- 1Laboratory of Microbiology and Infectious Diseases, Division of Special Projects, Institute of Scientific and Industrial Research, Osaka University, Ibaraki, Osaka, Japan
- 2Graduate School of Pharmaceutical Sciences, Osaka University, Suita, Japan
Honey has a complex chemistry, and its broad-spectrum antimicrobial activity varies with floral source, climate, and harvesting conditions. Methylglyoxal was identified as the dominant antibacterial component of manuka honey. Although it has been known that methylglyoxal has antibacterial activity against gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus, there is not much information describing its activity against gram-negative bacteria. In this study, we report the effect of methylglyoxal against multidrug-resistant Pseudomonas aeruginosa (MDRP) using 53 clinically isolated strains. We also assessed the effect of deleting the five multidrug efflux systems in P. aeruginosa, as well as the efflux systems in Escherichia coli and Salmonella enterica serovar Typhimurium, on MICs of methylglyoxal. Our results indicate that methylglyoxal inhibits the growth of MDRP at concentrations of 128–512 μg/ml (1.7–7.1 mM) and is not recognized by drug efflux systems.
Introduction
Pseudomonas aeruginosa is endemic among critically ill patients, and multidrug-resistant strains are increasingly being isolated in intensive care units (Ortega et al., 2004). Because P. aeruginosa is a virulent organism susceptible to a limited number of antibiotic agents, infections caused by this organism are difficult to cure and often require combination therapy. Multidrug-resistant P. aeruginosa (MDRP) has been defined as P. aeruginosa resistant to imipenem, amikacin, and ciprofloxacin (Sekiguchi et al., 2007). The increasing resistance of P. aeruginosa is a growing threat to the clinical management of such infections (Ortega et al., 2004).
In bacteria, resistance to bactericidal agents is often associated with multidrug efflux systems, which decrease cellular drug accumulation (Nikaido, 1996). In gram-negative bacteria, systems belonging to the resistance/nodulation/division (RND) family are particularly effective in generating resistance because they form a tripartite complex with the periplasmic proteins of the membrane fusion protein family and an outer membrane channel, ensuring that drugs are pumped out directly to the external medium (Nikaido and Pages, 2012). P. aeruginosa expresses several RND-type multidrug efflux systems, including MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY, which are significant determinants of multidrug resistance in laboratory and clinical isolates (Poole, 2004; Piddock, 2006; Lister et al., 2009). These systems are three-component systems comprising antiporters of the RND family driven by proton motive force (MexB, MexD, MexF, and MexY), outer membrane channels (OprM, OprJ, and OprN), and periplasmic membrane fusion proteins (MexA, MexC, MexF, and MexX). These pumps function in a manner similar to AcrAB–TolC, which is the best-studied RND-type multidrug pump of Escherichia coli (Nakashima et al., 2011; Nikaido, 2011). It is necessary to develop drugs that are not recognized by the efflux pumps to prevent multidrug resistance modulated by drug efflux systems.
Honey has several antibacterial features that are distinct from classical antibiotics, including high osmolarity, low pH, and generation of hydrogen peroxide by the bee-derived enzyme glucose oxidase (Allen et al., 1991). Antibacterial phenolic components have been identified in honey (Weston et al., 1999), and an antimicrobial peptide has been discovered in a Dutch medical-grade honey produced from an undisclosed floral source cultivated in greenhouses (Kwakman et al., 2010). Manuka honey is derived from nectar that has been collected by honey bees (Apis mellifera) foraging on a shrub known as manuka (Leptospermum scoparium) that is indigenous to New Zealand. Manuka honey is broad in spectrum, able to inhibit a diverse range of bacterial and yeast pathogens, and equally effective against multidrug-resistant bacteria (Blair et al., 2009; Henriques et al., 2010; Kwakman et al., 2011). It is used in modern wound-care formulations and has been shown to eradicate methicillin-resistant Staphylococcus aureus (MRSA) from wounds (Natarajan et al., 2001; Blaser et al., 2007; Gethin and Cowman, 2008; Visavadia et al., 2008). Clinically isolated strains of methicillin-susceptible and -resistant staphylococci were shown to be equally susceptible to manuka honey in vitro, with minimum inhibitory concentrations (MICs) reported to be <3% (v/v) [equivalent to 41,000 mg/L or 4.1% (w/v)] (Cooper et al., 1999, 2002b). Methylglyoxal was identified as the dominant active antibacterial component of manuka honey (Mavric et al., 2008; Adams et al., 2009b). Active manuka honey contains high levels of the reactive dicarbonyl methylglyoxal (Mavric et al., 2008; Adams et al., 2009a), which is formed nonenzymatically from nectar-derived dihydroxyacetone during ripening. Methylglyoxal was also found to be produced from dihydrocyacetone phosphate in E. coli, initiating a bypass of the glycolytic pathway (Cooper and Anderson, 1970). It was suggested that methylglyoxal inhibits protein synthesis by reacting with guanine residues in RNA and its precursors. It also inhibits DNA synthesis by reacting with guanine residues in DNA and its precursors (Krymkiewicz et al., 1971).
It has been known that methylglyoxal has antibacterial activity against gram-positive bacteria, including MRSA and vancomycin-resistant Enterococcus. It was also reported that methylglyoxal containing manuka honey is biocidal against S. aureus strains at a concentration of 33–66% w/v (equivalent methylglyoxal concentration, 260–530 μg/ml) (Jervis-Bardy et al., 2011). However, there is not much information describing methylglyoxal activity against gram-negative bacteria. Although it was previously reported that manuka honey is bactericidal against P. aeruginosa (Roberts et al., 2012), the effect of methylglyoxal on MDRP has been unknown. In this study, we report the antibacterial effect of methylglyoxal on MDRP using 53 clinically isolated strains. We also demonstrate that methylglyoxal is not recognized by drug efflux systems in P. aeruginosa, Salmonella enterica, and E. coli.
Materials and Methods
Bacterial Strains and Growth Conditions
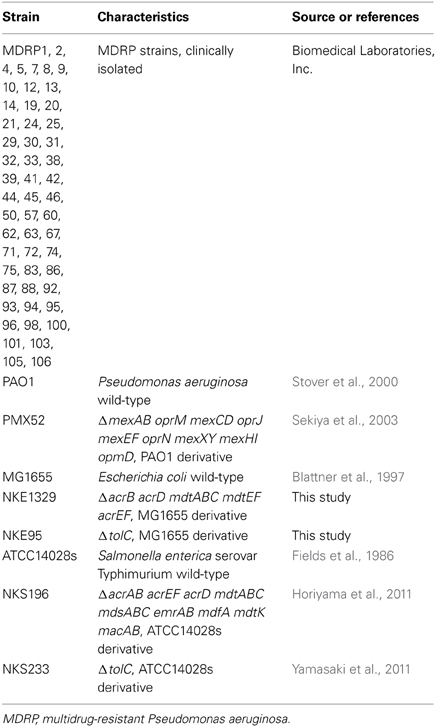
The bacterial strains used in this study are listed in Table 1. We used P. aeruginosa PAO1 (Stover et al., 2000), S. enterica serovar Typhimurium ATCC14028s (Fields et al., 1986), and E. coli MG1655 (Blattner et al., 1997) as wild-type strains. All clinically isolated MDRP strains, which showed resistance to imipenem, amikacin, and ciprofloxacin, were kindly provided by Biomedical Laboratories, Inc. (Tokyo, Japan).
Construction of Gene Deletion Mutants
P. aeruginosa PMX52 (Sekiya et al., 2003), a PAO1-derived strain lacking the genes encoding the MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY, and MexHI-OpmD drug efflux systems, was kindly provided by Tomofusa Tsuchiya of Ritsumeikan University, Japan. S. enterica serovar Typhimurium strains NKS196 (ΔacrAB acrEF acrD mdtABC mdsABC emrAB mdfA mdtK macAB) and NKS233 (ΔtolC) were constructed as described previously (Horiyama et al., 2011; Yamasaki et al., 2011).
To construct E. coli strains NKE1329 (ΔacrB acrD mdtABC mdtEF acrEF) and NKE95 (ΔtolC), we performed gene disruption using procedures described previously (Datsenko and Wanner, 2000). The following oligonucleotide primers were used for the construction of the mutants: acrB-P1 (AAAAAGGCCGCTTACGCGGCCTTAGTGAT
TACACGTTGTAGTGTAGGCTGGAGCTGCTTC); acrB-P2 (GAACAGTCCAAGTCTTAACTTAAACAGGAGCCGTT
AAGACCATATGAATATCCTCCTTAG); acrD-P1 (TGAAAAAGGCGACACATTGGCATGTCGCCTTTTTTATT
GCGTGTAGGCTGGAGCTGCTTC); acrD-P2 (AAGCCTACAACGATACGCAGAAACACGAGG
TCCTCTTTTACATATGAATATCCTCCTTAG); mdtA-P1 (ATCATTCCGCGAAACGTTTCAGGAAGAGAA
ACTCTTAACGGTGTAGGCTGGAGCTGCTTC); mdtC-P2 (GAGATACACCACCGGCGTGGTAT
ACAGCGTAAGGAGCTGGCATATGAATATCCTCCTTAG); mdtE-P1 (TTAAAGAACCGTTATTTCTCAAGAATTTT
CAGGGACTAAAGTGTAGGCTGGAGCTGCTTC); mdtF-P2 (AGGCTGAACCTTCATGTTCAACCTTACTCTCA
TTTACACGCATATGAATATCCTCCTTAG); acrE-P1 (TTGGGTAAATAACGCGCTTTTGGTTTTTT
GAGGAATAGTAGTGTAGGCTGGAGCTGCTTC); acrF-P2 (AAATAATAAAGGCACCCGAAAGCGCCTTTAT
GTTTCTGATCATATGAATATCCTCCTTAG); tolC-P1 (ACTGGTGCCGGGCTATCAGGCGCATAACCAT
CAGCAATAGGTGTAGGCTGGAGCTGCTTC); and tolC-P2 (TTACAGTTTGATCGCGCTAAATACTGCTTCACC
ACAAGGACATATGAATATCCTCCTTAG). The chloramphenicol resistance gene cat or the kanamycin resistance gene aph, flanked by Flp recognition sites, was amplified by PCR using the primers listed above. The resulting PCR products were used to transform E. coli MG1655, which harbors plasmid pKD46, expressing Red recombinase. The chromosomal structures of the mutated loci were verified by PCR; cat and aph were further eliminated using the plasmid pCP20, as described previously (Datsenko and Wanner, 2000). To construct the NKE1329 strain, the deletions were transferred to strains by P22 transduction, as described previously Davis et al. (1980).
Determination of MICs of Antimicrobial Compounds
Antibacterial activities were determined on Muller Hinton II agar (Becton Dickinson & Co., Franklin Lakes, NJ, USA) plates containing methylglyoxal (32–2048 μg/ml), imipenem (0.0625–2048 μg/ml), amikacin (0.125–4096 μg/ml), or ciprofloxacin (0.0078–2048 μg/ml) (Sigma, St. Louis, MO, USA). Agar plates were prepared using the two-fold agar dilution technique. Bacteria were grown at 37°C overnight and then tested at a final inoculum volume of 1 × 105 cfu/μl using a multipoint inoculator (Sakuma Seisakusyo, Tokyo, Japan). The inoculated agar plates were examined after incubation at 37°C for 16 h. MIC was the lowest concentration of a compound that inhibited cell growth.
Measurement of Bacterial Growth in the Presence of Methylglyoxal
E. coli (MG1655, NKE1329, and NKE95) and S. enterica (ATCC14028s, NKS196, and NKS233) strains were grown in Luria–Bertani broth (Becton Dickinson & Co., Franklin Lakes, NJ, USA), and P. aeruginosa (PAO1 and PMX52) strains were grown in Muller Hinton II (MHII) broth (Becton Dickinson & Co., Franklin Lakes, NJ, USA). Bacterial cells were cultured overnight at 37°C, and then 100 μl of cell cultures were diluted in 5 ml of the same medium. The diluted bacterial cells were incubated at 37°C until OD600 reached 0.5. Then, the bacterial cells were diluted in the same medium to an OD600 of 0.05. This diluted bacterial cells were incubated in NUNC Edge 96-well plates (Thermo scientific, MA, USA) with shaking at 37°C for 7 h. Bacterial growth was monitored using an Infinite M200 Pro plate reader (Tecan, Männedorf, Switzerland).
Results and Discussion
MICs of Imipenem, Amikacin, or Ciprofloxacin for Clinically Isolated MDRP
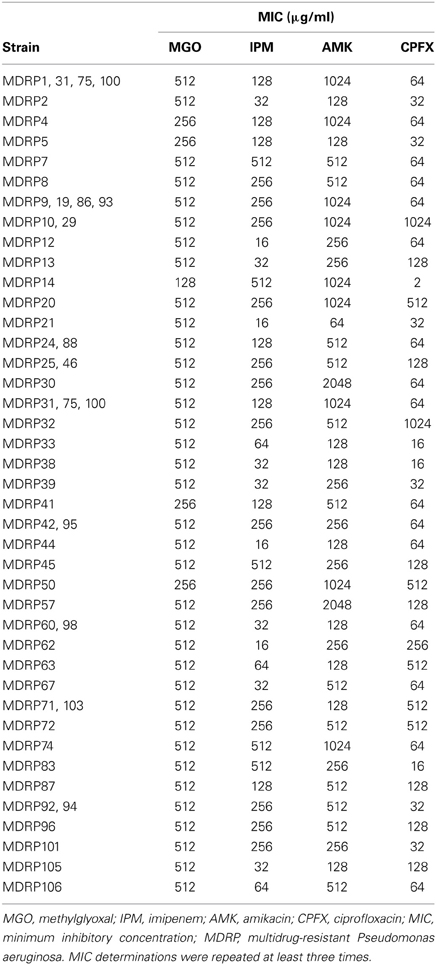
MDRP has been defined as P. aeruginosa resistant to imipenem (MIC, ≥16 μg/ml), amikacin (≥32 μg/ml), and ciprofloxacin (≥4 μg/ml) (Sekiguchi et al., 2007). Using this criterion, we determined that all 53 clinical isolates were MDRP (Table 2). The highest MIC of imipenem for strains MDRP7, 14, 45, 74, and 83 was 512 μg/ml. The highest MIC of amikacin for strains MDRP30 and 57 was 2048 μg/ml. The highest MIC of ciprofloxacin for strains MDRP10, 29, and 32 was 1024 μg/ml.
Susceptibilities of MDRP Strains to Methylglyoxal
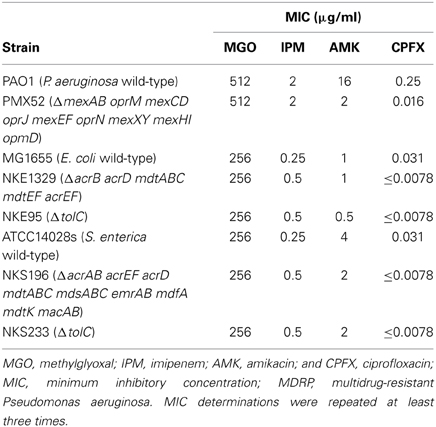
To evaluate the antibacterial activity of methylglyoxal against clinically isolated MDRP strains, we determined MICs using the 53 confirmed MDRP strains. The MIC of methylglyoxal for most of the MDRP strains was 512 μg/ml (Table 2), whereas the susceptibilities of these strains to imipenem, amikacin, and ciprofloxacin were different. The methylglyoxal concentration at which MDRP14 was susceptible was 128 μg/ml and that at which MDRP4, 5, 41, and 50 were susceptible was 256 μg/ml. We also tested the methylglyoxal susceptibility of the drug-sensitive wild-type strain P. aeruginosa PAO1. The MIC of methylglyoxal for PAO1 was 512 μg/ml (Table 3), which was the same that for most of the MDRP strains.
Effect of Drug Efflux Systems in P. aeruginosa, E. coli, and S. enterica to Methylglyoxal
Multidrug efflux pumps in P. aeruginosa, such as MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY, have been shown to be significant determinants of multidrug resistance in laboratory and clinical isolates (Poole, 2004; Piddock, 2006; Lister et al., 2009). The existence of an additional multidrug efflux system, MexHI-OpmD, was also previously reported (Sekiya et al., 2003) in this organism. Because multidrug efflux systems display the ability to transport various structurally unrelated drugs, we investigated whether methylglyoxal is exported by these drug efflux systems in P. aeruginosa. For this purpose, we measured MIC of methylglyoxal for the wild-type P. aeruginosa strain PAO1 and its efflux-deficient mutant strain PMX52 (ΔmexAB oprM mexCD oprJ mexEF oprN mexXY mexHI opmD). Although PMX52 was more susceptible to amikacin and ciprofloxacin than PAO1, MIC of methylglyoxal for these strains was the same. This suggests that methylglyoxal is not recognized by drug efflux systems in P. aeruginosa. To confirm whether same phenomenon could be observed in other gram-negative bacteria, we determined MICs of methylglyoxal for the efflux-deficient mutants of E. coli and S. enterica serovar Typhimurium. There are five RND-type drug efflux systems (AcrAB, AcrD, MdtABC, MdtEF, and AcrEF) in E. coli, and all of them require the TolC outer membrane channel for their function (Nishino et al., 2003). To investigate the defect of these drug efflux systems in E. coli, we measured MICs of methylglyoxal for MG1655 (wild-type), NKE1329 (ΔacrB acrD mdtABC mdtEF acrEF), and NKE95 (ΔtolC) strains. The susceptibility of NKE1329 and NKE95 to methylglyoxal was same as that of the wild-type strain, although they were more susceptible to ciprofloxacin than the wild-type strain. S. enterica serovar Typhimurium harbors at least nine drug efflux systems belonging to RND, multidrug and toxic compound extrusion, and ATP-binding cassette (ABC) superfamilies (Nishino et al., 2006). Seven of them (AcrAB, AcrEF, AcrD, MdtABC, MdsAbC, EmrAB, and MacAB) require TolC for their function (Horiyama et al., 2010). For S. enterica, we used ATCC14028s (wild-type), NKS196 (ΔacrAB acrEF acrD mdtABC mdsABC emrAB mdfA mdtK macAB), and NKS233 (ΔtolC) strains. Although NKS196 and NKS233 were more sensitive to ciprofloxacin than the wild-type strain ATCC14028s, MICs of methylglyoxal for ATCC14028s, NKS196, and NKS233 were the same. In addition to MIC determination using agar plates, we tested the effect of methylglyoxal on bacterial growth in liquid medium. The growth of E. coli (MG1655, NKE1329, and NKE9) and Salmonella (ATCC14028s, NKS196, and NKS233) strains was inhibited by methylglyoxal at a concentration of 256 μg/ml, and the growth of P. aeruginosa (PAO1 and PMX52) strains was inhibited at 512 μg/ml, which is consistent with MICs determined (Figure 1). These data suggest that methylglyoxal is not recognized by drug efflux systems in E. coli or S. enterica.
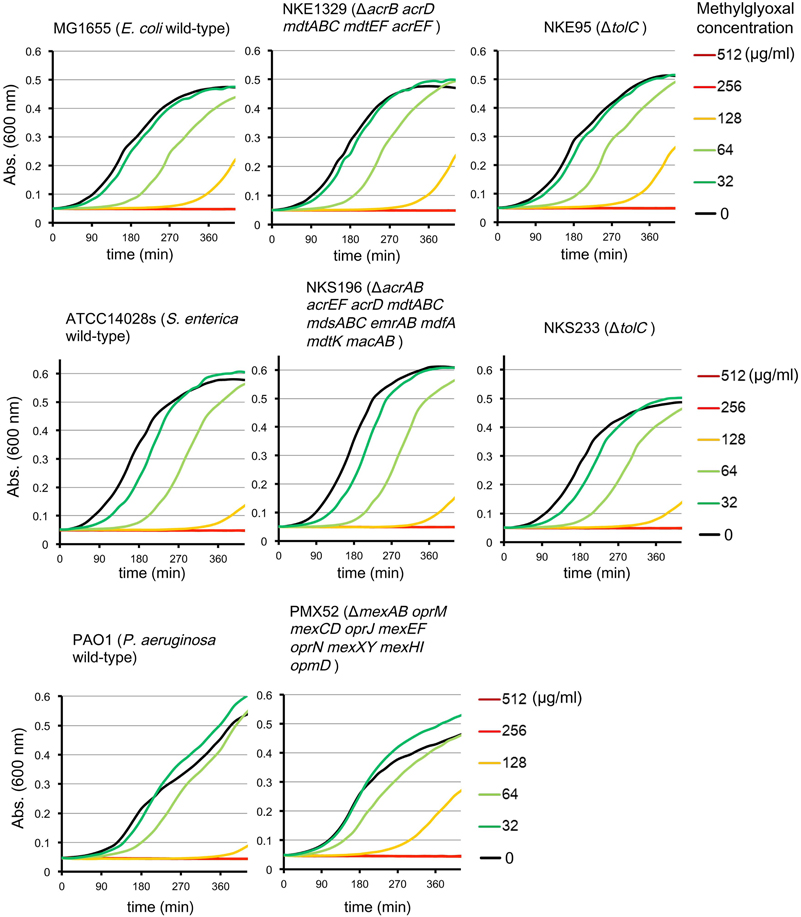
Figure 1. Effects of methylglyoxal on the growth of E. coli, S. enterica, and P. aeruginosa. Growth of E. coli (MG1655, NKE1329, and NKE95), S. enterica (ATCC14028s, NKS196, and NKS233), and P. aeruginosa (PAO1 and PMX52) strains were measured in liquid medium with or without methylglyoxal.
In this study, we showed that methylglyoxal equally inhibits drug-susceptible P. aeruginosa and MDRP at concentrations of 128–512 μg/ml (1.7–7.1 mM). Methylglyoxal is a key antimicrobial component of manuka honey, and manuka honey has previously been suggested as a topical treatment option for burn patients infected with P. aeruginosa (Cooper et al., 2002a). Jenkins and Cooper reported that MICs of manuka honey for MRSA and methicillin-resistant P. aeruginosa were 6–7% w/v (Jenkins and Cooper, 2012). This corresponds to 50–100 μg/ml methylglyoxal when manuka honey contains 7% of methylglyoxal. Cooper et al. also reported that MIC for E. coli is 16% w/v (Cooper et al., 2010), which corresponds to approximately 200 μg/ml methylglyoxal. It was previously reported that methylglyoxal is the dominant antibacterial constituent of manuka honey and that MIC of methylglyoxal for E. coli and S. aureus, determined using the agar well diffusion assay, is 1.1 mM (79.3 μg/ml) (Mavric et al., 2008). Our data showed that methylglyoxal itself inhibits the growth of MDRP strains at high concentrations, suggesting that methylglyoxal activity might be enhanced when in honey solution. Further research is required to demonstrate whether methylglyoxal and manuka honey exert their antibacterial effects through a common mechanism. We also showed that methylglyoxal is not recognized by drug efflux systems in P. aeruginosa, E. coli, and S. enterica.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tomofusa Tsuchiya of Ritsumeikan University and Yoshimi Matsumoto of Osaka University for providing strains of Pseudomonas aeruginosa. Katsuhiko Hayashi was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science. This study was supported by Grants-in-Aid for Scientific Research (Mitsuko Hayashi-Nishino) and Young Scientists from the Japan Society for the Promotion of Science (to Mitsuko Hayashi-Nishino and Kunihiko Nishino), the Kanae Foundation for the Promotion of Medical Science (to Kunihiko Nishino), the Senri Life Science Foundation (to Kunihiko Nishino), and the NEXT Program from the Cabinet Office, Government of Japan (to Kunihiko Nishino).
References
Adams, C. J., Boult, C. H., Deadman, B. J., Farr, J. M., Grainger, M. N. C., Manley-Harris, M., et al. (2009a). Isolation by HPLC and characterization of the bioactive fraction of New Zealand manuka (Leptospermum scoparium) honey. Carbohydr. Res. 344, 2609–2609. doi: 10.1016/j.carres.2009.08.008
Adams, C. J., Manley-Harris, M., and Molan, P. C. (2009b). The origin of methylglyoxal in New Zealand manuka (Leptospermum scoparium) honey. Carbohydr. Res. 344, 1050–1053. doi: 10.1016/j.carres.2009.03.020
Allen, K. L., Molan, P. C., and Reid, G. M. (1991). A survey of the antibacterial activity of some New Zealand honeys. J. Pharm. Pharmacol. 43, 817–822. doi: 10.1111/j.2042-7158.1991.tb03186.x
Blair, S. E., Cokcetin, N. N., Harry, E. J., and Carter, D. A. (2009). The unusual antibacterial activity of medical-grade Leptospermum honey: antibacterial spectrum, resistance and transcriptome analysis. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1199–1208. doi: 10.1007/s10096-009-0763-z
Blaser, G., Santos, K., Bode, U., Vetter, H., and Simon, A. (2007). Effect of medical honey on wounds colonised or infected with MRSA. J. Wound Care 16, 325–328.
Blattner, F. R., Plunkett, G. 3rd., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi: 10.1126/science.277.5331.1453
Cooper, R. A., and Anderson, A. (1970). The formation and catabolism of methylglyoxal during glycolysis in Escherichia coli. FEBS Lett. 11, 273–276. doi: 10.1016/0014-5793(70)80546-4
Cooper, R. A., Halas, E., and Molan, P. C. (2002a). The efficacy of honey in inhibiting strains of Pseudomonas aeruginosa from infected burns. J. Burn Care Rehabil. 23, 366–370. doi: 10.1097/00004630-200211000-00002
Cooper, R. A., Jenkins, L., Henriques, A. F., Duggan, R. S., and Burton, N. F. (2010). Absence of bacterial resistance to medical-grade manuka honey. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1237–1241. doi: 10.1007/s10096-010-0992-1
Cooper, R. A., Molan, P. C., and Harding, K. G. (1999). Antibacterial activity of honey against strains of Staphylococcus aureus from infected wounds. J. R. Soc. Med. 92, 283–285.
Cooper, R. A., Molan, P. C., and Harding, K. G. (2002b). The sensitivity to honey of Gram-positive cocci of clinical significance isolated from wounds. J. Appl. Microbiol. 93, 857–863. doi: 10.1046/j.1365-2672.2002.01761.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Davis, R. W., Bolstein, D., and Roth, J. R. (1980). Advanced Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986). Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193. doi: 10.1073/pnas.83.14.5189
Gethin, G., and Cowman, S. (2008). Bacteriological changes in sloughy venous leg ulcers treated with manuka honey or hydrogel: an RCT. J. Wound Care 17, 241–244, 246–247.
Henriques, A. F., Jenkins, R. E., Burton, N. F., and Cooper, R. A. (2010). The intracellular effects of manuka honey on Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 29, 45–50. doi: 10.1007/s10096-009-0817-2
Horiyama, T., Nikaido, E., Yamaguchi, A., and Nishino, K. (2011). Roles of Salmonella multidrug efflux pumps in tigecycline resistance. J. Antimicrob. Chemother. 66, 105–110. doi: 10.1093/jac/dkq421
Horiyama, T., Yamaguchi, A., and Nishino, K. (2010). TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 65, 1372–1376. doi: 10.1093/jac/dkq160
Jenkins, R., and Cooper, R. (2012). Improving antibiotic activity against wound pathogens with manuka honey in vitro. PLoS ONE 7:e45600. doi: 10.1371/journal.pone.0045600
Jervis-Bardy, J., Foreman, A., Bray, S., Tan, L., and Wormald, P. J. (2011). Methylglyoxal-infused honey mimics the anti-Staphylococcus aureus biofilm activity of manuka honey: potential implication in chronic rhinosinusitis. Laryngoscope 121, 1104–1107. doi: 10.1002/lary.21717
Krymkiewicz, N., Dieguez, E., Rekarte, U. D., and Zwaig, N. (1971). Properties and mode of action of a bactericidal compound (=methylglyoxal) produced by a mutant of Escherichia coli. J. Bacteriol. 108, 1338–1347.
Kwakman, P. H., de Boer, L., Ruyter-Spira, C. P., Creemers-Molenaar, T., Helsper, J. P., Vandenbroucke-Grauls, C. M., et al. (2011). Medical-grade honey enriched with antimicrobial peptides has enhanced activity against antibiotic-resistant pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 30, 251–257. doi: 10.1007/s10096-010-1077-x
Kwakman, P. H., te Velde, A. A., de Boer, L., Speijer, D., Vandenbroucke-Grauls, C. M., and Zaat, S. A. (2010). How honey kills bacteria. FASEB J. 24, 2576–2582. doi: 10.1096/fj.09-150789
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Mavric, E., Wittmann, S., Barth, G., and Henle, T. (2008). Identification and quantification of methylglyoxal as the dominant antibacterial constituent of Manuka (Leptospermum scoparium) honeys from New Zealand. Mol. Nutr. Food Res. 52, 483–489. doi: 10.1002/mnfr.200700282
Nakashima, R., Sakurai, K., Yamasaki, S., Nishino, K., and Yamaguchi, A. (2011). Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569. doi: 10.1038/nature10641
Natarajan, S., Williamson, D., Grey, J., Harding, K. G., and Cooper, R. A. (2001). Healing of an MRSA-colonized, hydroxyurea-induced leg ulcer with honey. J. Dermatol. Treat. 12, 33–36. doi: 10.1080/095466301750163563
Nikaido, H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77, 1–60. doi: 10.1002/9780470920541.ch1
Nikaido, H., and Pages, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Nishino, K., Latifi, T., and Groisman, E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141. doi: 10.1111/j.1365-2958.2005.04940.x
Nishino, K., Yamada, J., Hirakawa, H., Hirata, T., and Yamaguchi, A. (2003). Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to beta-lactams. Antimicrob. Agents Chemother. 47, 3030–3033. doi: 10.1128/AAC.47.9.3030-3033.2003
Ortega, B., Groeneveld, A. B., and Schultsz, C. (2004). Endemic multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Infect. Control Hosp. Epidemiol. 25, 825–831. doi: 10.1086/502303
Piddock, L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/CMR.19.2.382-402.2006
Poole, K. (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10, 12–26. doi: 10.1111/j.1469-0691.2004.00763.x
Roberts, A. E. L., Maddocks, S. E., and Cooper, R. A. (2012). Manuka honey is bactericidal against Pseudomonas aeruginosa and results in differential expression of oprF and algD. Microbiol. 158, 3005–3013. doi: 10.1099/mic.0.062794-0
Sekiguchi, J. I., Asagi, T., Miyoshi-Akiyama, T., Kasai, A., Mizuguchi, Y., Araake, M., et al. (2007). Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J. Clin. Microbiol. 45, 979–989. doi: 10.1128/JCM.01772-06
Sekiya, H., Mima, T., Morita, Y., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2003). Functional cloning and characterization of a multidrug efflux pump, mexHI-opmD, from a Pseudomonas aeruginosa mutant. Antimicrob. Agents Chemother. 47, 2990–2992. doi: 10.1128/AAC.47.9.2990-2992.2003
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Visavadia, B. G., Honeysett, J., and Danford, M. (2008). Manuka honey dressing: an effective treatment for chronic wound infections. Br. J. Oral Maxillofac. Surg. 46, 696–697. doi: 10.1016/j.bjoms.2007.12.014
Weston, R. J., Mitchell, K. R., and Allen, K. L. (1999). Antibacterial phenolic components of New Zealand manuka honey. Food Chem. 64, 295–301. doi: 10.1016/S0308-8146(98)00100-9
Keywords: manuka honey, methylglyoxal, drug efflux system, multidrug resistance, Pseudomonas aeruginosa
Citation: Hayashi K, Fukushima A, Hayashi-Nishino M and Nishino K (2014) Effect of methylglyoxal on multidrug-resistant Pseudomonas aeruginosa. Front. Microbiol. 5:180. doi: 10.3389/fmicb.2014.00180
Received: 11 April 2013; Accepted: 01 April 2014;
Published online: 17 April 2014.
Edited by:
Fiona Walsh, National University of Ireland Maynooth, IrelandReviewed by:
Agnese Lupo, Technical University of Dresden, GermanyEddie Cytryn, ARO, Volcani Agriculture Research Center, Israel
Copyright © 2014 Hayashi, Fukushima, Hayashi-Nishino and Nishino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunihiko Nishino, Laboratory of Microbiology and Infectious Diseases, Division of Special Projects, Institute of Scientific and Industrial Research, Osaka University, 8-1 Mihogaoka, Ibaraki, Osaka 567-0047, Japan e-mail: nishino@sanken.osaka-u.ac.jp
 Katsuhiko Hayashi
Katsuhiko Hayashi Aiko Fukushima
Aiko Fukushima Mitsuko Hayashi-Nishino
Mitsuko Hayashi-Nishino Kunihiko Nishino
Kunihiko Nishino