MicroRNAs in the pathophysiology and treatment of status epilepticus
- Department of Physiology and Medical Physics, Royal College of Surgeons in Ireland, Dublin, Ireland
MicroRNA (miRNA) are an important class of non-coding RNA which function as post-transcriptional regulators of gene expression in cells, repressing and fine-tuning protein output. Prolonged seizures (status epilepticus, SE) can cause damage to brain regions such as the hippocampus and result in cognitive deficits and the pathogenesis of epilepsy. Emerging work in animal models has found that SE produces select changes to miRNAs within the brain. Similar changes in over 20 miRNAs have been found in the hippocampus in two or more studies, suggesting conserved miRNA responses after SE. The miRNA changes that accompany SE are predicted to impact levels of multiple proteins involved in neuronal morphology and function, gliosis, neuroinflammation, and cell death. miRNA expression also displays select changes in the blood after SE, supporting blood genomic profiling as potential molecular biomarkers of seizure-damage or epileptogenesis. Intracerebral delivery of chemically modified antisense oligonucleotides (antagomirs) has been shown to have potent, specific and long-lasting effects on brain levels of miRNAs. Targeting miR-34a, miR-132 and miR-184 has been reported to alter seizure-induced neuronal death, whereas targeting miR-134 was neuroprotective, reduced seizure severity during status epilepticus and reduced the later emergence of recurrent spontaneous seizures. These studies support roles for miRNAs in the pathophysiology of status epilepticus and miRNAs may represent novel therapeutic targets to reduce brain injury and epileptogenesis.
Introduction
A prolonged, non-terminating seizure (status epilepticus, SE) is a neurological emergency that has potential to cause irreversible brain damage. Uncovering the molecular mechanisms by which seizures transition into an uninterrupted state and elucidating the downstream consequence of such seizures on the brain are important if we are to understand and improve treatment of this devastating condition. MicroRNA (miRNA) have recently been implicated in the pathophysiology of SE and their expressional responses, targets and mechanisms represent a new focus of research in this field with potential to better understand the condition, identify novel therapeutics, and develop diagnostic biomarkers.
Status Epilepticus
Seizures are the result of abnormal, synchronous discharges of groups of neurons in the brain. Most epileptic seizures are self-terminating, often ending within a minute or less (Chen and Wasterlain, 2006). This is thought to be due to homeostatic mechanisms including inactivation of ion channels, build up of the anticonvulsant adenosine within the extracellular space, the anti-excitatory effect of tissue acidosis, and other changes (Lado and Moshe, 2008; Loscher and Kohling, 2010). However, some seizures do not self-terminate. This can result in the development of SE, which is variously defined by duration, often as 30 min of continuous seizure activity or two or more seizures without complete recovery in between. SE can follow drug withdrawal in patients with epilepsy but also occurs due to a myriad of other factors including CNS infection (Tatum Iv et al., 2001). The molecular mechanisms underlying the transition from seizure to SE are poorly understood, but may involve loss of surface receptors for the inhibitory neurotransmitter γ-amino butyric acid (GABA; Wasterlain et al., 2009).
The threshold of impending SE is defined operationally as over 5 min of continuous seizure activity (Chen and Wasterlain, 2006). Such patients require urgent care. Current treatment is with anticonvulsants such as lorazepam or midazolam (intravenous or intramuscular) or certain anti-epileptic drugs including phenytoin (Rossetti and Lowenstein, 2011; Silbergleit et al., 2012). If SE persists, additional combinations may be necessary including intravenous pentobarbital or the anesthetic propofol (Rossetti and Lowenstein, 2011). There is recent clinical evidence supporting the use of the N-methyl-D-aspartate (NMDA) receptor antagonist ketamine (Gaspard et al., 2013; Synowiec et al., 2013).
Status epilepticus has the capability of causing profound brain damage. The central mechanism of seizure-induced neuronal injury is glutamate-mediated excitotoxicity but there is also an important contribution from apoptosis-associated signaling pathways (Meldrum, 1991; Fujikawa, 2006; Engel and Henshall, 2009). Status epilepticus also produces synaptic reorganization, gliosis, inflammation, blood-brain barrier (BBB) damage, and lasting changes to excitability (Coulter, 1999; Vezzani et al., 2011). Despite major progress, there remains a need to further improve our understanding of the pathophysiologic mechanisms of SE and explore novel approaches to treatment that may better interrupt SE (particularly pharmacoresistant SE) and prevent long-term deleterious consequences (e.g., provide neuroprotection and anti-epileptogenesis).
SE Triggers Large-Scale Changes in Expression of Protein-Coding Genes
Status epilepticus results in large-scale changes to expression of genes within affected brain regions such as the hippocampus. The most recent microarray analyses in animal models that featured genome-wide coverage found changes to over 1000 genes after SE (Gorter et al., 2006; Jimenez-Mateos et al., 2008; Lauren et al., 2010). Affected biological processes include metabolism, signaling, transport, immune response, transcriptional regulation, cytoskeleton, glial function, neuronal death, and extracellular matrix organization (Lukasiuk and Pitkanen, 2007; Wang et al., 2010; Pitkanen and Lukasiuk, 2011a). There has been recent progress in identifying transcription factors driving up- and down-regulation of protein-coding genes after SE, including Activating transcription factor 5 (ATF5; Torres-Peraza et al., 2013), CCAAT/enhancer-binding protein homologous protein (CHOP; Engel et al., 2013b), Neuron restrictive silencing factor (NRSF/RE1-silencing transcription factor; McClelland et al., 2011) and Nuclear factor erythroid 2-related factor 2 (Nrf2; Mazzuferi et al., 2013). Uncovering the regulatory mechanisms controlling translation of mRNA transcripts represents a largely unexplored aspect of the molecular pathophysiology of SE.
MicroRNA
MicroRNA represents a potentially critical post-transcriptional mechanism regulating protein levels after SE. miRNA are an endogenous class of small (~23 nt) non-coding RNA that function to regulate gene expression at a post-transcriptional level by targeting mRNAs and reducing protein production (Bartel, 2004). Biogenesis of miRNAs is a highly conserved process which begins with RNA pol II or III-dependent transcription of a primary transcript (pri-miRNA; Lee et al., 2004; Borchert et al., 2006). miRNAs can be transcribed as single units or as part of poly-cistronic miRNA “clusters”, such as miR-17~92 (He et al., 2005) and miR-379~410 (Seitz et al., 2004). The pri-miRNA is processed in the nucleus to a shorter hairpin by the Drosha microprocessor complex (Lee et al., 2003; Gregory et al., 2004). The resulting pre-miRNA is exported to the cytoplasm for further processing by the RNAase III enzyme Dicer. Cleavage of pre-miRNA by Dicer produces the mature miRNA duplex. One strand is selected and incorporated into the RNA-induced silencing complex (RISC) while the other strand is typically degraded.
How miRNAs Work
MiRNAs control protein output by binding to specific, complementary sequences in target mRNAs of protein-coding genes. MiRNA binding sites are most often found in the 3′untranslated region (UTR) but have also been identified at the 5′end and within the open reading frame (ORF; Bartel, 2009). In mammals, miRNAs usually do not have complete complementarity to the mRNA sequence and therefore do not trigger direct cleavage of the mRNA as occurs with the RNA interference pathway activated by short interfering RNAs (Krol et al., 2010). However, mRNA levels of targets are often reduced by miRNA targeting (Lim et al., 2005; Guo et al., 2010). Targeting involves a 7–8 nucleotide “seed” region within the 5′ end of the miRNA binding to the mRNA via Watson–Crick base-pairing, followed by a variable number of further binding sites (Bartel, 2009; Figure 1A). The molecular machinery driving this process is the RISC which is a multi-protein complex, comprising members of the argonaute family as well as GW182 proteins (Fabian et al., 2010). Ago2 is critical in loading the miRNA and bringing it together with the mRNA target. The effect of miRNA targeting of a mRNA can be inhibition of translation or deadenylation and subsequent degradation, or both (Fabian et al., 2010). RISCs containing miRNA and their targets may also be sequestered in processing (P) bodies, including at synapses, which is reversible, enabling later release of the mRNA for translation (Cougot et al., 2008; Saugstad, 2010).
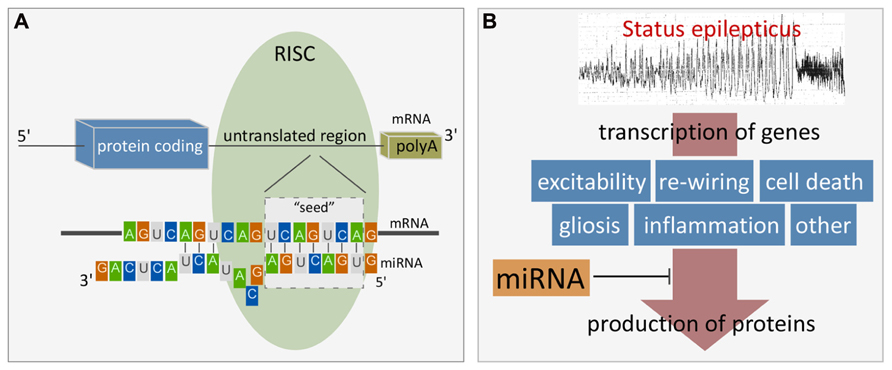
FIGURE 1. (A) Cartoon showing the site and mechanism of miRNA targeting to mRNA. Figure highlights the seven nucleotide “seed” region critical for miRNA binding to the mRNA target. Additional binding also determines specificity and potency. Alignment is facilitated by initial miRNA loading into the RISC which contains Ago2 and GW182 proteins. Binding typically occurs within the 3′UTR. The result is either degradation of the mRNA target or inhibition of translation. (B) Cartoon depicting scheme whereby miRNAs influence gene expression after SE. Status epilepticus results in transcriptional up-regulation of 100s of genes. miRNAs lie downstream of this and exert influence over protein production and as a result, influence post-injury outcomes such as repair, cell death and reorganization. Such miRNAs represent potential treatment targets to interrupt pathogenesis of damage and long-term consequences (e.g., hyperexcitability).
Impact of miRNAs On Protein Levels
There are over 1500 miRNAs in the human genome (miRBase v19). These are predicted to regulate the levels of at least one third of translated proteins, although over 60% of protein-coding genes are predicted to have miRNA regulatory sites (Friedman et al., 2009). Such extensive control is possible because a single miRNA may be capable of targeting perhaps 200 mRNAs. Not all mRNAs are targets for miRNA, however, and mRNA sequences with short 3′UTRs often lack miRNA binding sites meaning they are probably not significantly regulated in this manner. Conversely, mRNAs with tissue-specific expression or involved in developmental transitions tend to have longer 3′UTRs with more potential miRNA regulatory sites and these transcripts may be under potent miRNA control (Ebert and Sharp, 2012). MiRNAs also display cell and tissue-specific distribution (Lagos-Quintana et al., 2002; Sempere et al., 2004; Shao et al., 2010; He et al., 2012). In the brain, a large number of miRNAs display cell-specific enrichment that contributes to differentiation and distinguishes neurons from astrocytes, oligodendrocytes, and microglia (Jovicic et al., 2013).
The impact of a miRNA on protein levels of its targets is often only within the twofold range (Baek et al., 2008; Selbach et al., 2008). This may fall below the level capable of producing a phenotype, although under conditions of cell stress limited targeting may be enough to produce a larger effect. Multi-targeting of a single mRNA can produce much stronger effects, in the 10-fold range or targeting of multiple mRNAs within the same pathway (Ebert and Sharp, 2012). Nevertheless, for a given miRNA-mRNA pairing, these variables must be determined experimentally and not assumed based on bioinformatics predictions alone. A miRNA strongly predicted to target a particular mRNA may or may not be in a position to influence its translation. This is particularly important when considering the significance of miRNA changes reported in SE studies that analyzed pooled brain regions (e.g., whole hippocampus) containing multiple cell types expressing diverse transcripts and displaying varying degrees of vulnerability to damage after seizures.
Neuronal Activity and miRNA Expression
A key function of neuronal miRNAs is in regulating synaptic plasticity in response to neuronal activity (Schratt, 2009; McNeill and Van Vactor, 2012). Dendritic spines are major sites of excitatory communication in the brain and the molecular mechanisms regulating spine size are important determinants of learning, memory, and perhaps, seizure thresholds (Rochefort and Konnerth, 2012). In situ hybridization and other visualization techniques reveal enrichment of several miRNAs within dendrites, including miR-134 (Schratt et al., 2006) and miR-138 (Siegel et al., 2009) and neuronal depolarization or glutamate receptor activation results in changes to the expression of these miRNAs. Components of the RISC are also present within the synapto-dendritic compartment (Lugli et al., 2005) and certain dendritically localized miRNAs contain sequences within the precursor form that are identified by chaperone proteins which then shuttle them to the dendrite for later processing to the active mature form (Bicker et al., 2013). The targets of synapto-dendritic miRNAs are involved in dendritic morphogenesis and include p250GAP (by miR-132; Vo et al., 2005; Wayman et al., 2008), Lim kinase (Limk1), and Pumilio2 (by miR-134; Schratt et al., 2006; Fiore et al., 2009) and acyl protein thioesterase 1 (by miR-138; Siegel et al., 2009). This system enables prompt, localized control of neuronal morphology that is responsive to excitatory input (McNeill and Van Vactor, 2012).
Altered microRNA Expression Following SE
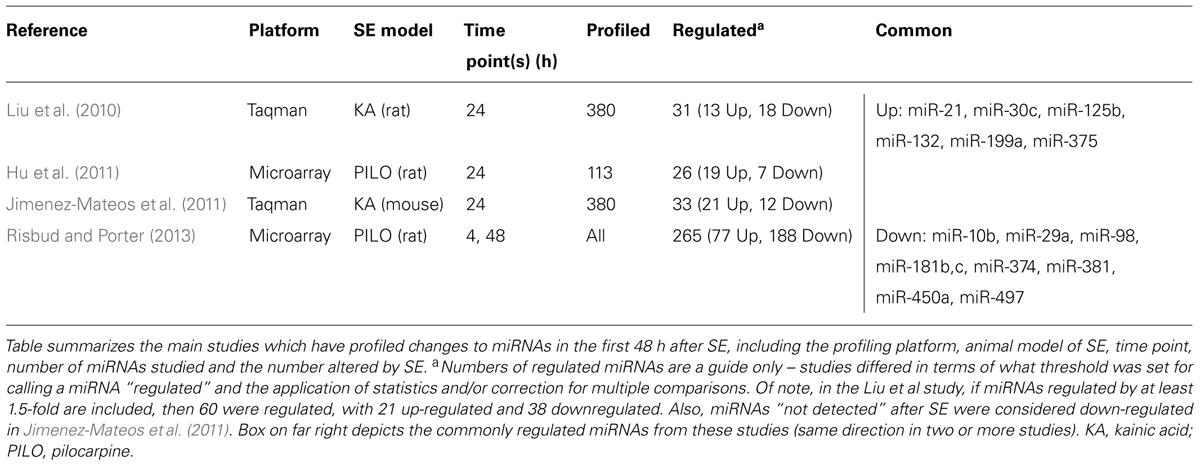
MiRNAs may serve important roles in SE by regulating protein production during and after seizures, thereby influencing hyperexcitability, injury, and repair responses (Figure 1B). Several studies have profiled miRNA responses in the hippocampus in the acute wake (≤48 h) of SE (Liu et al., 2010; Hu et al., 2011; Jimenez-Mateos et al., 2011; Risbud and Porter, 2013). Table 1 provides a summary of these studies, including the SE model and profiling platform. Combined, the four studies reveal increased expression levels of approximately 100 different miRNAs after SE while levels of about 200 miRNAs decreased. Although there are difficulties with cross-comparing data derived from different platforms and inter-study differences in the models, seizure severity and timing of sampling, a sub-set of miRNAs changed expression in at least two profiling studies. This includes six up-regulated miRNAs (miRNAs-21, -30c, -125b, -132, -199a, and -375) and nine down-regulated miRNAs (miRNAs-10b, -29a, -98, -181b,c, -374, -381, -450a, and -497; Table 1). In another study, miR-10b levels were found to be up-regulated after SE (Jimenez-Mateos et al., 2011). If studies that performed analyses of single miRNAs are also included then this list expands to include up-regulation of miR-34a, miR-134, and miR-155, and down-regulation of miR-9, miR-125a, miR-145, and miR-150 (Hu et al., 2012; Pichardo-Casas et al., 2012; Sano et al., 2012; Ashhab et al., 2013; Peng et al., 2013). Also, miR-146a has been consistently found to be upregulated after SE, albeit at later time points (Aronica et al., 2010; Hu et al., 2012; Omran et al., 2012). Thus, over 20 miRNAs appear to show conserved responses to SE. This is likely an under-estimate, however, since most profiling studies had limited spatio-temporal sampling and incomplete coverage of the rodent miRNAome. The roles of a selection of the profiling-identified miRNAs potentially relevant to seizures and brain function are reviewed briefly below. A potential caveat is that in most cases we do not have direct evidence that these miRNAs are functional – uploaded to the RISC – afterSE.
Conserved miRNAs Detected After SE in Profiling Studies
Among the four studies to have profiled miRNA responses to SE, miR-132 is the most consistently up-regulated miRNA, being identified in 3/4 of the studies (Hu et al., 2011; Jimenez-Mateos et al., 2011; Risbud and Porter, 2013). Up-regulation of miR-132 was also reported in two other studies that looked at individual miRNA responses to SE (Nudelman et al., 2010; Peng et al., 2013) and miR-132 is over-expressed in human temporal lobe epilepsy (Peng et al., 2013). Increased levels of miR-132 are also present in Ago2-eluted samples from the hippocampus after experimental SE, implying it is functional (Jimenez-Mateos et al., 2011).
The role of miR-132 in the brain is increasingly well understood. Expression of miR-132 is associated with synaptogenesis and a number of miR-132 targets are of potential relevance to the pathophysiology of SE (see Table 2). Overexpression of miR-132 in hippocampal neurons in culture was shown to cause neurite (Vo et al., 2005) and dendritic (Wayman et al., 2008) sprouting, and increase excitatory currents (Edbauer et al., 2010). Over-expression of miR-132 in vivo (~fivefold was achieved) resulted in an increase in spine density and was associated with a deficit in novel object recognition task (Hansen et al., 2010). In contrast, deletion of miR-132 in vivo is associated with decreased dendritic length and branching (Magill et al., 2010) and select defects in synaptic transmission (Pathania et al., 2012). Other predicted targets of miR-132 include MeCP2 (Lusardi et al., 2010), loss of which promotes cognitive deficits, hyperexcitability and seizures (Shahbazian et al., 2002). Targeting of acetylcholinesterase by miR-132 may increase cholinergic tone and influence excitability, hippocampal function, or inflammation (Friedman et al., 2007; Shaked et al., 2009; Shaltiel et al., 2013). The increased hippocampal miR-132 levels that accompany SE and are present in epilepsy may, therefore, influence neuronal morphology and contribute to hyperexcitability or cognitive dysfunction.
Much less is known about the remaining conserved miRNAs from the profiling studies. Most of the functional studies have been in cancer, where their targets have been linked to controlling apoptosis, invasiveness, and cell division. While this may fit with pathways expected to be regulated after SE, it is likely that some (or perhaps most) of the brain targets of these miRNAs in the setting of SE will be different.
Based on available functional studies and known targets, including members of the Bcl-2 family and p53 pathway, increased levels of miR-21, miR-125b, and down-regulation of miR-29a and miR-497 in SE would be expected to have an anti-apoptotic effect (Chan et al., 2005; Mott et al., 2007; Sathyan et al., 2007; Le et al., 2009; Park et al., 2009; Yin et al., 2010; Yadav et al., 2011; Amir et al., 2013). In contrast, down-regulation of miR-10b and miR-98 in SE would be predicted to have a pro-apoptotic effect based on their roles as “oncomirs” (Ma et al., 2007; Ozsait et al., 2010; Foley et al., 2011; Wang et al., 2011). These miRNAs may therefore be involved in the control of apoptosis-associated signaling and regulation of seizure-induced neuronal death (Bozzi et al., 2011; Henshall and Engel, 2013).
Members of the miR-181 family have been linked to promoting cell death (Shi et al., 2008) and may control expression of Bcl-2 family proteins (Ouyang et al., 2012). Astrocytes are particularly enriched in miR-181c, and reduced miR-181b and miR-181c levels promote astrocyte-derived cytokine responses (Hutchison et al., 2013). Thus, miR-181, like miR-146a (Iyer et al., 2012), may negatively regulate inflammatory responses in astrocytes after SE.
For the remaining miRNAs down-regulated in at least two profiling studies – miR-374, miR-381, miR-450a – there is little or no relevant experimental data beyond detection in some cancer models. Whether these represent novel miRNAs with roles in the pathogenesis of SE is uncertain but could be explored in future studies.
Control of microRNA Expression After SE
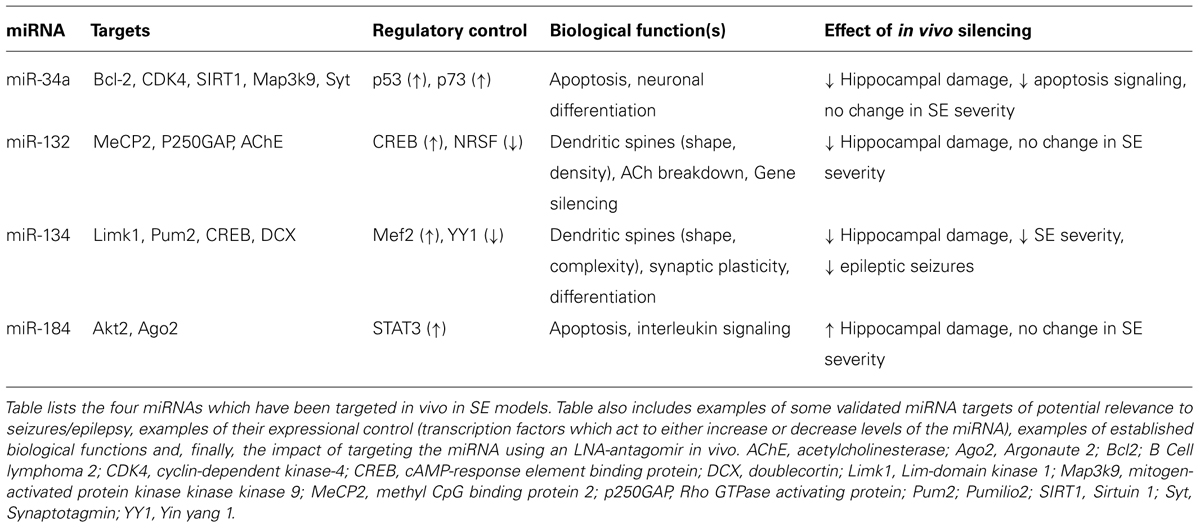
Although we have an increasingly expansive picture of which miRNAs change after SE and in what direction, we know little about the mechanisms controlling miRNA expression itself. No studies have directly explored miRNA regulatory control in SE, however the transcriptional control mechanism for some SE-regulated miRNAs is understood (Table 2). Expression of miR-34a is controlled by p53 (Chang et al., 2007; Raver-Shapira et al., 2007) as well as p73 (Agostini et al., 2011), and an inhibitor of p53 prevented miR-34a upregulation after SE (Sano et al., 2012). Expression of miR-132 is regulated by CREB, a stress-activated transcription factor that promotes neuronal survival (Lee et al., 2009). Some CREB-mediated effects may be pro-epileptogenic. Consistent with this, mice with decreased CREB levels develop fewer spontaneous seizures following pilocarpine-induced SE (Zhu et al., 2012). For miR-134, regulatory control is activity-dependent and driven by Mef2 (Fiore et al., 2009). miRNAs have also been identified under control of the transcriptional repressor NRSF. NRSF is implicated in the epigenetic silencing of multiple genes after SE and interference in NRSF function can recover expression and function of genes whose down-regulation is implicated in epileptogenesis (McClelland et al., 2011). MiR-124 is a known NRSF target that is involved in defining the neuronal phenotype. Levels of miR-124 have been reported to both increase (Peng et al., 2013) and decrease (Risbud and Porter, 2013) following SE in rats. These transcription factors and others may exert their influence on the post-SE molecular environment by modulating expression of miRNAs under their control. Their targeting represents potential approaches for modulating miRNA expression in SE.
Time-course studies have noted abrupt increases in miRNA expression, with rapid turn-on followed by restitution to baseline or lower levels after SE (McKiernan et al., 2012; Sano et al., 2012). This type of precise, dynamic response is reminiscent of miRNA responses during brain development (Krichevsky et al., 2003) and after other CNS insults (Lusardi et al., 2010), and supports tight transcriptional control of the spatiotemporal induction of miRNAs. This has implications for studies where only a single time point has been used because in the absence of a complete time course, erroneous conclusions may be drawn about the full response of a miRNA to SE.
Therapeutic miRNA Targeting in SE
Targeting miRNAs for therapeutic benefit is gaining increasing attention in multiple fields (Brown and Naldini, 2009; Stenvang et al., 2012). If miRNAs exert significant influence over processes involved in either seizure generation or the pathophysiological consequences of SE then miRNA targeting may have therapeutic potential. Delivery of a miRNA inhibitor or replenishment of an otherwise lost miRNA (e.g., via a miRNA mimic) could alter the excitability of the brain leading to less severe seizures or mitigate the downstream consequences leading to neuroprotection.
Brain-expressed miRNAs represent challenging targets for experimental and therapeutic modulation in vivo. First, miRNAs display cell-specific expression and tight transcriptional regulation along with their potential for multi-targeting, the control of which is still poorly understood. This means that delivery of an inhibitor or mimic may require careful timing and the means to control where it goes in the brain. Second, small molecules (e.g., <1000 Da) do not yet exist that selectively target miRNAs, although small molecules have been identified which alter miRNA biogenesis (Shan et al., 2008; Melo et al., 2011). A leading approach is to use antisense oligonucleotides (antagomirs; Stenvang et al., 2012). Modifications such as locked nucleic acid (LNA; Wahlestedt et al., 2000) make these potent and selective miRNA inhibitors and further modifications such as placement of cholesterol (Krutzfeldt et al., 2005) or other tags [e.g., penetratin peptide (Schratt et al., 2006)] facilitate cell entry. Studies show that for a miRNA to be inhibited the antagomir must be in several fold excess (Ebert et al., 2007). The mechanism by which antagomirs reduce miRNA function appears to differ depending on the chemistry of the molecules, and includes activation of degradation mechanisms and sequestration as a heteroduplex (Stenvang et al., 2012). Shorter sequences, so-called “tiny LNAs” which share common seed regions of miRNA families may enable blockade of multiple miRNA members and further potentiate targeting efficacy (Obad et al., 2011). A potentially attractive quality of antagomir targeting of miRNAs is prolonged suppression of the miRNA. Silencing of miRNAs by antagomirs has been reported to last several weeks in the periphery (Krutzfeldt et al., 2005; Elmen et al., 2008) and after injection into the brain (Jimenez-Mateos et al., 2012). Another challenge is that antagomirs do not cross an intact BBB (Krutzfeldt et al., 2005). To date, this has been overcome by direct intracerebroventricular microinjection of antagomirs (Jimenez-Mateos et al., 2011; Hu et al., 2012; Jimenez-Mateos et al., 2012; McKiernan et al., 2012; Sano et al., 2012). However, BBB integrity is disrupted by seizures (Marchi et al., 2012) therefore systemic injection may be sufficient to deliver antagomirs into the brain after SE. If the site of BBB disruption is limited to the area of pathologic activity then brain penetration may restrict delivery to the site of injury with minimal effects elsewhere in the brain. Alternatively, strategies can be used to temporarily breach the BBB (Campbell et al., 2008), or antagomirs can be given via intra-nasal delivery (Jimenez-Mateos et al., 2012) or encapsulated in a nanoparticle or exosome (Alvarez-Erviti et al., 2011).
Clinical trials are now underway using antagomirs for non-CNS conditions. Miravirsen targets miR-122, which is involved in hepatitis C virus replication and miravirsen was shown to be safe and effective in patients (Janssen et al., 2013). This raises the possibility of using miRNA-based therapeutics for other diseases, including CNS applications (Brown and Naldini, 2009; Stenvang et al., 2012).
miRNA Targeting In SE
Four miRNAs have been targeted in vivo in experimental models of SE using antagomirs (Table 2). The first to be targeted was miR-132 (Jimenez-Mateos et al., 2011). Intracerebroventricular injection of an LNA-modified antagomir targeting miR-132 reduced hippocampal levels of miR-132 when measured 24 h later in mice. Animals in which miR-132 had been silenced and then subjected to SE were found to have significantly less damage to the CA3 subfield of the hippocampus (Jimenez-Mateos et al., 2011). No effects of the antagomirs were reported on seizure severity. The mechanism of the protection is unknown and while miR-132 was confirmed in other experiments to be increased in the RISC, the mRNA targets in the RISC were not explored (Jimenez-Mateos et al., 2011). Whether the neuroprotection has any functional effects is unknown as cognitive tests have not yet been performed. However, a similar degree of neuroprotection in the same model was associated with fewer spontaneous seizures in long-term EEG monitoring studies (Jimenez-Mateos et al., 2008; Engel et al., 2010).
Silencing of two other miRNAs has been reported to alter seizure-induced neuronal death without affecting the severity of SE (Table 2). MiR-34a is another miRNA whose up-regulation has been reported in multiple models (Hu et al., 2012; Sano et al., 2012). Increased miR-34a levels promote apoptosis via suppressing anti-apoptotic proteins including Bcl-2 (Hermeking, 2010). However, the pro-apoptotic effect of miR-34a in neurons has been questioned (Agostini et al., 2011). More recently, miR-34a was shown to be a positive and negative regulator of neuronal differentiation, targeting synaptotagmin-1 (Agostini et al., 2011), and Numb1 (Fineberg et al., 2012). Inhibition of miR-34a using antagomir infusions into the ventricle of rodents was reported to reduce seizure-induced neuronal death in one study (Hu et al., 2012), but not in another (Sano et al., 2012). In contrast, targeting miR-184, a miRNA up-regulated by a protective episode of brief, non-harmful seizures, resulted in increased susceptibility to seizure-induced neuronal death in mice (McKiernan et al., 2012). Again, seizure severity was not affected. This supports miR-184 having protective effects against seizure-damage, although no candidate targets of this miRNA are obvious to explain this action (Table 2). Together with results of miR-132 and miR-34a, these studies reveal miRNAs as potential targets for modulating cell death after SE.
MiR-134 is another activity-regulated miRNA that has been found to be upregulated after SE in kainate and pilocarpine models of SE (Jimenez-Mateos et al., 2011, 2012; Peng et al., 2013). Levels of miR-134 were also confirmed to be increased in the RISC in Ago2 pull down experiments after SE and there were lower protein levels of two validated targets (Jimenez-Mateos et al., 2012). Targeting miR-134 using intracerebroventricular injections of LNA-modified antagomirs produced silencing of the miRNA lasting several weeks. When mice were injected with the antagomirs 24 h before SE, the resulting seizure severity was strongly reduced. Indeed, the seizure suppression was qualitatively similar to the effect of the anticonvulsant lorazepam in the same model (Jimenez-Mateos et al., 2012). Hippocampal damage in these antagomir pre-treated mice was also strongly reduced, although this may have been secondary to the anticonvulsant effect rather than a direct neuroprotective action.
In further experiments, the authors tested the effect of the antagomirs on the development of epilepsy. Antagomirs were injected 1 h after SE, ensuring the initial brain insult was similar between antagomir and scrambled-control SE mice. In EEG and video monitoring of the mice the antagomir-injected animals displayed ~90% fewer spontaneous seizures during the next month (Jimenez-Mateos et al., 2012). Seizure frequency remained reduced 2 months later indicating, presumably, a permanent protective effect. Chronic pathologic changes to the hippocampus including progressive neuronal loss, gliosis, and synaptic reorganization were also reduced (Jimenez-Mateos et al., 2012). The mechanism by which silencing miR-134 produces these strong anti-seizure effects is unknown, although in vitro experiments suggested they may be Limk1-dependent (Jimenez-Mateos et al., 2012). These findings suggest antagomirs targeting this miRNA could have neuroprotective and disease-modifying effects which might be a new therapeutic strategy for SE.
miRNAs as Biomarkers of Seizure-Damage and Epileptogenesis
MiRNAs have been recognized as having potential as non-invasive biomarkers (Scholer et al., 2010; de Planell-Saguer and Rodicio, 2011). Unique expression profiles of miRNAs have been reported in blood and other biofluids in animal models and patients and these may be useful as diagnostics, helping to discriminate between diseases with a similar clinical presentation, provide better stratification of patients, predicting disease course, and responses to therapy. Biofluid miRNA profiles could also have applications in toxicology and as markers of tissue damage. Part of their attraction lies with the conserved and widespread function of miRNAs in cell physiology and disease but there are also physico-chemical properties of miRNAs that make them suitable biomarkers. Unlike most other forms of RNA, miRNAs are remarkably stable in biofluids, remaining detectable in serum for weeks and they are also resistant to freeze-thaw and pH changes (Chen et al., 2008; McDonald et al., 2011; Blondal et al., 2013). The stability is attributable, at least in part, to binding to Ago2 (Arroyo et al., 2011; Turchinovich et al., 2011) and presence in membrane-enclosed circulating microvesicles such as exosomes (Hunter et al., 2008; Gallo et al., 2012).
There is an emerging consensus that biomarkers would be useful diagnostics in epilepsy (Pitkanen and Lukasiuk, 2011b; Simonato et al., 2012; Engel et al., 2013a). Molecular biomarkers of SE could be used to gauge insult severity, prognosis, and inform the choice of anticonvulsants or administration of anti-epileptogenic treatments, were they to become available. Evidence is emerging that miRNA signatures in biofluids can distinguish between different forms of neurological disease or acute brain injuries (Rong et al., 2011; Balakathiresan et al., 2012; Baraniskin et al., 2012; Haghikia et al., 2012). To date only a single study has looked at miRNA changes in the blood following SE (Liu et al., 2010). This revealed that kainate-induced seizures in rats produce unique miRNA expression profiles in blood that are different from those produced by other acute neurological injuries, including stroke, and hemorrhage (Liu et al., 2010). The study reported up-regulation of 15 miRNAs and decreased levels of 43 miRNAs in blood, although none passed correction for multiple comparisons (Liu et al., 2010). Nevertheless, this supports biofluid miRNA changes as a source of molecular biomarkers of SE. Notably, a number of the commonly regulated miRNAs identified in the hippocampus in profiling studies are found in serum and plasma, including miR-29a, miR-125b, and miR-375 (Blondal et al., 2013). Just as significantly, several miRNAs increased by SE in the hippocampus are not normally present in these biofluids, including miR-132 and miR-134 (Blondal et al., 2013), supporting their detection post-SE as a potential biomarker of seizures or injury.
Remaining Challenges
What are some of the future challenges? There is a need to identify the targets of seizure-regulated miRNAs. This could be achieved using techniques such as HITS-CLIP, whereby RISC-loaded RNAs are cross-linked to proteins followed by Ago2-immunoprecipitation and sequencing (Chi et al., 2009). Knowledge of the in vivo targets of miRNAs in SE models would also lead to better understanding of the mechanisms by which antagomirs produce their effects. The specificity of antagomirs in the brain has yet to be established, although some studies have looked at potential off-target effects (Jimenez-Mateos et al., 2011, 2012). Future studies should explore ways to deliver antagomirs via systemic routes while also including assessment of cognitive effects of miRNA silencing. This is particularly relevant for miR-132 and miR-134 because these directly regulate dendritic spines and small changes to levels of miRNA regulating dendritic spines have been found to produce behavioral phenotypes (Hansen et al., 2010).
There are several directions that could be taken to explore the potential of miRNAs as biomarkers in SE. For example, comparing profiles in different biofluids or between different models, and identifying miRNAs with predictive value for epileptogenesis. More clinical data are needed. For example, are miRNAs profiles in the brain or biofluids altered following SE in patients?
Several of the other commonly regulated miRNAs have yet to be targeted in animal models but likely represent focuses of the future. Combinations of miRNA targeting or delivery could offer ways to more completely block deleterious consequences of SE such as epileptogenesis. Experiments could also explore whether antagomirs can have effects on already established epilepsy. Can a disease-modifying effect be produced once epilepsy is established?
Clearly, miRNA functions are directly relevant to seizure thresholds, but clinical applications of miRNA-based therapeutics for SE would most likely be as disease-modifying post-treatments rather than acute anticonvulsants. This is because antagomirs take time to produce miRNA knockdown and measurable effects on the de-repressed targets. This means they are not realistic prospects for stopping SE, although perhaps there would be an application in super-refractory SE (Shorvon, 2011). Nevertheless, faster or more efficient targeting tools may emerge or indeed we may simply identify better miRNA targets or find ways to target the proteins under their control.
Summary
MiRNAs represent a major additional layer of gene expression control in SE, regulating protein levels within cells in the seizure-damaged brain. As functional studies begin to explore the importance of individual miRNAs in SE we are seeing influences on neuronal death, excitability, gliosis, and neuroinflammation. Many or even most processes dysregulated after SE may be controlled to some degree by miRNA expression. The arrival of miRNA-based inhibitors in clinical trials in other diseases herald translation to the clinic that may eventually also be possible for SE. Translation will be facilitated by focusing on the most critical miRNAs, identifying the molecular targets of miRNAs altered by SE and exploring antagomir delivery routes. Last, miRNAs represent an interesting class of biomarker that may have applications for tracking the severity of injury after SE and whether or not a patient is at risk of long-term consequences such as development or exacerbation of epilepsy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank Eva Jimenez-Mateos and Roger P. Simon for advice and helpful comments and would like to apologize to those authors whose relevant work was not cited here. The author also gratefully acknowledges funding from NINDS (R56 073714), Science Foundation Ireland (11/TIDA/B1988 and 08/IN1/B1875) and the Health Research Board (HRA-POR-2013-325).
References
Agostini, M., Tucci, P., Killick, R., Candi, E., Sayan, B. S., Rivetti Di Val Cervo, P., et al. (2011). Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. U.S.A. 108, 21093–21098. doi: 10.1073/pnas.1112061109
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345. doi: 10.1038/nbt.1807
Amir, S., Ma, A. H., Shi, X. B., Xue, L., Kung, H. J., and Devere White, R. W. (2013). Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS ONE 8:e61064. doi: 10.1371/journal.pone.0061064
Aronica, E., Fluiter, K., Iyer, A., Zurolo, E., Vreijling, J., Van Vliet, E. A., et al. (2010). Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 31, 1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x
Arroyo, J. D., Chevillet, J. R., Kroh, E. M., Ruf, I. K., Pritchard, C. C., Gibson, D. F., et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008. doi: 10.1073/pnas.1019055108
Ashhab, M. U., Omran, A., Kong, H., Gan, N., He, F., Peng, J., et al. (2013). Expressions of tumor necrosis factor alpha and microRNA-155 in immature rat model of status epilepticus and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 51, 950–958. doi: 10.1007/s12031-013-0013-9
Baek, D., Villen, J., Shin, C., Camargo, F. D., Gygi, S. P., and Bartel, D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64–71. doi: 10.1038/nature07242
Balakathiresan, N., Bhomia, M., Chandran, R., Chavko, M., Mccarron, R. M., and Maheshwari, R. K. (2012). MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J. Neurotrauma 29, 1379–1387. doi: 10.1089/neu.2011.2146
Baraniskin, A., Kuhnhenn, J., Schlegel, U., Schmiegel, W., Hahn, S., and Schroers, R. (2012). MicroRNAs in cerebrospinal fluid as biomarker for disease course monitoring in primary central nervous system lymphoma. J. Neurooncol. 109, 239–244. doi: 10.1007/s11060-012-0908-2
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bicker, S., Khudayberdiev, S., Weiss, K., Zocher, K., Baumeister, S., and Schratt, G. (2013). The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 27, 991–996. doi: 10.1101/gad.211243.112
Blondal, T., Jensby Nielsen, S., Baker, A., Andreasen, D., Mouritzen, P., Wrang Teilum, M., et al. (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. doi: 10.1016/j.ymeth.2012.09.015
Borchert, G. M., Lanier, W., and Davidson, B. L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13, 1097–1101. doi: 10.1038/nsmb1167
Bozzi, Y., Dunleavy, M., and Henshall, D. C. (2011). Cell signaling underlying epileptic behavior. Front. Behav. Neurosci. 5:45. doi: 10.3389/fnbeh.2011.00045
Brown, B. D., and Naldini, L. (2009). Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat. Rev. Genet. 10, 578–585. doi: 10.1038/nrg2628
Campbell, M., Kiang, A. S., Kenna, P. F., Kerskens, C., Blau, C., O’Dwyer, L., et al. (2008). RNAi-mediated reversible opening of the blood-brain barrier. J. Gene Med. 10, 930–947. doi: 10.1002/jgm.1211
Chan, J. A., Krichevsky, A. M., and Kosik, K. S. (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65, 6029–6033. doi: 10.1158/0008-5472.CAN-05-0137
Chang, T. C., Wentzel, E. A., Kent, O. A., Ramachandran, K., Mullendore, M., Lee, K. H., et al. (2007). Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 26, 745–752. doi: 10.1016/j.molcel.2007.05.010
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. doi: 10.1038/cr.2008.282
Chen, J. W., and Wasterlain, C. G. (2006). Status epilepticus: pathophysiology and management in adults. Lancet Neurol. 5, 246–256. doi: 10.1016/S1474-4422(06)70374-X
Chi, S. W., Zang, J. B., Mele, A., and Darnell, R. B. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. doi: 10.1038/nature08170
Cougot, N., Bhattacharyya, S. N., Tapia-Arancibia, L., Bordonne, R., Filipowicz, W., Bertrand, E., et al. (2008). Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 28, 13793–13804. doi: 10.1523/JNEUROSCI.4155-08.2008
Coulter, D. A. (1999). Chronic epileptogenic cellular alterations in the limbic system after status epilepticus. Epilepsia 40, S23–S33. doi: 10.1111/j.1528-1157.1999.tb00875.x
de Planell-Saguer, M., and Rodicio, M. C. (2011). Analytical aspects of microRNA in diagnostics: a review. Anal. Chim. Acta 699, 134–152. doi: 10.1016/j.aca.2011.05.025
Ebert, M. S., Neilson, J. R., and Sharp, P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726. doi: 10.1038/nmeth1079
Ebert, M. S., and Sharp, P. A. (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524. doi: 10.1016/j.cell.2012.04.005
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C. F., Seeburg, D. P., Batterton, M. N., et al. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384. doi: 10.1016/j.neuron.2010.01.005
Elmen, J., Lindow, M., Schutz, S., Lawrence, M., Petri, A., Obad, S., et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899. doi: 10.1038/nature06783
Engel, T., and Henshall, D. C. (2009). Apoptosis, Bcl-2 family proteins and caspases: the ABCs of seizure-damage and epileptogenesis? Int. J. Physiol. Pathophysiol. Pharmacol. 1, 97–115.
Engel, T., Murphy, B. M., Hatazaki, S., Jimenez-Mateos, E. M., Concannon, C. G., Woods, I., et al. (2010). Reduced hippocampal damage and epileptic seizures after status epilepticus in mice lacking proapoptotic Puma. FASEB J. 24, 853–861. doi: 10.1096/fj.09-145870
Engel, J. Jr., Pitkanen, A., Loeb, J. A., Edward Dudek, F., Bertram, E. H., Cole, A. J., et al. (2013a). Epilepsy biomarkers. Epilepsia 54(Suppl. 4), 61–69. doi: 10.1111/epi.12299
Engel, T., Sanz-Rodgriguez, A., Jimenez-Mateos, E. M., Concannon, C. G., Jimenez-Pacheco, A., Moran, C., et al. (2013b). CHOP regulates the p53-MDM2 axis and is required for neuronal survival after seizures. Brain 136, 577–592. doi: 10.1093/brain/aws337
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. doi: 10.1146/annurev-biochem-060308-103103
Fineberg, S. K., Datta, P., Stein, C. S., and Davidson, B. L. (2012). MiR-34a represses Numbl in murine neural progenitor cells and antagonizes neuronal differentiation. PLoS ONE 7:e38562. doi: 10.1371/journal.pone.0038562
Fiore, R., Khudayberdiev, S., Christensen, M., Siegel, G., Flavell, S. W., Kim, T. K., et al. (2009). Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 28, 697–710. doi: 10.1038/emboj.2009.10
Foley, N. H., Bray, I., Watters, K. M., Das, S., Bryan, K., Bernas, T., et al. (2011). MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 18, 1089–1098. doi: 10.1038/cdd.2010.172
Friedman, A., Behrens, C. J., and Heinemann, U. (2007). Cholinergic dysfunction in temporal lobe epilepsy. Epilepsia 48(Suppl. 5), 126–130. doi: 10.1111/j.1528-1167.2007.01300.x
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Fujikawa, D. G. (2006). “Neuroprotective strategies in status epilepticus,” in Status Epilepticus: Mechanisms and Management, eds C. G. Wasterlain and D. M. Treiman (Cambridge: MIT Press), 463–480.
Gallo, A., Tandon, M., Alevizos, I., and Illei, G. G. (2012). The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 7:e30679. doi: 10.1371/journal.pone.0030679
Gaspard, N., Foreman, B., Judd, L. M., Brenton, J. N., Nathan, B. R., Mccoy, B. M., et al. (2013). Intravenous ketamine for the treatment of refractory status epilepticus: a retrospective multicenter study. Epilepsia 54, 1498–1503. doi: 10.1111/epi.12247
Gorter, J. A., Van Vliet, E. A., Aronica, E., Breit, T., Rauwerda, H., Lopes Da Silva, F. H., et al. (2006). Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J. Neurosci. 26, 11083–11110. doi: 10.1523/JNEUROSCI.2766-06.2006
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240. doi: 10.1038/nature03120
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Haghikia, A., Hellwig, K., Baraniskin, A., Holzmann, A., Decard, B. F., Thum, T., et al. (2012). Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology 79, 2166–2170. doi: 10.1212/WNL.0b013e3182759621
Hansen, K. F., Sakamoto, K., Wayman, G. A., Impey, S., and Obrietan, K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5:e15497. doi: 10.1371/journal.pone.0015497
He, M., Liu, Y., Wang, X., Zhang, M. Q., Hannon, G. J., and Huang, Z. J. (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73, 35–48. doi: 10.1016/j.neuron.2011.11.010
He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828–833. doi: 10.1038/nature03552
Henshall, D. C., and Engel, T. (2013). Contribution of apoptosis-associated signaling pathways to epileptogenesis: lessons from Bcl-2 family knockouts. Front. Cell Neurosci. 7:110. doi: 10.3389/fncel.2013.00110.
Hermeking, H. (2010). The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199. doi: 10.1038/cdd.2009.56
Hu, K., Xie, Y. Y., Zhang, C., Ouyang, D. S., Long, H. Y., Sun, D. N., et al. (2012). MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 13:115. doi: 10.1186/1471-2202-13-115
Hu, K., Zhang, C., Long, L., Long, X., Feng, L., Li, Y., et al. (2011). Expression profile of microRNAs in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neurosci. Lett. 488, 252–257. doi: 10.1016/j.neulet.2010.11.040
Hunter, M. P., Ismail, N., Zhang, X., Aguda, B. D., Lee, E. J., Yu, L., et al. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3:e3694. doi: 10.1371/journal.pone.0003694
Hutchison, E. R., Kawamoto, E. M., Taub, D. D., Lal, A., Abdelmohsen, K., Zhang, Y., et al. (2013). Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 61, 1018–28. doi: 10.1002/glia.22483
Iyer, A., Zurolo, E., Prabowo, A., Fluiter, K., Spliet, W. G., Van Rijen, P. C., et al. (2012). MicroRNA-146a: a key regulator of astrocyte-mediated inflammatory response. PLoS ONE 7:e44789. doi: 10.1371/journal.pone.0044789
Janssen, H. L., Reesink, H. W., Lawitz, E. J., Zeuzem, S., Rodriguez-Torres, M., Patel, K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694. doi: 10.1056/NEJMoa1209026
Jimenez-Mateos, E. M., Bray, I., Sanz-Rodriguez, A., Engel, T., Mckiernan, R. C., Mouri, G., et al. (2011). miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 179, 2519–2532. doi: 10.1016/j.ajpath.2011.07.036
Jimenez-Mateos, E. M., Engel, T., Merino-Serrais, P., Mckiernan, R. C., Tanaka, K., Mouri, G., et al. (2012). Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 18, 1087–1094. doi: 10.1038/nm.2834
Jimenez-Mateos, E. M., Hatazaki, S., Johnson, M. B., Bellver-Estelles, C., Mouri, G., Bonner, C., et al. (2008). Hippocampal transcriptome after status epilepticus in mice rendered seizure damage-tolerant by epileptic preconditioning features suppressed calcium and neuronal excitability pathways. Neurobiol. Dis. 32, 442–453. doi: 10.1016/j.nbd.2008.08.008
Jovicic, A., Roshan, R., Moisoi, N., Pradervand, S., Moser, R., Pillai, B., et al. (2013). Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J. Neurosci. 33, 5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013
Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K., and Kosik, K. S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281. doi: 10.1261/rna.5980303
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. doi: 10.1038/nature04303
Lado, F. A., and Moshe, S. L. (2008). How do seizures stop? Epilepsia 49, 1651–1664. doi: 10.1111/j.1528-1167.2008.01669.x
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. doi: 10.1016/S0960-9822(02)00809-6
Lauren, H. B., Lopez-Picon, F. R., Brandt, A. M., Rios-Rojas, C. J., and Holopainen, I. E. (2010). Transcriptome analysis of the hippocampal CA1 pyramidal cell region after kainic acid-induced status epilepticus in juvenile rats. PLoS ONE 5:e10733. doi: 10.1371/journal.pone.0010733
Le, M. T., Teh, C., Shyh-Chang, N., Xie, H., Zhou, B., Korzh, V., et al. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862–876. doi: 10.1101/gad.1767609
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. doi: 10.1038/nature01957
Lee, B., Cao, R., Choi, Y. S., Cho, H. Y., Rhee, A. D., Hah, C. K., et al. (2009). The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J. Neurochem. 108, 1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. doi: 10.1038/nature03315
Liu, D. Z., Tian, Y., Ander, B. P., Xu, H., Stamova, B. S., Zhan, X., et al. (2010). Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 30, 92–101. doi: 10.1038/jcbfm.2009.186
Loscher, W., and Kohling, R. (2010). Functional, metabolic, and synaptic changes after seizures as potential targets for antiepileptic therapy. Epilepsy Behav. 19, 105–113. doi: 10.1016/j.yebeh.2010.06.035
Lugli, G., Larson, J., Martone, M. E., Jones, Y., and Smalheiser, N. R. (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905. doi: 10.1111/j.1471-4159.2005.03224.x
Lukasiuk, K., and Pitkanen, A. (2007). Gene and protein expression in experimental status epilepticus. Epilepsia 48(Suppl. 8), 28–32. doi: 10.1111/j.1528-1167.2007.01342.x
Lusardi, T. A., Farr, C. D., Faulkner, C. L., Pignataro, G., Yang, T., Lan, J., et al. (2010). Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J. Cereb. Blood Flow Metab. 30, 744–756. doi: 10.1038/jcbfm.2009.253
Ma, L., Teruya-Feldstein, J., and Weinberg, R. A. (2007). Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688. doi: 10.1038/nature06174
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387. doi: 10.1073/pnas.1015691107
Marchi, N., Granata, T., Ghosh, C., and Janigro, D. (2012). Blood-brain barrier dysfunction and epilepsy: pathophysiologic role and therapeutic approaches. Epilepsia 53, 1877–1886. doi: 10.1111/j.1528-1167.2012.03637.x
Mazzuferi, M., Kumar, G., Van Eyll, J., Danis, B., Foerch, P., and Kaminski, R. M. (2013). Nrf2 defense pathway: experimental evidence for its protective role in epilepsy. Ann. Neurol. doi: 10.1002/ana.23940 [Epub ahead of print].
McClelland, S., Flynn, C., Dube, C., Richichi, C., Zha, Q., Ghestem, A., et al. (2011). Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann. Neurol. 70, 454–464. doi: 10.1002/ana.22479
McDonald, J. S., Milosevic, D., Reddi, H. V., Grebe, S. K., and Algeciras-Schimnich, A. (2011). Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 57, 833–840. doi: 10.1373/clinchem.2010.157198
McKiernan, R. C., Jimenez-Mateos, E. M., Sano, T., Bray, I., Stallings, R. L., Simon, R. P., et al. (2012). Expression profiling the microRNA response to epileptic preconditioning identifies miR-184 as a modulator of seizure-induced neuronal death. Exp. Neurol. 237, 346–354. doi: 10.1016/j.expneurol.2012.06.029
McNeill, E., and Van Vactor, D. (2012). MicroRNAs shape the neuronal landscape. Neuron 75, 363–379. doi: 10.1016/j.neuron.2012.07.005
Meldrum, B. (1991). Excitotoxicity and epileptic brain damage. Epilepsy Res. 10, 55–61. doi: 10.1016/0920-1211(91)90095-W
Melo, S., Villanueva, A., Moutinho, C., Davalos, V., Spizzo, R., Ivan, C., et al. (2011). Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U.S.A. 108, 4394–4399. doi: 10.1073/pnas.1014720108
Mott, J. L., Kobayashi, S., Bronk, S. F., and Gores, G. J. (2007). mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26, 6133–6140. doi: 10.1038/sj.onc.1210436
Nudelman, A. S., Dirocco, D. P., Lambert, T. J., Garelick, M. G., Le, J., Nathanson, N. M., et al. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20, 492–498. doi: 10.1002/hipo.20646
Obad, S., Dos Santos, C. O., Petri, A., Heidenblad, M., Broom, O., Ruse, C., et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371–378. doi: 10.1038/ng.786
Omran, A., Peng, J., Zhang, C., Xiang, Q. L., Xue, J., Gan, N., et al. (2012). Interleukin-1beta and microRNA-146a in an immature rat model and children with mesial temporal lobe epilepsy. Epilepsia 53, 1215–1224. doi: 10.1111/j.1528-1167.2012.03540.x
Ouyang, Y. B., Lu, Y., Yue, S., and Giffard, R. G. (2012). miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 12, 213–219. doi: 10.1016/j.mito.2011.09.001
Ozsait, B., Komurcu-Bayrak, E., Levula, M., Erginel-Unaltuna, N., Kahonen, M., Rai, M., et al. (2010). Niemann-Pick type C fibroblasts have a distinct microRNA profile related to lipid metabolism and certain cellular components. Biochem. Biophys. Res. Commun. 403, 316–321. doi: 10.1016/j.bbrc.2010.11.026
Park, S. Y., Lee, J. H., Ha, M., Nam, J. W., and Kim, V. N. (2009). miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 16, 23–29. doi: 10.1038/nsmb.1533
Pathania, M., Torres-Reveron, J., Yan, L., Kimura, T., Lin, T. V., Gordon, V., et al. (2012). miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PLoS ONE 7:e38174. doi: 10.1371/journal.pone.0038174
Peng, J., Omran, A., Ashhab, M. U., Kong, H., Gan, N., He, F., et al. (2013). Expression Patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 50, 291–297. doi: 10.1007/s12031-013-9953-3
Pichardo-Casas, I., Goff, L. A., Swerdel, M. R., Athie, A., Davila, J., Ramos-Brossier, M., et al. (2012). Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Res. 1436, 20–33. doi: 10.1016/j.brainres.2011.12.001
Pitkanen, A., and Lukasiuk, K. (2011a). Mechanisms of epileptogenesis and potential treatment targets. Lancet Neurol. 10, 173–186. doi: 10.1016/S1474-4422(10)70310-0
Pitkanen, A., and Lukasiuk, K. (2011b). Molecular biomarkers of epileptogenesis. Biomark. Med. 5, 629–633. doi: 10.2217/bmm.11.67
Raver-Shapira, N., Marciano, E., Meiri, E., Spector, Y., Rosenfeld, N., Moskovits, N., et al. (2007). Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 26, 731–743. doi: 10.1016/j.molcel.2007.05.017
Risbud, R. M., and Porter, B. E. (2013). Changes in microRNA expression in the whole hippocampus and hippocampal synaptoneurosome fraction following pilocarpine induced status epilepticus. PLoS ONE 8:e53464. doi: 10.1371/journal.pone.0053464
Rochefort, N. L., and Konnerth, A. (2012). Dendritic spines: from structure to in vivo function. EMBO Rep. 13, 699–708. doi: 10.1038/embor.2012.102
Rong, H., Liu, T. B., Yang, K. J., Yang, H. C., Wu, D. H., Liao, C. P., et al. (2011). MicroRNA-134 plasma levels before and after treatment for bipolar mania. J. Psychiatr. Res. 45, 92–95. doi: 10.1016/j.jpsychires.2010.04.028
Rossetti, A. O., and Lowenstein, D. H. (2011). Management of refractory status epilepticus in adults: still more questions than answers. Lancet Neurol. 10, 922–930. doi: 10.1016/S1474-4422(11)70187-9
Sano, T., Reynolds, J. P., Jimenez-Mateos, E. M., Matsushima, S., Taki, W., and Henshall, D. C. (2012). MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 3, e287. doi: 10.1038/cddis.2012.23
Sathyan, P., Golden, H. B., and Miranda, R. C. (2007). Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 27, 8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007
Saugstad, J. A. (2010). MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J. Cereb. Blood Flow Metab. 30, 1564–1576. doi: 10.1038/jcbfm.2010.101
Scholer, N., Langer, C., Dohner, H., Buske, C., and Kuchenbauer, F. (2010). Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp. Hematol. 38, 1126–1130. doi: 10.1016/j.exphem.2010.10.004
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. doi: 10.1038/nature04367
Seitz, H., Royo, H., Bortolin, M. L., Lin, S. P., Ferguson-Smith, A. C., and Cavaille, J. (2004). A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14, 1741–1748. doi: 10.1101/gr.2743304
Selbach, M., Schwanhausser, B., Thierfelder, N., Fang, Z., Khanin, R., and Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63. doi: 10.1038/nature07228
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13. doi: 10.1186/gb-2004-5-3-r13
Shahbazian, M., Young, J., Yuva-Paylor, L., Spencer, C., Antalffy, B., Noebels, J., et al. (2002). Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35, 243–254. doi: 10.1016/S0896-6273(02)00768-7
Shaked, I., Meerson, A., Wolf, Y., Avni, R., Greenberg, D., Gilboa-Geffen, A., et al. (2009). MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973. doi: 10.1016/j.immuni.2009.09.019
Shaltiel, G., Hanan, M., Wolf, Y., Barbash, S., Kovalev, E., Shoham, S., et al. (2013). Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct. Funct. 218, 59–72. doi: 10.1007/s00429-011-0376-z
Shan, G., Li, Y., Zhang, J., Li, W., Szulwach, K. E., Duan, R., et al. (2008). A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 26, 933–940. doi: 10.1038/nbt.1481
Shao, N. Y., Hu, H. Y., Yan, Z., Xu, Y., Hu, H., Menzel, C., et al. (2010). Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics 11:409. doi: 10.1186/1471-2164-11-409
Shi, L., Cheng, Z., Zhang, J., Li, R., Zhao, P., Fu, Z., et al. (2008). hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 1236, 185–193. doi: 10.1016/j.brainres.2008.07.085
Shorvon, S. (2011). Super-refractory status epilepticus: an approach to therapy in this difficult clinical situation. Epilepsia 52(Suppl. 8), 53–56. doi: 10.1111/j.1528-1167.2011.03238.x
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716. doi: 10.1038/ncb1876
Silbergleit, R., Durkalski, V., Lowenstein, D., Conwit, R., Pancioli, A., Palesch, Y., et al. (2012). Intramuscular versus intravenous therapy for prehospital status epilepticus. N. Engl. J. Med. 366, 591–600. doi: 10.1056/NEJMoa1107494
Simonato, M., Loscher, W., Cole, A. J., Dudek, F. E., Engel, J. Jr., Kaminski, R. M., et al. (2012). Finding a better drug for epilepsy: preclinical screening strategies and experimental trial design. Epilepsia 53, 1860–1867. doi: 10.1111/j.1528-1167.2012.03541.x
Stenvang, J., Petri, A., Lindow, M., Obad, S., and Kauppinen, S. (2012). Inhibition of microRNA function by antimiR oligonucleotides. Silence 3, 1. doi: 10.1186/1758-907X-3-1
Synowiec, A. S., Singh, D. S., Yenugadhati, V., Valeriano, J. P., Schramke, C. J., and Kelly, K. M. (2013). Ketamine use in the treatment of refractory status epilepticus. Epilepsy Res. 105, 183–188. doi: 10.1016/j.eplepsyres.2013.01.007
Tatum Iv, W. O., French, J. A., Benbadis, S. R., and Kaplan, P. W. (2001). The etiology and diagnosis of status epilepticus. Epilepsy Behav. 2, 311–317. doi: 10.1006/ebeh.2001.0195
Torres-Peraza, J. F., Engel, T., Martin-Ibanez, R., Sanz-Rodriguez, A., Fernandez-Fernandez, M. R., Esgleas, M., et al. (2013). Protective neuronal induction of ATF5 in endoplasmic reticulum stress induced by status epilepticus. Brain 136, 1161–1176. doi: 10.1093/brain/awt044
Turchinovich, A., Weiz, L., Langheinz, A., and Burwinkel, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233. doi: 10.1093/nar/gkr254
Vezzani, A., French, J., Bartfai, T., and Baram, T. Z. (2011). The role of inflammation in epilepsy. Nat. Rev. Neurol. 7, 31–40. doi: 10.1038/nrneurol.2010.178
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431. doi: 10.1073/pnas.0508448102
Wahlestedt, C., Salmi, P., Good, L., Kela, J., Johnsson, T., Hokfelt, T., et al. (2000). Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 97, 5633–5638. doi: 10.1073/pnas.97.10.5633
Wang, Y. Y., Smith, P., Murphy, M., and Cook, M. (2010). Global expression profiling in epileptogenesis: does it add to the confusion? Brain Pathol. 20, 1–16. doi: 10.1111/j.1750-3639.2008.00254.x
Wang, S., Tang, Y., Cui, H., Zhao, X., Luo, X., Pan, W., et al. (2011). Let-7/miR-98 regulate Fas and Fas-mediated apoptosis. Genes Immun. 12, 149–154. doi: 10.1038/gene.2010.53
Wasterlain, C. G., Liu, H., Naylor, D. E., Thompson, K. W., Suchomelova, L., Niquet, J., et al. (2009). Molecular basis of self-sustaining seizures and pharmacoresistance during status epilepticus: the receptor trafficking hypothesis revisited. Epilepsia 50(Suppl. 12), 16–18. doi: 10.1111/j.1528-1167.2009.02375.x
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H. Y., et al. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098. doi: 10.1073/pnas.0803072105
Yadav, S., Pandey, A., Shukla, A., Talwelkar, S. S., Kumar, A., Pant, A. B., et al. (2011). miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J. Biol. Chem. 286, 37347–37357. doi: 10.1074/jbc.M111.235531
Yin, K. J., Deng, Z., Huang, H., Hamblin, M., Xie, C., Zhang, J., et al. (2010). miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol. Dis. 38, 17–26. doi: 10.1016/j.nbd.2009.12.021
Keywords: argonuate, dicer, epilepsy, epileptogenesis, hippocampal sclerosis, miRNA, non-coding RNA, RNA induced silencing complex
Citation: Henshall DC (2013) MicroRNAs in the pathophysiology and treatment of status epilepticus. Front. Mol. Neurosci. 6:37. doi: 10.3389/fnmol.2013.00037
Received: 28 June 2013; Accepted: 17 October 2013;
Published online: 12 November 2013.
Edited by:
Hermona Soreq, The Hebrew University of Jerusalem, IsraelReviewed by:
Kei Cho, University of Bristol, UKBjörn Spittau, Albert-Ludwigs-University Freiburg, Germany
Copyright © 2013 Henshall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David C. Henshall, Department of Physiology and Medical Physics, Royal College of Surgeons in Ireland, 123 St. Stephen’s Green, Dublin 2, Ireland e-mail: dhenshall@rcsi.ie
 David C. Henshall
David C. Henshall