- 1St. John’s Institute of Dermatology, Division of Genetics and Molecular Medicine, King’s College London, London, UK
- 2NIHR Biomedical Research Centre, Guy’s and St. Thomas’ Hospital, King’s College London, Guy’s Hospital, London, UK
- 3Clinical Oncology, Guy’s and St. Thomas’s NHS Foundation Trust, London, UK
The incidence of cutaneous melanoma has more than doubled over the last decades making it one of the fastest rising cancers worldwide. Improved awareness and early detection of malignant moles now permit earlier diagnosis aiming to decrease the likelihood of recurrence. However, it is difficult to identify those patients initially diagnosed with localized melanoma who subsequently develop metastatic disease. For this group, prognosis remains poor and clinical outcomes are variable and challenging to predict. Considerable efforts have focused on the search for novel prognostic tools, with numerous markers evaluated in the circulation and in tumor lesions. The most reliable predictors of patient outcome are the clinical and histological features of the primary tumor such as Breslow thickness, ulceration status, and mitotic rate. Elevated serum levels of the enzyme lactate dehydrogenase, likely to indicate active metastatic disease, are also routinely used to monitor patients. The emergence of novel immune and checkpoint antibody treatments for melanoma and increasing appreciation of key roles of the immune system in promoting or halting cancer progression have focused attention to immunological biomarkers. Validation of the most promising of these may have clinical applications in assisting prognosis, assessing endpoints in therapy, and monitoring responses during treatment.
Introduction
The incidence rates of melanoma are rising constantly, faster than for any other malignancy over the last two decades (1). Currently, melanoma has an incidence rate of 12.4/100,000 and a mortality rate of 3/100,000 in the UK (worldwide ASR 3.1/10000 and 0.8/10000, respectively) (2). Higher reported incidence rates partly reflect better surveillance and early diagnosis programs. Improvements in early detection could help identify patients at earlier, more curable disease stages, which may translate to enhanced overall survival (OS) rates (3). Currently, however, the number of patients dying of the disease is significantly greater than 1.3% diagnosed with incurable metastatic disease at presentation, implying a significant rate of disease progression in patients originally diagnosed with local disease (4–6). Treatment options for these patients are limited despite recently approved and emerging molecular targeted and immune therapies (7–9).
Classification and Staging
Cutaneous melanoma is classified into four types: (1) lentigomaligna melanomas with a papular or nodular structure; (2) superficial spreading (malignant) melanomas with large flat irregular pigmented lesions that grow laterally before invading the dermis; (3) nodular (malignant) melanomas with rapidly growing nodules that tend to ulcerate and bleed; (4) acral lentigo (malignant) melanomas mainly present at sites of friction, such as the sole, palm, or under nails.
Similarly, staging of melanoma is determined according to defined criteria (T: local extent of tumor; N: regional lymph-node involvement; M: distance metastasis-classification, Table 1). These parameters include the extent of dermal invasion (Breslow thickness and Clark’s level); the presence of ulceration; the mitotic rate of melanoma cells; and the presence of loco-regional or distant metastases. Although more advanced disease stage correlates with a worse prognosis, there have been reported cases of spontaneous melanoma lesion regressions and remissions of systemic disease, and this is perhaps attributed to immunological responses in these patients (4).
Loco-Regional Management
Localized disease in melanoma is currently treated by surgical excision with a 0.5 cm surrounding margin for tumor in situ, 1 cm margin for invasive melanoma of <2 mm Breslow thickness, and 2 cm margin for melanomas of ≥2 mm thickness (10, 11). Routine elective loco-regional lymphadenectomy has not been proven to be superior to observation alone (12). However, sentinel lymph-node biopsy has been suggested by several groups to be of benefit for patients with stage IB disease or higher, allowing for more accurate staging and therefore clearer information on prognosis (12). Radiotherapy could be used for primary lesions only if unresectable and for dissected lymph-node basins in patients at high risk of recurrence (13, 14).
Systemic Therapy
Conventional cytotoxic chemotherapy
Conventional cytotoxic drugs are largely ineffective in melanoma. Dacarbazine (DTIC), until 2011 the only standard therapy, has a low objective response rate (<20%) in metastatic disease (7). Numerous other chemotherapeutic agents including temozolomide, taxanes, fotemustine, and platin derivatives have been unsuccessfully trialed as single agents, combinations, or in conjunction with immune-modulators such as IFNα-2b or IL-2 at the cost of significant toxicity (14, 15).
Molecular targeted therapies
An activating mutation in the BRAFV600 gene on the long arm of chromosome 7 is found in approximately 50% of melanomas (16, 17). This change results in the constitutive activation of BRAF kinase, likely to promote RAS–RAF–MEK–ERK pathway-induced melanoma cell survival and proliferation. Vemurafinib is a pathway inhibitor drug that inhibits the mutated form of the BRAF kinase. Although originally designed to block the V600E (glutamate for valine) mutated form of BRAF, it can also inhibit other mutant forms such as the V600K (lysine for valine) (7, 18–20). In stage IV disease, vemurafenib has been shown to prolong progression-free survival (12.5 vs. 9.5 months) and OS (13.6 vs. 9.7 months) in comparison to dacarbazine (DTIC) (21). Vemurafenib, the first pathway inhibitor therapy approved for melanoma in 2011, represented the first significant change in outlook for stage IV disease in decades. However, following treatment, the majority of treated patients develop secondary resistance to the drug, thought to be associated with mechanisms overcoming the blockade of BRAF kinase inhibitors and with alternative activation of the MAPK kinase pathway (22). Treatment has also been linked to an enhanced risk of keratoacanthomas, squamous cell carcinomas (SCC), and the development of new melanomas (23–25) implying manipulation of other signaling cascades. Clinical trials are ongoing for BRAF inhibitors in the adjuvant setting and for combination therapies such as with BRAF and MEK inhibitor drugs. Recently, two trials reported that the combination of a BRAF inhibitor and a MEK inhibitor significantly improve progression-free survival (26, 27). Standard clinical evaluation now includes screening for the presence of the BRAF mutation in excised high risk and metastatic melanoma deposits. This test, now used for selecting patients to receive pathway inhibitor treatment, may be considered as a co-diagnostic tool for this treatment.
The identification of new mutations such as alternative BRAF, NRAS, c-kit, GNA11, and GNAQ mutations has led to the development of further kinase inhibitors and a number of these are being assessed pre-clinically and in clinical trials.
Immune therapies
The efficacy of immune therapies such as pegylated IFNα-2b and high dose IFNα-2b has been known for a number of years. These adjuvant treatments have shown improved disease-free survival (DFS), relapse-free survival (RFS) but only marginal improvements for OS (14, 28, 29). Their cost and toxicity, however, mean that their routine use is extremely limited. Likewise, high dose IL-2 was, until recently, the only effective therapy in stage IV disease, capable in rare cases of inducing prolonged remission (15). High toxicity confined treatment to a small subset of selected patients and in specialist clinical centers.
Ipilimumab is an IgG1 monoclonal antibody that recognizes the immune checkpoint cell surface molecule CTLA-4 on T cells. The antibody blocks the binding of the immunomodulatory molecule CTLA-4 to CD80/CD86 on the surface of antigen-presenting cells, preventing negative CTLA-4-mediated signals. This can result in universal activation of T cells, including some capable of specifically recognizing melanoma cells. Ipilimumab has shown efficacy in several clinical trials in untreated and treated metastatic patients in combination with a range of therapies such as peptide vaccines and as a mono-therapy (8, 30, 31). Some long-term responses are observed, albeit in a minority of patients and use of this agent is associated with autoimmune-like toxicities, most likely due to universal activation of CTLA-4-expressing T cells. The agent was approved by regulatory authorities in metastatic melanoma in 2011.
Based on the promise demonstrated by anti-CTLA-4 treatment, antibodies against other immune regulatory proteins such as PD-1, PD-L1, CD137, OX40, and CD40 are being investigated (32, 33). Administration of the anti-PD-1 antibody nivolumab demonstrated impressive objective responses in approximately 30% of individuals treated in a phase I trial (34). Combination therapy with nivolumab and ipilimumab in a dosing study demonstrated clinical safety and showed enhanced efficacy that appeared to be superior compared to data published on either antibody alone (35). Hamid et al. reported that the monoclonal antibody lambrolizumab (anti-PD-1) can induce sustained tumor regression in patients that had refractory disease despite ipilimumab treatment (36). Based on early clinical trial data, the anti-PD-1 antibody pembrolizumab was approved for the treatment of advanced melanoma in 2014 (37, 38).
The emergence of these new targeted and immune therapies, their success in only subsets of patient groups, as well as the need to assess patient benefit with combination treatments, all highlight the need to develop a range of tools, not only for prognosis but also to assist prediction and monitoring of treatment responses (39).
The Search for Biomarkers in Melanoma
The rapidly evolving clinical landscape of melanoma management and therapy mandates the search for new candidate biomarkers. Clinical trials traditionally rely on broad clinical groups and long-term objective outcomes, for example, assessment of OS in patients with previously treated stage IV melanoma. Patient stratification is partly served by co-diagnostic tools, such as in the case of the BRAF mutation test, which can help stratify patients who may receive pathway inhibitor drugs; however, these co-diagnostics do not always provide prognostic or predictive information. Biomarkers could revolutionize the process of drug development and those predictive of response could rationalize entry to trials for those patients most likely to benefit (40). New reliable biomarkers are thus required for all stages of melanoma management to assist with early detection, diagnosis, staging, prognosis as well as prediction, and monitoring of treatment response.
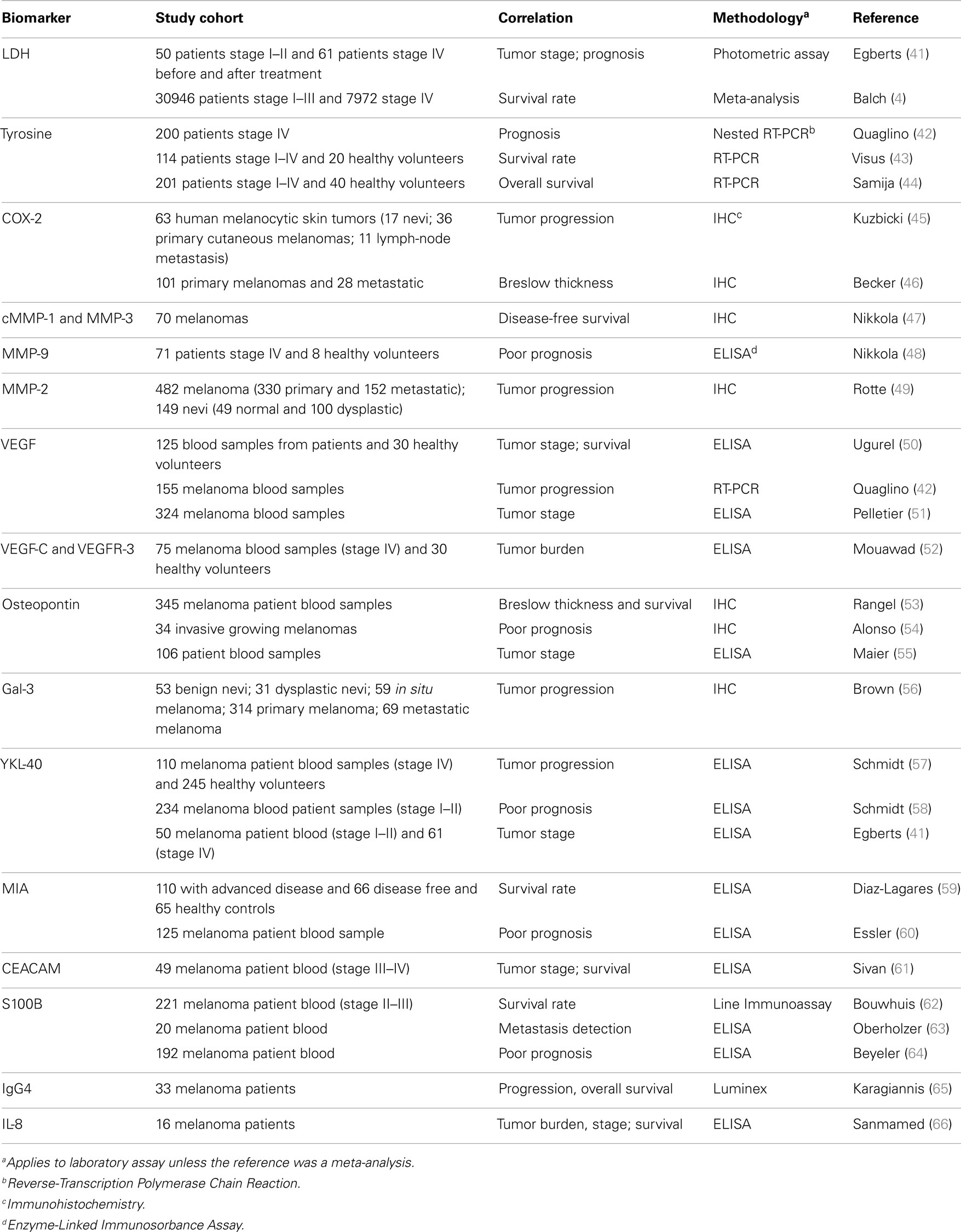
Disease-relevant candidate markers for melanoma have traditionally been derived from dissecting melanoma disease pathways. A number of candidates have been studied in sera and tumor specimens (Table 2) and some of these are discussed here. For this, a literature search using the search engine “Pubmed” was conducted with the following keywords and phrases: melanoma, malignant melanoma, metastatic melanoma, biomarker, serum/tissue biomarker in melanoma, melanoma therapy, melanoma immunotherapy, targeted therapy in melanoma, S100 melanoma, lactate dehydrogenase (LDH) melanoma, Braf melanoma, and diagnostic markers. All articles used in this review were peer reviewed. Markers only reported in a single article (with the exception of the immune inflammatory IL-8 and IgG4) were excluded, and markers reported in the literature only before the year 2000 were excluded.
Serum Biomarkers for Melanoma
Lactate dehydrogenase
The enzyme LDH is a clinical serological biomarker in melanoma. It remains to date the strongest prognostic indicator found to be elevated with tumor burden (67). LDH is mostly released upon cell damage or death, with both phenomena indicating higher tumor burden and disease progression. An increase in serum LDH is, however, not specific to malignancy, but also occurs in many other settings such as hemolysis, infection, infarction, and inflammation. Therefore, the positive predictive value in melanoma is limited by this false-positive rate (68). Recent studies have shown that LDH is less sensitive in early stage disease, but has negative predictive value for metastatic relapse (69–71). It is nonetheless at present a useful and clinically available tool for indication of tumor progression in patients at later disease stages and it is therefore incorporated in the TNM classification (Table 1).
Tyrosinase
Tyrosinase is part of the biosynthesis process of melanin and is constitutively expressed in melanocytes and melanoma cells. Tyrosinase mRNA levels are detectable in the blood of melanoma patients with advanced metastatic disease detected by nested RT-PCR (42, 43). Initial evaluations revealed that tyrosinase is an independent prognostic marker for tumor progression (42, 43, 72). Samija et al. demonstrated that tyrosine mRNA is associated with a decrease in OS (44). However, high variability in the levels of serum tyrosinase have been reported, most likely due to difficulties in sample processing and the transient presence of metastasizing tumor cells in the blood (43). Therefore, it is not surprising that several studies could not confirm a significant prognostic utility for tyrosinase (73). This includes a recent study that showed no differences in tyrosinase serum levels when comparing patients to healthy volunteers (74).
Vascular endothelial growth factor
Growth factors in combination with interleukins are major regulators of inflammatory conditions in the tumor microenvironment. Vascular endothelial growth factor (VEGF) is known to support tumor-associated angiogenesis, and to contribute to inflammatory conditions, which promote immunosuppression and redirection of effective anti-tumoral immunity. Ugurel et al. reported that VEGF was an independent prognostic marker for overall and progression-free survival in their cohort (125 stage I–IV patients) (50). Unfortunately, several subsequent studies could not confirm this finding. Although associations between VEGF and the disease were found, no correlation with disease progression was attained (51, 75).
Osteopontin
Osteopontin, a secreted integrin-binding glycol-phosphoprotein, has been described to reduce apoptosis, enhance tumor growth, and be a major component for the recruitment of tumor-promoting stromal cells from the bone marrow (76, 77). In a recent study, Maier et al. demonstrated that osteopontin in combination with S100B can help to differentiate patients who are likely to subsequently relapse and develop metastatic disease from those who do not (55). Yet, the presence of osteopontin is also associated with other medical conditions such as autoimmune diseases and this may translate to false-positive readouts in patients with melanoma.
YKL-40
YKL-40 is a glycoprotein secreted by many cells including cancer cells and by immune cells such as macrophages and activated neutrophils (78–80). The physiological functions of YKL-40 are yet not fully understood and reports of its functionality as an independent prognostic marker vary (41, 57–59). More importantly, immunomodulatory drugs such as IL-2 and IFN-α2b stimulate YKL-40 expression and therefore its use as a biomarker can yield false-negative readouts during treatment (81).
Melanoma-inhibitory activity protein
The small protein melanoma-inhibitory activity (MIA) is secreted by melanoma cells and is involved in cell–cell contact by interacting with the extracellular matrix. It is also thought that MIA promotes tumor cell invasion and metastasis (82). Equally, it was reported that MIA has a high sensitivity and specificity compared to other clinically relevant biomarkers such as LDH (59, 69). Elevated levels of MIA correlated with more advanced disease stages, poorer prognosis, and decreased DFS (59, 69, 77, 83). However, different studies using multivariate analysis demonstrated higher rates of false-positive readouts in women, suggesting an alternative source of MIA may exist and that this protein may participate in and be influenced by other biological processes unrelated to malignancy (60, 69, 84).
S100
The family of S100 proteins has been of special interest as diagnostic markers in melanoma over the last decade, since the expression and secretion of these proteins is much higher in malignant compared to healthy tissues (85–87). S100 proteins and in particular S100B were recently found to be elevated in the serum of melanoma patients with these elevated levels being associated with poorer prognosis (63, 64), DFS, and OS (88). In a more recent prognostic study of patients treated with IFN-α2b versus observation in stage II and III patients, investigators showed that S100B is associated with worse OS and distant metastasis-free survival (DMFS). Interestingly, S100B in patients’ sera correlated over time with disease progression, increasing further in parallel to increases in disease burden (62). False-positive results can occur after brain, liver, or renal injury as well as during infectious diseases (89–91). However, data acquired to date point to the merits of further evaluation of S100 as a potential clinical tool.
IL-8
Interleukin-8 (IL-8), also known as CXCL8, is a chemokine produced by malignant cells and associated with inflammatory responses; it can induce neutrophil chemotaxis and is also thought to be a potent promoter of angiogenesis. A recent study has shown that IL-8 correlated with tumor burden, stage, survival, and response to therapy such as BRAF inhibitors (66). These data show that IL-8, perhaps in combination with other chemokines and cytokines that participate in tumor inflammation, has potential as a prognostic marker and warrants further investigation.
Tissue-Specific Biomarkers for Melanoma
Tissue specific biomarkers are molecules demonstrated to be over-represented in cancer lesions, and may facilitate diagnosis, early detection, prognosis of disease progression, and patient stratification.
Cyclooxygenase-2
Cyclooxygenases (COX1–3) are a group of proteins that are important modulators in the human body, affecting essential pathways such as the catabolic metabolism. COX1–3 also converts arachidonic acid into prostaglandin. Of this group, COX-2 can be induced in tumor cells (92, 93). Becker et al. showed a correlation between COX-2 staining intensity and Breslow thickness in melanoma (46). Furthermore, Kuzbicki et al. reported a higher COX-2 staining intensity in melanoma lesions compared to benign nevi (45).
Galectin-3
The Galectin-3 molecule is mainly secreted by inflammatory cells and is associated with both tumor progression and metastasis in melanoma (94). However, using a multifactorial Cox regression analysis, Brown et al. showed an inverse association between galectin-3 with tumor size (thin tumors had more galectin-3) and improved OS (56).
Matrix metalloproteinases
Matrix metalloproteinases play an important role in remodeling the tumor tissue microenvironment as they are responsible for proteolytically breaking components of the local tissue architecture, promoting tissue remodeling, and facilitating tumor cell migration (95, 96). For these reasons, the proteins are over-represented in tumor tissues. Nikkola and colleagues showed that MMP-1- and MMP-3-positive melanoma metastases correlate with decreased DFS (47). Furthermore, Rotte et al. confirmed a higher expression of MMP-2 in melanoma when compared to normal and dysplastic nevi. In this study, the levels of MMP-2 expression positively correlated with tumor progression and worse survival (49). It is, however, noteworthy that this study was conducted using tumor tissue microarrays through the application of peroxidase developed with 3,3’-diaminobenzidine (DAB – a brown substrate), making the assessment of MMP-2 difficult to distinguish from melanoma cells in pigmented lesions.
Cell adhesion molecules
Cancer-associated cell adhesion proteins are proteins that adhere to different cells, show altered expression levels in cancer, and may participate in tumor cell migration, immune cell evasion, and angiogenesis (97). Molecules such as the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM-1) are known to foster interactions between somatic cells and immune cells. It was demonstrated that expression of CEACAM on tumor cells inhibits immune responses and leads to tumor progression (61). Investigation of CEACAM-1 expression suggested that it may be an independent factor to predict the risk of metastasis (98). Studies have also suggested associations between serum levels of cell adhesion molecules with the development of metastasis (99–101).
The chondroitin sulfate proteoglycan 4 as a biomarker
The chondroitin sulfate proteoglycan 4 (CSPG4), also known as the high molecular weight melanoma-associated antigen (HMW-MAA) or the melanoma-associated chondroitin sulfate proteoglycan (MCSP) is a glycoprotein–proteoglycan complex expressed on the surface of melanoma, glioma, neuroblastoma, certain breast carcinomas, and acute leukemias (102). CSPG4 is thought to play crucial roles in cell adhesion, melanoma migration, and metastasis (103). Although CSPG4 is over-expressed in over 80% of all melanomas, it is found at all disease stages and data so far have not yielded any correlations with disease progression (104). However, a recent study reported that a cytoplasmic form of the protein appears to correspond with response to treatment with a melanoma vaccine, indicating that CSPG4 may potentially be evaluated as a marker of response or progression (105).
The Immune Response as a Source of Biomarkers
Cancer development and progression is associated with dysregulated molecular pathways in tumor cells, but it is now accepted that it may also be affected by cross-talk with the immune response. The latter could lead either to immune cell activation and destruction of cancer cells or to immune response redirection and suppression to support cancer cell survival and metastasis (106). Different immune sentinels and molecules associated with tumor inflammation with either possible tumor-promoting or tumor-eradicating effects may be evaluated as potential biomarkers to predict clinical course or to monitor how patients respond to treatments (107). For example, the immune checkpoint molecule programed death-ligand 1 (PD-L1), which is also expressed in melanoma and other cancers has been described in preliminary studies to identify enhanced aggressive and invasive disease and to be an independent risk factor for worse clinical outcome (108). Furthermore, gene expression profiling analyses suggest that high expression of immunological parameters may predict responses to anti-CTLA-4 antibody therapies (109).
The potential value of immune sentinels in cancer could be exemplified by reports that monitoring tumor infiltrating lymphocytes (TIL) (immune score) may be more accurate prognostically than standard methods such as TNM stage (110, 111). Further analysis into the phenotype of the TIL revealed that while cytotoxic T cells may predict better patient outcomes and responses to checkpoint blockade therapies, high numbers of regulatory T cells (Treg: CD4+FoxP3+CD25high) among TIL, or the presence of myeloid-derived suppressor cells (MDSC) may correlate with poorer survival in many cancers and with lower clinical response rates to anti-CTLA-4 antibodies (109, 112–114). Although most clinical studies are focused on T cells, emerging evidence also points to a potential prognostic value for B cells in cancer. For instance, the presence of CD20+ cells among TIL correlated with a more favorable prognosis in ovarian cancer, while other studies point to immune regulatory roles for B cells (115). These findings suggest that the specific immune status of patients may be indicative of capacity to respond to and possibly restrict tumor progression.
Immunoglobulins: Antibody Class Bias and Tumor-Reactive Responses
Early disease stages are characterized by mostly minimal disease burden with no more than 1 × 106 tumor cells, which translate to very low or undetectable serum levels of potential biomarkers. Unless actively secreted, molecules released from these few cells may not be easily detectable in the circulation, thus monitoring such markers could be challenging.
B cells can undergo class switching to produce affinity matured antibodies in response to tissue damage and to the presence of antigens, including cancer antigens, and this may happen at early stages of cancer development. An individual B cell can produce 5,000–20,000 antibodies/min, and B cells can go through mitosis every 3 days, which can maintain or further enhance antibody production (116–118). The potential specificity of B cell and antibody responses to tumor antigens and amplification of the “tumor signal” may fulfill all three desirable attributes of a biomarker, namely detectability, sensitivity, and specificity. In support of this notion, the immunoglobulin G kappa chain has been shown to be a positive prognostic biomarker in breast cancer, non-small lung cancer, and colorectal cancer, not only for monitoring disease progression but also for predicting the response to neoadjuvant chemotherapy (115, 119, 120).
Until recently, the nature and functional significance of the humoral response in cancer were poorly understood. Moreover, whether and how the humoral response may be “immunoedited” by tumors has remained unknown. We have reported a tumor-reactive mature memory B cell compartment in the circulation of patients with melanoma. By isolating tumor-reactive antibody clones from patients’ mature B cells, we found that patient-derived antibodies can activate effector cells to destroy tumor cells ex vivo by recognizing tumor antigens on the surface of these cells (121). These findings suggest that host humoral immunity recognizes the presence of cancer and may potentially be activated. Antibodies, whether administered as therapeutics or expressed by host B cells, have the potential to induce potent anti-tumoral responses by sequestering effector cells to destroy tumor cells. However, since it is known that immunoediting in the tumor microenvironment may suppress the anti-tumoral functions of T cells, it would be reasonable to hypothesize that tumor-induced inflammatory conditions triggering suppression of T cell functions can lead to immunoediting of humoral responses as well and may substantially impair the potency of B cell and antibody functions against cancer.
Hints that immune escape mechanisms may operate to suppress tumor-reactive humoral responses came from our findings that this tumor-reactive memory B cell compartment appears more prominent at earlier disease stages and is reduced with disease progression. It has also been demonstrated that both the broad and the tumor-reactive mature memory B cell compartments are reduced in patients with advanced disease (122).
In melanoma, early studies indicated dysregulation and polarization of IgG antibody subclasses in the serum of patients with melanoma, but the biological relevance and functional implications of these findings were unclear (123). More recently, the presence of infiltrating IgG4+ plasma cells in hepatic cholangiocarcinomas (124) has been reported, pointing to diverted immunoglobulin class/subclass distribution in cancer. In support of these findings, we demonstrated that in melanoma tumor microenvironments B cells are polarized to favor production of IgG4, an antibody subclass with substantially restricted effector functions compared to well-known potent IgG1 antibodies and with capacity to impair effector functions of tumor-reactive IgG1 antibodies (65). This bias may occur in “alternative” Th2 cytokine environments in melanoma, where the presence of the immunoregulatory cytokine IL-10 can favor class switching to IgG4 and can enhance IgG4 production by class-switched B cells in the presence of IL-4 (125, 126). This biological relevance of IgG4 subclass antibodies in disease pathogenesis adds to the relatively newly described roles of IgG4 subclass in some inflammatory diseases (IgG4-related disease) (127).
Tumor antigen recognition may be another attribute of antibodies that could render them promising potential biomarkers. This may be facilitated by emerging screening platforms such as “immunosignaturing,” which is designed to identify reactive antibody signatures against panels of antigens, including those found in cancers. The “signatures” resulting from this process could allow for the identification of antibody reactivity patterns that may predict disease course. The potential for this approach has been demonstrated in the diagnosis of Alzheimer’s disease (128) and to detect antibodies against brain tumor antigens (129), but it may also be evaluated as a clinical biomarker tool in other cancers including melanoma.
Different facets of antibodies, including tumor specificity and reactivity, as well as antibody class and subclass bias may therefore be examined in the context of cancer including; as measures of the potency of adaptive immunity in cancer, as readouts of tumor-induced immunoediting and as powerful therapeutic agents to activate patient immune cells against cancer (35). Host humoral responses associated with tumor-immune escape may be linked to a higher risk of disease relapse and could point to readily detectable biomarkers, possibly at early disease stages.
Concluding Thoughts
Biomarkers that move beyond the current clinical pathological and radiological parameters, helping to identify those patients with early disease at high risk of relapse and guiding therapy choices for patients with metastatic disease, are still needed. A number of potential candidate biomarkers, including immunological markers warrant further evaluation in melanoma. Circulating or tumor-resident immune cells, including those associated with immunosuppressive forces in melanoma such as Treg MDSC, IgG4+ B cells, and also cytokines, chemokines, checkpoint molecules, and antibodies may point to yet unexplored biomarker signatures associated with particular clinical outcomes. Despite the considerable progress made in immune monitoring technologies, it has been challenging to draw accurate correlations between immunological parameters and clinical outcomes or patient responses to therapeutic agents. The reasons might include complex interactions between immune and tumor cells and the variable patient immune responses, making it difficult to account for all the interactions required for adequate prognostic readouts. Even when associations with melanoma are demonstrated, there is significant variability among patients, possibly reflecting the heterogeneity of individual tumors and of individual patient immune responses.
Other important challenges for biomarker discovery and validation remain. As most biomarkers are normal cellular or immunological proteins, they can be detected in both healthy and disease states, reducing their specificity. Additionally, many studies report relatively small sizes of patient samples, and this may indicate a limitation potentially preventing biomarker selection and routine use. Future directions may include larger cohorts assessed in multi-center studies and also assessing combinations of multiple putative markers to identify specific prognostic footprints. Finally, complex statistical analyses and comprehensive algorithms will be needed in order to integrate these multiple lower specificity biomarkers.
In conclusion, the potential of biomarkers to contribute toward better clinical monitoring and management will most likely be aided through further refinement of emerging design, statistical, bioinformatics, and analytical methods (130), as well as via our enhanced understanding of disease pathways and tumor-immune cell cross-talk.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London (Sophia N. Karagiannis and Panagiotis Karagiannis). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. Matthew Fittall is supported by an NIHR funded Integrated Academic Training Clinical Fellowship at King’s College London, UK. The authors acknowledge support from Cancer Research UK (C30122/A11527; C30122/A15774; Sophia N. Karagiannis); KCL Experimental Cancer Medicine Centre jointly funded by Cancer Research UK, the National Institute for Health Research, Welsh Assembly Government, HSC R&D Office for Northern Ireland and Chief Scientist Office, Scotland (Sophia N. Karagiannis); The Medical Research Council (MR/L023091/1; Sophia N. Karagiannis); and the CRUK/EPSRC/MRC/NIHRKCL/UCL Comprehensive Cancer Imaging Centre (C1519/A10331; Sophia N. Karagiannis).
References
1. Giblin AV, Thomas JM. Incidence, mortality and survival in cutaneous melanoma. J Plast Reconstr Aesthet Surg (2007) 60(1):32–40. doi:10.1016/j.bjps.2006.05.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. CRUK. Melanoma Statistics and Outlook. (2011). Available from: http://www.cancerhelp.org.uk/type/melanoma/treatment/melanoma-statistics-and-outlook
3. Verdecchia P, Angeli F, Repaci S, Mazzotta G, Gentile G, Reboldi G. Comparative assessment of angiotensin receptor blockers in different clinical settings. Vasc Health Risk Manag (2009) 5:939–48. doi:10.2147/VHRM.S7263
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol (2009) 27(36):6199–206. doi:10.1200/JCO.2009.23.4799
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Balch CM, Balch GC, Sharma RR. Identifying early melanomas at higher risk for metastases. J Clin Oncol (2012) 30(13):1406–7. doi:10.1200/JCO.2011.40.6983
6. Green AC, Baade P, Coory M, Aitken JF, Smithers M. Population-based 20-year survival among people diagnosed with thin melanomas in Queensland, Australia. J Clin Oncol (2012) 30(13):1462–7. doi:10.1200/JCO.2011.38.8561
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med (2011) 364(26):2507–16. doi:10.1056/NEJMoa1103782
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. doi:10.1056/NEJMoa1003466
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Hodi FS, Oble DA, Drappatz J, Velazquez EF, Ramaiya N, Ramakrishna N, et al. CTLA-4 blockade with ipilimumab induces significant clinical benefit in a female with melanoma metastases to the CNS. Nat Clin Pract Oncol (2008) 5(9):557–61. doi:10.1038/ncponc1183
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet (2005) 365(9460):687–701. doi:10.1016/S0140-6736(05)17951-3
11. Marsden JR, Newton-Bishop JA, Burrows L, Cook M, Corrie PG, Cox NH, et al. Revised U.K. guidelines for the management of cutaneous melanoma 2010. Br J Dermatol (2010) 163(2):238–56. doi:10.1111/j.1365-2133.2010.09883.x
12. Morton DL, Thompson JF, Cochran AJ, Mozzillo N, Elashoff R, Essner R, et al. Sentinel-node biopsy or nodal observation in melanoma. N Engl J Med (2006) 355(13):1307–17. doi:10.1056/NEJMoa060992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Burmeister BH, Henderson MA, Ainslie J, Fisher R, Di Iulio J, Smithers BM, et al. Adjuvant radiotherapy versus observation alone for patients at risk of lymph-node field relapse after therapeutic lymphadenectomy for melanoma: a randomised trial. Lancet Oncol (2012) 13(6):589–97. doi:10.1016/S1470-2045(12)70138-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Dummer R, Hauschild A, Guggenheim M, Keilholz U, Pentheroudakis G, Group EGW. Cutaneous melanoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol (2012) 23(Suppl 7):vii86–91. doi:10.1093/annonc/mds229
15. Petrella T, Quirt I, Verma S, Haynes AE, Charette M, Bak K, et al. Single-agent interleukin-2 in the treatment of metastatic melanoma: a systematic review. Cancer Treat Rev (2007) 33(5):484–96. doi:10.1016/j.ctrv.2007.04.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature (2002) 417(6892):949–54. doi:10.1038/nature00766
17. Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol (2011) 29(10):1239–46. doi:10.1200/JCO.2010.32.4327
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med (2012) 367(18):1694–703. doi:10.1056/NEJMoa1210093
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med (2012) 367(2):107–14. doi:10.1056/NEJMoa1203421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA (2008) 105(8):3041–6. doi:10.1073/pnas.0711741105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol (2014) 15(3):323–32. doi:10.1016/S1470-2045(14)70012-9
22. Khattak M, Fisher R, Turajlic S, Larkin J. Targeted therapy and immunotherapy in advanced melanoma: an evolving paradigm. Ther Adv Med Oncol (2013) 5(2):105–18. doi:10.1177/1758834012466280
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol (2012) 30(3):316–21. doi:10.1200/JCO.2011.36.7680
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med (2012) 366(3):207–15. doi:10.1056/NEJMoa1105358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Zimmer L, Hillen U, Livingstone E, Lacouture ME, Busam K, Carvajal RD, et al. Atypical melanocytic proliferations and new primary melanomas in patients with advanced melanoma undergoing selective BRAF inhibition. J Clin Oncol (2012) 30(19):2375–83. doi:10.1200/JCO.2011.41.1660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med (2014) 371(20):1877–88. doi:10.1056/NEJMoa1406037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med (2014) 371(20):1867–76. doi:10.1056/NEJMoa1408868
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Eggermont AM, Suciu S, Testori A, Santinami M, Kruit WH, Marsden J, et al. Long-term results of the randomized phase III trial EORTC 18991 of adjuvant therapy with pegylated interferon alfa-2b versus observation in resected stage III melanoma. J Clin Oncol (2012) 30(31):3810–8. doi:10.1200/JCO.2011.41.3799
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Mocellin S, Pasquali S, Rossi CR, Nitti D. Interferon alpha adjuvant therapy in patients with high-risk melanoma: a systematic review and meta-analysis. J Natl Cancer Inst (2010) 102(7):493–501. doi:10.1093/jnci/djq009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Robert C, Schadendorf D, Messina M, Hodi FS, O’Day S; Investigators MDX. Efficacy and safety of retreatment with ipilimumab in patients with pretreated advanced melanoma who progressed after initially achieving disease control. Clin Cancer Res (2013) 19(8):2232–9. doi:10.1158/1078-0432.CCR-12-3080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med (2011) 364(26):2517–26. doi:10.1056/NEJMoa1104621
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. McArthur GA, Ribas A. Targeting oncogenic drivers and the immune system in melanoma. J Clin Oncol (2013) 31(4):499–506. doi:10.1200/JCO.2012.45.5568
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Malas S, Harrasser M, Lacy KE, Karagiannis SN. Antibody therapies for melanoma: new and emerging opportunities to activate immunity (Review). Oncol Rep (2014) 32(3):875–86. doi:10.3892/or.2014.3275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med (2012) 366(26):2443–54. doi:10.1056/NEJMoa1200690
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med (2013) 369(2):122–33. doi:10.1056/NEJMoa1302369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med (2013) 369(2):134–44. doi:10.1056/NEJMoa1305133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Robert C, Ribas A, Wolchok JD, Hodi FS, Hamid O, Kefford R, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet (2014) 384(9948):1109–17. doi:10.1016/S0140-6736(14)60958-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Poole RM. Pembrolizumab: first global approval. Drugs (2014) 74(16):1973–81. doi:10.1007/s40265-014-0314-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Ilieva KM, Correa I, Josephs DH, Karagiannis P, Egbuniwe IU, Cafferkey MJ, et al. Effects of BRAF mutations and BRAF inhibition on immune responses to melanoma. Mol Cancer Ther (2014) 13(12):2769–83. doi:10.1158/1535-7163.MCT-14-0290
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Yap TA, Sandhu SK, Workman P, de Bono JS. Envisioning the future of early anticancer drug development. Nat Rev Cancer (2010) 10(7):514–23. doi:10.1038/nrc2870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Egberts F, Kotthoff EM, Gerdes S, Egberts JH, Weichenthal M, Hauschild A. Comparative study of YKL-40, S-100B and LDH as monitoring tools for Stage IV melanoma. Eur J Cancer (2012) 48(5):695–702. doi:10.1016/j.ejca.2011.08.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Quaglino P, Osella-Abate S, Cappello N, Ortoncelli M, Nardo T, Fierro MT, et al. Prognostic relevance of baseline and sequential peripheral blood tyrosinase expression in 200 consecutive advanced metastatic melanoma patients. Melanoma Res (2007) 17(2):75–82. doi:10.1097/CMR.0b013e328054c667
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Visus C, Andres R, Mayordomo JI, Martinez-Lorenzo MJ, Murillo L, Saez-Gutierrez B, et al. Prognostic role of circulating melanoma cells detected by reverse transcriptase-polymerase chain reaction for tyrosinase mRNA in patients with melanoma. Melanoma Res (2007) 17(2):83–9. doi:10.1097/CMR.0b013e3280a60878
44. Samija I, Lukac J, Maric-Brozic J, Buljan M, Alajbeg I, Kovacevic D, et al. Prognostic value of microphthalmia-associated transcription factor and tyrosinase as markers for circulating tumor cells detection in patients with melanoma. Melanoma Res (2010) 20(4):293–302. doi:10.1097/CMR.0b013e32833906b6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Kuzbicki L, Lange D, Chwirot BW. Cyclooxygenase-2 immunohistochemistry in human melanoma: differences between results obtained with different antibodies. Melanoma Res (2009) 19(5):294–300. doi:10.1097/CMR.0b013e32832e0bde
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Becker MR, Siegelin MD, Rompel R, Enk AH, Gaiser T. COX-2 expression in malignant melanoma: a novel prognostic marker? Melanoma Res (2009) 19(1):8–16. doi:10.1097/CMR.0b013e32831d7f52
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Kahari VM, Pyrhonen S. High expression levels of collagenase-1 and stromelysin-1 correlate with shorter disease-free survival in human metastatic melanoma. Int J Cancer (2002) 97(4):432–8. doi:10.1002/ijc.1636
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Nikkola J, Vihinen P, Vuoristo MS, Kellokumpu-Lehtinen P, Kahari VM, Pyrhonen S. High serum levels of matrix metalloproteinase-9 and matrix metalloproteinase-1 are associated with rapid progression in patients with metastatic melanoma. Clin Cancer Res (2005) 11(14):5158–66. doi:10.1158/1078-0432.CCR-04-2478
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Rotte A, Martinka M, Li G. MMP2 expression is a prognostic marker for primary melanoma patients. Cell Oncol (Dordr) (2012) 35(3):207–16. doi:10.1007/s13402-012-0080-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Ugurel S, Rappl G, Tilgen W, Reinhold U. Increased serum concentration of angiogenic factors in malignant melanoma patients correlates with tumor progression and survival. J Clin Oncol (2001) 19(2):577–83.
51. Pelletier F, Bermont L, Puzenat E, Blanc D, Cairey-Remonnay S, Mougin C, et al. Circulating vascular endothelial growth factor in cutaneous malignant melanoma. Br J Dermatol (2005) 152(4):685–9. doi:10.1111/j.1365-2133.2005.06507.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Mouawad RSC, Soubrane C, Khayat D. Prognostic relevance of pretreatment soluble vascular endothelial growth factors (A,C,D) and their receptors (R1, R2 and R3) in advanced melanoma patients. J Clin Oncol (2007) 25(18s):8540.
53. Rangel J, Nosrati M, Torabian S, Shaikh L, Leong SP, Haqq C, et al. Osteopontin as a molecular prognostic marker for melanoma. Cancer (2008) 112(1):144–50. doi:10.1002/cncr.23147
54. Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res (2007) 67(7):3450–60. doi:10.1158/0008-5472.CAN-06-3481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Maier T, Laubender RP, Sturm RA, Klingenstein A, Korting HC, Ruzicka T, et al. Osteopontin expression in plasma of melanoma patients and in melanocytic tumours. J Eur Acad Dermatol Venereol (2012) 26(9):1084–91. doi:10.1111/j.1468-3083.2011.04210.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Brown ER, Doig T, Anderson N, Brenn T, Doherty V, Xu Y, et al. Association of galectin-3 expression with melanoma progression and prognosis. Eur J Cancer (2012) 48(6):865–74. doi:10.1016/j.ejca.2011.09.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Schmidt H, Johansen JS, Gehl J, Geertsen PF, Fode K, von der Maase H. Elevated serum level of YKL-40 is an independent prognostic factor for poor survival in patients with metastatic melanoma. Cancer (2006) 106(5):1130–9. doi:10.1002/cncr.21678
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Schmidt H, Johansen JS, Sjoegren P, Christensen IJ, Sorensen BS, Fode K, et al. Serum YKL-40 predicts relapse-free and overall survival in patients with American joint committee on cancer stage I and II melanoma. J Clin Oncol (2006) 24(5):798–804. doi:10.1200/JCO.2005.03.7960
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Diaz-Lagares A, Alegre E, Arroyo A, Gonzalez-Cao M, Zudaire ME, Viteri S, et al. Evaluation of multiple serum markers in advanced melanoma. Tumour Biol (2011) 32(6):1155–61. doi:10.1007/s13277-011-0218-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Essler M, Link A, Belloni B, Mirceva V, Souvatzoglou M, Thaler M, et al. Prognostic value of [18F]-fluoro-deoxy-glucose PET/CT, S100 or MIA for assessment of cancer-associated mortality in patients with high risk melanoma. PLoS One (2011) 6(9):e24632. doi:10.1371/journal.pone.0024632
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Sivan S, Suzan F, Rona O, Tamar H, Vivian B, Tamar P, et al. Serum CEACAM1 correlates with disease progression and survival in malignant melanoma patients. Clin Dev Immunol (2012) 2012:290536. doi:10.1155/2012/290536
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Bouwhuis MG, Suciu S, Kruit W, Sales F, Stoitchkov K, Patel P, et al. Prognostic value of serial blood S100B determinations in stage IIB-III melanoma patients: a corollary study to EORTC trial 18952. Eur J Cancer (2011) 47(3):361–8. doi:10.1016/j.ejca.2010.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Oberholzer PA, Urosevic M, Steinert HC, Dummer R. Baseline staging of melanoma with unknown primary site: the value of serum s100 protein and positron emission tomography. Dermatology (2008) 217(4):351–5. doi:10.1159/000155878
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Beyeler M, Waldispuhl S, Strobel K, Joller-Jemelka HI, Burg G, Dummer R. Detection of melanoma relapse: first comparative analysis on imaging techniques versus S100 protein. Dermatology (2006) 213(3):187–91. doi:10.1159/000095034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Karagiannis P, Gilbert AE, Josephs DH, Ali N, Dodev T, Saul L, et al. IgG4 subclass antibodies impair antitumor immunity in melanoma. J Clin Invest (2013) 123(4):1457–74. doi:10.1172/JCI65579
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Sanmamed MF, Carranza-Rua O, Alfaro C, Onate C, Martin-Algarra S, Perez G, et al. Serum interleukin-8 reflects tumor burden and treatment response across malignancies of multiple tissue origins. Clin Cancer Res (2014) 20(22):5697–707. doi:10.1158/1078-0432.CCR-13-3203
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Solassol J, Du-Thanh A, Maudelonde T, Guillot B. Serum proteomic profiling reveals potential biomarkers for cutaneous malignant melanoma. Int J Biol Markers (2011) 26(2):82–7. doi:10.5301/JBM.2011.8344
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Vereecken P, Cornelis F, Van Baren N, Vandersleyen V, Baurain JF. A synopsis of serum biomarkers in cutaneous melanoma patients. Dermatol Res Pract (2012) 2012:260643. doi:10.1155/2012/260643
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Hofmann MA, Gussmann F, Fritsche A, Biesold S, Schicke B, Kuchler I, et al. Diagnostic value of melanoma inhibitory activity serum marker in the follow-up of patients with stage I or II cutaneous melanoma. Melanoma Res (2009) 19(1):17–23. doi:10.1097/CMR.0b013e32831bc78c
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Hofmann MA, Schicke B, Fritsch A, Biesold S, Gussmann F, Kuchler I, et al. Impact of lymph node metastases on serum level of melanoma inhibitory activity in stage III melanoma patients. J Dermatol (2011) 38(9):880–6. doi:10.1111/j.1346-8138.2011.01219.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Kluger HM, Hoyt K, Bacchiocchi A, Mayer T, Kirsch J, Kluger Y, et al. Plasma markers for identifying patients with metastatic melanoma. Clin Cancer Res (2011) 17(8):2417–25. doi:10.1158/1078-0432.CCR-10-2402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Schmidt H, Sorensen BS, Fode K, Nexo E, von der Maase H. Tyrosinase messenger RNA in peripheral blood is related to poor survival in patients with metastatic melanoma following interleukin-2-based immunotherapy. Melanoma Res (2005) 15(5):409–16. doi:10.1097/00008390-200510000-00009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Garbe C, Leiter U, Ellwanger U, Blaheta HJ, Meier F, Rassner G, et al. Diagnostic value and prognostic significance of protein S-100beta, melanoma-inhibitory activity, and tyrosinase/MART-1 reverse transcription-polymerase chain reaction in the follow-up of high-risk melanoma patients. Cancer (2003) 97(7):1737–45. doi:10.1002/cncr.11250
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Zhang H, Fu T, McGettigan S, Kumar S, Liu S, Speicher D, et al. IL-8 and cathepsin B as melanoma serum biomarkers. Int J Mol Sci (2011) 12(3):1505–18. doi:10.3390/ijms12031505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Vihinen PP, Hilli J, Vuoristo MS, Syrjanen KJ, Kahari VM, Pyrhonen SO. Serum VEGF-C is associated with metastatic site in patients with malignant melanoma. Acta Oncol (2007) 46(5):678–84. doi:10.1080/02841860600965020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. McAllister SS, Gifford AM, Greiner AL, Kelleher SP, Saelzler MP, Ince TA, et al. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell (2008) 133(6):994–1005. doi:10.1016/j.cell.2008.04.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Perrotta R, Bevelacqua Y, Malaguarnera G, Paladina I, Giordano M, Malaguarnera M. Serum markers of cutaneous melanoma. Front Biosci (2010) 2:1115–22. doi:10.2741/E170
78. Volck B, Price PA, Johansen JS, Sorensen O, Benfield TL, Nielsen HJ, et al. YKL-40, a mammalian member of the chitinase family, is a matrix protein of specific granules in human neutrophils. Proc Assoc Am Physicians (1998) 110(4):351–60.
79. Schultz NA, Johansen JS. YKL-40—a protein in the field of translational medicine: a role as a biomarker in cancer patients? Cancers (Basel) (2010) 2(3):1453–91. doi:10.3390/cancers2031453
80. Johansen JS, Jensen BV, Roslind A, Nielsen D, Price PA. Serum YKL-40, a new prognostic biomarker in cancer patients? Cancer Epidemiol Biomarkers Prev (2006) 15(2):194–202. doi:10.1158/1055-9965.EPI-05-0011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Krogh M, Christensen IJ, Bouwhuis M, Johansen JS, Schmidt H, Hansson J, et al. Prognostic value of serum YKL-40 in stage IIB-III melanoma patients receiving adjuvant interferon therapy. J Clin Oncol (2010) 28(15s):8587.
82. Palmer SR, Erickson LA, Ichetovkin I, Knauer DJ, Markovic SN. Circulating serologic and molecular biomarkers in malignant melanoma. Mayo Clin Proc (2011) 86(10):981–90. doi:10.4065/mcp.2011.0287
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Juergensen A, Holzapfel U, Hein R, Stolz W, Buettner R, Bosserhoff A. Comparison of two prognostic markers for malignant melanoma: MIA and S100 beta. Tumour Biol (2001) 22(1):54–8. doi:10.1159/000030147
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Bosserhoff AK, Kuster H, Hein R. Elevated MIA levels in the serum of pregnant women and of children. Clin Exp Dermatol (2004) 29(6):628–9. doi:10.1111/j.1365-2230.2004.01623.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Davey GE, Murmann P, Hoechli M, Tanaka T, Heizmann CW. Calcium-dependent translocation of S100A11 requires tubulin filaments. Biochim Biophys Acta (2000) 1498(2–3):220–32. doi:10.1016/S0167-4889(00)00098-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Hsieh HL, Schafer BW, Cox JA, Heizmann CW. S100A13 and S100A6 exhibit distinct translocation pathways in endothelial cells. J Cell Sci (2002) 115(Pt 15):3149–58.
87. Petersson S, Shubbar E, Enerback L, Enerback C. Expression patterns of S100 proteins in melanocytes and melanocytic lesions. Melanoma Res (2009) 19(4):215–25. doi:10.1097/CMR.0b013e32832c6358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Kruijff S, Bastiaannet E, Kobold AC, van Ginkel RJ, Suurmeijer AJ, Hoekstra HJ. S-100B concentrations predict disease-free survival in stage III melanoma patients. Ann Surg Oncol (2009) 16(12):3455–62. doi:10.1245/s10434-009-0629-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Molina R, Navarro J, Filella X, Castel T, Ballesta AM. S-100 protein serum levels in patients with benign and malignant diseases: false-positive results related to liver and renal function. Tumour Biol (2002) 23(1):39–44. doi:10.1159/000048687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Michetti F, Corvino V, Geloso MC, Lattanzi W, Bernardini C, Serpero L, et al. The S100B protein in biological fluids: more than a lifelong biomarker of brain distress. J Neurochem (2012) 120(5):644–59. doi:10.1111/j.1471-4159.2011.07612.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Tsoporis JN, Mohammadzadeh F, Parker TG. S100B: a multifunctional role in cardiovascular pathophysiology. Amino Acids (2011) 41(4):843–7. doi:10.1007/s00726-010-0527-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Meyer S, Vogt T, Landthaler M, Berand A, Reichle A, Bataille F, et al. Cyclooxygenase 2 (COX2) and peroxisome proliferator-activated receptor gamma (PPARG) are stage-dependent prognostic markers of malignant melanoma. PPAR Res (2009) 2009:848645. doi:10.1155/2010/848645
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Bosserhoff AK. Novel biomarkers in malignant melanoma. Clin Chim Acta (2006) 367(1–2):28–35. doi:10.1016/j.cca.2005.10.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Buljan M, Situm M, Tomas D, Milosevic M, Kruslin B. Prognostic value of galectin-3 in primary cutaneous melanoma. J Eur Acad Dermatol Venereol (2011) 25(10):1174–81. doi:10.1111/j.1468-3083.2010.03943.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Stamenkovic I. Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol (2003) 200(4):448–64. doi:10.1002/path.1400
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol (2000) 10(6):415–33. doi:10.1006/scbi.2000.0379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Sapoznik S, Ortenberg R, Schachter J, Markel G. CEACAM1 in malignant melanoma: a diagnostic and therapeutic target. Curr Top Med Chem (2012) 12(1):3–10. doi:10.2174/156802612798919259
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Thies A, Moll I, Berger J, Wagener C, Brummer J, Schulze HJ, et al. CEACAM1 expression in cutaneous malignant melanoma predicts the development of metastatic disease. J Clin Oncol (2002) 20(10):2530–6. doi:10.1200/JCO.2002.05.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Ebrahimnejad A, Streichert T, Nollau P, Horst AK, Wagener C, Bamberger AM, et al. CEACAM1 enhances invasion and migration of melanocytic and melanoma cells. Am J Pathol (2004) 165(5):1781–7. doi:10.1016/S0002-9440(10)63433-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Franzke A, Probst-Kepper M, Buer J, Duensing S, Hoffmann R, Wittke F, et al. Elevated pretreatment serum levels of soluble vascular cell adhesion molecule 1 and lactate dehydrogenase as predictors of survival in cutaneous metastatic malignant melanoma. Br J Cancer (1998) 78(1):40–5. doi:10.1038/bjc.1998.439
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Hirai S, Kageshita T, Kimura T, Tsujisaki M, Imai K, Wakamatsu K, et al. Serum levels of sICAM-1 and 5-S-cysteinyldopa as markers of melanoma progression. Melanoma Res (1997) 7(1):58–62. doi:10.1097/00008390-199702000-00009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Ross AH, Cossu G, Herlyn M, Bell JR, Steplewski Z, Koprowski H. Isolation and chemical characterization of a melanoma-associated proteoglycan antigen. Arch Biochem Biophys (1983) 225(1):370–83. doi:10.1016/0003-9861(83)90042-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Campoli M, Ferrone S, Wang X. Functional and clinical relevance of chondroitin sulfate proteoglycan 4. Adv Cancer Res (2010) 109:73–121. doi:10.1016/B978-0-12-380890-5.00003-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Vergilis IJ, Szarek M, Ferrone S, Reynolds SR. Presence and prognostic significance of melanoma-associated antigens CYT-MAA and HMW-MAA in serum of patients with melanoma. J Invest Dermatol (2005) 125(3):526–31. doi:10.1111/j.0022-202X.2005.23798.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Reynolds SR, Vergilis IJ, Szarek M, Ferrone S, Bystryn JC. Cytoplasmic melanoma-associated antigen (CYT-MAA) serum level in patients with melanoma: a potential marker of response to immunotherapy? Int J Cancer (2006) 119(1):157–61. doi:10.1002/ijc.21820
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol (2007) 19(2):209–16. doi:10.1016/j.coi.2007.01.001
107. Whiteside TL. Immune responses to cancer: are they potential biomarkers of prognosis? Front Oncol (2013) 3:107. doi:10.3389/fonc.2013.00107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Massi D, Brusa D, Merelli B, Ciano M, Audrito V, Serra S, et al. PD-L1 marks a subset of melanomas with a shorter overall survival and distinct genetic and morphological characteristics. Ann Oncol (2014) 25(12):2433–42. doi:10.1093/annonc/mdu452
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother (2012) 61(7):1019–31. doi:10.1007/s00262-011-1172-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (2006) 313(5795):1960–4. doi:10.1126/science.1129139
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med (2005) 353(25):2654–66. doi:10.1056/NEJMoa051424
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Whiteside TL, Mandapathil M, Szczepanski M, Szajnik M. Mechanisms of tumor escape from the immune system: adenosine-producing Treg, exosomes and tumor-associated TLRs. Bull Cancer (2011) 98(2):E25–31. doi:10.1684/bdc.2010.1294
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Lanca T, Silva-Santos B. The split nature of tumor-infiltrating leukocytes: implications for cancer surveillance and immunotherapy. Oncoimmunology (2012) 1(5):717–25. doi:10.4161/onci.20068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother (2014) 63(3):247–57. doi:10.1007/s00262-013-1508-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Nielsen JS, Sahota RA, Milne K, Kost SE, Nesslinger NJ, Watson PH, et al. CD20+ tumor-infiltrating lymphocytes have an atypical CD27− memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res (2012) 18(12):3281–92. doi:10.1158/1078-0432.CCR-12-0234
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett (2007) 581(19):3652–7. doi:10.1016/j.febslet.2007.04.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Forster I, Rajewsky K. The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc Natl Acad Sci USA (1990) 87(12):4781–4. doi:10.1073/pnas.87.12.4781
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Cooperman J, Neely R, Teachey DT, Grupp S, Choi JK. Cell division rates of primary human precursor B cells in culture reflect in vivo rates. Stem Cells (2004) 22(6):1111–20. doi:10.1634/stemcells.22-6-1111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
119. Schmidt M, Hellwig B, Hammad S, Othman A, Lohr M, Chen Z, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res (2012) 18(9):2695–703. doi:10.1158/1078-0432.CCR-11-2210
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res (2008) 68(13):5405–13. doi:10.1158/0008-5472.CAN-07-5206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Gilbert AE, Karagiannis P, Dodev T, Koers A, Lacy K, Josephs DH, et al. Monitoring the systemic human memory B cell compartment of melanoma patients for anti-tumor IgG antibodies. PLoS One (2011) 6(4):e19330. doi:10.1371/journal.pone.0019330
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Carpenter EL, Mick R, Rech AJ, Beatty GL, Colligon TA, Rosenfeld MR, et al. Collapse of the CD27+ B-cell compartment associated with systemic plasmacytosis in patients with advanced melanoma and other cancers. Clin Cancer Res (2009) 15(13):4277–87. doi:10.1158/1078-0432.CCR-09-0537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Daveau M, Pavie-Fischer J, Rivat L, Rivat C, Ropartz C, Peter HH, et al. IgG4 subclass in malignant melanoma. J Natl Cancer Inst (1977) 58(2):189–92.
124. Harada K, Shimoda S, Kimura Y, Sato Y, Ikeda H, Igarashi S, et al. Significance of immunoglobulin G4 (IgG4)-positive cells in extrahepatic cholangiocarcinoma: molecular mechanism of IgG4 reaction in cancer tissue. Hepatology (2012) 56(1):157–64. doi:10.1002/hep.25627
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
125. Jeannin P, Lecoanet S, Delneste Y, Gauchat JF, Bonnefoy JY. IgE versus IgG4 production can be differentially regulated by IL-10. J Immunol (1998) 160(7):3555–61.
126. Satoguina JS, Weyand E, Larbi J, Hoerauf A. T regulatory-1 cells induce IgG4 production by B cells: role of IL-10. J Immunol (2005) 174(8):4718–26. doi:10.4049/jimmunol.174.8.4718
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Stone JH, Zen Y, Deshpande V. IgG4-related disease. N Engl J Med (2012) 366(6):539–51. doi:10.1056/NEJMra1104650
128. Restrepo L, Stafford P, Johnston SA. Feasibility of an early Alzheimer’s disease immunosignature diagnostic test. J Neuroimmunol (2013) 254(1–2):154–60. doi:10.1016/j.jneuroim.2012.09.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Hughes AK, Cichacz Z, Scheck A, Coons SW, Johnston SA, Stafford P. Immunosignaturing can detect products from molecular markers in brain cancer. PLoS One (2012) 7(7):e40201. doi:10.1371/journal.pone.0040201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Tsoka S, Ainali C, Karagiannis P, Josephs DH, Saul L, Nestle FO, et al. Toward prediction of immune mechanisms and design of immunotherapies in melanoma. Crit Rev Biomed Eng (2012) 40(4):279–94. doi:10.1615/CritRevBiomedEng.v40.i4.40
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: biomarkers, melanoma, humoral immunity, antibodies, cancer, prognosis, immune response, inflammation
Citation: Karagiannis P, Fittall M and Karagiannis SN (2015) Evaluating biomarkers in melanoma. Front. Oncol. 4:383. doi: 10.3389/fonc.2014.00383
Received: 07 October 2014; Accepted: 22 December 2014;
Published online: 23 January 2015.
Edited by:
Adetunji T. Toriola, Washington University School of Medicine and Siteman Cancer Center, USAReviewed by:
Soldano Ferrone, University of Pittsburgh Cancer Institute, USAAnne E. Cust, The University of Sydney, Australia
Copyright: © 2015 Karagiannis, Fittall and Karagiannis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophia N. Karagiannis, St. John’s Institute of Dermatology, Division of Genetics and Molecular Medicine, King’s College London & NIHR Biomedical Research Centre at Guy’s and St. Thomas’s Hospitals and King’s College London, 9th Floor, Tower Wing, Guy’s Hospital, London, SE1 9RT, UK e-mail: sophia.karagiannis@kcl.ac.uk
 Panagiotis Karagiannis
Panagiotis Karagiannis Matthew Fittall
Matthew Fittall Sophia N. Karagiannis
Sophia N. Karagiannis