Coexisting protist-bacterial community accelerates protein transformation in microcosm experiments
- 1Center for Marine Environmental Studies, Ehime University, Matsuyama, Japan
- 2Faculty of Environment and Resources, Nong Lam University, Ho Chi Minh City, Vietnam
- 3Faculty of Engineering, Yokohama National University, Yokohama, Japan
Proteins constitute the major portion of labile substances in the marine environment and are an important source of organic matter supporting marine ecosystems. However, previous studies have revealed that specific bacterial membrane proteins are refractory in the oceans. We here show by kinetic analyses of protease degradation activity using inactivated Pseudomonas aeruginosa (Pa) cells as a proteinaceous substrate that bacterial proteases are insufficient to completely hydrolyze proteins, which may partially cause the protein accumulation in seawater. Protease activity was monitored simultaneously in 8 microcosms subjected to differing conditions. Some Pa proteins were retained for 30 days in the presence of bacteria without protists, whereas the Pa proteins were completely disappeared in the presence of both, indicating that these proteins were substantially incorporated into protist biomass. Our result suggests that protists play an important role in the transformation of bacterial proteins in seawater. Our experiments also imply that the functional/taxonomic diversity should be taken into account when considering decomposition activity in marine environments.
Introduction
Proteins make up more than 50% of the organic matter found in marine organisms (Romankevich, 1984), suggesting a large input from proteins to the dissolved combined amino acid (DCAA) pool, which is comprised of those amino acids released after hydrolysis of macromolecules (Keil and Kirchman, 1993). A majority of DCAA pool, indeed, consists of proteins (Keil and Kirchman, 1993), and can support a substantial fraction of bacterial growth (Keil and Kirchman, 1993; Rosenstock and Simon, 1993; Middelboe et al., 1995). Proteins and their precursors, amino acids, have also been detected at high enough concentrations to be considered an important contributor to the carbon and nitrogen pools (Keil et al., 1994; McCarthy et al., 1998; Horiuchi et al., 2004). The wide distribution and biological importance of proteins, therefore, have attracted much attention to their fates in aquatic environments. It is generally assumed that proteins are labile in seawater and thus are not readily preserved (Hollibaugh and Azam, 1983). However, research has indicated that proteinaceous materials are preserved in freshwater, estuarine, and marine waters and sediments (Tanoue, 1995; Tanoue et al., 1996; Nguyen and Harvey, 1997, 1998; Fogel and Tuross, 1999; Pantoja and Lee, 1999a; Moore et al., 2012). Tanoue et al. (1995), found that approximately 30 polypeptides with molecular masses ranging from 30 to 48 kDa were detected in seawater samples collected from the Arctic to the Antarctic. Among these polypeptides, the 48-kDa protein was found to be the major dissolved protein, with a concentration greater than 0.1 μg l−1. This protein was shown to be an intact homolog of porin P (OprP), an outer membrane porin protein induced by phosphate depletion in Pseudomonas aeruginosa (Hancock et al., 1982; Tanoue et al., 1995). Porins derived from diverse bacterial groups were later shown to be a major source of dissolved protein (Suzuki et al., 1997, 2000; Yamada and Tanoue, 2009). One explanation for the detection of membrane vesicle-associated proteins in the oceans is that they are degraded slowly due to the low collision frequency between vesicles and bacteria resulting from the low protein concentration of seawater (Nagata and Kirchman, 1996). Steric protection of bacterial membrane proteins by the membrane itself as well as cell wall components also impedes bacterial degradation of these proteins (Nagata et al., 1998) and represent important mechanisms enhancing the preservation of proteins (Moore et al., 2012). In addition, the chemical forms of proteins (i.e., “fresh” proteins, and “modified” proteins) also influence both their degradation and utilization by bacterial communities (Keil and Kirchman, 1993; Azúa et al., 2003; McKee et al., 2014).
Marine heterotrophic bacteria use extracellular to assimilate protein, thus transforming dissolved organic matter into particulate matter available to higher trophic levels. The transformation process composes of two steps. Polymers and particles are first hydrolyzed by extracellular enzymes into smaller oligo-sized substances, which then diffuse into the bacterial periplasmic space through porins (Hancock and Brinkman, 2002) and are incorporated into the cell biomass interacting with permeases (Azam and Cho, 1987). The aminopeptidases have been thought to be the major proteolytic enzymes in seawater (Chróst, 1990; Bochdansky et al., 1995; Crump et al., 1998; Murrell et al., 1999); however, more recent work indicates that trypsin-type enzymes are also detectable at different levels in seawater (Obayashi and Suzuki, 2005; Bong et al., 2013; Steen and Arnosti, 2013). Although the source(s) of proteolytic enzymes could be direct secretion from marine bacteria (Arnosti, 2011) and mesophilic archaea (Lloyd et al., 2013) into the surrounding water, sources other than bacteria and archaea can be hypothesized (Mohapatra and Fukami, 2004; Salerno and Stoecker, 2009).
Proteins and proteases can be an important model for use in studying the role of microbial communities in determining the fate of organic matter. A number of proteases of both marine bacteria (Arnosti et al., 2005; Obayashi and Suzuki, 2005; Obayashi et al., 2010; Kiersztyn et al., 2012; Bong et al., 2013) and protists (Richardot et al., 1999; Sala and Güde, 1999; Mohapatra and Fukami, 2004) have been detected in seawater. However, with few efforts to study the effect of grazers on protein hydrolysis (Pérez et al., 2003; Kawasaki and Benner, 2006), the role of proteolytic enzymes in protein degradation in communities composed of both bacteria and protists is poorly understood. Previous studies have sought to determine the role of microbial enzymes in the protein degradation process in seawater (Suzuki et al., 1997, 1999; Obayashi and Suzuki, 2008). Here, we expand on previous research by examining the role of protists as well through monitoring of both enzyme activity and substrate fate in seawater microcosms. By comparing proteolytic activity and protein degradation profiles in samples of fractionated seawater, we elucidated some of the mechanistic basis of variability on the protein degradation process within different microbial communities. The inclusion of protists in our analyses provides a more comprehensive understanding of the processing of proteins in seawater because previous research has focused primarily on bacterial communities.
Materials and Methods
Sample Collection
Seawater was collected at Mori port, Matsuyama, Ehime, Japan (33°48'39.30″ N, 132°41'40.16″ E) in August 2011. The seawater parameters were: water temperature 26.5°C, pH 7.8, and salinity 31 (psu). Seawater was pre-filtered through a 50-μm nylon mesh to remove large particles and phytoplankton, contained and kept cold in 10-l plastic buckets until being processed within 2–3 h. Portions of collected seawater were then sequentially filtered through a GF/F filter (Whatman, 0.7-μm nominal mesh size) and 0.2-μm disposable capsule filter (Advantec). Treated seawater was stored at 4°C and used in experiments within 12 h.
Model Protein Substrate Preparation
Marine Pseudomonas aeruginosa strain eco3 was employed as a substrate source. Strain eco3 was isolated from a coastal site off Ehime, Japan (Nonaka et al., 2010). The bacterium was cultured in a medium with phosphate-deficient artificial seawater (Hancock et al., 1982) with 0.1 M sodium N-2-hydroxyethyl piperazine-N'-2-ethanesulfonate (HEPES) (pH 7.0), 7 mM (NH4)2SO4, 20 mM sodium succinate (pH 7.0), 0.1% (v/v) trace ion mixture described in Hancock et al. (1981), 0.2 mM sodium phosphate buffer (pH 7.0), and 1% proteose peptone No. 3 (Becton, Dickinson). Cells were incubated at 37°C with shaking at 105 rpm in the dark, which allowed the production of OprP. After 24 h of incubation, the bacterium was harvested by centrifugation at 3000 × g for 30 min at 4°C (Himac CF16RX, Hitachi). The cells were resuspended in phosphate buffered saline (PBS) and then disrupted by sonicating on ice 6 times for 10 s each at a power of 50 W using an ultrasonic disruptor (UD-201, TOMY). The sonicated-P. aeruginosa cells are hereafter referred to by the abbreviation “Pa” to distinguish them from natural bacteria.
Microcosm Experiment
Eight microcosms were prepared in duplicate to assess the degradation of Pa in seawater.
Approximately 0.4 g wet weight of Pa was suspended in 0.8 l of treated water in 1-l polycarbonate bottles (Nalgene, Nunc). The microcosm bottles were: natural seawater plus Pa cells (bottle name: SW+Pa), GF/F-filtered seawater (nominal mesh size 0.7 μm) plus Pa cells (SW-0.7+Pa), 0.2-μm-filtered seawater plus Pa cells (SW-0.2+Pa), and autoclaved seawater plus Pa cells (Auto-SW+Pa). Microcosms without Pa were designated as SW, SW-0.7, SW-0.2, and Auto-SW. All microcosms were incubated at 26°C in the dark and shaken by hand once a day. A subsample was taken from each bottle at the start of the microcosm experiment (day 0) and on days 3, 5, 10, 15, and 30 to assess the abundance of bacteria and protists, measure enzyme (protease) activity, and determine the Pa protein profile. Although the use of GF/F traps almost half of bacteria (Lee and Fuhrman, 1987), this filtration method is applicable to remove protist and retain natural bacteria.
Bacterial and Protist Cell Counts
The abundances of bacteria and protists were determined by direct cell counting using epifluorescence microscopy. Subsamples were fixed with formaldehyde (final concentration = 2%). For bacterial cell counts, fixed subsamples were appropriately diluted with PBS and then stained with DAPI (final concentration = 0.3 μg ml−1) for 7 min in the dark (Sherr et al., 2001). Stained samples were filtered onto 0.2-μm black nucleopore filters (Advantec), washed several times with PBS, and observed using epifluorescence microscopy (×1000, Olympus BX51, Olympus). Samples were enumerated in duplicate for the Pa-containing microcosms and in triplicate for the Pa-free microcosms. Approximately 200 cells or 40 views were counted for each subsample. The coefficient of variation (CV) of bacterial counting was 10.8 ± 9.6% (average ± SD; n = 120).
Protist cells were counted according to the method reported by Sherr and Sherr (1983). Briefly, fixed samples were stained with DAPI for 7 min and then filtered onto black 0.8-μm nuclepore filters (Whatman). The filters were then flooded with fluorescein iso-thiocyanate (FITC) solution for 10 min while in the filter holders. The FITC was then allowed to pass through and the filters were rinsed several times with ice-cold 0.5 M sodium carbonate buffer (pH 9.5) and viewed under an epifluorescence microscope (×1000, Olympus BX51). Samples were analyzed in duplicate. The CV of protist counting was 47.4 ± 34.4% (average ± SD; n = 24).
Protease Activity
Protease activity was measured fluorometrically by monitoring the hydrolytic product 7-amino-4-methylcoumarin (AMC), which results from the hydrolysis of synthetic peptide analog methylcoumarylamide (MCA) substrates (Peptide Institute, Osaka). The MCA substrates employed were Leu-MCA, Boc-Phe-Ser-Arg-MCA, and Suc-Ala-Ala-Pro-Phe-MCA for determination of leucine-aminopeptidase, trypsin-type, and chymotrypsin-type activities, respectively (Obayashi and Suzuki, 2005). Each substrate was dissolved in dimethyl sulfoxide (DMSO, final concentration = 10%) and mixed with samples at a final concentration of 100 μM in a total volume of 300 μl. The concentration of AMC in each assay mixture was measured using a microplate reader (SH8100, Corona) at 380/460 nm (excitation/emission) both before and after incubation at 26°C for 1 h. All samples were measured in duplicate. By comparing the enzyme activity of Auto-SW+Pa microcosm with the other Pa-added microcosms, it was possible for us to be sure that we were tracking the enzyme activity of the natural community vs. Pa cells. The activity in Auto-SW+Pa showed the potential activity of Pa cells (injured and surviving Pa cells) because the community composed of only Pa. In SW-0.2+Pa, SW-0.7+Pa, and SW+Pa microcosms, there were other microbes besides Pa cells. The usage of fluorogenic substrates (MCA-substrates) could also affect the results by introducing substrate competiveness between Pa proteins and MCA-proteins in mixtures of seawater and DMSO because the peptide bonds attached to the aromatic fluorescent tags are difficult to cleave (Stevenson, 1994; Pantoja and Lee, 1999b). Nevertheless, by carrying out the same procedure at every measurement, the comparison of protease activity to obtain the changing profiles over time was possible. Although this precludes the extrapolation of the results to the functioning of full natural communities, our findings can still be regarded as insights into the potential of the combination between bacteria and protists to degrade proteins in seawater.
Protein Degradation Profiling
Degradation of the crude protein fraction of Pa cells was monitored by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Sambrook and Russell, 2001). Subsamples of microcosm water (40 or 80 ml) were mixed with trichloroacetic acid (TCA, final concentration = 10% [w/v]) and left on ice for 30 min. The resulting precipitates were centrifuged at 10,000 × g for 15 min at 4°C, after which 1 ml of ice-cold ethanol was added. The precipitates were recovered by centrifugation and sequentially washed with 1 ml of diethyl ether and then air-dried. The dried pellets were dissolved in SDS-PAGE sample buffer (50 mM Tris-HCl pH 6.8, 100 mM dithiothreitol, 2% [w/v] SDS, 10% [v/v] glycerol, and 0.1% [w/v] bromophenol blue), heated at 95°C for 3 min, then loaded onto a 15% polyacrylamide gel with a 5% stacking gel. The amount of sample loaded into each lane was equivalent to 2 ml of microcosm seawater. Electrophoresis was performed at a constant current of 20 mA for 40 min. After electrophoresis, the gel was stained with Coomassie brilliant blue-R250 (CBB-R250). Protein band intensities were scanned for semi-quantifying by densitometry with ImageJ software version 1.48 k. The protein intensity for each microcosm was expressed as a percent of the initially added Pa in the same gel, in order to eliminate variation due to variable staining intensity among gels. We initially added Pa cells in each microcosm as 0.4 g wet cells 800 ml−1, which gave 0.02 μg protein ml−1 lane−1 in SDS-PAGE analysis.
Results
Changes in the Abundance of Bacteria and Protists
Changes in the number of bacteria over time are summarized in Figure 1. The initial cell densities were: 2.0–3.5 × 107 cells ml−1 in the Pa-containing microcosms (indicating the sum of natural bacteria and Pa cells), 6.9–10.0 × 105 cells ml−1 in unfiltered (SW) and GF/F-filtered (SW-0.7) microcosms (indicating natural bacteria), and 2.0 × 104 cells ml−1 in 0.2-μm-filtered (SW-0.2) and autoclaved (Auto-SW) microcosms (indicating the effects of filtration and autoclaving). Among the microcosms without Pa addition, there was no apparent change in the cell density in the SW, Auto-SW, and SW-0.7 microcosms during the incubation period, whereas the density in the SW-0.2 microcosm rapidly increased from 2.5 × 104 to 5.1 × 105 cells ml−1 by day 5 and remained at this level throughout the study period. In the Pa-containing microcosms, the density of cells in the Auto-SW+Pa, SW-0.7+Pa, and SW-0.2+Pa samples remained fairly stable; however, the density of cells in the SW+Pa microcosm decreased between days 5 and 10. Because the seawater used for the SW and SW+Pa microcosms was not filtered, bacterial predators were not removed. Microscopic observation confirmed that the SW and SW+Pa microcosms contained protists, and it was determined that 1.7 × 103 and 2.1 × 103 protist cells ml−1 were present on day 0 in the SW and SW+Pa microcosms, respectively (Figure 2). The numbers of protists in the SW+Pa and SW microcosms increased slightly to 8.1 × 103 and 3.2 × 103 cells ml−1 until day 5, respectively and then returned to the initial level. The protist cell size was determined to be ca. 40 μm by epifluorescence microscopy. No protists were detected in other microcosms with filtered seawater, indicating that sequential filtration through 0.7- and 0.2-μm filters removed these organisms.
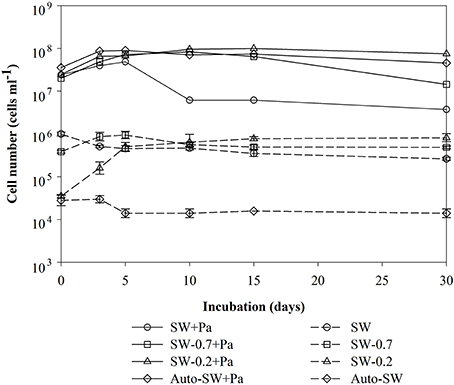
Figure 1. Time-course analysis of changes in the number of bacteria in each microcosm over time. Each value is an average of duplicate (samples from Pa-containing microcosms) or triplicate (samples from Pa-free microcosms) counts. Error bars representing the standard deviations for the triplicate.
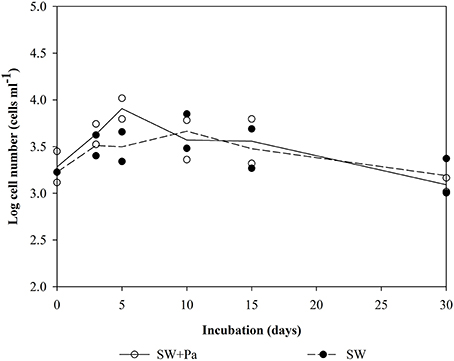
Figure 2. Number of protist cells in the SW and SW+Pa microcosms over the course of the study. Each value is an average of duplicate measurements. <DL, below detectable level. Protists were not detected in other micrososms.
Changes in Protease Activity
Three types of protease activity (aminopeptidase, trypsin-type, and chymotrypsin-type) were measured during the experimental period (Figure 3). The level of each type of protease activity was much higher in microcosms with Pa addition (left panels) than those in microcosms without Pa addition (right panels). SW+Pa (Figure 3A) showed higher activity than 1000 nmol−1 l−1 h−1 of trypsin on day 0, which increased on day 3. This increase should be derived from survived P. aeruginosa cells. In the unfiltered microcosm (SW, Figure 3B), trypsin-type activity was higher (200 nmol−1 l−1 h−1) than aminopeptidase and chymotrypsin, whose activities decreased over time. Protease activities in the SW-0.7+Pa microcosm (Figure 3C) increased greatly, which was not observed in SW-0.7 without Pa (Figure 3D). The trypsin activity in SW-0.7+Pa elevated over 8000 nmol−1 l−1 h−1 on day 10 and aminopeptidase elevated over 5000 nmol−1 l−1 h−1 at day 5. Because protists were removed by the GF/F filtration, the increase in the activities is derived from natural bacteria. The protease activity profile of SW-0.2+Pa (Figure 3E) was different from that of SW-0.7. Drastic increase in trypsin and aminopeptidase until day 10 was not observed. SW-0.2 (Figure 3F) showed the increase in trypsin activity initially (Figure 3F) and reached the highest value on day 3, although the value was low (500 nmol l−1 h−1). The SW-0.2 microcosm (Figure 3F) contained free enzymes derived from bulk seawater and/or those released from the cell lysis on filtration, and a small number of bacteria passing through 0.2-μm filter.
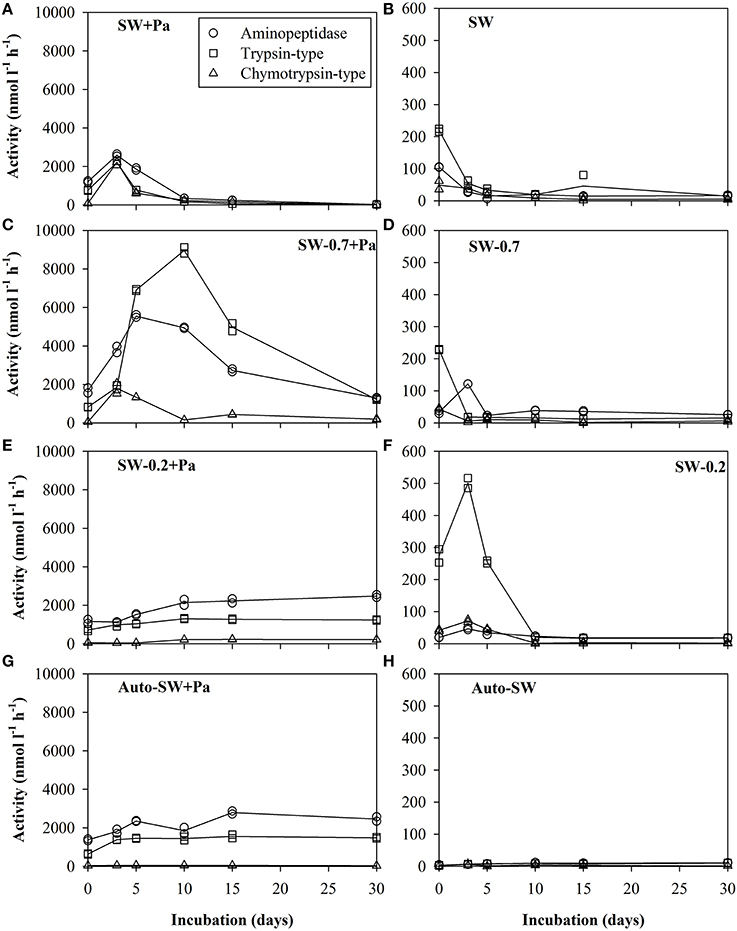
Figure 3. Proteolytic activity in the (A) SW+Pa, (B) SW, (C) SW-0.7+Pa, (D) SW, (E) SW-0.2+Pa, (F) SW-0.2, (G) Auto-SW+Pa, and (H) Auto-SW microcosms over the course of the study. Solid lines show the average of duplicate measurements and plots are of data obtained from duplicate samples. Left panels are microcosms with addition of Pa cells and right panels are those without Pa cells. Aminopeptidase, trypsin-type, and chymotrypsin-type activities were assayed using the substrates Leu-MCA, Boc-Phe-Ser-Arg-MCA, and Suc-Leu-Leu-Val-Tyr-MCA, respectively. Note that the scale of the y-axis in the left panels differs from that in the right panels.
The protease activities in the negative control (Auto-SW+Pa, Figure 3G) showed only Pa-derived enzyme activity. Blank control (Auto-SW, Figure 3H) did not show enzyme activity at all.
Changes in Microcosm Protein Profiles
We examined changes in the protein profile in seawater microcosms under different conditions using SDS-PAGE. Because natural seawater did not contain detectable amounts of protein, as determined by CBB staining (Figure 4D), protein bands detected on the gels were derived from the added Pa cells. Proteins in the SW+Pa microcosm disappeared completely by day 15 (Figure 4A). However, 28% of protein was retained in the SW-0.7+Pa microcosm on day 30 (Figure 4B). The SW-0.2+Pa showed 41% retaining of the protein on day 30 (Figure 4C). Pa proteins were preserved on day 30 in Auto-SW+Pa (Figure 4E), although it decreased to 59%.
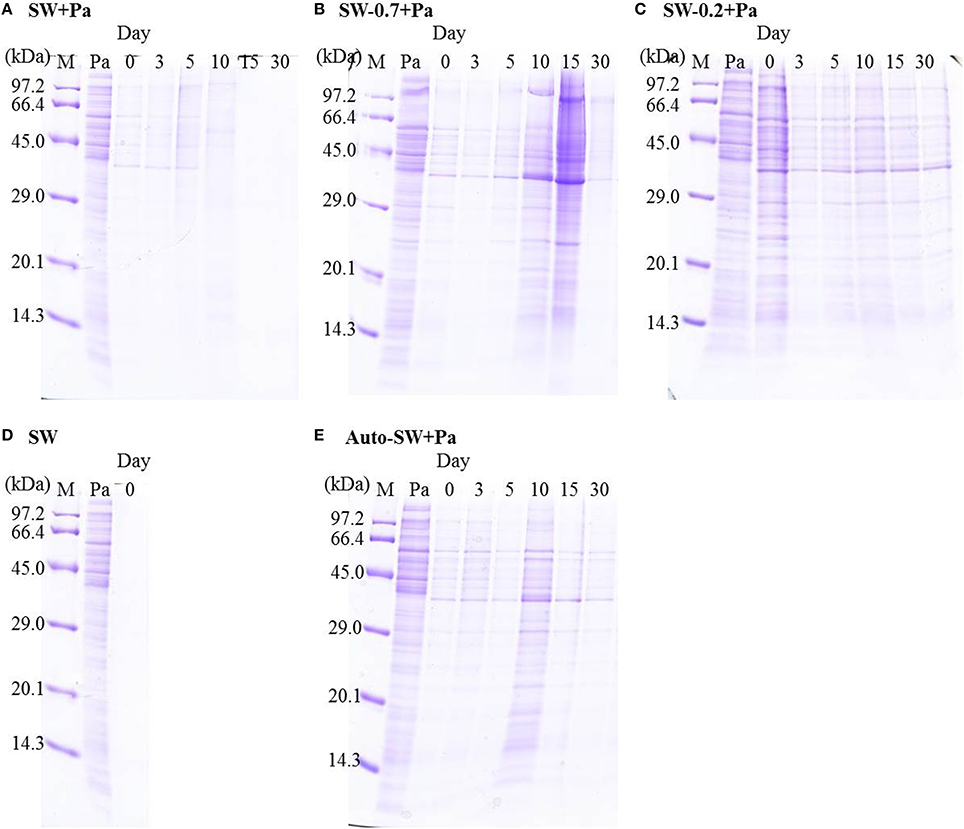
Figure 4. SDS-PAGE profiles of proteins in the (A) SW+Pa, (B) SW-0.7+Pa, (C) SW-0.2+Pa, (D)SW, and (E) Auto-SW+Pa microcosms. M, molecular size marker; Pa, proteins from P. aeruginosa.
Discussion
Changes in cell abundance of natural bacteria, injured Pa, and surviving Pa in the Pa-containing microcosms and of natural bacteria in the Pa-free microcosms were observed. The numbers of bacteria in both microcosms remained generally stable; however, the bacterial abundance in the SW+Pa microcosm was markedly lower on day 10 due to the effect of grazing by bacterivorous protists. The steep decrease in bacterial abundance in SW+Pa could be caused by the decrease in the injured Pa not being able to recover and grazed by protists. Weber et al. (2012), also points out that protists prefer to graze inactive bacteria. The protist abundance in SW+Pa was higher than that in SW because the protists in SW+Pa had more available food not only natural bacteria, but also injured Pa and surviving Pa and more quantity of food to take up. The relatively constant bacterial abundance after day 10 in the SW+Pa microcosm (Figure 1) suggests that the protists could not effectively graze on the bacteria because the bacteria may have developed grazing-resistance mechanisms such as size elongation, aggregation, and flagellum development (Alonso-Sáez et al., 2009). Obayashi et al. (2010), reported that marine microbes degrade, utilize, and grow on proteins derived from P. aeruginosa.
The proteolytic activity differed significantly between microcosms (Figure 3). The protease activity levels in the Auto-SW+Pa microcosm were higher compared with the A microcosm, suggesting that the increases in aminopeptidase and trypsin-type activities in the Auto-SW+Pa microcosm are derived from the temporary activation of surviving P. aeruginosa cells and/or the release of intracellular enzymes brought about by sonication in the process of preparing the Pa protein fraction.
Among the Pa-free microcosms, the SW-0.2 microcosm showed the most elevation in trypsin-type activity, with the highest value observed on day 3 (Figure 3F). On day 0, the SW-0.2 microcosm contained free enzymes derived from bulk seawater and/or those released from cell lysis by virus and filtration, and a small number of bacteria passing through 0.2-μm filter, whereas SW-0.2+Pa contained enzymes from Pa in addition to SW-0.2. This suggests that natural bacteria produce more protease activity when their initial abundance is low. The effort of bacteria to degrade a large amount of substrates irrelative to their initial abundance, which may cause their robust growth (Figure 1). This could explain the high enzyme activity in the SW-0.2 microcosm.
The SW-0.7+Pa microcosm showed the highest protease activity among those containing Pa (Figure 3C). The high activity can be attributed to the combination of microorganisms derived from natural seawater and surviving P. aeruginosa, as evidenced by the observation that the enzyme activity associated with surviving and active P. aeruginosa in the Auto-SW+Pa microcosm (Figure 3G) was lower than in the SW-0.7+Pa microcosm. Arnosti et al. (2011), suggested that in seawater the diversity of enzymes that degrade complex substrates is positively correlated with the diversity of the microbial community. Obayashi and Suzuki (2008), also reported that each bacterial isolate shows substrate specificity, whereas a variety of substrates are hydrolyzed by the natural bacterial community in bulk seawater. Hence, the greater variety of microorganisms and their respective capability to degrade organic matter might have made the community in the SW-0.7+Pa microcosm more active and robust. These results show the importance of various microbes and their coupled degradation process in protein degradationas well as organic matter recycling.
Studies of enzyme production by various bacterial strains (Priest, 1984) or mixed populations (Chróst, 1991) indicated that the production of specific enzymes is induced by their substrates and inhibited by their end products. Boetius (1995), showed that hydrolytic activity reflects the respective enzyme substrate rather than microbial biomass. Here, the highest protease activity observed was trypsin-type activity in the SW-0.7+Pa microcosm, suggesting that proteins from sonicated-Pa cells are favored substrates for proteases exhibiting this type of activity. On the other hand, the fact that bacteria preferentially take up high-molecular-weight DOM (Amon and Benner, 1994) could also have caused the observed elevation in trypsin-type enzyme activity. However, higher endohydrolase activity (i.e., trypsin-type enzymes) relative to exohydrolase activity (i.e., aminopeptidases) could lead to the unexpected accumulation of polymer and/or oligomer intermediates if these poly/oligomers are too large to be assimilated by the prokaryotes and no other microorganisms are capable of utilizing them.
Despite some increase of bacteria in SW-0.2+Pa, the proteolytic activity was similar to Auto-SW+Pa (Figures 3E,G), which could have been due to the unmarked differences in cell abundance since the amount of Pa added to the SW-0.2+Pa and Auto-SW+Pa microcosms was identical and represented a much greater number of cells compared with the number of natural indigenous bacteria.
Protein degradation profiling revealed that some proteins remained until the end of the incubation in the Auto-SW+Pa, SW-0.7+Pa, and SW-0.2+Pa microcosms (Figures 4B,C,E), even in the presence of high levels of protease activity (Figure 3). This result indicates that living natural bacteria and surviving Pa did not sufficiently incorporate substrates into their biomass in these microcosms. Although the protease activity in the SW+Pa microcosm was the lowest among those containing Pa, all of the protein bands completely disappeared by day 15 (Figure 4A), implying that the protein transformation process is highly efficient in the presence of both protists and bacteria. Since enzymes have substrate specificity, the result in enzyme activity in SW-0.7+Pa only showed the availability of enzymes that existed, but not their intrinsic ability to hydrolyze the natural substrates. Enzymes from protists and bacteria should be different from each other. Despite the low proteolytic activity in SW+Pa, various enzymes actively degraded proteins into substrates which were promptly incorporated into microbes. The major difference between those 2 microcosms is the presence of protists. Consequently, these results lead us to propose a new hypothesis, which holds that protein transformation in the marine environment involves uptake of bacterial proteins by not only bacteria but also protists and/or proteolytic degradation by proteases originating from various microbes, including protists. In other words, efficient proteolysis in seawater may require the diversity at functional/taxonomic level rather than simply diversity within bacterial communities. Recent studies have also supported our hypothesis that the extent to which a high-molecular-weight substrate is bioavailable depends on microbial community capabilities (D'Ambrosio et al., 2014) and the organisms using polymers are quite distinct from those utilizing monomer constituents (Kirchman et al., 2007; Alonso-Sáez et al., 2012). The transfer of organic matter can also be enhanced by virus infection in natural environment (Middelboe et al., 2003). However, in the present experiment, Pa-protein degradation by virus in our experiment can be ignored due to the preparation method by sonication treatment.
In conclusion, bacteria produce high levels of proteolytic activity in the absence of protists. However, bacteria alone cannot efficiently transform proteins in seawater. Proteins could be rapidly transformed in a coexisting protist-bacterial community, in which protists might work as not only predators but also sources of proteases, which are required for complete transformation of proteins in seawater. Further hypotheses are that protists may play a role as protein consumers and/or the excretion of protists' proteases should assist DOM decomposition, which accelerates production of usable molecules for bacterial growth. Bacterial growth can be stimulated by the increase in amino acids and dipeptides, followed by enhancing protists growth. Thus, protists may play an important role in the protein transformation by means of enzyme production and substrate utilization in seawater. The results from the present study provide new evidence demonstrating interactions between protists and bacteria in relation to protein transformation in aquatic ecosystems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partly supported by KAKENHI, JSPS, Japan. Ngo V. Thao was financially supported by the scholarship from Ehime University.
References
Alonso-Sáez, L., Sánchez, O., and Gasol, J. M. (2012). Bacterial uptake of low molecular weight organics in the subtropical Atlantic: are major phylogenetic groups functionally different? Limnol. Oceanogr. 57, 798–808. doi: 10.4319/lo.2012.57.3.0798
Alonso-Sáez, L., Unanue, M., Latatu, A., Azua, I., Ayo, B., Artolozaga, I., et al. (2009). Changes in marine prokaryotic community induced by varying types of dissolved organic matter and subsequent grazing pressure. J. Plank. Res. 31, 1373–1383. doi: 10.1093/plankt/fbp081
Amon, R. M. W., and Benner, R. (1994). Rapid cycling of high-molecular-weight dissolved organic matter in the ocean. Nature 369, 549–552. doi: 10.1038/369549a0
Arnosti, C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arnosti, C., Durkin, A. S., and Jeffrey, W. H. (2005). Patterns of extracellular enzyme activities among pelagic marine microbial communities: implication for cycling of dissolved organic carbon. Aquat. Microb. Ecol. 38, 135–145. doi: 10.3354/ame038135
Arnosti, C., Steen, A. D., Ziervogel, K., Ghobrial, S., and Jeffrey, W. H. (2011). Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS ONE 6:e28900. doi: 10.1371/journal.pone.0028900
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Azam, F., and Cho, B. (1987). “Bacterial utilization of organic matter in the sea,” in Ecology of Microbial Communities, eds M. M. Fletcher, T. R. Gray, and J. G. Jones (Cambridge: Cambridge University Press), 261–281.
Azúa, I., Unanue, M., Ayo, B., Artolozaga, I., Arrieta, J. M., and Iriberri, J. (2003). Influence of organic matter quality in the cleavage of polymers by marine bacterial communities. J. Plank. Res. 25, 1451–1460. doi: 10.1093/plankt/fbg105
Bochdansky, A. B., Puskaric, S., and Herndl, G. J. (1995). Influence of zooplankton grazing on free dissolved enzymes in the sea. Mar. Ecol. Prog. Ser. 121, 53–63. doi: 10.3354/meps121053
Boetius, A. (1995). Microbial hydrolytic enzyme activities in deep-sea sediments. Helgoländer Meeresunters. 49, 177–1187. doi: 10.1007/BF02368348
Bong, C. W., Obayashi, Y., and Suzuki, S. (2013). Succession of protease activity in seawater and bacterial isolates during starvation in a mesocosm experiment. Aquat. Microb. Ecol. 69, 33–46. doi: 10.3354/ame01618
Chróst, R. J. (1990). “Microbial ectoenzymes in aquatic environments,” in Aquatic Microbial Ecology: Biochemical and Molecular Approaches, eds J. Overbeck and R. J. Chróst (New York, NY: Brock/Springer), 47–78.
Chróst, R. J. (1991). “Environmental control of the synthesis and activity of aquatic microbial ectoenzymes,” in Microbial Enzymes in the Aquatic Environment, ed R. J. Chróst (New York, NY: Springer), 29–59.
Crump, B. C., Baross, J. A., and Simenstad, C. A. (1998). Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat. Microb. Ecol. 14, 7–18. doi: 10.3354/ame014007
D'Ambrosio, L., Ziervogel, K., Macgregor, B., Teske, A., and Arnosti, C. (2014). Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J. 8, 2167–2179. doi: 10.1038/ismej.2014.67
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fogel, M. L., and Tuross, N. (1999). Transformation of plant biochemicals to geological macromolecules during early diagenesis. Oecologia 120, 336–346. doi: 10.1007/s004420050867
Hancock, R. E., and Brinkman, F. S. (2002). Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56, 17–38. doi: 10.1146/annurev.micro.56.012302.160310
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hancock, R. E. W., Poodle, K., and Benz, R. (1982). Outer membrane protein P of Pseudomonas aeruginosa: regulation by phosphate deficiency and formation of small anion-specific channels in lipid bilayer membranes. J. Bacteriol. 150, 730–738.
Hancock, R. E. W., Raffle, V. J., and Nicas, T. I. (1981). Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 19, 777–785. doi: 10.1128/AAC.19.5.777
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hollibaugh, J. T., and Azam, F. (1983). Microbial-degradation of dissolved proteins in seawater. Limnol. Oceanogr. 28, 1104–1116. doi: 10.4319/lo.1983.28.6.1104
Horiuchi, T., Takano, Y., Ishibashi, J.-I., Marumo, K., Urabe, T., and Kobayashi, K. (2004). Amino acids in water samples from deep sea hydrothermal vents at Suiyo Seamount, Izu-Bonin Arc, Pacific Ocean. Org. Geochem. 35, 1121–1128. doi: 10.1016/j.orggeochem.2004.06.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawasaki, N., and Benner, R. (2006). Bacterial release of dissolved organic matter during cell growth and decline: molecular origin and composition. Limnol. Oceanogr. 51, 2170–2180. doi: 10.4319/lo.2006.51.5.2170
Keil, R. G., and Kirchman, D. L. (1993). Dissolved combined amino acids: chemical form and utilization by marine bacteria. Limnol. Oceanogr. 38, 1256–1270. doi: 10.4319/lo.1993.38.6.1256
Keil, R. G., Montluçon, D. B., Prahl, F. G., and Hedges, J. I. (1994). Sorptive preservation of labile organic matter in marine sediments. Nature 370, 549–551. doi: 10.1038/370549a0
Kiersztyn, B., Siuda, W., and Chróst, R. J. (2012). Persistence of bacterial proteolytic enzymes in lake ecosystems. FEMS Microbiol. Ecol. 80, 124–134. doi: 10.1111/j.1574-6941.2011.01276.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kirchman, D. L., Elifantz, H., Dittel, A. I., Malmstrom, R. R., and Cottrell, M. T. (2007). Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52, 495–507. doi: 10.4319/lo.2007.52.2.0495
Lee, S., and Fuhrman, J. A. (1987). Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53, 1298–1303.
Lloyd, K. G., Schreiber, L., Petersen, D. G., Kjeldsen, K. U., Lever, M. A., Steen, A. D., et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218. doi: 10.1038/nature12033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCarthy, M. D., Hedges, J. I., and Benner, R. (1998). Major bacterial contribution to marine dissolved organic nitrogen. Science 281, 231–234. doi: 10.1126/science.281.5374.231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKee, G. A., Kobiela, M. E., and Hatcher, P. (2014). Effect of Michael adduction on peptide preservation in natural waters. Environ. Sci. Processes Impacts 16, 2087–2097. doi: 10.1039/C4EM00075G
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Middelboe, M., Borch, N., and Kirchman, D. (1995). Bacterial utilization of dissolved free amino acids, dissolved combined amino acids and ammonium in the Delaware Bay estuary: effects of carbon and nitrogen limitation. Mar. Ecol. Prog. Ser. 128, 109–120. doi: 10.3354/meps128109
Middelboe, M., Riemann, L., Steward, G. F., Hansen, V., and Nybroe, O. (2003). Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat. Microb. Ecol. 33, 1–10. doi: 10.3354/ame033001
Mohapatra, B. R., and Fukami, K. (2004). Production of aminopeptidase by marine heterotrophic nanoflagellates. Aquat. Microb. Ecol. 34, 129–137. doi: 10.3354/ame034129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, E. K., Nunn, B. L., Goodlett, D. R., and Harvey, H. R. (2012). Identifying and tracking proteins through the marine water column: insights into the inputs and preservation mechanisms of protein in sediments. Geochim. Cosmochim. Acta 83, 324–359. doi: 10.1016/j.gca.2012.01.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murrell, M. C., Hollibaugh, J. T., Silver, M. W., and Wong, P. S. (1999). Bacterioplankton dynamics in northern San Francisco Bay: role of particle association and seasonal freshwater flow. Limnol. Oceanogr. 44, 295–308. doi: 10.4319/lo.1999.44.2.0295
Nagata, T., Fukuda, R., Koike, I., Kogure, K., and Kirchman, D. L. (1998). Degradation by bacteria of membrane and soluble protein in seawater. Aquat. Microb. Ecol. 14, 29–37. doi: 10.3354/ame014029
Nagata, T., and Kirchman, D. L. (1996). Bacterial degradation of protein adsorbed to model submicron particles in seawater. Mar. Ecol. Prog. Ser. 132, 241–248. doi: 10.3354/meps132241
Nguyen, R. T., and Harvey, H. R. (1997). Protein and amino acid cycling during phytoplankton decomposition in oxic and anoxic waters. Org. Geochem. 27, 115–128. doi: 10.1016/S0146-6380(97)00076-4
Nguyen, R. T., and Harvey, H. R. (1998). “Protein preservation during early diagenesis in marine waters and sediments,” in Nitrogen-Containing Macromolecules in the Bio- and Geosphere, ACS Symposium Series 707, eds B. A. Stankiewicz and P. F. van Bergen (Washington, DC: American Chemical Society), 88–112.
Nonaka, L., Inubushi, A., Shinomiya, H., Murase, M., and Suzuki, S. (2010). Differences of genetic diversity and antibiotics susceptibility of Pseudomonas aeruginosa isolated from hospital, river and coastal seawater. Environ. Microbiol. Rep. 2, 465–472. doi: 10.1111/j.1758-2229.2010.00178.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Obayashi, Y., and Suzuki, S. (2005). Proteolytic enzymes in coastal surface seawater: significant activity of endopeptidases and exopeptidases. Limnol. Oceanogr. 50, 722–726. doi: 10.4319/lo.2005.50.2.0722
Obayashi, Y., and Suzuki, S. (2008). Occurrence of exo- and endopeptidases in dissolved and particulate fractions of coastal seawater. Aquat. Microb. Ecol. 50, 231–237. doi: 10.3354/ame01169
Obayashi, Y., Ueoka, N., and Suzuki, S. (2010). Degradation and utilization of protein derived from Pseudomonas aeruginosa by marine microbial community. J. Oceanogr. 66, 513–521. doi: 10.1007/s10872-010-0043-7
Pantoja, S., and Lee, C. (1999a). Molecular weight distribution of proteinaceous material in Long Island Sound sediments. Limnol. Oceanogr. 44, 1323–1330. doi: 10.4319/lo.1999.44.5.1323
Pantoja, S., and Lee, C. (1999b). Peptide decomposition by extracellular hydrolysis in coastal seawater and salt marsh sediment. Mar. Chem. 63, 273–291. doi: 10.1016/S0304-4203(98)00067-X
Pérez, M. T., Pausz, C., and Herndl, G. J. (2003). Major shift in bacterioplankton utilization of enantiomeric amino acids between surface waters and the ocean's interior. Limnol. Oceanogr. 48, 755–763. doi: 10.4319/lo.2003.48.2.0755
Richardot, M., Debroas, D., Thouvenot, A., Romagoux, J. C., Berthon, J. L., and Devaux, J. (1999). Proteolytic and glycolytic activities in size-fractionated surface water samples from an oligotrophic reservoir in relation to plankton communities. Aquat. Sci. 61, 279–292. doi: 10.1007/s000270050066
Rosenstock, B., and Simon, M. (1993). Use of dissolved combined and free amino acids by planktonic bacteria in Lake Constance. Limnol. Oceanogr. 38, 1521–1531. doi: 10.4319/lo.1993.38.7.1521
Sala, M. M., and Güde, H. (1999). Role of protozoans on the microbial ectoenzymatic activity during the degradation of macrophytes. Aquat. Microb. Ecol. 20, 75–82. doi: 10.3354/ame020075
Salerno, M., and Stoecker, D. K. (2009). Ectocellular glucosidase and peptidase activity of the mixotrophic dinoflagellate Prorocentrum Minimum (Dinophyceae). J. Phycol. 45, 34–45. doi: 10.1111/j.1529-8817.2008.00622.x
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning. A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Sherr, B., and Sherr, E. (1983). Enumeration of heterotrophic microprotozoa by epifluorescence microscopy. Estuar. Coast. Shelf. S 16, 1–7. doi: 10.1016/0272-7714(83)90089-6
Sherr, B., Sherr, E., and del Giorgio, P. D. (2001). “Enumeration of total and highly active bacteria,” in Methods in Microbiology, Marine Microbiology, Vol. 30, ed J. H. Paul (London: Academic Press), 129–159.
Steen, A. D., and Arnosti, C. (2013). Extracellular peptidase and carbohydrate hydrolase activities in an Arctic fjord (Smeerenburgfjord, Svalbard). Aquat. Microb. Ecol. 69, 93–99. doi: 10.3354/ame01625
Stevenson, F. J. (1994). Humus Chemistry: Genesis, Composition, Reactions. New York, NY: bJohn Wiley & Sons.
Suzuki, S., Fujita, T., Kimata, N., Kogure, K., and Tanoue, E. (2000). Isolation of bacteria with membrane proteins homologous to Vibrio anguillarum porin Omp35La. Microbes Environ. 15, 185–189. doi: 10.1264/jsme2.2000.189
Suzuki, S., Kogure, K., and Tanoue, E. (1997). Immunochemical detection of dissolved proteins and their source bacteria in marine environments. Mar. Ecol. Prog. Ser. 158, 1–9. doi: 10.3354/meps158001
Suzuki, S., Kogure, K., and Tanoue, E. (1999). “Bacterial porin in the ocean and its ecological implications,” in Microbial Biosystems: New Frontiers, Proceedings of the 8th International Symposium on Microbial Ecology, eds C. R. Bell, M. Brylinsky, and P. Johnson-Green (Halifax, NS: Atlantic Canada Society for Microbial Ecology), 175–178.
Tanoue, E. (1995). Detection of dissolved protein molecules in oceanic waters. Mar. Chem. 51, 239–252. doi: 10.1016/0304-4203(95)00061-5
Tanoue, E., Ishii, M., and Midorikawa, T. (1996). Discrete dissolved and particulate proteins in oceanic waters. Limnol. Oceanogr. 41, 1334–1343. doi: 10.4319/lo.1996.41.6.1334
Tanoue, E., Nishiyama, S., Kamo, M., and Tsugita, A. (1995). Bacterial membranes: possible source of a major dissolved protein in seawater. Geochim. Cosmochim. Acta 59, 2643–2648. doi: 10.1016/0016-7037(95)00134-4
Weber, F., Campo, J. D., Wylezich, C., Massana, R., and Jügens, K. (2012). Unveiling trophic functions of uncultured protist taxa by incubation experiments in the brackish Baltic Sea. PLoS ONE 7:e41970. doi: 10.1371/journal.pone.0041970
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: protease, Pseudomonas aeruginosa, protein transformation, protist, seawater
Citation: Thao NV, Obayashi Y, Yokokawa T and Suzuki S (2014) Coexisting protist-bacterial community accelerates protein transformation in microcosm experiments. Front. Mar. Sci. 1:69. doi: 10.3389/fmars.2014.00069
Received: 06 October 2014; Paper pending published: 03 November 2014;
Accepted: 17 November 2014; Published online: 03 December 2014.
Edited by:
Benjamin Van Mooy, Woods Hole Oceanographic Institution, USAReviewed by:
Andrew Decker Steen, University of Tennessee, Knoxville, USAKarl Kaiser, Texas A&M University, USA
Copyright © 2014 Thao, Obayashi, Yokokawa and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoru Suzuki, Center for Marine Environmental Studies, Ehime University, Bunkyo-3, Matsuyama, Ehime 790-8577, Japan e-mail: ssuzuki@ehime-u.ac.jp
 Ngo V. Thao
Ngo V. Thao Yumiko Obayashi
Yumiko Obayashi Taichi Yokokawa1
Taichi Yokokawa1  Satoru Suzuki
Satoru Suzuki