- 1Molecular Immunology Group, Department of Pathology and Pathogen Biology, Royal Veterinary College, Hatfield, UK
- 2Leibniz Institute for Farm Animal Biology, Dummerstorf, Germany
- 3School of Veterinary Medicine and Sciences, Faculty of Medicine and Health Sciences, University of Nottingham, Sutton Bonington, UK
- 4Department of Pathobiological Sciences, University of Wisconsin–Madison, Madison, WI, USA
- 5Department of Microbiology and Immunology, Faculty of Medicine, Dentistry and Health Sciences, University of Melbourne, Melbourne, VIC, Australia
Staphylococcus aureus, sequence type (ST) 398, is an emerging pathogen and the leading cause of livestock-associated methicillin-resistant S. aureus infections in Europe and North America. This strain is characterized by high promiscuity in terms of host-species and also lacks several traditional S. aureus virulence factors. This does not, however, explain the apparent ease with which it crosses species-barriers. Recently, TIR-domain containing proteins (Tcps) which inhibit the innate immune response were identified in some Gram-negative bacteria. Here we report the presence of two proteins, S. aureus TIR-like Protein 1 (SaTlp1) and S. aureus TIR-like Protein 2 (SaTlp2), expressed by ST398 which contain domain of unknown function 1863 (DUF1863), similar to the Toll/IL-1 receptor (TIR) domain. In contrast to the Tcps in Gram-negative bacteria, our data suggest that SaTlp1 and SaTlp2 increase activation of the transcription factor NF-κB as well as downstream pro-inflammatory cytokines and immune effectors. To assess the role of both proteins as potential virulence factors knock-out mutants were created. These showed a slightly enhanced survival rate in a murine infectious model compared to the wild-type strain at one dose. Our data suggest that both proteins may act as factors contributing to the enhanced ability of ST398 to cross species-barriers.
Introduction
Staphylococcus aureus is an important commensal and pathogen of animal species causing a variety of diseases ranging from skin and soft tissue infections to more severe, life threatening infections such as toxic shock syndrome in both human and animal species (Lowy, 1998). In addition, S. aureus is a major pathogen causing bovine mastitis, a disease of high economic importance (Holmes and Zadoks, 2011). Here, infections with S. aureus mainly causes subclinical infections of the udder parenchyma, which is accompanied by the absent of the expression of early innate immune defense genes when compared to infection with Escherichia coli (Petzl et al., 2008). Indeed, comparative kinetic analysis between E. coli and S. aureus infected primary bovine mammary epithelial cells (pbMECs) showed that S. aureus infection failed to induce tumor necrosis factor (TNF), interleukin (IL)-1 and IL-8 (CXCL8), which did not depend on the lack of recognition by bovine Toll-like receptor (TLR) 2 or 4 (Yang et al., 2008; Gunther et al., 2011). Treatment of S. aureus infections is complicated by its propensity to acquire antibiotic resistance determinants, most notably the emergence of methicillin-resistant S. aureus (MRSA). Human MRSA infections can be classified as being community-associated (CA-MRSA), healthcare-associated (HA-MRSA), or livestock-associated (LA-MRSA; Lindsay, 2010). Although most S. aureus strains are considered to be host specific (Fitzgerald, 2012), interest in livestock-associated S. aureus was renewed with the discovery of MRSA sequence type (ST) 398, initially in pigs and more recently in calves, chickens, horses, and pets (Armand-Lefevre et al., 2005; Graveland et al., 2011). ST398, as determined by multilocus sequence typing (MLST), is considered a newly emerging, pathogenic and zoonotic strain and the major cause of LA-MRSA in Europe and North America (Smith et al., 2009; Graveland et al., 2010), displaying significant diversity and high content of antimicrobial resistance genes, but so far, no significant virulence genes compared to other ST-lineages have been identified (Jamrozy et al., 2012). Current data indicates a low incidence of human to human transmission of this strain suggesting that the majority of infections are of a zoonotic origin (Wassenberg et al., 2011). Interestingly, it has been shown that ST398 can be either MRSA or methicillin-sensitive S. aureus (MSSA), both having been isolated from infections of a range of domestic animals including pigs, horses, chickens, and cows (Witte et al., 2007). Additionally, an increase in the incidence rate of clinical mastitis cases due to ST398 has been reported in recent years, adding to the zoonotic risk (Holmes and Zadoks, 2011). In this regard, it is also worth mentioning that infection of the udder by S. aureus does not seem to induce an appropriate innate immune response (Yang et al., 2008), potentially indicating some immune evasion strategies. Given these observations, the relative ease with which ST398 crosses the species-barrier and the lack of certain important S. aureus virulence factors such as panton-valentine leukocidin (PVL), toxic shock syndrome toxin-1 (TSST-1), and leukotoxin M (LukM; Vanderhaeghen et al., 2010) it is possible that ST398 may possesses additional and non-traditional virulence factors. One such group of virulence factors could be represented by Toll/IL-1 receptor (TIR)-domain containing proteins (Tcps), which have been identified in various bacteria (Spear et al., 2009). Whereas Tcps may have a normal function in protein–protein interaction (Spear et al., 2009), the presence of some Tcps in Gram-negative bacteria has been shown to shut-down TLR signaling (Spear et al., 2009). TLRs are vital components of the innate immune system, recognizing microbe-associated molecular patterns (MAMPs), leading to activation of transcription factors important for initiating the immune response (Akira et al., 2006). The intracellular TIR domain of TLRs is responsible for recruitment and activation of TIR-domain containing adaptor molecules such as myeloid differentiation factor 88 (MyD88), TIR-domain containing adaptor inducing interferon-β (TRIF), TIR-domain containing adaptor protein (TIRAP), and sterile-α and HEAT/Armadillo motifs-containing protein (SARM; McGettrick and O’Neill, 2004). Tcps share low sequence identity to their mammalian counterparts, and both are characterized by the presence of three boxes of conserved residues set in a core sequence ranging from 135 to 160 amino acids (Rana et al., 2011). In addition to Tcps, several additional protein classes have recently been identified that may interact and interfere with TLR signaling. A conserved region has been identified in transmembrane receptors including similar expression to fibroblast-growth factor (SEF) and IL17Rs in eukaryotes and bacteria. This region shows structural homology to TIR domains, the key functional subdomain used by the Toll/IL-1 family receptors. A similar domain is also found in Act1/CIKS, an essential adaptor downstream of IL-17R family members that is required for activation of NF-κB and other signals. Thus, the SEF/IL17R/CIKS/Act1 homology (SEFIR) domain is observed in both transmembrane and cytoplasmic proteins (Onishi et al., 2010). Sequence and structure-based fold prediction server results support the distant similarity of the SEFIR domain to the TIR structure, except for the most C-terminal region. SEFIR and TIR domains are related, belonging to a new STIR domain superfamily consisting of SEFIR, TIR and the recently identified TIR-like domain of unknown function (DUF) 1863 (Novatchkova et al., 2003). In Toll/IL-1R-like pathways, the cytoplasmically localized TIR domain of a receptor and the TIR domain of a soluble adaptor interact physically and activate signaling. The similarity between the SEFIR and TIR domains involves the above mentioned conserved boxes 1 and 2 of the TIR domain that are implicated in homotypic dimerization, but there is no sequence similarity between SEFIR domains and the TIR sequence box 3. By analogy, it was suggested that SEFIR-domain proteins function as signaling components of Toll/IL-1R-similar pathways and that their SEFIR domain mediates physical protein–protein interactions between pathway components (Wu et al., 2012).
Several studies have revealed that a diverse range of both pathogenic and non-pathogenic microorganisms express proteins containing STIR domains (Newman et al., 2006; Cirl et al., 2008; Spear et al., 2012). Despite low sequence identity, the crystal structure of Paracoccus denitrificans (PdTIR) is structurally similar to the TIR domain of human TLR1. PdTIR interacts with both MyD88 and the TLR4 TIR domains, however, no inhibitory function has been demonstrated (Low et al., 2007; Chan et al., 2009). In contrast, only Tcps identified in Gram-negative bacteria have been shown to inhibit MyD88-dependent TLR signaling. TlpA from Salmonella enterica inhibits activation of the transcription factor NF-κB (Newman et al., 2006). Similarly TcpB, present in a range of Brucella species including B. melitensis, B. ovis and B. abortus, and TcpC from E. coli inhibits NF-κB activation by direct interaction with MyD88, subsequently aiding bacterial macrophage entry and enhancing degradation of MAL (Radhakrishnan et al., 2009, 2011; Radhakrishnan and Splitter, 2010). In contrast, the Yersinia pestis TIR-domain protein (YpTdp) has been shown to interact with MyD88 and down-regulate activation of IL-1β; although the presence of this protein did not affect the phenotype of deletion mutants in a mouse infection model (Rana et al., 2011; Spear et al., 2012). There is less information available regarding SEFIR- (Wu et al., 2012) and DUF1863-domain containing proteins within the STIR superfamily.
The hypothesis of the current work was to assess whether proteins belonging to these families are in general present in Gram-positive bacteria, and specifically in ST398, which may explain the ease with that this specific S. aureus crosses into other species. Here we present data identifying two DUF1863-domain containing proteins, named S. aureus TIR-Like Proteins 1 and 2 (SaTlp1 and SaTlp2) in ST398 S. aureus, which affect TLR-dependent NF-κB activation in HEK293 and pbMECs. Initial experiments showed that a knock-out (KO) mutation of ST398 lacking both proteins seemed to enhanced survival of mice in an in vivo infection model using a defined colony forming unit (CFU), although the data did not reach a level of significance. These proteins may be important virulence factors and contribute to the ability of ST398 to cross species-barriers.
Materials and Methods
Cells Culture Conditions
HEK293 cells do not express TLR2 or TLR5 on their own, and do not respond to the appropriate TLR ligands (Willcocks et al., 2013; Metcalfe et al., 2014). HEK293 cells stably transfected with bovine TLR2 (GenBAnk Acc. No AY634629; HEK-boTLR2) were generated in house (Willcocks et al., 2013). HEK cells expressing human TLR5 were purchased from Invivogen (293-hTLR5), and have been used before (Metcalfe et al., 2014). Expression of TLRs was maintained using Geneticin (Gibco) at 1 mg ml-1 for boTLR2 and Blasticidin (Lifetech) at 10 μg ml-1 for 293-hTLR5. Both cell types were grown in DMEM (Gibco) supplemented with 10% fetal calf serum (PAA). The preparation of pbMECs as cells that have been shown to be affected by S. aureus expression, their culture in RPMI 1640 (Gibco) on collagen-coated plates, and general challenge conditions were previously described (Gunther et al., 2011).
Bacterial Strains, Growth Conditions, and MLST
S. aureus strains isolated from various species and disease presentations (Table 1), were grown on 5% Sheep blood agar (Oxoid) and cultured in brain heart infusion medium (Oxoid) at 37°C. Bacterial genomic DNA was isolated using a Wizard® Genomic DNA Purification Kit (Promega) following manufacturer’s guidelines. Polymerase chain reaction (PCR) was performed with Easy-A High-Fidelity PCR Mastermix (Agilent) using the primers stated on http://saureus.mlst.net/ (Aanensen and Spratt, 2005) for the genes arcC, aroE, glpF, gmk, pta, tpi, and tpi. Each primer pair amplified an internal fragment of the housekeeping gene and allowed accurate sequencing of ∼450-bp fragments of each gene on both strands. For each locus, the sequences obtained from all isolates were compared and the different sequences were assigned allele numbers. For each isolate, the alleles at each of the seven loci defined the allelic profile that corresponded to a ST.
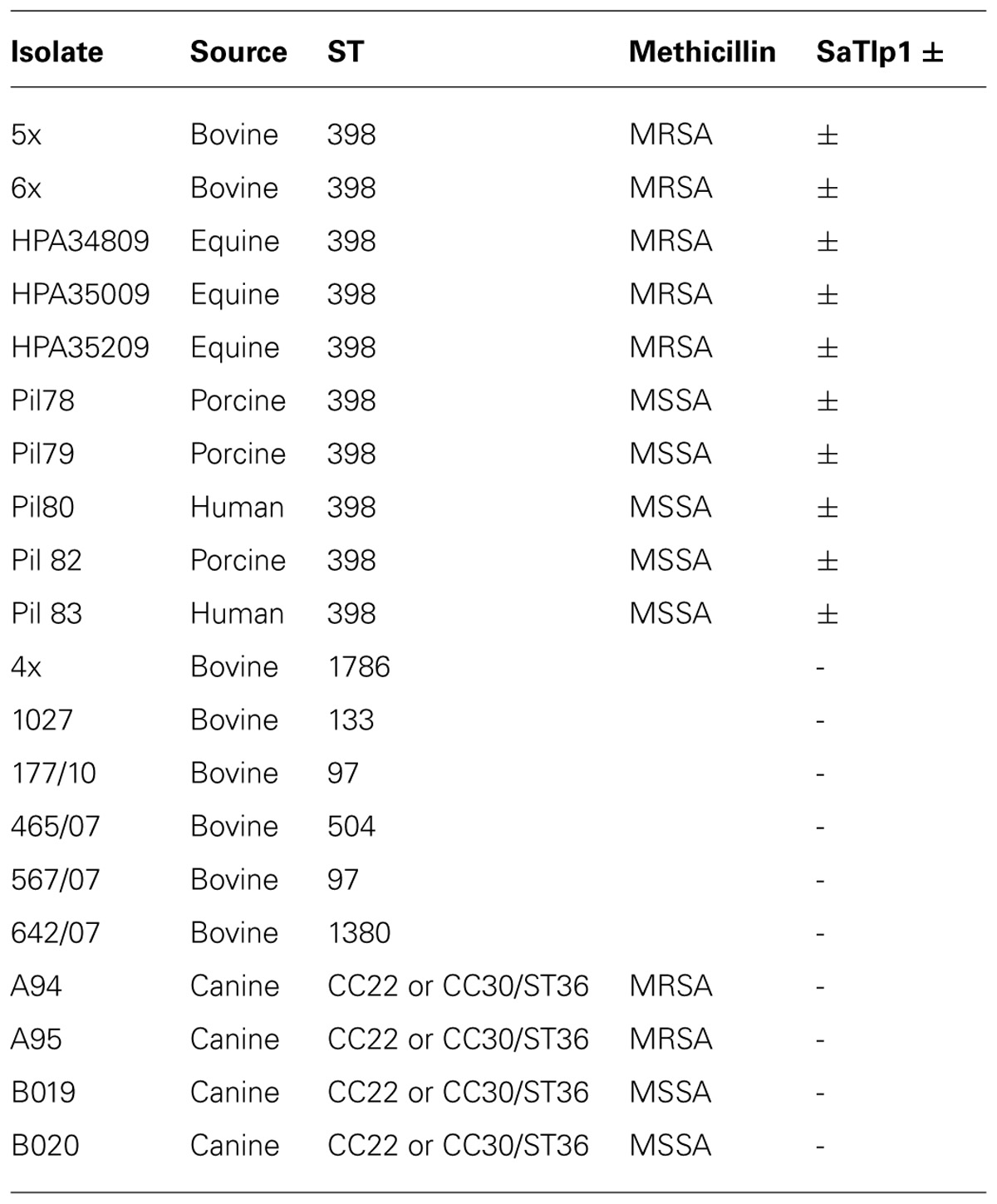
TABLE 1. Names, species isolated from, MLST group, methicillin-sensitivity (when relevant) and presence of SaTlp1 of the Staphylococcus aureus strains.
Identification and Cloning of Bacterial Tcps
Identification of Tcp-like proteins within S. aureus sequences were performed by searching for homologs of mammalian and bacterial STIR domains in the non-redundant protein sequences database1; using the PSI-BLAST algorithm (Altschul et al., 1997). Identified proteins were screened initially for structural similarities to known Tcp using SMART (Schultz et al., 19982) and the similarity of their tertiary structure compared to other proteins using the BioInfoBank Meta Server3. Subsequently, primers (Table 2) were used to amplify the sequences of SaTlp1 and SaTlp2 (GenBank Acc. No. CAQ50581.1 and CAQ50581.2, respectively) which also introduced a mutation to alter the start codons to ATG, allowing transcription in eukaryotic cells. PCR products were excised from 1% agarose gels using a GenElute Gel Extraction Kit (Sigma), cloned into pGEM-T Easy (Promega) and inserts sequenced. Subsequently, SaTlp1, SaTlp2, and TcpB were cloned into the pcDNA3.3 TOPO plasmid (Invitrogen). All plasmids were propagated in TOP10 E. coli (Invitrogen) bacteria, grown in Luria Broth supplemented with 100 μg ml-1 ampicillin (Invivogen), purified using a PureYield Plasmid Miniprep kit (Promega) and subsequently used for transfection into HEK-boTLR2, 293-hTLR5, or pbMEC cells.
TABLE 2. Primers used for cloning and subsequent screening for SaTlp1 and SaTlp2, and subsequent cloning into pcDNA3.3 and pIMAY.
Computational Modeling of SaTlp1
The secondary structure prediction and 3D template suggestions for structural SaTlp1, TcpB, and TLR2 were performed by HHpred (Soding et al., 2005) and used to generate a manually adjusted structural alignment. The PdTIR structure is comprised of four chains with differing structures which were manually aligned with an Arabidopsis thaliana TIR-like protein and human TIR domains from TLR2 (1FYW), TLR10 (1FYX), and MyD88 (2JS7) to the target sequences. The PDB structure 3LD8 was used to span the BB loop insertion which was not present in any of the TIR protein template sequences. MODELER version 9.10 (Fiser and Sali, 2003a) was used to generate 100 models and the poorly defined loop region (residues 43–49) of the highest ranking model refined by MODLOOP (Fiser and Sali, 2003b). Models were validated using PROCHECK (Morris et al., 1992), Verify3D (Eisenberg et al., 1997), and ProQ (Wallner et al., 2003). Sequence and structural similarity were analyzed by PDBeFold (Krissinel and Henrick, 2007).
NF-κB Luciferase Assay
The generation of HEK cells expressing TLR2, the main receptor for recognition of Gram-positive bacteria as well as the NF-κB reporter gene assay have been described recently (Willcocks et al., 2013). HEK cells expressing either boTLR2 or TLR5 (293-hTLR5; Invivogen) were seeded at a density of 2.5 × 105 and were transfected after 24 h with 250 ng NF-kB-Luc (Firefly luciferase gene under the control of NF-κB promoter; Promega) and 1250 ng pSaTlp1-3.3, pSaTlp2-3.3, or pTcpB-3.3, using TurboFectTM in vitro Transfection Reagent (Fermentas) or Lipofectamine 2000 (Invitrogen) following manufacturers’ guidelines and returned to the incubator for 24 h. A plasmid where TcpB had been ligated into pcDNA3.3 TOPO in the reverse orientation (pTcpB3.3INV) was used as a control. Cells were then split into six wells of a 24 well plate with three unstimulated wells and three wells of cells stimulated with either 100 ng ml-1 of the TLR2/6 ligand FSL-1 (Invivogen), 500 ng ml-1 of the TLR5 ligand flagellin (Invivogen) or 30 μg ml-1 heat-killed E. coli. (FBI Dummerstorf). After 24 h gene activation was analyzed using the Luciferase® Reporter Assay System (Promega) following manufacturer’s guidelines. The cell lysates were made using passive lysis buffer, centrifuged at 16,000×g for 5 min and the OD 280 nm measured using a ND-1000 spectrophotometer (Nanodrop) to allow for normalization, as previously described (Liu et al., 2011).
Assessment Cytokine Production by Quantitative PCR
To measure the effect of intracellular SaTlp1 or TcpB proteins on the production of inflammatory response genes in pbMEC, their mRNA concentration was determined for those cells transfected with the respective expression vector or the negative control plasmid. The pbMECs were grown to 80% confluence into 9 cm plates and were subsequently transfected with 1000 ng of the respective expression vector using Lipofectamine as described above. After an overnight recovery the transfected cells of each 9 cm plate were split into two wells of a six well plate (in order to obtain duplicates for the following inflammatory challenge). Subsequent to an additional recovery of 1 day the pbMEC were stimulated with 30 μg ml-1 heat-killed E. coli for three hours. After the stimulation total RNA was extracted using Trizol (Invitrogen) and RNA quality was determined as previously described (Gunther et al., 2011). For cDNA synthesis, 1.5 μg RNA was primed with a mixture of gene specific reverse oligonucleotide primers (Table 3) as described (Goldammer et al., 2004). After cDNA purification with HiPure columns (Roche) relative mRNA concentrations were determined with quantitative real-time PCR (RT-qPCR) using the LightCycler instrument and the SYBR green Plus kit (both from Roche), essentially as described previously (Goldammer et al., 2004; Gunther et al., 2011). Sequences of the oligonucleotide primers for the Chloride intracellular channel protein 1 (CLIC1; house-keeping gene), the proinflammatory cytokines TNF-α, IL-1β, IL-6, and CXCL8, inducible Nitric oxide synthase 2 (iNOS2), the bactericidal β-defensin lingual antimicrobial peptide (LAP) and the acute phase protein Serum amyloid A (SAA3) are given in Table 3.
Assessment of Cytokine Production by Elisa
To measure the effect of intracellular SaTlp1 or TcpB proteins on the production of inflammatory response genes in pbMEC, the corresponding supernatants from pbMEC used to generate mRNA were analyzed for the presence of CXCL8 and TNF using ELISA systems as described recently (Metcalfe et al., 2014). IL-1β was analyzed using a bovine IL-1β ELISA kit (Pierce Protein Biology Products) as per manufacture’s recommendation.
Generation of ΔSaTlp1/ΔSaTlp2 Mutant
In order to delete SaTlp1 and SaTlp2 from the ST398 genome the pIMAY plasmid and protocols were used as previously described (Monk et al., 2012). Deletion constructs were produced by amplifying regions 500-bp upstream and downstream of the genes using primer pairs A/B and C/D respectively. These PCR products were used as templates for the spliced overlap extension (SOE) PCR using A/D primers. The PCR product was purified and cleaved at endonuclease sites added by the A and D primers and cloned into pIMAY using standard protocols. Allelic exchange was achieved by transforming electrocompetent ST398 with the pIMAY construct and plating on brain heart infusion agar (BHIA) with chloramphenicol (Cm) at 28° C (Monk et al., 2012). Potential mutants were identified after antisense secY induction by colony PCR with OUT-F/OUT-R primers. The double mutant was confirmed by sequencing the deleted region from isolated genomic DNA.
Murine Infection Model
The murine peritoneal S. aureus infection is generally considered by be a good model to assess invasiveness and subsequently infectivity of S. aureus. The study was performed at the AAALAC-accredited Michigan State University In vivo Facility (East Lansing, MI, USA, study number 14IVF-IF-1003, IACUC approval No. 01/13-013-004). The study was conducted in accordance with the current guidelines for animal welfare (Guide for the Care and Use of Laboratory Animals, 8th Edn, 2011). The procedures used in this study were reviewed and approved by the Institutional Animal Care and Use Committee. Six- to eight-week-old female CD-1 mice (Charles River, Portage, MI, USA) with an average weight of 23.8 gm were randomly assigned into 13 groups (n = 6 per group). Animals were housed three per cage and provided food and water ad libitum. Animals were acclimated 8 days prior to use. One day prior to infection a parent culture of S. aureus ST398, and the KO strain ΔSaTlp1/ΔSaTlp2 were removed from frozen storage and streaked out onto trypticase soy agar supplemented with 5% lysed sheep blood cells (TSA-5% SB). Cultures were grown overnight at 37°C, at ambient atmosphere. On the day of infection, the cultures were removed from the incubator and used to prepare an inoculum of each organism in trypticase soy broth (TSB). Bacterial concentrations were measured in the broth by forward light scatter measurement at 600 nm (O.D.600) and adjusted to provide a target value of 0.600. A 1:100 dilution was prepared, using TSB supplemented with 10% gastric hog mucin (GHM) as the diluent, to generate a high dose concentration for challenge of 7.0 log10 bacteria in a dose volume of 0.5 mL. Serial 1:10 dilutions of the high concentration were made into TSB-10% GHM to prepare 6.0 log10, 5.0 log10, 4.0 log10, 3.0 log10, and 2.0 log10 challenge doses of each strain, and the inoculum concentration of each dosing preparation was confirmed by plating serial dilutions for CFU enumeration. Animals received mucin control, wild-type S. aureus, or KO S. aureus as a single intraperitoneal injections. Animals were monitored at least twice daily for signs of morbidity. During periods of expected high mortality, mice were monitored a minimum of four times per day. Animals exhibiting clinical signs listed as criteria for implementation of early euthanasia in AUF 01/13-013-00 were euthanized by CO2 asphyxiation, followed by cervical dislocation. Deaths were recorded as the number “died” or “euthanized” on a daily basis, and the study was terminated on Day 6. Any remaining animals were euthanized at that time.
Statistical Analysis
All in vitro experiments were performed at least three times, with samples run in at least duplicates. Data are presented as averages of all experiments, and results were assessed for statistical significance by a two-way ANOVA followed by a Bonferroni t-test between treatment groups using Excel and GraphPad Prism software packages (version 5; GraphPad Software, Inc., La Jolla, CA, USA).
For the in vivo experiment, data were collected on an Excel spreadsheet. Survival curves were compared statistically using Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxson tests (GraphPad Prism 5.03). Differences were considered significant at p < 0.05. LD50 curves were generated using GraphPad Prism 5.03. A sigmoidal Emax dose response model derived from the Hill equation (four-parameter logistic equation with variable slope using GraphPad Prism v5.03) was used to determine the relationship between the antibacterial drug dose and the efficacy, as determined by bacterial burden: Y = D +(A-D)/(1 +10(log C–X)*α). For the four-parameter logistic equation, Y is the observed effect, D is the bottom, A is the top, C is the LD50 or 50% of the observed maximum effect, X is the log of the challenge concentration and α is the Hill slope. The bottom value (D) was constrained to 0 and the top value (C) was left unconstrained. Data are shown as Kaplan–Meier Survival Curves.
Results
SaTlp1 AND SaTlp2 Share Structural Homology to Known TIR Domains
Initially we tried to identify Tcp in S. aureus using a bioinformatics approach. As Tcps in other bacteria have been shown to show similarities to mammalian TLR TIR domains, we compared different TLR TIR domains against all available S. aureus genomes (taxid:1280) using BLAST. This resulted in the identification of two immediately adjacent proteins (e = 8 × 10-6 and e = 3 × 10-5, respectively) containing a DUF1863 domain, located within a recently identified putative transposon. SMART annotation confirmed that these proteins, named S. aureus Tcp-like protein 1 and 2 (SaTlp1 and SaTlp2) contain DUF1863 domains. SaTlp1 and SaTlp2 share less than 20% sequence identity with other members of the TIR superfamily. However, despite low similarity at the sequence level, secondary structure prediction suggests that at least four of the central beta strands common to TIR-like domains are present in these bacterial proteins. The closest structural homologs identified by HHpred are matches to the DUF1863 (PF08937) and TIR or TIR_2 (PF13676 and PF01582) domain-containing families and proteins sharing a Flavodoxin-like fold (SCOP 31128 and 31130). The closest secondary structure match to both SaTlp1 and TcpB was PdTIR (PDB: 3H16) while the Eubacterium rectale putative signal transduction protein containing a Flavodoxin-like fold (PDB: 3HYN) was the top ranked hit for SaTlp2 with probabilities of 99.9, 100, and 99.7%, respectively. Within eukaryotic TIR domains, three conserved regions called boxes 1, 2, and 3 are proposed as putative adaptor binding sites, essential for signaling. The box 1 motif can be clearly identified in all of the TIR-like domains included in the alignment (Figure 1A). However, both SaTlp1 and SaTlp2 appear to have a helical insertion within the BB loop (box 2) and, unlike the other bacterial proteins shown, seem to entirely lack box 3. Computational modeling of SaTlp1 predicts the conserved TIR-like tertiary structure of four/five central beta strands surrounded by helices (Figure 1B). While the modeled structure of TcpB is most similar to PdTIR (∼0.5 Å), SaTlp1 shows closer structural homology to TLR TIR domains (∼1.9 Å).
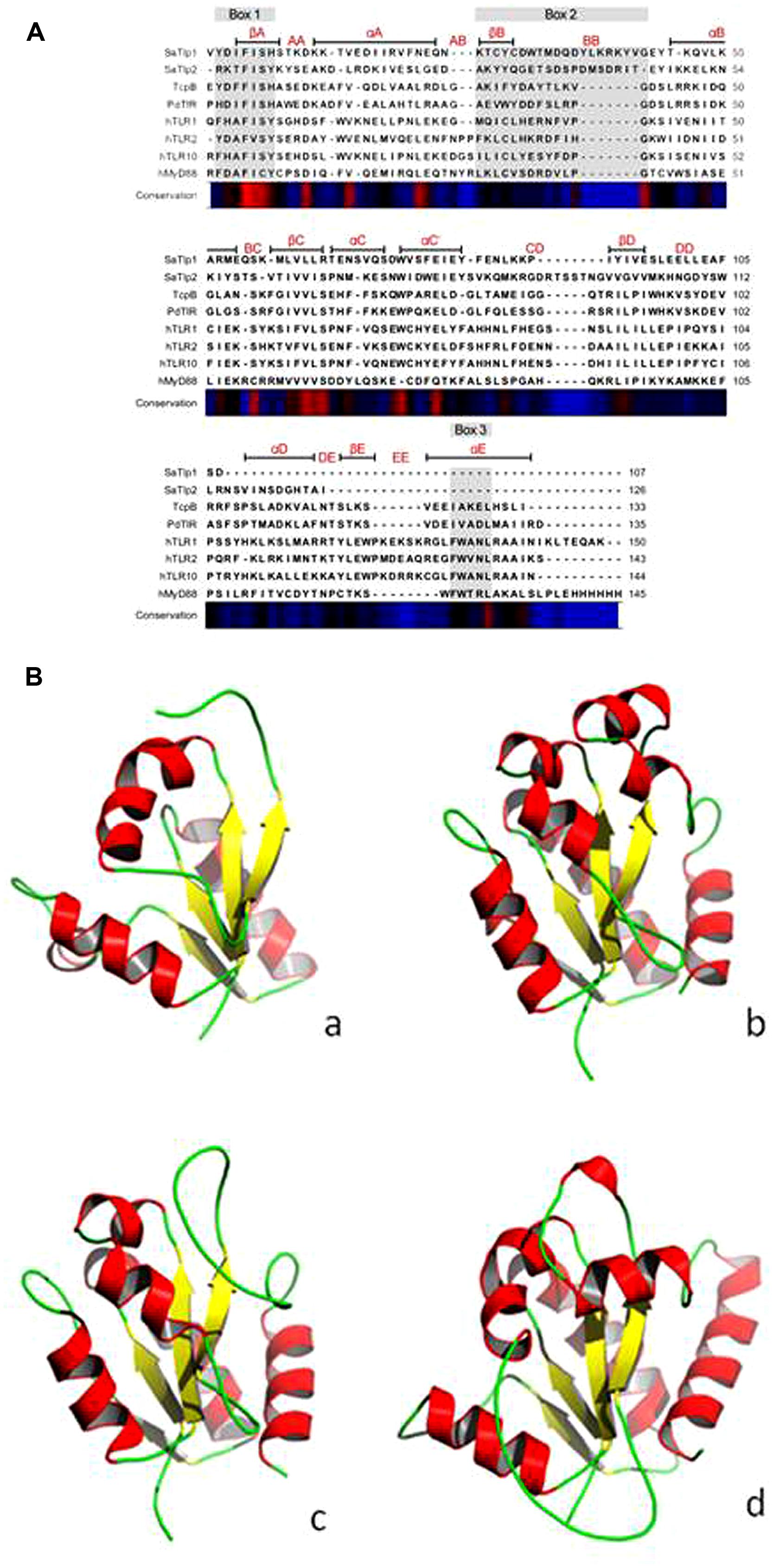
FIGURE 1. (A) Alignment of the DUF1863 domains of SaTlp1, SaTlp2, and TcpB with TIR domains of known structure. The regions corresponding to the TIR motifs boxes 1, 2, and 3 are indicated by the rectangles above the alignment, highlighted in gray. Secondary structure elements corresponding to the known proteins are annotated above with conservation from high (red) to low (blue) shown below. (B) Predicted tertiary structure of (a) SaTlp1, (b) TcpB and (d) boTLR2 and crystal structure of (c) PdTIR.
SaTlp1 and SaTlp2 are Present in All ST398 Isolates and CC75 Staphyloccoccus aureus
Having identified two putative Tcp-like proteins, we next assessed the occurrence of these genes in different S. aureus strains. To do so, a phylogenetic tree of all S. aureus strains used in this study was created by MLST analysis, and the presence of SaTlp1 and SaTlp2 in these strains determined (Table 1). Interestingly, SaTlp1 and SaTlp2 were identified in all ST398 isolates tested, regardless of the species of isolation, clinical symptoms of the host-species or methicillin-sensitivity status. Additionally, BLAST searches on 89 ST398 isolates were performed which revealed that all of these contained these genes. In addition to ST398, BLAST searches revealed these genes were present in S. aureus clonal complex (CC) 75 (also known as S. argenteus), however, they were not detected in any other S. aureus lineage. ST398 and CC75 are only distantly related (Figure 2) and published work on CC75 states that it is a highly divergent lineage, showing an average seven fold greater nucleotide divergence in orthologous genes with typical S. aureus strains than other strains (Holt et al., 2011). Considering this divergent relationship but the similarity of these genes it seems highly likely that the transposon containing them was transferred between the strains post-dating their divergence.
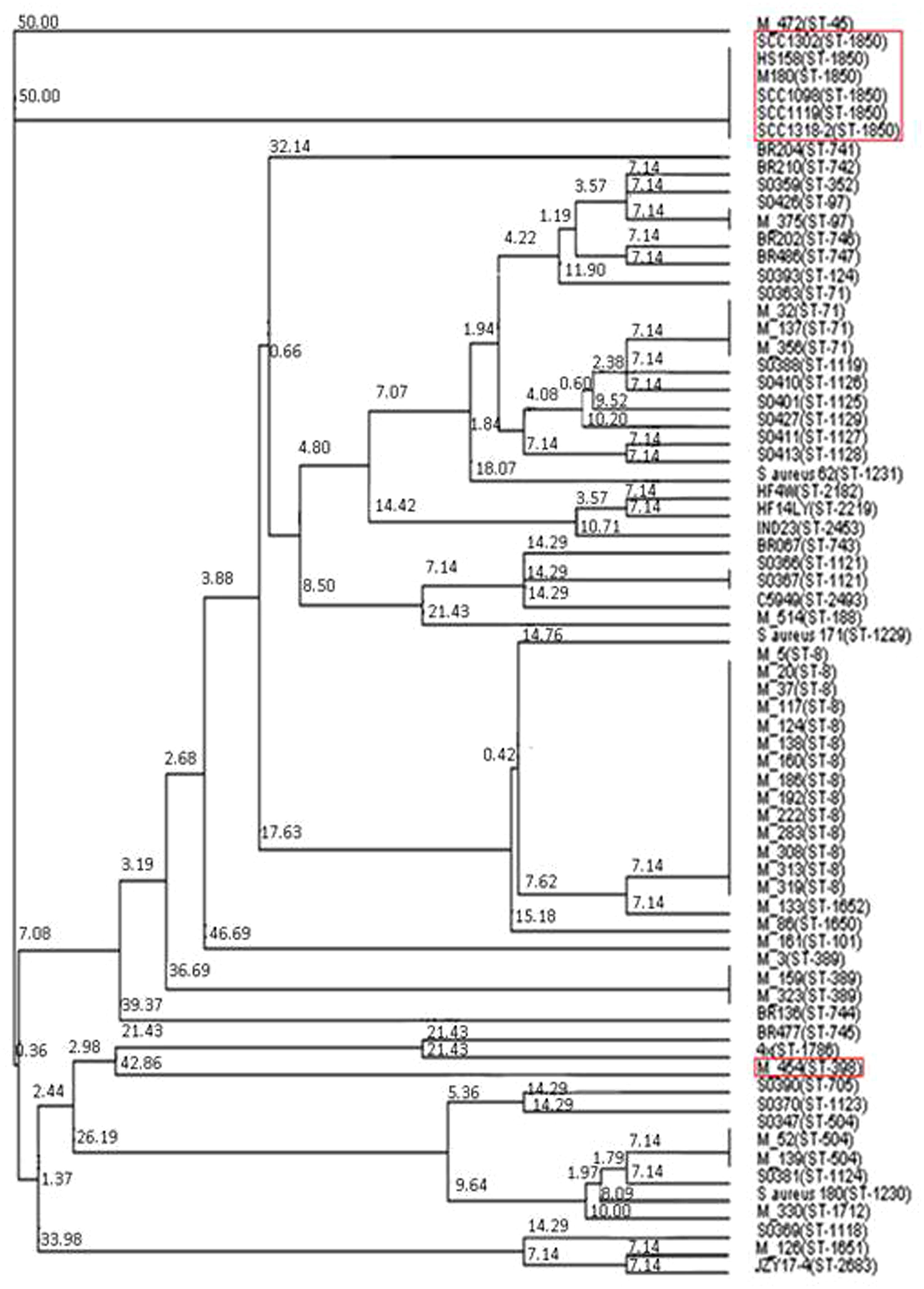
FIGURE 2. Neighbor joining tree of S. aureus isolates. Strains falling under “Bovine Mastitis” disease classification on MLST.net and ST398 and ST1850. Red boxes indicate STs in which SaTlp1 and SaTlp2 have been identified. The tree was computed by using concatenated nucleotide sequences of seven housekeeping genes and was constructed using the tool provided by the MLST database (http://saureus.mlst.net/sql/uniquetree.asp?). Bootstrap values are indicated above the branches.
Both SaTlp1 and SaTlp2 Up-Regulate NF-κB Activation in HEK-boTLR2 and pbMEC
To assess whether the presence of the Tcp-like proteins in this S. aureus strain interferes with the activation of the innate immune response, we tested whether the identified proteins would exhibit similar effects as described for Tcps of other bacteria which seem to inhibit TLR signaling. We analyzed whether transfection of HEK cells expressing boTLR2 with SaTlp1 and SaTlp2 affected the downstream activity of the transcription factor NF-κB after stimulation with the TLR2 ligand FSL-1 (Figure 3A). The levels of NF-κB-induced luciferase were significantly up-regulated following transfection with SaTlp1 and SaTlp2 plasmids (both p < 0.001) when compared to the negative control. Simultaneous transfection of both plasmids also up-regulated NF-κB activation (p < 0.001). In the HEK-boTLR2 system these plasmids up-regulated NF-κB-dependent luciferase between 4.1 (SaTlp1) and 6.1 (SaTlp1/2) fold following stimulation compared to the negative control. In contrast, transfection of HEK-boTLR2 cells with TcpB which had been shown before to block TLR-dependent NF-κB activation (Radhakrishnan et al., 2009) did not induce significant NF-κB stimulation, confirming these data. FSL-1 failed to activate NF-κB in HEK293 not expressing TLR2 (Data not shown).
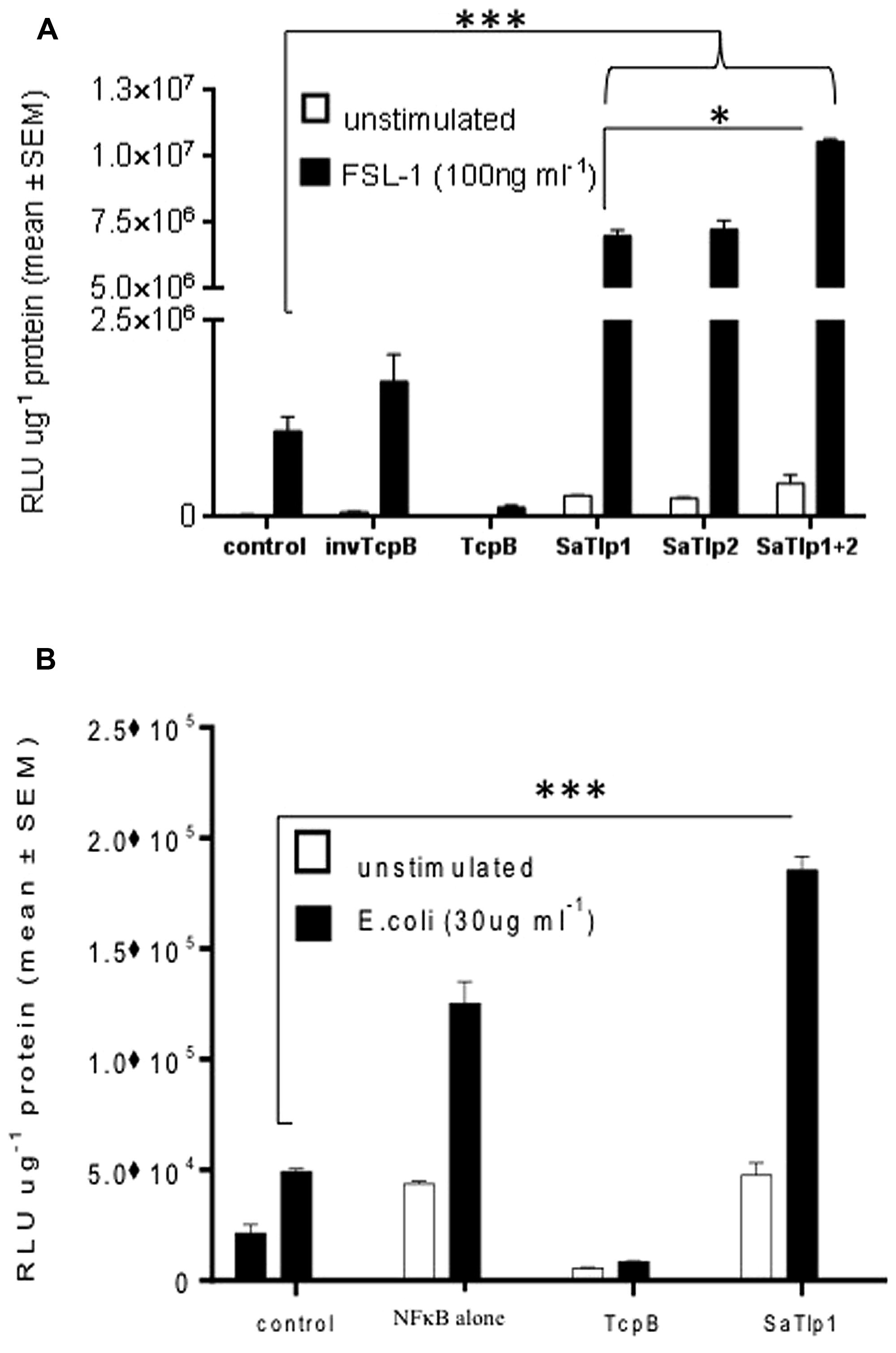
FIGURE 3. Impact of SaTlp1 and SaTlp2 on NF-κB activation in two different cell types. Cells were transfected as described, and relative luciferase units were analyzed. Graphs showing the RLU per μg protein in lysates from (A) HEK293-boTLR2 cells stimulated with 100 ng ml-1 FSL-1 and (B) primary bovine mammary epithelial cells (pbMECs) stimulated with 30 μg ml-1 heat-killed E. coli. Cells were transfected with plasmids indicated. Error bars represent the standard error of the mean for each condition (±SEM) of three independent repeats performed in triplicates. Control: medium alone; NFkB alone: cells transfected with NFkB coding plasmid only; invTcpB: cells transfected with a plasmid in which the TcpB sequence was inserted in reverse direct; SaTlp1: cels transfected with plasmid coding for SaTlp1; SaTlp2: cells transfected with plasmid coding for SaTlp2; SaTlp1+2: cells transfected with both plasmids. Significant fold changes are denoted by asterisks in the figure (*p < 0.05; ***p < 0.001).
As the HEK cell system is a somewhat artificial system, we next assessed the effects of SaTlp1 using pbMECs which express TLR2 and have been used before to assess the effect of mastitis causing pathogens on the innate immune response (Yang et al., 2008; Gunther et al., 2011). As seen in HEK293 cells, transfection of SaTlp1 (p = 0.006) caused increased NF-κB activation in pbMEC in response to heat-killed E. coli. As before transfection of pbMECs with TcpB before E. coli stimulation did not activate NF-κB signaling (p < 0.001). However, this up-regulation was overall lower compared to HEK293 cells with SaTlp1 up-regulating NF-κB by 1.4 fold compared to the negative control (Figure 3B).
The Effect of SaTlp1 and SaTlp2 is Not Limited to One Species or a Specific TLR
To assess whether the responses seen in both cell types would be unique to the interaction with boTLR2, we examined the effect of SaTlp1 and SaTlp2 in HEK cells transfected with hTLR5 cells stimulated with flagellin (Figure 4). Transfection of SaTlp1 SaTlp2 (both p < 0.001) and SaTlp1/2 (p < 0.001) led to an up-regulation of NF-κB activity of between 2.0 (SaTlp2) and 2.5 (SaTlp1) fold. In contrast, transfection of HEK cells expressing hTLR5 with TcpB down-regulated its activation in response to flagellin (p < 0.001). These results are therefore similar to those for boTLR2 expressing cells, suggesting the action of both proteins may not be limited to a specific TLR.
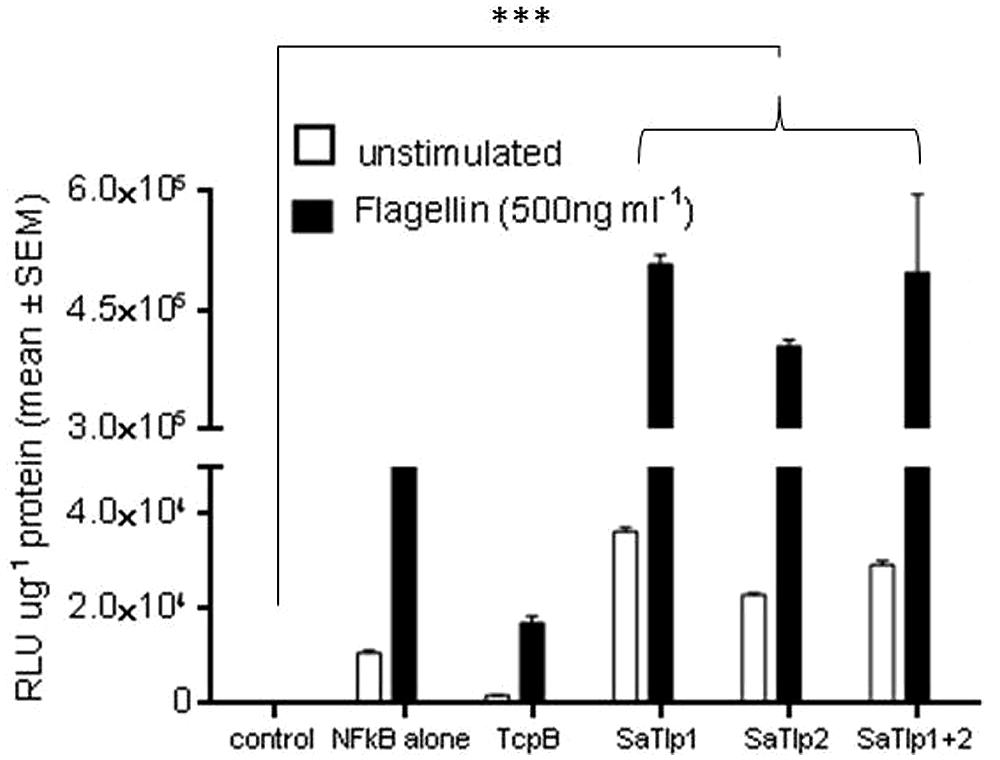
FIGURE 4. Impact of SaTlp1 and SaTlp2 on NF-κB activation in HEK293-hTLR5 cells. Graphs showing the RLU per μg protein in lysates of cells stimulated with stimulated with 500 ng ml-1 flagellin. Cells were transfected with plasmids indicated. Control: medium alone; NFkB alone: cells transfected with NFkB coding plasmid only; SaTlp1: cells transfected with plasmid coding for SaTlp1; SaTlp2: cells transfected with plasmid coding for SaTlp2; SaTlp1+2: cells transfected with both plasmids. Error bars represent the standard error of the mean for each condition (±SEM) of three independent repeats performed in triplicates. Significant fold changes are denoted by asterisks in the figure (***p < 0.001).
Up-Regulation of NF-κB Activation by SaTlp1 Increases mRNA and Protein Expression of Primary and Secondary Response Genes
Having established that SaTlp1 stimulated NF-κB activation, we investigated whether this was also reflected at the transcriptional level for NF-κB-dependent cytokine production. We analyzed expression of the primary response genes IL-1β, IL-6, CXCL8, and TNFα as well as the secondary response genes iNOS-2, LAP, and SAA3 by qPCR, similar to that described before (Gunther et al., 2011). Stimulation of pbMEC transfected with either empty plasmids or plasmids containing SaTlp1 followed by E. coli stimulation resulted in the up-regulation of IL-1β, IL-6, CXCL8, and TNF-α mRNA, levels to various extents (Figure 5). In contrast, transfection of pbMEC with a plasmid coding for TcpB in general reduced expression of these genes after E. coli stimulation (Figure 5). Transfection of pbMEC with plasmids coding for SaTlp1 and TcpB followed by subsequent stimulation with E. coli increased mRNA expression for the secondary response genes iNOS-2, LAP and SAA3 (Figure 6), with LAP expression being effected more by TcpB.
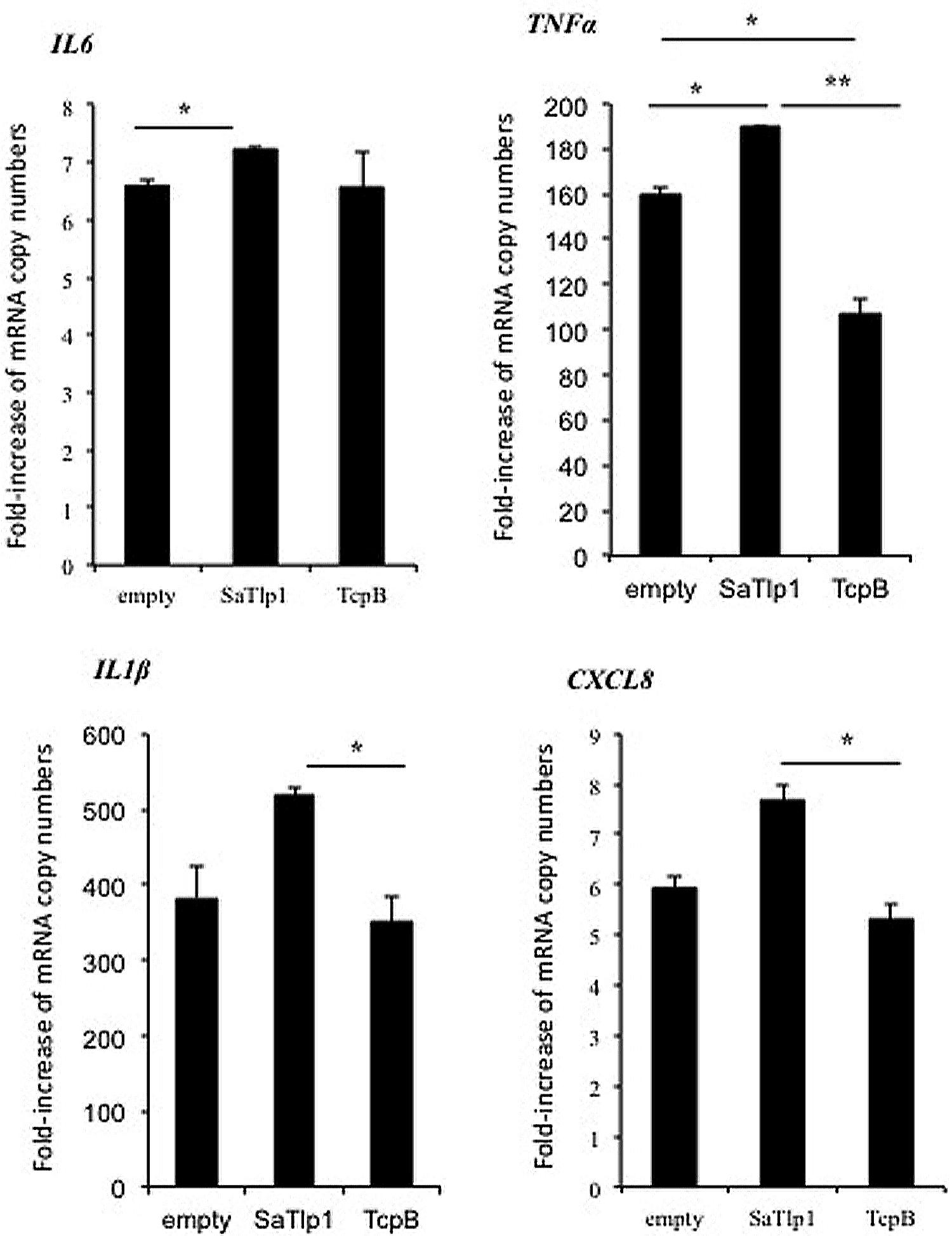
FIGURE 5. Average fold increase of mRNA levels of the pro-inflammatory genes IL-1β, IL-6, TNF, CXCL8 in pbMEC transfected with the empty plasmid, or plasmids coding for TcpB and SaTlp1 and stimulated with 30 μg ml–1 heat-killed E. coli for three hours. Error bars represent the standard error of the mean for each condition (±SEM) of three independent repeats performed in triplicates. Significant fold changes are denoted by asterisks in the figure (*p < 0.05; **p < 0.01).
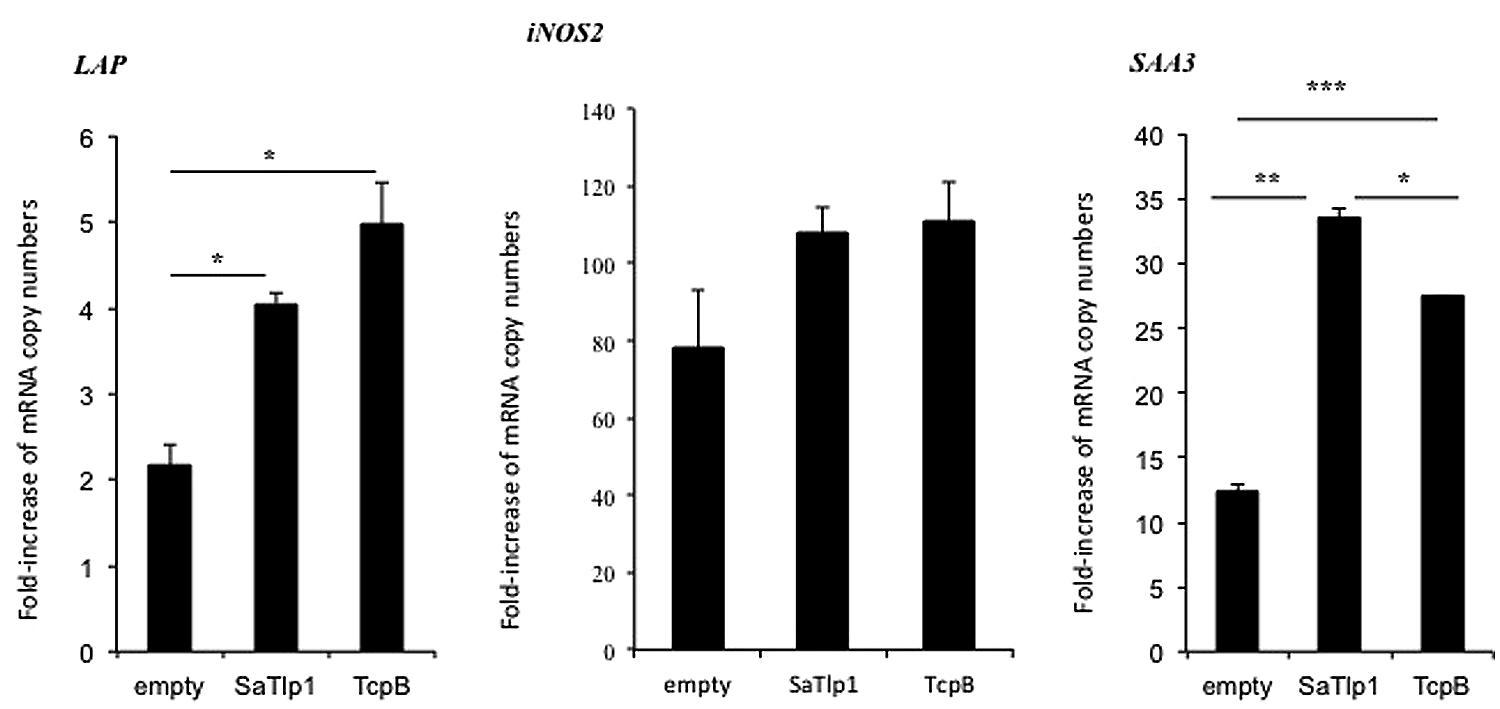
FIGURE 6. Average fold increase of mRNA levels of the secondary response genes iNOS-2 LAP and SAA3 in pbMEC transfected with the empty plasmid or plasmids coding for TcpB and SaTlp1 and stimulated with 30 μg ml–1 heat-killed E. coli for three hours. Error bars represent the standard error of the mean for each condition (±SEM) of three independent repeats performed in triplicates. Significant fold changes are denoted by asterisks in the figure (*p < 0.05; **p < 0.01; ***p < 0.001).
Corresponding supernatants were analyzed for the presence of CXCL8, TNF-α, and IL-1β. The secretion pattern followed in general the results obtained for qPCR, with values obtained for all three cytokines being in generally low. Similar as to the data obtained by qPCR, supernatants of pbMEC transfected with SaTlp1 before stimulation with E. coli contained the largest amount of all three cytokines (Figures 7A–C), whereas no differences were seen between untransfected pbMEC and pbMEC transfected with TcpB.
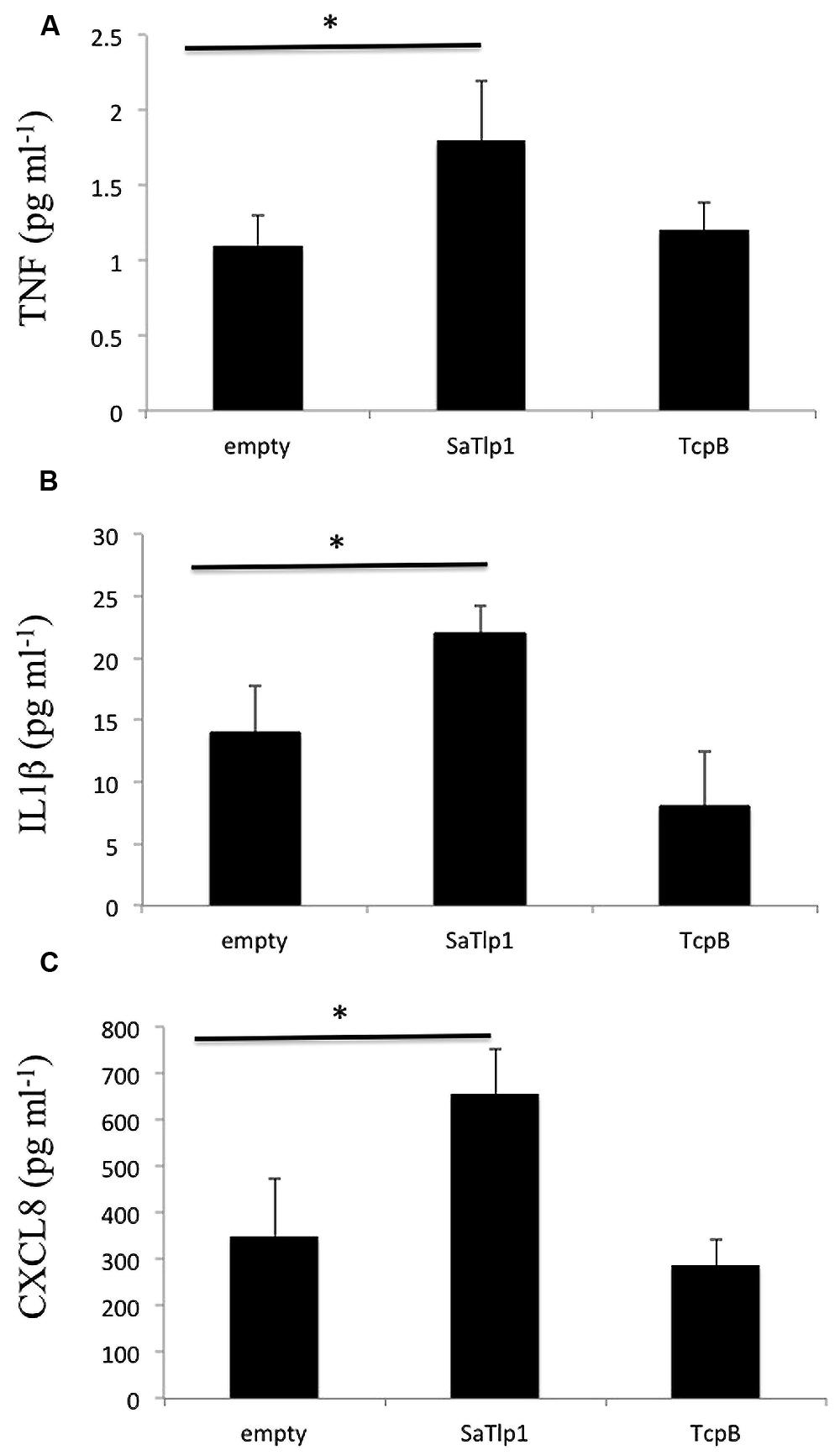
FIGURE 7. Values of the pro-inflammatory TNF (A), IL-1β (B), and CXCL8 (C) analyzed in the supernatants of pbMEC transfected with the empty plasmid, or plasmids coding for TcpB and SaTlp1 and stimulated with 30 μg ml–1 heat-killed E. coli for three hours. Error bars represent the standard error of the mean for each condition (±SEM) of three independent repeats performed in triplicates. Significant fold changes are denoted by asterisks in the figure (*p < 0.05).
A ΔsaTlp1/ΔsaTlp2 ST398 Potentially Impacts On Survival Rate in a Staphylococcus aureus Mice Challenge Model
As the presence of SaTlp1 and SaTlp2 seemed to increase NF-κB activation and pro-inflammatory cytokine production in response to subsequent stimulation, we next assessed whether the absence of the genes has an impact on the virulence of S. aureus ST398 in vivo. After creation of a ΔsaTlp1/ΔsaTlp2 ST398 KO mutant, the growth rate or colony appearance of both wild-type and mutant S. aureus were compared and showed no difference (data not shown). In order to determine whether there are differences in the in vivo pathogenicity of wild-type and ΔsaTlp1/ΔsaTlp2 ST398 a murine sepsis model was used in order to calculate the median lethal dose (MLD) of the bacteria. CD-1 mice were challenged via i.p. injection with 102–107 CFU of either wild-type or mutant S. aureus after which the time until their death was monitored. The data showed a trend for increased survival rates for mice infected with the ΔsaTlp1/ΔsaTlp2 mutant, most noticeably in the case of infection with 105 CFU when 2 of 6 mice challenged with mutant bacteria survived until day 6 of the experiment whereas all mice infected with wild-type had died by day 2 (Figure 8). Despite the fact that none of the observed differences reach level of significance, the lowest p-value (p = 0.13) was obtained for 105 CFU. Considering the data from all challenge concentrations, the MLD of the strains was calculated using the Reed-Meunch equation. This showed the MLD of ΔsaTlp1/ΔsaTlp2 ST398 to be 3630 CFU compared to one of 575 for the wild-type ST398 indicating that in order for 50% of challenged mice to be killed by the mutant, an infectious dose of over 6.3 times that of wild-type ST398 is required. This suggests that although differences between wild-type and ΔsaTlp1/ΔsaTlp2 ST398 are not significant at individual concentrations, when is considered together there does appear to be genuine differences in their pathogenicity.
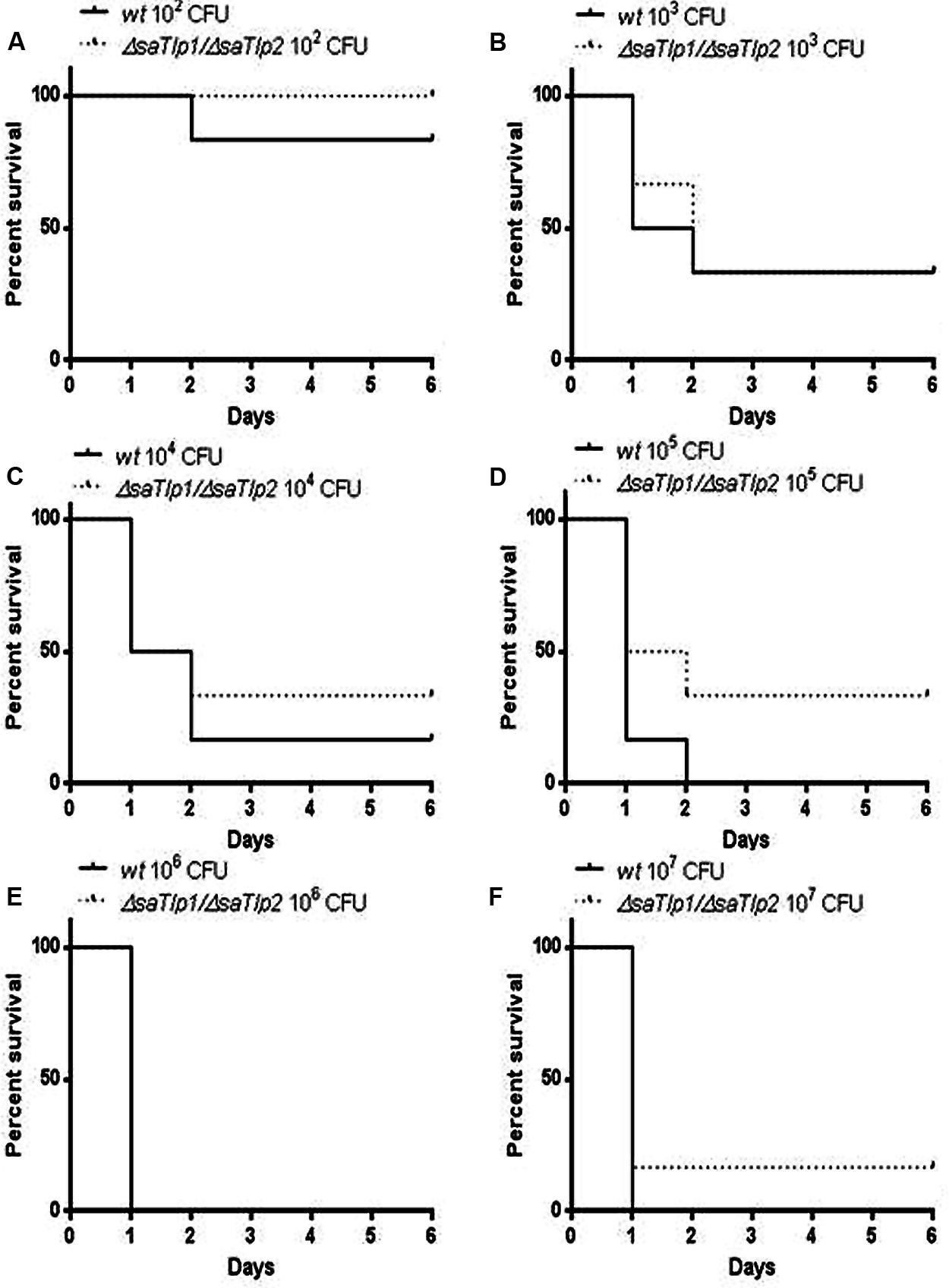
FIGURE 8. Kaplan–Meier survival curves for groups of mice infected with different CFU of either wild-type or ΔsaTlp1/ΔsaTlp2 ST398 S. aureus. Each figure represent six mice challenged by intra-peritoneal injection with 0.5 mL containing wild-type ST398 and six mice challenged with ΔsaTlp1/ΔsaTlp2 KO mutant which were then monitored for 6 days and deaths recorded. Figures show survival rates for mice challenged with (A) 102 CFU (B) 103 CFU (C) 104 CFU (D) 105 CFU (E) 106 CFU and (F) 107 CFU.
Discussion
Staphylococci have evolved numerous mechanisms to evade immune recognition (Foster, 2005). In the present study, we describe the identification of two DUF1863 domain-containing proteins in the emerging zoonotic S. aureus ST398 which show structural similarity to TLR TIR domains. As described for other proteins of the STIR-superfamily, both proteins show only a weak sequence homology, with the mentioned boxes 1 and 2 being identifiable but not box 3. Furthermore our data suggests that expression of these proteins results in significantly increased activity of the transcription factor NF-κB in HEK-boTLR2, HEK-hTLR5, and pbMEC cells, as well as mRNA expression levels of pro-inflammatory primary and secondary response genes. However, in contrast to work published on the function of Tcps identified in Gram-negative bacteria, which were shown to down-regulate NF-κB activation as well as subverting the ensuing innate immune response (Radhakrishnan et al., 2009; Yadav et al., 2010; Rana et al., 2011), our data seems to suggest that these proteins activate NF-κB signaling and the production of inflammatory mediators. Although there is still a certain amount of debate as to whether TIR, SEFIR, and DUF1863 domains should be classified separately or as a single domain, our data suggests that they may have distinct functions within the same signaling pathways.
In addition to the differences seen in activation of NF-κB, this study is the first to investigate these presence of such proteins from Gram-positive bacteria, as all published work on bacterial Tcps has been in Gram-negative bacteria such and B. melitensis (TcpB), E. coli (TcpC), and P. denitrificans (PdTIR). It is unclear whether NF-κB down-regulation is associated with Gram-negative and up-regulation with Gram-positive bacteria or if there are examples of up- and down-regulation for Tcp present in both. Further to this it must be considered that TIR domains are most likely to have more general protein/protein interaction functions, as reviewed recently (Spear et al., 2009), and modulation of TLR signaling by these proteins may not be the rule.
Although the identified proteins are likely to be ubiquitous within ST398, they appear to be rare within the S. aureus species as a whole. A recently published genome, however, showed that they are present in a very distantly related, early branching S. aureus strain CC75, isolated in Northern Australia, also known a S. argenteus (Holt et al., 2011). There is conflicting evidence as to whether the genetic element containing these genes is ancestral or acquired and further study would determine if there are any notable pathogenic similarities between these two strains, particularly in the context of ST398. Strains of this group are notable for their high promiscuity, with the ability to easily cross species-barriers, having been isolated from a range of livestock animals and also humans. Furthermore ST398 S. aureus are often missing several traditional staphylococcal virulence factors such as PVL, suggesting that these bacteria may have acquired alternative factors that allow for their successful host colonization. It could be hypothesized that these Tlps may represent novel staphylococcal virulence factors, which may be unique to this specific MLST. Indeed, and in contrast to our data, the interference of S. aureus with MyD88-dependent TLR signaling was recently suggested (Gunther et al., 2011), with the authors demonstrating an inhibitory effect of S. aureus on TNF-α and IL-1 mRNA production. The fact that our data show that the identified proteins are influencing the pro-inflammatory response system on both, mRNA and protein level indicates that both proteins are potentially interesting virulence factors that warrant further investigation. This is of even more interest as not only SaTlp1, but also TcpB from B. melitiensis seemed to affect mRNA expression levels of the secondary response genes LAP and SAA3. These require de novo synthesis and have been shown to need chromatin-remodeling (Liu et al., 2011) or the transcription factor C/EBPδ (Litvak et al., 2009), and were differently expressed compared to the pro-inflammatory genes.
We are fully aware that the in vivo model used may not be the most appropriate one to assess the function of the two identified S. aureus proteins. However, murine models for S. aureus are still considered the appropriate standard for invasiveness/survival (Kim et al., 2014; Zhang et al., 2014). The in vivo infection data from a murine sepsis model provided data which suggests a trend toward increased survival time for mice infected with the ΔsaTlp1/ΔsaTlp2 mutant compared to the wild-type ST398 strain although differences between these were not statistically significant at any individual S. aureus concentration. The level where the greatest differences are apparent was when mice were infected with 105 CFU. This is with a p-value of 0.1292 according to the Log-rank test which, although not significant, would certainly merit further investigation considering the relatively small sample size at each CFU concentration (n = 6). It is possible that at CFU concentrations lower than 105 there may be less effect as the mice may be more able to fight the infection regardless, whereas at higher concentrations the effects of these proteins may be negated due to the sheer number of bacteria present. Around the 105 mark may represent something of a sweet spot where effect of these proteins can be seen. Additionally the small sample size is clearly susceptible to natural variation between mice and challenge doses. This is especially important as while large differences in survival of infected mice were not observed there may be more subtle in vivo effects of these proteins similar to those proposed for YpTlp (Spear et al., 2012). Subtle in vivo effects would very likely require relatively large sample sizes in order to be able to produce statistically significant results. The exact mechanisms used by these proteins are yet to be elucidated, as a direct interaction with host proteins has not been identified. It should also be noted that there is still no clear picture as to how these bacterial proteins would reach any cellular target, as they do not contain domains associated with export from the bacterium. However, these findings are similar to TcpB of Brucella. Despite this, TcpB exhibits lipid-binding properties (Radhakrishnan and Splitter, 2010), and interacts with phosphoinositides through its N-terminal domain, leading to co-localization with the plasma membrane and components of the cytoskeleton (Radhakrishnan et al., 2009). Furthermore this Brucella TIR protein has the property of cell permeability which would provide a mechanism for virulence and could mean that it influences nearby cells not infected with the bacteria, as well as contributing to trafficking within the cell to influence host cell machinery.
In conclusion, SaTlp1 and SaTlp2 are potential novel virulence factors in ST398 S. aureus which appear to interact with the innate immune signaling machinery of host cells in novel ways. The presence of these proteins may account for some of the unusual characteristics of strains of S. aureus within ST398 and are therefore interesting targets for understanding these characteristics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank W. Petzl and H. Zerbe (Centre of Clinical Veterinary Medicine of the Ludwig-Maximilians-University Munich) and E. Statis (RVC) for providing the different Staphylococci strains, and G. Splitter for TcpB and control plasmids. The authors declare no conflict of interest. This project was supported by a BBSRC iCASE grant (BB/G018073/1) with Zoetis Animal Health to Dirk Werling. This manuscript represents publication No PID_00483 of the RVC.
Footnotes
- ^http://blast.ncbi.nlm.nih.gov
- ^http://smart.embl-heidelberg.de/
- ^http://bioinfo.pl/
- ^http://www.aaalac.org/accreditedorgsdirectorysearch/aaalacprgms.cfm
References
Aanensen, D. M., and Spratt, B. G. (2005). The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 33, W728–W733. doi: 10.1093/nar/gki415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W.,et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Armand-Lefevre, L., Ruimy, R., and Andremont, A. (2005). Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11, 711–714. doi: 10.3201/eid1105.040866
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chan, S. L., Low, L. Y., Hsu, S., Li, S., Liu, T., Santelli, E.,et al. (2009). Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J. Biol. Chem. 284, 21386–21392. doi: 10.1074/jbc.C109.007591
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cirl, C., Wieser, A., Yadav, M., Duerr, S., Schubert, S., Fischer, H.,et al. (2008). Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14, 399–406. doi: 10.1038/nm1734
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenberg, D., Luthy, R., and Bowie, J. U. (1997). VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277, 396–404. doi: 10.1016/S0076-6879(97)77022-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fiser, A., and Sali, A. (2003a). Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491. doi: 10.1016/S0076-6879(03)74020-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fiser, A., and Sali, A. (2003b). ModLoop: automated modeling of loops in protein structures. Bioinformatics 19, 2500–2501. doi: 10.1093/bioinformatics/btg362
Fitzgerald, J. R. (2012). Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20, 192–198. doi: 10.1016/j.tim.2012.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Foster, T. J. (2005). Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958. doi: 10.1038/nrmicro1289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goldammer, T., Zerbe, H., Molenaar, A., Schuberth, H. J., Brunner, R. M., Kata, S. R.,et al. (2004). Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin. Diagn. Lab. Immunol. 11, 174–185.
Graveland, H., Duim, B., Van Duijkeren, E., Heederik, D., and Wagenaar, J. A. (2011). Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 301, 630–634. doi: 10.1016/j.ijmm.2011.09.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Graveland, H., Wagenaar, J. A., Heesterbeek, H., Mevius, D., Van Duijkeren, E., and Heederik, D. (2010). Methicillin resistant Staphylococcus aureus ST398 in veal calf farming: human MRSA carriage related with animal antimicrobial usage and farm hygiene. PLoS ONE 5:e10990. doi: 10.1371/journal.pone.0010990
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gunther, J., Esch, K., Poschadel, N., Petzl, W., Zerbe, H., Mitterhuemer, S.,et al. (2011). Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect. Immun. 79, 695–707. doi: 10.1128/IAI.01071-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Holmes, M. A., and Zadoks, R. N. (2011). Methicillin resistant S. aureus in human and bovine mastitis. J. Mammary Gland Biol. Neoplasia 16, 373–382. doi: 10.1007/s10911-011-9237-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Holt, D. C., Holden, M. T., Tong, S. Y., Castillo-Ramirez, S., Clarke, L., Quail, M. A.,et al. (2011). A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 3, 881–895. doi: 10.1093/gbe/evr078
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jamrozy, D. M., Fielder, M. D., Butaye, P., and Coldham, N. G. (2012). Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS ONE 7:e40458. doi: 10.1371/journal.pone.0040458
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, H. K., Missiakas, D., and Schneewind, O. (2014). Mouse models for infectious diseases caused by Staphylococcus aureus. J. Immunol. Methods 410, 88–99. doi: 10.1016/j.jim.2014.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krissinel, E., and Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. doi: 10.1016/j.jmb.2007.05.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lindsay, J. A. (2010). Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 98–103. doi: 10.1016/j.ijmm.2009.08.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Litvak, V., Ramsey, S. A., Rust, A. G., Zak, D. E., Kennedy, K. A., Lampano, A. E.,et al. (2009). Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat. Immunol. 10, 437–443. doi: 10.1038/ni.1721
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, S., Shi, X., Bauer, I., Gunther, J., and Seyfert, H. M. (2011). Lingual antimicrobial peptide and IL-8 expression are oppositely regulated by the antagonistic effects of NF-kappaB p65 and C/EBPbeta in mammary epithelial cells. Mol. Immunol. 48, 895–908. doi: 10.1016/j.molimm.2010.12.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Low, L. Y., Mukasa, T., Reed, J. C., and Pascual, J. (2007). Characterization of a TIR-like protein from Paracoccus denitrificans. Biochem. Biophys. Res. Commun. 356, 481–486. doi: 10.1016/j.bbrc.2007.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McGettrick, A. F., and O’Neill, L. A. (2004). The expanding family of MyD88-like adaptors in Toll-like receptor signal transduction. Mol. Immunol. 41, 577–582. doi: 10.1016/j.molimm.2004.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Metcalfe, H. J., La Ragione, R. M., Smith, D. G., and Werling, D. (2014). Functional characterisation of bovine TLR5 indicates species-specific recognition of flagellin. Vet. Immunol. Immunopathol. 157, 197–205. doi: 10.1016/j.vetimm.2013.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monk, I. R., Shah, I. M., Xu, M., Tan, M.-W., and Foster, T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3:e00277–11. doi: 10.1128/mBio.00277-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morris, A. L., Macarthur, M. W., Hutchinson, E. G., and Thornton, J. M. (1992). Stereochemical quality of protein structure coordinates. Proteins 12, 345–364. doi: 10.1002/prot.340120407
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Newman, R. M., Salunkhe, P., Godzik, A., and Reed, J. C. (2006). Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immunol. 74, 594–601. doi: 10.1128/IAI.74.1.594-601.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Novatchkova, M., Leibbrandt, A., Werzowa, J., Neubuser, A., and Eisenhaber, F. (2003). The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 28, 226–229. doi: 10.1016/S0968-0004(03)00067-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Onishi, R. M., Park, S. J., Hanel, W., Ho, A. W., Maitra, A., and Gaffen, S. L. (2010). SEF/IL-17R (SEFIR) is not enough: an extended SEFIR domain is required for il-17RA-mediated signal transduction. J. Biol. Chem. 285, 32751–32759. doi: 10.1074/jbc.M110.121418
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Petzl, W., Zerbe, H., Gunther, J., Yang, W., Seyfert, H. M., Nurnberg, G.,et al. (2008). Escherichia coli, but not Staphylococcus aureus triggers an early increased expression of factors contributing to the innate immune defense in the udder of the cow. Vet. Res. 39, 18. doi: 10.1051/vetres:2007057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radhakrishnan, G. K., Harms, J. S., and Splitter, G. A. (2011). Modulation of microtubule dynamics by a TIR domain protein from the intracellular pathogen Brucella melitensis. Biochem. J. 439, 79–83. doi: 10.1042/BJ20110577
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radhakrishnan, G. K., and Splitter, G. A. (2010). Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem. Biophys. Res. Commun. 397, 59–63. doi: 10.1016/j.bbrc.2010.05.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radhakrishnan, G. K., Yu, Q., Harms, J. S., and Splitter, G. A. (2009). Brucella TIR domain-containing protein mimics properties of the Toll-like receptor adaptor protein TIRAP. J. Biol. Chem. 284, 9892–9898. doi: 10.1074/jbc.M805458200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rana, R. R., Simpson, P., Zhang, M., Jennions, M., Ukegbu, C., Spear, A. M.,et al. (2011). Yersinia pestis TIR-domain protein forms dimers that interact with the human adaptor protein MyD88. Microb. Pathog. 51, 89–95. doi: 10.1016/j.micpath.2011.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Smith, T. C., Male, M. J., Harper, A. L., Kroeger, J. S., Tinkler, G. P., Moritz, E. D.,et al. (2009). Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS ONE 4:e4258. doi: 10.1371/journal.pone.0004258
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soding, J., Biegert, A., and Lupas, A. N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248. doi: 10.1093/nar/gki408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spear, A. M., Loman, N. J., Atkins, H. S., and Pallen, M. J. (2009). Microbial TIR domains: not necessarily agents of subversion? Trends Microbiol. 17, 393–398. doi: 10.1016/j.tim.2009.06.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spear, A. M., Rana, R. R., Jenner, D. C., Flick-Smith, H. C., Oyston, P. C., Simpson, P.,et al. (2012). A Toll/interleukin (IL)-1 receptor domain protein from Yersinia pestis interacts with mammalian IL-1/Toll-like receptor pathways but does not play a central role in the virulence of Y. pestis in a mouse model of bubonic plague. Microbiology 158, 1593–1606. doi: 10.1099/mic.0.055012-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vanderhaeghen, W., Hermans, K., Haesebrouck, F., and Butaye, P. (2010). Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 138, 606–625. doi: 10.1017/S0950268809991567
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wallner, B., Fang, H., and Elofsson, A. (2003). Automatic consensus-based fold recognition using Pcons, ProQ, and Pmodeller. Proteins 53(Suppl. 6), 534–541. doi: 10.1002/prot.10536
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wassenberg, M. W., Bootsma, M. C., Troelstra, A., Kluytmans, J. A., and Bonten, M. J. (2011). Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin. Microbiol. Infect. 17, 316–319. doi: 10.1111/j.1469-0691.2010.03260.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Willcocks, S., Offord, V., Seyfert, H. M., Coffey, T. J., and Werling, D. (2013). Species-specific PAMP recognition by TLR2 and evidence for species-restricted interaction with Dectin-1. J. Leukoc. Biol. 94, 449–458. doi: 10.1189/jlb.0812390
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Witte, W., Strommenger, B., Stanek, C., and Cuny, C. (2007). Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13, 255–258. doi: 10.3201/eid1302.060924
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, B., Gong, J., Liu, L., Li, T., Wei, T., and Bai, Z. (2012). Evolution of prokaryotic homologues of the eukaryotic SEFIR protein domain. Gene 492, 160–166. doi: 10.1016/j.gene.2011.10.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yadav, M., Zhang, J., Fischer, H., Huang, W., Lutay, N., Cirl, C.,et al. (2010). Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS Pathog. 6:e1001120. doi: 10.1371/journal.ppat.1001120
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, W., Zerbe, H., Petzl, W., Brunner, R. M., Gunther, J., Draing, C.,et al. (2008). Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-kappaB in mammary epithelial cells and to quickly induce TNFalpha and interleukin-8 (CXCL8) expression in the udder. Mol. Immunol. 45, 1385–1397. doi: 10.1016/j.molimm.2007.09.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, B., Hua, Y., Yu, B., Lau, C. C., Cai, J., Zheng, S.,et al. (2014). Recombinant ESAT-6-like proteins provoke protective immune responses against invasive Staphylococcus aureus disease in a murine model. Infect. Immun. doi: 10.1128/IAI.02498-14 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: TLR signaling, Staphylococcus aureus, bacterial proteins, TCP, innate immune response, mouse model
Citation: Patterson NJ, Günther J, Gibson AJ, Offord V, Coffey TJ, Splitter G, Monk I, Seyfert H-M and Werling D (2014) Two TIR-like domain containing proteins in a newly emerging zoonotic Staphylococcus aureus strain sequence type 398 are potential virulence factors by impacting on the host innate immune response. Front. Microbiol. 5:662. doi: 10.3389/fmicb.2014.00662
Received: 23 June 2014; Accepted: 14 November 2014;
Published online: 09 December 2014.
Edited by:
Hao Shen, University of Pennsylvania, USAReviewed by:
Lauren A. Zenewicz, The University of Oklahoma, USAHans-Joachim Schuberth, University of Veterinary Medicine Hanover, Germany
Copyright © 2014 Patterson, Günther, Gibson, Offord, Coffey, Splitter, Monk, Seyfert and Werling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dirk Werling, Molecular Immunology Group, Department of Pathology and Pathogen Biology, Royal Veterinary College, Hawkshead Lane, Hatfield, AL9 7TA, UK e-mail: dwerling@rvc.ac.uk
 Nicholas J. Patterson
Nicholas J. Patterson Juliane Günther
Juliane Günther Amanda J. Gibson
Amanda J. Gibson Victoria Offord1
Victoria Offord1 Tracey J. Coffey
Tracey J. Coffey Gary Splitter
Gary Splitter Ian Monk
Ian Monk Hans-Martin Seyfert
Hans-Martin Seyfert Dirk Werling
Dirk Werling