Comparing miRNAs and viroids; highly conserved molecular mechanisms for the transmission of genetic information
- 1LSU Neuroscience Center, Louisiana State University Health Sciences Center, New Orleans, LA, USA
- 2Department of Microbiology, Louisiana State University Health Sciences Center, New Orleans, LA, USA
- 3Department of Ophthalmology, Louisiana State University Health Sciences Center, New Orleans, LA, USA
- 4Department of Pharmacology, Louisiana State University Health Sciences Center, New Orleans, LA, USA
- 5Department of Neurology, Louisiana State University Health Sciences Center, New Orleans, LA, USA
Introduction
All forms of life on earth, from the most ancient Archaebacteria and Lycopodiaceae to higher eukaryotes, display phenotypes based on genetic information transfer contained within, and regulated by, hierarchic orders of RNA. While DNA is capable of storing large amounts of genetic information over long periods of time, RNA-based signaling represents a transient, highly selective, ancient, and still evolving information transfer mechanism. The information-storage capability of RNA, its solubility and the mobility of small non-coding RNA (ncRNA) species, ability to self-replicate, to catalyze chemical reactions, to direct the amino acid polymerization into peptides, and to variably regulate the expression of highly selective genetic information underscores RNA as an important, and perhaps pre-eminent, driver in the evolution of genetic complexity and information transfer (Crick, 1968; Orgel, 1968; Ambros, 2004; Sempere et al., 2004; Lukiw, 2007; Mehler and Mattick, 2007; Lukiw et al., 2008, 2012; Bartel, 2009; Kosik, 2009; Cech, 2012). This opinion paper will highlight some intriguing similarities in evolutionary ancient, molecular, and mechanistic features shared by small, non-protein coding single-stranded RNA (ssRNA) entities known as microRNAs (miRNAs), and infective, self-replicating circular ssRNA plant pathogens known as viroids. We will further highlight the intrinsic potential of miRNA and viroids to induce pathogenetic signaling in widely diverse species.
microRNAs (miRNAs)
micro RNAs (miRNAs) are 18–25 nucleotide, conserved, non-coding ssRNAs that represent the smallest known carriers of highly selective genetic regulatory information in plants and animals. Mature miRNAs are typically derived from larger, double-stranded pre-miRNAs transcribed by RNA polymerase II (RNA Pol II) and processed by at least 2 RNA cleavage steps in which the activities of the RNAses Drosha and Dicer are involved, resulting in the generation of mature miRNA species (Ambros, 2004; Lukiw, 2007; Bartel, 2009; Lukiw et al., 2012). While our perceptions on the mechanism and relevance of miRNA signaling continues to evolve, it is now generally accepted that the primary mode of miRNA action is to recognize and bind to complementary RNA sequences in the 3′ un-translated region (3′-UTR) of target messenger RNAs (mRNAs) and in doing so, down-regulate their expression (Lukiw et al., 2008; Guo et al., 2010; Witkos et al., 2011; Lukiw, 2012a). While miRNAs are generally considered to be vitally essential, post-transcriptional regulators of gene expression, it is not often appreciated that these ssRNAs: (i) are very highly selected in their ribonucleotide sequence; (ii) exhibit remarkable cell and tissue specificity; (iii) represent a nucleotide signaling system that is evolutionarily ancient; (iv) are the smallest yet identified ribonucleic acid carriers of specific genetic regulatory information; (v) possess interesting viroid-like properties (see below); (vi) are the most abundant extracellular nucleic acids contained in human circulatory fluids including the cerebrospinal fluid (CSF) and blood serum; and (vii) as such may spread genetic information and gene signaling information, both homeostatic and pathogenic, amongst neighboring cells, tissues and perhaps between individual species (Arteaga-Vazquez et al., 2006; Lukiw, 2007, 2013; Yuva-Aydemir et al., 2011; Zhao et al., 2013). Rudimentary ribonucleic acid sequence analysis indicates that a “typical” 22 nucleotide ssRNA composed of the 4 different ribonucleotides (A,G,C,U) could have over 1013 possible sequence combinations. However, the fact that there typically only about 2 × 103 different miRNAs so far identified, suggests an extremely high evolutionary selection pressure to utilize only specific ribonucleotide sequences in miRNA that will yield biologically useful miRNA–mRNA interactions (Orgel, 1968; Lukiw, 2007, 2013; Bartel, 2009). Diverse investigative methods that include miRNA array-, RNA-sequencing-, Northern dot blot-, and RT-PCR-based analysis suggest that abundant miRNAs in specialized cells such as human brain cells probably number less than ~50, and only a significantly smaller fraction of these are utilized in pathological signaling of, for example, neurological disorders (Burmistrova et al., 2007; Lukiw and Pogue, 2007; Lukiw, 2012a). Interestingly, the abundance, speciation, and complexity of these highly selected miRNAs may vary amongst different human populations in health and disease (Lukiw, 2012b, 2013). Similar to messenger RNA (mRNA), miRNAs appear to follow the same stability rules involving adenine-uridine (AU) rich elements (AREs) in their primary sequence; a higher content of AREs in miRNAs are generally correlated with short miRNA half-lives, and absence of AU or UA dinucleotides may confer miRNA stability (Chen and Shyu, 1995; Cui et al., 2005; Sethi and Lukiw, 2009). Interestingly, while mammalian brain and retinal miRNAs in particular may have in general a relatively short half-life, miRNA half-lives may be considerably extended by miRNA-binding proteins, extensive miRNA secondary and tertiary structures, circularization or interaction with RNA circles, containment within vesicles, or by combinations of these and other factors (Krol et al., 2010; Boon and Vickers, 2013; Hansen et al., 2013; Kosaka et al., 2013; Memczak et al., 2013; Perkel, 2013). In double stranded miRNA precursors, virtually all of the miRNA sequence base-pairs with complementary sequences in other parts of the same molecule to form a highly structured pre-miRNA considerably more resistant to degradation than ssRNA alone (Figure 1).
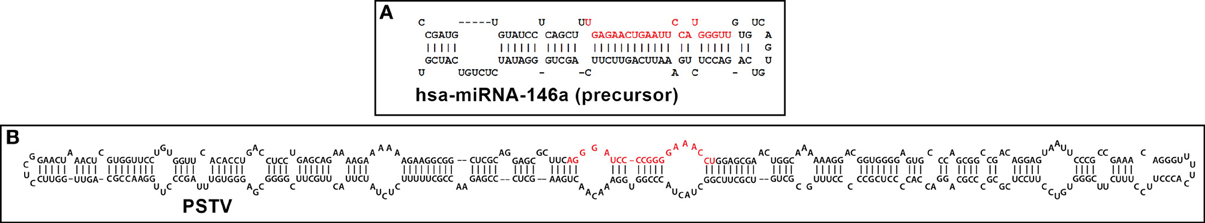
Figure 1. Typical representative structures of (A) a 99 nucleotide “hairpin”-shaped precursor to a 22 nucleotide homo sapien microRNA 146a (hsa-miRNA-146a) and (B) a 359 nucleotide circular potato spindle tuber viroid (PSTV) indicate extensive intra-strand base pairing and formation of doubled stranded RNA (dsRNA), imperfect base-pairing resulting in “bulges” and stem-loop structures (Krol and Krzyzosiak, 2006; Ritchie et al., 2007; Ding, 2009; Triboulet and Gregory, 2010). In both cases these RNA precursors are further processed by an RNase III of the family of Dicer-like proteins to generate smaller ssRNA species 18–25 nucleotides in length; these “mature” miRNA and viroid specific RNA (vsRNA) sequences are highlighted in red; note that these sizes are similar to endogenous small interfering RNA (as miRNA or vsRNA) and thus might alter the normal cultivar- and viroid-dependent gene expression in the host plant by viroids, or of mRNA in other plant and animal species, including humans (Arteaga-Vazquez et al., 2006; Krol and Krzyzosiak, 2006; Ritchie et al., 2007; Lukiw et al., 2008; Ding, 2009; Triboulet and Gregory, 2010; Hammann and Steger, 2012; Navarro et al., 2012). PSTV is the smallest known pathogen of all living species; it is interesting that both the plant and animal kingdoms have adopted similar ssRNA strategies to convey only the most essential genetic regulatory information in miRNA or vsRNA translocation and/or signal propagation, some of which may convey, in whole or part, pathological effects. Human miRNA-146a and PSTV ssRNA sequences and/or precursor structures were derived from GenBank accession NR_029701.1; GI:262205399 (http://www.ncbi.nlm.nih.gov/nuccore/NR_029701.1); miRBase (http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000477); or M36163.1; GI:333356 (http://www.ncbi.nlm.nih.gov/nuccore/M36163.1); secondary structures were predicted using online web servers such as http://mfold.rna.albany.edu/?q=mfold.
The conservation amongst specific miRNAs and ribonucleotide sequence preservation is remarkable. Using RNA-sequencing and novel genome-wide computational approaches to detect miRNAs based on nucleotide sequence structure and alignment, the miRNA-854 family has been shown to be expressed in Arabidopsis thaliana, Caenorhabditis elegans, Mus musculus, and Homo sapiens (Arteaga-Vazquez et al., 2006; unpublished observations). In these widely diverse species (Arabidopsis—Homo sapiens divergence about 1.5 billion years) miRNA-854 commonly targets an oligouridylate binding protein 1b mRNA 3′-UTR that normally encodes a member of a heterogeneous nuclear RNA binding protein family, suggesting a common origin of miRNA-854 as an ancient regulator of basal eukaryotic transcription (Wang et al., 1999; Hedges, 2002; Arteaga-Vazquez et al., 2006; Taft et al., 2010; unpublished). Moreover, secondary and/or tertiary structures may also be conserved between miRNAs, and internal stems, loops and mis-paired “bulges” commonly appear in specific positions in many pre-miRNA sequences (Figure 1) (Saetrom et al., 2006). Unique ssRNA sequences, some of which are conserved across both plant and animal species, further define and regulate the expression a relatively discrete subset of mRNAs with which they may interact, thus defining a complex, interwoven miRNA-mRNA regulatory network (Orgel, 1968; Lukiw et al., 1992; Ambros, 2004; Taganov et al., 2006; Bartel, 2009; Lukiw, 2012a,c). ssRNAs presumably also carry a signal encoded within the sequence of their RNA which may be transmitted via a unique molecular shape, molecular topology, and/or RNA charge density across the entire molecule. Intriguingly miRNAs enriched in certain foods may pass horizontally between species, carrying genetic regulatory information well outside of the cells in which they were initially generated (Alexandrov et al., 2012; Sarkies and Miska, 2013; Zhang et al., 2013).
Viroids
The smallest known ssRNA viruses are retroviruses, such as Rous Sarcoma virus (RSV), with a 3.5 × 104 nucleotide genome (Adams and Carstens, 2012; http://www.ictvonline.org/). Smaller than any known ssRNA viruses are minimalist plant pathogens known as viroids. It remains debatable whether viroids are a biological oddity, an evolutionary fossil of pre-cellular evolution, and/or a highly novel and evolving, self-perpetuating pathogenic entity (Diener, 1991, 2003; Rocheleau and Pelchat, 2006; Ding, 2009; Diermann et al., 2010; Sano et al., 2010; Wang and Ding, 2010; Navarro et al., 2012). The host range of many viroids is currently expanding, essentially as a result of a fast and continuing evolution of sequences/structures that gain new biological functions (Diener, 1991, 2003; Ding, 2009; Navarro et al., 2012). Comprising a small family of about 35 different, non-coding, un-encapsulated, autonomously infectious circular ssRNA plant pathogens ranging in size from 246 to 401 nucleotides, viroids possess the highest in vivo mutation rate among all known nucleic acids (Ding, 2009). Viroids are not only of evolutionary, virological and biological interest but are also of agricultural and economic concern since viroid infections compromise the yield of several important food crops worldwide (Diener, 1991, 2003; Sano et al., 2010; Wang and Ding, 2010). The first discovered viroid was the 359 nucleotide potato spindle tuber viroid (PSTV) of the viroid family Pospiviroidae, a circular ssRNA which primarily causes an infectious disease of potato plants (chiefly Solanum tuberosum) (Figure 1) (Ding, 2009; Adams and Carstens, 2012; Hammann and Steger, 2012). Viroids are transcribed using a unidirectional rolling-circle mechanism in the infected plant nuclei (for the family Pospiviroidae; involving stunting diseases of potato, apple, and coconut) or the chloroplast (for the viroid family Avsunviroidae; involving stunting diseases of avocado, eggplant, and peach). After replication, viroid precursors exit the nucleus or chloroplast via an Exportin-5 or related transport mechanism (in a fashion similar to miRNA translocation); viroids then traffic from cell to cell to establish systemic infection. Recent findings indicate that like miRNAs, viroid infection and pathogenic activity is associated with the appearance of a small viroid-specific ssRNA (vsRNA), 21–24 nucleotides in size, processed by an RNaseIII of the family of Dicer-like proteins from a double stranded RNA (dsRNA) viroid precursor (Figure 1) (Diener, 2003; Ding, 2009; Adams and Carstens, 2012; Hammann and Steger, 2012). Mature vsRNA is therefore highly analogous to miRNA in their mechanism of generation and translocation, and both have potential to not only alter the normal gene expression patterns in the host to cause disease, but also to spread these pathogenic signals to other cells and tissues via diffusion or through circulating fluids (Diener, 2003; Diermann et al., 2010; Sano et al., 2010; Wang and Ding, 2010; Adams and Carstens, 2012; Hammann and Steger, 2012). Indeed vsRNAs have sizes, structures and actions strikingly similar to both naturally occurring endogenous miRNAs, and each are known to have considerable potential to transiently or permanently alter the ssRNA-dependent gene expression patterns in the host (Diener, 2003; Ritchie et al., 2007; Ding, 2009; Hammann and Steger, 2012).
miRNAs and Viroids; Extensive Similarity in Structure, Processing, and Biological Function
That 21–24 nucleotide viroid-generated vsRNA are sufficient to induce diseases in plants is highly reminiscent of the finding that miRNAs, through analogous genetic mechanisms, have capabilities to contribute to disease processes in both plant and animal species (Diener, 1991, 2003; Lukiw and Pogue, 2007; Ding, 2009; Pogue et al., 2010; Hammann and Steger, 2012). Very much like miRNAs, pre-viroid ssRNAs and vsRNAs encode no proteins, have no naturally protective protein coat, do not reverse-transcribe into DNA when they replicate, and are significantly inducible by external stress or environmental factors (Diener, 2003; Ritchie et al., 2007; Adams and Carstens, 2012). Naked, infectious viroids are initially generated as a precursor dsRNA (pre-viroid) structure from which a mature ssRNA is excised by RNAse III-type nuclease activities (see Figure 1) (Ritchie et al., 2007; Lukiw, 2012b; Navarro et al., 2012; Perkel, 2013). As far as is known, miRNAs do not replicate in vivo; in fact they are probably physically too small to do so, and in in vitro experiments usually require nucleotide-linkers to effectively copy miRNAs using RT-PCR-based systems. Interestingly, circularization of pre-viroids, pri-miRNAs or miRNAs and/or the formation of complex higher order secondary and/or tertiary structures may further stabilize them (Rocheleau and Pelchat, 2006; Sethi and Lukiw, 2009; Perkel, 2013). The potential transmission and spreading of miRNA and viroid information-carrying signals from cell to cell, tissue to tissue and perhaps between species has a tremendous bearing on our understanding on the complex genetic interactions between diverse forms of life in the plant and animal kingdoms, and their symbiotic exchanges of biological information in natural environments (Hedges, 2002; Arteaga-Vazquez et al., 2006; Hammann and Steger, 2012; Sarkies and Miska, 2013).
Concluding Remarks
Intriguing similarities between the structure and function of miRNAs and viroids underscore the idea that during evolution, once nature has found and tested a successful molecular design and “information transfer mechanism” it is highly preserved, and this design is used repeatedly in diverse biological applications across diverse species. These biological applications range from the post-transcriptional modulation of gene expression to the potential for disease induction in both the plant and animal kingdom. Such conservation appears to lie in the molecular-genetic mechanism of miRNA and viroid ribonucleotide sequence structure and ensuing complementarity-based ssRNA-mRNA interaction. Intriguingly, ssRNA miRNA and viroid ribonucleotide sequences contain fingerprints for conservation across many diverse species, and these fingerprints represent some of the most highly conserved nucleic acid sequences known (Triboulet and Gregory, 2010; Witkos et al., 2011; Figure 1). It has recently been shown that infection of human brain cells with a high phenotypic re-activator strain of the double-stranded linear DNA herpes simplex-1 (HSV-1) induces, and then utilizes a host-specific miRNA-146a to maintain and support HSV-1 invasiveness to propagate successful infection (Figure 1; Hill et al., 2009; Lukiw et al., 2010; Ball et al., 2013). It will be interesting to ascertain if other RNA—or DNA-based “helper viruses” promote or intensify miRNA or viroid re-activities, if miRNA effects can be moderated by vsRNA or other ssRNA or dsRNA, if miRNA and viroid activities are equally affected by the presence of circular RNA (circRNA), if miRNAs are able to interact with helper viruses to modulate or amplify miRNA effects, and what potential roles other environmental factors might play in miRNA—or vsRNA-mediated gene activity and pathogenicity (Krol and Krzyzosiak, 2006; Lukiw, 2012a; Hansen et al., 2013; Memczak et al., 2013).
Acknowledgments
The work in this paper was presented in part at the Alzheimer Association International Conference 2013 (AAIC 2013) held in Boston MA 13–18 July 2013. Sincere thanks are extended to S. Bhattacharjee and Y. Zhao for miRNA array work and initial data interpretation, and to D Guillot and AI Pogue for expert technical assistance. Research on miRNA in the Lukiw laboratory involving the miRNA signaling in the innate-immune response was supported by a COBRE III pilot grant, INBRE-NIH 5P20RR016456-11, NIA AG038834 and NEI EY006311.
References
Adams, M. J., and Carstens, E. B. (2012). Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch. Virol. 157, 1411–1422. doi: 10.1007/s00705-012-1299-6
Alexandrov, P. N., Dua, P., Hill, J. M., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2012). microRNA (miRNA) speciation in Alzheimer's disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int. J. Biochem. Mol. Biol. 3, 365–373.
Arteaga-Vazquez, M., Caballero-Pérez, J., and Vielle-Calzada, J. P. (2006). A family of miRNAs present in plants and animals. Plant Cell 18, 3355–3369. doi: 10.1105/tpc.106.044420
Ball, M. J., Lukiw, W. J., Kammerman, E. M., and Hill, J. M. (2013). Intracerebral propagation of Alzheimer's disease: strengthening evidence of a herpes simplex virus etiology. Alzheimers Dement. 9, 169–175. doi: 10.1016/j.jalz.2012.07.005
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Boon, R. A., and Vickers, K. C. (2013). Intercellular transport of microRNAs. Arterioscler. Thromb. Vasc. Biol. 33, 186–192. doi: 10.1161/ATVBAHA.112.300139
Burmistrova, O. A., Goltsov, A. Y., Abramova, L. I., Kaleda, V. G., Orlova, V. A., and Rogaev, E. I. (2007). MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11). Biochemistry (Mosc.) 72, 578–582. doi: 10.1134/S0006297907050161
Cech, T. R. (2012). The RNA worlds in context. Cold Spring Harb. Perspect. Biol. 4:a006742. doi: 10.1101/cshperspect.a006742
Chen, C. Y., and Shyu, A. B. (1995). AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470. doi: 10.1016/S0968-0004(00)89102-1
Crick, F. H. (1968). “The origin of the genetic code.” J. Mol. Biol. 38, 367–379. doi: 10.1016/0022-2836(68)90392-6
Cui, J. G., Zhao, Y., and Lukiw, W. J. (2005). Isolation of high spectral quality RNA using run-on gene transcription; application to gene expression profiling of human brain. Cell. Mol. Neurobiol. 25, 789–794. doi: 10.1007/s10571-005-4035-x
Diener, T. O. (1991). Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J. 5, 2808–2813.
Diener, T. O. (2003). Discovering viroids—a personal perspective. Nat. Rev. Microbiol. 1, 75–80. doi: 10.1038/nrmicro736
Diermann, N., Matoušek, J., Junge, M., Riesner, D., and Steger, G. (2010). Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 391, 1379–1390. doi: 10.1515/BC.2010.148
Ding, B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47, 105–131. doi: 10.1146/annurev-phyto-080508-081927
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Hammann, C., and Steger, G. (2012). Viroid-specific small RNA in plant disease. RNA Biol. 9, 809–819. doi: 10.4161/rna.19810
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hedges, S. B. (2002). The origin and evolution of model organisms. Nat. Rev. Genet. 3, 838–849. doi: 10.1038/nrg929
Hill, J. M., Zhao, Y., Clement, C., Neumann, D. M., and Lukiw, W. J. (2009). HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport 20, 1500–1505. doi: 10.1097/WNR.0b013e3283329c05
Kosaka, N., Yoshioka, Y., Hagiwara, K., Tominaga, N., Katsuda, T., and Ochiya, T. (2013). Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front. Genet. 4:173. doi: 10.3389/fgene.2013.00173
Kosik, K. S. (2009). MicroRNAs tell an evo-devo story. Nat. Rev. Neurosci. 10, 754–759. doi: 10.1038/nrn2713
Krol, J., Busskamp, V., Markiewicz, I., Stadler, M. B., Ribi, S., Richter, J., et al. (2010). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631. doi: 10.1016/j.cell.2010.03.039
Krol, J., and Krzyzosiak, W. J. (2006). Structure analysis of microRNA precursors. Methods Mol. Biol. 342, 19–32. doi: 10.1385/1-59745-123-1:19
Lukiw, W. J. (2007). Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport 18, 297–300. doi: 10.1097/WNR.0b013e3280148e8b
Lukiw, W. J. (2012a). NF-κB-regulated, proinflammatory miRNAs in Alzheimer's disease. Alzheimers Res. Ther. 4, 47–54. doi: 10.1186/alzrt150
Lukiw, W. J. (2012b). Evolution and complexity of micro RNA in the human brain. Front. Genet. 3:166. doi: 10.3389/fgene.2012.00166
Lukiw, W. J. (2012c). NF-κB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp. Neurol. 235, 484–490. doi: 10.1016/j.expneurol.2011.11.022
Lukiw, W. J. (2013). Variability in micro RNA (miRNA) abundance, speciation and complexity amongst different human populations and potential relevance to Alzheimer's disease (AD). Front. Cell. Neurosci. 7:133. doi: 10.3389/fncel.2013.00133
Lukiw, W. J., Alexandrov, P. N., Zhao, Y., Hill, J. M., and Bhattacharjee, S. (2012). Spreading of Alzheimer's disease inflammatory signaling through soluble micro-RNA. Neuroreport 23, 621–626. doi: 10.1097/WNR.0b013e32835542b0
Lukiw, W. J., Cui, J. G., Yuan, L. Y., Bhattacharjee, P. S., Corkern, M., Clement, C., et al. (2010). Acyclovir or Aβ42 peptides attenuate HSV-1-induced miRNA-146a levels in human primary brain cells. Neuroreport 21, 922–927. doi: 10.1097/WNR.0b013e32833da51a
Lukiw, W. J., Handley, P., Wong, L., and McLachlan, D. R. C. (1992). BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem. Res. 17, 591–597. doi: 10.1007/BF00968788
Lukiw, W. J., and Pogue, A. I. (2007). Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J. Inorg. Biochem. 101, 1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004
Lukiw, W. J., Zhao, Y., and Cui, J. G. (2008). An NF-κB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 283, 31315–31322. doi: 10.1074/jbc.M805371200
Mehler, M. F., and Mattick, J. S. (2007). Noncoding RNAs and RNA editing in brain development, functional diversification and neurological disease. Physiol. Rev. 87, 799–823. doi: 10.1152/physrev.00036.2006
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012). Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 94, 1474–1480. doi: 10.1016/j.biochi.2012.02.020
Orgel, L. E. (1968). Evolution of the genetic apparatus. J. Mol. Biol. 38, 381–393. doi: 10.1016/0022-2836(68)90393-8
Perkel, J. M. (2013). Assume nothing: the tale of circular RNA. Biotechniques 55, 55–57. doi: 10.2144/000114061
Pogue, A. I., Cui, J. G., Li, Y. Y., Zhao, Y., Culicchia, F., and Lukiw, W. J. (2010). Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett. 476, 18–22. doi: 10.1016/j.neulet.2010.03.054
Ritchie, W., Legendre, M., and Gautheret, D. (2007). RNA stem-loops: to be or not be cleaved by RNAse III. RNA 13, 457–462. doi: 10.1261/rna.366507
Rocheleau, L., and Pelchat, M. (2006). The subviral RNA database: a toolbox for viroids, the hepatitis delta virus and satellite RNAs research. BMC Microbiol. 6:24. doi: 10.1186/1471-2180-6-24
Saetrom, P., Snøve, O., Nedland, M., Grünfeld, T. B., Lin, Y., Bass, M. B., et al. (2006). Conserved microRNA characteristics in mammals. Oligonucleotides 16, 115–144. doi: 10.1089/oli.2006.16.115
Sano, T., Barba, M., Li, S. F., and Hadidi, A. (2010). Viroids and RNA silencing: mechanism, role in viroid pathogenicity and development of viroid-resistant plants. GM Crops 1, 80–86. doi: 10.4161/gmcr.1.2.11871
Sarkies, P., and Miska, E. A. (2013). Molecular biology. Is there social RNA? Science 341, 467–468. doi: 10.1126/science.1243175
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. doi: 10.1186/gb-2004-5-3-r13
Sethi, P., and Lukiw, W. J. (2009). Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci. Lett. 459, 100–104. doi: 10.1016/j.neulet.2009.04.052
Taft, R. J., Pang, K. C., Mercer, T. R., Dinger, M., and Mattick, J. S. (2010). Non-coding RNAs: regulators of disease. J. Pathol. 220, 126–139. doi: 10.1002/path.2638
Taganov, K. D., Boldin, M. P., Chang, K. J., and Baltimore, D. (2006). NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486. doi: 10.1073/pnas.0605298103
Triboulet, R., and Gregory, R. I. (2010). Autoregulatory mechanisms controlling the microprocessor. Adv. Exp. Med. Biol. 700, 56–66. doi: 10.1007/978-1-4419-7823-3_6
Wang, D. Y., Kumar, S., and Hedges, S. B. (1999). Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. Biol. Sci. 266, 163–171. doi: 10.1098/rspb.1999.0617
Wang, Y., and Ding, B. (2010). Viroids: small probes for exploring the vast universe of RNA trafficking in plants. J. Integr. Plant Biol. 52, 28–39. doi: 10.1111/j.1744-7909.2010.00900.x
Witkos, T. M., Koscianska, E., and Krzyzosiak, W. J. (2011). Practical aspects of miRNA target prediction. Curr. Mol. Med. 11, 93–109. doi: 10.3791/2034
Yuva-Aydemir, Y., Simkin, A., Gascon, E., and Gao, F. B. (2011). MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 8, 557–564. doi: 10.4161/rna.8.4.16019
Zhang, R., Zhang, Q., Niu, J., Lu, K., Xie, B., Cui, D., et al. (2013). Screening of microRNAs associated with Alzheimer's disease using oxidative stress cell model and different strains of senescence accelerated mice. J. Neurol. Sci. doi: 10.1016/j.jns.2013.12.017. [Epub ahead of print].
Zhao, Y., Bhattacharjee, S., Jones, B. M., Hill, J., Dua, P., and Lukiw, W. J. (2013). Regulation of neurotropic signaling by the inducible, NF-kB-sensitive miRNA-125b in Alzheimer's disease (AD) and in primary human neuronal-glial (HNG) cells. Mol. Neurobiol. doi: 10.1007/s12035-013-8595-3. [Epub ahead of print].
Keywords: micro RNAs (miRNAs), viroids, evolution, molecular speciation and complexity, miRNA-146a, genetic variation, PSTV, potato spindle tuber viroid
Citation: Hill JM and Lukiw WJ (2014) Comparing miRNAs and viroids; highly conserved molecular mechanisms for the transmission of genetic information. Front. Cell. Neurosci. 8:45. doi: 10.3389/fncel.2014.00045
Received: 03 January 2014; Accepted: 01 February 2014;
Published online: 20 February 2014.
Edited by:
Lawrence Rajendran, University Zurich, SwitzerlandReviewed by:
Andrew Francis Hill, The University of Melbourne, AustraliaLeonora Balaj, Massachusetts General Hospital, USA
Maria Mittelbrunn, National Center of Cardiovascular Research, Spain
Copyright © 2014 Hill and Lukiw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: wlukiw@lsuhsc.edu
 James M. Hill1,2,3,4
James M. Hill1,2,3,4  Walter J. Lukiw
Walter J. Lukiw