Not all that glitters is gold: off-target recombination in the somatostatin–IRES-Cre mouse line labels a subset of fast-spiking interneurons
- Department of Neurobiology and Anatomy and the Sensory Neuroscience Research Center, West Virginia University, Morgantown, WV, USA
Neurons in the mammalian brain are a highly diverse population with a complex assortment of electrophysiological, morphological and molecular properties, which has hindered efforts to classify them into genetically and functionally meaningful subtypes and to understand their various roles in the normal or pathological brain. Nowhere is this issue more acutely felt than in the study of inhibitory cortical interneurons, whose classification is still a source of contention and confusion (Markram et al., 2004; Petilla Interneuron Nomenclature et al., 2008; Defelipe et al., 2013). A major boon to investigators has been the development of mouse lines in which genetically defined subsets of interneurons express fluorescent proteins, allowing their identification and targeting during electrophysiological recordings or imaging experiments (Oliva et al., 2000; Meyer et al., 2002; Chattopadhyaya et al., 2004; Ma et al., 2006). More recently, investigators funded by the NIH Neuroscience Blueprint project have developed a toolbox of “driver” lines in which the Cre recombinase gene is inserted immediately downstream to genes that are known markers of specific interneuron subsets (Taniguchi et al., 2011). These Cre lines can be bred with mice carrying floxed genes, to generate cell type-specific knockouts of genes of interest. In addition, by breeding these lines with mice from a parallel toolbox of Cre reporter lines in which a gene coding sequence is inserted after a lox-STOP-lox cassette, or by transfecting them with viral vectors carrying similar constructs, investigators can induce cell-type specific expression of any gene of interest, from inert fluorescent proteins to calcium probes and light-activated ion channels or pumps (Madisen et al., 2010, 2012; Zariwala et al., 2012). While these technologies carry great promise and have already enabled some important findings, the rush to use them also carries considerable risk, if the relevant expression patterns are not fully characterized. A case in point is the somatostatin–IRES-Cre (SOM-Cre) mouse line (Taniguchi et al., 2011), in which Cre expression was targeted to cells containing the neuropeptide somatostatin (SOM). In the cerebral cortex, SOM-containing neurons are a well-studied population of dendritic-targeting inhibitory interneurons (Ma et al., 2006; Silberberg and Markram, 2007; Fanselow et al., 2008; Tan et al., 2008; Ma et al., 2010; Fino and Yuste, 2011). The SOM-Cre line has already been used in several high-profile studies, and in most of these the authors tacitly assumed—but did not validate—that Cre-mediated recombination was restricted to SOM interneurons (Adesnik et al., 2012; Gentet et al., 2012; Lee et al., 2012; Wilson et al., 2012; Chiu et al., 2013; Kvitsiani et al., 2013; Xu et al., 2013). We found, however, that 6–10% of neurons expressing a Cre-dependent reporter in any given cortical layer were fast-spiking/ parvalbumin-expressing (FS/PV) interneurons, a subtype quite distinct from SOM interneurons in electrophysiological etc., morphological and molecular properties (Rudy et al., 2011) [Note that there is another SOM-Cre line reported in the literature (Lovett-Barron et al., 2012), which we did not test].
Our experiments complied with all relevant institutional and federal animal use guidelines and regulations and were approved by the West Virginia University Institutional Animal Care and Use Committee, and the methods have been previously published (Hu et al., 2011). We crossed SOM-Cre males with females of the Ai14 reporter line (Madisen et al., 2010), to generate double transgenic progeny in which SOM interneurons express td-Tomato, a red fluorescent protein (RFP). We refer to such double transgenics as “SOM-RFP mice.” We conducted whole-cell recordings in brain slices prepared from the somatosensory cortex of SOM-RFP mice, and were surprised to find that 18% (20/112) of RFP-expressing neurons in cortical layers 3 through 5 exhibited an electrophysiological “fingerprint” typical of FS interneurons and clearly distinct from that of SOM interneurons (Beierlein et al., 2003; Hu et al., 2011) (Figure 1A). To verify the subclass identity objectively, we developed a simple non-linear classifier based on three electrophysiological parameters: spike width at half height (SWHH), after-hyperpolarization (AHP) and adaptation ratio (AR), measured as previously described (Ma et al., 2006). Each cell was tested for three conditions: SWHH < 0.26 ms, AHP ≥ 14.5 mV and AR > 0.56, and was classified as FS if at least 2 conditions were true and as SOM otherwise. We first tested this classifier on a “ground truth” dataset of 91 GFP-expressing interneurons from the X94 line (Ma et al., 2006); all but one were classified correctly as SOM interneurons. Since in the X94 line GFP-expressing SOM interneurons have quasi fast-spiking firing properties, separating them correctly from FS interneurons was a stringent test of the classifier. We then tested the classifier on a sample of 15 GFP-expressing interneurons from the G42 line (Chattopadhyaya et al., 2004) and 96 RFP-expressing interneurons from progeny of a PV-Cre line (Hippenmeyer et al., 2005); all 111 interneurons were classified correctly as FS. Finally, we applied the classifier to our sample of SOM-RFP interneurons; 21 interneurons were classified as FS, including the 20 initially identified. Recordings from progeny of SOM-Cre mice bred with a different reporter line [Ai39 (Madisen et al., 2012)] yielded a similar, though smaller percentage of FS interneurons (2 out of 19).
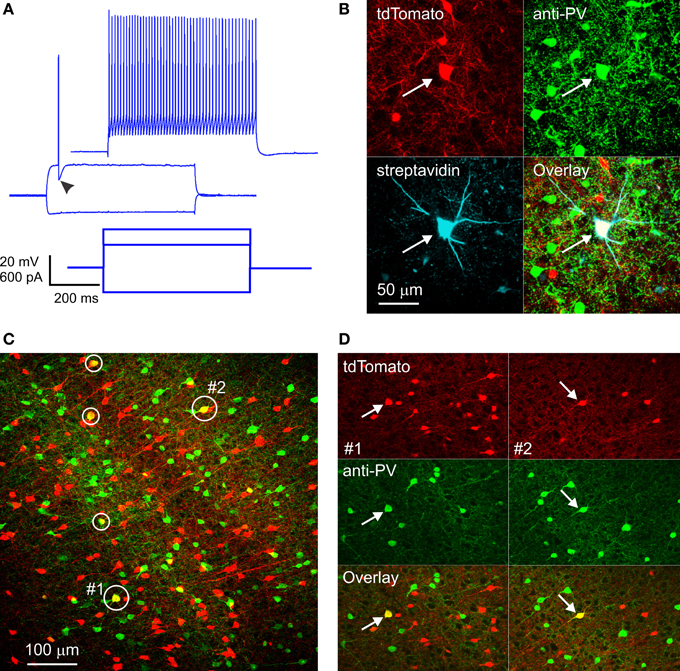
Figure 1. RFP-expressing fast-spiking interneurons in SOM-RFP mice. (A) Voltage responses (upper panels) of an RFP-expressing layer 5 barrel cortex neuron to the intracellular current steps shown in the lower panel. Note the pronounced after-hyperpolarization (arrowhead) and the non-adapting spike train, typical of FS interneurons. (B) The same neuron was found post-hoc to contain biocytin and RFP and to be immunopositive for PV. (C) A projection of a confocal stack from cortical area A1, showing five RFP-expressing cells immunopositive for PV (circled). The cells labeled #1 and #2 are shown in (D). (D) Single optical sections from subregions of the same field of view as in (C), in the vicinity of cell #1 (left panels, arrows) and cell #2 (right panels, arrows), separated into red and green channels.
FS interneurons are uniquely characterized by their PV expression (Kawaguchi and Kubota, 1993); we fixed a subset of slices in which we recorded RFP-expressing FS interneurons, stained them with an antibody to PV and with fluorescent streptavidin (to label the biocytin-filled neurons recorded from), and imaged them on a confocal microscope. Out of 7 RFP-expressing FS neurons recovered, five were immunopositive for PV (Figure 1B), substantiating their electrophysiological identification. The remaining two neurons were likely false negatives, due to wash-out of the cytoplasmic PV protein during the whole-cell recording.
Since electrophysiological sampling can be biased (for example, FS interneurons may be more likely to be targeted for recordings because they are typically larger than SOM cells), we sectioned fixed brains from four SOM-RFP mice and dually immunostained them against PV and SOM. Two brains were from third postnatal week pups (the age range used in our recording experiments), and two were from ~1 month old animals. We imaged sections representing five cortical areas (cingulate, M1, S1, A1, and V1) on a confocal microscope and counted RFP+, PV+, and RFP+/PV+ double-labeled cells in confocal stacks, verifying that double labeled cells were indeed so in single optical sections (Figures 1C,D). In total, about 18,000 RFP-expressing neurons were examined. Of these, on average 6% were immunopositive for PV. This number is a lower estimate, because weakly double-labeled cells (cells with fluorescence intensity in either channel weaker than the average intensity of single-labeled cells in the same optical section) were not counted as double-labeled. When averaged by layer, cortical area and age group, percentage of RFP+/PV+ double-labeled cells was highest in layer 4 of S1, reaching 14% in one animal and averaging 11.5 and 10% in the younger and older animals, respectively. Percentage of double-labeled cells was somewhat lower in other cortical areas and layers, with the lowest (2%) found in layers 2/3 of V1 of the younger animals, but in the older animals we observed 6–10% double labeled cells in all areas and layers except layers 2/3 of V1 (3%). We carefully examined all RFP+/PV+ double-labeled neurons for potential SOM expression, but only 0.4% of them appeared to be immunopositive for SOM. Finally, in the two brains with the best SOM immunostaining (one from each age range) we also examined RFP+/PV− cells for SOM immunolabeling, and found that 8% of RFP-expressing neurons, on average, were immunonegative for both SOM and PV. To what neuronal subtype these immunonegative neurons belonged remains to be determined; however, some of them could have been false-negative for PV, suggesting that the fraction of FS/PV interneurons in these animals could have been higher than our estimate, closer to the percentage observed in our electrophysiological recordings.
Our observation of RFP expression in FS interneurons cannot be explained by leaky expression of the RFP gene (i.e., expression in cells that did not undergo recombination) or by non-specific expression of the Cre gene (i.e., expression not under the control of the endogenous SOM promoter), because we would then expect to see widespread RFP expression in excitatory neurons, which are the majority cortical cell type. Thus, RFP expression must be under the same genetic control as the endogenous SOM gene; however, we found no SOM expression in the subset of double-labeled RFP+/PV+ neurons, consistent with previous studies which observed no overlap between SOM and PV protein expression in mouse and rat cortical interneurons (Gonchar and Burkhalter, 1997; Xu et al., 2010). This apparent paradox can be explained in two different ways. First, both SOM and FS/PV interneurons are born from embryonic progenitors in the medial ganglionic eminence (Batista-Brito and Fishell, 2009), and it is possible that a subset of progenitors (or of post-mitotic neuroblasts) destined to become FS/PV interneurons transiently express SOM. In the SOM-Cre mice these cells will also transiently express Cre recombinase, undergo Cre-mediated recombination and then express RFP for life, even after losing their SOM expression and attaining their mature FS phenotype. Alternatively, it has been reported that a subset of adult mouse cortical interneurons co-express PV and SOM mRNA (Lee et al., 2010). In the SOM-Cre line these neurons will co-express PV mRNA and the bicistronic SOM-IRES-Cre transcript. It is possible that the Cre transcript is translated into protein even though the SOM transcript is not, or that both transcripts are translated but at very low levels, sufficient for Cre-mediated recombination but not for detection of SOM protein.
The two alternative mechanisms above imply slightly different risks to investigators: the first implies that recombination in FS interneurons is expected when breeding SOM-Cre mice with Cre reporter lines, but not necessarily when transfecting adult neurons with viral vectors, while the second mechanism would result in off-target recombination regardless of the mode of delivery of the reporter construct. Either way, our findings underscore an important caveat for researchers using “subtype specific” mouse driver lines, including those in which the Cre coding sequence is presumed to be under the control of the endogenous gene promoter—these lines should be used with caution and with proper validation. Most studies using the SOM-Cre line appear to use it without validation; an exception is a recent study that tested recombination specificity and found that 5% of recombined neurons in the visual cortex were immunopositive for PV (Pfeffer et al., 2013). We found, by a lower estimate, 6–10% off-target recombination in FS interneurons in most cortical areas and layers. When using the SOM-Cre line to express optogenetic constructs for activation or silencing of SOM interneurons, this degree of contamination by FS interneurons can potentially affect the results and lead to erroneous conclusions. For example, a recent study Kvitsiani et al. (2013) observed that about one third of SOM-Cre interneurons tagged by Cre-dependent channelrhodopsin (ChR2) exhibited fast spike waveforms and high firing rates reminiscent of FS interneurons; but the possibility that some of these were actually ChR2-expressing FS interneurons was not considered. We submit that investigators ignoring the potential for off-target recombination when using the SOM-Cre line, or indeed any other Cre driver line that has not been fully characterized, are doing so at their own risk.
Acknowledgments
We thank Drs. Eric Tucker and Peter Mathers for helpful discussions and comments on the manuscript, Qingyan Wang and Cary Johnson for excellent technical support, and Kevin Hao for help with confocal imaging. SOM-Cre mice were generously provided by Dr. Josh Huang. The work was supported by NIH grant NS050437 from the National Institutes of Health. The authors of this manuscript are fully and solely responsible for its content.
References
Adesnik, H., Bruns, W., Taniguchi, H., Huang, Z. J., and Scanziani, M. (2012). A neural circuit for spatial summation in visual cortex. Nature 490, 226–231. doi: 10.1038/nature11526
Batista-Brito, R., and Fishell, G. (2009). Chapter 3 the developmental integration of cortical interneurons into a functional network. Curr. Top. Dev. Biol. 87, 81–118. doi: 10.1016/S0070-2153(09)01203-4
Beierlein, M., Gibson, J. R., and Connors, B. W. (2003). Two dynamically distinct inhibitory networks in layer 4 of the neocortex. J. Neurophysiol. 90, 2987–3000. doi: 10.1152/jn.00283.2003
Chattopadhyaya, B., Di Cristo, G., Higashiyama, H., Knott, G. W., Kuhlman, S. J., Welker, E., et al. (2004). Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. J. Neurosci. 24, 9598–9611. doi: 10.1523/JNEUROSCI.1851-04.2004
Chiu, C. Q., Lur, G., Morse, T. M., Carnevale, N. T., Ellis-Davies, G. C., and Higley, M. J. (2013). Compartmentalization of GABAergic inhibition by dendritic spines. Science 340, 759–762. doi: 10.1126/science.1234274
Defelipe, J., Lopez-Cruz, P. L., Benavides-Piccione, R., Bielza, C., Larranaga, P., Anderson, S., et al. (2013). New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 14, 202–216. doi: 10.1038/nrn3444
Fanselow, E. E., Richardson, K. A., and Connors, B. W. (2008). Selective, state-dependent activation of somatostatin-expressing inhibitory interneurons in mouse neocortex. J. Neurophysiol. 100, 2640–2652. doi: 10.1152/jn.90691.2008
Fino, E., and Yuste, R. (2011). Dense inhibitory connectivity in neocortex. Neuron 69, 1188–1203. doi: 10.1016/j.neuron.2011.02.025
Gentet, L. J., Kremer, Y., Taniguchi, H., Huang, Z. J., Staiger, J. F., and Petersen, C. C. (2012). Unique functional properties of somatostatin-expressing GABAergic neurons in mouse barrel cortex. Nat. Neurosci. 15, 607–612. doi: 10.1038/nn.3051
Gonchar, Y., and Burkhalter, A. (1997). Three distinct families of GABAergic neurons in rat visual cortex. Cereb. Cortex 7, 347–358. doi: 10.1093/cercor/7.4.347
Hippenmeyer, S., Vrieseling, E., Sigrist, M., Portmann, T., Laengle, C., Ladle, D. R., et al. (2005). A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 3:e159. doi: 10.1371/journal.pbio.0030159
Hu, H., Ma, Y., and Agmon, A. (2011). Submillisecond firing synchrony between different subtypes of cortical interneurons connected chemically but not electrically. J. Neurosci. 31, 3351–3361. doi: 10.1523/JNEUROSCI.4881-10.2011
Kawaguchi, Y., and Kubota, Y. (1993). Correlation of physiological subgroupings of nonpyramidal cells with parvalbumin- and calbindinD28k-immunoreactive neurons in layer V of rat frontal cortex. J. Neurophysiol. 70, 387–396.
Kvitsiani, D., Ranade, S., Hangya, B., Taniguchi, H., Huang, J. Z., and Kepecs, A. (2013). Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature 498, 363–366. doi: 10.1038/nature12176
Lee, S., Hjerling-Leffler, J., Zagha, E., Fishell, G., and Rudy, B. (2010). The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J. Neurosci. 30, 16796–16808. doi: 10.1523/JNEUROSCI.1869-10.2010
Lee, S. H., Kwan, A. C., Zhang, S., Phoumthipphavong, V., Flannery, J. G., Masmanidis, S. C., et al. (2012). Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature 488, 379–383. doi: 10.1038/nature11312
Lovett-Barron, M., Turi, G. F., Kaifosh, P., Lee, P. H., Bolze, F., Sun, X. H., et al. (2012). Regulation of neuronal input transformations by tunable dendritic inhibition. Nat. Neurosci. 15, 423–430. doi: 10.1038/nn.3024
Ma, W. P., Liu, B. H., Li, Y. T., Huang, Z. J., Zhang, L. I., and Tao, H. W. (2010). Visual representations by cortical somatostatin inhibitory neurons–selective but with weak and delayed responses. J. Neurosci. 30, 14371–14379. doi: 10.1523/JNEUROSCI.3248-10.2010
Ma, Y., Hu, H., Berrebi, A. S., Mathers, P. H., and Agmon, A. (2006). Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J. Neurosci. 26, 5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006
Madisen, L., Mao, T., Koch, H., Zhuo, J. M., Berenyi, A., Fujisawa, S., et al. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 15, 793–802. doi: 10.1038/nn.3078
Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. doi: 10.1038/nn.2467
Markram, H., Toledo-Rodriguez, M., Wang, Y., Gupta, A., Silberberg, G., and Wu, C. (2004). Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 5, 793–807. doi: 10.1038/nrn1519
Meyer, A. H., Katona, I., Blatow, M., Rozov, A., and Monyer, H. (2002). In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J. Neurosci. 22, 7055–7064.
Oliva, A. A. Jr., Jiang, M., Lam, T., Smith, K. L., and Swann, J. W. (2000). Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J. Neurosci. 20, 3354–3368.
Petilla Interneuron Nomenclature, G., Ascoli, G. A., Alonso-Nanclares, L., Anderson, S. A., Barrionuevo, G., Benavides-Piccione, R., et al. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568. doi: 10.1038/nrn2402
Pfeffer, C. K., Xue, M., He, M., Huang, Z. J., and Scanziani, M. (2013). Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat. Neurosci. 16, 1068–1076. doi: 10.1038/nn.3446
Rudy, B., Fishell, G., Lee, S., and Hjerling-Leffler, J. (2011). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev. Neurobiol. 71, 45–61. doi: 10.1002/dneu.20853
Silberberg, G., and Markram, H. (2007). Disynaptic inhibition between neocortical pyramidal cells mediated by Martinotti cells. Neuron 53, 735–746. doi: 10.1016/j.neuron.2007.02.012
Tan, Z., Hu, H., Huang, Z. J., and Agmon, A. (2008). Robust but delayed thalamocortical activation of dendritic-targeting inhibitory interneurons. Proc. Natl. Acad. Sci. U.S.A. 105, 2187–2192. doi: 10.1073/pnas.0710628105
Taniguchi, H., He, M., Wu, P., Kim, S., Paik, R., Sugino, K., et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. doi: 10.1016/j.neuron.2011.07.026
Wilson, N. R., Runyan, C. A., Wang, F. L., and Sur, M. (2012). Division and subtraction by distinct cortical inhibitory networks in vivo. Nature 488, 343–348. doi: 10.1038/nature11347
Xu, H., Jeong, H. Y., Tremblay, R., and Rudy, B. (2013). Neocortical somatostatin-expressing GABAergic interneurons disinhibit the thalamorecipient layer 4. Neuron 77, 155–167. doi: 10.1016/j.neuron.2012.11.004
Xu, X., Roby, K. D., and Callaway, E. M. (2010). Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J. Comp. Neurol. 518, 389–404. doi: 10.1002/cne.22229
Keywords: Cre mice, inhibitory interneurons, somatostatin, parvalbumin, fast spiking, Cre reporters
Citation: Hu H, Cavendish JZ and Agmon A (2013) Not all that glitters is gold: off-target recombination in the somatostatin–IRES-Cre mouse line labels a subset of fast-spiking interneurons. Front. Neural Circuits 7:195. doi: 10.3389/fncir.2013.00195
Received: 02 November 2013; Accepted: 25 November 2013;
Published online: 10 December 2013.
Edited by:
Yoshiyuki Kubota, National Institute for Physiological Sciences, JapanReviewed by:
Barry W. Connors, Brown University, USACopyright © 2013 Hu, Cavendish and Agmon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: aric.agmon@gmail.com
 Hang Hu
Hang Hu John Z. Cavendish
John Z. Cavendish Ariel Agmon
Ariel Agmon