as discussed below.
Long Stress Induced Non-Coding Transcript 5 (LSINCT5)
Long stress induced non-coding transcript 5 (LSINCT5) is a nuclear localized lncRNA that was discovered in screens to identify lncRNAs upregulated upon treatment with stress inducing chemicals. In normal tissues, LSINCT5 is highly expressed in proliferative and stress responsive tissues such as the colon and spleen compared to non-proliferative tissues such as brain. Expression of LSINCT5 is increased in several cancers, including breast and ovarian cancer. Because of its high expression in proliferative tissues and cancer, LSINCT5 could potentially function as a regulator of proliferation. This is supported by LSINCT5 knockdown in breast and ovarian cancer cell lines, which impairs proliferation (Silva et al., 2011). Additionally, knockdown of LSINCT5 leads to altered expression of several genes including suppression of CXCR4, a breast cancer marker associated with metastasis, the lncRNA NEAT1 and the protein coding gene PSPC1 (paraspeckle component 1; Silva et al., 2011). LSINCT co-localizes with NEAT1 in the nucleus (Smith and Jessica, 2012), suggesting LSINCT5 regulates nuclear paraspeckle maintenance. How LSINCT5 contributes to paraspeckle function is yet to be determined, and it is possible that it acts to regulate proliferative genes in specific nuclear bodies.
Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1)
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved, abundant lncRNA whose expression is localized to the nucleus. It was discovered in non-small cell lung cancer as a prognostic marker of later metastasis (Ji et al., 2003) but since then has been found in several other types of human cancer, including breast cancer. MALAT1 expression is typically upregulated in cancer and has been associated with several neoplastic phenotypic features including proliferation, cell death, migration, invasion, and metastasis (reviewed in Gutschner et al., 2013b).
Metastasis associated lung adenocarcinoma transcript 1 is a core component of nuclear speckles, which are involved in mRNA processing, splicing, and export (Hutchinson et al., 2007). Blocking transcription causes a redistribution of MALAT1 from the nuclear speckles to the nucleoplasm (Bernard et al., 2010), indicating that MALAT1 regulates active transcription or processing. MALAT1 also localizes with several splicing factors, including SRSF1–3 (Anko et al., 2012). MALAT1 depletion decreases the association of these splicing factors with the nuclear speckles and differential alternative splicing of pre-mRNA (Tripathi et al., 2013). It is proposed that MALAT1 regulates the recruitment of splicing factors to these pre-mRNAs. MALAT1 also regulates other nuclear factors, such as polycomb group proteins. This lncRNA binds to unmethylated Polycomb 2 protein (PC2) in response to growth signals, shifting its location from repressive polycomb bodies on growth control gene loci to active interchromatin granules, activating their expression. MALAT1 acts as a switch for polycomb target genes from repressive nuclear domains to active nuclear domains, influencing gene expression (Gutschner et al., 2013b).
Metastasis associated lung adenocarcinoma transcript 1 also regulates expression of cell cycle genes. It is required for G1/S transition and mitotic progression, and depletion of MALAT1 leads to cell cycle arrest and activates p53 and p53 target genes. Additionally, cells depleted in MALAT1 show reduced expression of B-MYB, an oncogenic transcription factor involved in G2/M progression. MALAT1 regulates the cell cycle by modulating key transcription factors which, when deregulated, could promote cancer progression (Tripathi et al., 2013).
In breast cancer, MALAT1 is also upregulated (Guffanti et al., 2009), although its function remains to be determined. MALAT1 harbors mutations and deletions in luminal breast cancer, in a region which modulates interaction with the splicing factors 1–3 (SRSF1–3; Gutschner et al., 2013b). These mutations could alter the function of MALAT1, leading to deregulated splicing events. Treatment of breast cancer cells with high levels of estradiol reduces MALAT1 transcription, reducing proliferation, migration, and invasion (Zhao et al., 2014). Although higher expression of MALAT1 is associated with poorer outcomes in a number of cancers, Sun et al. (2009) have shown that MALAT1 is decreased in proliferating mammary glands of c-myc transgenic mice and further decreased to undetectable levels in mammary tumors. MALAT1 may regulate many genes, and its regulation is context specific.
Ribosome Associated lncRNAs
The advent of high throughput technologies is uncovering unexpected roles for lncRNAs. Ingolia et al. (2011) performed genome wide ribosome profiling in mouse embryonic stem cells, a recently developed technique that involves digesting RNA and sequencing the portion bound to the 80S ribosome to give a profile of ribosome occupancy. Unexpectedly, they discovered that a large set of intergenic lncRNAs was bound to ribosomes, raising the question of whether these lncRNAs were translated to small proteins. In light of these findings, Guttman et al. (2013) revisited these experiments with a new metric to discriminate between transcripts that were being actively translated and those just bound to the ribosome. During translation, ribosomes release their transcript upon encountering a stop codon, which is seen as a drop in 3′UTR occupancy. As this should not occur for non-coding transcripts, Guttman et al. (2013) developed a metric based on ribosome release of these transcripts. What they found was that even though a large number of long intergenic ncRNAs (lincRNA) bind, they are not actively translated. This is not just restricted to lincRNAs, as independent studies have also found a large proportion of other classes of lncRNAs bound to actively translating ribosomes (van Heesch et al., 2014).
As these lncRNAs have been confirmed to not encode for proteins, this raises the question of why so many lncRNAs are bound to ribosomes. Few of these lncRNAs have been studied in detail, but those that have been investigated have been found to regulate translation by associating with polysomes during stress conditions and either upregulating specific proteins, as is the case for Uchl1AS (Carrieri et al., 2012), or globally inhibiting translation as for mlonRNAs found in yeast (Galipon et al., 2013). Their roles in disease processes have not been established. Here, we highlight ribosome associated lncRNAs and discuss their putative roles in regulating ribosome function in breast cancer.
Translational Regulatory lncRNA (TreRNA)
Translational regulatory lncRNA (TreRNA) was identified through genome wide computational analysis (Orom et al., 2010) and found to be upregulated in cellular invasion and tumor metastasis, as well as upregulated in lymph node metastatic breast cancer samples when compared to matched primary tumors. TreRNA is also overexpressed in primary colon cancer samples compared to healthy controls, indicating an association between treRNA expression and cancer progression, including metastatic development (Gumireddy et al., 2013).
Further evidence that treRNA promotes invasion and metastasis has been obtained from in vitro and in vivo studies. Enforced expression of treRNA in breast cancer cell lines and mouse models significantly promoted invasion and lung metastases, respectively, whereas knockdown suppressed cellular migration, invasion, and metastasis in mouse (Gumireddy et al., 2013). TreRNA also leads to suppression of epithelial markers, which is associated with metastasis and transition to a mesenchymal state. Enhanced treRNA expression suppresses E-cadherin, and to a lesser extent zonula occludens-1 (ZO-1) and β-catenin, and increases expression of fibronectin and vimentin (Gumireddy et al., 2013). This change in cellular markers indicates a partial epithelial to mesenchymal transition.
This lncRNA employs different mechanisms of action depending on its cellular location. In the nucleus, treRNA acts as an enhancer for neighboring genes such as Snail (Orom et al., 2010). Cytoplasmic treRNA on the other hand acts to regulate E-cadherin by modulating its translation. Expression of treRNA reduces the level of E-cadherin protein without affecting mRNA abundance, and redistributes E-cadherin mRNA from high molecular weight polysomes to low molecular weight polysomes, strongly indicating that it suppresses translation of E-cadherin. How treRNA functions to inhibit translation is yet to be known, but it is proposed that treRNA promotes the formation of a ribonucleoprotein complex with hnRNP K, FXR1, FXR2, PUF60, and SF3B. This treRNA–protein complex may interact directly or indirectly with E-cadherin mRNA to inhibit its translation (Gumireddy et al., 2013).
LncRNAs in Estrogen Signaling
Estrogens are mitogens that promote cellular growth and proliferation, and development of the mammary gland. Estrogens signal by binding to nuclear receptors that regulate gene transcription. Mice lacking estrogen receptors cannot undergo pubertal mammary gland development. Aberrant estrogen signaling participates in breast cancer. Increased estrogen exposure may stimulate excess proliferation and has been linked to higher rates of breast cancer. 60–70% of breast cancers overexpress the estrogen receptor and show increased responses to the mitogenic effects of estrogen (reviewed in Manavathi et al., 2013). Estrogen directly modifies the expression of protein coding genes and also interacts with certain lncRNAs to modify gene expression, as discussed below.
H19
H19 is derived from a large imprinted locus on human chromosome 11p15.5. This lncRNA is abundantly expressed in the developing embryo, highly expressed in the mammary bud and adjacent tissues and differentially regulated during mammary gland development. H19 is expressed exclusively from the maternal allele where it acts to regulate the expression of IGF2, the paternally expressed growth factor from the same locus, by repressing both its transcription (Wilkin et al., 2000) and translation (Li et al., 1998). H19 is transcriptionally induced by the transcription factor E2F1, which promotes cell cycle progression. Its expression is repressed by pRB and E2F6, both of which are E2F-dependent transcriptional inhibitors (Ratajczak, 2012). c-Myc, which binds to E-box regions, also promotes transcription of H19 (Barsyte-Lovejoy et al., 2006). As H19 is under the transcriptional control of two well-known oncoproteins, E2F1 and c-myc, it is likely that it also participates in a network that promotes tumor progression.
H19 expression is normally downregulated in normal adult tissue, with only a few tissues, including the mammary gland, showing basal expression. In mouse mammary glands H19 expression is repressed during prepubertal development, upregulated during both puberty and pregnancy, repressed during lactation and then once again upregulated during involution indicating that it is not necessary for terminal differentiation but may instead function in proliferation, migration, and pre-terminal differentiation (Ratajczak, 2012). H19 is upregulated in breast cancer, with over 70% of breast adenomas showing overexpression, and is significantly correlated with ER+ and PR+ breast cancers (Adriaenssens et al., 1998). Estradiol transcriptionally regulates H19, as treatment with estradiol of ovariectomized and adrenalectomized mice increases H19 expression in the uterus, as well as increasing its expression in breast cancer cell lines (Adriaenssens et al., 1999). Transfection of H19 in breast cancer cells also increases their ability to form tumors when injected into mice (Lottin et al., 2002) and its overexpression leads to an increase in breast cancer proliferation by increasing entry into S phase, whereas knockdown prevented cell cycle progression through S phase (Berteaux et al., 2005).
SOX2 Overlapping Transcript (SOX2OT)
SRY (sex determining region Y)-box 2 (SOX2) is a transcription factor that acts a key regulator of pluripotency and lies within the intron of a lncRNA, called SOX2OT. SOX2 is required for embryonic stem cell self-renewal and pluripotency and is often deregulated in cancer (Liu et al., 2013). SOX2OT is a stable transcript, which, like SOX2, is expressed in embryonic stem cells and downregulated upon induction of embryoid body differentiation. SOX2OT is dynamically regulated during embryogenesis of several vertebrates, and restricted to the brain in adult mouse and humans (Amaral et al., 2009).
In breast cancer, SOX2 overexpression is observed in aggressive tumors and associated with a stem cell like phenotype and SOX2OT plays a key role in regulating SOX2. Expression of both SOX2 and SOX2OT are positively correlated in breast cancer samples, with ER+ tumors showing greater expression of both genes than ER- tumors, suggesting that this lncRNA is sensitive to steroid hormones such as estrogens. Ectopic expression of SOX2OT induced SOX2 expression, suggesting a positive regulatory role for SOX2OT. Orthotopic xenografts of breast cancer cells also show upregulation of both SOX2 and SOX2OT (Askarian-Amiri et al., 2014). SOX2OT not only regulates SOX2 in vertebrate development, but also regulates SOX2 during disease processes such as cancer.
LncRNAs and Drug Resistance
Survival rates of patients with breast cancer have significantly improved over the past several decades. While many factors are credited for this more positive outlook, including early detection methods, this improvement can also be attributed to new treatment programs as well as the development of new drugs. However, continuous chemotherapy often leads to drug resistance, hindering the long-term control of cancers. Chemoresistance is often reported to be a result of increase in efflux transporters or deregulation of signaling pathways, and there is increasing evidence that epigenetic factors contribute to drug resistance. In the following section lncRNAs that contribute to drug resistance in breast cancer will be discussed.
Adriamycin Resistance Associated (ARA)
Doxorubicin, also known as Adriamycin, is a chemotherapeutic drug that acts as a topoisomerase II poison. It is used to treat a variety of cancers, including breast cancer (Cortes-Funes and Coronado, 2007). However, prolonged exposure to doxorubicin induces drug resistance, making the cancer harder to treat. Jiang et al. (2014) recently identified several lncRNAs including ARA that showed differential expression in doxorubicin resistant breast and liver cancer cell lines. This lncRNA is derived from the intronic region of the PAK3 gene, and is upregulated in doxorubicin resistant MCF7 cells compared to the parental cell line. Treatment of sensitive breast cancer cell lines with doxorubicin led to an increase in ARA expression, and thus prolonged exposure to doxorubicin may lead to constant upregulation of ARA during the initiation of drug resistance. ARA may also be involved in self-sufficiency in estrogen signaling, as estrogen receptor negative breast cancer cell lines expressed greater levels of ARA than estrogen receptor positive cell lines.
Silencing of ARA in doxorubicin resistant breast cancer cell lines restored sensitivity to doxorubicin, as well as inhibiting cellular proliferation, inducing apoptosis and the accumulation of cells in the G2/M phase and reducing cell motility. Genes regulated by ARA are enriched in several pathways, including MAPK, focal adhesion, PPAR, and metabolism signaling pathways. ARA may contribute a survival and proliferative advantage to doxorubicin resistant cells, and inhibition of ARA returns the breast cancer cell to the parental drug sensitive phenotype (Jiang et al., 2014).
Urothelial Carcinoma-Associated 1 (UCA1)
Urothelial carcinoma-associated 1 (UCA1) was originally identified as being dramatically upregulated in bladder cancer, and is known as a highly specific and sensitive biomarker for diagnosing bladder carcinoma (Wang et al., 2008). UCA1 expression is elevated in embryonic tissues, then turned off in most tissues after birth. It is reactivated in cancer, indicating that UCA1 is an oncofetal gene (Wang et al., 2008). In breast cancer, UCA1 is upregulated and displays oncogenic properties. In vitro, ectopic expression of UCA1 enhances breast cancer cell growth, while UCA1-siRNA suppresses growth, leading to the accumulation of cells in G1 phase, as well as decreased Ki67 staining in tumors from mice (Huang et al., 2014). An isoform of UCA1 is associated with doxorubicin resistance in a squamous cell carcinoma line (Tsang et al., 2007), but it is not known whether this applies to breast cancer.
Urothelial carcinoma-associated 1 forms a ribonucleoprotein complex with heterogeneous nuclear ribonucleoprotein 1 (hnRNP-1). Its expression is mainly restricted to the nucleus, where it plays a key role in the splicing of pre-mRNA (Izquierdo et al., 2005), but low levels are also expressed in the cytoplasm. In the cytoplasm, hnRNP1 acts as an activator of translation. By binding to the 5′ UTR of mRNA, hnRNP recruits ribosomal proteins to initiate translation of several mRNAs such as that of p27, a well-known tumor suppressor protein that inhibits cyclin-dependent kinases. UCA1 inhibits the interaction between the p27 mRNA and hnRNP, preventing the translation of p27. Thus, there exists a competition between p27 and UCA1, as indicated by a decrease in p27 mRNA and hnRNP complex when UCA1 is overexpressed. Additionally, although both UCA1 and p27 mRNA interact with hnRNP1, they are present as separate hnRNP complexes, perhaps indicating that they compete for the same binding site, or that binding of either RNA in an allosteric site induces a conformational change that prevents binding of the other (Huang et al., 2014).
LncRNAs as Potential Therapeutic Targets
As discussed in this review, several lncRNAs have been implicated in cancer and modulation of these lncRNAs could lead to new therapeutic advances for cancer treatment.
Long ncRNAs can be targeted by RNA interference technologies such as siRNAs and shRNAs. These molecules show a high degree of selectivity and knockdown efficiency and could therefore be used to silence lncRNAs for treatment of disease. Although the use of siRNA and shRNA for therapeutic use is still in development, a few siRNAs have been used in clinical trials for a number of diseases, including cancer (Burnett et al., 2011). Various chemical modifications can also improve the stability and delivery of these RNA targeting molecules, which would be valuable if they were to be used in a clinical setting to target critical lncRNAs (Lorenz et al., 2004).
Other strategies that could be used to target lncRNAs include antisense oligonucleotides (ASO), single stranded DNA or RNA molecules with a specific sequence designed to target a particular RNA. ASOs bind to their target RNAs with high sequence specificity, and induce their degradation by RNAse H1. They have been used experimentally to inhibit MALAT1 in cancer cells and subcutaneous tumors in nude mice (Gutschner et al., 2013a) with success. ASOs could be designed to target other lncRNAs which act as oncoRNAs so as to inhibit their expression.
Long ncRNAs have also been proposed as potential biomarkers for cancer progression. As discussed previously, HOTAIR and BC200 are upregulated in breast cancer as well as in other cancers. High expression of HOTAIR is correlated with poor survival in breast cancer, increased recurrence of hepatocellular carcinoma following surgical hepatectomy, and high grades of gastrointestinal stromal tumor (GIST; Gupta et al., 2010; Niinuma et al., 2012; Ishibashi et al., 2013). HOTAIR and BC200 (Iacoangeli et al., 2004) may be useful to predict patient outcome and tumor aggressiveness in breast cancer.
However, therapeutic use of lncRNAs is not without its limitations. New tools will be needed to study their function, of which RNA interference technologies will be vital. Nevertheless, identifying the mechanism of lncRNAs will require extensive characterization of individual cases. These studies will also need to address the biological context of lncRNAs, as lncRNAs might regulate different functions in different tissues or disease processes.
Long ncRNAs are expressed at low abundance compared to protein coding genes, which can make functional studies challenging. However, this may be due to their expression in only small subpopulations of cells, providing highly specific targets for disease therapy (Djebali et al., 2012). LncRNAs show conserved function and structure in different species, and despite their poor sequence conservation could potentially be targeted for drug therapy. However, extrapolating results in animal models to humans must be done with care, especially for those lncRNAs whose roles depend on their sequence conservation such as miRNA decoys.
LncRNAs are active in multiple levels of gene regulation. They play important roles in the human mammary gland development and breast carcinogenesis. Several studies have examined their role as therapeutic targets and it is possible that lncRNAs could be targeted clinically in the future.
Concluding Remarks
Long ncRNAs are important regulators of gene expression and have a wide variety of functions that enable them to interact with multiple pathways that modulate breast cancer progressions. These lncRNAs have been shown to act as oncoRNAs, often upregulated in breast cancer and facilitating tumor growth and progression, or acting as tumor suppressors, showing downregulation in breast cancer and inhibiting tumorigenesis (Table 1). Their role as therapeutic agents is currently being explored, and highly specific lncRNA expression signatures make them attractive markers for accurate disease diagnostics. As well as their use for diagnosis and prognosis, the advancement of RNA-based therapeutics opens up lncRNAs as new targets for therapy. The functional role of the plethora of lncRNAs is only beginning to be elucidated, and provides an untapped resource for the development of cancer therapies and biomarkers.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Marjan E. Askarian-Amiri was supported by a Rodney and Elaine Davies Repatriation Fellowship and an Auckland Medical Research Foundation project grant. Herah Hansji holds PhD scholarships from the University of Auckland and the Cancer Society of New Zealand. Euphemia Y. Leung was supported by the New Zealand Breast Cancer Society.
References
Adriaenssens, E., Dumont, L., Lottin, S., Bolle, D., Lepretre, A., Delobelle, A.,et al. (1998). H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 153, 1597–1607. doi: 10.1016/S0002-9440(10)65748-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Adriaenssens, E., Lottin, S., Dugimont, T., Fauquette, W., Coll, J., Dupouy, J. P.,et al. (1999). Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene 18, 4460. doi: 10.1038/sj.onc.1202819
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alaei-Mahabadi, B., and Larsson, E. (2013). Limited evidence for evolutionarily conserved targeting of long non-coding RNAs by microRNAs. Silence 4, 4. doi: 10.1186/1758-907X-4-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amaral, P., Dinger, M. E., Mercer, T. R., and Mattick, J. S. (2008). The eukaryotic genome as an RNA machine. Science 319, 1787–1789. doi: 10.1126/science.1155472
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Amaral, P. P., Neyt, C., Wilkins, S. J., Askarian-Amiri, M. E., Sunkin, S. M., Perkins, A. C.,et al. (2009). Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15, 2013–2027. doi: 10.1261/rna.1705309
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anderson, S., Rudolph, M., Mcmanaman, J., and Neville, M. (2007). Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 9, 204. doi: 10.1186/bcr1653
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anko, M. L., Muller-Mcnicoll, M., Brandl, H., Curk, T., Gorup, C., Henry, I.,et al. (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17. doi: 10.1186/gb-2012-13-3-r17
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Askarian-Amiri, M. E., Crawford, J., French, J. D., Smart, C. E., Smith, M. A., Clark, M. B.,et al. (2011). SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17, 878–891. doi: 10.1261/rna.2528811
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Askarian-Amiri, M. E., Seyfoddin, V., Smart, C. E., Wang, J., Kim, J. E., Hansji, H.,et al. (2014). Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS ONE 9:e102140. doi: 10.1371/journal.pone.0102140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bak, R. O., and Mikkelsen, J. G. (2014). miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev. RNA 5, 317–333. doi: 10.1002/wrna.1213
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barsyte-Lovejoy, D., Lau, S. K., Boutros, P. C., Khosravi, F., Jurisica, I., Andrulis, I. L.,et al. (2006). The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 66, 5330–5337. doi: 10.1158/0008-5472.CAN-06-0037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergmann, J. H., and Spector, D. L. (2014). Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18. doi: 10.1016/j.ceb.2013.08.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z.,et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bertani, S., Sauer, S., Bolotin, E., and Sauer, F. (2011). The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell. 43, 1040–1046. doi: 10.1016/j.molcel.2011.08.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berteaux, N., Lottin, S., Monte, D., Pinte, S., Quatannens, B., Coll, J.,et al. (2005). H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 280, 29625–29636. doi: 10.1074/jbc.M504033200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bertone, P., Stolc, V., Royce, T. E., Rozowsky, J. S., Urban, A. E., Zhu, X.,et al. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246. doi: 10.1126/science.1103388
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhan, A., Hussain, I., Ansari, K. I., Kasiri, S., Bashyal, A., and Mandal, S. S. (2013). Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 425, 3707–3722. doi: 10.1016/j.jmb.2013.01.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H.,et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, C., Ballabio, A., Rupert, J. L., Lafreniere, R. G., Grompe, M., Tonlorenzi, R.,et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. doi: 10.1038/349038a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burnett, J. C., Rossi, J. J., and Tiemann, K. (2011). Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 6, 1130–1146. doi: 10.1002/biot.201100054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N.,et al. (2005). The transcriptional landscape of the mammalian genome. Science 311, 1559–1563.
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S.,et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. doi: 10.1016/j.cell.2009.01.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, W., Böcker, W., Brosius, J., and Tiedge, H. (1997). Expression of neural BC200 RNA in human tumours. J. Pathol. 183, 345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Clark, P. M., Loher, P., Quann, K., Brody, J., Londin, E. R., and Rigoutsos, I. (2014). Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci. Rep. 4, 5947. doi: 10.1038/srep05947
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Colley, S. M., and Leedman, P. J. (2011). Steroid receptor RNA activator – a nuclear receptor coregulator with multiple partners: insights and challenges. Biochimie 93, 1966–1972. doi: 10.1016/j.biochi.2011.07.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cortes-Funes, H., and Coronado, C. (2007). Role of anthracyclines in the era of targeted therapy. Cardiovasc. Toxicol. 7, 56–60. doi: 10.1007/s12012-007-0015-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H.,et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dinger, M. E., Amaral, P. P., Mercer, T. R., Pang, K. C., Bruce, S. J., Gardiner, B. B.,et al. (2008). Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445. doi: 10.1101/gr.078378.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Djebali, S., Davis, C., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A.,et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, Z., Fei, T., Verhaak, R. G., Su, Z., Zhang, Y., Brown, M.,et al. (2013). Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913. doi: 10.1038/nsmb.2591
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Froberg, J. E., Yang, L., and Lee, J. T. (2013). Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 425, 3698–3706. doi: 10.1016/j.jmb.2013.06.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Galipon, J., Miki, A., Oda, A., Inada, T., and Ohta, K. (2013). Stress-induced lncRNAs evade nuclear degradation and enter the translational machinery. Genes Cells 18, 353–368. doi: 10.1111/gtc.12042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ganesan, S., Silver, D. P., Drapkin, R., Greenberg, R., Feunteun, J., and Livingston, D. M. (2004). Association of BRCA1 with the inactive X chromosome and XIST RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 123–128. doi: 10.1098/rstb.2003.1371
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ganesan, S., Silver, D. P., Greenberg, R. A., Avni, D., Drapkin, R., Miron, A.,et al. (2002). BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111, 393–405. doi: 10.1016/S0092-8674(02)01052-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gee, H. E., Buffa, F. M., Camps, C., Ramachandran, A., Leek, R., Taylor, M.,et al. (2011). The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br. J. Cancer 104, 1168–1177. doi: 10.1038/sj.bjc.6606076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geng, Y. J., Xie, S. L., Li, Q., Ma, J., and Wang, G. Y. (2011). Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 39, 2119–2128. doi: 10.1177/147323001103900608
Gibb, E., Brown, C. J., and Lam, W. L. (2011). The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10, 38. doi: 10.1186/1476-4598-10-38
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ginger, M. R., Shore, A. N., Contreras, A., Rijnkels, M., Miller, J., Gonzalez-Rimbau, M. F.,et al. (2006). A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc. Natl. Acad. Sci. U.S.A. 103, 5781–5786. doi: 10.1073/pnas.0600745103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glinsky, G. V. (2009). Human genome connectivity code links disease-associated SNPs, microRNAs and pyknons. Cell Cycle 8, 925–930. doi: 10.4161/cc.8.6.7937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guffanti, A., Iacono, M., Pelucchi, P., Kim, N., Soldà, G., Croft, L. J.,et al. (2009). A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10:163. doi: 10.1186/1471-2164-10-163
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gumireddy, K., Li, A., Yan, J., Setoyama, T., Johannes, G. J., Orom, U. A.,et al. (2013). Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 32, 2672–2684. doi: 10.1038/emboj.2013.188
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gupta, R., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J.,et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi: 10.1038/nature08975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gutschner, T., and Diederichs, S. (2012). The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719. doi: 10.4161/rna.20481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gutschner, T., Hämmerle, M., Eissmann, M., Hsu, J., Kim, Y., Hung, G.,et al. (2013a). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189. doi: 10.1158/0008-5472.CAN-12-2850
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gutschner, T., Hämmerle, M., and Diederichs, S. (2013b). MALAT1: a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 91, 791–801. doi: 10.1007/s00109-013-1028-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G.,et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346. doi: 10.1038/nature10887
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guttman, M., Russell, P., Ingolia, N. T., Weissman, J. S., and Lander, E. S. (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. doi: 10.1016/j.cell.2013.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hennighausen, L., and Robinson, G. (2001). Signaling pathways in mammary gland development. Dev. Cell 1, 467–475. doi: 10.1016/S1534-5807(01)00064-8
Huang, J., Zhou, N., Watabe, K., Lu, Z., Wu, F., Xu, M.,et al. (2014). Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 5, e1008. doi: 10.1038/cddis.2013.541
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D.,et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. doi: 10.1016/j.cell.2010.06.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hung, T., Wang, Y., Lin, M. F., Koegel, A. K., Kotake, Y., Grant, G. D.,et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629. doi: 10.1038/ng.848
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hussein-Fikret, S., and Fuller, P. J. (2005). Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol. Cell. Endocrinol. 229, 149–160. doi: 10.1016/j.mce.2004.08.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi: 10.1186/1471-2164-8-39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iacoangeli, A., Lin, Y., Morley, E. J., Muslimov, I. A., Bianchi, R., Reilly, J.,et al. (2004). BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis 25, 2125–2133. doi: 10.1093/carcin/bgh228
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ingolia, N. T., Lareau, L. F., and Weissman, J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. doi: 10.1016/j.cell.2011.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ishibashi, M., Kogo, R., Shibata, K., Sawada, G., Takahashi, Y., Kurashige, J.,et al. (2013). Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol. Rep. 29, 946–950. doi: 10.3892/or.2012.2219
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Izquierdo, J. M., Majos, N., Bonnal, S., Martinez, C., Castelo, R., Guigo, R.,et al. (2005). Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 19, 475–484. doi: 10.1016/j.molcel.2005.06.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jackson, A. L., and Levin, A. A. (2012). Developing microRNA therapeutics: approaching the unique complexities. Nucleic Acid Ther. 22, 213–225. doi: 10.1089/nat.2012.0356
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ji, P., Diederichs, S., Wang, W., Boing, S., Metzger, R., Schneider, P. M.,et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041. doi: 10.1038/sj.onc.1206928
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, M., Huang, O., Xie, Z., Wu, S., Zhang, X., Shen, A.,et al. (2014). A novel long non-coding RNA-ARA: adriamycin resistance associated. Biochem. Pharmacol. 87, 254–283. doi: 10.1016/j.bcp.2013.10.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jobe, E. M., Mcquate, A. L., and Zhao, X. (2012). Crosstalk among epigenetic pathways regulates neurogenesis. Front. Neurosci. 6:59. doi: 10.3389/fnins.2012.00059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, P. A., and Baylin, S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. doi: 10.1016/j.cell.2007.01.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kapranov, P., St Laurent, G., Raz, T., Ozsolak, F., Reynolds, C. P., Sorensen, P.,et al. (2010). The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 8:149. doi: 10.1186/1741-7007-8-149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kapusta, A., Kronenberg, Z., Lynch, V. J., Zhuo, X., Ramsay, L., Bourque, G.,et al. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. doi: 10.1371/journal.pgen.1003470
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D.,et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 3, ra8. doi: 10.1126/scisignal.2000568
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T.,et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326. doi: 10.1158/0008-5472.CAN-11-1021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kondrashov, A. V., Kiefmann, M., Ebnet, K., Khanam, T., Muddashetty, R. S., and Brosius, J. R. (2005). Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353, 88–103. doi: 10.1016/j.jmb.2005.07.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kurisu, T., Tanaka, T., Ishii, J., Matsumura, K., Sugimura, K., Nakatani, T.,et al. (2006). Expression and function of human steroid receptor RNA activator in prostate cancer cells: role of endogenous hSRA protein in androgen receptor-mediated transcription. Prostate Cancer Prostatic Dis. 9, 173–178. doi: 10.1038/sj.pcan.4500867
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lanz, R. B., Chua, S. S., Barron, N., Soder, B. M., Demayo, F., and O’Malley, B. W. (2003). Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell Biol. 23, 7163–7176. doi: 10.1128/MCB.23.20.7163-7176.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lanz, R. B., Mckenna, N. J., Onate, S. A., Albrecht, U., Wong, J., Tsai, S. Y.,et al. (1999). A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97, 17–27. doi: 10.1016/S0092-8674(00)80711-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leeb, M., Steffen, P. A., and Wutz, A. (2009). X chromosome inactivation sparked by non-coding RNAs. RNA Biol. 6, 94–99. doi: 10.4161/rna.6.2.7716
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leygue, E., Dotzlaw, H., Watson, P. H., and Murphy, L. C. (1999). Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 59, 4190–4193.
Li, Y. M., Franklin, G., Cui, H. M., Svensson, K., He, X. B., Adam, G.,et al. (1998). The H19 transcript is associated with polysomes and may regulate IGF2 expression intrans. J. Biol. Chem. 273, 28247–28252. doi: 10.1074/jbc.273.43.28247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lim, E., Wu, D., Pal, B., Bouras, T., Asselin-Labat, M., Vaillant, F.,et al. (2010). Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 12, R21. doi: 10.1186/bcr2560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, K., Lin, B., Zhao, M., Yang, X., Chen, M., Gao, A.,et al. (2013). The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell. Signal. 25, 1264–1271. doi: 10.1016/j.cellsig.2013.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lorenz, C., Hadwiger, P., John, M., Vornlocher, H. P., and Unverzagt, C. (2004). Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 14, 4975–4977. doi: 10.1016/j.bmcl.2004.07.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lottin, S., Adriaenssens, E., Dupressoir, T., Berteaux, N., Montpellier, C., Coll, J.,et al. (2002). Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23, 1885–1895. doi: 10.1093/carcin/23.11.1885
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Manavathi, B., Dey, O., Gajulapalli, V. N. R., Bhatia, R. S., Bugide, S., and Kumar, R. (2013). Derailed estrogen signaling and breast cancer: an authentic couple. Endocr. Rev. 34, 1–32. doi: 10.1210/er.2011-1057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maruyama, R., Shipitsin, M., Choudhury, S., Wu, Z., Protopopov, A., Yao, J.,et al. (2012). Altered antisense-to-sense transcript ratios in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 2820–2824. doi: 10.1073/pnas.1010559107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mattick, J. S. (2007). A new paradigm for developmental biology. J. Exp. Biol. 210, 1526–1547. doi: 10.1242/jeb.005017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mattick, J. S., and Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15 Spec No. 1, R17–R29. doi: 10.1093/hmg/ddl046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morey, L., and Helin, K. (2010). Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332. doi: 10.1016/j.tibs.2010.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mourtada-Maarabouni, M., Pickard, M. R., Hedge, V. L., Farzaneh, F., and Williams, G. T. (2008). GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28, 195–208. doi: 10.1038/onc.2008.373
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murphy, L. C., Simon, S. L., Parkes, A., Leygue, E., Dotzlaw, H., Snell, L.,et al. (2000). Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 60, 6266–6271.
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R.,et al. (2008). The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720. doi: 10.1126/science.1163802
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nallar, S. C., and Kalvakolanu, D. V. (2013). Regulation of snoRNAs in cancer: close encounters with interferon. J. Interferon Cytokine Res. 33, 189–198. doi: 10.1089/jir.2012.0106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U.,et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. doi: 10.1038/nature12943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niinuma, T., Suzuki, H., Nojima, M., Nosho, K., Yamamoto, H., Takamaru, H.,et al. (2012). Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72, 1126–1136. doi: 10.1158/0008-5472.CAN-11-1803
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Odom, D. T., Dowell, R. D., Jacobsen, E. S., Gordon, W., Danford, T. W., Macisaac, K. D.,et al. (2007). Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 39, 730–732. doi: 10.1038/ng2047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G.,et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58. doi: 10.1016/j.cell.2010.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orom, U. A., and Shiekhattar, R. (2013). Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154, 1190–1193. doi: 10.1016/j.cell.2013.08.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osato, N., Suzuki, Y., Ikeo, K., and Gojobori, T. (2007). Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 176, 1299–1306. doi: 10.1534/genetics.106.069484
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paci, P., Colombo, T., and Farina, L. (2014). Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 8:83. doi: 10.1186/1752-0509-8-83
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J.,et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 32, 232–246. doi: 10.1016/j.molcel.2008.08.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pauli, A., Valen, E., Lin, M. F., Garber, M., Vastenhouw, N. L., Levin, J. Z.,et al. (2012). Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22, 577–591. doi: 10.1101/gr.133009.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perez, D., Hoage, T. R., Pritchett, J. R., Ducharme-Smith, A. L., Halling, M. L., Ganapathiraju, S. C.,et al. (2007). Long, abundantly-expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 17, 642–655. doi: 10.1093/hmg/ddm336
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perez, D. S., Hoage, T. R., Pritchett, J. R., Ducharme-Smith, A. L., Halling, M. L., Ganapathiraju, S. C.,et al. (2008). Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 17, 642–655. doi: 10.1093/hmg/ddm336
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poliseno, L., Salmena, L., Zhang, J., Carver, B. Haveman, W. J., and Pandolf, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qiu, M. T., Hu, J. W., Yin, R., and Xu, L. (2013). Long noncoding RNA: an emerging paradigm of cancer research. Tumor Biol. 34, 613–620. doi: 10.1007/s13277-013-0658-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raho, G., Barone, V., Rossi, D., Philipson, L., and Sorrentino, V. (2000). The gas 5 gene shows four alternative splicing patterns without coding for a protein. Gene 256, 13–17. doi: 10.1016/S0378-1119(00)00363-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ratajczak, M. Z. (2012). Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem. Cytobiol. 50, 171–179. doi: 10.5603/FHC.2012.0026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richardson, A. L., Wang, Z. C., De Nicolo, A., Lu, X., Brown, M., Miron, A.,et al. (2006). X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132. doi: 10.1016/j.ccr.2006.01.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rigoutsos, I., Huynh, T., Miranda, K., Tsirigos, A., McHardy, A., and Platt, D. (2006). Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc. Natl. Acad. Sci. U.S.A. 103, 6605–6610. doi: 10.1073/pnas.0601688103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A.,et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schneider, C., King, R. M., and Philipson, L. (1988). Genes specifically expressed at growth arrest of mammalian cells. Cell 54, 787–793. doi: 10.1016/S0092-8674(88)91065-3
Shore, A. N., Kabotyanski, E. B., Roarty, K., Smith, M. A., Zhang, Y., Creighton, C. J.,et al. (2012). Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation. PloS Genet. 8:e1002840. doi: 10.1371/journal.pgen.1002840
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Silva, J. M., Boczek, N. J., Berres, M. W., Ma, X., and Smith, D. I. (2011). LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 8, 496–505. doi: 10.4161/rna.8.3.14800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, B., Welm, A., and Welm, B. (2012). On the shoulders of giants: a historical perspective of unique experimental methods in mammary gland research. Semin. Cell Dev. Biol. 23, 583–590. doi: 10.1016/j.semcdb.2012.03.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, C., and Steitz, J. A. (1998). Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell Biol. 18, 6897–6909.
Smith, D. I., and Jessica, S. M. (2012). Abstract A1: long stress-induced non-coding transcripts (LSINCTs) and cancer. Cancer Res. 72, A1. doi: 10.1158/1538-7445.nonrna12-a1
Sun, Y., Wu, J., Wu, S. H., Thakur, A., Bollig, A., Huang, Y.,et al. (2009). Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Res. Treat. 118, 185–196. doi: 10.1007/s10549-008-0171-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tay, Y., Zhang, J., Thomson, A. M., Lim, B., and Rigoutsos, I. (2008). MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455, 1124–1128. doi: 10.1038/nature07299
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tian, D., Sun, S., and Lee, J. T. (2010). The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403. doi: 10.1016/j.cell.2010.09.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Torarinsson, E., Sawera, M., Havgaard, J. H., Fredholm, M., and Gorodkin, J. (2006). Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 16, 885–889. doi: 10.1101/gr.5226606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tripathi, V., Shen, Z., Chakraborty, A., Giri, S., Freier, S. M., Wu, X.,et al. (2013). Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PloS Genet. 9:e1003368. doi: 10.1371/journal.pgen.1003368
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsai, M., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F.,et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsang, W. P., Wong, T. W., Cheung, A. H., Co, C. N., and Kwok, T. T. (2007). Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 13, 890–898. doi: 10.1261/rna.359007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Heesch, S., Van Iterson, M., Jacobi, J., Boymans, S., Essers, P. B., De Bruijn, E.,et al. (2014). Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15, R6. doi: 10.1186/gb-2014-15-1-r6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vincent-Salomon, A., Ganem-Elbaz, C., Manie, E., Raynal, V., Sastre-Garau, X., Stoppa-Lyonnet, D.,et al. (2007). X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 67, 5134–5140. doi: 10.1158/0008-5472.CAN-07-0465
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, F., Li, X., Xie, X., Zhao, L., and Chen, W. (2008). UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 582, 1919–1927. doi: 10.1016/j.febslet.2008.05.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Liu, X., Wu, H., Ni, P., Gu, Z., Qiao, Y.,et al. (2010). CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38, 5366–5383. doi: 10.1093/nar/gkq285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, K., and Chang, H. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell. 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y.,et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, L., Zeng, X., Chen, S., Ding, L., Zhong, J., Zhao, J. C.,et al. (2013). BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 32, 1584–1597. doi: 10.1038/emboj.2013.95
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watson, C. J., and Khaled, W. T. (2008). Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 135, 995–1003. doi: 10.1242/dev.005439
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilkin, F., Paquette, J., Ledru, E., Mamelin, C., Pollak, M., and Deal, C. L. (2000). H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. Eur. J. Biochem. 267, 4020–4027. doi: 10.1046/j.1432-1327.2000.01438.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, G. T., and Farzaneh, F. (2012). Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev.Cancer 12, 84–88. doi: 10.1038/nrc3195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, S., Yuan, Q., Hao, H., Guo, Y., Liu, S., Zhang, Y.,et al. (2011). Expression of OCT4 pseudogenes in human tumours: lessons from glioma and breast carcinoma. J. Pathol. 223, 672–682. doi: 10.1002/path.2827
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Z., Chen, C., Liu, Y., and Wu, C. (2014). 17β-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 445, 388–393. doi: 10.1016/j.bbrc.2014.02.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Y., Zhong, Y., Wang, Y., Zhang, X., Batista, D. L., Gejman, R.,et al. (2007). Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 282, 24731–24742. doi: 10.1074/jbc.M702029200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: long non-coding RNA, breast cancer, mammary gland development, gene regulation, epigenetics
Citation: Hansji H, Leung EY, Baguley BC, Finlay GJ and Askarian-Amiri ME (2014) Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front. Genet. 5:379. doi: 10.3389/fgene.2014.00379
Received: 24 July 2014; Accepted: 13 October 2014;
Published online: 31 October 2014.
Edited by:
Muller Fabbri, University of Southern California, USAReviewed by:
Antonio Sorrentino, Exiqon A/S, DenmarkIvan Vannini, Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori, Italy
Isidore Rigoutsos, Thomas Jefferson University, USA
Copyright © 2014 Hansji, Leung, Baguley, Finlay and Askarian-Amiri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Graeme J. Finlay and Marjan E. Askarian-Amiri, Auckland Cancer Society Research Centre, University of Auckland, 85 Park Road, Grafton, Auckland, New Zealand e-mail: g.finlay@auckland.ac.nz; m.askarian-amiri@auckland.ac.nz
 Herah Hansji
Herah Hansji Euphemia Y. Leung
Euphemia Y. Leung Bruce C. Baguley
Bruce C. Baguley Graeme J. Finlay
Graeme J. Finlay Marjan E. Askarian-Amiri
Marjan E. Askarian-Amiri