- 1Department of Molecular Cellular and Developmental Biology, University of Michigan, Ann Arbor, MI, USA
- 2Department of Microbiology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA, USA
Many bacteria produce cytolytic toxins that target host cells or other competing microbes. It is well known that environmental factors control toxin expression, however, recent work suggests that some bacteria manipulate the fold of these protein toxins to control their function. The β-sheet rich amyloid fold is a highly stable ordered aggregate that many toxins form in response to specific environmental conditions. When in the amyloid state, toxins become inert, losing the cytolytic activity they display in the soluble form. Emerging evidence suggest that some amyloids function as toxin storage systems until they are again needed, while other bacteria utilize amyloids as a structural matrix component of biofilms. This amyloid matrix component facilitates resistance to biofilm disruptive challenges. The bacterial amyloids discussed in this review reveal an elegant system where changes in protein fold and solubility dictate the function of proteins in response to the environment.
Amyloids
The secondary structure of the amyloid fold is one that is seen throughout life. Amyloids have long been studied because of their importance in human neurodegenerative diseases such as Alzheimer’s and Huntington’s diseases. There have been many reviews published on disease associated amyloids and they will not be discussed in this review (Chiti and Dobson, 2006; Eisenberg and Jucker, 2012). Amyloids were initially described in the 1850s as deposits in human tissues that stained with iodine, which was a characteristic of starch. A few years later though, it was shown that there were no carbohydrates in amyloid deposits but they consisted of proteins. Since then, amyloids have been found to be produced in many organisms and the biophysical and chemical properties of amyloids have been significantly investigated.
Amyloids are composed of fibrous oligomers of proteins that are characterized by a cross β-sheet structure running perpendicular to the fiber axis. The core of the amyloid fiber is made of protein backbones that form many hydrogen bonds between them leading to strong molecular forces. Because the strength of the amyloid is mainly independent of the side chains, proteins that can fold into amyloids do not have any sequence motifs, making it difficult to predict amyloid forming proteins. Even though they do not contain sequence homology, amyloids can be identified using biophysical properties including SDS-insolubility, protease resistance, and binding to the amyloid specific dyes Thioflavin T and Congo red (CR).
Amyloids have long been thought to be a result of protein misfolding, but over the past decade, this view has evolved to the understanding that some organisms utilize the amyloid fold for various functions aptly named Functional Amyloids (Table 1; Chapman et al., 2002). The most well studied functional amyloids are those made by bacteria that help form microbial communities called biofilms. These biofilms contain bacterial cells as well as matrix containing carbohydrates and proteins that hold them together affixed to a surface. Increasingly, it is being found that bacteria utilize the strength of the amyloid fold to make the strong biofilm matrix that resists disruption from stressors. As will be discuss below, many bacteria have developed toxin systems that are able to attack niche competitors or the host that can be abrogated by sequestering these toxins as amyloids where some have a second function in biofilm stability.
Amyloids as Structural Molecules
Curli
Curli are the most well studied bacterial functional amyloid. Through a dedicated pathway, curli form amyloids on the surface of Enterobacteria, such as Escherichia coli, and Salmonella, that aid bacteria in attaching to surfaces as well as defending the population from stress (Saldaña et al., 2009; Goulter-Thorsen et al., 2011; Zhou et al., 2012). Curli are made through a highly controlled master regulator CsgD, which induces the transcription of other curli specific genes (csg) to produce these amyloids (Figure 1; Brombacher et al., 2003). The major functional subunit of curli, CsgA, is secreted from the cell in a soluble form, leaving the outer-membrane through the pore formed by a hexamer of CsgG (Figure 1; Chapman et al., 2002; Robinson et al., 2006; Epstein et al., 2009). The minor fiber subunit CsgB is linked with the membrane and facilitates the nucleation of CsgA into amyloid fibers (Figure 1; Bian and Normark, 1997; Hammer et al., 2007). Proper assembly, localization, and regulation of curli fibers are modulated by CsgC, CsgE, and CsgF (Gibson et al., 2007; Nenninger et al., 2009, 2011; Evans and Chapman, 2014). Not only are the curli genes under strict genetic regulation, but it has been shown that cellular chaperones can modulate the fold of CsgA to prevent improper folding in the cells (Evans et al., 2011).
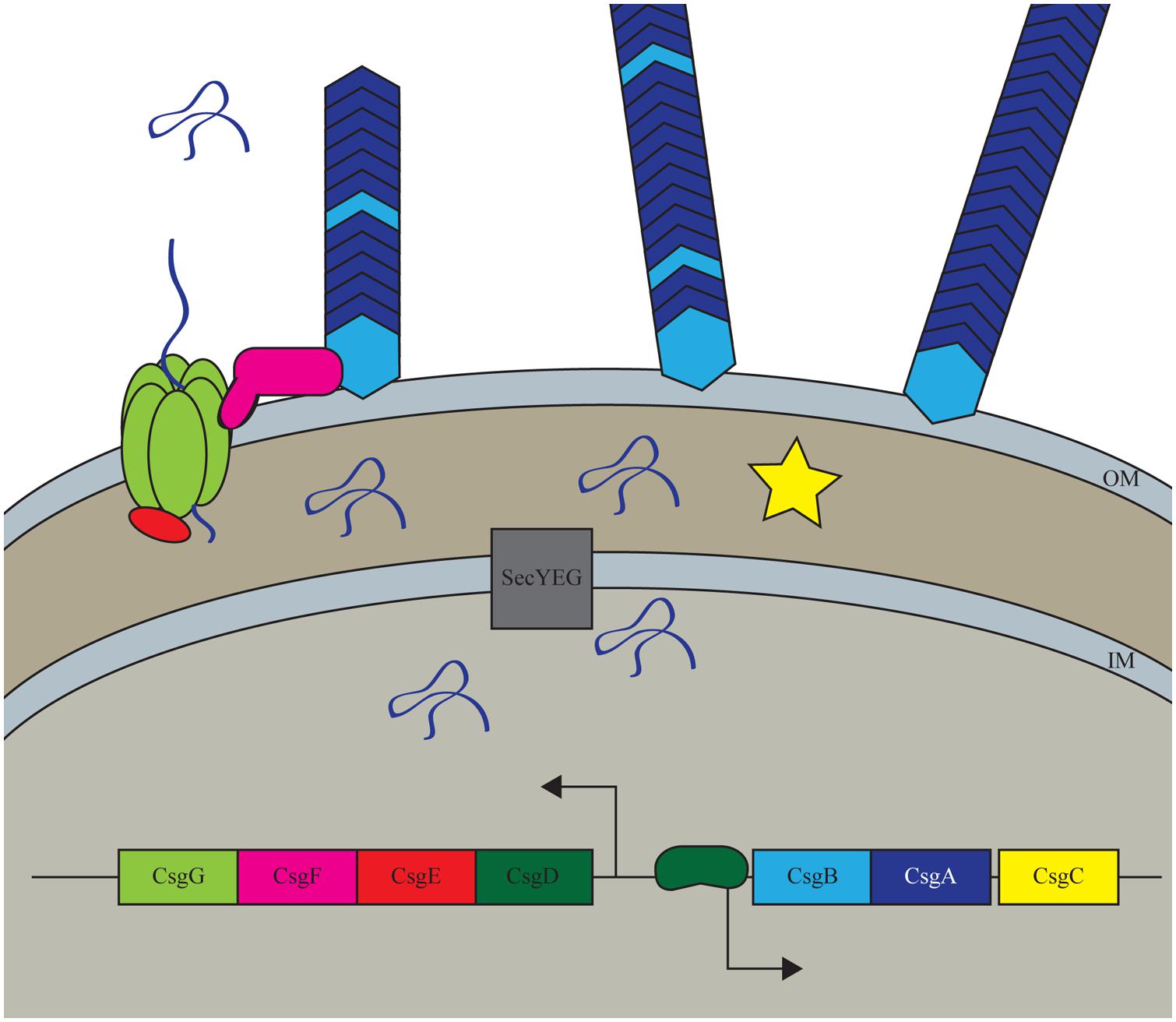
FIGURE 1. Curli biogenesis model. The curli system in Escherichia coli is a highly controlled process that only expressed the curli amyloid under conditions that promote biofilm formation. The system is transcriptionally controlled by the master regulator CsgD which increases the transcription of the major and minor subunits CsgA and CsgB. All Csg proteins other than CsgD are secreted through the Sec secretion pathway into the periplasm where CsgA, CsgB, and CsgF are then transclocated outside of the cell through the CsgG pore complex. CsgE and CsgF aid in proper export and localization of the structural components while CsgC has a less well understood role in the periplasm.
Curli fibers are important for E. coli surface colonization and biofilm formation (Chapman et al., 2002; Saldaña et al., 2009; Crémet et al., 2013; DePas et al., 2013; Giaouris et al., 2013). The expression of curli is a tightly regulated process in regards to the environment around the bacteria as well as within a biofilms community. Recently, it has been shown that there is spatial regulation within an E. coli rugose biofilms where curli producing cells are localized to the exterior of the biofilms, whereas cells on the interior of the community were not producing curli fibers (DePas et al., 2013; Serra et al., 2013). This bimodal growth allows for a protective shell of matrix-encased cells that contain a population of cells that ready to disperse and disseminate when conditions become favorable.
Other Functional Amyloids Produced by Bacteria
Emerging evidence suggest that amyloids likely play a structural role in some naturally occurring environmental biofilms. Recent work utilizing conformational antibodies that specifically bind to the amyloid fold, and the amyloid-specific dye thioflavin-T, provide evidence of amyloids being present in biofilm samples for fresh water lakes, drinking water, and activated sludge from a water treatment facility (Larsen et al., 2007). The bacteria present in these biofilms include representatives from Actinobacteria, Bacteroidetes, Chloroflexi, and Proteobacteria. Further studies revealed one member of this community, Pseudomonas fluorescens, was able to produce an amyloid found in the biofilm matrix (Dueholm et al., 2010). Proteomic analysis revealed the major subunit of the amyloid to consist of a protein named FapC (Dueholm et al., 2010). The genes necessary for formation of this amyloid were traced to the fapA-F operon, which is conserved in many Pseudomonas species. FapC contains repeat motifs and conserved Asn/Gln consensus residues similar to curli and the prion and spider silk amyloid proteins (Dueholm et al., 2010). Further studies have demonstrated that other Pseudomonads also form Fap fibrils that result in biofilm formation (Dueholm et al., 2013). These finding suggest functional amyloids are likely abundant in naturally occurring biofilms consisting of diverse microbial members.
The pathogens Mycobacterium tuberculosis and Streptococcus mutans have also been found to produce functional amyloids. In the case of M. tuberculosis, thin, aggregative flexible pili, named MTP, were observed during human infection (Alteri et al., 2007). These pili possess biophysical and morphological characteristics of amyloids and bind to the human extracellular matrix component, laminin. Proteomic analysis suggests the structural subunit of MTP is a proteolytically processed version of a 10.5 kDa protein encoded by the open reading frame Rv3312A (mtp) in M. tuberculosis strain H37Rv (Alteri et al., 2007). In addition, serum from tuberculosis patients contained antibodies that specifically recognized MTP, suggesting a role for MTP during infection (Alteri et al., 2007). MTP was also found to be important in the formation of biofilms by M. tuberculosis (Ramsugit et al., 2013). S. mutans is a member of the oral microbiome and is linked to the disease dental caries because of it’s ability to produce acid from the utilization of dietary sugars. Recent work suggests that the S. mutans adhesin P1 (antigen I/II, PAc) is an amyloid-forming protein (Oli et al., 2012). During biofilm growth S. mutans displayed amyloid fibers as evidenced by transmission electron microscopy, bound the amyloidophilic dyes CR and Thioflavin T (ThT), and possessed green birefringent properties of CR-stained protein aggregates when viewed under cross-polarized light (Oli et al., 2012). Importantly, human dental plaques contain microbial amyloids, suggesting a role for this protein fold in dental carries (Oli et al., 2012).
Chaplins are a class of hydrophobic proteins that spontaneously self-assemble into amyloid fibrils (Claessen et al., 2003). The spore-forming filamentous bacterium S. coelicolor uses chaplin amyloids to complete its lifecycle progression (Claessen et al., 2003). Under starvation conditions S. coelicolor produces aerial hyphae that extend upward out of the soil. Spores are produced in these hyphae and they are release once the soil surface has been breached. Vegetative S. coelicolor cell surfaces are hydrophilic, so to break the soil/air interface, the cells must first develop a hydrophobic coat. To this end, S. coelicolor secretes monomeric chaplin proteins (encoded by chpA-H; Claessen et al., 2003; Elliot et al., 2003). These hydrophobic proteins have been shown to form β-sheet rich amyloid fibers on contact with air, therefore chaplin amyloids are essential for S. coelicolor to complete its lifecycle from vegetative cells to spore containing hyphae.
Bifunctional Proteins
Phenol Soluble Modulins
Phenol soluble modulins (PSMs) are a family of proteins that are found in Staphylococci, most notably the significant human pathogen Staphylococcus aureus and the human commensal Staphylococcus epidermidis (Mehlin et al., 1999; Wang et al., 2007). S. aureus has nine characterized PSM peptides that are all regulated by the accessory gene regulator (AGR) quorum sensing system (Janzon et al., 1989; Wang et al., 2007). There are four PSMα, two PSMβ, and δ-toxin that are present in three separate regions of the chromosome. The newest member to this family is the N-terminal signal sequence of the AgrD molecule N-AgrD (Schwartz et al., 2014). This sequence is critical for localization of the propeptide to the membrane and once cleaved from the rest of the AgrD molecule has many structural and functional similarities to the other PSMs (Schwartz et al., 2014). In addition, some stains of S. aureus contain a pathogenicity island that harbors an ninth PSM called PSM-mec (Queck et al., 2009). The PSMs are secreted from the cells by a dedicated, essential secretion system called phenol-soluble modulin transporter (PMT; Chatterjee et al., 2013). These PSM peptides are amphipathic α-helices, meaning that one face of the helix in hydrophobic while the other is hydrophilic (Wang et al., 2007). This shared property is thought to allow for them to form pores in the membranes of competing microbes and host cells to invade tissues and evade immune cells.
Phenol soluble modulins have been shown to be critical determinants of S. aureus to cause skin abscesses and wounds in murine models as well as aiding in the survival of S. aureus in murine bacteremia models (Wang et al., 2007). PSMs stimulate neutrophil chemotaxis through the human formyl peptide receptor 2 (FPR2), at nanomolar concentrations, independent of the formylation state of the peptides (Figure 2; Kretschmer et al., 2010). Once the neutrophils are in close proximity, PSMs are able to infiltrate cells and cause cell death (Figure 2; Wang et al., 2007). Recently though, the field has shifted toward the hypothesis that in the host, PSMs may be important in virulence once S. aureus is phagocytosed by neutrophils (Surewaard et al., 2012). This hypothesis is supported by data demonstrating the serum lipoproteins are able to bind to and inactivate PSMs, meaning that they would be unable to function in the presence of serum in the host (Surewaard et al., 2012). Secondly, once phagocytosed by neutrophils, S. aureus cells highly upregulate the production of PSM peptides which aid in escaping from the phagolysosome (Figure 2; Surewaard et al., 2012).
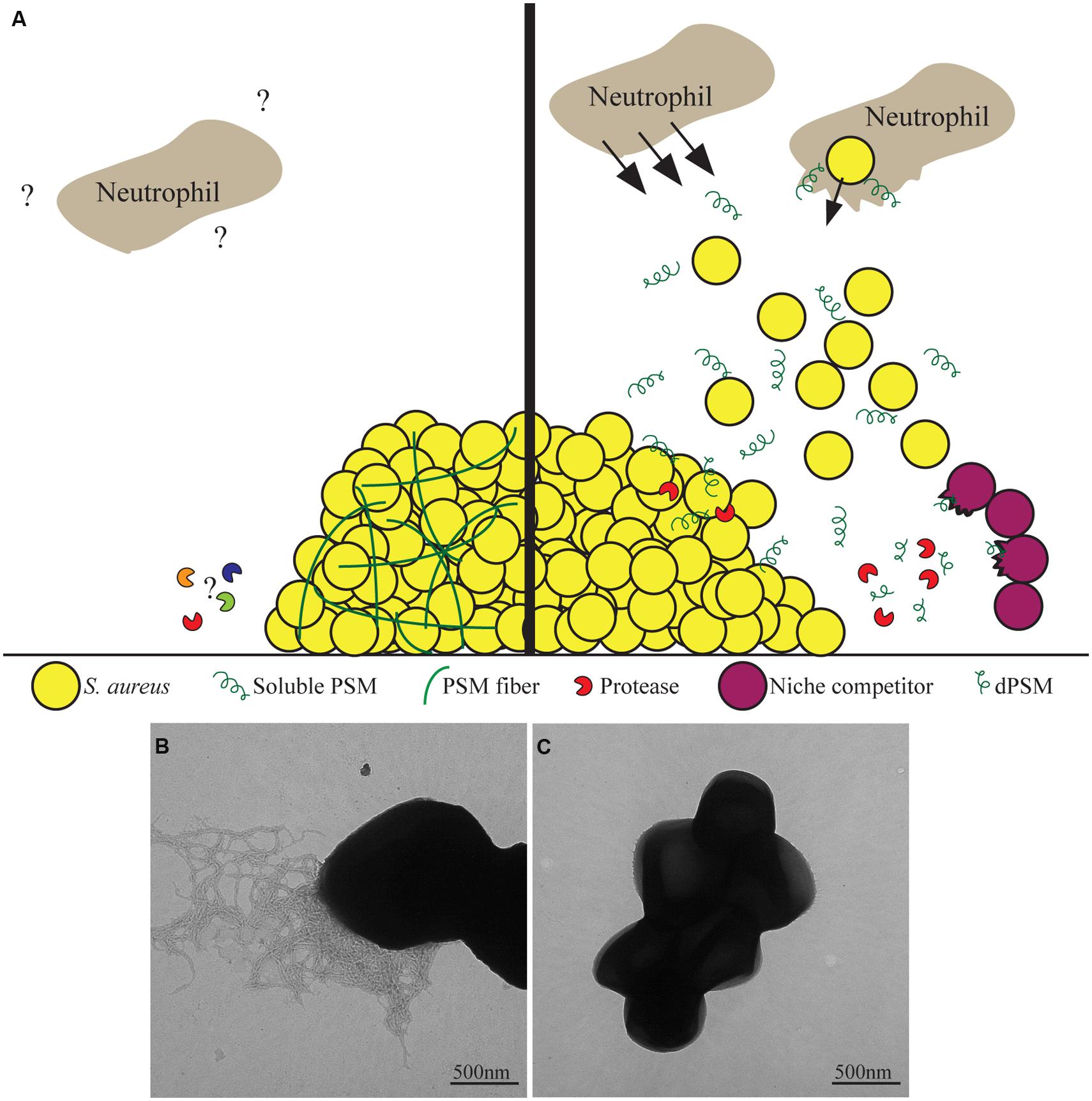
FIGURE 2. Phenol soluble modulins as bifunctional proteins. (A) Staphylococcus aureus produces Phenol soluble modulins (PSM) proteins that have been found to have many diverse functions. As soluble molecules, they are able to stimulate chemotaxis of neutrophils as well as aid S. aureus escape from the phagolysosome upon phagocytosis. Additionally, soluble PSMs can disperse biofilms as well as be proteolytically processed into PSM derivatives of phenol-soluble modulins (dPSMs) that can kill niche competing bacteria. Once polymerized into amyloid fibers, PSMs provide functional support to the biofilm community that prevents degradation by biofilm dispersing enzymes. The interactions between PSM fibers and immune cells have yet to be characterized. (B) Transmission electron microscopy of a S. aureus biofilm that is producing PSM amyloids fibers. (C) S. aureus biofilm cells under conditions where amyloid fibers are not detected.
Phenol soluble modulins are not only reported to be important for S. aureus pathogenesis and virulence against the host. PSMs have also been shown to be antimicrobial against potential competitors. PSMs were first determined to have antimicrobial effects from S. epidermidis (Cogen et al., 2010). PSMs share structural similarity to mammalian antimicrobial peptides such as LL-37, thus it was tested to determine if S. epidermidis PSMs were able to kill mammalian pathogens (Cogen et al., 2010). Two PSMs from S. epidermidis were found to have antimicrobial effects against S. aureus as well as group A Streptococcus (GAS) and worked in conjunction with LL-37 to have synergy (Cogen et al., 2010). Focus then turned to determine if and how S. aureus PSMs may act against niche competing bacteria. It was found that full length PSM peptides possessed little antimicrobial activity but derivatives of PSMs (dPSMs), PSMs that have been proteolytically processed to be missing the first few amino acids, have strong antimicrobial properties against S. pyrogenes, S. epidermidis, and GAS (Figure 2; Joo et al., 2011; Gonzalez et al., 2012). Furthermore, when a colony of S. aureus is grown in close proximity to S. epidermidis or GAS, dPSMs are localized to the zone of inhibition of the competing bacteria (Gonzalez et al., 2012).
Along with their role as toxins, the biophysical properties of PSMs give them several unique properties in modulating S. aureus communities. First these are the ability of S. aureus PSMα and PSMβ to facilitate dissemination and spreading of a colony over soft agar plates (Tsompanidou et al., 2011). This suggests that some PSMs are able to act with surfactant-like properties lowering hydropathy and allowing for S. aureus to spread (Tsompanidou et al., 2013). Additionally, PSMs have been shown to be important for the formation of biofilms. The PSMs are important biosurfactants that aid in the characteristic waves of dissemination of parts of the biofilms to colonize other areas (Figure 2; Periasamy et al., 2012).
Along with these properties attributed to the soluble PSM peptides, they are able to form amyloid fibers which stabilize S. aureus biofilms (Schwartz et al., 2012). This switch changes the soluble α-helical peptides into β-sheet rich protein aggregates (Schwartz et al., 2012). This aggregation, like other amyloids, is through a self-templating mechanism that facilitates the transformation of other nearby proteins to adopt this amyloid fold. The PSMs formed amyloid fibers in biofilms that were grown in a non-standard rich media containing peptone, glucose, and NaCl. These biofilms were completely resistant to known biofilm dispersing enzymes Proteinase K, DNase, and Dispersin B, suggesting that the PSM amyloids are able to structurally stabilize the biofilm against enzymatic targeting of the previously characterized matrix components (Figure 2; Schwartz et al., 2012). Importantly, PSMs were demonstrated to have bifunctional abilities to either strengthen biofilms or disperse them dependent on their secondary structure (Figure 2). If monomeric PSMs were added to an established biofilm they exhibits surfactant like properties, dispersing the biofilms in a concentration dependent manner, whereas PSM fiber addition does not disperse biofilms (Schwartz et al., 2012).
Further studies are needed to investigate the flexibility of the PSM peptides to switch from soluble peptides to amyloid fibers and if this change is irreversible or only temporary. Interestingly, where PSMα and PSMβ peptides were shown to be essential for S. aureus colony spreading, it has been shown that colony spreading can be inhibited by δ-toxin (Omae et al., 2012). This may suggest a role in amyloid nucleation by δ-toxin on the other PSMs that inhibit their ability to act as surfactants. It is tempting to speculate that these fibers may be reservoirs of toxins that S. aureus can utilize to both defend itself while also causing the population to disseminate and escape. It would also be interesting to see if the aggregation of these peptides into amyloids fibers is able to abrogate neutrophil chemotaxis thus acting as a way to hide from the immune system when forming biofilms in the host. Much more work is needed to fully understand how S. aureus and other Staphylococcal species utilize the fold of these PSM peptides to modulate their function.
Microcin E492
Microcin E492 (MccE492) is part of a large family of bacteriocins that are antimicrobials secreted by bacteria to kill niche competitors. In general, bacteriocins are pore-forming proteins that kill competitor microbes by forming pores in their membranes, decreasing membrane potential (de Lorenzo and Pugsley, 1985). MccE492 is produced by Klebsiella pnumoniae that is able to target many Enterobacteria species such as E. coli, and Salmonella (de Lorenzo and Pugsley, 1985; Destoumieux-Garzón et al., 2003). MccE492 is found as both an unmodified peptide as well as a posttranslationally modified molecule with a catechol-type siderophore molecule (Thomas et al., 2004). This post translational modification allows for microcin to be recognized by siderophore catecholate receptors of target organisms that cause an uptake of the mature MccE492 molecule into the periplasm (Destoumieux-Garzón et al., 2003). The exact mechanism of cell death is unknown due to the fact that there needs to be much more MccE492 present for antimicrobial activity compared to the amount needed for membrane permeabilization (Destoumieux-Garzón et al., 2003).
Microcin E492 is unique to known microcins in that it is produced through exponential and stationary phase, whereas other microcins are only produced in stationary phase. Interestingly, MccE492 loses its antimicrobial activity when the population enters stationary phase even though the protein is still present at high levels (Corsini et al., 2002). This observation led to the discovery that in stationary phase, MccE492 aggregates to form amyloid fibers (Biéler et al., 2005). Aggregation into an amyloid abolishes the toxic effects of this peptide (Biéler et al., 2005). The aggregation of the peptide is modulated by many environmental factors as well as the state of posttranslational modification of the peptides.
MccE492 is produced in both the unmodified and modified forms in culture. The modified, antimicrobial MccE492 is the predominant form while the bacteria are growing in exponential phase of the culture (Marcoleta et al., 2013). When the bacteria begin to enter stationary phase, they decrease the production of the modified form making the unmodified MccE492 more prevalent in the population (Marcoleta et al., 2013). Unmodified MccE492 polymerizes faster than the modified peptide in forming amyloid fibers leading them to hypothesis that the bacteria may begin to produce more unmodified peptide in stationary phase is to begin to detoxify the environment by sequestering these peptides in inert amyloids (Marcoleta et al., 2013). Even though the unmodified form is more efficient in polymerizing, the modified MccE492 is found in fibers with the unmodified form (Marcoleta et al., 2013). Moreover, polymerization of the modified MccE492 is accelerated in the presence of unmodified seeds, small amyloids that can nucleate amyloid elongation (Marcoleta et al., 2013).
Apart from the influence of posttranslational modification on MccE492 polymerization, the environment has a profound effect of the rate of polymerization and can even cause disassembly of MccE492 amyloids (Shahnawaz and Soto, 2012). Basic pH, low ionic strength, and dilution of the fibers all led to fiber disassembly in two hours (Shahnawaz and Soto, 2012). These disassembled fibers regained their antimicrobial activity that was absent while in amyloid fibers (Shahnawaz and Soto, 2012). Even more striking was the ability of these disassembled toxins to reform amyloids when the environment was again changed (Shahnawaz and Soto, 2012). These data demonstrate that in the case of MccE492, there are many factors that influence the toxicity of the peptides suggesting that the Klebsiella pneumonia has evolved a mechanism of efficiently modulating the toxicity of MccE942. This data also leads to the exciting hypothesis that other amyloids may have conditions that lead to the disassembly of the amyloid fiber.
TasA
TasA was originally described as a protein in Bacillus subtilis that was involved in sporulation but was later found to be expressed in stationary phase cells (Stover and Driks, 1999b,c). It has been demonstrated to have widespread antimicrobial activities against both plant and animal pathogens and commensal bacteria (Stover and Driks, 1999c). Recently though, TasA has been shown to produce amyloid fibers in B. subtilis biofilms that contributes heavily to the formation of complex community architecture similar to curli in E. coli (Romero et al., 2010). TasA is part of an operon that also encodes for TapA, the minor amyloid subunit and fiber anchor, and SipW, the signal peptidase that processes TapA and TasA (Stover and Driks, 1999a,c). It has been proposed that the antimicrobial effects of TasA may be due to the formation of toxic oligomers that many amyloidogenic proteins generate, but to our knowledge, this has not yet been investigated (Romero et al., 2010).
Listeriolysin O
Listeriolysin O (LLO) of Listeria monocytogenes is a cholesterol-dependent cytolysin whose activity is dependent on pH. LLO formed pores in the membrane of phagolysosome allowing for L. monocytogenes to escape and carry out its replicative phase in the host cell cytosol (Portnoy et al., 1988; Cossart et al., 1989). The pH dependent activity of LLO was shown to be due to an irreversible structural change in the protein that lead to a decreased ability in hemolytic activity (Schuerch et al., 2005). Later, it was appreciated that this structural change was actually the result of LLO forming an amyloid (Bavdek et al., 2012). This pH change of the protein into an amyloid suggests that this proteins has evolved to form pores while the bacteria is trapped in a phagolysosome, but once it escapes into the cytosol, the higher pH inactivates the proteins by triggering amyloidogenesis (Bavdek et al., 2012). This amyloidogenesis may prevent the LLO toxin from lysing the infected cell while L. monocytogenes replicates in the host cell. It is unknown if LLO amyloids demonstrate any activity intracellularly.
Harpins
Harpins are a class of proteins that are produced by gram-negative plant pathogens. They are characterized by being glycine rich, heat stable proteins that are secreted by a type III secretion system that can trigger a hypersensitive response (HR) in plants and are predicted to have α-helical regions (Wei et al., 1992). Harpins trigger this HR when they are present in the intercellular space. The plant cells detect these proteins and respond using the early defense response through an apoptosis-like cell death. Pathogens lacking harpins, such as HpaG of Xanthomonas axonopodis, have decreased virulence (Kim et al., 2003, 2004). The mechanism by which harpins trigger HR in plants in not fully understood, but there is some data supporting harpins interacting with and disrupting cell membranes leading to depolarization (Lee et al., 2001).
In 2007, a harpin from Xanthamonas, HpaG, was characterized biochemically (Oh et al., 2007). This group found that under conditions that mimic plant apoplasts, HpaG formed amyloid fibers (Oh et al., 2007). A mutant of the proteins (L50P) that did not trigger HR in plants and was also unable to form amyloid fibers (Oh et al., 2007). From this, the authors suggest that the transition to amyloid fibers is an important step in triggering HR. Furthermore, harpins from other plant pathogens, Ercinia amylovora, and Pseudomonas syringae also formed amyloid fibers in plant apoplast-like conditions (Oh et al., 2007). More detailed studies on harpins are needed to determine exactly what properties can be attributed to soluble and amyloid forms of these proteins.
Amyloid Inhibitors
An exciting field is emerging that is trying to speed up or slow down the formation of amyloid fibers using small molecules. The idea is that for many amyloidogenic proteins, the toxicity is due to the formation of intermediate oligomers that can disrupt membranes. By accelerating the polymerization of amyloid subunits, we may be able to bypass the toxic, degenerative affect of amyloids slowing the progression of neurodegenerative disease. Along similar lines, since many bacterial amyloids have been shown to aid in adherence to surfaces, if we use small molecules to interfere with the polymerization, bacteria may not be able to anchor themselves to the host in infections leading to faster clearance of pathogens. Conversely, in cases like the PSMs of S. aureus, since the soluble form is a toxin that is able to disrupt the immune response, by causing polymerization of the monomers could prevent their toxic function to cells allowing the immune system to clear the infection.
Curlicides and pilicides are ring-fused 2-pyridone molecules that are designed to look like peptide backbones. They have been designed to mimic a proteins backbone and interact with proteins that form amyloids by either disrupting their ability to polymerize, or nucleating and accelerating the amyloid maturation (Andersson et al., 2013). They have so far been characterized with their interaction with E. coli curli and type 1 pili (Andersson et al., 2013). These molecules are not only able to influence the in vitro polymerization of CsgA, but they are able to affect the biogenesis of curli and pili in E. coli biofilms (Andersson et al., 2013). Additionally, uropathogenic E. coli treated with curlicides were attenuated in a murine model of a urinary tract infection (Cegelski et al., 2009). Other groups have taken the approach of designing small, non-natural peptides that can disrupt amyloid formation (Sievers et al., 2011). This has been shown to be successful in vitro with disease associated amyloids (Sievers et al., 2011). Recently, TasA has been proposed to be model amyloid for screening molecules with widespread activity against amyloids necessary for biofilm formation (Romero et al., 2013). Ongoing research is increasing the efficacy of these molecules as well as characterizing their ability to modulate the biogenesis of other amyloids.
Functional Amyloids in Other Kingdoms of Life
Eukaryotic amyloids have been the subject of many recent reviews (Liebman and Chernoff, 2012; Watt et al., 2013; Wickner et al., 2013). Importantly, functional amyloids are not exclusive to bacteria. They have been found to be important in diverse eukaryotes from yeast to humans. It has recently become appreciated that humans have several functional amyloids. The protein Pmel17, a protein made in melanosomes for mammalian pigmentation, forms amyloids fibers (Fowler et al., 2005). The production of this amyloids is highly regulated with cells utilizing proteolytic processing to mature the proteins into a form that can form amyloids (Berson et al., 2001; Kummer et al., 2009). These processing steps prevent the proteins from forming amyloids in other cellular compartments preventing the toxicity that is associated with disease-associated amyloids. Additionally, it has been found that peptide hormones that are stored in the secretory granules of the mammalian pituitary form amyloids (Maji et al., 2009). It was found that in vitro, 31 of 42 studied hormones were able to form amyloids fibers (Maji et al., 2009). Mouse pituitary’s also contained amyloid fibers using various approaches to identify amyloids (Maji et al., 2009). It is thought that these peptide hormones are stored in secretary granules as amyloids until they are needed. The cells can then secrete the granules and dilution of the amyloids causes disassociation and activation of the peptide hormones (Maji et al., 2009).
Many yeast form prions, self-propagating amyloids that are heritable elements. This method of non-mendelian inheritance was first proposed for [URE3] (Wickner, 1994). It was shown that the cytoplasmically inherited [URE3] element has the opposite effect on ureidosuccinate metabolism as the Ure2 protein and that cells cured of [URE3] were able to regain the element when Ure2 is overexpressed (Aigle and Lacroute, 1975; Wickner, 1994). This lead Wickner to the hypothesis that cytoplasmically inherited elements in yeast were prions (Wickner, 1994). Since then, several yeast proteins have been shown to form prions that, in most cases, abrogate the function of proteins.
Concluding Remarks
It is becoming ever more appreciated that the amyloid fold is not just a product of protein misfolding, but it is a ubiquitously used protein fold throughout the kingdoms of life. Amyloids provide structure or control availability of proteins such as toxins or signaling molecules. Even more exciting is the discovery that some of these proteins have been found to have different functions when they are in their soluble or insoluble forms. The production of functional amyloids is a highly controlled and regulated process that is controlled on several levels including transcriptional, translational, and posttranslational. The difficulty associated with breaking up these proteins is the property that has made them so valuable for many organisms. In the case of many bacteria, these amyloids provide a structural component that keep the community protected against mechanical and enzymatic disruption (Schwartz et al., 2012). In others, they are used as reservoirs of toxins that are ready to become active once the environment changes (Shahnawaz and Soto, 2012). This field is only beginning to look at the effect that these functional amyloids play in the dynamic relationships between bacterial species as well as how these proteins may be involved in bacterial interactions with the host at a commensal or pathogenic level. It will be exciting to see where this field of bifunctional bacterial proteins goes as well as targeting these proteins to disperse biofilms or to sequester toxins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the members of the laboratories of Blaise Boles and Matt Chapman at the University of Michigan for insightful conversation. This work was funded by a NIH grant NIAID AI081748 to Blaise R. Boles and American Heart Association Fellowship 13PRE13810001 to Adnan K. Syed.
References
Aigle, M., and Lacroute, F. (1975). Genetical aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136, 327–335. doi: 10.1007/BF00341717
Alteri, C. J., Xicohténcatl-Cortes, J., Hess, S., Caballero-Olín, G., Giron, J. A., and Friedman, R. L. (2007). Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. U.S.A. 104, 5145–5150. doi: 10.1073/pnas.0602304104
Andersson, E. K., Bengtsson, C., Evans, M. L., Chorell, E., Sellstedt, M., Lindgren, A. E. G.,et al. (2013). Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem. Biol. 20, 1245–1254. doi: 10.1016/j.chembiol.2013.07.017
Bavdek, A., Kostanjšek, R., Antonini, V., Lakey, J. H., Dalla Serra, M., Gilbert, R. J. C.,et al. (2012). pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 279, 126–141. doi: 10.1111/j.1742-4658.2011.08405.x
Berson, J. F., Harper, D. C., Tenza, D., Raposo, G., and Marks, M. S. (2001). Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol. Biol. Cell 12, 3451–3464. doi: 10.1091/mbc.12.11.3451
Bian, Z., and Normark, S. (1997). Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 16, 5827–5836. doi: 10.1093/emboj/16.19.5827
Biéler, S., Estrada, L., Lagos, R., Baeza, M., Castilla, J., and Soto, C. (2005). Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 280, 26880–26885. doi: 10.1074/jbc.M502031200
Brombacher, E., Dorel, C., Zehnder, A. J., and Landini, P. (2003). The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149, 2847–2857. doi: 10.1099/mic.0.26306-0
Cegelski, L., Pinkner, J. S., Hammer, N. D., Cusumano, C. K., Hung, C. S., Chorell, E.,et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5, 913–919. doi: 10.1038/nchembio.242
Chapman, M. R., Robinson, L. S., Pinkner, J. S., Roth, R., Heuser, J., Hammar, M.,et al. (2002). Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855. doi: 10.1126/science.1067484
Chatterjee, S. S., Joo, H.-S., Duong, A. C., Dieringer, T. D., Tan, V. Y., Song, Y.,et al. (2013). Essential Staphylococcus aureus toxin export system. Nat. Med. 19, 364–367. doi: 10.1038/nm.3047
Chiti, F., and Dobson, C. M. (2006). Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366. doi: 10.1146/annurev.biochem.75.101304.123901
Claessen, D., Rink, R., de Jong, W., Siebring, J., de Vreugd, P., Boersma, F. G. H.,et al. (2003). A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726. doi: 10.1101/gad.264303
Cogen, A. L., Yamasaki, K., Sanchez, K. M., Dorschner, R. A., Lai, Y., MacLeod, D. T.,et al. (2010). Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 130, 192–200. doi: 10.1038/jid.2009.243
Corsini, G., Baeza, M., Monasterio, O., and Lagos, R. (2002). The expression of genes involved in microcin maturation regulates the production of active microcin E492. Biochimie 84, 539–544. doi: 10.1016/S0300-9084(02)01415-3
Cossart, P., Vicente, M. F., Mengaud, J., Baquero, F., Perez-Diaz, J. C., and Berche, P. (1989). Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57, 3629–3636.
Crémet, L., Corvec, S., Batard, E., Auger, M., Lopez, I., Pagniez, F.,et al. (2013). Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli. Diagn. Microbiol. Infect. Dis. 75, 252–255. doi: 10.1016/j.diagmicrobio.2012.11.019
de Jong, W., Wösten, H. A. B., Dijkhuizen, L., and Claessen, D. (2009). Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol. Microbiol. 73, 1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x
de Lorenzo, V., and Pugsley, A. P. (1985). Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27, 666–669. doi: 10.1128/AAC.27.4.666
DePas, W. H., Hufnagel, D. A., Lee, J. S., Blanco, L. P., Bernstein, H. C., Fisher, S. T.,et al. (2013). Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 2629–2634. doi: 10.1073/pnas.1218703110
Destoumieux-Garzón, D., Thomas, X., Santamaria, M., Goulard, C., Barthélémy, M., Boscher, B.,et al. (2003). Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49, 1031–1041. doi: 10.1046/j.1365-2958.2003.03610.x
Dueholm, M. S., Petersen, S. V., Sønderkaer, M., Larsen, P., Christiansen, G., Hein, K. L.,et al. (2010). Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x
Dueholm, M. S., Søndergaard, M. T., Nilsson, M., Christiansen, G., Stensballe, A., Overgaard, M. T.,et al. (2013). Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2, 365–382. doi: 10.1002/mbo3.81
Eisenberg, D., and Jucker, M. (2012). The amyloid state of proteins in human diseases. Cell 148, 1188–1203. doi: 10.1016/j.cell.2012.02.022
Elliot, M. A., Karoonuthaisiri, N., Huang, J., Bibb, M. J., Cohen, S. N., Kao, C. M.,et al. (2003). The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17, 1727–1740. doi: 10.1101/gad.264403
Epstein, E. A., Reizian, M. A., and Chapman, M. R. (2009). Spatial clustering of the curlin secretion lipoprotein requires curli fiber assembly. J. Bacteriol. 191, 608–615. doi: 10.1128/JB.01244-08
Evans, M. L., and Chapman, M. R. (2014). Curli biogenesis: order out of disorder. Biochim. Biophys. Acta 1844, 1551–1558. doi: 10.1016/j.bbamcr.2013.09.010
Evans, M. L., Schmidt, J. C., Ilbert, M., Doyle, S. M., Quan, S., Bardwell, J. C. A.,et al. (2011). E. coli chaperones DnaK, Hsp33 and Spy inhibit bacterial functional amyloid assembly. Prion 5, 323–334. doi: 10.4161/pri.18555
Fowler, D. M., Koulov, A. V., Alory-Jost, C., Marks, M. S., Balch, W. E., and Kelly, J. W. (2005). Functional amyloid formation within mammalian tissue. PLoS Biol. 4:e6. doi: 10.1371/journal.pbio.0040006.sg001
Giaouris, E., Samoilis, G., Chorianopoulos, N., Ercolini, D., and Nychas, G.-J. (2013). Differential protein expression patterns between planktonic and biofilm cells of Salmonella enterica serovar Enteritidis PT4 on stainless steel surface. Int. J. Food Microbiol. 162, 105–113. doi: 10.1016/j.ijfoodmicro.2012.12.023
Gibson, D. L., White, A. P., Rajotte, C. M., and Kay, W. W. (2007). AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella enteritidis. Microbiology 153, 1131–1140. doi: 10.1099/mic.0.2006/000935-0
Gonzalez, D. J., Okumura, C. Y., Hollands, A., Kersten, R., Akong-Moore, K., Pence, M. A.,et al. (2012). Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry. J. Biol. Chem. 287, 13889–13898. doi: 10.1074/jbc.M112.349860
Goulter-Thorsen, R. M., Taran, E., Gentle, I. R., Gobius, K. S., and Dykes, G. A. (2011). CsgA production by Escherichia coli O157:H7 alters attachment to abiotic surfaces in some growth environments. Appl. Environ. Microbiol. 77, 7339–7344. doi: 10.1128/AEM.00277-11
Hammer, N. D., Schmidt, J. C., and Chapman, M. R. (2007). The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U.S.A. 104, 12494–12499. doi: 10.1073/pnas.0703310104
Janzon, L., Löfdahl, S., and Arvidson, S. (1989). Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 219, 480–485. doi: 10.1007/BF00259623
Joo, H.-S., Cheung, G. Y. C., and Otto, M. (2011). Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J. Biol. Chem. 286, 8933–8940. doi: 10.1074/jbc.M111.221382
Kim, J.-G., Jeon, E., Oh, J., Moon, J. S., and Hwang, I. (2004). Mutational analysis of Xanthomonas harpin HpaG identifies a key functional region that elicits the hypersensitive response in nonhost plants. J. Bacteriol. 186, 6239–6247. doi: 10.1128/JB.186.18.6239-6247.2004
Kim, J.-G., Park, B. K., Yoo, C.-H., Jeon, E., Oh, J., and Hwang, I. (2003). Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166. doi: 10.1128/JB.185.10.3155-3166.2003
Kretschmer, D., Gleske, A.-K., Rautenberg, M., Wang, R., Köberle, M., Bohn, E.,et al. (2010). Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe 7, 463–473. doi: 10.1016/j.chom.2010.05.012
Kummer, M. P., Maruyama, H., Huelsmann, C., Baches, S., Weggen, S., and Koo, E. H. (2009). Formation of Pmel17 amyloid is regulated by juxtamembrane metalloproteinase cleavage, and the resulting C-terminal fragment is a substrate for γ-secretase. J. Biol. Chem. 284, 2296–2306. doi: 10.1074/jbc.M808904200
Larsen, P., Nielsen, J. L., Dueholm, M. S., Wetzel, R., Otzen, D., and Nielsen, P. H. (2007). Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x
Lee, J., Klessig, D. F., and Nürnberger, T. (2001). A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13, 1079–1093. doi: 10.1105/tpc.13.5.1079
Liebman, S. W., and Chernoff, Y. O. (2012). Prions in yeast. Genetics 191, 1041–1072. doi: 10.1534/genetics.111.137760
Maji, S. K., Perrin, M. H., Sawaya, M. R., Jessberger, S., Vadodaria, K., Rissman, R. A.,et al. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332. doi: 10.1126/science.1173155
Marcoleta, A., Marín, M., Mercado, G., Valpuesta, J. M., Monasterio, O., and Lagos, R. (2013). Microcin e492 amyloid formation is retarded by posttranslational modification. J. Bacteriol. 195, 3995–4004. doi: 10.1128/JB.00564-13
Mehlin, C., Headley, C. M., and Klebanoff, S. J. (1999). An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J. Exp. Med. 189, 907–918. doi: 10.1084/jem.189.6.907
Nenninger, A. A., Robinson, L. S., Hammer, N. D., Epstein, E. A., Badtke, M. P., Hultgren, S. J.,et al. (2011). CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol. Microbiol. 81, 486–499. doi: 10.1111/j.1365-2958.2011.07706.x
Nenninger, A. A., Robinson, L. S., and Hultgren, S. J. (2009). Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U.S.A. 106, 900–905. doi: 10.1073/pnas.0812143106
Oh, J., Kim, J.-G., Jeon, E., Yoo, C.-H., Moon, J. S., Rhee, S.,et al. (2007). Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J. Biol. Chem. 282, 13601–13609. doi: 10.1074/jbc.M602576200
Oli, M. W., Otoo, H. N., Crowley, P. J., Heim, K. P., Nascimento, M. M., Ramsook, C. B.,et al. (2012). Functional amyloid formation by Streptococcus mutans. Microbiology 158, 2903–2916. doi: 10.1099/mic.0.060855-0
Omae, Y., Sekimizu, K., and Kaito, C. (2012). Inhibition of colony-spreading activity of Staphylococcus aureus by secretion of δ-hemolysin. J. Biol. Chem. 287, 15570–15579. doi: 10.1074/jbc.M112.357848
Periasamy, S., Joo, H.-S., Duong, A. C., Bach, T.-H. L., Tan, V. Y., Chatterjee, S. S.,et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286. doi: 10.1073/pnas.1115006109
Portnoy, D. A., Jacks, P. S., and Hinrichs, D. J. (1988). Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167, 1459–1471. doi: 10.1084/jem.167.4.1459
Queck, S. Y., Khan, B. A., Wang, R., Bach, T.-H. L., Kretschmer, D., Chen, L.,et al. (2009). Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 5:e1000533. doi: 10.1371/journal.ppat.1000533.g010
Ramsugit, S., Guma, S., Pillay, B., Jain, P., Larsen, M. H., Danaviah, S.,et al. (2013). Pili contribute to biofilm formation in vitro in Mycobacterium tuberculosis. Antonie Van Leeuwenhoek 104, 725–735. doi: 10.1007/s10482-013-9981-6
Robinson, L. S., Ashman, E. M., Hultgren, S. J., and Chapman, M. R. (2006). Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59, 870–881. doi: 10.1111/j.1365-2958.2005.04997.x
Romero, D., Aguilar, C., Losick, R., and Kolter, R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234. doi: 10.1073/pnas.0910560107
Romero, D., Sanabria-Valentín, E., Vlamakis, H., and Kolter, R. (2013). Biofilm inhibitors that target amyloid proteins. Chem. Biol. 20, 102–110. doi: 10.1016/j.chembiol.2012.10.021
Saldaña, Z., Xicohténcatl-Cortes, J., Avelino, F., Phillips, A. D., Kaper, J. B., Puente, J. L.,et al. (2009). Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11, 992–1006. doi: 10.1111/j.1462-2920.2008.01824.x
Schuerch, D. W., Wilson-Kubalek, E. M., and Tweten, R. K. (2005). Molecular basis of listeriolysin O pH dependence. Proc. Natl. Acad. Sci. U.S.A. 102, 12537–12542. doi: 10.1073/pnas.0500558102
Schwartz, K., Sekedat, M. D., Syed, A. K., O’Hara, B., Payne, D. E., Lamb, A.,et al. (2014). The AgrD N-terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. Infect. Immun. doi: 10.1128/IAI.02111-14
Schwartz, K., Syed, A. K., Stephenson, R. E., Rickard, A. H., and Boles, B. R. (2012). Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 8:e1002744. doi: 10.1371/journal.ppat.1002744.t001
Serra, D. O., Richter, A. M., Klauck, G., Mika, F., and Hengge, R. (2013). Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 4:e00103–13. doi: 10.1128/mBio.00103-13
Shahnawaz, M., and Soto, C. (2012). Microcin amyloid fibrils A are reservoir of toxic oligomeric species. J. Biol. Chem. 287, 11665–11676. doi: 10.1074/jbc.M111.282533
Sievers, S. A., Karanicolas, J., Chang, H. W., Zhao, A., Jiang, L., Zirafi, O.,et al. (2011). Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475, 96–100. doi: 10.1038/nature10154
Stover, A. G., and Driks, A. (1999a). Control of synthesis and secretion of the Bacillus subtilis protein YqxM. J. Bacteriol. 181, 7065–7069.
Stover, A. G., and Driks, A. (1999b). Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. J. Bacteriol. 181, 5476–5481.
Stover, A. G., and Driks, A. (1999c). Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J. Bacteriol. 181, 1664–1672.
Surewaard, B. G., Nijland, R., Spaan, A. N., Kruijtzer, J. A., de Haas, C. J., and van Strijp, J. A. (2012). Inactivation of staphylococcal phenol soluble modulins by serum lipoprotein particles. PLoS Pathog. 8:e1002606. doi: 10.1371/journal.ppat.1002606.g007
Thomas, X., Destoumieux-Garzón, D., Peduzzi, J., Afonso, C., Blond, A., Birlirakis, N.,et al. (2004). Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279, 28233–28242. doi: 10.1074/jbc.M400228200
Tsompanidou, E., Denham, E. L., Becher, D., de Jong, A., Buist, G., van Oosten, M.,et al. (2013). Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 79, 886–895. doi: 10.1128/AEM.03157-12
Tsompanidou, E., Sibbald, M. J. J. B., Chlebowicz, M. A., Dreisbach, A., Back, J. W., van Dijl, J. M.,et al. (2011). Requirement of the agr locus for colony spreading of Staphylococcus aureus. J. Bacteriol. 193, 1267–1272. doi: 10.1128/JB.01276-10
Wang, R., Braughton, K. R., Kretschmer, D., Bach, T.-H. L., Queck, S. Y., Li, M.,et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514. doi: 10.1038/nm1656
Watt, B., van Niel, G., Raposo, G., and Marks, M. S. (2013). PMEL: a pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res. 26, 300–315. doi: 10.1111/pcmr.12067
Wei, Z. M., Laby, R. J., Zumoff, C. H., Bauer, D. W., He, S. Y., Collmer, A.,et al. (1992). Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88. doi: 10.1126/science.1621099
Wickner, R. B. (1994). [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569. doi: 10.1126/science.7909170
Wickner, R. B., Edskes, H. K., Bateman, D. A., Kelly, A. C., Gorkovskiy, A., Dayani, Y.,et al. (2013). Amyloids and yeast prion biology. Biochemistry 52, 1514–1527. doi: 10.1021/bi301686a
Keywords: functional amyloid, bacterial toxin, biofilm, bifunctional protein, aggregation
Citation: Syed AK and Boles BR (2014) Fold modulating function: bacterial toxins to functional amyloids. Front. Microbiol. 5:401. doi: 10.3389/fmicb.2014.00401
Received: 05 May 2014; Accepted: 16 July 2014;
Published online: 01 August 2014.
Edited by:
Salvador Ventura, Universitat Autonoma de Barcelona, SpainReviewed by:
Paras Jain, Albert Einstein College of Medicine, USAClaudio Soto, University of Texas, USA
Copyright © 2014 Syed and Boles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Blaise R. Boles, Department of Microbiology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, 51 Newton Road, Iowa City, IA, USA e-mail: blaise-boles@uiowa.edu
 Adnan K. Syed
Adnan K. Syed Blaise R. Boles
Blaise R. Boles