- 1Plant Physiology and Morphology Laboratory, Crop Science Department, Agricultural University of Athens, Athens, Greece
- 2General and Agricultural Microbiology Laboratory, Crop Science Department, Agricultural University of Athens, Athens, Greece
Nicotianamine is an essential molecule for Fe homeostasis in plants, its primary precursor is the S-containing compound methionine, and it is biosynthesized by the enzyme family of nicotianamine synthases (NASs). In maize, a graminaceous plant that follows Strategy II for Fe uptake, ZmNAS genes can be subgrouped into two classes, according to their roles and tissue specific expression profiles. In roots, the genes of class I provide NA for the production of deoxymugineic acid (DMA), which is secreted to the rhizosphere and chelates Fe(III). The Fe(III)-DMA complex is then inserted to the root via a ZmYS1 transporter. The genes of class II provide NA for local translocation and detoxification of Fe in the leaves. Due to the connection between S and Fe homeostasis, S deficiency causes Fe deprivation responses to graminaceous plants and when S is supplied, these responses are inverted. In this study, maize plants were grown in pots with sterile river sand containing FePO4 and were inoculated with the mycorrhizal fungus Rhizophagus irregularis. The plants were grown under S deficient conditions until day 60 from sowing and on that day sulfate was provided to the plants. In order to assess the impact of AM symbiosis on Fe homeostasis, the expression patterns of ZmNAS1, ZmNAS3 (representatives of ZmNAS class I and class II), and ZmYS1 were monitored before and after S supply by means of real time RT-PCR and they were used as indicators of the plant Fe status. In addition, total shoot Fe concentration was determined before and after S supply. AM symbiosis prevented Fe deprivation responses in the S deprived maize plants and iron was possibly provided directly to the mycorrhizal plants through the fungal network. Furthermore, sulfate possibly regulated the expression of all three genes revealing its potential role as signal molecule for Fe homeostasis.
Introduction
Iron is an essential micronutrient for plants. Graminaceous plants follow the Strategy II for iron acquisition from the rhizosphere. Iron homeostasis in maize involves a series of processes, including the biosynthesis of deoxymugineic acid (DMA) for iron uptake from the rhizosphere and the translocation of iron throughout the plant body toward the sink organs (Kobayashi et al., 2006). In this iron uptake pathway, three molecules of S-adenosyl-methionine are combined to form nicotianamine (NA) which is then used as the precursor for DMA biosynthesis. DMA is secreted to the rhizosphere where it chelates Fe(III) and the complex DMA-Fe(III) is inserted into the root cell via the YS1 transporter (Curie et al., 2001; Nozoye et al., 2013). In addition to its role in iron uptake, NA plays also a dominant role in iron transfer, being used for the intercellular and intracellular Fe transport in all plants' organs, as well as long distance transport through the phloem (Kobayashi et al., 2006; Zhou et al., 2013b).
The primary precursor of NA is methionine, a sulfur-containing amino acid, so sulfur deprivation has a strong effect on iron homeostasis; as a result, S deficiency causes Fe deprivation responses to the graminaceous plants, which can be inverted when S is provided (Astolfi et al., 2003, 2010; Bouranis et al., 2003). The strong connection between sulfur and iron is typified by the Fe-S clusters, where most of the metabolically active Fe is bound to S. In chloroplasts, the most abundant Fe-S proteins are ferredoxin, photosystem I and cytochrome b6f complex. In mitochondria, major Fe-S proteins are complex I, II and III of the respiratory chain and aconitase in the citric acid cycle. This connection between the two nutrients suggests coordination between the metabolisms of S and Fe (Forieri et al., 2013; Vigani et al., 2013).
The enzyme family of nicotianamine synthases (NASs) produces NA using S-adenosyl-methionine as substrate molecule. Recent studies revealed the evolutionary relationship and tissue specific expression profiles of ZmNAS genes in maize leading to their grouping into two classes. The ZmNASs of class I are mainly expressed in the roots when iron is sufficient (Zhou et al., 2013a,b). ZmNASs of class I are mostly responsible for providing the precursor for DMA synthesis as well as for the long distance translocation of Fe in stem (Mizuno et al., 2003; Zhou et al., 2013b). The ZmNASs of class II are commonly accumulated in meristems and mesophyll cells of the leaves (Zhou et al., 2013a,b). These NAS genes are important for local iron distribution in leaves and sheaths and play a key role in iron homeostasis and detoxification (Mizuno et al., 2003; Zhou et al., 2013b). All ZmNAS genes of class I are induced as a response to Fe deficiency, while in Fe excess they are downregulated. On the other hand, all class II ZmNASs are downregulated in Fe deficiency and retain their expression levels or get overexpressed in Fe excess (Mizuno et al., 2003; Zhou et al., 2013b).
ZmYS1 is a membrane protein and functions as a proton-coupled symporter that mediates iron uptake in maize. ZmYS1 expression at both the mRNA and protein levels responds rapidly to changes in iron availability, whilst it is not regulated by zinc or copper deficiency (Curie et al., 2001; Roberts et al., 2004; Schaaf et al., 2004; Nozoye et al., 2013).
Arbuscular mycorrhizal (AM) symbiosis improves plant nutrient uptake under low nutrient availability (Bonfante and Genre, 2008). The role of AM symbiosis on phosphate has been extensively studied (Smith et al., 1994, 2003; Javot et al., 2007) and there seems to be a specific symbiotic phosphorus acquisition pathway (Harrison et al., 2002). Moreover, other nutrients such as nitrogen and sulfur are shown to be translocated from the fungal to the plant partner (Ames et al., 1983; Allen and Shachar-Hill, 2009; Leigh et al., 2009; Sieh et al., 2013). Few studies have been conducted in order to reveal the impact of AM symbiosis on iron uptake and the role of this symbiosis to plant iron homeostasis is still unclear. Studies on sorghum revealed that AM fungi can mobilize and/or take up Fe from soil and translocate it to the plant (Caris et al., 1998). However, other studies on maize propose that AM fungi increase total Fe in the shoot in the absence of other micronutrients and only in low P levels (Liu et al., 2000).
In this study, mycorrhizal and non-mycorrhizal plants were grown under prolonged sulfur deficiency in the presence of insoluble iron, as FePO4 salt. This salt has been used in previous studies, as practically insoluble form of phosphate, for the investigation of the impact of AM fungi on plant growth or P nutrition (Bolan et al., 1987; Virant-Klun and Gogala, 1995). In our study FePO4 had a dual role: it was the only source for Fe and the main source for P as the nutrient solution provided was P insufficient in order to promote the establishment of mycorrhizal symbiosis. Two months after sowing, sulfur was supplied to the plants, in the form of sulfate, to promote iron acquisition. The period before S supply was used for the determination of the impact of AM symbiosis on Fe status under long-term S deficient conditions while after S supply, components of the iron acquisition pathway were mainly investigated. The expression patterns of two ZmNASs were monitored in roots and leaves and ZmYS1 was monitored in roots before and after sulfur repletion. Taking into account the expression profiles and tissue specific localization of ZmNASs, ZmNAS1 was selected as representative of class I and ZmNAS3 as representative of class II. In order to monitor the influence of AM symbiosis on Fe homeostasis, the expression patterns of ZmNAS1 and ZmYS1 in the roots, ZmNAS3 in the leaves as well as total Fe concentrations in the shoots were used as indicators of the plants' iron status before and immediately after (24 and 48 h) sulfur supply.
Materials and Methods
Plant Material and Growth Conditions
Maize (Zea mays L., “Cisko,” Syngenta Hellas) seeds were thoroughly washed and placed on wet filter paper, in the dark at 28°C to germinate for 4 days. Then, the seedlings were transferred to batch culture boxes and grew hydroponically in well-aerated distilled H2O for the next 4 days. On day eight from sowing, all parts below the crown (i.e., mesocotyl and embryonic root system) as well as the seed were detached from the seedlings, which were thereafter, hydroponically grown in well-aerated nutrient solution, completely deprived of Fe and S and containing a low P concentration (10 μM), for 2 days. On day 10 from sowing, the seedlings were transferred to individual pots with sterile river sand (121°C for 1 h, 250 ml per pot) and the addition of practically insoluble FePO4(500 mg per pot). For the mycorrhizal treatment, 300 mg of Rhizophagus irregularis inoculum (synonym: Glomus irregulare DAOM197198, SYMPLANTA-001 standard grade, Symplanta) were added to each pot. The plants were watered two times a week with the Fe and S deficient nutrient solution until day 60 from sowing. Sulfur was provided in the form of sulfate on day 60, with the use of a nutrient solution only deprived of Fe. The nutrient solution deprived of Fe and S contained 5 mM KNO3, 10 μM KH2PO4, 2 mM Mg(NO3)2 6H2O, 4 mM Ca(NO3)2 4H2O, 0.86 mM CaCl2 2H2O, 0.9 μM ZnCl2, 30 μM H3BO3, 0.9 μM CuCl2 2H2O, 0.5 μM MoO3 85%, and 20 μM MnCl2 4H2O. The iron-deficient nutrient solution applied on day 60 contained 5 mM KNO3, 10 μM KH2PO4, 2 mM Mg(NO3)2 6H2O, 2.5 mM CaSO4 2H2O, 1 mM MgSO4 7H2O, 4 mM Ca(NO3)2 4H2O, 0.9 μM ZnCl2, 30 μM H3BO3, 0.9 μM CuCl2 2H2O, 0.5 μM MoO3 85%, and 20 μM MnCl2 4H2O. A controlled environment of 250 μmol photons m−2 s−1 photosynthetic photon flux density and a 14-h light photoperiod with day/night growth conditions at shoot base 28/23°C and RH 36/40% was used.
Plant Samplings for Gene Expression Analysis
Samplings were performed on days 30, 45, 60, 61 (24 h after sulfur supply), and 62 (48 h after sulfur supply) from sowing and 3 h after the onset of light. The sampling of day 60 took place before the addition of sulfur. A schematic illustration of the experimental design, indicating also the days of the samplings, is presented in Figure 1. Lateral roots as well as two young expanding leaves were immediately frozen in liquid nitrogen and stored at −80°C until use. On each experiment, plant material from at least three biological replicates per treatment and sampling day was used. Lateral roots were chosen as root samples for the gene expression analysis because, according to Gutjahr and Paszkowski (2013), in roots of monocotyledon plants AM fungi preferentially colonize lateral roots. The young expanding leaves were chosen as strong sinks of Fe.
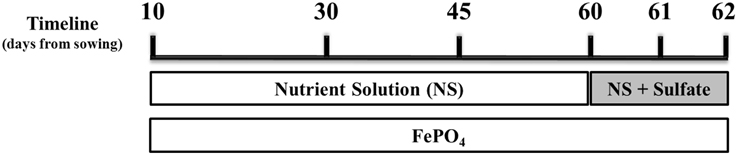
Figure 1. Schematic illustration of the experimental design. Mycorrhizal and non-mycorrhizal plants were grown under S deficient conditions until day 60; all plants were watered with a nutrient solution deprived of Fe and S and containing a low P concentration (10 μM). On day 60 S was provided to the plants as sulfate. Fe was provided to the plants in the form of sparingly soluble FePO4, throughout the experiment. Samplings took place on days 30, 45, 60 (before S supply), 61 and 62.
Primer Design
Performing Blast searches we identified the cDNA sequences of all recorded ZmNAS and ZmYS genes. Primers were designed for a representative of each class of the ZmNAS gene family as well as for ZmYS1 (MaizeGDB: GRMZM2G156599). ZmNAS1;1 (MaizeGDB: GRMZM2G385200) and ZmNAS3 (MaizeGDB: GRMZM2G478568) were chosen for class I and II respectively. Real-time PCR primers were designed to amplify 100–200 bp fragments in the 3′untranslated regions, using the Primer Blast tool of NCBI. All primers were designed for 60°C annealing temperature and their sequences are as follows: ZmNAS1;1: Forward 5′-GGAACTTTTGAGCACCTATGCG-3′ and Reverse 5′-CACTTCACAATGCATAGCATCGAAT-3′; ZmNAS3: Forward 5′-CGTGTCTACACCACATGCGT-3′ and Reverse 5′-TCGGACTTCGACTTCTACCCT-3′; ZmYS1: Forward 5′-GTCTTCCATTCTCGCTCTGG-3′ and Reverse 5′-CAACCAACCACAGTTGATGC-3′. The gene of ubiquitin was used as an internal control and the target was detected by the following primers-pair: ZmUBQ (NCBI: NM_001138130): Forward 5′-TGTCTTCATGGCCAACCACT-3′ and Reverse 5′-GCTTGATAGGTAGGCGGGTG-3′.
RNA Extraction and Real-Time RT-PCR
Total nucleic acids were extracted from root and shoot samples using the Phenol-Chloroform protocol (Brusslan and Tobin, 1992) and were treated thereafter with Recombinant DNase I (RNase-free, Takara Bio Inc) in order to get the total RNA of each sample. An amount of 500 ng of RNA was reverse-transcribed using the PrimeScript RT reagent (Perfect Real Time, Takara Bio Inc).
Measurements of real-time RT-PCR were performed using KAPA SYBR FAST Master Mix (KAPA Biosystems) in the MxPro Mx3005P thermocycler (Stratagene, USA). The real-time RT-PCR was performed according to the respective protocol of the kit, using optical 96-well plates with the following PCR program: 95°C for 10 min, 40 cycles of 95°C for 30 s and 60°C for 1 min and the final cycle of 95°C for 1 min and 60°C for 30 s and 95°C for 30 s. Melting curve analysis was carried out after each amplification to exclude unspecific amplifications from the analysis. PCR amplification efficiencies were obtained using the LinRegPCR software (Ruijter et al., 2009). The relative expression ratios were calculated with the mathematical formula of Pfaffl (2001), using as reference the gene of ubiquitin and as targets the genes of ZmNAS1, ZmNAS3, and ZmYS1. As control the samples of day 30 (for the samplings before sulfur supply) or day 60 (for the samplings after sulfur supply) of the respective treatment were used.
Verification of Mycorrhizal Colonization
In order to confirm the mycorrhizal colonization in the root samples two tests were performed, using both a histological and a molecular approach. Large and fine lateral roots were cleared in 2.5% KOH and stained with Trypan blue (Koske and Gemma, 1989) for fungal detection. For the molecular approach, using the total pre-DNase-treated nucleic acids extract, a PCR was performed with primers specific for the internal transcribed spacer 1 of the ribosomal RNA gene of R. irregularis (NCBI: JF820567). The sequences of the primers were the following ones: Forward 5′-TGATCTTTGATCATGGTTTCGC-3′ and Reverse 5′-TCGCACTTCGCTACGTTCTT-3′.
Total Fe Determination
Samples of maize shoots were used for the total Fe determination and were harvested on days 45, 60, and 65. The sampling of day 60 was conducted before sulfur supply. Samples were oven-dried at 80°C, the dry weight was recorded and the appropriate dry mass was ground to pass a 40 mesh screen using an analytical mill (IKA, model A10) prior to chemical analysis. Samples were digested with hot H2SO4 and repeated additions of 30% H2O2 until the digestion was complete, and thereafter, total Fe was determined in the diluted digests by atomic absorption spectrophotometry (GBC, Model Avanta spectrophotometer) (Mills and Jones, 1996).
Statistical Analysis
The experiment was performed two times under the same conditions and during two distinct time periods: autumn 2013 and spring 2014. Data were analyzed by t-test variance analysis with two-tailed distribution and two-sample unequal variance to determine the significance of differences among samplings.
Results
Verification of Mycorrhizal Colonization
All mycorrhizal plants used were verified for the presence of R. irregularis prior to further analysis. Staining with Trypan blue showed that the roots of the maize plants were already colonized by the fungus on day 45. Arbuscules, vesicles and fungal hyphae were present in all large as well as fine lateral root samples examined. The molecular approach used to verify the mycorrhizal colonization revealed fungal presence in the root samples of mycorrhizal plants from day 45 until the end of the experiment (Figure 2). As depicted on Figure 2 the amplification products of the internal transcribed spacer 1 of the ribosomal RNA of R. irregularis have been comparable between the two repetitions of the experiment.
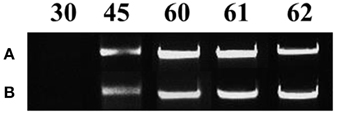
Figure 2. Verification of mycorrhizal colonization via PCR amplification of the internal transcribed spacer 1 of the ribosomal RNA gene of Rhizophagus irregularis in the root samples of the mycorrhizal treatment. Each amplification product illustrates a representative biological replicate of every sampling. Numbers indicate the respective sampling days. A: first repetition of the experiment (autumn 2013), B: second repetition of the experiment (spring 2014).
Expression Levels of ZmNAS1 in the Roots and ZmNAS3 in the Leaves
The expression profiles of ZmNAS1 and ZmNAS3 were monitored in both roots and leaves. The corresponding profiles of ZmNAS1 in the leaves and ZmNAS3 in the roots are provided as supplemental data (Figures S1, S2).
On day 30 there was no significant difference in the expression levels of each gene between mycorrhizal and non-mycorrhizal plants (Figure 3, insets). Before sulfur supply, the roots of non-mycorrhizal plants revealed no significant change in the expression of ZmNAS1 on day 45 which was followed by a significant upregulation on day 60 (Figure 3A). On the other hand, mycorrhizal roots showed a differential response; ZmNAS1 was downregulated on both days 45 and 60 (Figure 3A). In the leaves of non-mycorrhizal plants, ZmNAS3 was downregulated on both days before S repletion while the leaves of mycorrhizal plants showed an overexpression of ZmNAS3 on day 60 (Figure 3B).
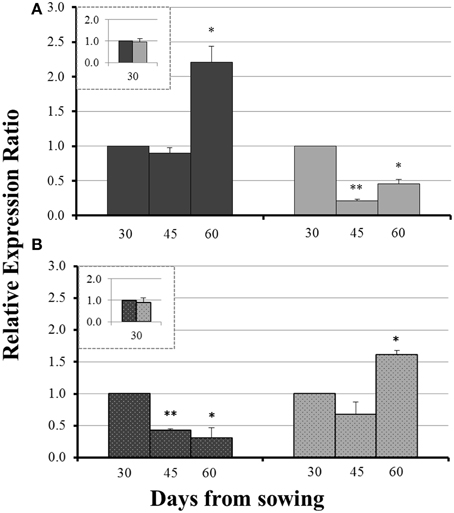
Figure 3. Expression of ZmNAS1 in the roots (A) and ZmNAS3 in the leaves (B) of non-mycorrhizal (black columns) and mycorrhizal (gray columns) maize plants, before sulfur supply, relative to the expression of ubiquitin. Day 30 of each treatment was used as control for the calculation of the relative expression ratios. The inset provides the relative expression ratio of each gene in the mycorrhizal plants on day 30, using the respective sample of non-mycorrhizal plants as control. Bars show the mean of the biological replicates ± SE, */** indicated when the difference between the sampling and the respective control is statistically significant at p < 0.05/0.005 respectively.
The influx of sulfate, 24 h after sulfur supply (day 61), resulted in a common response of both ZmNAS1 in the roots and ZmNAS3 in the leaves between mycorrhizal and non-mycorrhizal plants. Both genes were downregulated, 24 h after sulfur repletion, in all plants irrespective of the fungal presence (Figure 4). However, 48 h after sulfur supply (day 62), mycorrhizal plants showed again a differential response in relation to non-mycorrhizal plants. While in non-mycorrhizal plants both ZmNAS1 in roots and ZmNAS3 in leaves were upregulated, these genes presented a strong downregulation in the corresponding organs of mycorrhizal plants (Figure 4).
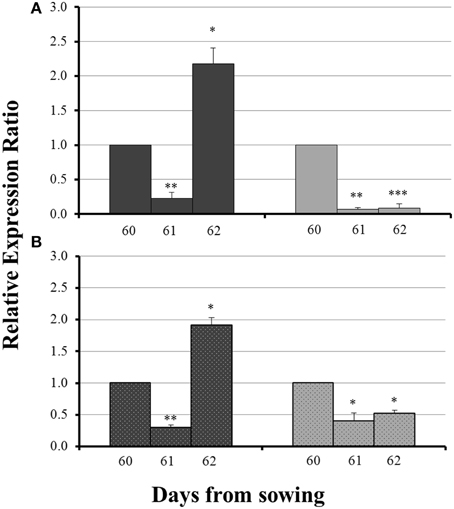
Figure 4. Expression of ZmNAS1 in the roots (A) and ZmNAS3 in the leaves (B) of non-mycorrhizal (black columns) and mycorrhizal (gray columns) maize plants, after sulfur supply, relative to the expression of ubiquitin. Day 60 of each treatment was used as control for the calculation of the relative expression ratios. Bars show the mean of the biological replicates ± SE, */**/*** indicated when the difference between the sampling and the respective control is statistically significant at p < 0.05/0.005/0.0005 respectively.
Expression Levels of ZmYS1 in the Roots
No significant difference in the expression levels of ZmYS1 between mycorrhizal and non-mycorrhizal roots was monitored on day 30 (Figure 5, inset). Before sulfur supply, ZmYS1 was significantly overexpressed in the roots of non-mycorrhizal plants (Figure 5A). On the other hand, mycorrhizal roots did not show any considerable response and the expression ratios of ZmYS1 remained stable until day 60 (Figure 5A). The influx of sulfate, 24 h after sulfur supply (day 61), resulted again in a downregulation of ZmYS1 in the roots of all plants (Figure 5B). However, 48 h after sulfur supply (day 62), mycorrhizal plants revealed once more a differential response against non-mycorrhizal plants. ZmYS1 was strongly upregulated in non-mycorrhizal roots while mycorrhizal roots presented an intense downregulation (Figure 5B).
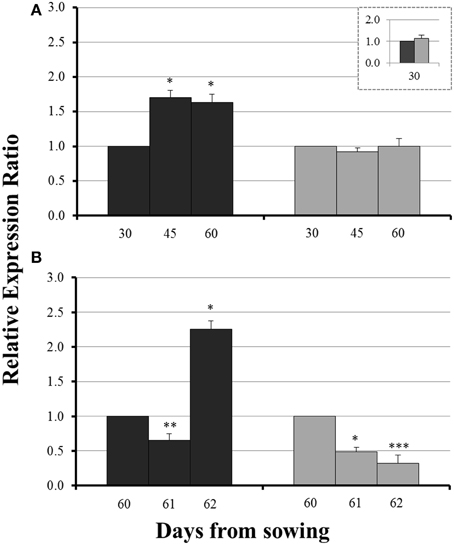
Figure 5. Expression of ZmYS1 in the roots of non-mycorrhizal (black columns) and mycorrhizal (gray columns) maize plants, before (A) and after (B) sulfur supply, relative to the expression of ubiquitin. Day 30 or day 60 of each treatment was used accordingly as control for the calculation of the relative expression ratios. Bars show the mean of the biological replicates ± SE, */**/*** indicated when the difference between the sampling and the respective control is statistically significant at p < 0.05/0.005/0.0005 respectively.
Total Fe Concentrations in the Shoots
Total Fe concentration of the aerial part of non-mycorrhizal plants decreased significantly from day 45 to day 60. In the mycorrhizal plants, the corresponding concentrations of total Fe presented no significant change, although the dry mass was increasing (data not shown). On day 65, i.e., 5 days after the addition of sulfate, total Fe concentration increased in the shoots of all plants. Interestingly, Fe concentration in mycorrhizal plants on that day was found to be 3.4 times higher than the corresponding one of non-mycorrhizal plants (Table 1).
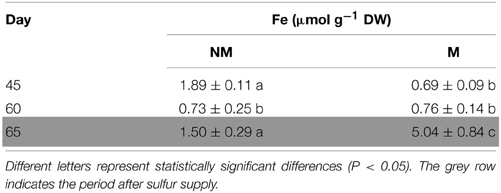
Table 1. Time course of total Fe concentrations (mean values ± SE) in the shoots of mycorrhizal (M) and non-mycorrhizal (NM) maize plants.
Discussion
Mycorrhizal Colonization Prevents Fe Deprivation Responses in S Deprived Maize Plants
AM symbiosis modifies the nutrient status of the plants; however the impact of AM colonization on the plants' iron status has not been clarified, yet. Only a few studies on graminaceous plants have shown a potential contribution of AM fungi in Fe uptake and they aimed to examine whether and/or how much Fe is transferred to the mycorrhizal plants in contrast to non-mycorrhizal plants (Caris et al., 1998; Liu et al., 2000). In our work, three key Fe homeostasis genes were used for the assessment of the effect of AM symbiosis on Fe homeostasis in maize plants.
Class I ZmNAS genes are mainly expressed in the roots and play a crucial role in the Strategy II Fe acquisition pathway, providing the essential NA molecules for the formation of DMA. The genes of class II are mostly expressed in the leaves and are involved in the short distance translocation and/or detoxification of Fe (Mizuno et al., 2003; Zhou et al., 2013a,b). When Fe is depleted, class I genes are induced, promoting the Strategy II Fe acquisition pathway and class II genes are downregulated, possibly because under such conditions there are lower needs of Fe transport in the leaves. Moreover, the gene of the Fe(III)-DMA chelate transporter, ZmYS1, is strongly and rapidly induced in Fe deprived conditions (Roberts et al., 2004). However, when Fe is in excess, class I ZmNAS genes are downregulated and the genes of class II either retain their expression levels or get overexpressed (Zhou et al., 2013b). ZmNAS1 and ZmNAS3 are members of class I and class II, respectively, so their expression patterns follow the expression profiles mentioned above, when Fe is either depleted or in excess (Table 2).
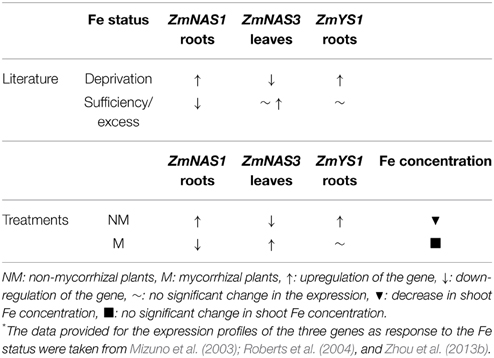
Table 2. Alterations in the expression profiles of ZmNAS1, ZmNAS3, and ZmYS1 as response to Fe status (according to the literature*) and the treatments conducted in this study before S supply.
In this study, sulfur deficient conditions had a strong, negative impact on the Strategy II Fe acquisition pathway. As proposed by previous studies, S deprivation generates Fe deficiency in graminaceous plants (Astolfi et al., 2003; Bouranis et al., 2003). Before S supply, non-mycorrhizal plants showed an anticipated response to S depletion; the concentration of total Fe in the shoots reduced from day 45 to day 60 (Table 2), probably due to a dilution effect. In addition, ZmNAS1 was induced on day 60 in the roots and ZmNAS3 was downregulated in the leaves (Table 2). The expression profiles of these genes in the non-mycorrhizal plants suggest that these plants sensed Fe deprivation and the reducing Fe concentration confirms that the plants had experienced a difficulty in taking up Fe from the provided sparingly soluble FePO4 salt. On the other hand, mycorrhizal plants revealed a completely diverse response in the sulfur deficient conditions. ZmNAS1 was downregulated in the roots and ZmNAS3 was upregulated in the leaves on day 60, which in turn suggests that mycorrhizal plants probably sensed Fe sufficient or excessive conditions (Table 2). Moreover, total shoot Fe concentrations remained at the same levels from day 45 to day 60 (Table 2), suggesting that mycorrhizal plants took up Fe. Especially on day 60, although shoot Fe concentrations were equal between mycorrhizal and non-mycorrhizal plants, the expression profiles of the two genes were exactly the opposite between the treatments, showing that the same iron concentration is sensed as sufficient for the mycorrhizal and insufficient for the non-mycorrhizal plants.
The above divergent observations can be supported by the expression profiles of ZmYS1. The expression levels of this gene respond rapidly to the changes of the plant Fe status (Roberts et al., 2004). An upregulation of ZmYS1, observed under Fe deprived conditions, was also observed in the roots of non-mycorrhizal plants before S supply, clearly suggesting that these plants sensed Fe insufficiency (Table 2). In contrast, mycorrhizal roots sustained the expression levels of ZmYS1 until day 60 a fact that endorses the expression profiles of ZmNAS1 and ZmNAS3. The expected Fe deprivation responses in the S deprived conditions were prevented because of the AM symbiosis, whilst S deprivation obstructed Fe absorption and/or translocation in non-mycorrhizal plants leading them to sense Fe deficient conditions. Consequently, it is suggested that the combined expression patterns of ZmNAS1 and ZmYS1 in the roots and ZmNAS3 in the leaves can be used as indicators of the plant Fe status (Table 2).
Mycorrhizal Symbiosis Alters the Fe Uptake Pathway in Maize Plants
AM symbiosis severely alters the processes of nutrient uptake according to the needs of the mycorrhizal plants. Phosphorus, for instance, is provided to the mycorrhizal plants through the fungus and there was recorded loss of function of the direct P uptake pathway in the colonized roots (Smith et al., 2003). In this study, ZmNAS1 and ZmYS1, key genes of the Strategy II Fe uptake pathway, were used in order to determine the impact of AM symbiosis on the plant Fe acquisition pathway.
Forty-eight hours after sulfur supply (day 62), mycorrhizal and non-mycorrhizal plants showed again completely diverse responses. The overexpression of ZmNAS1 in the roots of non-mycorrhizal plants, 48 h after S supply (Figure 4A), was anticipated as S supply contributes in the production of DMA by promoting the Strategy II Fe acquisition pathway (Astolfi et al., 2010). The severe overexpression of ZmYS1 in the roots (day 62) as well as the increase of total Fe concentration in the shoots of non-mycorrhizal plants (day 65) sustain this hypothesis (Figure 5B, Table 1). On the other hand, mycorrhizal plants did not show an intention to enhance the Strategy II pathway, even if S was provided. The downregulation of both ZmNAS1 and ZmYS1 in the roots (Figures 4A, 5B) as well as the increased shoot Fe levels on day 65 (Table 1), suggest that Fe was mainly transported directly to them by the AM fungus in a special, symbiotic Fe uptake pathway. Further analyses should be conducted in order to confirm the existence of such a pathway.
Sulfate Probably Regulates the Expression of ZmNAS1, ZmNAS3, and ZmYS1
Sulfur and iron metabolisms are highly interrelated and there is a co-regulation between their uptake pathways (Forieri et al., 2013; Vigani et al., 2013). As depicted in Figures 4, 5, 24 h after sulfate supply (day 61), there was a common response in the expression of ZmNAS1, ZmNAS3, and ZmYS1 of all plants. All genes were suppressed and such suppression, 24 h after S supply, is difficult to be explained. We assume that the incoming sulfate probably played a role as signal molecule for Fe homeostasis throughout the plant body, a role which has been previously given to sulfate by Forieri et al. (2013). The exact way by which the expressions of these genes are regulated by sulfate, as well as the potential impact of this signaling procedure remains unknown and if this is the case, it is of great interest for future analyses.
Concluding Remarks
AM symbiosis managed to prevent Fe deprivation responses in S deprived maize plants. It is suggested that Fe was provided directly to the mycorrhizal plants through the fungal network. Moreover, sulfate probably regulates the expression of three key Fe homeostasis genes in maize providing a hint of its role as a signal molecule.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to express our appreciation to Syngenta Hellas for the supply of maize seeds. We are also grateful for the contribution of V. Petris to the determination of Fe content. Many thanks to A. Venieraki and E. Vezyri for their support and helpful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00257/abstract
Abbreviations
AM, arbuscular mycorrhiza; AMF, arbuscular mycorrhizal fungi; DMA, deoxymugineic acid; NA, nicotianamine; NAS, nicotianamine synthase; YS, yellow stripe.
References
Allen, J. W., and Shachar-Hill, Y. (2009). Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol. 149, 549–560. doi: 10.1104/pp.108.129866
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ames, R. N., Reid, C. P. P., Porter, L. K., and Cambardella, C. (1983). Hyphal uptake and transport of nitrogen from two 15N-labelled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol. 95, 381–396. doi: 10.1111/j.1469-8137.1983.tb03506.x
Astolfi, S., Zuchi, S., Hubberten, H.-M., Pinton, R., and Hoefgen, R. (2010). Supply of sulphur to S-deficient young barley seedlings restores their capability to cope with iron shortage. J. Exp. Bot. 61, 799–806. doi: 10.1093/jxb/erp346
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Astolfi, S., Zuchi, S., Passera, C., and Cesco, S. (2003). Does the sulfur assimilation pathway play a role in the response to Fe deficiency in maize (Zea mays L.) plants? J. Plant Nutr. 26, 2111–2121. doi: 10.1081/PLN-120024268
Bolan, N. S., Robson, A. D., and Barrow, N. J. (1987). Effects of vesicular-arbuscular mycorrhiza on the availability of iron phosphates to plants. Plant Soil 99, 401–410. doi: 10.1007/BF02370885
Bonfante, P., and Genre, A. (2008). Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13, 492–498. doi: 10.1016/j.tplants.2008.07.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bouranis, D. L., Chorianopoulou, S. N., Protonotarios, V. E., Siyiannis, V. F., Hopkins, L., and Hawkesford, M. J. (2003). Leaf responses of young iron−inefficient maize plants to sulfur deprivation. J. Plant Nutr. 26, 1189–1202. doi: 10.1081/PLN-120020364
Brusslan, J. A., and Tobin, E. M. (1992). Light-independent developmental regulation of cab gene expression in Arabidopsis thaliana seedlings. Proc. Natl. Acad. Sci. U.S.A. 89, 7791–7795. doi: 10.1073/pnas.89.16.7791
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Caris, C., Hördt, W., Hawkins, H.-J., Römheld, V., and George, E. (1998). Studies of iron transport by arbuscular mycorrhizal hyphae from soil to peanut and sorghum plants. Mycorrhiza 8, 35–39. doi: 10.1007/s005720050208
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Forieri, I., Wirtz, M., and Hell, R. (2013). Toward new perspectives on the interaction of iron and sulfur metabolism in plants. Front. Plant Sci. 4:357. doi: 10.3389/fpls.2013.00357
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gutjahr, C., and Paszkowski, U. (2013). Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 4:204. doi: 10.3389/fpls.2013.00204
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Javot, H., Penmetsa, R. V., Terzaghi, N., Cook, D. R., and Harrison, M. J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 1720–1725. doi: 10.1073/pnas.0608136104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kobayashi, T., Nishizawa, N. K., and Mori, S. (2006). “Molecular analysis of iron-deficient graminaceous plants,” in Iron Nutrition in Plants and Rhizospheric Microorganisms, eds. L. L. Barton and J. Abadía (Dordrecht: Springer), 395–435. doi: 10.1007/1-4020-4743-6_20
Koske, R. E., and Gemma, J. N. (1989). A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 92, 486–489. doi: 10.1016/S0953-7562(89)80195-9
Leigh, J., Hodge, A., and Fitter, A. H. (2009). Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol. 181, 199–207. doi: 10.1111/j.1469-8137.2008.02630.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, A., Hamel, C., Hamilton, R. I., Ma, B. L., and Smith, B. L. (2000). Acquisition of Cu, Zn, Mn and Fe by mycorrhizal maize (Zea mays L.) grown in soil at different P and micronutrient levels. Mycorrhiza 9, 331–336. doi: 10.1007/s005720050277
Mills, H. A., and Jones, J. B. (1996). Plant Analysis Handbook II. Athens: MicroMacro Publishing, Inc.
Mizuno, D., Higuchi, K., Sakamoto, T., Nakanishi, H., Mori, S., and Nishizawa, N. K. (2003). Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol. 132, 1989–1997. doi: 10.1104/pp.102.019869
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nozoye, T., Nakanishi, H., and Nishizawa, N. K. (2013). Characterizing crucial components of iron homeostasis in the maize mutants ys1 and ys3. PLOS ONE 8:e62567. doi: 10.1371/journal.pone.0062567
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2002–2007. doi: 10.1093/nar/29.9.e45
Roberts, L. A., Pierson, A. J., Panaviene, Z., and Walker, E. L. (2004). Yellow stripe1. Expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol. 135, 112–120. doi: 10.1104/pp.103.037572
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ruijter, J. M., Ramakers, C., Hoogaars, W., Bakker, O., van den Hoff, M. J. B., Karlen, Y., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schaaf, G., Ludewig, U., Erenoglu, B. E., Mori, S., Kitahara, T., and von Wirén, N. (2004). ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 279, 9091–9096. doi: 10.1074/jbc.M311799200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sieh, D., Watanabe, M., Devers, E. A., Brueckner, F., Hoefgen, R., and Krajinski, F. (2013). The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol. 197, 606–616. doi: 10.1111/nph.12034
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smith, S. E., Dickson, S., Morris, C., and Smith, F. A. (1994). Transfer of phosphate from fungus to plant in VA mycorrhizas: calculation of the area of symbiotic interface and of fluxes of P from different fungi to Allium porrum L. New Phytol. 127, 93–99. doi: 10.1111/j.1469-8137.1994.tb04262.x
Smith, S. E., Smith, F. A., and Jakobsen, I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133, 16–20. doi: 10.1104/pp.103.024380
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vigani, G., Zocchi, G., Bashir, K., Philippar, K., and Briat, J.-F. (2013). Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci. 18, 305–311. doi: 10.1016/j.tplants.2013.01.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Virant-Klun, I., and Gogala, N. (1995). Impact of VAM on phosphorus nutrition of maize with low soluble phosphate fertilization. J. Plant Nutr. 18, 1815–1823. doi: 10.1080/01904169509365025
Zhou, M.-L., Qi, L.-P., Pang, J.-F., Zhang, Q., Lei, Z., Tang, Y.-X., et al. (2013a). Nicotianamine synthase gene family as central components in heavy metal and phytohormone response in maize. Funct. Integr. Genomics 13, 229–239. doi: 10.1007/s10142-013-0315-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, X., Li, S., Zhao, Q., Liu, X., Zhang, S., Sun, C., et al. (2013b). Genome-wide identification, classification and expression profiling of nicotianamine synthase (NAS) gene family in maize. BMC Genomics 14:238. doi: 10.1186/1471-2164-14-238
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: maize, arbuscular mycorrhizal symbiosis, sulfur, iron homeostasis, nicotianamine synthase, yellow stripe
Citation: Chorianopoulou SN, Saridis YI, Dimou M, Katinakis P and Bouranis DL (2015) Arbuscular mycorrhizal symbiosis alters the expression patterns of three key iron homeostasis genes, ZmNAS1, ZmNAS3, and ZmYS1, in S deprived maize plants. Front. Plant Sci. 6:257. doi: 10.3389/fpls.2015.00257
Received: 22 September 2014; Accepted: 01 April 2015;
Published: 20 April 2015.
Edited by:
Stanislaus Francis D'Souza, BMG Bhabha Atomic Research Centre, IndiaReviewed by:
Ruediger Hell, University of Heidelberg, GermanyThomas Leustek, Rutgers, The State University of New Jersey, USA
Copyright © 2015 Chorianopoulou, Saridis, Dimou, Katinakis and Bouranis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Styliani N. Chorianopoulou, Plant Physiology and Morphology Laboratory, Crop Science Department, Agricultural University of Athens, Iera Odos 75, 11855 Athens, Greece s.chorianopoulou@aua.gr
 Styliani N. Chorianopoulou
Styliani N. Chorianopoulou Yiorgos I. Saridis1
Yiorgos I. Saridis1 Dimitris L. Bouranis
Dimitris L. Bouranis