- 1Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE, USA
- 2Department of Pediatrics, Philipps-University Marburg, Marburg, Germany
- 3The Eppley Cancer Institute, University of Nebraska Medical Center, Omaha, NE, USA
- 4Department of Internal Medicine, University of Nebraska Medical Center, Omaha, NE, USA
VH replacement refers to RAG-mediated secondary recombination of the IgH genes, which renews almost the entire VH gene coding region but retains a short stretch of nucleotides as a VH replacement footprint at the newly generated VH–DH junction. To explore the biological significance of VH replacement to the antibody repertoire, we developed a Java-based VH replacement footprint analyzer program and analyzed the distribution of VH replacement products in 61,851 human IgH gene sequences downloaded from the NCBI database. The initial assignment of the VH, DH, and JH gene segments provided a comprehensive view of the human IgH repertoire. To our interest, the overall frequency of VH replacement products is 12.1%; the frequencies of VH replacement products in IgH genes using different VH germline genes vary significantly. Importantly, the frequencies of VH replacement products are significantly elevated in IgH genes derived from different autoimmune diseases, including rheumatoid arthritis, systemic lupus erythematosus, and allergic rhinitis, and in IgH genes encoding various autoantibodies or anti-viral antibodies. The identified VH replacement footprints preferentially encoded charged amino acids to elongate IgH CDR3 regions, which may contribute to their autoreactivities or anti-viral functions. Analyses of the mutation status of the identified VH replacement products suggested that they had been actively involved in immune responses. These results provide a global view of the distribution of VH replacement products in human IgH genes, especially in IgH genes derived from autoimmune diseases and anti-viral immune responses.
Introduction
To protect our body from various infectious agents, the adaptive immune system has evolved the capability to generate a vast number of antibody (Ab) specificities through somatic rearrangement of previously separated variable (V), diversity (D) (for heavy chain only), and joining (J) gene segments to form the variable domain exons of immunoglobulin genes (1–3). V(D)J recombination is catalyzed by a pair of recombination activating gene products (RAG1 and RAG2) (4–6). Specific joining of the V, D, and J gene segments is directed by the recombination signal sequences (RSS) flanking each rearranging gene segment (7). The RSS is composed of a highly conserved heptamer (5’-CACTGTG-3’) and a nonamer (5’-ACAAAAACC-3’) separated by a non-conserved spacer region with either 12 or 23 bp in length (7–9). There are 44 functional VH genes, 27 DH genes, and 6 JH genes within the human IgH locus. The diversified IgH repertoire is generated at different levels, including the random recombination of V, D, and J genes segments, imprecise processing of the coding-ends, addition of non-template nucleotides by terminal deoxynucleotidyl transferase (TdT), random pairing of IgH with Igκ or Igλ light chains, and later through somatic hypermutation and class switch recombination during antigen dependent germinal center reaction (2). Previous analyses of the IgH repertoire have provided important information regarding the developmental process and function of B lineage cells (10, 11). For examples, earlier studies on the expression and rearrangement status of IgH genes demonstrated that IgH gene are rearranged sequentially during early B lineage cell development, in which DH to JH rearrangements occurs prior to VH to DJH rearrangements followed by rearrangement of the Igκ and then Igλ light chain genes (12, 13). Analyses of the Ig gene repertoires of different autoimmune diseases such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) revealed skewed usages of specific germline VH genes (14–16), unusually long CDR3 regions within the IgH and IgL genes (17, 18), and accumulation of somatic hypermutation in the variable regions of IgH and IgL genes (15, 19).
The random process of V(D)J recombination is essential for generating a diverse IgH repertoire, however, it also produces non-functional IgH genes or IgH genes encoding autoreactive antigen receptors (2, 20). Early B lineage cells carrying non-functional IgH rearrangements must re-initiate the V(D)J recombination process to generate functional B-cell receptors (BCRs) for subsequent development; on the other hand, B-cells expressing autoreactive receptors will be removed from the repertoire through receptor editing, clonal deletion, or anergy to establish central tolerance (1, 21, 22). Receptor editing refers to RAG-mediated secondary recombination of previously rearranged IgH or IgL genes (1, 21, 22). The organizations of the Igκ and Igλ loci allow continuous secondary recombination by joining an upstream VL gene with a downstream JL gene segment. The previously formed VLJL joints are deleted during secondary recombination leaving no trace in the newly formed VLJL junctions; the only indication of extensive light chain gene editing is the elevated usage of the 3′ Jκ or Jλ genes and the deletion of the Igκ locus (23, 24).
The unwanted IgH genes can also be changed through a RAG-mediated VH replacement process using the cryptic recombination signal sequences (cRSSs) embedded within the framework-3 regions of previously rearranged VH genes (21, 22, 25). The concept of VH replacement was originally proposed to explain the observation that functional IgH genes were generated in mouse pre-B-cell leukemia lines initially harboring non-functional IgH rearrangements (26–28). Comparison of the functional IgH genes versus the non-functional IgH rearrangements suggested a VH to VHDJH recombination process mediated by the cRSS sites (26, 27). Subsequently, the occurrence of VH replacement had been demonstrated in mouse models carrying knocked-in IgH genes encoding anti-DNA Abs, anti-NP Abs, or non-functional IgH genes in both alleles (29–34). Despite these findings, the natural occurrence of VH replacement during early B-cell development in mouse remains to be determined (35, 36).
Ongoing VH replacement in human B-cells had been found in a human leukemia cell line, EU12, by detection of RAG-mediated cRSS double stranded DNA breaks (DSBs) and by amplification of different VH replacement excision circles (37). The detection of DSBs at the VH3–cRSS borders in human bone marrow immature B-cells provided the first evidence for the natural occurrence of VH replacement during normal B-cell development in humans (37). The occurrence of VH replacement in bone marrow immature B-cells is consistent with the observation that RAG1 and RAG2 genes can be reinduced in these cells to catalyze IgL gene editing (24, 38, 39). Our recent studies showed that VH replacement occurs in the newly immigrated immature B-cells in the peripheral blood of healthy donors, which can be further induced through BCR-mediated signaling in Ref. (40). The cRSS-mediated VH replacement was of particular interest because the cRSS motifs are found in 40 out of 44 human VH germline genes and in the majority of mouse VH germline genes (22, 41). VH replacement renews almost the entire VH gene coding region but retains a short stretch of nucleotides as a VH replacement footprint at the VH–DH junction (37). Such footprints can be used to identify VH replacement products through analysis of IgH gene sequences. The initial analyses of 417 human IgH gene sequences estimated that VH replacement products contribute to about 5% of the normal IgH repertoire (37). Interestingly, analyses of the amino acids encoded by the VH replacement footprints revealed that these footprints preferentially contribute charged amino acids into the IgH CDR3 regions, which is different from the low frequency of charged amino acids encoded by human germline DH genes or N region sequences added by TdT (37).
To explore the biological significance of VH replacement, we developed a Java-based computer program and analyzed 61,851 human IgH gene sequences from the NCBI database to determine the distribution of VH replacement products.
Materials and Methods
Development of the VH Replacement Footprint Analyzer Program
The VH replacement footprint analyzer (VHRFA) program was developed using the NetBeans 7.01 IDE with Java development kit (JDK) and tested under Windows, Mac OS X, and Ubuntu Linux (42). The reference human VH germline gene sequences were downloaded from the IMGT database to generate the library of VH replacement footprints with different lengths. For the initial test of the VHRFA program, we used 417 IgH sequences that had been analyzed in our previous study to manually identify potential VH replacement products (37, 43). The 61,851 human IgH gene sequences were downloaded from the NCBI database on April 20, 2011.
Analysis of IgH Gene Sequences and Identification of Potential VH Replacement Products Using the VHRFA Program
The IgH gene sequence files from NCBI database were first converted into FASTA files and uploaded to the VHRFA program. The VH, DH, and JH germline gene usages were assigned by automatic submission of sequences in batches to the IMGT/V-Quest program (http://www.imgt.org/IMGT_vquest/share/textes/) (44) and the results were exported as Microsoft Excel files to a local computer. Identical IgH gene sequences in the original NCBI database were removed based on their VH–DH–JH junctions and the remaining 39,438 unique human IgH gene sequences with identifiable VH, DH, and JH genes were further analyzed to identify potential VH replacement products and calculate the frequencies of VH replacement products in subsequent analyses. Briefly, the IgH gene sequences with clear identifiable VH, DH, and JH genes were analyzed to identify VH replacement footprints with 7, 6, 5, 4, and 3-mer VH replacement footprint motifs at their VH–DH junction (N1) regions and DH–JH junction (N2) regions. The frequency of VH replacement products was calculated by dividing the number of IgH genes with VH replacement footprints in the N1 regions with the total number of unique IgH gene sequences. IgH genes with 7, 6, 4, and 3-mer VH replacement footprint motifs within their N1 regions were also analyzed and discussed. The positive prediction value with 95% confidence interval using the 6, 5, 4, and 3-mer VH replacement footprint motifs to assign VH replacement products are 68, 59, 54, and 52%, respectively. In the following comparison, the VH replacement products mainly refer to IgH genes with 5-mer VH replacement footprint within their N1 regions.
The distribution of VH replacement products in IgH genes derived from different keyword sub-categories were analyzed based on the information linked to each sequence in the NCBI GenBank files. The frequencies of VH replacement products with pentameric footprints were used for all these comparisons. For mutational analysis the IgH gene sequences had a minimum of ≥80% nucleotide similarity to the assigned germline VH gene sequences.
Statistical Analysis
Statistical significance was determined by using either the two-tailed Chi square test with Yates’ correction or the unpaired t-test. p < 0.05 is considered statistically significant and p < 0.0001 is considered extremely statistically significant.
Results
Differential Usage of Germline VH, DH, and JH Genes in Human IgH Gene Sequences
We have developed a Java-based VHRFA computer program to analyze large number of IgH gene sequences and to identify potential VH replacement products (42). In the current study, the 61,851 human IgH gene sequences were downloaded from the NCBI database. The initial analysis showed that 54,970 IgH genes have identifiable VH, DH, and JH gene segments. After removal of duplicate IgH sequences, the remaining 39,438 unique IgH genes with identifiable VH, JH, and DH genes were further analyzed. The usages of the VH, JH, and DH germline genes in these sequences represent a combinatorial view of the human IgH repertoire from many studies (Figure 1). The usages of all the 44 functional human germline VH genes were confirmed in this dataset (Figure 1A); the frequencies of individual VH germline gene usage varied considerably. For different families of VH genes, the VH3 family of genes was predominantly utilized, followed by the VH4 and VH1 families of genes (Figure 1A). Such results are consistent with previous analyses of small groups of IgH gene sequences, Among individual VH genes, the VH3–23 gene was used the most frequently in 9536 IgH genes (25%). The VH4–28 gene was used less frequently, which was only found in 13 IgH rearrangements (0.03%). The differential usages of individual VH germline genes did not seem to correlate with their relative location within the IgH locus (Figure 1A). Within the IgH locus, the VH1–24, VH2–26, and VH3–30 genes are located very close to the VH3–23 and VH4–28 genes. However, the frequency of the VH3–23 gene usage is only 4-fold higher than those of the VH3–30 gene, but is 50- and 80-fold higher than that of the VH1–24 and VH2–26 genes, respectively (Figure 1A).
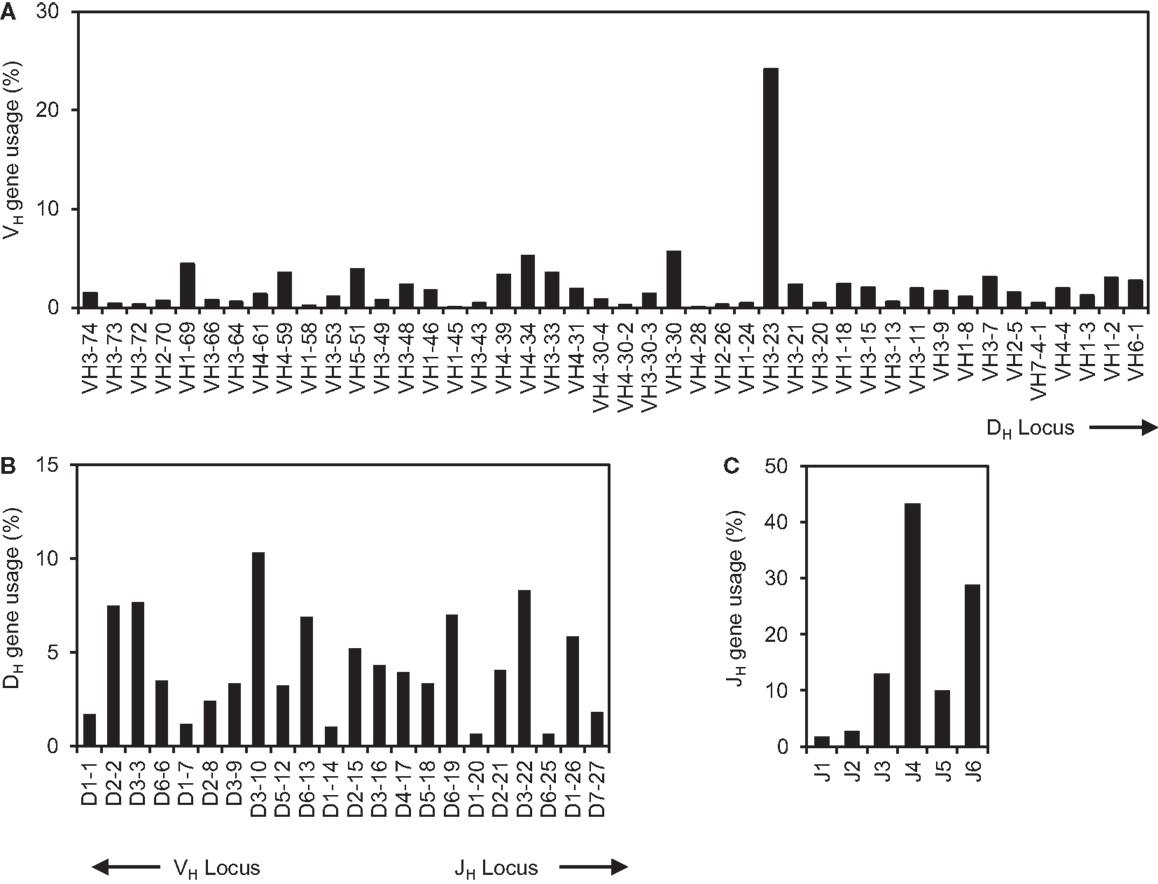
Figure 1. The comprehensive analysis of human IgH repertoire. The 61,851 human IgH gene sequences were downloaded from the NCBI database on May, 2012. The sequences were first analyzed for their VH, DH, and JH gene usage using the IMGT/V-Quest program and the identical sequences were removed. The frequencies of VH (A), DH (B), and JH (C) germline gene usages in the 39,438 unique human IgH gene sequences were shown.
Among different DH genes, the DH3 gene family was predominantly used in 35% of IgH genes, in which the DH3–10, DH3–3, and DH3–22 genes were used frequently; The DH1 gene family was used less frequently (Figure 1B). Among JH germline genes, the JH4 gene was predominantly used followed by the JH6 gene (Figure 1C). These results are consistent with previous individual reports with small number of IgH sequences. Taken together, this analysis provides a comprehensive view of the existing human IgH gene sequences in the NCBI database.
Identification of VH Replacement Products Using the VHRFA Program
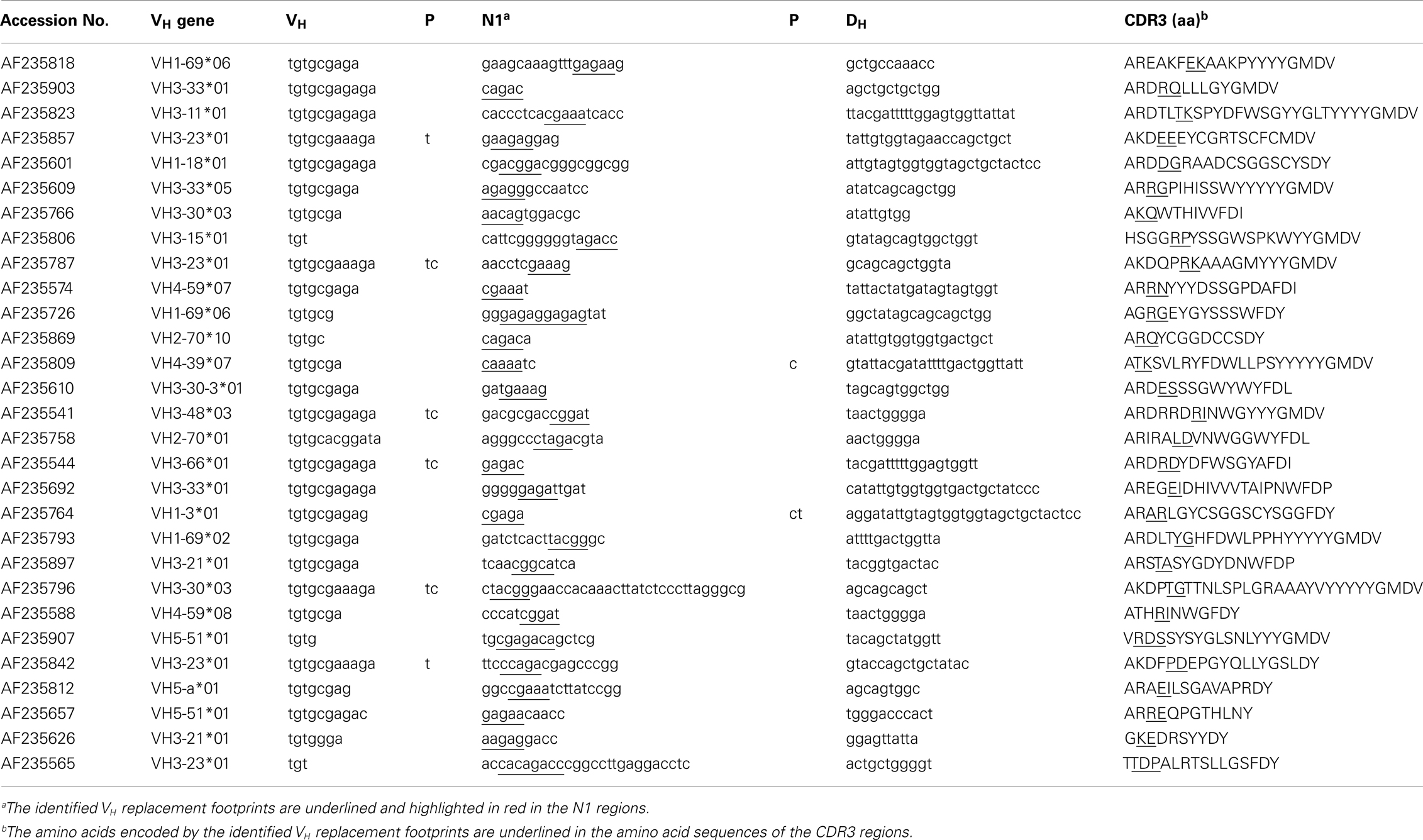
To identify potential VH replacement products in a large number of IgH gene sequences, the VHRFA program first generated libraries of potential VH replacement footprint database with different length based on the VH gene 3′ ending sequences following the conserved cRSS sites of all the functional human VH germline genes (Tables S1 and S2 in Supplementary Material). Then, the VHRFA program uses these libraries to search for the presence of VH replacement footprint motifs with specified lengths at the VH–DH junction (N1) regions or the DH–JH junction (N2) regions of IgH genes. As an initial test of the newly developed VHRFA program, we reanalyzed the 417 human IgH gene sequences that had been to manually identify potential VH replacement products analyzed in a previous study (37). The VHRFA program efficiently identified VH replacement footprint motifs with 3, 4, 5, 6, or 7 nucleotides in both the N1 and N2 regions (Table 1, top). The frequencies of VH replacement footprint motifs with 3, 4, or 5-mer in the N1 regions are significantly higher than those in the N2 regions (Table 1, top), indicating that the addition of such motifs in the N1 region is not a random event. Based on the identification of 5-mer VH replacement footprints, 7.3% of the IgH gene sequences can be assigned as potential VH replacement products. Further review of these IgH genes confirmed the identified pentameric VH replacement motifs within the VH–DH junctions (Table 2, N1 regions). If we consider the 4- or 3-mer VH replacement footprints within the N1 regions, 25 or 54.7% of IgH genes can be assigned as potential VH replacement products, respectively (Table 1; Table S3 in Supplementary Material). These results are consistent with our previously manual assignment of VH replacement products in this group of IgH genes and provide the first validation of the VHRFA program.
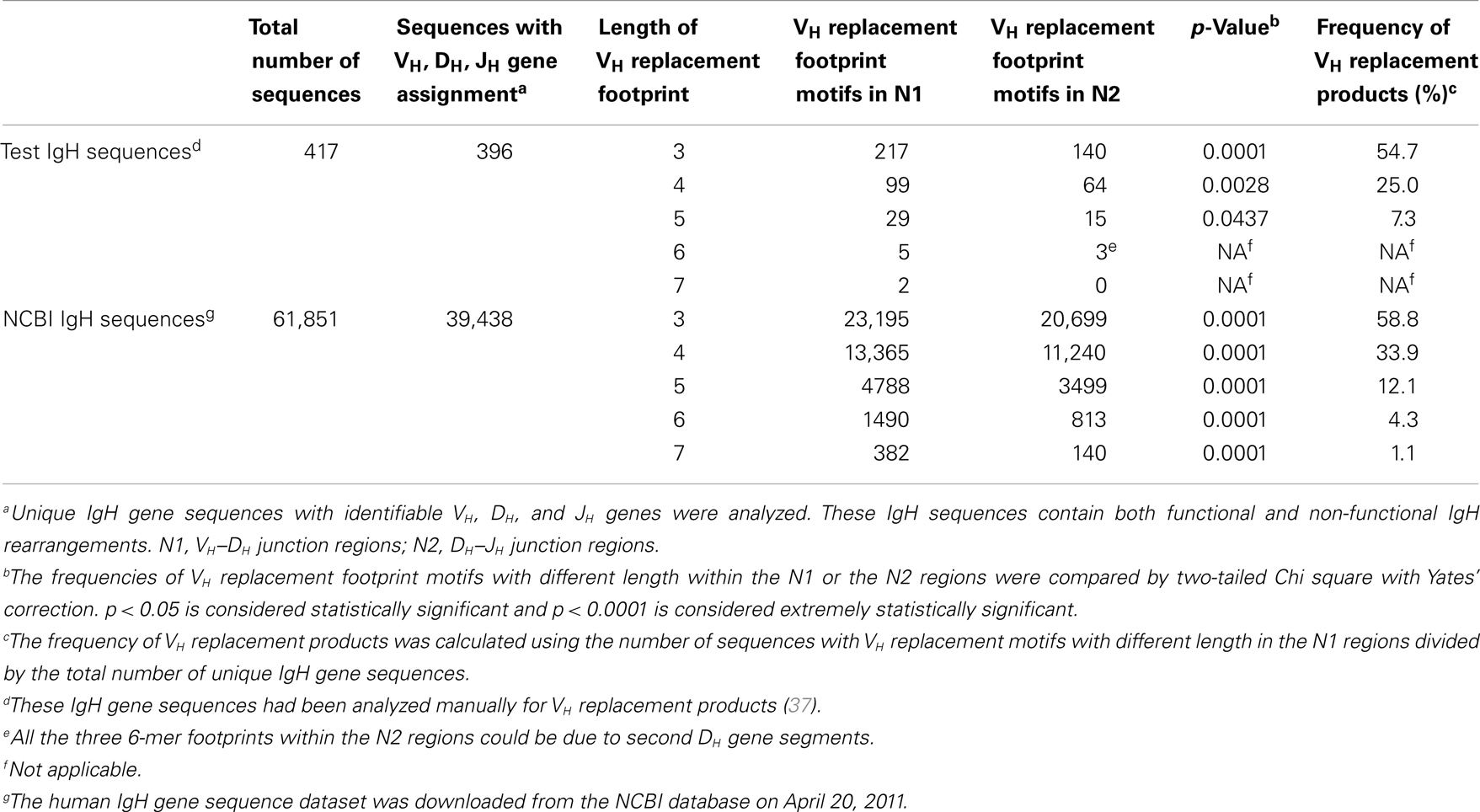
Table 1. Frequencies of VH replacement footprint motifs in the N1 and N2 regions of human IgH genes.
Contribution of VH Replacement Products to the Human IgH Repertoire
With the help of the VHRFA program, we searched for potential VH replacement products in the 39,438 unique human IgH sequences with identifiable VH, DH, and JH genes from the NCBI database. We first compared the frequencies of VH replacement footprint motifs with 3, 4, 5, 6, or 7 nucleotides within the N1 and N2 regions (Table 1, bottom). The frequencies of 3, 4, 5, 6, and 7-mer VH replacement footprint motifs in the N1 regions are extremely statistically significantly higher than those in the N2 regions (Table 1, bottom, p < 0.0001), indicating that the presence of such motifs at the N1 region is likely contributed by VH replacement rather than random nucleotide addition. Among these IgH gene sequences, 12.1% of them contain the 5-mer VH replacement footprint motifs and can be assigned as potential VH replacement products (Table 1, bottom). This number indicates a significant contribution of VH replacement products to the diversification of the human IgH repertoire. If we consider the 4- and 3-mer VH replacement footprints motifs, 33.9 and 55.8% of IgH genes can be assigned as potential VH replacement products (Table 1, bottom).
Within the large number of IgH genes, there are 3818 non-functional IgH gene sequences and 687 of them contain the 5-mer VH replacement footprint motifs in their N1 regions, which can be assigned as potential VH replacement products. The frequency of VH replacement products in non-functional IgH genes (18%) is extremely statistically significantly higher than that in the overall functional IgH genes (p < 0.0001, two-tailed Chi square test with Yates’ correction). Identification of VH replacement products in non-functional IgH genes fulfills the prediction that VH replacement is a random process that can generate both functional and non-functional IgH rearrangement products. Taken together, these results uncovered a previously unrealized contribution of VH replacement products to the diversification of human IgH repertoire.
Distribution of VH Replacement Products in IgH Genes Using Different VH Genes
Using the VHRFA program, we further analyzed the distribution of VH replacement products in IgH genes using different VH genes. The frequencies of VH replacement products in IgH genes using different VH germline genes are different (Figure 2). For examples, the frequencies of VH replacement products in IgH genes using the VH2–5, VH3–30, VH3-30-3, VH1–69, and VH3–34 genes are 23.88, 19.12, 16.64, 14.28, and 13.13%, which are extremely statistically significantly higher than that in IgH genes using the VH6-1 gene (p < 0.0001, two-tailed Fisher’s exact test) (Figure 2). As an internal control, 7.56% of IgH genes using the VH6-1 gene have 5-mer VH replacement footprints within their N1 regions, which is statistically significantly lower than that in the overall IgH gene sequences (p = 0.0004, two-tailed Fisher’s exact test).
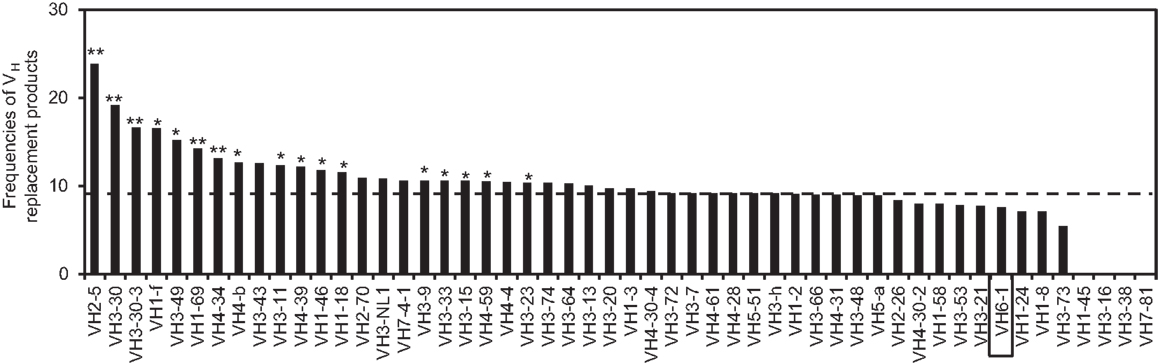
Figure 2. Distribution of VH replacement products in IgH genes using different VH genes. The frequencies of VH replacement products in functional IgH genes using each VH germline genes are compared with that in IgH genes using the VH6-1 gene. **p < 0.0001, *p < 0.05. The result for IgH genes using the VH6-1 gene is highlighted in the box and the frequency of VH replacement products in all the IgH genes is indicated by the dashed line.
VH Replacement Products are Highly Enriched in IgH Genes Derived from Patients with Autoimmune Diseases or Viral Infections
The overall frequency of VH replacement products in the 39,438 unique IgH genes from the NCBI database (12.1%) is much higher than what was estimated in the 417 IgH genes obtained from healthy donors. We reasoned that the majority of IgH gene sequences deposited at the NCBI database was derived from diseased subjects, which may have higher frequencies of VH replacement products. Next, we investigated the distribution of VH replacement products in IgH genes derived from different disease sub-categories. Using the keyword analysis function within the VHRFA program, we can correlate the frequencies of VH replacement products with different sub-categories of IgH gene sequences from the NCBI database. For examples, the frequency of VH replacement products in 558 IgH genes derived from healthy donors is 8.6% (Figure 3), which is similar to the result obtained from previous analysis of the 417 IgH gene sequences from healthy donors. Interestingly, the frequencies of VH replacement products in IgH genes derived from subjects with different autoimmune diseases, such as allergic rhinitis, RA, and SLE are statistically significantly higher than that in the healthy donors (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction; Table S4 in Supplementary Material). The frequencies of VH replacement products are further enriched in IgH genes derived from RA synovium and in IgH genes encoding rheumatoid factors, suggesting that B-cells expressing VH replacement products are positively selected in the RA synovium to encode rheumatoid factors (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction; Table S4 in Supplementary Material). Similarly, VH replacement products are highly enriched in IgH genes derived from SLE plasmablasts (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction; Table S4 in Supplementary Material), suggesting that these enriched VH replacement products contribute to the production of autoAbs in SLE.
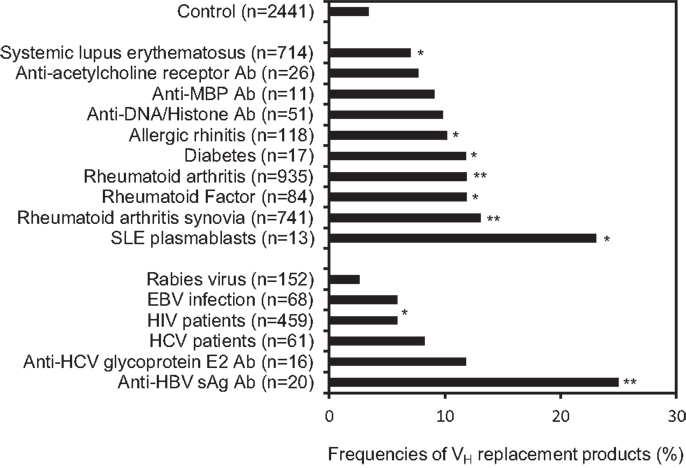
Figure 3. VH replacement products are significantly enriched in IgH genes derived from autoimmune diseases or viral infections and in IgH genes encoding autoreactive or anti-viral Abs. Frequencies of VH replacement products in IgH gene sequences derived from different sub-categories were analyzed based on the identification of pentameric VH replacement footprints within their V–D junctions. The frequencies of VH replacement products in IgH genes derived from different autoimmune diseases and viral infections, or in IgH genes encoding auto Abs, anti-viral Abs, or anti-bacterial Abs were compared with that from healthy controls. The number of analyzed IgH gene sequences in each subcategory are indicated (n). The arrow head indicates the overall frequency of VH replacement products (12.1%) in the 39,438 unique human IgH sequences. Statistical significance was determined using a two-tailed Chi square test with Yate’s correction. *p < 0.05 is considered statistically significant and **p < 0.0001 is considered extremely statistically significant.
The accumulation of VH replacement in IgH genes derived from patients with different autoimmune diseases suggested that VH replacement products may contribute to the production of autoAbs. Indeed, further analyses showed that VH replacement products are statistically significantly enriched in IgH genes encoding rheumatoid factors, anti-Rh (D) Abs, and anti-acetylcholine receptor Abs (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction; Table S4 in Supplementary Material).
To our surprise, the frequencies of VH replacement products are significantly elevated in IgH genes derived from different viral infections. For examples, the frequencies of VH replacement products in IgH genes derived from HIV and HCV infected patients are statistically significantly higher than that in healthy donors (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction; Table S4 in Supplementary Material). Further analyses showed that the VH replacement products contribute to about 30% of IgH genes encoding anti-HCV glycoprotein E2 Abs or anti-HBVsAg Abs. Such frequencies are statistically significantly higher than that in healthy donors (Figure 3, p < 0.05, two-tailed Chi square test with Yates’ correction). Taken together, these results showed that VH replacement products are highly enriched in IgH genes derived from patients with different autoimmune diseases and viral infections.
VH Replacement Elongates the IgH CDR3
VH replacement renews almost the entire VH coding region. Due to the location of the cRSS site, a short stretch of nucleotides is remained as a VH replacement footprint at the newly formed VH–DH junction after the VH replacement process (37). Such VH replacement footprints can contribute up to two amino acids into the IgH CDR3 to elongate the CDR3. The average CDR3 length of the identified VH replacement products is 18.2 ± 5.0 aa (n = 4417), which is extremely statistically significantly longer than that of the non-VH replacement products (15.4 ± 4.4 aa, Figure 3, p < 0.0001, unpaired t-test) (Figure 4). This result confirmed that VH replacement elongates the IgH CDR3 region.
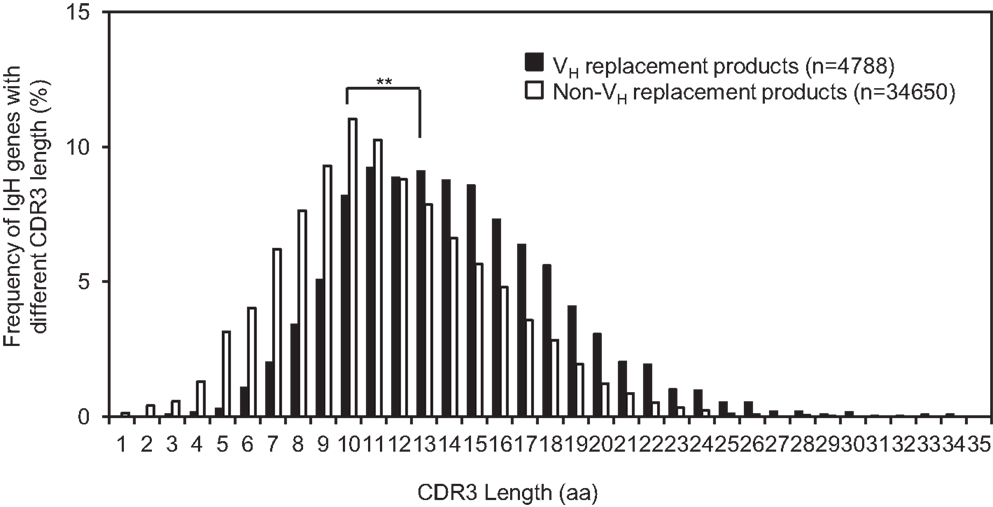
Figure 4. The average CDR3 length of identified VH replacement products is significantly longer than that of non-VH replacement products. The distribution of IgH genes with different CDR3 lengths is shown in the bar graph. The average CDR3 length of VH replacement products (black bars) was compared to that of non-VH replacement products (white bars). Statistical significance was determined by using an unpaired t-test. **p < 0.0001 is considered extremely statistically significant.
The VH Replacement Footprints Preferentially Encode Charged Amino Acids
Our previous analysis showed that the VH replacement footprints preferentially encoded charged amino acids in the IgH CDR3 regions (37, 45). This is likely predetermined by the conservation of amino acid sequence at the 3′ ends of VH germline genes. Here, analysis of the amino acids encoded by the identified pentameric VH replacement footprints in the 4417 VH replacement products showed that 57% of them are charged amino acids. Such frequency is extremely statistically significantly higher than that in the N1 regions of non-VH replacement products (25%) (Figure 5A, p < 0.0001, two-tailed Chi square test with Yates’ correction). Detailed analyses showed that the frequencies of K, R, D, and E residues encoded by the VH replacement footprints are statistically significantly higher than their usage in the N1 regions of non-VH replacement products (Figure 5B, p < 0.05, two-tailed Chi square test with Yates’ correction). These results confirmed our previous prediction that VH replacement footprints preferentially contribute charged amino acids to the IgH CDR3 regions.
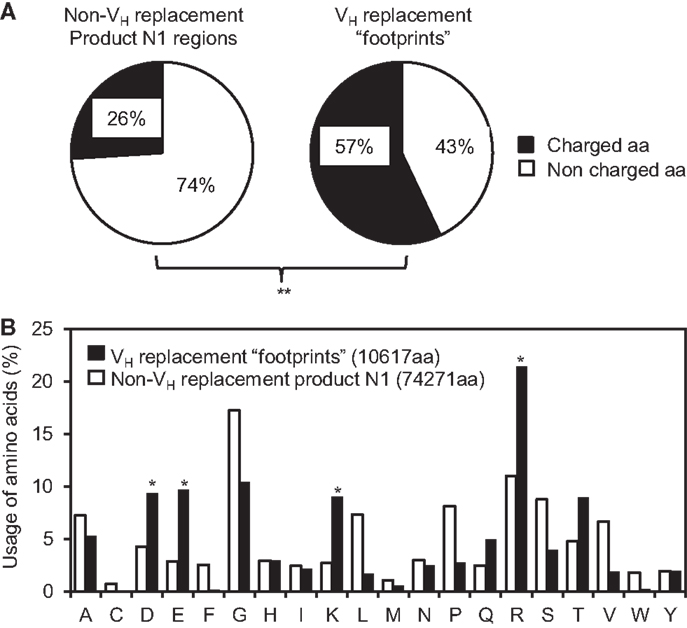
Figure 5. VH replacement footprints preferentially contribute charged amino acids into the IgH CDR3 regions. (A) Frequencies of charged and uncharged amino acids (aa) in the N1 regions of non-VH replacement products were compared with those encoded by the VH replacement footprints. (B) The usages of different amino acids in the N1 regions of non-VH replacement products (white bars) or encoded by the VH replacement footprints (black bars) were analyzed and shown in the bar graph. The total number of amino acids analyzed for each population is indicated. Statistical significance was determined using a two-tailed Chi square test with Yate’s correction. *p < 0.05 is considered statistically significant. **p < 0.0001 is considered extremely statistically significant.
VH Replacement Products are Positively Selected during Autoimmune or Anti-Viral Responses
Charged amino acids within IgH CDR3 are not well tolerated during Ab repertoire development, they are frequently found within the IgH CDR3 regions of autoreactive or anti-viral Abs, which may play important roles in binding charged self or viral antigens, respectively. Further analyses of VH replacement products derived from different autoimmune diseases or viral infections showed that the identified VH replacement footprints predominantly encode charged amino acids (Figure 6A). Detailed analyses showed that the identified VH replacement footprints in IgH genes encoding anti-DNA/histone Abs or rheumatoid factors encoded significantly lower frequencies of negatively charged residues, including D, E, N, and Q residues (Figure 6B, p < 0.05, two-tailed Chi square test with Yates’ correction).
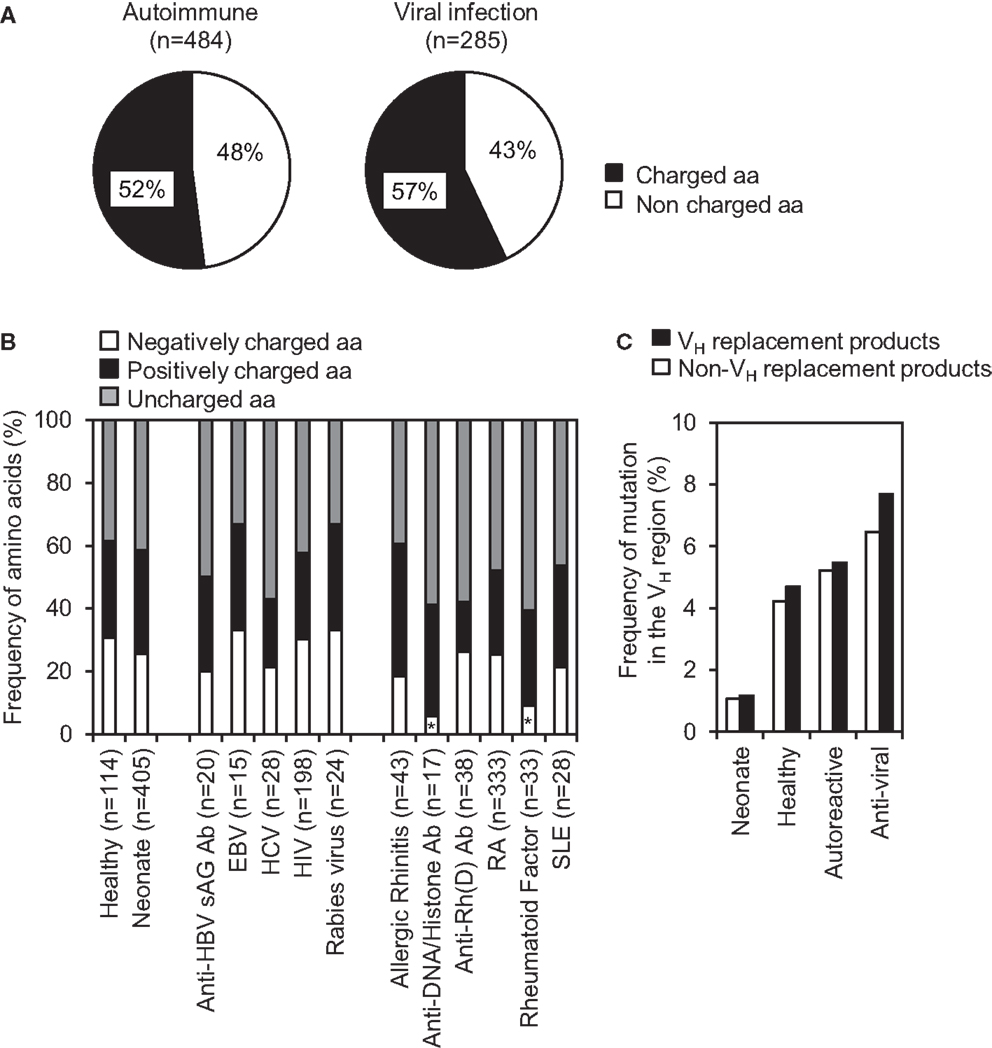
Figure 6. VH replacement footprints preferentially contribute charged amino acids into the CDR3 regions of IgH genes derived from autoimmune diseases and viral infections. (A) Frequencies of charged and uncharged amino acids encoded by the VH replacement footprints in IgH genes derived from autoimmune diseases and viral infections. n, Total number of amino acids analyzed in each subcategory. (B) Frequencies of negatively and positively charged residues encoded by the VH replacement footprints in IgH genes derived from autoimmune diseases or viral infections. Statistical significance was determined using a two-tailed Chi square test with Yate’s correction. *p < 0.05 is considered statistically significant. (C) Comparison of overall somatic mutation rates (%) within VH region of VH replacement products versus non-VH replacement products in different sub-categories.
The identified VH replacement products have similar mutation rate when compared with the non-VH replacement product derived from healthy donors, patients with autoimmune diseases or viral infections (Figure 6C). As negative controls, VH replacement products or non-VH replacement products in neonatal IgH gene sequences have much lower mutation rates (Figure 6C). The accumulation of mutations within these VH replacement products indicates that these enriched VH replacement products in autoimmune diseases or viral infections had been positively selected.
Discussion
In order to determine the distribution of VH replacement products in these IgH genes and explore the biological significance of VH replacement products in human antibody diversification and diseases, we developed a Java based computer program VHRFA to analyze large number of IgH gene sequences and to identify potential VH replacement products (42). Previous analyses of the IgH gene repertoire have provided important insights regarding the developmental process and function of B lineage cells. Due to the tremendous diversity, the complete human IgH repertoire cannot be experimentally determined. Within the NCBI database, there are 61,851 human IgH gene sequences (May, 2012 version). The initial analysis of the VH, DH, and JH gene usages in the 61,851 human IgH gene sequences provides a comprehensive view of the human IgH repertoire. In this dataset, the usage of every functional VH germline gene was confirmed, although their usages differ dramatically.
Using the VHRFA program, we identified VH replacement products and analyzed their distributions in the 39,438 unique IgH sequences. Based on the identification of pentameric VH replacement footprint motifs within the VH–DH junctions, 12.1% of the IgH genes can be assigned as potential VH replacement products. These results confirmed our previous estimation that VH replacement products contribute to the diversification of the human IgH repertoire. Interestingly, the frequencies of VH replacement products in IgH genes using the VH2–5, VH3–30, VH3-30-3, VH3–49, VH1–69, and VH3–34 are statistically significantly higher than that in the overall IgH genes. In contrast, the frequency of VH replacement products in IgH genes using the VH6-1 gene is statistically significantly lower than that in the overall IgH genes. Among the non-functional IgH genes, 18% of them contain the pentameric VH replacement footprints and can be assigned as potential VH replacement products. These results confirmed the prediction that VH replacement is a random process that can generate both functional and non-functional IgH rearrangements. Moreover, the high frequency of VH replacement products in non-functional IgH genes suggested that VH replacement products were negatively selected during B-cell development. Based on this reasoning, the frequency of VH replacement products in the non-functional IgH genes may represent the true frequency of VH replacement during early stages of B-cell development, because these non-functional IgH rearrangements cannot encode BCRs and had not been selected during B-cell development.
Due to the location of the cRSS site, a short stretch of nucleotides has the potential to remain as a VH replacement footprint at the VH–DH junctions following the VH replacement process (25, 37, 46). The leftover VH replacement footprints will elongate the IgH CDR3 regions (25, 37, 46). Analyses of the identified 4788 VH replacement products showed that the average CDR3 length of the identified VH replacement products is 2.8 aa longer than that of non-VH replacement products. Previously, it surprised us that the identified VH replacement footprints preferentially encode charged amino acids within the IgH CDR3 regions (22, 37, 46). Recent analyses showed that the positions of the cRSS and high frequencies of charged amino acids encoded by the following nucleotides are highly conserved in IgH genes from different vertebrates (47). In the current study, 57% of the identified VH replacement footprints encoded charged amino acids in the IgH CDR3 regions. Normally, charged amino acids within IgH CDR3 are not well tolerated during antibody repertoire development, probably due to charged residues may generate autoAbs. Indeed, our analysis revealed that VH replacement products are significantly enriched in IgH genes derived from patients with different autoimmune diseases, including RA, allergic rhinitis, and SLE or in IgH genes encoding different autoAbs such as rheumatoid factor, anti-rhesus D antigen, and anti-acetylcholine receptor Abs. Our recent analyses of large number of mouse IgH genes also showed that the frequencies of VH replacement products are enriched in IgH genes derived from autoimmune prone mice (48). These results suggested that VH replacement products contribute to the generation of autoantibodies in both human and mouse.
Another important and interesting finding from this analysis of large number of IgH gene sequences is that the frequencies of VH replacement products are significantly elevated in IgH genes derived from various viral infections, including HIV, HCV, and in IgH genes encoding Abs against HCV glycoprotein E2 or HBV surface antigens. Our recent studies showed that VH replacement products are highly enriched in IgH genes encoding different subgroups of anti-HIV antibodies, especially in CD4i and PGT antibodies (49). These results suggested that VH replacement products may contribute to the generation of anti-viral Abs. The majority of the VH replacement footprints identified from anti-viral Abs also encode charged amino acids, which may be important for binding charged viral antigens. Moreover, the accumulation of mutations in these VH replacement products indicated that these enriched VH replacement products in patients with viral infections are positively selected during anti-viral responses. The identification of VH replacement products in autoimmune diseases and anti-viral responses suggested a potential link between viral infections and the pathogenesis of autoimmune diseases. It has long been postulated that chronic viral infections contribute to autoimmunity. However, clear examples that Abs against viral antigens cross-react with self-antigens have only been found in a few cases (50, 51). Here, our results reveal a shared pattern of accumulation of VH replacement products in IgH genes derived from autoimmune diseases and anti-viral responses.
VH replacement was originally proposed as a receptor editing mechanism to change unwanted IgH genes that are either non-functional or encoding autoreactive Abs. The enrichment of VH replacement products in IgH genes derived from autoimmune diseases or encoding autoAbs is particular puzzling. There are several possible mechanisms to explain this finding. First, we have recently shown that crosslinking cell surface BCRs induces VH replacement in human immature B-cells (40). Thus, the levels of VH replacement recombination might be induced in the immature B-cells during either the anti-viral immune response or autoimmune disease due to persistent antigen stimulation or chronic inflammation. In supporting of this assumption, the number of newly emigrated immature B-cells in the peripheral blood is increased during inflammatory response; and these mobilized immature B-cells may continue to undergo VH replacement recombination ectopically. Second, the intrinsic feature of VH replacement is elongating the IgH CDR3 with charged amino acid. VH replacement products may frequently encode autoAbs and they are efficiently deleted during normal B-cell development. The observed elevated frequencies of VH replacement products in different autoimmune diseases may reflect the defective negative selection in these diseased subjects. Moreover, ectopically occurred VH replacement may bypass the stringent negative selection in the bone marrow and release VH replacement products in the periphery. Last, due to the special features of VH replacement products in generating IgH genes with long and charged CDR3, it is possible that VH replacement products are positively selected by viral antigens during anti-viral responses to produce specific anti-viral Abs. In supporting of this notion, the identified potential VH replacement products encoding anti-HIV antibodies all have very long CDR3 regions with multiple charged amino acid residues (49). The accumulated mutations within the VH genes of the identified VH replacement products in the current study also indicated the positive selection. However, the leftover VH replacement products generated during a chronic viral infection may encode Abs that cross-react with self-antigens and later contribute to autoimmunity. In fact, many cell surface antigens and viral antigens are negatively charged, which may be a reason for the selection of VH replacement products with long and charged CDR3 regions.
In our sequence based analysis, the assignment of VH replacement is dependent on the identification of VH replacement footprints within the VH–DH junctions. Any deletion at the 3′ of VH genes or the 5′ of VH replacement footprint motifs during the primary or secondary IgH gene recombination, respectively, may destroy the pentameric VH replacement footprints. Therefore, it is possible that the sequence analysis based study still under-estimates the frequency of VH replacement products. Using the VHRFA program, we extended our analysis our VH replacement products to include potential VH replacement footprint motifs with different lengths. For examples, 33.9% of the IgH genes contain the tetrameric VH replacement footprint motifs and 58.8% of IgH genes contain the trimeric VH replacement footprint motifs. These results revealed a significant contribution of VH replacement products to the IgH repertoire. Recent studies in mice carrying non-functional IgH genes on both IgH alleles demonstrated that VH replacement occurs efficiently to generate almost normal numbers of B-cells with diversified IgH repertoires (52). However, only about 20% of the IgH gene sequences from this study contained residual VH replacement footprints. Therefore, the majority IgH genes generated through VH replacement recombination have no leftover VH replacement footprints. Theoretically, 66% of IgH rearrangements will be out of reading frame and 44% of developing B-cells may carry non-functional IgH rearrangements on both alleles. If all of these B-cells are rescued by VH replacement, a minimum of 44% of the IgH genes might be generated through VH replacement recombination. Under this assumption, IgH genes containing the tetrameric or the trimeric VH replacement footprint motifs at their N1 regions should also be considered as potential VH replacement products.
Like any sequence based analysis program, the VHRFA program also has its limitation. Although sequence motifs assemble the VH gene 3′ ending sequences can be identified in the N1 regions, such motifs can also be identified within the N2 regions at relative lower frequencies. Theoretically, VH replacement can only leave footprint within the N1 region; the existence of VH replacement footprint like motifs within the N2 regions can only be generated by random nucleotide addition. For IgH genes using the VH6-1 gene, which is the first VH germline gene 5′ to the DH locus, there should have no VH replacement footprint like motifs within the VH–DH junctions, but the VHRFA program still identifies 7.56% of the sequences contains VH replacement footprint like motifs within the VH–DH junctions. We can only refer such motifs as the contribution of random nucleotide addition.
In summary, analyses of a large number of human IgH gene sequences from the NCBI database uncovered a significant contribution of VH replacement products to human Ab repertoire, especially in IgH genes derived from autoimmune diseases or anti-viral responses. Understanding how VH replacement is regulated and how VH replacement products are positively or negatively selected during normal or diseased conditions will be the focus of future studies, because modulation of the level of VH replacement may offer unique approaches to treat different human diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Miles D. Lange, Lin Huang, and Zhixin Zhang conceived and designed the study. Lin Huang developed the Java-based VHRFA software. Miles D. Lange and Lin Huang analyzed the raw data and generated figures and tables. Miles D. Lange, Lin Huang, and Zhixin Zhang validated the results. All authors contributed to the development of the project and final writing of the manuscript. This study was supported in part by NIH grants AI074948 (Zhixin Zhang), AI076475 (Zhixin Zhang), and AR059351 (Kaihong Su). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fimmu.2014.00345/abstract
Abbreviations
aa, amino acid; cRSS, cryptic recombination signal sequence; EBV, Epstein–Barr virus; HBV, hepatitis virus B; HCV, hepatitis virus C; HIV, human immunodeficiency virus; RA, rheumatoid arthritis; RA, rheumatoid arthritis; RAG, recombination activating gene products; SLE, systemic lupus erythematosus; VHRFA, VH replacement footprint analyzer.
References
1. Meffre E, Casellas R, Nussenzweig MC. Antibody regulation of B cell development. Nat Immunol (2000) 1:379–85. doi: 10.1038/80816
2. Rajewsky K. Clonal selection and learning in the antibody system. Nature (1996) 381:751–8. doi:10.1038/381751a0
3. Tonegawa S. Somatic generation of antibody diversity. Nature (1983) 302:575–81. doi:10.1038/302575a0
4. Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science (1990) 248:1517–23. doi:10.1126/science.2360047
5. Schatz DG, Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell (1988) 53:107–15. doi:10.1016/0092-8674(88)90492-8
6. Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell (1989) 59:1035–48. doi:10.1016/0092-8674(89)90760-5
7. Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol (1994) 56:27–150. doi:10.1016/S0065-2776(08)60450-2
8. Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res (1994) 22:1785–96. doi:10.1093/nar/22.10.1785
9. Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity (1998) 9:115–25. doi:10.1016/S1074-7613(00)80593-2
10. ten Boekel E, Melchers F, Rolink AG. Changes in the V(H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity (1997) 7:357–68. doi:10.1016/S1074-7613(00)80357-X
11. ten Boekel E, Melchers F, Rolink AG. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity (1998) 8:199–207. doi:10.1016/S1074-7613(00)80472-0
12. Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J (1984) 3:1209–19.
13. Burrows P, LeJeune M, Kearney JF. Evidence that murine pre-B cells synthesise μ heavy chains but no light chains. Nature (1979) 280:838–40. doi:10.1038/280838a0
14. Cappione AJ, Pugh-Bernard AE, Anolik JH, Sanz I. Lupus IgG VH4.34 antibodies bind to a 220-kDa glycoform of CD45/B220 on the surface of human B lymphocytes. J Immunol (2004) 172:4298–307. doi:10.4049/jimmunol.172.7.4298
15. Odendahl M, Jacobi A, Hansen A, Feist E, Hiepe F, Burmester GR, et al. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. J Immunol (2000) 165:5970–9. doi:10.4049/jimmunol.165.10.5970
16. Pugh-Bernard AE, Silverman GJ, Cappione AJ, Villano ME, Ryan DH, Insel RA, et al. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J Clin Invest (2001) 108:1061–70. doi:10.1172/JCI12462
17. Bridges SL, Lavelle JC, Lee SK, Byer S, Schroeder HW. CDR3 fingerprinting of immunoglobulin kappa light chains expressed in rheumatoid arthritis. Evidence of antigenic selection or dysregulation of gene rearrangement in B cells. Ann N Y Acad Sci (1997) 815:423–6. doi:10.1111/j.1749-6632.1997.tb52093.x
18. Bridges SL, Lee SK, Koopman WJ, Schroeder HW. Analysis of immunoglobulin gamma heavy chain expression in synovial tissue of a patient with rheumatoid arthritis. Arthritis Rheum (1993) 36:631–41. doi:10.1002/art.1780360509
19. Grimaldi CM, Hicks R, Diamond B. B cell selection and susceptibility to autoimmunity. J Immunol (2005) 174:1775–81. doi:10.4049/jimmunol.174.4.1775
20. Rajewsky K, Forster I, Cumano A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science (1987) 238:1088–94. doi:10.1126/science.3317826
21. Nemazee D, Weigert M. Revising B cell receptors. J Exp Med (2000) 191:1813–7. doi:10.1084/jem.191.11.1813
22. Zhang Z, Burrows PD, Cooper MD. The molecular basis and biological significance of VH replacement. Immunol Rev (2004) 197:231–42. doi:10.1111/j.0105-2896.2004.0107.x
23. Casellas R, Shih TA, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, et al. Contribution of receptor editing to the antibody repertoire. Science (2001) 291:1541–4. doi:10.1126/science.1056600
24. Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell (1998) 92:173–82. doi:10.1016/S0092-8674(00)80912-5
25. Zhang Z. VH replacement in mice and humans. Trends Immunol (2007) 28:132–7. doi:10.1016/j.it.2007.01.003
26. Kleinfield R, Hardy RR, Tarlinton D, Dangl J, Herzenberg LA, Weigert M. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature (1986) 322:843–6. doi:10.1038/322843a0
27. Reth M, Gehrmann P, Petrac E, Wiese P. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature (1986) 322:840–2. doi:10.1038/322840a0
28. Usuda S, Takemori T, Matsuoka M, Shirasawa T, Yoshida K, Mori A, et al. Immunoglobulin V gene replacement is caused by the intramolecular DNA deletion mechanism. EMBO J (1992) 11:611–8.
29. Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity (1995) 3:747–55. doi:10.1016/1074-7613(95)90064-0
30. Chen C, Nagy Z, Radic MZ, Hardy RR, Huszar D, Camper SA, et al. The site and stage of anti-DNA B-cell deletion. Nature (1995) 373:252–5. doi:10.1038/373252a0
31. Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity (1997) 6:97–105. doi:10.1016/S1074-7613(00)80673-1
32. Cascalho M, Ma A, Lee S, Masat L, Wabl M. A quasi-monoclonal mouse. Science (1996) 272:1649–52. doi:10.1126/science.272.5268.1649
33. Cascalho M, Wong J, Wabl M. VH gene replacement in hyperselected B cells of the quasimonoclonal mouse. J Immunol (1997) 159:5795–801.
34. Lutz J, Muller W, Jack HM. VH replacement rescues progenitor B cells with two nonproductive VDJ alleles. J Immunol (2006) 177:7007–14. doi:10.4049/jimmunol.177.10.7007
35. Davila M, Liu F, Cowell LG, Lieberman AE, Heikamp E, Patel A, et al. Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells. J Exp Med (2007) 204:3195–208. doi:10.1084/jem.20071224
36. Watson LC, Moffatt-Blue CS, McDonald RZ, Kompfner E, it-Azzouzene D, Nemazee D, et al. Paucity of V-D-D-J rearrangements and VH replacement events in lupus prone and nonautoimmune TdT-/- and TdT+/+ mice. J Immunol (2006) 177:1120–8. doi:10.4049/jimmunol.177.2.1120
37. Zhang Z, Zemlin M, Wang Y-H, Munfus D, Huye LE, Findley HW, et al. Contribution of VH gene replacement to the primary B cell repertoire. Immunity (2003) 19:21–31. doi:10.1016/S1074-7613(03)00170-5
38. Sandel PC, Monroe JG. Negative selection of immature B cells by receptor editing or deletion is determined by site of antigen encounter. Immunity (1999) 10:289–99. doi:10.1016/S1074-7613(00)80029-1
39. Verkoczy L, it-Azzouzene D, Skog P, Martensson A, Lang J, Duong B, et al. A role for nuclear factor kappa B/rel transcription factors in the regulation of the recombinase activator genes. Immunity (2005) 22:519–31. doi:10.1016/j.immuni.2005.03.006
40. Liu J, Lange MD, Hong SY, Xie W, Xu K, Huang L, et al. Regulation of VH replacement by B cell receptor-mediated signaling in human immature B cells. J Immunol (2013) 190:5559–66. doi:10.4049/jimmunol.1102503
41. Radic M, Zouali M. Receptor editing, immune diversification and self-tolerance. Immunity (1996) 5:505–11. doi:10.1016/S1074-7613(00)80266-6
42. Huang L, Lange MD, Zhang Z. VH replacement footprint analyzer-I, a Java-based computer program for analyses of immunoglobulin heavy chain genes and potential VH replacement products in human and mouse. Front Immunol (2014) 5:40. doi:10.3389/fimmu.2014.00040
43. Zemlin M, Bauer K, Hummel M, Pfeiffer S, Devers S, Zemlin C, et al. The diversity of rearranged immunoglobulin heavy chain variable region genes in peripheral blood B cells of preterm infants is restricted by short third complementarity-determining regions but not by limited gene segment usage. Blood (2001) 97:1511–3. doi:10.1182/blood.V97.5.1511
44. Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res (2008) 36:W503–8. doi:10.1093/nar/gkn316
45. Liu Y, Fan R, Zhou S, Yu Z, Zhang Z. Potential contribution of VH gene replacement in immunity and disease. Ann N Y Acad Sci (2005) 1062:175–81. doi:10.1196/annals.1358.020
46. Zhang Z, Wang YH, Zemlin M, Findley HW, Bridges SL, Burrows PD, et al. Molecular mechanism of serial VH gene replacement. Ann N Y Acad Sci (2003) 987:270–3. doi:10.1111/j.1749-6632.2003.tb06060.x
47. Sun Y, Liu Z, Li Z, Lian Z, Zhao Y. Phylogenetic conservation of the 3’ cryptic recombination signal sequence (3’cRSS) in the VH genes of jawed vertebrates. Front Immunol (2012) 3:392. doi:10.3389/fimmu.2012.00392
48. Huang L, Lange MD, Yu Y, Li S, Su K, Zhang Z. Contribution of VH replacement products in mouse antibody repertoire. PLoS One (2013) 8:e57877. doi:10.1371/journal.pone.0057877
49. Liao H, Guo JT, Lange MD, Fan R, Zemlin M, Su K, et al. Contribution of VH replacement products to the generation of anti-HIV antibodies. Clin Immunol (2013) 146:46–55. doi:10.1016/j.clim.2012.11.003
50. Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. EBV and systemic lupus erythematosus: a new perspective. J Immunol (2005) 174:6599–607. doi:10.4049/jimmunol.174.11.6599
51. McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med (2005) 11:85–9. doi:10.1038/nm1167
Keywords: B-cell, antibody, IgH genes, cryptic RSS, VH replacement, VH replacement footprint, autoimmune disease, anti-viral response
Citation: Lange MD, Huang L, Yu Y, Li S, Liao H, Zemlin M, Su K and Zhang Z (2014) Accumulation of VH replacement products in IgH genes derived from autoimmune diseases and anti-viral responses in human. Front. Immunol. 5:345. doi: 10.3389/fimmu.2014.00345
Received: 23 May 2014; Accepted: 06 July 2014;
Published online: 22 July 2014.
Edited by:
Harry W. Schroeder, University of Alabama at Birmingham, USAReviewed by:
Biao Zheng, Baylor College of Medicine, USA; University of Texas Medical School, USAHarry W. Schroeder, University of Alabama at Birmingham, USA
Copyright: © 2014 Lange, Huang, Yu, Li, Liao, Zemlin, Su and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixin Zhang, LTC 11706A, Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE 68198-7660, USA e-mail: zhangj@unmc.edu
 Miles D. Lange
Miles D. Lange Lin Huang1
Lin Huang1 Yangsheng Yu
Yangsheng Yu Hongyan Liao
Hongyan Liao Michael Zemlin
Michael Zemlin Kaihong Su
Kaihong Su Zhixin Zhang
Zhixin Zhang