Brain-wide imaging of neurons in action
- Department of Psychiatry, University of California San Diego, La Jolla, CA, USA
A commentary on
Whole-brain functional imaging at cellular resolution using light-sheet
by Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M., and Keller, P. J. (2013). Nat. Methods. 10, 413–420. doi: 10.1038/nmeth.2434
Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light
by Schrödel, T., Prevedel, R., Aumayr, K., Zimmer, M., and Vaziri, A. (2013). Nat. Methods. 10, 1013–1020. doi: 10.1038/nmeth.2637
Fully comprehending the brain as an information-processing organ will involve analysis of both the physical and algorithmic components of the system. The physical substrates comprising the brain, including neurophysiology, ion gradients, neurotransmitter release, and the molecules involved in action potential firing have been studied since Hodgkin and Huxley developed the squid giant axon model in 1952 Hodgkin et al. (1952). In order to understand how complex properties like response to stimulus, cognition, and emotion are derived from simple physical processes like an action potential, the algorithms of neural circuitry need to be described (Power et al., 2011). Mapping whole brain neural networks (Figure 1) and describing dynamic interactions during behavior is a daunting technological challenge that neuroimaging researchers are tackling with greater precision. In the March and September 2013 issues of Nature Methods, Ahrens et al. (2013) and Schrödel et al. (2013) describe whole-brain functional imaging at the single-neuron level in two model organisms adapting two fluorescence microscopy techniques.
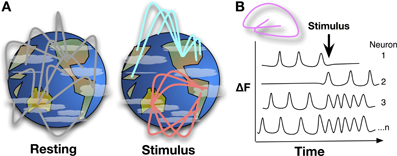
Figure 1. (A) Mapping whole brain neural activity is a daunting task, it involves determining how interacting networks respond to stimuli, encode information, and change with development, experience, and aging. Global network interactions of the brain can be described like linkages across continents, with background signaling occurring during resting state, e.g., asynchronous networks as described in Ahrens et al. Upon stimulation, a pathway or node may be activated while suppressing others, changing the global signaling network during and shortly after stimulation. Whole-brain, single-cell resolution, monitoring of activity over time will help to map these networks and nodes. (B) Detecting Ca2+ bursts via fluorescence (δF) in individual neurons brain-wide shows how neuron actions are correlated and anti-correlated at resting state or during stimulus and behavior.
At the core of both reports is the use of genetically encoded Ca2+ reporters that fluoresce in the presence of high Ca2+ concentration. During an action potential of a neuron, Ca2+ is released into the cytoplasm and the Ca2+ is quickly returned to its compartments in the endoplasmic reticulum and extracellular space as the neuron restores resting potential. Thus a Ca2+ “bursts” marks a firing neuron. The genetically encoded reporters are derived from fusing the Ca2+-binding domain from calmodulin, another peptide domain, and a circularly permuted green fluorescent protein (GFP) whose fluorescence properties are modulated by the Ca2+ sensing interaction. These are termed GFP CalModulin proteins (GCaMP), which have been modified and optimized for Ca2+ responsiveness and fluorescence properties (Akerboom et al., 2012). While the GCaMPs have been used since 2001 for detecting Ca2+ activity in the roundworm (Nakai et al., 2001; Chen et al., 2013), Caenorhabditis elegans, Schrödel et al. (2013) developed a sculpted light technique for recording activity of the majority of head ganglion neurons in response to chemical stimulus with high temporal and spatial resolution. Ahrens et al. (2013) used laser scanning light-sheet microscopy of the albino zebrafish, species Danio rerio, to capture activity of over 80% of the neurons in the brain every 1.3 seconds and identified anticorrelated hindbrain oscillations (Ahrens et al., 2013).
The light-sculpted technique involves two-photon excitation in which the spectrum of a femtosecond laser pulse is temporally separated by a grating. Thus, the light is separated into its component wavelengths (hence, “sculpted”), and pulsed, rather than continuously illuminated, to the sample in such a manner that the spectral components of the pulse overlap in time and space only at the focus region with near diffraction-limited axial (i.e., z-plane) confinement. With this technique, they achieved a wide-field excitation area of 60 μm diameter with 1.9 μm axial confinement. Imaging a depth of 30 μm, they were able to essentially detect neuronal Ca2+ pulses in the entire volume of an 85,000 μm3 cylinder of the C. elegans brain 4–6 times per second. By further adapting the GCaMP with a nuclear localization signal, they were able to increase resolution to single-cell without sacrificing the ability to detect fast Ca2+ spikes.
The light-sheet microscopy technique described in Ahrens et al. (2013) allows for larger volume of detection in the zebrafish brain but with diminished time resolution. Using a laser to scan a 4 μm excitation beam vertically across the head of a live zebrafish, and the objective capturing emitted fluorescence from the active neurons. In 5 μm steps, with 30 ms scans per section, the Ca2+ dynamics of a 9.6 × 107 μm3 box is captured in 1.6 s.
Both technologies have the ability to nearly simultaneously detect neuronal firing across the whole brain of a model organism using genetically encoded, neuronally expressed, calcium detectors. The advantage of C. elegans and the sculpted light technique is the ability to demonstrate neuronal response to chemical stimulus with high time resolution and that all 302 neurons and 8000 synaptic connections have been mapped (White et al., 1986). On the other hand, the light-sheet technique records a larger volume and two orders of magnitude more neurons without the necessity of nuclear localized GCaMP to achieve single-cell resolution. Computational methods to analyze the activity of a large number of neurons (100,000 for the zebrafish) are being developed to derive meaning from these studies. Ahrens et al. (2013) did show anticorrelated oscillations during resting state. With modification and specialization of the GCaMP genetics, it will be possible to describe the functional networks involved in resting state activity, stimulus, information integration, and behavior. GCaMP expression limited to specific neural types like inhibitory, excitatory, or neurotransmitter-specific cells will begin to identify how regulatory networks and activation pathways are encoded in the brain architecture.
Using these techniques to probe behavior will help to establish basic principles of neural circuit function. They may also be used to analyze activity-dependent growth and function across the brain, or brain-wide neuronal activity through development and aging. Understanding how the activity of neural circuits are altered when exposed to drugs of abuse will also help us understand the dynamic processes of addiction and habit. We are beginning to understand that behavior, consciousness, and information processing function of the brain are derived from the retrieval and integration of information encoded in neurons and describing the network properties as they happen is an important first-step in mapping the brain. As future work is prompted by the international efforts of Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative (Insel et al., 2013), announced in April 2013, we should expect variations on these technologies to begin decoding algorithms of the complex network properties of brain function and mental health.
Erick T. Tatro is supported by the US National Institutes of Health grants R01MH94159, R03DA033849, P50DA026306, and R21DA036423.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M., and Keller, P. J. (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420. doi: 10.1038/nmeth.2434
Akerboom, J., Chen, T.-W., Wardill, T. J., Tian, L., Marvin, J. S., Mutlu, S., et al. (2012). Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012
Chen, T.-W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Hodgkin, A. L., Huxley, A. F., and Katz, B. (1952). Measurement of current-voltage relations in the membrane of the giant axon of loligo. J. Physiol. 116, 424–448.
Insel, T. R., Landis, S. C., and Collins, F. S. (2013). Research priorities. the NIH BRAIN initiative. Science 340, 687–688. doi: 10.1126/science.1239276
Nakai, J., Ohkura, M., and Imoto, K. (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141. doi: 10.1038/84397
Power, J. D., Cohen, A. L., Nelson, S. M., Wig, G. S., Barnes, K. A., Church, J. A., et al. (2011). Functional network organization of the human brain. Neuron 72, 665–678. doi: 10.1016/j.neuron.2011.09.006
Schrödel, T., Prevedel, R., Aumayr, K., Zimmer, M., and Vaziri, A. (2013). Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat. Methods 10, 1013–1020. doi: 10.1038/nmeth.2637
Keywords: whole brain imaging, C. elegans, R. danio, calcium sensor, two-photon imaging, GFP CalModulin proteins, light-sheet imaging, sculpted light imaging
Citation: Tatro ET (2014) Brain-wide imaging of neurons in action. Front. Neural Circuits 8:31. doi: 10.3389/fncir.2014.00031
Received: 27 February 2014; Paper pending published: 13 March 2014;
Accepted: 15 March 2014; Published online: 03 April 2014.
Edited by:
Mark A. Frye, University of California, Los Angeles, USAReviewed by:
Jonathan T. Pierce-Shimomura, University of Texas, USACopyright © 2014 Tatro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: etatro@ucsd.edu
 Erick T. Tatro
Erick T. Tatro