Gene regulatory network analysis reveals differences in site-specific cell fate determination in mammalian brain
- 1Computational Biology, Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belval, Luxembourg
- 2Developmental and Cellular Biology, Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belval, Luxembourg
Neurogenesis—the generation of new neurons—is an ongoing process that persists in the adult mammalian brain of several species, including humans. In this work we analyze two discrete brain regions: the subventricular zone (SVZ) lining the walls of the lateral ventricles; and the subgranular zone (SGZ) of the dentate gyrus (DG) of the hippocampus in mice and shed light on the SVZ and SGZ specific neurogenesis. We propose a computational model that relies on the construction and analysis of region specific gene regulatory networks (GRNs) from the publicly available data on these two regions. Using this model a number of putative factors involved in neuronal stem cell (NSC) identity and maintenance were identified. We also demonstrate potential gender and niche-derived differences based on cell surface and nuclear receptors via Ar, Hif1a, and Nr3c1. We have also conducted cell fate determinant analysis for SVZ NSC populations to Olfactory Bulb interneurons and SGZ NSC populations to the granule cells of the Granular Cell Layer. We report 31 candidate cell fate determinant gene pairs, ready to be validated. We focus on Ar—Pax6 in SVZ and Sox2—Ncor1 in SGZ. Both pairs are expressed and localized in the suggested anatomical structures as shown by in situ hybridization and found to physically interact. Finally, we conclude that there are fundamental differences between SGZ and SVZ neurogenesis. We argue that these regulatory mechanisms are linked to the observed differential neurogenic potential of these regions. The presence of nuclear and cell surface receptors in the region specific regulatory circuits indicate the significance of niche derived extracellular factors, hormones and region specific factors such as the oxygen sensitivity, dictating SGZ and SVZ specific neurogenesis.
Introduction
Neurons constitute a specific cell lineage in mammals. Vast majority of them arise during embryonic development, they do not undergo replicative aging and new neurons are generated from neural stem cells (NSCs) through a process called neurogenesis only in specific regions in the adult mammalian brain (Altman and Das, 1965; Lois and Alvarez-Buylla, 1993; Lois et al., 1996; Gage et al., 1998). Cell divisions during neurogenesis can be either symmetric or asymmetric. Symmetric division can increase the pool of stem cells when they lead to two new stem cells, while terminal symmetric division leads to the generation of two neurons and a depletion of the stem cell pool (Doetsch et al., 1997, 1999; Palmer et al., 2000). Asymmetric mitosis of NSCs produces one NSC and one neural progenitor cell (NPC), daughter cell with limited differentiation capacity for neuronal or glial lineages (Gage, 2000; Temple, 2001). Extracellular factors and brain region specific transcription factors (TFs) are believed to be essential for NSC maintenance and proliferation which include epidermal growth factor (EGF) (Gritti et al., 1999; Tropepe et al., 1999), fibroblast growth factor (FGF) (Kalyani et al., 1997; Raballo et al., 2000), Sonic Hedgehog (Shh) (Wechsler-Reya and Scott, 1999) and Wnt signaling (Ikeya et al., 1997).
Two major sites of adult neurogenesis reported to date are the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG) and the subventricular zone (SVZ) lining the lateral ventricles (Lois and Alvarez-Buylla, 1993; Gage et al., 1998). Although both sites harbor NSCs, the differentiation process is site specific and results in different types of cells. The SVZ has the ability to produce neurons, glia and oligodendrocytes (Rao and Mayer-Proschel, 1997; Rao, 1999; Alvarez-Buylla et al., 2001), while the SGZ specific stem and progenitor cells are able to produce granule neurons and astrocytes (Seri et al., 2001; Fukuda et al., 2003). Given the paramount importance of natural reservoirs of NSCs in neurodegenerative diseases, the site-specific regulatory mechanisms acting in SGZ and SVZ are surprisingly not well understood.
The niche of the SGZ in the DG of the hippocampus consists of populations of stem and progenitor cell types, which have distinct and variable cell division and survival rates. Quiescent NSCs have a single radial process that extends through the granule cell layer (GCL) and express markers such as glial fibrillary acidic protein (GFAP) and Nestin (Seri et al., 2001; Fukuda et al., 2003). Another cell type is the horizontal cell, which divides more rapidly; however, the exact lineage association between radial and horizontal cells is still not established. Quiescent NSCs leave their dormant state and proliferate; they divide to produce transit-amplifying progenitors (TAPs) that have the potential to differentiate into neurons and astrocytes. The immature neurons produced from TAPs have the potential to differentiate to give rise to mature glutamatergic granule neurons in the GCL (Cameron et al., 1993; Kempermann et al., 2004). During this process intra- and intercellular signals arising from the microenvironment, including cellular components (vascular and glial cells, and granule neurons in the environment) and non-cellular components (extracellular matrix proteins and secreted molecules), have an effect on the activity of SGZ NSCs as well as the differentiation and survival capacity of the immature neurons along the SGZ GCL axis (Palmer et al., 2000; Ma et al., 2005, 2009; Morrens et al., 2012). Furthermore, both astrocytes and neurons differentiated from the TAPs are reported to play an instructive role to promote NSC self-renewal and differentiation. This naturally creates the feedback loop to maintain homeostasis in the microenvironment. Another lines of evidence indicate that neural progenitor cells respond to neuronal activity in the form of glutamate and GABA secretion as part of their differentiation program (Song et al., 2002; Deisseroth et al., 2004; Tozuka et al., 2005). In addition to these neurotransmitters, astrocytes are described also a potential source of niche factors such as Notch, Shh, bone morphogenetic proteins (BMPs), and Wnts (Ahn and Joyner, 2005; Lie et al., 2005; Ables et al., 2010; Mira et al., 2010).
The second region in which neuronal production persists throughout life in mammals is within the SVZ of the lateral ventricle. Earlier studies have shown that a subset of radial glial cells subsequently give rise to astrocyte-like NSCs that can serve as quiescent stem cells of the SVZ during postnatal and adult stages (Merkle et al., 2004; Spassky et al., 2005). In addition to their reported morphological resemblance to astrocytes, type B1 cells also express markers like GFAP, glutamate transporter (GLAST) and brain–lipid-binding protein (BLBP) (Doetsch et al., 1999; Nomura et al., 2010). Type C cells, the progeny of type B cells, rapidly proliferate and are often reside in clusters near blood vessels indicating possible niche-provided factors for their survival (Kriegstein and Alvarez-Buylla, 2009). Similar to type 2 TAPs of the SGZ, type C cells have elevated levels of Dlx2, Ascl1, and Pax6 (Brill et al., 2008). Finally, those type C cells give rise to neuroblasts, which form migratory pathways enclosed within astrocyte tubes, the so called rostral migratory stream reaching from the SVZ to the olfactory bulb (Lois et al., 1996). These multiple progenitor cell types ultimately give rise to a diverse array of olfactory bulb (OB) interneurons, including deep granule interneurons and superficial granule interneurons (Merkle et al., 2007; Lledo et al., 2008). Neurons of the OB are mostly GABAergic interneurons; however, fate-mapping study suggested the existence of glutamateric- and dopaminergic neurons (Brill et al., 2009) as well. Together, current studies support the model of “microenvironment supported adult neurogenesis,” in which NSCs are regulated by microenvironment derived signals, allowing their proliferative expansion and differentiation into mature cell types in SGZ and in SVZ. Ultimately, niche-derived signals are relayed to the NSC genome to control transcription of genes involved in self-renewal, differentiation and survival. The detailed review including of SGZ and SVZ neurogenesis is found elsewhere (Hsieh, 2012).
At the molecular level, region specific roles of the TFs, cell surface markers as well as extracellular factors constitute a distinct microenvironment in SGZ and SVZ resulting in reported differences in cell lineage commitment such as the propensity to differentiate into granule cells of the GCL and interneurons of the OB respectively.
The role of TFs determining cell fates, known as lineage specifiers, is one of the important concepts emerged from studying stem cell differentiation. A simple model considers that two lineage specifiers, which repress each other and activate themselves, are responsible for binary cell fate decisions. Several examples of these binary cell fate choice mechanisms have emerged in the last decade. A more general view postulates that stem/progenitor states, corresponding to metastable states, are maintained by the balanced expression of two rival lineage specifiers, which compete for differentiation into mutually exclusive cell fates (Huang et al., 2006; Roeder and Glauche, 2006). Moreover, disturbance of this equilibrium leads to cell fate differentiation.
In this manuscript we performed a comparative transcriptomics analysis between the mouse SGZ and SVZ in order to investigate region specific gene expression profiles, including TFs, receptors and extracellular factors. Moreover, gene regulatory networks (GRNs) of differentially expressed TFs between these two regions were constructed using a method that we have previously introduced (Crespo et al., 2013). Further, key strongly connected components (SCC) belonging to each region-specific GRN were identified. Indeed, SCCs have been shown to be important GRN stability motifs that contain genes responsible for maintaining cell identity (Crespo and Del Sol, 2013). Finally, based on the notion that two rival cell fate determinants exhibit a significantly disbalanced expression pattern in the differentiated cells in comparison to the stem/progenitor cell, we identified several pairs of these TFs belonging to SCCs as candidates of cell fate determinants in these regions.
Our results allow us to highlight potential cell fate determinant pair candidates and their site specificity in regions of neurogenesis. These pairs constitute a good starting point for future studies addressing cellular reprogramming and transdifferentiation for neurodegenerative disorders.
Materials and Methods
Gene Expression Data
The gene expression microarray data for SGZ and GCL are obtained from NCBI Gene Expression Omnibus (GEO) under accession number GSE39697 (Edgar et al., 2002). In this study authors explain that brains from 10 to 11-week-old C57BL/6 male mice (n = 9) were quickly removed, flash-frozen in OCT (Sakura Finetek) and stored at −80°C. Cryostat sections (12 μm) were stained with Cresyl Violet and rapidly dehydrated through graded xylenes. An Arcturus PixCell II machine (Arcturus) was used to isolate two- to three-cell thick bands from the outer and inner (SGZ, and specifically excluding cells of the hilus) portions of the dentate GCL (Miller et al., 2013). The RNA purification, amplification and microarray hybridization (to Affymetrix MG-U74Av2 arrays) experiments are performed according to the protocols in house. Explanation of the animal procedures, RNA purification, amplification and microarray hybridization can be found in detailed elsewhere (Miller et al., 2013).
The RNASeq data for SVZ is obtained from NCBI GEO under accession number GSE45282 (Ramos et al., 2013). In this study the brain from adult (older than postnatal day 60) male C57BL/6 mice was removed from the skull and placed in the ice-cold L15 media and a coronal slab (0.5-mm-thick) was obtained. The lateral SVZ and striatum were then micro dissected, avoiding contamination from the corpus callosum. FACS sorting of the SVZ cells was performed as described earlier (Pastrana et al., 2009). DG region was microdissected with a Vibratome and 300-mm-thick coronal sections obtained are placed in ice-cold L15 media. The high-throughput sequencing (Illumina) and sequencing data processing and mouse genome mapping is performed according to protocols as explained in detail in Ramos et al. (2013).
The background gene expression signal for detecting over/under expression for predicting pairs has been obtained from GEO under accession number GSE2882 (Sugino et al., 2006). In this study authors have characterized 12 populations of neurons from the mouse forebrain. These regions include hippocampus, cingulate cortex, somatosensory cortex, amygdala and thalamus. The data consists of 36 samples from 12 different cell types with three biological replicates (each sample from a different animal). The animals were all adult male mice, 57–106 day old with varying genetic background (The contribution of C57BL/6J ranged from 31 to 100%, with an average of 67 ± 26% (mean ± SD) (Sugino et al., 2006). The median expression per gene calculated from this data has served as the basal expression level per gene for the mouse forebrain.
In situ Hybridization
The ISH data presented herein is obtained from Allen Brain Atlas portal (http://www.brain-map.org/). The in situ hybridization shown in this manuscript is from the Allen Mouse Brain Atlas. All technical details describing this project is available under the “Documentation” tabs in the atlas (http://mouse.brain-map.org/static/atlas).
Microarray Data Normalization, Discretization and Analysis
Gene expression data from GSE3997, GSE45282, and GSE2882 have been merged and used for downstream analysis. The in-group normalization of the data has been performed in R. BioConductor “affy” package is used for reading Affymetrix microarray data and summarizing the probe level data into Robust Multichip Analysis (RMA) expression measures (Bolstad et al., 2003). The FPKM converted values of RNASeq data from GSE45282 has been used for downstream analysis.
Quartile expression
Equal Frequency Discretization (EFD) divides the sorted E(n,:) into k intervals so that each interval contains approximately the same number of expression values. We have extended the Equal Frequency Discretization method (EFD) to discretize and calculate the differential expression between SGZ, GCL, SVZ, and OB expression data. This discretization allows us to assign expression values to each gene between 1 and 4 depending on the relative abundance (Quartiles 1–4) of the transcripts (mRNA, lncRNA, miRNA etc.) in each dataset. We have used the median values for summarizing different probes for the same gene product. We have used the median values also per gene when there are multiple replicates were available (in SGZ, GCL, and SGZ). The choice of the median values was motivated by choosing a robust discretization to avoid the outlier effects. Hence increase the sensitivity of the differential expression calculation.
We have calculated the Quartile Expression (QE) for each gene by evaluating the quartile change of the gene in two experimental conditions. Therefore, in this comparison differentially expressed is defined as the change in the relative abundance (or quartile expression) of the gene in two different regions. We have defined minimum two (±2) difference as statistically significant to increase sensitivity to signal ratio by avoiding the possible boundary conditions around the quartiles. We have calculated QE for SGZ vs. SVZ, SGZ vs. GCL, and SVZ vs. OB (Supplementary Table).
We have also calculated the distribution of the gene expression skewness (SGZ: 6.4–7.18 and SVZ: 2.87–2.89) and kurtosis (SGZ: 64.1–76.9 and SVZ: 10.6–10.8) per region (in SGZ and SVZ) for examining the suitability of our approach. The quartile expression values for the previously associated genes, and violin plots including all experiments for both SGZ and SVZ are shown in Figure 1B.
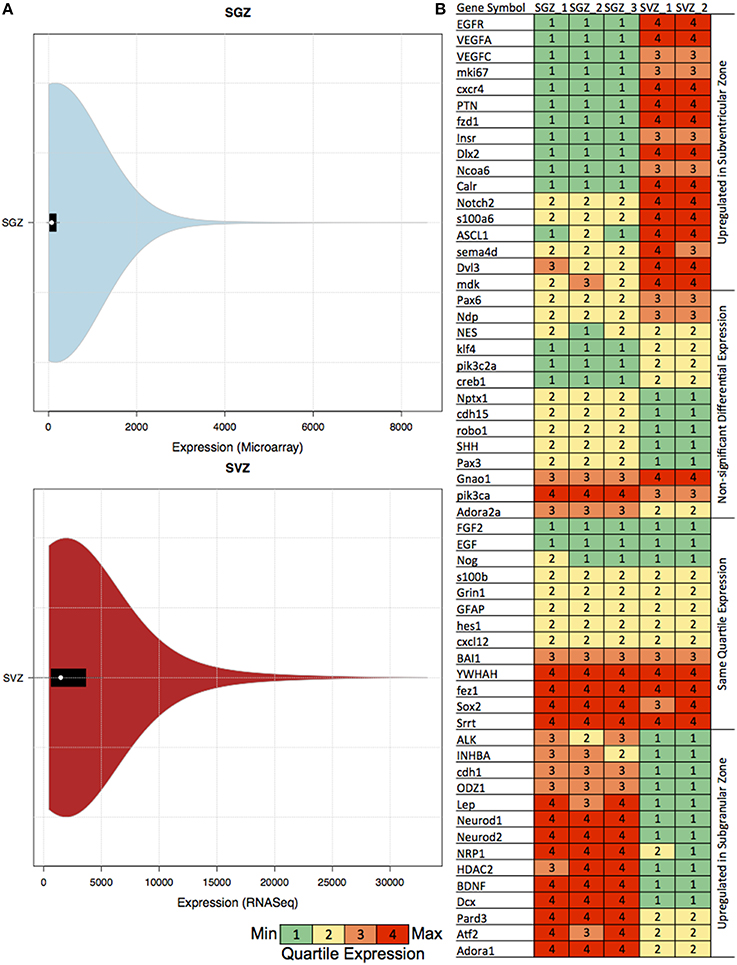
Figure 1. Table showing the quartile expression of genes associated with neurogenesis and the distribution of gene expression in SGZ and SVZ. (A) Violin plots showing the density trace with the box plot of gene expression in SGZ (microarray expression intensity) and SVZ (gene-level signal intensity) values. The black region in each plot is the box plot with white dot with the mean and the upper and lower bounds of the plots are the upper and lower quartiles. As shown in the figures, even though the absolute values are different, the violin plot distribution of the expression in each region is very similar, hence rendering the comparison via quartile expression viable. (B) The genes associated with neurogenesis are clustered in four clusters according to their differential expression in SGZ and SVZ: Upregulated in Subventricular Zone, Non-significant Differential Expression, Same Quartile Expression and Upregulated in Subgranular Zone. The quartile expression is color coded as shown below green, yellow, orange, and red indicating low, low-medium, high-medium and high expression respectively.
Gene Ontology Analysis
For Gene Ontology (GO) analysis we have used funcassociate (Berriz et al., 2009) to investigate key factors, pathways and regulatory mechanisms that are responsible for the maintenance and differentiation of NSC identities in mammalian brain.
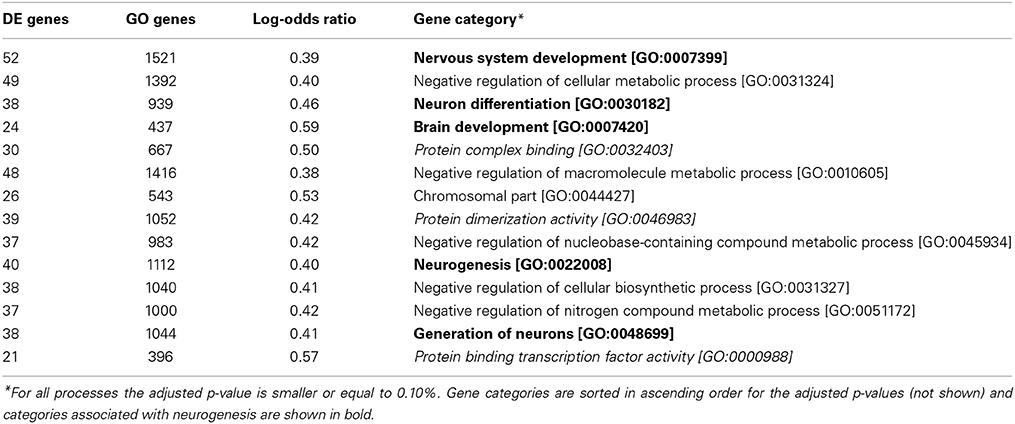
We have employed a strict criterion for performing GO analysis for differentially expresses genes between SGZ and SVZ. Only GO processes with adjusted P-value below 0.1% (P_adj ≤ 0.001), log-odds ratio over 0.3 (LOD ≥ 0.3) are considered significant. We have sorted the results for number of genes in each associated GO term (X). For presentation in in Table 1, we selected top 14 GO terms that have minimum number of genes per GO term for specificity.
We have used the package funcassociate (Berriz et al., 2009) that statistically identifies the terms that were associated with differentially expressed genes (defined by adjusted p ≤ 0.1 and LOD ≥ 0.3).
Gene Regulatory Network Construction, Contextualization and Analysis
Human direct gene interactions were retrieved from MetaCore from Thompson Reuters [GeneGo Inc. (https://portal.genego.com/)] to infer biological relationships and associations that have been extracted from the biomedical literature relevant for these sites.
The interaction types “Transcription regulation” and “Binding” were kept for the subsequent analyses. Pruning of network edges (interactions) was performed using the method developed by Crespo et al. (2013), which was implemented in MATLAB using the genetic algorithm (ga) function. This algorithm is designed with the assumption that each cellular phenotype is a stable steady state (attractor) of the GRN. The algorithm removes edges that are inconsistent with the Booleanized gene expression data, thereby obtaining a network not only based on previous knowledge but also customized for a given biological condition. The Boolean simulation was carried out using the pbn-matlab-toolbox (http://code.google.com/p/pbn-matlab-toolbox/downloads/list) using the synchronous updating scheme. The logic rule was defined, so that the number of activating edges and inhibiting edges acting on a gene were compared and the one with a higher number dominated. For example, if a gene had three activating edges and two inhibiting edges acting on it, the activation logic was used in the next update. If both numbers were the same, the inhibition logic was set to dominate. During this process, “unassigned” interactions (i.e., interactions without knowledge of activation or inhibition) were randomly assigned “activation” or “inhibition” and the one that yielded a better result was taken. The contextualized network was loaded in Cytoscape (version 2.7.0) (Shannon et al., 2003). The network stability motif, strongly connected component (SCC), was identified using the Cytoscape BiNoM (version 2.5) plugin (Zinovyev et al., 2008).
Cell Fate Determinant Pairs Analysis
The difference in the expression ratio of a pair of genes between the stem/progenitor cell and differentiated cell was computed by
where gene1 and gene2 indicate the log2 median gene expression value over biological replicates. The suffix “differentiated” indicates the differentiated cell types (i.e., GCL or OB) and “stem” indicates the stem/progenitor cell types (i.e., SGZ and SVZ). This value was calculated for all pairs of TFs and statistically significant pairs were detected by a robust z-test. The values were first normalized by their expression ratios of the stem/progenitor cells and the z-score was computed after trimming the 2.5% outliers on both sides. The z-score of the trimmed data points was extrapolated from the z-score function. The z-score was then converted into the p-value. The p-value was corrected for multiple testing by the Benjamini-Hochberg method. Pairs with adjusted p-value below 0.05 were kept. Additional criteria were set to narrow down candidate cell fate determinants [Supplementary Figure S3 (flowchart)]. These criteria were (1) one of the genes in a pair has to be differentially up-regulated in the differentiated cell type (i.e., GCL and OB), (2) both genes in a pair are not differentially down-regulated in the stem/progenitor cells (i.e., SGZ and SVZ), and either (3) both genes in a pair are directly connected in the SCC or (4) one of the genes in a pair is present in the SCC and the other gene is not present in the SCC but is known to directly interact with the other gene.
Results
The neurogenic potential of the SVZ along the walls of the lateral ventricle and the SGZ of the DG is well established. Each region has been studied in isolation that resulted in our understanding of the mammalian neurogenesis in-vivo. Naturally gene ontologies (such as neurogenesis; GO:0022008, nervous system development; GO:0007399, regulation of nervous system development; GO:0051960) have been created from these studies. This equipped us with tools to identify neurogenic gene expression signatures from other processes of the neural system.
Recent studies indicate the differences between the SGZ and SVZ, site-specific regulation of these regions by molecular cues arising from their complex heterogeneous cellular environment and their differential contribution in generating various cell types in the adult mammalian brain.
For the molecular annotation and functional analysis of NSC in two regions in the adult mammalian brain we have merged and analyzed genome-wide gene expression of SGZ and GCL from Miller et al. (2013) and SVZ and OB from Ramos et al. (2013).
SGZ and SVZ Differ in Expression of Factors Associated with Neurogenesis
We have composed the specific markers and genes previously associated with neurogenesis to the best of our knowledge. The list of associated factors with their references are shown in the Supplementary Tables under Genes associated and Present tab.
To be able to compare GRNs we have extended the Equal Frequency Discretization method (EFD) to calculate the Quartile Expression (QE) for each gene. This discretization allows us to assign expression values to each gene between 1 and 4 depending on the relative abundance in the experiment, therefore in this comparison differentially expressed is defined as the change in the relative abundance (or Quartile Expression) of the gene in two different regions. We have also calculated the distribution of the gene expression, skewness and kurtosis per region (SGZ and SVZ) for characterizing our method (See Materials and Methods). The distributions of gene expression for each region show very similar patterns as shown by the violin plots in Figure 1A, hence allowing our method to capture the essential differences in gene expression between SGZ and SVZ.
There were 1578 genes differentially expressed (having a differential quartile expression QDE ≤ −2 or QDE ≥ 2) between two regions among which 828 of them were up-regulated in SVZ and 750 of them were up-regulated in SGZ.
The quartile expression values for the previously associated genes for each region are shown in Figure 1B. In depth analysis of the quartile expression of these genes indicates four clusters: (i) Up-regulated in SGZ, (ii) Up-regulated in SVZ, (iii) Same quartile expression, and (iv) Non-significant differential expression.
One of the stark differences is observed in the Vascular Endothelial Growth Factor (VEGF) genes (Vegfa and Vegfc). Vegf over-expression is associated with increased neurogenesis earlier with SVZ and SGZ neurogenesis (Jin et al., 2002; Warner-Schmidt and Duman, 2007) and reviewed in detail (Fournier and Duman, 2012; Mackenzie and Ruhrberg, 2012). Although the link with Vegf genes and proliferation-linked neurogenesis is established, region specific effects and the possible role in determining cell fate is not explored.
The overexpression of the homeobox protein Dlx2 and the EGF receptor (Egfr) are observed to be SVZ specific in our analysis. This finding overlaps with the reported SVZ specific interaction between two genes, Dlx2 promoting the lineage transition from NSCs to TAPs and at the same time enhancing the proliferative response of neuronal progenitors to Egf (Suh et al., 2009).
Another important factor Notch2, whose down-regulation is implicated in subsequent NSC differentiation to astroglial lineage (Tchorz et al., 2012), is observed to be upregulated in SVZ with respect to SGZ. This is in line with the astrocytic features associated to SVZ NSCs.
Neurod is very well established as a transcription factor required for maintenance of neurogenesis and differentiation of the granule cells in the cerebellum and hippocampus (Miyata et al., 1999). The essential role of NeuroD1 in the differentiation and survival of neuronal precursors is stressed by Gao et al. (2009). The elevated Neurod1 and Neurod2 expression in SGZ is possibly due to the different spatial scales of neurogenesis and differentiation for SGZ and SVZ (SGZ to GCL being more compact and SVZ to OB being spread out including RMS).
Sox2 is essential for maintenance of neural stem cell fate and as expected is expressed in the last (4th) quartile in both SGZ and SVZ regions. GFAP, another widely used marker of quiescent NSC, is expressed in both SGZ and SVZ.
SGZ and SVZ Differ in Classes of Differentially Expressed Genes
We next sought to measure the extent to which the gene expression profiles of the neurogenic niche are conserved between SGZ and SVZ and also determine the functional classes of the genes distinguishing different regions. We have performed an over-representation analysis using the Gene Ontology (GO) associations. GO terms are nested functional categories that summarize the known molecular functions, biological processes and cellular compartments associated with each gene. Enriched GO terms for genes that are differentially expressed between SGZ and SVZ are shown in Table 1 (Adjusted p ≤ 0.1%).
Given the neurogenic potential of both regions, we expect to find only marginal differences in GO analysis, possibly related to the specific differentiation potential of each zone. However, over represented biological processes that differentiate gene expression of both regions include five categories associated with neurogenesis, namely: Nervous system development, Neuron differentiation, Brain development, Neurogenesis and Generation of Neurons. This is striking, because significant portion of stem cell maintenance and cellular proliferation markers (such as Sox2 and Gfap) are not differentially expressed between SGZ and SVZ. However, GO enrichment shows strong association with neurogenesis and neuron differentiation indicating possible involvement of niche specific factors. This leads to the conclusion that in addition to the mutual and comparable expression of stem cell factors such as Sox2, Oct4, Klf4, Myc, and Nanog, the genes associated with neurogenesis are not expressed uniformly in SGZ and SVZ, leading to diverse differentiation potentials in each region.
It is also important to note that three molecular functions, protein complex binding, protein dimerization activity and protein binding transcription factor activity, are over represented together suggesting potential differences on the protein function level.
SGZ and SVZ have Distinct Regulatory Mechanisms
In order to identify differences between the SGZ and SVZ, we discretized their genome-wide expression patterns, and calculated differentially expressed genes (DEGs) based on our criteria as explained in materials and methods. We then constructed two region specific GRNs from differentially expressed TFs using a method that we previously implemented (Crespo et al., 2013). This method removes interactions which are inconsistent with expression data and generates networks that explain cell specific gene expression programme. We previously used network strongly connected components (SCCs) as gene regulatory network (GRN) stability motifs and identified candidate genes that are responsible for the stabilization of cellular identities (Crespo and Del Sol, 2013). Therefore, we next identified SCCs in the GRNs (Figures 2A,D). The SCC for the SGZ had 58 genes (17 are up and 41 are down-regulated in SGZ), whereas the SCC from the SVZ had 63 genes (31 are up and 32 are down regulated in SVZ). Both SCCs had 37 genes in common with the same predicted gene expression state including Ar, Hif1a, Foxo3, Lef1, Jun, Jund, Fos, Snai2, Nr3c1, Sp3, and Atf2. Additionally 21 and 26 of those genes are unique to the SGZ and SVZ, respectively, revealing the similar and distinct gene regulatory mechanisms that stabilize the NSCs in these regions. We have also focused on highly connected genes and genes that had the highest number of out-going edges within each SCC, as these genes are likely to have a high regulatory influence on the SCC. Indeed, this criterion called “out-degree interface” was previously used in our study (Crespo and Del Sol, 2013) for identifying master regulatory TFs within SCCs. Here, we have identified Nr3c1, Hif1a, and Ar as master regulators in this system with the three highest out-degree interfaces in both SGZ and SVZ.
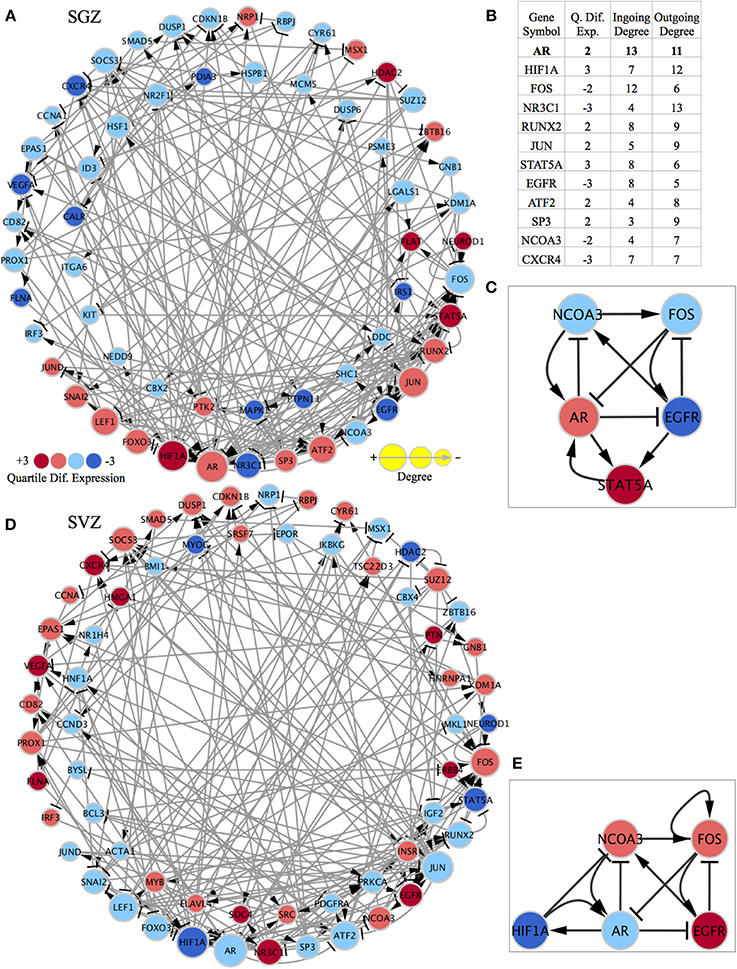
Figure 2. Region specific gene regulatory networks and sub-networks and gene list showing the importance of highly connected genes. The quartile differential expression of each gene in the network is color-coded below showing the difference in quartile expression with respect to the other region. Red indicates significantly higher (+3), pink indicates higher (+2), light blue indicates lower (−2), dark blue indicates significantly lower differential quartile expression in the region of interest vs. the other region. The size of each node is size coded to indicate the degree of this gene in the network as displayed below. The standard graphical notation for biological networks is used arrow heads indicating activation/up-regulating and bar-headed arrows indicating inhibition/down-regulating interactions. (A) SCC of the SGZ specific regulatory network: The outer ring consists of genes that are shared with the SCC of the SVZ specific network and have differential quartile expression. The inner ring consists of genes only present in the SCC of the SGZ regulatory network (their expression is important only in SGZ regulatory network and not in SVZ). (B) The table showing the quartile expression, ingoing & outgoing degrees for highly connected genes in the shared genes (outside ring genes) in both networks. The list of genes and their order of importance is very similar in SGZ and SVZ specific networks (the degrees in the table are from SGZ specific network and are almost identical in the SVZ specific network). (C) The AR contextualized SGZ specific SCC sub-network showing the interconnected nature and the regulation of factors around AR. (D) SCC of the SVZ specific regulatory network: The outer ring consists of genes that are shared with the SCC of the SGZ specific network and have differential quartile expression. The inner ring consists of genes only present in the SCC of the SVZ regulatory network (their expression is important only in SVZ regulatory network and not in SGZ). (E) The AR contextualized SVZ specific SCC sub-network showing the interconnected nature and the regulation of factors around AR. Note that FOS has a self-activation and STAT5A is replaced by HIF1A in this network for representation purposes.
To further elucidate the network dynamics in the proximity of these master regulators we have chosen Ar (androgen receptor) as our focus due to its highest total degree (3rd highest outgoing and 1st ingoing) and its gender specific biological implications (Figure 2B).
Subsequently, we have built SGZ and SVZ specific GRNs from AR (and all 1st neighbor genes) as explained above and the SCCs of these GRNs were identified (Supplementary Figures S1, S2 and Supplementary Networks). This has accomplished by while pruning of network edges, instead of global optimization, we have implemented optimization for edges connected to Ar for obtaining perfect consistency around Ar. The SGZ specific SCC sub-network around Ar is shown in Figure 2C. Network dynamics analysis in this sub-network indicates the “dominance” of AR in this subnetwork. Ar directly regulates all three out-degree connected nodes; inhibit Ncoa3, Egfr and forms a reinforcing feedback loop with Stat5. Moreover, the expression pattern observed in SGZ vs. SVZ for this sub-network corresponds to an attractor state presenting the stability of the observed expression pattern.
Ar is present in many of the body's tissue, where it binds to androgens (such as testosterone). Its expression is highly gender specific and the resulting androgen-receptor complex binds to DNA and regulates the activity of androgen responsive genes.
An inverse correlation between EGF and androgen signaling is reported in epithelial cells (Léotoing et al., 2007). It has been demonstrated that, cell cycle exit at G0 phase and the setting of the differentiation process is a prerequisite for having an active androgen signaling. Once cells have completed this process, androgen signaling is essential for the maintenance of the differentiated functions and for repressing the EGF-dependent signaling in epithelial cells.
Here we report that the balance between AR and EGFR signaling is skewed toward Ar up-regulation in SGZ and Egfr up-regulation in SVZ, indicating a possible mechanism for distinct differentiation phenotypes in those regions.
Ncoa3 encodes for a co-activator that interacts with nuclear hormone receptors to enhance their transcriptional activity. It has histone acetyltransferase activity and considered to act via remodeling of chromatin. Involved in the co-activation of different nuclear receptors, such as for steroids (GR and ER), retinoids (RARs and RXRs), thyroid hormone (TRs), vitamin D3 (VDR) and prostanoids (PPARs). In SGZ sub-network, the up-regulation of Ar combined with the scarcity in EGF signaling results in down-regulation of Ncoa3, in turn results in the impeded support for the Ar expression.
The SVZ specific SCC-sub-network around Ar is shown in Figure 2E. Analysis of this sub-network indicates Fos dominance with self-activation and a negative feedback loop via Egfr. Furthermore, Fos inhibits Ar expression, practically rendering the Ar dependent inhibition of Ncoa3 and Egfr obsolete.
Hif1a constitutes another remarkable finding significantly differentially expressed between SGZ and SVZ. Hif1a encodes the alpha subunit of transcription factor Hif-1, which functions as a master regulator of cellular homeostatic response to hypoxia by activating transcription of many genes. These include genes involved in energy metabolism, angiogenesis, apoptosis, and others whose protein products increase oxygen delivery or facilitate metabolic adaptation to hypoxia.
Mazumdar et al., have demonstrated that Hif1a modulates Wnt/β-catenin signaling in hypoxic embryonic stem (ES) cells by enhancing β-catenin activation and expression of the downstream effectors Lef-1 and Tcf-1. They have extended this finding to primary cells, including isolated NSCs, and NOT to differentiated cells. They also report that in vivo, Wnt/β-catenin activity was closely associated with low O2 regions in the SGZ of the hippocampus. The deletion of Hif1a impaired hippocampal Wnt-dependent processes, including NSC proliferation, differentiation and neuronal maturation. This decline correlated with reduced Wnt/β-catenin signaling in the SGZ (Mazumdar et al., 2010).
Differential expression and regulation of Hif1a (up-regulated in SGZ) in our analysis is in perfect agreement and suggests diverse niches with varying O2 sensitivity and Wnt/β-catenin activity in SGZ and SVZ.
Region Specific Cell Fate Determinant Pairs
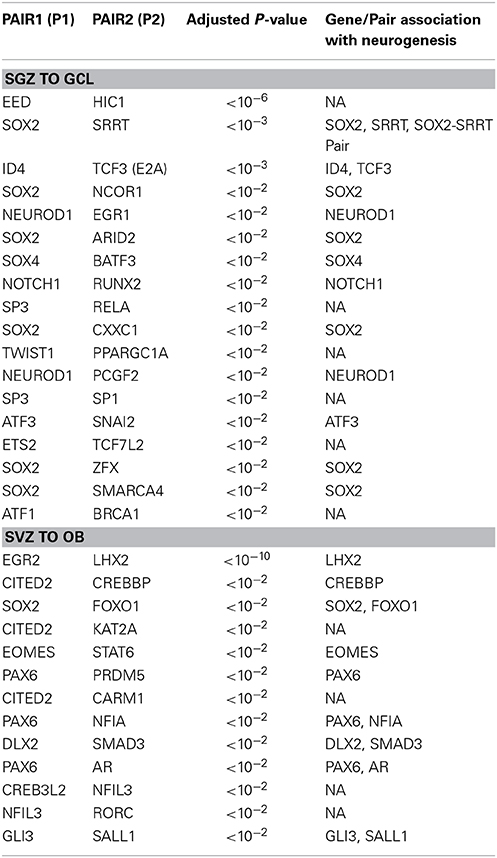
Previous theoretical and experimental studies indicated that two rival cell fate determinants show a balanced expression pattern in the stem/progenitor cell and breaking this balance would lead to differentiation (Huang et al., 2006; Roeder and Glauche, 2006). Assuming that this notion also holds true for neurogenesis, we next aimed to identify pairs of TFs that exhibited this expression pattern between the SGZ and GCL, and between the SVZ and OB. The goal here is to find region specific cell fate determinants, which give rise to the granule cells of the GCL and interneurons of the OB from the SGZ and SVZ, respectively. In addition, we set other criteria to narrow down candidate cell fate determinants [Supplementary Figure S3 (flowchart)]. These criteria were (1) one of the genes in a pair has to be differentially up-regulated in the differentiated cell type (i.e., GCL and OB), (2) both genes in a pair are not differentially down-regulated in the stem/progenitor cells (i.e., SGZ and SVZ), and either (3) both genes in a pair are directly connected in the SCC or (4) one of the genes in a pair is present in the SCC and the other gene is not present in the SCC but is known to directly interact with the other gene.
The results of the cell fate determinant pair analysis are shown in Table 2. For the SGZ to GCL predictions, out of 18 predicted pairs, 12 of them has at least one of the pair is associated earlier with neurogenesis while 6 pairs are novel predictions. From the SVZ to OL region predictions 9 out of 13 predictions have one or both of the pair genes have a reported link to neurogenesis.
A necessary condition for the cell fate determinant pairs is that they are colocalized around the regions of neurogenesis (SGZ and SVZ) in adult male mice. Hence, we have included in-situ hybridization images for two of our candidate pairs (Sox2—Ncor1 and Ar—Pax6) from Allen Mouse Brain Atlas as shown in Figures 3A,B. The details of the experiments and imaging can be found elsewhere (See Materials and Methods, In-situ Hybridization).
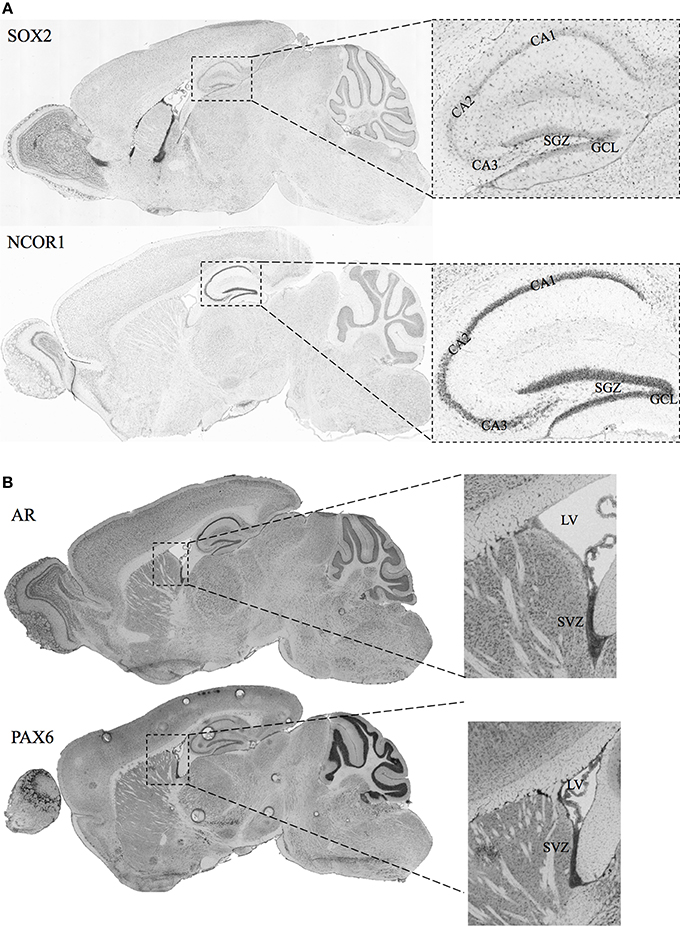
Figure 3. In-situ Hybridization images for selected candidate cell fate determinant pairs from Allen Brain Atlas. (A) Images showing the staining for SOX2 and NCOR1 in the sagittal section of mice brain. The right panels show the zoomed view for the Lateral Ventricle (LV) and the SVZ. (B) Images showing the staining for AR and PAX6 in the sagittal section of adult male mice brain. The right panels show the zoomed view for the Lateral Ventricle (LV) and the SVZ. Original images can be accessed freely on http://mouse.brain-map.org/. (SOX2: http://mouse.brain-map.org/experiment/show/79677365, NCOR1: http://mouse.brain-map.org/experiment/show/687, AR: http://mouse.brain-map.org/experiment/show/100142554, PAX6: http://mouse.brain-map.org/experiment/show/79677341).
It is not surprising that stem cell marker Sox2 is identified as a potential candidate in 6 out of 18 candidate pairs in SGZ to GCL region. Our candidate pair Sox2—Srrt predicted as cell fate determinant in SGZ is reported to function together as a fundamental mechanism to maintain neural stem cell identity in SVZ (Andreu-Agullo et al., 2011). The only Sox2 candidate from the SVZ—OB analysis is the Sox2—Foxo1 pair which is well characterized for having a pivotal role in maintaining stem cell identity (Zhang et al., 2011).
Ncor1 is another factor predicted together with Sox2 as a cell fate determinant in SGZ to GCL analysis. NCOR1 acts as a transcriptional co-regulatory protein, which recruits histone deacetylases to DNA promoter regions hence assisting nuclear receptors in the down regulation of DNA expression. Its expression in the mouse brain appears to be highly specific and localized around DG as observed from the in-situ-hybridization images from the Allen Mouse Brain Atlas (Data not shown). The interaction between NCOR1 and SOX2 is reported via the SMRT/NcoR complex during neurogenesis in NSC by Engelen et al. but requires functional identification on the cellular level (Engelen et al., 2011). The ISH of Sox2 and Ncor1 in Figure 3A indicates a highly specific Ncor1 localization around DG whereas Sox2 is localized around SGZ as well as in the SVZ, Lateral Ventricles (LV) and the Rostral Migratory Stream (RMS). These suggest significant colocalization of Sox2 and Ncor1 expression around SGZ and GCL in-vivo.
Our candidate pair Eed—Hic1, consists of Embryonic Ectoderm Development gene (Eed), a member of the epigenetic modifier complex Polycomb Repressive Complex 2 (PRC2), and an upstream transcription factor of Wnt signaling pathway, Hc1 (Valenta et al., 2006). The canonical PRC2 comprises of Eed, Suz12, and Ezh2 and controls stem cell development into different cell and tissue types of the body (Kaneko et al., 2010). PRC2 functions to methylate lysine 27 of histone 3 (H3K27me3) via interactions with the methyl transferase enhancer-of-zest 2 (Ezh2). PRC2 and Ezh2 serve also in balancing self-renewal vs. differentiation and neuron vs. glial fate choices in early and late embryogenesis respectively (Pereira et al., 2010). Lately, HIC1 is demonstrated to recruit PRC2 for regulating the expression of a subset of its targets (Boulay et al., 2012) strongly suggesting that the HIC1—EED pair can function as a cell fate determinant via Wnt dependent manner.
Among the SVZ—OB cell fate determinant pairs we focus on Ar—Pax6 pair. The transcription factor Pax6 is reported to be essential for generating specific subpopulation of granule and periglomerular neurons in the olfactory bulb and neuronal and glial populations in the SGZ (Kohwi et al., 2005; Klempin et al., 2012). In Figure 3B, we demonstrate the localization of both Ar and Pax6 around the SVZ. Furthermore, Pax6, and not Ar, expression is localized in OB.
AR has been associated as a potent regulator of adult neurogenesis only very recently. It has been reported by Hamson et al., that androgens, such as testosterone (T) and dihydrotestosterone (DHT), but not estradiol, increased the survival of new neurons in the DG and regulate hippocampal neurogenesis in an Ar dependent manner (Hamson et al., 2013). Here we identify a possible link between the Pax6 and Ar in determining the balance between self-renewal and differentiation in the adult SVZ.
Discussion
In this work we have focused on identifying region specific molecular mechanisms and cell fate determining factors in two main areas of neurogenesis (SGZ and SVZ) in the adult mammalian brain. To our knowledge, this is the first study attempting to identify genome wide differential regulatory mechanisms between two discrete regions of adult neurogenesis (SGZ and SVZ).
We have combined matching strain, sex and age mice gene expression profiles of (i) SGZ and GCL (Miller et al., 2013) with SVZ and OB (Ramos et al., 2013). We have transformed these region specific expression profiles to discrete values and integrated them forming the complete dataset.
Subsequently, we have constructed region specific GRNs via reported interactions in MetaCore and by carrying out network contextualization to each region. This allowed us to pinpoint region specific regulatory factors within these region specific GRNs, which are differentially active in each region. We have characterized Hif1a, Nr3c1, and Ar as three essential factors that are responsible for the differential regulation observed in these regions.
Hif1a is demonstrated to promote neurogenesis in a Wnt pathway dependent manner. HIF-1 is heterodimer of HIF-1α and HIF-1β/ARNT that regulates the response to hypoxia. Hif1a knockout, which encodes the HIF-1α subunit, reduces Wnt pathway target gene expression, actively reducing BrdU incorporation and the number of newborn Dcx positive neurons in the SGZ of mammalian hippocampus. These effects were rescued by the inhibition of GSK-3 and by expression of stabilized β-catenin, suggesting the HIF-1 involvement in the upstream of the Wnt pathway to promote neurogenesis (Mazumdar et al., 2010).
Furthermore, differential regulation of Hif1a (up-regulated in SGZ) arguably has unexpected implications such as promoting diverse niches within SGZ and SVZ with varying oxygen sensitivity and Wnt/β-catenin activity. The recent finding from Christen et al., demonstrate that the oxygen saturation in the adult rat brain is highly heterogeneous and can be detected with high spatial resolution via magnetic resonance imaging (MRI) (Christen et al., 2014). This brings us to the conclusion that oxygen saturation/sensitivity is one of the factors giving rise to the differences observed between SGZ and SVZ neurogenesis. Therefore, further in-vivo studies addressing oxygen dependency of SGZ and SVZ neurogenesis are required.
Ar (androgen receptor) and Nr3c1 (glucocorticoid receptor) belong to a family of nuclear hormone receptors of the NR3C class, which also includes mineralocorticoid and progesterone receptors. They are ubiquitously expressed in various tissues and regulate downstream gene expression by binding to the nuclear response elements of the genome or direct interaction with other TFs such as NF-kappaB, Ap-1 or Stat.
Hamson et al. recently demonstrated that gonadal steroids [testosterone (T) and dihydrotestosterone (DHT)] are regulating adult neurogenesis in adult male rats. They have provided evidence that androgens promote neurogenesis in the adult hippocampus by increasing the survival of newborn neurons via an Ar-dependent mechanism (Hamson et al., 2013). Nr3c1 is a novel prediction resulted from our analysis and awaiting to be experimentally validated with adult neurogenesis.
In addition to region specific differences, an important conclusion from our analysis indicates the gender specificity of adult neurogenesis. Our analyses have been performed on male adult mice. The pivotal role of Ar in SGZ and Nr3c1 in SGZ sub-networks highlight the significance of niche specific expression of nuclear hormone receptors in the neurogenesis context. Considering gender dependent expression of Ar and Nr3c1, our results strongly indicate gender as well as region specificity of neurogenesis in the adult mammalian brain. Further studies are required to address gender and region specificity of adult neurogenesis. Focusing on region specific spatial structures with quantification of hormones in each niche would unveil the intrinsic properties of adult SGZ and SVZ NSCs, their impact on aging and their potential to regenerate lost cell populations in neurodegenerative diseases or brain injuries.
Lastly, we have performed an in-depth analysis aiming to elucidate the region specific cell fate determinant pairs in SGZ and SVZ. A significant and unique contribution in this work is the developed methodology that systematically determines TF combinations (pairs) of cell fate determinants following an unbiased in-silico analysis. The methodology we have implemented considers NSCs (in this study) as the source and the differentiated cell as the target phenotype without prior knowledge to determine a list of cell fate determinant pairs with statistical significance. Henceforth, the scope of this study can easily be extended to other neural cell lineage differentiations such as the differentiation of astrocytes from the TAPs of the SGZ or the dopaminergic neurons of the OB from the SVZ.
Finally, using our methodology we have identified 18 TF pairs in SGZ to GCL and 13 TF pairs in SVZ to OB as cell fate determinant candidates. We present all 31 pairs in two NSCs regions; most of them having at least one gene associated with neurogenesis. In all but one (Sox2—Srrt) of the predicted pairs the interaction between the genes in the pair and the function of the predicted pair as a cell fate determinant remains to be experimentally verified.
In SGZ to GCL, 13 out of 18 pairs have at least one gene associated with neurogenesis/differentiation whereas; one pair is reported to maintain neural stem-cell identity in SVZ (Sox2—Srrt) (Andreu-Agullo et al., 2011). The remaining five pairs constitute novel predictions resulted from our study. Nine out of thirteen pairs predicted in SVZ to OB, have at least one of the genes (two pairs have both genes) is linked to neurogenesis/differentiation. The remaining four pairs are novel predictions that are not yet associated with neurogenesis. We would like to specifically emphasize the pairs (Sox2—Ncor1) and (Id4—Tcf3) in SGZ to GCL and (Ar—Pax6) and (Egr2—Lhx2) in SVZ to OB that have been predicted to function as cell fate determinants.
In summary, we characterized two aspects of adult neurogenesis: site-specific regulatory mechanisms in SGZ vs. SVZ and candidate cell fate determinant factors for each region. We identified putative factors and target genes, many of which are involved in NSC identity and maintenance. We demonstrate here possible gender and niche-derived differences based on cell surface and nuclear receptors via Ar, Hif1a and Nr3c1. We have reported 31 candidate cell fate determinant pairs, ready to be validated, for SGZ and SVZ together. We focused on cell fate determination by AR—Pax6 in SVZ and Sox2—Ncor1 in SGZ. Both pairs are expressed and localized in the suggested anatomical structures as shown by the ISH and found to physically interact. An extrapolation of our results suggests that the common features of neurogenesis observed in the NSC populations in SGZ and SVZ should be inspected very carefully. The fact that many cell surface and nuclear receptors have differentially regulatory roles indicates strong involvement of the niche-derived factors. Hence, to be able to understand neurogenesis in depth we will have to develop gender and region specific animal models of neurogenesis which constitute the prerequisite for studying gender bias observed in neurodegenerative diseases and aging.
Author Contributions
Gökhan Ertaylan contributed to the study design, collected data, performed the analysis and drafted the manuscript. Satoshi Okawa helped perform the analysis and helped draft the manuscript. Jens Christian Schwamborn discussed findings and interpretation of the results and helped drafting the manuscript. Antonio del Sol designed the study, discussed findings and interpretation of the results and drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Gökhan Ertaylan was supported by AFR Postdoctoral Grant from the Luxembourg National Research Fund (FNR) with project ANEMO with reference number 4001584/PDR 2012-1. Authors also would like to thank the reviewers of this manuscript for their invaluable feedback.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fncel.2014.00437/abstract
Abbreviations
BrdU, 5′-bromodeoxy-uridine; BG, Background; BMP, Bone Morphogenetic Protein; CNS, Central Nervous System; DNA, Deoxyribonucleic Acid; EGF, Epidermal Growth Factor; GCL, Granule Cell Layer; GFAP, Glial Fibrillary Acidic Protein; IL, Interleukin; LIF, Leukemia Inhibitory Factor; INF, Interferon; LPS, Lipopolysacharide; LV, Lateral Ventricles; NF-κB, Nuclear Factor kappa B; NPC, Neural Progenitor Cell; OB, Olfactory Bulb; RMS, Rostral Migratory Stream; RNA, Ribonucleic Acid; SCC, Strongly Connected Component; SGZ, Subgranular Zone; Shh, Sonic Hedgehog; SOX, Sry-containing; SVZ, Subventricular Zone; TNF, Tumor Necrosis Factor; Wnt, Wingless-type; MMTV, integration site family member.
References
Ables, J. L., Decarolis, N. A., Johnson, M. A., Rivera, P. D., Gao, Z., Cooper, D. C., et al. (2010). Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J. Neurosci. 30, 10484–10492. doi: 10.1523/JNEUROSCI.4721-09.2010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ahn, S., and Joyner, A. L. (2005). In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437, 894–897. doi: 10.1038/nature03994
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Altman, J., and Das, G. D. (1965). Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335. doi: 10.1002/cne.901240303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alvarez-Buylla, A., Garcia-Verdugo, J. M., and Tramontin, A. D. (2001). A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2, 287–293. doi: 10.1038/35067582
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Andreu-Agullo, C., Maurin, T., Thompson, C. B., and Lai, E. C. (2011). Ars2 maintains neural stem-cell identity through direct transcriptional activation of Sox2. Nature 481, 195–198. doi: 10.1038/nature10712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berriz, G. F., Beaver, J. E., Cenik, C., Tasan, M., and Roth, F. P. (2009). Next generation software for functional trend analysis. J. Gerontol. 25, 3043–3044. doi: 10.1093/bioinformatics/btp498
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bolstad, B. M., Irizarry, R. A., Astrand, M., and Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on varianceand bias. Bioinformatics 19, 185–193. doi: 10.1093/bioinformatics/19.2.185
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boulay, G., Dubuissez, M., Van Rechem, C., Forget, A., Helin, K., Ayrault, O., et al. (2012). Hypermethylated in Cancer 1 (HIC1) Recruits Polycomb Repressive Complex 2 (PRC2) to a subset of its target genes through interaction with Human Polycomb-like (hPCL) proteins. J. Biol. Chem. 287, 10509–10524. doi: 10.1074/jbc.M111.320234
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brill, M. S., Ninkovic, J., Winpenny, E., Hodge, R. D., Ozen, I., Yang, R., et al. (2009). Adult generation of glutamatergic olfactory bulb interneurons. Nat. Neurosci. 12, 1524–1533. doi: 10.1038/nn.2416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brill, M. S., Snapyan, M., Wohlfrom, H., Ninkovic, J., Jawerka, M., Mastick, G. S., et al. (2008). A Dlx2- and Pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J. Neurosci. 28, 6439–6452. doi: 10.1523/JNEUROSCI.0700-08.2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cameron, H. A., Woolley, C. S., McEwen, B. S., and Gould, E. (1993). Differ- entiation of newly born neurons and glia in the dentate gyrus of the adult rat. Neuroscience 56, 337–344. doi: 10.1016/0306-4522(93)90335-D
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christen, T., Bouzat, P., Pannetier, N., Coquery, N., Moisan, A., Lemasson, B., et al. (2014). Tissue oxygen saturation mapping with magnetic resonance imaging. J. Cereb. Blood Flow Metab. 34, 1550–1557. doi: 10.1038/jcbfm.2014.116
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Crespo, I., and Del Sol, A. (2013). A general strategy for cellular reprogramming: the importance of transcription factor cross-repression. Stem Cells 31, 2127–2135. doi: 10.1002/stem.1473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Crespo, I., Krishna, A., Le Béchec, A., and del Sol, A. (2013). Predicting missing expression values in gene regulatory networks using a discrete logic modeling optimization guided by network stable states. Nucleic Acids Res. 41:e8. doi: 10.1093/nar/gks785
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deisseroth, K., Singla, S., Toda, H., Monje, M., Palmer, T. D., and Malenka, RC. (2004). Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron 42, 535–552. doi: 10.1016/S0896-6273(04)00266-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doetsch, F., Caille, I., Lim, D. A., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (1999). Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716. doi: 10.1016/S0092-8674(00)80783-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Doetsch, F., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (1997). Cellular composition and three-dimensional organization of the sub- ventricular germinal zone in the adult mammalian brain. J. Neurosci. 17, 5046–5061.
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Engelen, E., Akinci, U., Bryne, J. C., Hou, J., Gontan, C., Moen, M., et al. (2011). Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 43, 607–611. doi: 10.1038/ng.825
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fournier, N. M., and Duman, R. S. (2012). Role of vascular endothelial growth factor in adult hippocampal neurogenesis: implications for the pathophysiology and treatment of depression. Behav. Brain Res. 227, 440–449. doi: 10.1016/j.bbr.2011.04.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fukuda, S., Kato, F., Tozuka, Y., Yamaguchi, M., Miyamoto, Y., and Hisatsune, T. (2003). Two distinct subpopulations of nestin-positive cells in adult mouse dentate gyrus. J. Neurosci. 23, 9357–9366.
Gage, F. H. (2000). Mammalian neural stem cells. Science 287, 1433–1438. doi: 10.1126/science.287.5457.1433
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gage, F. H., Kempermann, G., Palmer, T. D., Peterson, D. A., and Ray, J. (1998). Multipotent progenitor cells in the adult dentate gyrus. J. Neurobiol. 36, 249–266.
Gao, Z., Ure, K., Ables, J. L., Lagace, D. C., Nave, K. A., Goebbels, S., et al. (2009). Neurod1 is essential for the survival and maturation of adult-born neurons. Nat. Neurosci. 12, 1090–1092. doi: 10.1038/nn.2385
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gritti, A., Frolichsthal-Schoeller, P., Galli, R., Parati, E. A., Cova, L., Pagano, S. F., et al. (1999). Epidermal and fibroblast growth factors behave as mitogenic regulators for a single multipotent stem cell-like population from the subventricular region of the adult mouse forebrain. J. Neurosci. 19, 3287–3297.
Hamson, D. K., Wainwright, S. R., Taylor, J. R., Jones, B. A., Watson, N. V., and Galea, L. A. M. (2013). Androgens increase survival of adult-born neurons in the dentate gyrus by an androgen receptor-dependent mechanism in male rats. Endocrinology 154, 3294–3304. doi: 10.1210/en.2013-1129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, J. (2012). Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 26, 1010–1021. doi: 10.1101/gad.187336.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, S., Guo, Y.-P., May, G., and Enver, T. (2006). Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol. 305, 695–713. doi: 10.1016/j.ydbio.2007.02.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ikeya, M., Lee, S. M., Johnson, J. E., McMahon, A. P., and Takada, S. (1997). Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389, 966–970. doi: 10.1038/40146
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, K., Zhu, Y., Sun, Y., Mao, X. O., Xie, L., and Greenberg, D. A. (2002). Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 11946–11950. doi: 10.1073/pnas.182296499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kalyani, A., Hobson, K., and Rao, M. S. (1997). Neuroepithelial stem cells from the embryonic spinal cord: isolation, characterization, and clonal analysis. Dev. Biol. 186, 202–223. doi: 10.1006/dbio.1997.8592
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaneko, S., Li, G., Son, J., Xu, C.-F., Margueron, R., Neubert, T. A., et al. (2010). Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 24, 2615–2620. doi: 10.1101/gad.1983810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G., Jessberger, S., Steiner, B., and Kronenberg, G. (2004). Milestones of neuronal development in the adult hippocam- pus. Trends Neurosci. 27, 447–452. doi: 10.1016/j.tins.2004.05.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klempin, F., Marr, R. A., and Peterson, D. A. (2012). Modification of Pax6 and Olig2 expression in adult hippocampal neurogenesis selectively induces stem cell fate and alters both neuronal and glial populations. Stem Cells 30, 500–509. doi: 10.1002/stem.1005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohwi, M., Osumi, N., Rubenstein, J. L. R., and Alvarez-Buylla, A. (2005). Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J. Neurosci. 25, 6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kriegstein, A., and Alvarez-Buylla, A. (2009). The glial nature of em- bryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184. doi: 10.1146/annurev.neuro.051508.135600
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Léotoing, L., Manin, M., Monté, D., Baron, S., Communal, Y., Lours, C., et al. (2007). Crosstalk between androgen receptor and epidermal growth factor receptor-signalling pathways: a molecular switch for epithelial cell differentiation. J. Mol. Endocrinol. 39, 151–162. doi: 10.1677/JME-07-0021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lie, D. C., Colamarino, S. A., Song, H. J., Desire, L., Mira, H., Consiglio, A., et al. (2005). Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375. doi: 10.1038/nature04108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lledo, P. M., Merkle, F. T., and Alvarez-Buylla, A. (2008). Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 31, 392–400. doi: 10.1016/j.tins.2008.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lois, C., and Alvarez-Buylla, A. (1993). Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc. Natl. Acad. Sci. U.S.A. 90, 2074–2077. doi: 10.1073/pnas.90.5.2074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lois, C., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (1996). Chain migration of neuronal precursors. Science 271, 978–981. doi: 10.1126/science.271.5251.978
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, D. K., Jang, M. H., Guo, J. U., Kitabatake, Y., Chang, M. L., Pow-Anpongkul, N., et al. (2009). Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. doi: 10.1126/science.1166859
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, D. K., Ming, G. L., and Song, H. (2005). Glial influences on neural stem cell development: cellular niches for adult neurogenesis. Curr. Opin. Neurobiol. 15, 514–520. doi: 10.1016/j.conb.2005.08.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mackenzie, F., and Ruhrberg, C. (2012). Diverse roles for VEGF-A in the nervous system. Development 139, 1371–1380. doi: 10.1242/dev.072348
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mazumdar, J., O'Brien, W. T., Johnson, R. S., LaManna, J. C., Chavez, J. C., Klein, P. S., et al. (2010). O2 regulates stem cells through Wnt/β-catenin signalling. Nat. Cell Biol. 12, 1007–1013. doi: 10.1038/ncb2102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merkle, F. T., Mirzadeh, Z., and Alvarez-Buylla, A. (2007). Mosaic orga- nization of neural stem cells in the adult brain. Science 317, 381–384. doi: 10.1126/science.1144914
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merkle, F. T., Tramontin, A. D., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (2004). Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. U.S.A. 101, 17528–17532. doi: 10.1073/pnas.0407893101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, J. A., Nathanson, J., Franjic, D., Shim, S., Dalley, R. A., Shapouri, S., et al. (2013). Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development 140, 4633–4644. doi: 10.1242/dev.097212
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mira, H., Andreu, Z., Suh, H., Lie, D. C., Jessberger, S., Consiglio, A., et al. (2010). Signaling through BMPR-IA regulates quies- cence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89. doi: 10.1016/j.stem.2010.04.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miyata, T., Maeda, T., and Lee, J. E. (1999). NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13, 1647–1652. doi: 10.1101/gad.13.13.1647
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morrens, J., Van Den Broeck, W., and Kempermann, G. (2012). Glial cells in adult neurogenesis. Glia 60, 159–174. doi: 10.1002/glia.21247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nomura, T., Goritz, C., Catchpole, T., Henkemeyer, M., and Frisen, J. (2010). EphB signaling controls lineage plasticity of adult neural stem cell niche cells. Cell Stem Cell 7, 730–743. doi: 10.1016/j.stem.2010.11.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmer, T. D., Willhoite, A. R., and Gage, F. H. (2000). Vascular niche for adult hippocampal neurogenesis. J. Comp. Neurol. 425, 479–494. doi: 10.1002/1096-9861(20001002)425:4<479::AID-CNE2>3.0.CO;2-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pastrana, E., Cheng, L.-C., and Doetsch, F. (2009). Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc. Natl. Acad. Sci. U.S.A. 106, 6387–6392. doi: 10.1073/pnas.0810407106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M.-W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. doi: 10.1073/pnas.1002530107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raballo, R., Rhee, J., Lyn-Cook, R., Leckman, J. F., Schwartz, M. L., and Vaccarino, F. M. (2000). Basic fibroblast growth factor (FGF-2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012–5023.
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K.-Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncrnas of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rao, M. S. (1999). Multipotent and restricted precursors in the central nervous system. Anat. Rec. 257, 137–148.
Rao, M. S., and Mayer-Proschel, M. (1997). Glial-restricted precursors are derived from multipotent neuroepithelial stem cells. Dev. Biol. 188, 48–63. doi: 10.1006/dbio.1997.8597
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roeder, I., and Glauche, I. (2006). Towards an understanding of lineage specification in hematopoietic stem cells: a mathematical model for the interaction of transcription factors GATA-1 and PU.1. J. Theor. Biol. 241, 852–865. doi: 10.1016/j.jtbi.2006.01.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seri, B., Garciìa-Verdugo, J. M., McEwen, B. S., and Alvarez-Buylla, A. (2001). Astrocytes give rise to new neurons in the adult mammalian hippocampus. J. Neurosci. 21, 7153–7160.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, H., Stevens, C. F., and Gage, F. H. (2002). Astroglia induce neuro- genesis from adult neural stem cells. Nature 417, 39–44. doi: 10.1038/417039a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spassky, N., Merkle, F. T., Flames, N., Tramontin, A. D., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (2005). Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25, 10–18. doi: 10.1523/JNEUROSCI.1108-04.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sugino, K., Hempel, C. M., Miller, M. N., Hattox, A. M., Shapiro, P., Wu, C., et al. (2006). Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 9, 99–107. doi: 10.1038/nn1618
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suh, Y., Obernier, K., Hölzl-Wenig, G., Mandl, C., Herrmann, A., Wörner, K., et al. (2009). Interaction between DLX2 and EGFR regulates proliferation and neurogenesis of SVZ precursors. Mol. Cell. Neurosci. 42, 308–314. doi: 10.1016/j.mcn.2009.08.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tchorz, J. S., Tome, M., Cloëtta, D., Sivasankaran, B., Grzmil, M., Huber, R. M., et al. (2012). Constitutive Notch2 signaling in neural stem cells promotes tumorigenic features and astroglial lineage entry. Cell Death Dis. 3, e325. doi: 10.1038/cddis.2012.65
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Temple, S. (2001). The development of neural stem cells. Nature 414, 112–117. doi: 10.1038/35102174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tozuka, Y., Fukuda, S., Namba, T., Seki, T., and Hisatsune, T. (2005). GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron 47, 803–815. doi: 10.1016/j.neuron.2005.08.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tropepe, V., Sibilia, M., Ciruna, B. G., Rossant, J., Wagner, E. F., and van der Kooy, D. (1999). Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev. Biol. 208, 166–188. doi: 10.1006/dbio.1998.9192
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valenta, T., Lukas, J., Doubravska, L., Fafilek, B., and Korinek, V. (2006). HIC1 attenuates Wnt signaling by recruitment of TCF-4 and β-catenin to the nuclear bodies. EMBO J. 25, 2326–2337. doi: 10.1038/sj.emboj.7601147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warner-Schmidt, J. L., and Duman, R. S. (2007). VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc. Natl. Acad. Sci. U.S.A. 104, 4647–4652. doi: 10.1073/pnas.0610282104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wechsler-Reya, R. J., and Scott, M. P. (1999). Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 22, 103–114. doi: 10.1016/S0896-6273(00)80682-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Yalcin, S., Lee, D.-F., Yeh, T.-Y. J., Lee, S.-M., Su, J., et al. (2011). FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13, 1092–1099. doi: 10.1038/ncb2293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zinovyev, A., Viara, E., Calzone, L., and Barillot, E. (2008). BiNoM: a Cytoscape plugin for manipulating and analyzing biological networks. J. Gerontol. 24, 876–877. doi: 10.1093/bioinformatics/btm553
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: SGZ, SVZ, gene regulatory network, neurogenesis, neural stem cells, differentiation, NSC
Citation: Ertaylan G, Okawa S, Schwamborn JC and del Sol A (2014) Gene regulatory network analysis reveals differences in site-specific cell fate determination in mammalian brain. Front. Cell. Neurosci. 8:437. doi: 10.3389/fncel.2014.00437
Received: 13 October 2014; Accepted: 02 December 2014;
Published online: 18 December 2014.
Edited by:
Isabel Farinas, Universidad de Valencia, SpainReviewed by:
Alexander K. Murashov, East Carolina University, USATakumi Takizawa, Gunma University, Japan
Copyright © 2014 Ertaylan, Okawa, Schwamborn and del Sol. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio del Sol, Luxembourg Centre for Systems Biomedicine, 7 Avenue des Hauts Fourneaux, 4362 Belval, Luxembourg e-mail: antonio.delsol@uni.lu
 Gökhan Ertaylan
Gökhan Ertaylan Satoshi Okawa1
Satoshi Okawa1  Jens C. Schwamborn
Jens C. Schwamborn Antonio del Sol
Antonio del Sol