- Division of Marine Life Science, Genetics and Genomics, Faculty of Fisheries Sciences, Hokkaido University, Hakodate, Japan
Phosphatidylinositol (PtdIns) is involved not only in the structural composition of eukaryotic cellular membranes but also in the regulation of a wide variety of physiological processes influencing growth and development (Xue et al., 2009; Janda et al., 2013). Because molecules that participate in PtdIns signaling are generated via PtdIns metabolism, extensive attention has been paid to genes encoding enzymes involved in this metabolism in order to elucidate developmental and stress-response mechanisms. PtdIns is synthesized from CDP-diacylglycerol and cytoplasmic inositol by PtdIns synthase (PIS) and is phosphorylated sequentially on its inositol ring to produce PtdIns4P and PtdIns(4,5)P2 by PtdIns 4-kinase (PI4K) and PtdIns phosphate-kinase (PIPK), respectively (Xue et al., 2009; Janda et al., 2013). Subsequently, PtdIns(4,5)P2 is cleaved by phosphoinositide-specific phospholipase C (PI-PLC) into the second messengers diacylglycerol and inositol-1,4,5-trisphosphate [Ins(1,4,5)P3, IP3] (Xue et al., 2009; Janda et al., 2013), which then activate protein kinase C (PKC) and the IP3 receptor involved in release of Ca2+ from the ER into the cytoplasm, respectively, in animal cells (Rebecchi and Pentyala, 2000; Suh et al., 2008). In fact, genes encoding orthologs of PKC and the IP3 receptor are not found in terrestrial plant genomes, suggesting differences in second messenger systems between animals and plants.
Significant genomic information has been accumulated for unicellular algae including the green alga Chlamydomonas reinhardtii (Merchant et al., 2007), the red alga Cyanidioschyzon merolae (Matsuzaki et al., 2004), for the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Armbrust et al., 2004; Bowler et al., 2008), and also for multicellular seaweeds such as the red seaweeds Pyropia yezoensis (Nakamura et al., 2013) and Chondrus crispus (Collén et al., 2013), and brown seaweed Ectocarpus siliculosus (Cock et al., 2010). In addition, large-scale EST information has been accumulated for the red seaweeds Porphyra umbilicalis and Porphyra purpurea (Chan et al., 2012; Stiller et al., 2012). Because understanding the evolutionary aspects of PtdIns signaling can help us to understand the process of establishment of the plant PtdIns signaling system, algal PI-PLC and PIPK protein structures have been compared with those of terrestrial plants.
PI-PLC
The first record of the PI-PLC gene in plants was presented by Hirayama et al. (1995). Since then, information on PI-PLC genes from terrestrial plants and green algae has accumulated from results of molecular cloning and in silico analysis based on genome sequence data. In mammals, PI-PLC isozymes have been divided into six groups according to their domain structural characteristics (Rebecchi and Pentyala, 2000; Suh et al., 2008). For example, PLCδ consists the Pleckstrin homology (PH) domain, the EF hand, and X, Y, and C2 domains (Figure 1), although PLCγ and PLCε contain the SH2 and SH3 domains and the Ras-GEF and RA domains, respectively, in addition to domains found in PLCδ. The PH domain participates in membrane binding, whereas the X and Y domains are responsible for catalytic activity. Plant PI-PLCs reported so far, however, lack the PH domain, similar in that to the mammalian PI-PLCζ isoform (Pokotylo et al., 2014). They consist of an N-terminal EF-hand, a central X/Y catalytic domain, and a C-terminal C2 domain (Figure 1). The EF hand plays an important role in Ca2+-dependent activation of PI-PLCζ, suggesting a similar function in plant PI-PLCs. Indeed, it has been demonstrated that the activities of PI-PLCs from plants were stimulated by Ca2+ in a concentration dependent manner (for instance, Hirayama et al., 1995; Otterhag et al., 2001; Hunt et al., 2004; Mikami et al., 2004). Although the EF hand in mammalian PI-PLC has four repeats of helix-loop-helix structure, plant enzymes carry only two of these repeats due to truncation (Figure 1). The C2 domain in plant PI-PLCs was suggested to have evolved separately from animal PI-PLCs and is possibly involved in membrane localization (Rupwate and Rajasekharan, 2012).
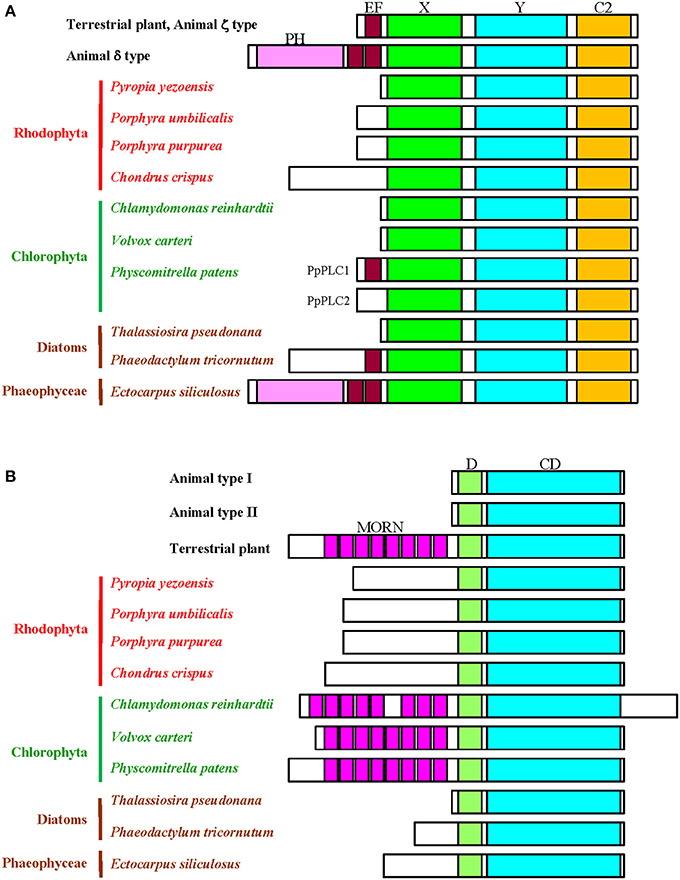
Figure 1. Schematic comparison of the domain structures of the PI-PLC signaling components. (A), PI-PLC; (B), PIPK. D, dimerization domain; CD, catalytic domain.
Genome analysis has identified nine copies of the PI-PLC genes (AtPLC1-AtPLC9) in A. thaliana, although AtPLC8 and AtPLC9 with amino acid substitutions in the Y domain do not group into the clade containing other PLCs during phylogenetic analysis (Mueller-Roeber and Pical, 2002; Hunt et al., 2004). However, only one and two copies of PI-PLC genes have been found in C. reinhardtii and Physcomitrella patens, respectively (Mikami et al., 2004; Awasthi et al., 2012). Moreover, because red and brown seaweeds as well as green microalgae have only a single copy of the PI-PLC gene, amplification and functional diversification of these genes likely occurred during evolution of streptophytes (land plants and charophytic algae) with establishment of multicellularity and vascular systems after the colonization of land.
It is worth noting that P. patens has a novel PI-PLC without PtdIns(4,5)P2-hydrolyzing activity, with an insertion at its N-terminal EF hand (Mikami et al., 2004). Figure 1 shows the absence of the EF hand in unicellular and multicellular algae from aqueous environments, except for the diatom P. tricornutum and the brown seaweed E. siliculosus (Figure 1). The EF hand might have appeared after the colonization of land by green algal lineages and, subsequently, PI-PLCs lacking the EF hand disappeared during the evolution of terrestrial green plants. Thus, the EF-hand-mediated regulatory mode of PI-PLC activation seems to have evolved in the green plant lineages after the colonization of land.
The evolutionary establishment of the domain structure of PI-PLC in brown seaweeds is highly complex. As shown in Figure 1, P. tricornutum and E. siliculosus have the EF hand motif. Thus, the EF hand was acquired during the evolution of diatoms and brown seaweeds. Surprisingly, PI-PLC in E. siliculosus contains the non-truncated EF hand and the PH domain, just as does the mammalian PI-PLCδ isoform (Figure 1). The reason that only the brown seaweed has an isoform of the PI-PLCδ type remains to be resolved.
PIPK
The first molecular characterization of the plant PIPK gene was performed by Mikami et al. (1998), following molecular cloning and in silico identification based on the genome sequences of land plant PIPKs. Due to their substrate specificity in animals, PIKs can be divided into three subfamilies, known as types I, II, and III. Types I and II enzymes convert PtdIns(4)P and PtdIns(5)P, respectively, to produce PtdIns(4,5)P2 (Anderson et al., 1999), whereas type III enzymes use PI(3)P to produce PI(3,5)P2 similar to yeast Fab1 (Mueller-Roeber and Pical, 2002). Despite such differences in substrate specificity, the structures of type I and II PIPKs are highly similar in animals; however, based on substrate specificity and structure, the plant enzymes are classified as type I/II PIPKs (Mueller-Roeber and Pical, 2002; Thole and Nielsen, 2008; Saavedra et al., 2012).
The genome of A. thaliana contains 11 PIPKs, subdivided into A and B subfamilies according to their structural differences. The domain structure of subfamily A (AtPIPK10 and AtPIPK11) shows similarity to animal PIPKs, whereas the domain structure of subfamily B members (AtPIP5K1-AtPIP5K9) includes a long N-terminal extension containing a repeat of the membrane occupation and recognition nexus (MORN) motif (Mueller-Roeber and Pical, 2002; Saavedra et al., 2012) Although the domain containing MORN motifs has been hypothesized to be a module involved in plasma membrane-localization and phosphatidic acid (PA)-dependent enzymatic activation (Im et al., 2007), it was recently demonstrated that the activation loop conserved in the lipid kinase domain is responsible for both plasma membrane localization and PA-dependent activation of A. thaliana and P. patens PIPKs (Mikami et al., 2010a,b; Saavedra et al., 2012). Plant genomes other than that of A. thaliana contain only the B subfamily and a small number of genes for PIPK orthologs exist in C. reinhardtii (single copy; Awasthi et al., 2012) and P. patens (two copies; Saavedra et al., 2009, 2011, 2012). Because red and brown seaweeds as well as green microalgae have only a single copy of the PIPK gene, amplification and functional diversification of PIPK genes also likely occurred during evolution, as in PI-PLC.
As shown in Figure 1, the MORN motifs are not found in PIPKs from red algae, diatoms, and brown E. siliculosus, indicating the presence of the MORN repeats is restricted into the green lineage, although PIPKs without the MORN motifs are conserved in A. thaliana, as mentioned above. Because the common origin of the red and green algae can be ascribed to a single symbiosis (Keeling, 2010), there are two possibilities for the presence of the MORN motifs in the green lineage. One is that although an ancestral plant cell has a MORN motif-containing PIPK, this motif disappeared from red algae after the divergence of red and green algae. The other is that an ancestral plant cell has a PIPK consisting of the dimerization and catalytic domains and the MORN motifs appeared only in green algae after the divergence of red and green algae. The absence of the MORN motifs in the brown seaweed can be explained by either possibility. According to these findings, the activation mode of PIPK differs between green plants and red and brown algae. Elucidation of the functions of the MORN motif is necessary to fully explain such a difference.
Conclusions
Comparative genomics provide an evolutionary insight into the taxonomic origin of enzymes and their isoforms, which is generally drawn based on the presence and number of gene family members. Here, it is demonstrated that analysis of domain structure can allow novel evolutionary conclusions. Because domain structure is involved in the regulation of the active state of each protein, comparison of plant genomes with a focus on the structure of and changes in protein domains could open new scenarios regarding the origin and evolution of signaling networks that regulate development and stress responses in plants.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported in part by a KAKENHI Grant (Number 2566016003) from the Japan Society for the Promotion of Science.
References
Anderson, R. A., Boronenkov, I. V., Doughman, S. D., Kunz, J., and Loijens, J. C. (1999). Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274, 9907–9910. doi: 10.1074/jbc.274.15.9907
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Awasthi, M., Batra, J., and Kateriya, S. (2012). Disulphide bridges of phospholipase C of Chlamydomonas reinhardtii modulates lipid interaction and dimer stability. PLoS ONE 7:e39258. doi: 10.1371/journal.pone.0039258
Bowler, C., Allen, A. E., Badger, J. H., Grimwood, J., Jabbari, K., Kuo, A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Chan, C. X., Zäuner, S., Wheeler, G., Grossman, A. R., Prochnik, S. E., Blouin, N. A., et al. (2012). Analysis of Porphyra membrane transporters demonstrates gene transfer among photosynthetic eukaryotes and numerous sodium-coupled transport systems. Plant Physiol. 158, 2001–2012. doi: 10.1104/pp.112.193896
Cock, J. M., Sterck, L., Rouzé, P., Scornet, D., Allen, A. E., Amoutzias, G., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465, 617–621. doi: 10.1038/nature09016
Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
Hirayama, T., Ohto, C., Mizoguchi, T., and Shinozaki, K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 92, 3903–3907. doi: 10.1073/pnas.92.9.3903
Hunt, L., Otterhag, L., Lee, J. C., Lasheen, T., Hunt, J., Seki, M., et al. (2004). Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytol. 162, 643–654. doi: 10.1111/j.1469-8137.2004.01069.x
Im, Y. J., Davis, A. J., Perera, I. Y., Johannes, E., Allen, N. S., and Boss, W. F. (2007). The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J. Biol. Chem. 282, 5443–5452. doi: 10.1074/jbc.M611342200
Janda, M., Planchais, S., Djafi, N., Martinec, J., Burketova, L., Valentova, O., et al. (2013). Phosphoglycerolipids are master players in plant hormone signal transduction. Plant Cell Rep. 32, 839–851. doi: 10.1007/s00299-013-1399-0
Keeling, P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748. doi: 10.1098/rstb.2009.0103
Matsuzaki, M., Misumi, O., Shin-i, T., Maruyama, S., Takahara, M., Miyagishima, S., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. doi: 10.1126/science.1143609
Mikami, K., Katagiri, T., Iuchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 15, 563–568. doi: 10.1046/j.1365-313X.1998.00227.x
Mikami, K., Repp, A., Graebe-Abts, E., and Hartmann, E. (2004). Isolation of cDNAs encoding typical and novel types of phosphoinositide-specific phospholipase C from the moss Physcomitrella patens. J. Exp. Bot. 55, 1437–1439. doi: 10.1093/jxb/erh140
Mikami, K., Saavedra, L., Hiwatashi, Y., Uji, T., Hasebe, M., and Sommarin, M. (2010b). A dibasic amino acid pair conserved in the activation loop directs plasma membrane localization and is necessary for activity of plant type I/II phosphatidylinositol phosphate kinase. Plant Physiol. 153, 1004–1015. doi: 10.1104/pp.109.152686
Mikami, K., Saavedra, L., and Sommarin, M. (2010a). Is membrane occupation and recognition nexus domain functional in plant phosphatidylinositol phosphate kinases? Plant Signal. Behav. 5, 1241–1244. doi: 10.4161/psb.5.10.12922
Mueller-Roeber, B., and Pical, C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46. doi: 10.1104/pp.004770
Nakamura, Y., Sasaki, N., Kobayashi, M., Ojima, N., Yasuike, M., Shigenobu, Y., et al. (2013). The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS ONE 8:e57122. doi: 10.1371/journal.pone.0057122
Otterhag, L., Sommarin, M., and Pical, C. (2001). N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 497, 165–170. doi: 10.1016/S0014-5793(01)02453-X
Pokotylo, I., Kolesnikov, Y., Kravets, V., Zachowski, A., and Ruelland, E. (2014). Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 96, 144–157. doi: 10.1016/j.biochi.2013.07.004
Rebecchi, M. J., and Pentyala, S. N. (2000). Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 80, 1291–1335.
Rupwate, S. D., and Rajasekharan, R. (2012). C2 domain is responsible for targeting rice phosphoinositide specific phospholipase C. Plant Mol. Biol. 78, 247–258. doi: 10.1007/s11103-011-9862-1
Saavedra, L., Balbi, V., Dove, S. K., Hiwatashi, Y., Mikami, K., and Sommarin, M. (2009). Characterization of phosphatidylinositol phosphate kinases from the moss Physcomitrella patens: PpPIPK1 and PpPIPK2. Plant Cell Physiol. 50, 595–609. doi: 10.1093/pcp/pcp018
Saavedra, L., Balbi, V., Lerche, J., Mikami, K., Heilmann, I., and Sommarin, M. (2011). PIPKs are essential for rhizoid elongation and caulonemal cell development in the moss Physcomitrella patens. Plant J. 67, 635–647. doi: 10.1111/j.1365-313X.2011.04623.x
Saavedra, L., Mikami, K., Malhó, R., and Sommarin, M. (2012). PIP kinases and their role in plant tip growing cells. Plant Signal Behav. 7, 1302–1305. doi: 10.4161/psb.21547
Stiller, J. W., Perry, J., Rymarquis, L. A., Accerbi, M., Green, P. J., Prochnik, S., et al. (2012). Major developmental regulators and their expression in two closely related species of Porphyra (Rhodophyta). J. Phycol. 48, 883–896. doi: 10.1111/j.1529-8817.2012.01138.x
Suh, P.-G., Park, J. I., Manzoli, L., Cocco, L., Peak, J. C., Katan, M., et al. (2008). Multiple roles of phosphoinositide-specific phospholipase C isozyme. BMB Rep. 41, 415–434. doi: 10.5483/BMBRep.2008.41.6.415
Thole, J. M., and Nielsen, E. (2008). Phosphoinositides in plants: novel functions in membrane trafficking. Curr. Opin. Plant Biol. 11, 620–631. doi: 10.1016/j.pbi.2008.10.010
Keywords: alga, phosphoinositide-specific phospholipase C, phosphatidylinositol phosphate-kinase, protein domain, genome
Citation: Mikami K (2014) Structural divergence and loss of phosphoinositide-specific phospholipase C signaling components during the evolution of the green plant lineage: implications from structural characteristics of algal components. Front. Plant Sci. 5:380. doi: 10.3389/fpls.2014.00380
Received: 26 June 2014; Accepted: 17 July 2014;
Published online: 05 August 2014.
Edited by:
Eric Ruelland, Centre National de la Recherche Scientifique, FranceReviewed by:
Imara Yasmin Perera, North Carolina State University, USAEric Ruelland, Centre National de la Recherche Scientifique, France
Copyright © 2014 Mikami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: komikami@fish.hokudai.ac.jp
 Koji Mikami
Koji Mikami