- Department of Life Sciences, University of Limerick, Limerick, Ireland
Plant growth is highly dependent on bacteria, saprophytic, and mycorrhizal fungi which facilitate the cycling and mobilization of nutrients. Over 95% of the sulfur (S) in soil is present in an organic form. Sulfate-esters and sulfonates, the major forms of organo-S in soils, arise through deposition of biological material and are transformed through subsequent humification. Fungi and bacteria release S from sulfate-esters using sulfatases, however, release of S from sulfonates is catalyzed by a bacterial multi-component mono-oxygenase system. The asfA gene is used as a key marker in this desulfonation process to study sulfonatase activity in soil bacteria identified as Variovorax, Polaromonas, Acidovorax, and Rhodococcus. The rhizosphere is regarded as a hot spot for microbial activity and recent studies indicate that this is also the case for the mycorrhizosphere where bacteria may attach to the fungal hyphae capable of mobilizing organo-S. While current evidence is not showing sulfatase and sulfonatase activity in arbuscular mycorrhiza, their effect on the expression of plant host sulfate transporters is documented. A revision of the role of bacteria, fungi and the interactions between soil bacteria and mycorrhiza in plant S supply was conducted.
Introduction
Sulfur (S), an essential macro-element required for growth, is increasingly becoming limiting to crop yield and quality as a result of a reduction in atmospheric S levels and crop varieties removing S from soil more rapidly (Fowler et al., 2005). S present in soil is approximately 95% organically bound largely in one of two major forms; sulfate-esters and sulfonates (Figure 1; Autry and Fitzgerald, 1990; Kertesz and Mirleau, 2004). These forms of organo-S are not directly available to plants which rely upon microbes in soil and rhizosphere for organo-S mobilization (Kertesz et al., 2007). Plant root activity impacts the physico-chemical properties of the soil through the release of organic compounds (rhizodeposition) which accounts for 15–30% of photosynthetically produced carbon (C; Russell, 1977). This process provides soil organisms with an energy source that enables them to fulfill their respective functional roles (Lynch and Whipps, 1990; Farrar et al., 2003).
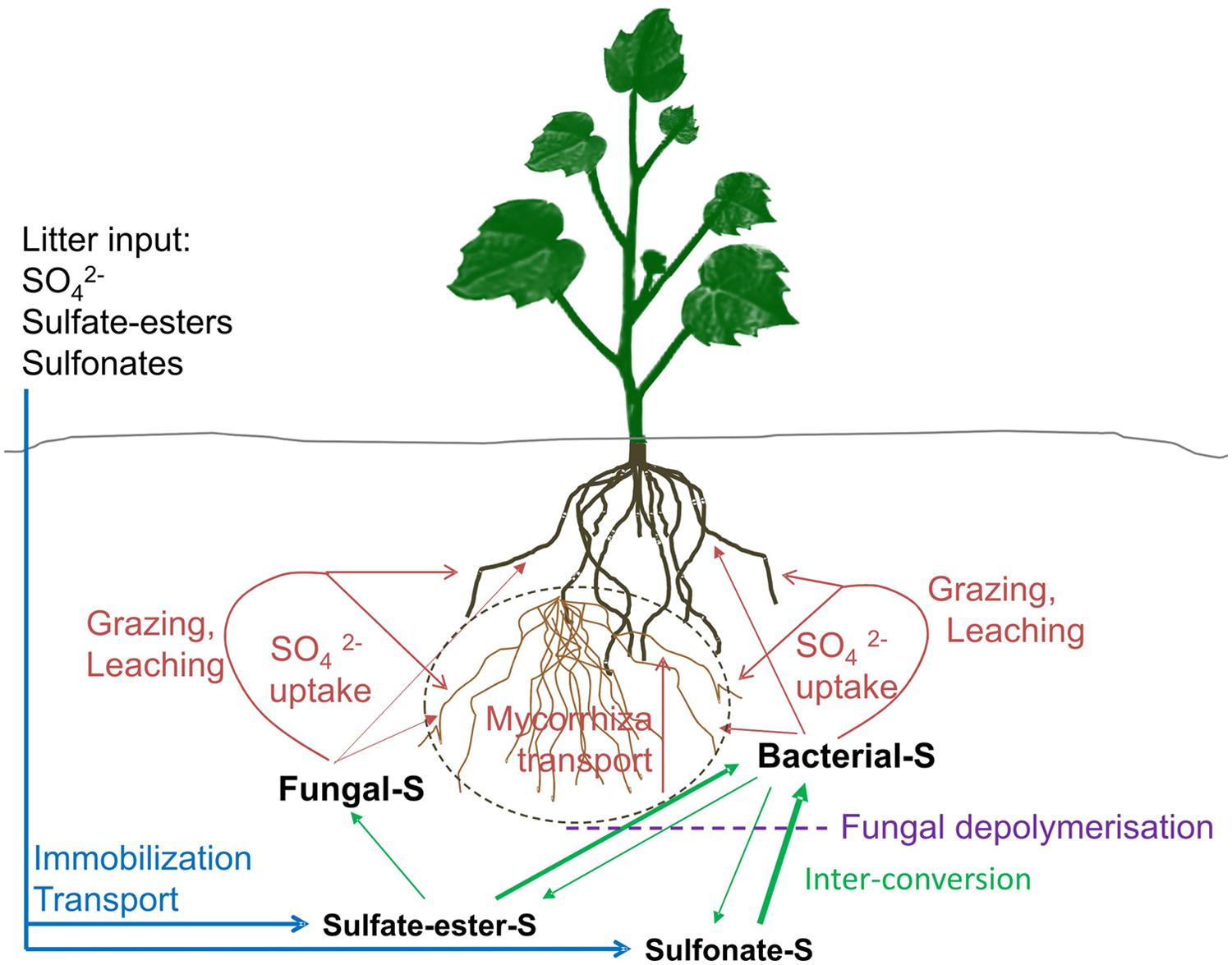
FIGURE 1. Illustration of the sulfur cycle in soil with plant cover. Major sulfur (S) inputs to soils originate from organic litter deposition and animal droppings (blue lines). Most of this deposited S is organically bound (organo-S). Atmospheric deposition of inorganic S has greatly declined in Europe, America and elsewhere, thus is often only a minor source for plants. Organo-S (sulfate-esters and sulfonates) can be transformed by soil microbes between the two major organo-S pools or mineralized to inorganic S (green lines, thickness suggests main direction of pathway). At the same time, inorganic S can be immobilized into organo-S (green lines). While the sulfate-ester pool is largely available to both fungi and bacteria, sulfonates are primarily accessible to bacteria only and aromatic sulfonates are only available to a particular functional clade of bacteria. Bacterial sulfonate desulfurization via the mono-oxygenase multi-enzyme pathway may occur intracellular, thus polymeric sulfonate may need depolymerisation, e.g., by saprophytic fungi prior to uptake (dotted purple line). Organo-S mineralised by fungi and bacteria need to be made available for plant uptake in the form of sulfate. This may happen via sulfate uptake by mycorrhizal fungal hyphae as an intermediate step (dashed gray line). In the absence of any direct evidence of a sulfate transport system from fungus or bacterium to the plant root or symbiotic mycorrhizal hyphae, release of mineralised S through autolysis and grazing by protists and microscopic nematodes may play an import role in inorganic sulfate release and plant sulfate uptake (red lines).
Many bacteria and fungi in soil are capable of mineralizing S from sulfate-esters (Klose et al., 1999). In contrast, an exclusively bacterial multicomponent mono-oxygenase enzyme complex is necessary to mobilize sulfonates, the dominant organo-S source in soil (Vermeij et al., 1999; Kertesz and Mirleau, 2004). In fact, soil S cycling may involve complex interactions between several free living and symbiotic root associated microbial populations. Arbuscular mycorrhizal (AM) fungi form symbiosis with 80% of land plant species which depend upon them for growth (Wang and Qiu, 2006). AM fungal symbiosis is characterized by fungal penetration of root cortical cells forming microscopic branched structures called arbuscules that increase efficiency of plant-fungus metabolite exchange (Smith and Read, 1997). Extra-radicular AM hyphae provide surfaces for functional bacterial populations to colonize. A number of studies have reported interactions between AM fungi and phosphorus (P) and nitrogen (N) mobilizing bacteria (Richardson et al., 2009; Hodge and Storer, 2014), and the impact of AM on bacterial community structures (Bianciotto and Bonfante, 2002; Toljander et al., 2007). Like S, both N, and P exist predominantly inaccessible to plants which rely on interactions with mycorrhizal fungi and associated microbes to facilitate their mobilization (Richardson et al., 2009).
Sulfur for Plant Growth
S owes its importance as a component of the (i) proteinaceous amino acids cysteine and methionine, (ii) non-protein amino acids including cystine, lanthionine, and ethionine (iii) tripeptide glutathione, and (iv) components including vitamins thiamine and biotin, phytochelatins, chlorophyll, coenzyme A, S-adenosyl-methionin and sulfolipids (Scherer, 2001). S plays critical structural roles in cells as disulphide bonds in proteins, is involved in enzyme regulation (redox control), provides protection from oxidative stress via glutathione, and its derivatives are involved in heavy metal stress mediation (Leustek and Saito, 1999). Plant S also plays an important role in disease protection and defense response as a component of glucosinolates and allin compounds (Jones et al., 2004; Brader et al., 2006). Various plant species prevent fungal infection via deposition of elemental S in the xylem parenchyma (Cooper and Williams, 2004).
Plant S demand is dependent on species and stage of development, with increased demand observed during periods of vegetative growth and seed development (Leustek and Saito, 1999). Inorganic sulfate () is the dominant plant available source of S, while to a lesser extent atmospheric reduced S may be utilized (Leustek et al., 2000). Regulation of uptake involves multiple transport steps and a large family of transporters have been characterized (Hawkesford, 2003). Assimilation of to cysteine occurs primarily in the chloroplasts of young leaves, while cysteine and methionine can also be synthesized in roots and seeds (Leustek and Saito, 1999). S starvation has been shown to negatively impact plant vitality when the P and N status is adequate (Sieh et al., 2013). During S limitation plant transporters are up-regulated for rapid up-take from the rhizosphere leading to a zone of depletion (Buchner et al., 2004). In this zone, bacterial desulfurization of organo-S is induced to mineralize organo-S, thus indirectly regulating plant S uptake (Kertesz and Mirleau, 2004). However, S-deficiency in plants can result in reduced root exudation (Alhendawi et al., 2005) or alteration of root exudates (Astolfi et al., 2010) which can influence bacterial communities seeking exudates as source of carbon.
X-ray absorption near edge structure (XANES) spectroscopy has revealed that sulfonates and sulfate-esters compose 30–70% and 20–60% of the organo-S in soil, respectively (Zhao et al., 2006). Directly plant available constitutes less than 5% of the total soil S (Autry and Fitzgerald, 1990). Organo-S compounds arise through deposition of biological material containing S, including plant and animal residues, and are subsequently incorporated into organic molecules through complex humification processes (Guggenberger, 2005). Animal residues are particularly high in organo-S with sheep dung comprising ∼80% of S as sulfonates, and while is rapidly leached from soil, organo-S can persist for longer time periods (Haynes and Williams, 1993). Additionally, soil-S pools are not static but rapidly interconverted between forms by soil microbial activity (Freney et al., 1975; Kertesz et al., 2007). Sulfonates were found to be mineralized more rapidly than other S-fractions and accounted for the majority of S released in short term incubation studies (Zhao et al., 2003, 2006). These findings indicate that C-bound S in soils may be of greatest importance (Ghani et al., 1992).
Microbial Mineralization of Organo-S
Microbial mineralization of organo-S is undertaken to access carbon, energy or S, with the latter also vital for plant growth (Ghani et al., 1992; Cook et al., 1998; Cook and Denger, 2002). Sulfate-ester mineralization is catalyzed by sulfatases of the esterase class (Deng and Tabatabai, 1997). Arylsulfatase enzymes act on aromatic sulfate-esters by splitting the O-S bond while alkylsulfatase enzymes act on aliphatic sulfate-esters by splitting the C-O bond (Kertesz, 1999). Both reactions release sulfate and are common in rhizospheric soil (Kertesz and Mirleau, 2004). Bacterial arylsulfatase activity is induced during S starvation and repressed in the presence of in Pseudomonas aeruginosa, while in a Streptomyces strain, a membrane bound sulfatase was also induced independently via substrate presence (Hummerjohann et al., 2000; Cregut et al., 2013). The ability to mobilize sulfate-esters has been observed in a range of bacteria including Pseudomonas, Klebsiella, Salmonella, Enterobacter, Serratia, and Comamonas (Hummerjohann et al., 2000). Additionally, arylsulfatase activity is influenced by various external factors including soil temperature, moisture content, vegetative cover, and crop rotation (Tabatabai and Bremner, 1970).
Fungi play an important role in the rhizosphere as plant symbionts or as free living saprotrophs. Soil filamentous fungi were reported to be important in mobilization of sulfate-esters (Omar and Abd-Alla, 2000; Baum and Hrynkiewicz, 2006), where enhanced arylsulfatase activity was found under S-limiting conditions (Fitzgerald, 1976; Marzluf, 1997). Likewise, wood-rotting fungi utilized sulfate-esters and thiols from wood (Schmalenberger et al., 2011).
The most abundant organo-S source in soil is present as aliphatic or aromatic sulfonates (Autry and Fitzgerald, 1990; Zhao et al., 2006). The ability to mobilize S from aliphatic sulfonates is widespread among soil bacteria with over 90% of morphologically distinct isolates capable of C2-sulfonate utilization (King and Quinn, 1997). However, aromatic sulfonates have been shown to be of greater importance for S nutrition and the ability to mobilize these sulfonates has been associated with plant growth promotion (PGP) of tomato (Kertesz and Mirleau, 2004) and Arabidopsis (Kertesz et al., 2007).
The desulfonating ability of the sewage sludge bacterial isolate Pseudomonas putida S-313 has been widely studied across a broad substrate range (Kertesz et al., 1994; Cook et al., 1998; Vermeij et al., 1999; Kahnert et al., 2000). Mobilization of from aromatic and aliphatic sulfonates is catalyzed by a FMNH2-dependent monooxygenase enzyme complex encoded in the ssu gene cluster (Eichhorn et al., 1999). The monooxygenase SsuD cleaves sulfonates to their corresponding aldehydes and the reduced flavin for this process is provided by the FMN-NADPH reductase SsuE. Although its function is unknown, ssuF from the ssu gene cluster was found to be essential for sulfonate desulfurization as well. For aromatic desulfonation the asfRABC gene cluster is required as an additional ‘tool-kit’ to complement ssu. The asf gene cluster includes a substrate binding protein, an ABC type transporter, a reductase/ferredoxin electron transport system involved in electron transfer and energy provision during oxygenation of the C-S bond, and a LysR-type regulatory protein, which activates the system during limitation (Vermeij et al., 1999). Transposon mutagenesis in the asfA gene of sewage isolate P. putida S-313 resulted in mutants without the capability to utilize aromatic sulfonates, while the utilization of aliphatic sulfonates was unchanged (Vermeij et al., 1999). This mutant was used in a plant growth experiment alongside its wild type, where the PGP effect was directly attributed to an functioning asfA gene (Kertesz and Mirleau, 2004). This particular type of bacterium has recently been isolated from the hyphae of symbiotic mycorrhizal fungi (Gahan and Schmalenberger, 2014). Various recent studies on the bacterial phylogeny of aromatic sulfonate mobilizing bacteria have expanded the diversity to the Beta-Proteobacteria; Variovorax, Polaromonas, Hydrogenophaga, Cupriavidus, Burkholderia, and Acidovorax, the Actinobacteria; Rhodococcus and the Gamma-Proteobacteria; Pseudomonas (Figure 2; Schmalenberger and Kertesz, 2007; Schmalenberger et al., 2008, 2009; Fox et al., 2014). Additionally, Stenotrophomonas and Williamsia species, isolated from hand-picked AM hyphae, have recently been added to these groups (Gahan and Schmalenberger, 2014).
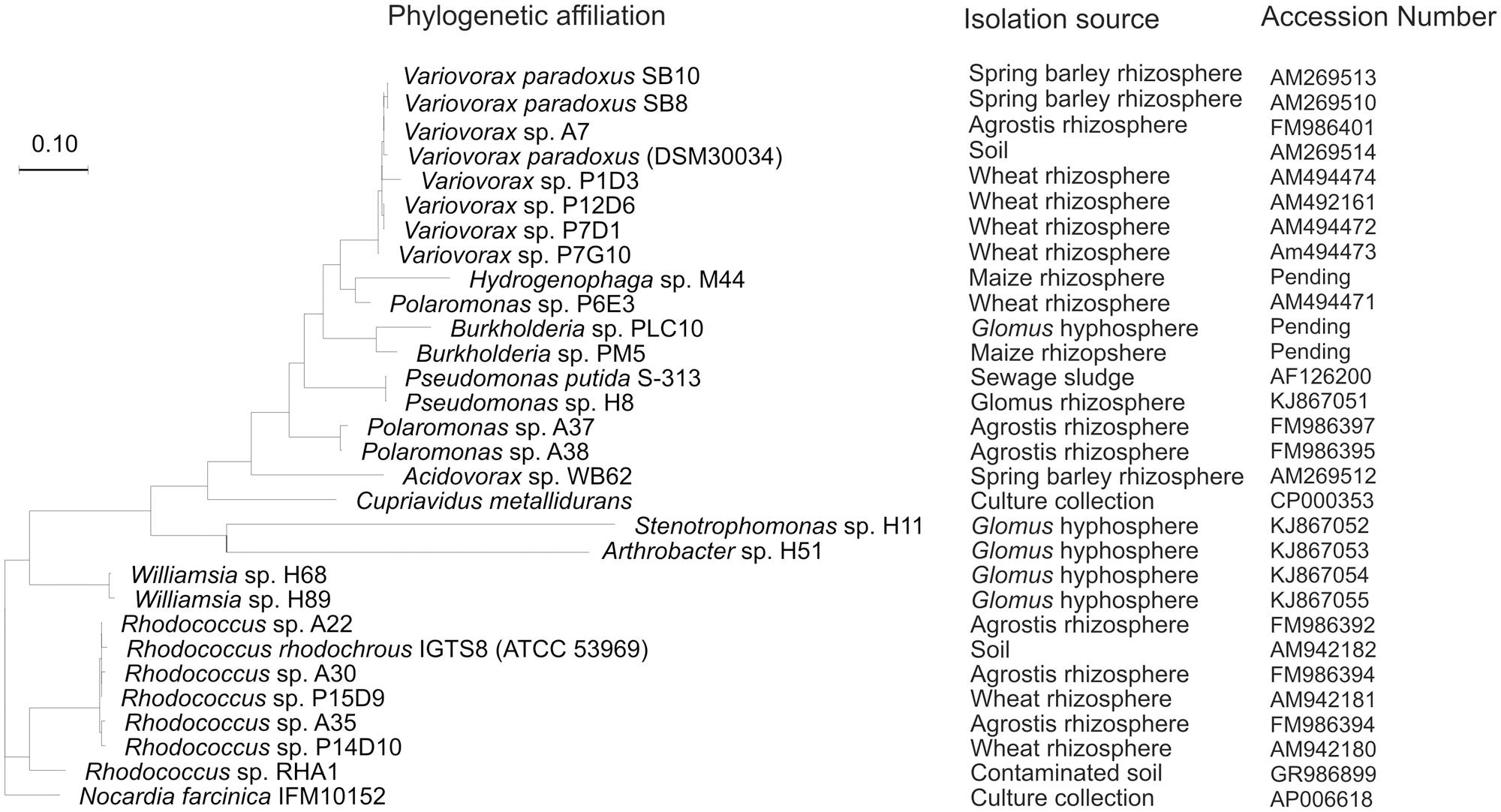
FIGURE 2. Randomized axelerated maximum likelihood tree from truncated AsfA sequences obtained from aromatic sulfonate desulfurizing bacteria isolated from soil, rhizosphere, or hyphosphere alongside strains from culture collections.
Until now, there has been little evidence to suggest fungal catalysis of sulfonate desulfurization (Kertesz et al., 2007; Schmalenberger et al., 2011). Indeed, while some saprotrophic fungi appear to breakdown some sulfonated molecules they do not release inorganic S in the process, for example, the white rot fungus Phanerochaete chrysporium transforms the aromatic alkylbenzene sulfonate but does so exclusively on its side chain without S-release (Yadav et al., 2001). Cultivation of fungi in vitro suggested that sulfonates could be utilized as an S source by wood degrading fungus Geophyllum trabeum, however, XANES spectra taken from wood accessible solely to the fungus displayed no evidence of sulfonate mobilization (Schmalenberger et al., 2011). Other cultivation experiments indicated a use of aliphatic sulfonates by various strains of yeasts via a putative 2-oxoglutarate dependent dioxygenase pathway (Uria-Nickelsen et al., 1993; Linder, 2012). However, this desulfurization capability may be limited to certain C4–C6 alkanesulfonates as this is the case for the taurine dioxygenase (Kertesz, 1999). Thus, the importance of bacteria and fungi with a dioxygenase pathway for sulfonate desulfurization is still somewhat unclear. As aforementioned, bacterial desulfonation based on the monooxygenase pathway occurs intracellularly and, as such, availability of sulfonates of different molecular size may be of importance. Therefore, saprotrophic fungi, including several genera of the Basidomycota, may play a role in sulfonate mobilization by secreting enzymes such as laccases and peroxidases in order to depolymerize large organic compounds in the soil (Figure 1; Muralikrishna and Renganathan, 1993; Tuor et al., 1995; Heinzkill et al., 1998). Lignolytic degradation of large organic complexes releases mono and oligomeric sulfonates which can be further mobilized by functional bacterial guilds as described above (Kertesz et al., 2007).
The Role of Arbuscular Mycorrhiza in Sulfur Supply
Arbuscular mycorrhizal fungi are the most common form of mycorrhizal association and their evolution can be dated back 460 million years (Smith and Read, 1997). They form symbiosis with 77% of angiosperms, 45% of 84 species of gymnosperms and 52% of 400 species of fern and lycopod (Wang and Qiu, 2006). The defining characteristic structure, the arbuscule, acts as an efficient site for plant-fungus metabolite exchange (Smith and Read, 1997). AM intra-radicular hyphae (IRH) provide the means for fungal extension within the host plant’s cortical region (Morton and Benny, 1990), while extra-radicular hyphae (ERH) have three primary functions – nutrient acquisition, infection of host plants, and production of fertile spores (Nagahashi and Douds, 2000).
Available studies on the effects of AM colonization on uptake of S have presented equivocal results (Gray and Gerdemann, 1973; Cooper and Tinker, 1978; Rhodes and Gerdemann, 1978). However, studies have shown that the presence of AM fungi enhances S uptake for maize, clover (Gray and Gerdemann, 1973) and tomato (Cavagnaro et al., 2006). More recently, AM fungus G. intraradices on transformed carrot roots demonstrated uptake of reduced forms of S in vitro (Allen and Shachar-Hill, 2009). Rates of this uptake and transfer of reduced S were comparable to that of when the latter was largely absent. Soil to root translocation is demand driven, with strongly induced absorption under conditions of S limitation. This rapid uptake of in the rhizosphere leads to a zone of depletion similar to that observed with P (Buchner et al., 2004). The AM fungal ERH could extend out past this zone of depletion and may play an important role in provision of S under conditions of S limitation (Kertesz et al., 2007). Recent investigations revealed that AM fungi can influence the expression of plant sulfate transporters and as a consequence improve the S nutritional status of the host plant (Giovannetti et al., 2014). This is important for all hyphospheric and rhizospheric soil microbes as lack of readily available sulfate in soil can lead to a reduction in plant exudates (Alhendawi et al., 2005) and as a consequence can affect soil microbial activity due to reduced availability of photosynthate as a source of carbon.
Extra-radicular hyphae are surrounded by complex bacterial and fungal communities that interact with the plant-mycorrhiza partnership and sustain its metabolic functioning (Frey-Klett and Garbaye, 2005). AM formation effects microbial communities in the rhizosphere via alteration of root exudates and translocation of energy rich C compounds to the extended soil environment for instance in the form of hyphal exudates (Barea et al., 2002; Boer et al., 2005). AM hyphae have a surface area several orders of magnitude greater than the plant roots which provides a niche for functional microbial interactions essential for nutrient cycling (Gryndler et al., 2000). Diverse soil microbial communities are essential for soil fertility and plant vitality (Gianinazzi and Schüepp, 1994; Siciliano et al., 2014) and AM hyphae have been shown to host a larger community of sulfonate desulfurizing bacteria than bulk soil (Gahan and Schmalenberger, 2014). Sulfonate desulfurization has been found to be characteristically rhizo- and hyphospheric in nature (Figure 2) and dominant sulfonate desulfurizing hyphospheric bacteria were found to be able to putatively attach and migrate with hyphae (Gahan and Schmalenberger, 2014). Inoculation of Lolium perenne soil microcosms with AM fungi significantly increased percentage root colonization and the quantity of cultivable sulfonate mobilizing bacteria (Gahan and Schmalenberger, 2013). Increased abundance of desulfonating bacteria as a result of elevated AM root colonization may be beneficial for plant-S supply. Likewise, addition of 2-(N-morpholine)-ethanesulfonic acid (MES) to soil putatively stimulated sulfonate mobilizing bacteria whose metabolites may have been responsible for the enhanced ERH growth of Glomus intraradices (Vilarino et al., 1997). This is important for maximizing S uptake as enhanced hyphal growth stemming from sulfonate mobilizing bacterial metabolites may further stimulate the proliferation of this community in a potential positive feedback loop. AM fungi may, therefore, play an increasingly important role in plant S metabolism not only through uptake and up-regulation of plant sulfate transporters but also through interaction with organo-S mobilizing microbes.
The hyphosphere of AM fungi can be regarded as a zone of increased bacterial abundance and activity, similar to the rhizosphere (Linderman, 1988; Andrade et al., 1998). Recent studies on the hyphosphere of ectomycorrhizae found that bacteria were co-migrating with the hyphae in vitro, putatively using a type III secretion system (T3SS) encoded infection needle for attachment (Warmink and van Elsas, 2008). This T3SS was also recently found to be present in aromatic sulfonate desulfurizing bacteria from the AM hyphosphere (Gahan and Schmalenberger, 2014), thus co-migration with ERH of AM fungi may be established via deployment of such an infection needle. While various pathogens are known to utilize T3SS for toxin injection into the host cells, nothing is known about any potential transfer of plant nutrients via such an infection needle to the mycorrhizal hyphae.
Currently, there is a profound knowledge gap when it comes to transfer of S from associated microbes to the plant host and its fungal symbiont. Extracellular sulfatases release S into soil solution which is then available to plant roots, mycorrhizal hyphae and various microbes, the release of S from sulfonates is potentially more complicated. While the possibility exists of a targeted transfer of S to the plant host via the ERH of AM fungi, there is currently no direct evidence provided in the literature. However, indirect release of S from sulfonate desulfurizing bacteria is a possibility. These bacteria may be turned over through grazing by microscopic predators such as nematodes and protozoa in the microbial loop (Bonkowski, 2004; Irshad et al., 2011). Indeed, soil amendments with biochar resulted not only in a significant increase in aromatic sulfonate desulfurizing bacteria but also in a significant increase in bacteria feeding nematodes (Fox et al., 2014), thus nematode activity may enhance the release of sulfonate desulfurized S in the rhizosphere and mycorrhizosphere/hyphosphere (Figure 1).
In conclusion, as a result of the limited nature of plant available S in soil it is increasingly necessary to understand the pathways and interactions required to mobilize the sulfate-esters and sulfonates that dominate the soil S pool. Saprotrophic fungi can depolymerize large humic material releasing sulfate-esters to bacteria and fungi, and sulfonates to specialist bacteria in possession of a monooxygenase enzyme complex. Desulfurizing microbial populations have been shown to be enriched in the rhizosphere and hyphosphere, however, released is quickly assimilated leaving an S depleted zone in the rhizosphere. AM fungi can extend past this zone, and indeed, are stimulated by organo-S mobilizing bacterial metabolites to expand their hyphal networks, increasing the area of soil and volume of S available to the plant. Additionally, inoculation with AM fungi has been shown to increase both percentage root colonization and the magnitude of the sulfonate mobilizing bacterial community. Inoculation practices, therefore, have huge potential to sustainably increase crop yield in areas where S is becoming a limiting factor to growth.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alhendawi, R. A., Kirkby, E. A., and Pilbeam, D. J. (2005). Evidence that sulfur deficiency enhances molybdenum transport in xylem sap of tomato plants. J. Plant Nutr. 28, 1347–1353. doi: 10.1081/PLN-200067449
Allen, J. W., and Shachar-Hill, Y. (2009). Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol. 149, 549–560. doi: 10.1104/pp.108.129866
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Andrade, G., Mihara, K., Linderman, R., and Bethlenfalvay, G. (1998). Soil aggregation status and rhizobacteria in the mycorrhizosphere. Plant Soil 202, 89–96. doi: 10.1023/A:1004301423150
Astolfi, S., Zuchi, S., Hubberten, H. M., Pinton, R., and Hoefgen, R. (2010). Supply of sulphur to S-deficient young barley seedlings restores their capability to cope with iron shortage. J. Exp. Bot. 61, 799–806. doi: 10.1093/jxb/erp346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Autry, A. R., and Fitzgerald, J. W. (1990). Sulfonate S: a major form of forest soil organic sulfur. Biol. Fertil. Soils 10, 50–56. doi: 10.1007/BF00336124
Barea, J.-M., Azcón, R., and Azcón-Aguilar, C. (2002). Mycorrhizosphere interactions to improve plant fitness and soil quality. Antonie Van Leeuwenhoek 81, 343–351. doi: 10.1023/A:1020588701325
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baum, C., and Hrynkiewicz, K. (2006). Clonal and seasonal shifts in communities of saprotrophic microfungi and soil enzyme activities in the mycorrhizosphere of Salix spp. J. Plant Nutr. Soil Sci. 169, 481–487. doi: 10.1002/jpln.200521922
Bianciotto, V., and Bonfante, P. (2002). Arbuscular mycorrhizal fungi: a specialised niche for rhizospheric and endocellular bacteria. Antonie Van Leeuwenhoek 81, 365–371. doi: 10.1023/A:1020544919072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boer, W. D., Folman, L. B., Summerbell, R. C., and Boddy, L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. doi: 10.1016/j.femsre.2004.11.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bonkowski, M. (2004). Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 162, 617–631. doi: 10.1111/j.1469-8137.2004.01066.x
Brader, G., Mikkelsen, M. D., Halkier, B. A., and Tapio Palva, E. (2006). Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 46, 758–767. doi: 10.1111/j.1365-313X.2006.02743.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buchner, P., Takahashi, H., and Hawkesford, M. J. (2004). Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55, 1765–1773. doi: 10.1093/jxb/erh206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cavagnaro, T. R., Jackson, L. E., Six, J., Ferris, H., Goyal, S., Asami, D.,et al. (2006). Arbuscular mycorrhizas, microbial communities, nutrient availability, and soil aggregates in organic tomato production. Plant Soil 282, 209–225. doi: 10.1007/s11104-005-5847-7
Cook, A. M., and Denger, K. (2002). Dissimilation of the C2 sulfonates. Arch. Microbiol. 179, 1–6. doi: 10.1007/s00203-002-0497-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cook, A. M., Laue, H., and Junker, F. (1998). Microbial desulfonation. FEMS Microbiol. Rev. 22, 399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x
Cooper, K. M., and Tinker, P. (1978). Translocation and transfer of nutrients in vesicular arbuscular mycorrhizas. New Phytol. 88, 327–339. doi: 10.1111/j.1469-8137.1981.tb01728.x
Cooper, R. M., and Williams, J. S. (2004). Elemental sulphur as an induced antifungal substance in plant defence. J. Exp. Bot. 55, 1947–1953. doi: 10.1093/jxb/erh179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cregut, M., Piutti, S., Slezack-Deschaumes, S., and Benizri, E. (2013). Compartmentalization and regulation of arylsulfatase activities in Streptomyces sp., Microbacterium sp. and Rhodococcus sp. soil isolates in response to inorganic sulfate limitation. Microbiol. Res. 168, 12–21. doi: 10.1016/j.micres.2012.08.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deng, S., and Tabatabai, M. (1997). Effect of tillage and residue management on enzyme activities in soils: III. Phosphatases and arylsulfatase. Biol. Fertil. Soils 24, 141–146. doi: 10.1007/s003740050222
Eichhorn, E., Van Der Ploeg, J. R., and Leisinger, T. (1999). Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274, 26639–26646. doi: 10.1074/jbc.274.38.26639
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Farrar, J., Hawes, M., Jones, D., and Lindow, S. (2003). How roots control the flux of carbon to the rhizosphere. Ecology 84, 827–837. doi: 10.1890/0012-9658(2003)084[0827:HRCTFO]2.0.CO;2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fitzgerald, J. W. (1976). Sulfate ester formation and hydrolysis: a potentially important yet often ignored aspect of the sulfur cycle of aerobic soils. Bacteriol. Rev. 40:698.
Fowler, D., Smith, R., Muller, J., Hayman, G., and Vincent, K. (2005). Changes in the atmospheric deposition of acidifying compounds in the UK between 1986 and 2001. Environ. Pollut. 137, 15–25. doi: 10.1016/j.envpol.2004.12.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fox, A., Kwapinski, W., Griffiths, B. S., and Schmalenberger, A. (2014). The role of sulfur and phosphorus mobilizing bacteria in biochar induced growth promotion of Lolium perenne. FEMS Microbiol. Ecol. 90, 78–91. doi: 10.1111/1574-6941.12374
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Freney, J., Melville, G., and Williams, C. (1975). Soil organic matter fractions as sources of plant-available sulphur. Soil Biol. Biochem. 7, 217–221. doi: 10.1016/0038-0717(75)90041-3
Frey-Klett, P., and Garbaye, J. (2005). Mycorrhiza helper bacteria: a promising model for the genomic analysis of fungal–bacterial interactions. New Phytol. 168, 4–8. doi: 10.1111/j.1469-8137.2005.01553.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gahan, J., and Schmalenberger, A. (2013). “Bacterial and fungal communities in the mycorrhizospheres of Agrostis, Lolium and Plantago respond to inoculation with arbuscular mycorrhizal fungi,” in Agricultural Research Forum 2014, ed. M. G. Diskin (Tullamore: Teagasc), 4.
Gahan, J., and Schmalenberger, A. (2014). Arbuscular mycorrhizal hyphae in grassland select for a diverse and abundant hyphospheric bacterial community involved in sulfonate desulfurization. Appl. Soil Ecol.
Ghani, A., Mclaren, R., and Swift, R. (1992). Sulphur mineralisation and transformations in soils as influenced by additions of carbon, nitrogen and sulphur. Soil Biol. Biochem. 24, 331–341. doi: 10.1016/0038-0717(92)90193-2
Gianinazzi, S., and Schüepp, H. (1994). Impact of Arbuscular Mycorrhizas on Sustainable Agriculture and Natural Ecosystems. Berlin: Birkhäuser (Springer). doi: 10.1007/978-3-0348-8504-1
Giovannetti, M., Tolosano, M., Volpe, V., Kopriva, S., and Bonfante, P. (2014). Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol. 204, 609–619. doi: 10.1111/nph.12949
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gray, L., and Gerdemann, J. (1973). Uptake of sulphur-35 by vesicular-arbuscular mycorrhizae. Plant Soil 39, 687–689. doi: 10.1007/BF00264184
Gryndler, M., Hršelová, H., and Stříteská, D. (2000). Effect of soil bacteria on hyphal growth of the arbuscular mycorrhizal fungus Glomus claroideum. Folia Microbiol. 45, 545–551. doi: 10.1007/BF02818724
Guggenberger, G. (2005). “Humification and mineralization in soils,” in Microorganisms in Soils: Roles in Genesis and Functions, eds F. Buscot and A. Varma (Berlin: Springer), 85–106. doi: 10.1007/3-540-26609-7_4
Hawkesford, M. J. (2003). Transporter gene families in plants: the sulphate transporter gene family—redundancy or specialization? Physiol. Plant. 117, 155–163. doi: 10.1034/j.1399-3054.2003.00034.x
Haynes, R., and Williams, P. (1993). Nutrient cycling and soil fertility in the grazed pasture ecosystem. Adv. Agron. 49, 119–199. doi: 10.1016/S0065-2113(08)60794-4
Heinzkill, M., Bech, L., Halkier, T., Schneider, P., and Anke, T. (1998). Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl. Environ. Microbiol. 64, 1601–1606.
Hodge, A., and Storer, K. (2014). Arbuscular mycorrhiza and nitrogen: implications for individual plants through to ecosystems. Plant Soil (in press). doi: 10.1007/s11104-014-2162-1
Hummerjohann, J., Laudenbach, S., Rétey, J., Leisinger, T., and Kertesz, M. A. (2000). The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol. 182, 2055–2058. doi: 10.1128/JB.182.7.2055-2058.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Irshad, U., Villenave, C., Brauman, A., and Plassard, C. (2011). Grazing by nematodes on rhizosphere bacteria enhances nitrate and phosphorus availability to Pinus pinaster seedlings. Soil Biol. Biochem. 43, 2121–2126. doi: 10.1016/j.soilbio.2011.06.015
Jones, M. G., Hughes, J., Tregova, A., Milne, J., Tomsett, A. B., and Collin, H. A. (2004). Biosynthesis of the flavour precursors of onion and garlic. J. Exp. Bot. 55, 1903–1918. doi: 10.1093/jxb/erh138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kahnert, A., Vermeij, P., Wietek, C., James, P., Leisinger, T., and Kertesz, M. A. (2000). The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol. 182, 2869–2878. doi: 10.1128/JB.182.10.2869-2878.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kertesz, M. A. (1999). Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24, 135–175.
Kertesz, M. A., Cook, A. M., and Leisinger, T. (1994). Microbial metabolism of sulfur and phosphorus-containing xenobiotics. FEMS Microbiol. Rev. 15, 195–215. doi: 10.1111/j.1574-6976.1994.tb00135.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kertesz, M. A., Fellows, E., and Schmalenberger, A. (2007). Rhizobacteria and plant sulfur supply. Adv. Appl. Microbiol. 62, 235–268. doi: 10.1016/S0065-2164(07)62008-5
Kertesz, M. A., and Mirleau, P. (2004). The role of microbes in plant sulphur supply. J. Exp. Bot. 55, 1939–1945. doi: 10.1093/jxb/erh176
King, J., and Quinn, J. (1997). The utilization of organosulphonates by soil and freshwater bacteria. Lett. Appl. Microbiol. 24, 474–478. doi: 10.1046/j.1472-765X.1997.00062.x
Klose, S., Moore, J. M., and Tabatabai, M. A. (1999). Arylsulfatase activity of microbial biomass in soils as affected by cropping systems. Biol. Fertil. Soils 29, 46–54. doi: 10.1007/s003740050523
Leustek, T., Martin, M. N., Bick, J. A., and Davies, J. P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Ann. Rev. Plant Biol. 51, 141–165. doi: 10.1146/annurev.arplant.51.1.141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leustek, T., and Saito, K. (1999). Sulfate transport and assimilation in plants. Plant Physiol. 120, 637–644. doi: 10.1104/pp.120.3.637
Linder, T. (2012). Genomics of alternative sulfur utilization in ascomycetous yeasts. Microbiology 158, 2585–2597. doi: 10.1099/mic.0.060285-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Linderman, R. (1988). Mycorrhizal interactions with the rhizosphere microflora: the mycorrhizosphere effect. Phytopathology 78, 366–371.
Lynch, J., and Whipps, J. (1990). Substrate flow in the rhizosphere. Plant Soil 129, 1–10. doi: 10.1007/BF00011685
Marzluf, G. A. (1997). Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 51, 73–96. doi: 10.1146/annurev.micro.51.1.73
Morton, J. B., and Benny, G. L. (1990). Revised classification of arbuscular mycorrhizal fungi (Zygomycetes): a new order, Glomales, two new suborders, Glomineae and Gigasporineae, and two new families, Acaulosporaceae and Gigasporaceae, with an emendation of Glomaceae. Mycotaxon 37, 471–491.
Muralikrishna, C., and Renganathan, V. (1993). Peroxidase-catalyzed desulfonation of 3, 5-dimethyl-4-hydroxy and 3, 5-dimethyl-4-aminobenzenesulfonic acids. Biochem. Biophys. Res. Commun. 197, 798–804. doi: 10.1006/bbrc.1993.2549
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nagahashi, G., and Douds, D. D. (2000). Partial separation of root exudate components and their effects upon the growth of germinated spores of AM fungi. Mycol. Res. 104, 1453–1464. doi: 10.1017/S0953756200002860
Omar, S., and Abd-Alla, M. (2000). Physiological aspects of fungi isolated from root nodules of faba bean (Vicia faba L.). Microbiol. Res. 154, 339–347. doi: 10.1016/S0944-5013(00)80008-7
Rhodes, L., and Gerdemann, J. (1978). Influence of phosphorus nutrition on sulfur uptake by vesicular-arbuscular mycorrhizae of onion. Soil Biol. Biochem. 10, 361–364. doi: 10.1016/0038-0717(78)90058-5
Richardson, A. E., Barea, J.-M., Mcneill, A. M., and Prigent-Combaret, C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. doi: 10.1007/s11104-009-9895-2
Russell, R. S. (1977). Plant Root Systems: Their Function and Interaction with Soil. London: McGraw-Hill Book Company.
Scherer, H. (2001). Sulphur in crop production—invited paper. Eur. J. Agron. 14, 81–111. doi: 10.1016/S1161-0301(00)00082-4
Schmalenberger, A., Hodge, S., Bryant, A., Hawkesford, M. J., Singh, B. K., and Kertesz, M. A. (2008). The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ. Microbiol. 10, 1486–1500. doi: 10.1111/j.1462-2920.2007.01564.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmalenberger, A., Hodge, S., Hawkesford, M. J., and Kertesz, M. A. (2009). Sulfonate desulfurization in Rhodococcus from wheat rhizosphere communities. FEMS Microbiol. Ecol. 67, 140–150. doi: 10.1111/j.1574-6941.2008.00602.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmalenberger, A., and Kertesz, M. A. (2007). Desulfurization of aromatic sulfonates by rhizosphere bacteria: high diversity of the asfA gene. Environ. Microbiol. 9, 535–545. doi: 10.1111/j.1462-2920.2006.01172.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmalenberger, A., Pritzkow, W., Ojeda, J. J., and Noll, M. (2011). Characterization of main sulfur source of wood-degrading Basidiomycetes by S K-edge X-ray absorption near edge spectroscopy (XANES). Int. Biodeterior. Biodegradation 65, 1215–1223. doi: 10.1016/j.ibiod.2011.08.013
Siciliano, S. D., Palmer, A. S., Winsley, T., Lamb, E., Bissett, A., Brown, M. V.,et al. (2014). Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol. Biochem. 78, 10–20. doi: 10.1016/j.soilbio.2014.07.005
Sieh, D., Watanabe, M., Devers, E. A., Brueckner, F., Hoefgen, R., and Krajinski, F. (2013). The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol. 197, 606–616. doi: 10.1111/nph.12034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tabatabai, M., and Bremner, J. (1970). Arylsulfatase activity of soils. Crop Sci. Soc. Am. J. 34, 225–229. doi: 10.2136/sssaj1970.03615995003400020016x
Toljander, J. F., Lindahl, B. D., Paul, L. R., Elfstrand, M., and Finlay, R. D. (2007). Influence of arbuscular mycorrhizal mycelial exudates on soil bacterial growth and community structure. FEMS Microbiol. Ecol. 61, 295–304. doi: 10.1111/j.1574-6941.2007.00337.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tuor, U., Winterhalter, K., and Fiechter, A. (1995). Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J. Biotechnol. 41, 1–17. doi: 10.1016/0168-1656(95)00042-O
Uria-Nickelsen, M. R., Leadbetter, E. R., and Godchaux, W. III. (1993). Sulfonate-sulfur assimilation by yeasts resembles that of bacteria. FEMS Microbiol. Lett. 114, 73–77. doi: 10.1111/j.1574-6968.1993.tb06553.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vermeij, P., Wietek, C., Kahnert, A., Wüest, T., and Kertesz, M. A. (1999). Genetic organization of sulphur-controlled aryldesulphonation in Pseudomonas putida S-313. Mol. Microbiol. 32, 913–926. doi: 10.1046/j.1365-2958.1999.01398.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vilarino, A., Frey, B., and Shüepp, H. (1997). MES [2-(N-morpholine)-ethane sulphonic acid] buffer promotes the growth of external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices in an alkaline sand. Biol. Fertil. Soils 25, 79–81. doi: 10.1007/s003740050284
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warmink, J. A., and van Elsas, J. D. (2008). Selection of bacterial populations in the mycosphere of Laccaria proxima: is type III secretion involved. ISME J. 2, 887–900. doi: 10.1038/ismej.2008.41
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yadav, J. S., Lawrence, D. L., Nuck, B. A., Federle, T. W., and Reddy, C. A. (2001). Biotransformation of linear alkylbenzene sulfonate (LAS) by Phanerochaete chrysosporium: oxidation of alkyl side-chain. Biodegradation 12, 443–453. doi: 10.1023/A:1015056530264
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, F., Knights, J., Hu, Z., and Mcgrath, S. (2003). Stable sulfur isotope ratio indicates long-term changes in sulfur deposition in the Broadbalk experiment since 1845. J. Environ. Qual. 32, 33–39. doi: 10.2134/jeq2003.3300
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: sulfonate desulfurization, sulfate esters, mycorrhizal fungi, plant–microbe interactions, asf gene cluster, sulfatases, mycorrhizosphere
Citation: Gahan J and Schmalenberger A (2014) The role of bacteria and mycorrhiza in plant sulfur supply. Front. Plant Sci. 5:723. doi: 10.3389/fpls.2014.00723
Received: 30 September 2014; Accepted: 01 December 2014;
Published online: 16 December 2014.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Tamara Gigolashvili, University of Cologne, GermanyMichael Kertesz, University of Sydney, Australia
Copyright © 2014 Gahan and Schmalenberger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Achim Schmalenberger, Department of Life Sciences, University of Limerick, Castletroy, Limerick, Ireland e-mail: achim.schmalenberger@ul.ie
 Jacinta Gahan
Jacinta Gahan Achim Schmalenberger
Achim Schmalenberger