- Psychiatric Epigenomics Laboratory, Queensland Brain Institute, The University of Queensland, Brisbane, QLD, Australia
Non-coding RNAs (ncRNAs) have emerged as critical regulators of transcription, epigenetic processes, and gene silencing, which make them ideal candidates for insight into molecular evolution and a better understanding of the molecular pathways of neuropsychiatric disease. Here, we provide an overview of the current state of knowledge regarding various classes of ncRNAs and their role in neural plasticity and cognitive function, and highlight the potential contribution they may make to the development of a variety of neuropsychiatric disorders, including schizophrenia, addiction, and fear-related anxiety disorders.
Introduction
From whence we came, whither we go: the rise of the RNA world. When Woese, Orgel, and Crick each wrote about the origin and evolution of the genetic code they laid the foundation for “The RNA world hypothesis,” which was later developed by Gilbert (Woese, 1967; Crick, 1968; Orgel, 1968; Gilbert, 1986). This hypothesis stemmed from the fact that ancient RNA had the ability to transmit heritable information as well as, in the case of ribozymes, being endowed with enzymatic capacity (Cech et al., 1981; Kruger et al., 1982; Guerrier-Takada et al., 1983; Joyce, 2002; Cech, 2011). With the advent of next-generation sequencing technology, coordinated efforts such as the ENCODE and FANTOM 3 projects have revealed that more than 90% of the genome is actively transcribed, whereas only ~2% codes for functional protein. These landmark observations brought into question the true value of RNA as a regulatory molecule and have since motivated scientists to embark on a deeper exploration of the biological function of so-called “junk DNA” or “genomic dark matter” (Johnson et al., 2005; Derrien et al., 2011). Many non-coding RNAs (ncRNAs) are highly conserved and correlate with the degree of eukaryotic complexity, further strengthening the argument that they are more than simple artifacts of evolution (Griffiths-Jones et al., 2005; Taft et al., 2007; Heimberg et al., 2008; Mattick, 2011). In fact, their ability to drive transcription, direct epigenetic processes, and silence gene expression make ncRNAs ideal candidates for insight into molecular evolution and a better understanding of the molecular pathways of neuropsychiatric disease. In this review, we provide an overview of the current state of knowledge regarding the involvement of various classes of ncRNAs in neural plasticity as well as their potential role in a variety of neuropsychiatric disorders, particularly those characterized by impairments in cognitive function.
Non-Coding RNA: A Key Driver of Gene Regulation
The broad potential of ncRNAs to critically regulate transcription and translation has been appreciated for some time with the discovery and characterization of various ncRNA families, the majority of which have been shown to be capable of directing epigenetic processes (Mattick and Makunin, 2006). A simplistic division of the different classes of ncRNAs can be made based on their size: small ncRNAs, between 20 and 200nt, and long ncRNAs, arbitrarily identified as any ncRNA over 200 nt. Small ncRNAs amazed the scientific world when they were identified as in Caenorhabditis elegans as being involved in the post-transcriptional regulation of genes during development (Lee et al., 1993; Wightman et al., 1993). Moreover, Fire and Mello were awarded a Nobel prize in 2006 for their discovery of RNA interference in C. elegans (Fire et al., 1998). Long ncRNAs have also recently entered the spotlight. For example, Rinn et al. (2007) recently discovered a long ncRNA called HOTAIR, which regulates epigenetic control over the HOXD locus, thereby providing novel evidence for a functionally relevant role for this enigmatic family of non-coding transcripts.
Small ncRNAs: miRNAs
MicroRNAs (miRNAs) are a class of endogenous, small ncRNAs that mediate post-transcriptional gene silencing by complementary binding to the 3′-untranslated region (3′ UTR) of their target mRNA, a key process for regulating gene expression in a tissue- and developmental stage-specific manner (Liu and Kohane, 2009). As illustrated in Figure 1A, the biogenesis of miRNAs begins in the nucleus through a canonical pathway as a large primary-RNA (pri-RNA) molecule, which folds into a stem loop transcribed by both RNA polymerase II (RNA Pol II) and RNA polymerase III (RNA Pol III; Kim, 2005; Borchert et al., 2006). This hairpin structure is then processed by an enzyme known as Drosha, which works with the RNA-binding protein DGCR8 (DiGeorge critical region 8, called Phasa in invertebrates; Kim, 2009) to produce a ~70 nt (precursor) pre-miRNA that is then shuttled to the cytoplasm via Exportin5 and Ran-GTP61. The pre-miRNA is cleaved in the cytoplasm by an enzyme called Dicer, together with double-stranded RNA binding proteins (dsRBPs) including TRBP (Yi et al., 2003). At this stage, a double-stranded mature miRNA can be detected; however, its inhibitory capacity depends on Dicer-mediated incorporation of one of its strands, together with the Argonaute (Ago) protein, into the RNA-induced silencing complex (RISC; Chendrimada et al., 2005). The RISC then directs the miRNA toward a complementary “seed” sequence within the 3′ UTR of its target mRNA (Wightman et al., 1993; Reinhart et al., 2000; Pillai et al., 2005). The level of complementarity between the miRNA and its target mRNA determines whether the mRNA is degraded or translation is disrupted (Sakurai et al., 2011). Recent findings suggest that these canonical processes have many variations (Volk and Shomron, 2011). For example, insertion at 5′ UTR and coding sequences has been shown to result in similar miRNA activity, and substrates for the formation of miRNA can found be within intronic regions (mirtrons), directly spliced by RNA Pol II, thereby eluding the activity of Drosha (Lee et al., 2004; Ruby et al., 2007). Furthermore, they may also be derived from small nucleolar RNAs, transfer RNAs or by tRNase Z activity (Yang and Lai, 2011).
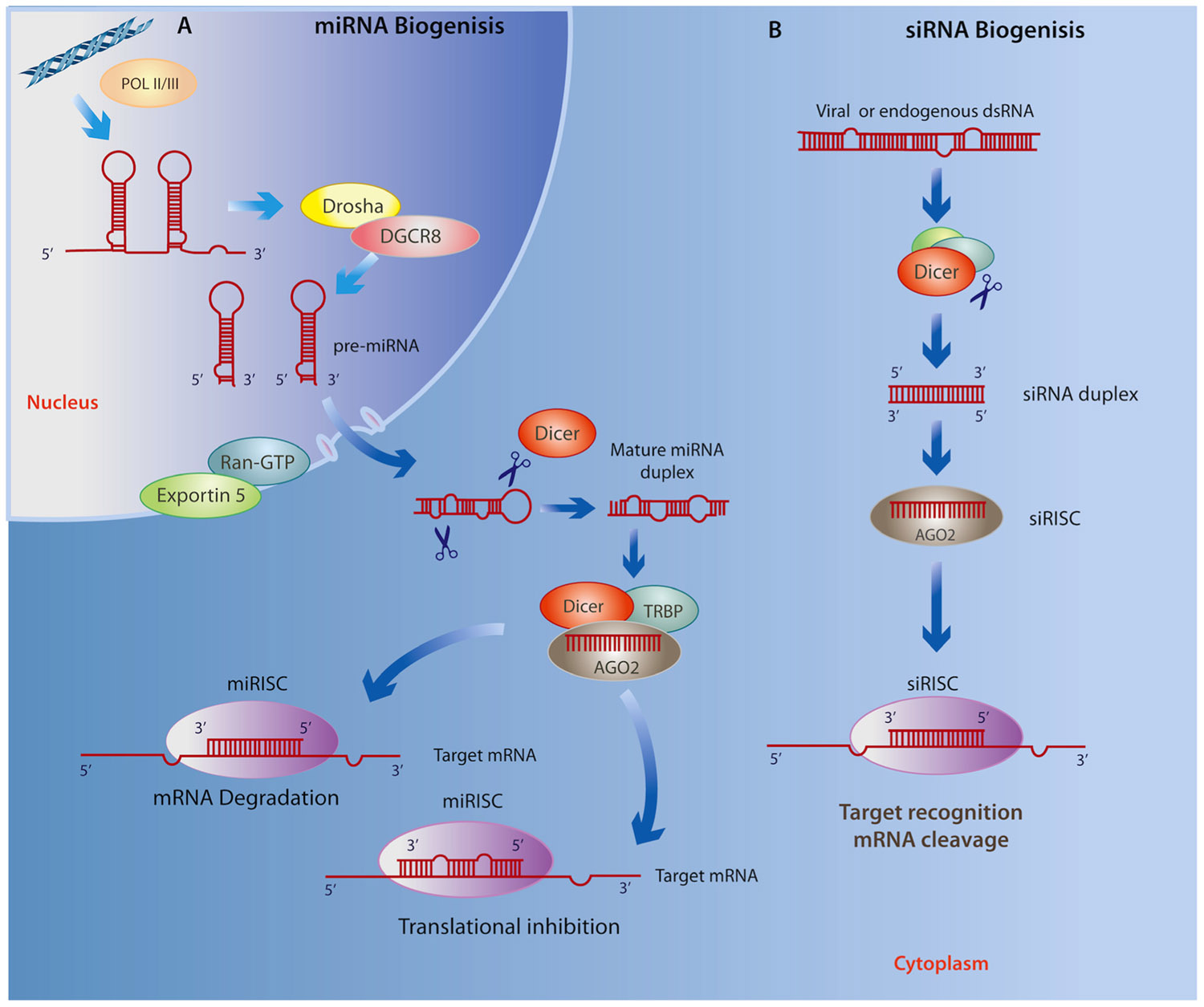
FIGURE 1.(A) miRNA biogenesis begins in the nucleus where Drosha and DGCR8 cleave the primary RNA resulting in a miRNA precursor that is exported to the cytoplasm by Exportin5 and Ran-GTP61 where it is then processed by Dicer and TRBP. Next, the mature miRNA duplex is incorporated to the RISC where it is cleaved Dicer, associates with AGO2, and subsequently binds to the 3′ UTR of a target RNA leading to either mRNA degradation or translational repression. (B) Similarly, siRNA biogenesis depends on Dicer activity and incorporation into the RISC, however siRNAs avoid cleavage by Drosha as they are derived from exogenous viral or endogenous double-stranded RNA molecules mostly within the cytoplasm. Given the specificity of base pairing, the final outcome of the siRNA interference is the cleavage and degradation of their target mRNAs.
MicroRNAs are predicted to affect up to 60% of protein-coding transcripts (Lewis et al., 2005; Friedman et al., 2009), and have been implicated in essential biological processes including stem cell division particularly in mammalian development (Hatfield et al., 2005; Shcherbata et al., 2006; Qi et al., 2009), differentiation and development (Reinhart et al., 2000; Foshay and Gallicano, 2009; Xu et al., 2009; Liu et al., 2012; Zhang et al., 2012b), as well as apoptosis and cancer (Wang et al., 2008b, 2009; Kim, 2009; Corthals et al., 2011; Rissland et al., 2011; Sreekumar et al., 2011; Nie et al., 2012; Yi et al., 2012). Recent investigations using Dicer knockout mice have also shown that miRNA dysregulation leads to abnormalities in brain development and neuronal stem cell differentiation (De Pietri Tonelli et al., 2008; Huang et al., 2010; Kawase-Koga et al., 2010), increased cortical neurodegeneration (Davis et al., 2008; Hebert et al., 2010), and altered learning and memory (Konopka et al., 2010). Although most miRNAs are expressed in the brain, their role in neurogenesis, synaptic plasticity, and cognitive function is only beginning to be elucidated, and considerable work is required in order to better understand their influence within the context of neuropsychiatric disorders (Bredy et al., 2011; O’Connor et al., 2011). For example, among an increasing number of miRNAs implicated in schizophrenia, miR-132 is a CREB-regulated miRNA driven by NMDA receptor signaling, which has been shown to be important for cognitive processing and is dysregulated in the brains of patients with the disorder (Kim et al., 2010; Moreau et al., 2011; Miller et al., 2012). Furthermore, miR-219, another NMDA-receptor-regulated miRNA, has been functionally linked to the behavioral impairments associated with schizophrenia (Kocerha et al., 2009). The progression of Alzheimer’s disease is characterized by cognitive decline and anxiety disorder related to dementia. Recently, Zovoilis et al. (2011) identified miR-34c as a negative regulator of memory consolidation in mice, with miR-34c also markedly increased in the brains of patients with Alzheimer’s disease. Interestingly, around the same time, it was demonstrated that the expression of miR-34c increases dramatically in response to acute stress, thereby providing a potential link between miRNA activity, cognitive function and anxiety in Alzheimer’s disease (Haramati et al., 2011).
More recently, we reported a role for miRNAs in regulating cognitive processes associated with fear-related anxiety disorder. Fear-extinction learning in C57/Bl6J mice led to increased expression of the brain-specific miRNA, miR-128b, which disrupted stability of several plasticity-related target genes and regulated formation of fear-extinction memory. We proposed that increased miR-128b activity might facilitate the transition from retrieval of the original fear memory toward the formation of a new fear-extinction memory, the disruption of which would have important implications for the development of fear-related anxiety disorder (Lin et al., 2011).
MicroRNA-mediated regulation of gene function has also been implicated in the development of addiction. The miRNAs miR-124, Let-7d, and miR-181 are up-regulated in the nucleus accumbens in response to cocaine, and knockdown of each of these miRNAs influences cocaine-seeking behavior (Chandrasekar and Dreyer, 2009, 2011). Using an elegant dopamine cell type-specific knockout approach, Schaefer et al. (2010) demonstrated that Ago2, a subunit of Argonaute, is critical for mediating the motivational aspects of cocaine self-administration. Importantly, there are specific Ago2-dependent miRNAs that are cocaine-inducible, including miR-181a, miR-324, and miR-369, which may therefore be involved in this effect. Indeed, miR-324 and miR-369 regulate MEF2 and FosB, two plasticity-related genes required for cognitive functioning associated with cocaine-seeking behavior (Pulipparacharuvil et al., 2008; Renthal et al., 2009). As MEF2 has been implicated in the suppression of excitatory synaptic density (Flavell et al., 2006), a miRNA-mediated negative regulation of MEF2 may remove this constraint and increase synaptogenesis in response to cocaine-associated learning. Furthermore, miR-212 expression is increased in the striatum after extended access to cocaine, and over-expression of this miRNA decreases cocaine-seeking behavior (Hollander et al., 2010). Recently, it has been demonstrated that the methyl CpG-binding protein 2 (MECP2) mediates the escalation of reward-seeking behavior in rats through a homeostatic interaction with miR-212 and subsequent regulation of brain-derived neurotrophic factor (BDNF) in the striatum (Im et al., 2010). This is a particularly interesting finding as it provides some of the first evidence to suggest that ncRNA-directed regulation of the epigenome occurs in an activity-dependent manner in vivo.
Short Interfering RNAs
Very little is known about the role of short interfering RNAs (siRNAs) in the context of neurodegenerative disease or neuropsychiatric disorders; however, we include these small ncRNAs because of their demonstrated ability for targeting gene silencing in a variety of contexts. Similar to miRNAs, siRNAs are ~22 nt non-coding transcripts that are derived from natural antisense transcripts, pseudogenes, and repetitive sequences within endogenous dsRNA, as well as by an exogenous viral replication mechanism that is able to initiate degradation of homologous mRNAs (Sharp, 2001; Tam et al., 2008; Cech, 2011; Zhang et al., 2012c). The biogenesis of siRNAs is similar to that of miRNA; however, unlike the nuclear origin of miRNAs, they may originate in both the cytoplasm and nucleus (Figure 1B; reviewed in Verdel et al., 2009). This class of ncRNAs is associated with the Ago family as well as the RISC complex, which transports the guide strand to a target mRNA. Importantly, siRNAs show significantly more specific base pairing relative to miRNAs, and use Slicer activity to cleave and degrade their target mRNAs (Valencia-Sanchez et al., 2006). The specific base complement of exogenous siRNAs to viral mRNAs, particularly in Drosophila and C. elegans, represents an efficient antiviral immune response (Wilkins et al., 2005; Umbach and Cullen, 2009), whereas endogenous siRNAs are important for controlling somatic transposable elements in Drosophila (Chung et al., 2008) as well as in plants (Ito, 2011).
Perhaps the most striking quality of siRNAs is their synthetic reproducibility, which makes them attractive tools to be used in gene therapy for a variety of diseases. The first successful attempt to treat a human disease through RNAi-mediated gene silencing was demonstrated with the use of siRNA against hepatitis C in mice (Song et al., 2003). SiRNA has since emerged as the next “new class of drugs” (Peer and Lieberman, 2011); however, several hurdles must be overcome before siRNA-mediated gene therapy will make its way to the clinic. The primary issue is to effectively deliver siRNAs without degradation by phagocytosis or the production of off-target effects. A second concern is the design of membrane-permeable siRNAs that readily cross the blood–brain barrier (Dominska and Dykxhoorn, 2010). Advances in siRNA delivery have begun to address these challenges through the development of labeled magnetic nanoparticles, PEGylated LPD (liposome-polycation-DNA), and cationic liposomes (Li et al., 2011). siRNA has been successfully used to treat viral infections including polio, human papilloma virus, echoviruses, and adenoviruses (Morrissey et al., 2005; Saulnier et al., 2006; Tan et al., 2007; Eckstein et al., 2010). Furthermore, siRNAs have also been demonstrated to be efficacious in the treatment of human pancreatic cancer (Zhong et al., 2012), lung carcinoma (Podesta et al., 2009), and fibrodysplasia ossificans progressiva (Kaplan et al., 2011; Takahashi et al., 2011).
Reports on the putative function of both endogenous and exogenous siRNAs in the brain and their regulation of behavior are emerging. Within the context of Parkinson’s disease, a neurodegenerative disease characterized by cognitive decline, Hommel et al. (2003) found that siRNA-mediated knockdown of Th, which is a gene responsible for the production of tyrosine hydroxylase and a key enzyme within the dopaminergic pathway, attenuated locomotor abnormalities in a mouse model of Parkinson’s disease. Neuronal dysfunction due to mutations in the human α-synuclein gene affect dopaminergic levels and have been associated with idiopathic Parkinson’s disease as well as dementia with Lewy bodies. Delivery of lentiviral-mediated siRNA has been shown to effectively diminish endogenous expression of this gene both in vitro and in vivo (Sapru et al., 2006). SiRNA designed to target human Htt, a gene involved in Huntington’s disease, which is also a neurodegenerative disorder characterized by cognitive impairment, attenuates striatal neuropathology, and motor deficits in a mouse model of Huntington’s disease (Difiglia et al., 2007). Moreover, these studies have been extended in both mice and non-human primates, in which administration of siRNAs selective for Huntingtin are effective in delaying neurodegeneration, and may also have some efficacy in preventing the cognitive decline observed in Huntington’s disease (Lombardi et al., 2009; Pfister et al., 2009; Zhang et al., 2009; McBride et al., 2011; Stiles et al., 2012).
It has been widely demonstrated that exogenous siRNA is also a useful approach for inducing loss of function in studies investigating genes involved in synaptic plasticity, and learning and memory. Early research found that siRNAs could be used to silence endogenous and heterologous genes within the rat hippocampus (Krichevsky and Kosik, 2002). The important role of GluR1 in the acquisition and consolidation of spatial memory has recently been demonstrated using siRNA (Das et al., 2012). Interestingly, knockdown of CREB by siRNA has elucidated the important role of O-GlcNAc-mediated glycolysation of CREB in regulating dendritic outgrowth, a process critical for the formation of long-term memory (Rexach et al., 2012). Concordantly, siRNA-mediated knockdown of the alpha 2 subunit of the GABAA receptor within the amygdala decreases binge drinking, thereby suggesting a potential role for endogenous siRNA in the regulation of addiction-like behavior (Liu et al., 2011). Furthermore, Panossian et al. (2012) employed an RNAi approach to demonstrate the essential role of the stress hormone neuropeptide Y (NPY) in regulating the stress response.
Remarkably, the stability and turnover of both miRNA and siRNA depend on the methylation status of their 3′ UTR, which potentially protects these small RNAs from urydination and exonucleolytic degradation (Ji and Chen, 2012; Zhao et al., 2012).This observation supports the prospective role of endogenous siRNAs, which is to control gene expression in mammalian oocytes (Tam et al., 2008; Watanabe et al., 2008; Nejepinska et al., 2012). These regulatory properties are also consistent with the findings of Song et al. (2011) who identified endogenous siRNAs in humans and mice, which are involved in developmental programming of male germ cells. Even more exciting is the fact that endogenous siRNAs may also direct transcriptional gene silencing in the metazoan genome through the regulation of chromatin modification. For example, in C. elegans, Schizosaccharomyces pombe, and Drosophila they direct H3K9 methylation to establish the formation of heterochromatin (Fagegaltier et al., 2009; Burkhart et al., 2011).
Experience-dependent expression of endogenous siRNAs has only recently been uncovered in the mammalian genome. For instance, siRNA associated with genes involved in synaptic plasticity are up-regulated in the hippocampus of mice subjected to an olfactory discrimination paradigm (Smalheiser et al., 2010). As mentioned, endogenous siRNAs are mostly derived from repetitive elements in germinal tissues, thus supporting the hypothesis that siRNAs might have evolved to sustain genomic stability during early development. However, considering the remarkable findings of Smalheiser et al. (2011), who identified an endogenous siRNA complementary to SynGAP1, and that dysregulation of SynGAP1 correlates with mental retardation as well as autism (Hamdan et al., 2009), these observations suggest a potentially important role for endogenous siRNA in cognitive disorders. Together, these early findings lay the foundation for the use of siRNA-mediated gene knockdown as a therapeutic intervention in a variety of disorders; however, it is not yet known whether manipulation of siRNAs will be applicable to the clinical treatment of cognitive deficits associated with neuropsychiatric disease.
PIWI-Interacting RNAs
P-element induced wimpy testis (PIWI)-interacting RNAs (PiRNAs) are the most distinct but least investigated of all the small ncRNAs, and appear to function by repressing target gene expression within the nucleus instead of the cytoplasm (Saito et al., 2006). They are associated with a subfamily of Ago proteins containing an N-terminus called PAZ (from PIWI homologous Argonaute and Zwillie) and a C-terminus PIWI domain (Parker et al., 2005). PIWI proteins are highly abundant in the germline, and play an essential role in normal gonadal development in mice, with mutations resulting in male infertility (Cox et al., 1998; Girard et al., 2006; Klattenhoff and Theurkauf, 2008; Beyret and Lin, 2011). In 2001, Aravin et al. discovered a novel small ncRNA involved in the silencing of the testis-expressed Stellate gene in Drosophila but they did not realize that they were looking at PIWI protein interacting RNAs (Aravin et al., 2001; Siomi et al., 2011). Later, Watanabe et al. (2006) revealed the existence of mouse germline small RNAs between ~26 and ~30 nt long, which they named gsRNAs. Although these investigators verified an expression pattern that correlated with MIWI and MILI, two members of the PIWI protein family, they did not uncover the extent of the association and the true nature of the piRNAs. This connection was later made in mice where it was discovered that MILI and MIWI murine PIWI proteins bind, respectively, to 26 and 30 nt RNAs, which subsequently became known as piRNAs (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006). Soon after, piRNAs were found in rat testes (Lau et al., 2006) as well as in Drosophila where they induced silencing of the Suppressor of Stellate locus found on the Y chromosome (Vagin et al., 2006).
Although it was initially suggested that piRNAs are only expressed in germinal cells (Carmell et al., 2007; Grimson et al., 2008; Beyret and Lin, 2011), they are also found in somatic tissue (Malone et al., 2009; Lee et al., 2011; Yan et al., 2011; Rajasethupathy et al., 2012). PiRNAs elude cleavage by Dicer and Drosha and are mostly derived from repetitive elements called piRNA clusters (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Saito et al., 2006; Brennecke et al., 2007; Malone et al., 2009; Malone and Hannon, 2009). Although these clusters retain interspecies conservation, piRNA sequences themselves show no evolutionary conservation (Aravin et al., 2006; Girard et al., 2006; Lau et al., 2006). This interesting characteristic suggests a locus-specific function of piRNAs whereby they inhibit only those transcripts derived from the genomic region in which they originate (Lau et al., 2006). PiRNAs can also be derived from the 3′ UTR of protein-coding genes and euchromatic transposable elements (Muerdter et al., 2012), as well as from non-repetitive genomic DNA (Kim et al., 2008). In addition, through the insertion and transcription of an exogenous sequence within an endogenous piRNA cluster locus, de novo piRNAs can be produced from non-repetitive protein-coding sequences (Kawaoka et al., 2012). This revelation paves the way for the exploration of the role of piRNAs in the regulation of protein-coding genes through sequence-specific recognition and a Dicer-independent siRNA pathway, perhaps by using PIWI nuclease activity.
PiRNAs are transcribed through two well-described mechanisms (Figure 2). The so called “ping pong” pathway is mainly observed in the cytoplasm of germ cells where pre-piRNAs derived from transposons are cleaved by unknown nucleases and form a complex with MILI proteins to direct MILI slicer activity against target mRNAs. This step results in a new secondary piRNA. The secondary piRNA is then coupled to a MIWI2 protein, which in turn exhibits endonuclease activity on the opposite strand and produces a new pre-piRNA, which again forms a complex with MILI in an amplification loop (Brennecke et al., 2007; Kim et al., 2009; Siomi et al., 2011; Pillai and Chuma, 2012). The MIWI2 interacting piRNA is also transported back to the nucleus where it represses transposable elements through the regulation of DNA methylation (Aravin and Bourc’his, 2008; Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008; Watanabe et al., 2011). A modification may occur where pre-piRNAs are associated with MIWI proteins instead of MILI and generate a secondary piRNA that appears to remain in the cytoplasm and drives the “primary pathway,” which is present in somatic cells (Kim et al., 2009; Siomi et al., 2011; Pillai and Chuma, 2012).
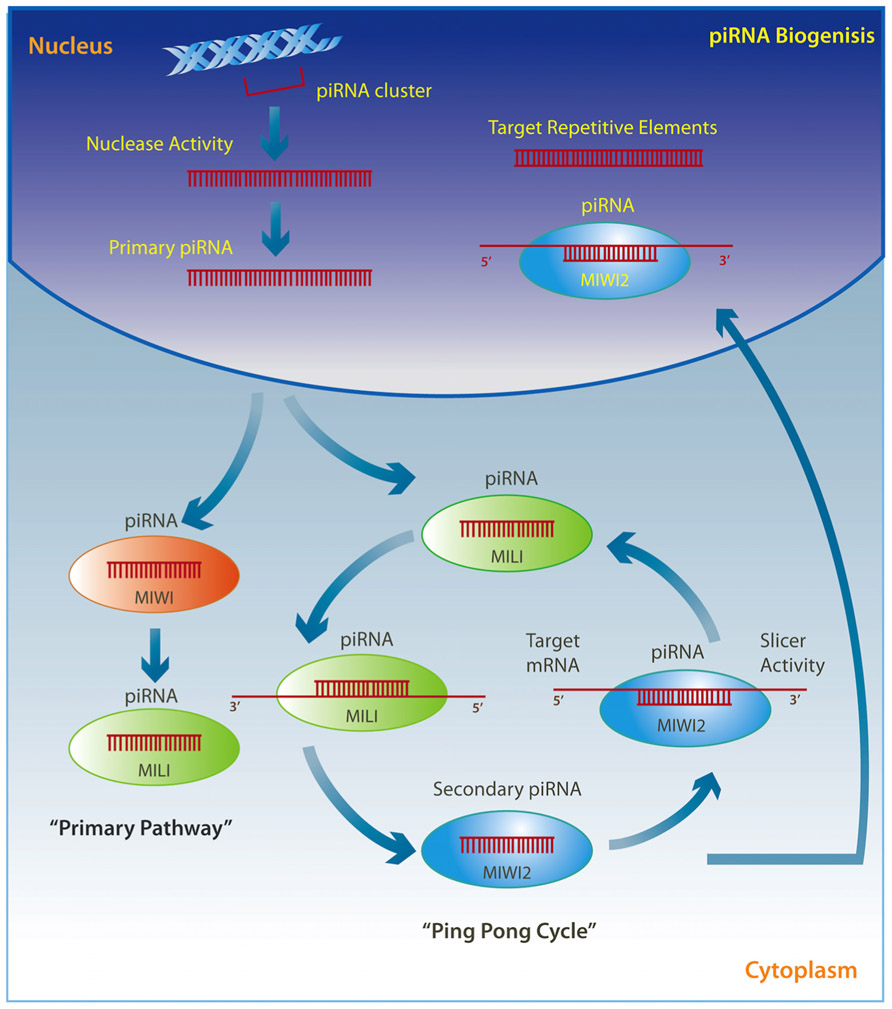
FIGURE 2. Primary single-stranded intergenic piRNAs are transcribed within the nucleus from well-conserved clusters of repetitive elements. Once in the cytoplasm they either interact with MIWI proteins and initiate the “primary pathway,” or directly with MILI proteins to trigger the “ping-pong cycle.” PiRNAs associated with MILI direct its slicer activity against target mRNAs, thereby producing a secondary piRNA, which is then coupled to MIWI2. MIWI2 piRNA can return to the nucleus where it targets repetitive elements, or remains within the cytoplasm where it exerts endonuclease activity on the opposite strand to reproduce a new pre-piRNA that interacts with MILI in a self-amplification loop.
Remarkably, piRNAs exploit DNA methylation and chromatin remodeling to serve their regulatory function in mammals. Mutations induced in murine MILI result in a loss of DNA methylation and a consequent up-regulation of long interspersed element-1 (LINE-1) and IAP (intracisternal A particle) elements (Aravin et al., 2007). Similarly, MIWI2 mutants also show increased expression of LINE-1 and IAP repetitive sequences, which correlate with higher DNA demethylation levels in the testes (Carmell et al., 2007). Initially, co-immunoprecipitation studies of MIWI2 failed to demonstrate a direct interaction with DNMT3a and DNMT3b, suggesting that PiRNAs do not directly regulate the activity of these enzymes (Aravin et al., 2008). However, additional investigations have shown impaired de novo DNA methylation and decreased piRNA levels in germline cells derived from MILI knockout mice, thus verifying a role for MILI-bound piRNAs in timing the methylation of transposons (Kuramochi-Miyagawa et al., 2008). Interestingly, Watanabe et al. (2011) proposed that piRNAs may interact with other small ncRNAs transcribed from differentially methylated genomic regions in order to drive de novo methylation of the locus corresponding to the associated ncRNA. PiRNAs seem to also participate in chromatin remodeling processes by recruiting major chromatin modifiers. These piRNAs, identified as piALU RNAs, have been shown to regulate chromatin organization within and around centromeres as well as by directing mechanisms of DNA repair and transcription (Blackwell et al., 2012).
Although the main role of piRNAs appears to be in the maintenance of genomic integrity and DNA stability through epigenetic silencing of transposable elements during early development (Vagin et al., 2006; Aravin et al., 2007; Brennecke et al., 2007; O’Donnell and Boeke, 2007; Dharap et al., 2011), the identification of ischemia-responsive piRNAs in the cortex of the adult rat (Dharap et al., 2011) suggests an important role for piRNAs across the lifespan. The recent demonstration that intergenic piRNAs are expressed in the mouse hippocampus, particularly within the dendritic compartment, indicates that piRNAs are functionally active in the adult central nervous system (CNS). Knockdown of piRNA QD541777 in hippocampal neurons leads to a decreased number of dendritic spines potentially via the regulation of mRNA derived from Cdk5rap1 and Mark 1/2, two genes with active roles in CNS development (Lee et al., 2011). Moreover, a cluster of four piRNAs was also found to be associated with other genes involved in dendrite formation and neuronal migration (Lee et al., 2011). Consistent with these findings, Kandel and colleagues recently discovered neuronally expressed piRNAs that promote DNA methylation of the CREB2 promoter thereby facilitating synaptic plasticity in Aplysia (Rajasethupathy et al., 2012). These findings suggest a potential role for piRNAs in transcriptional gene silencing and in the epigenetic facilitation of long-term memory.
Interestingly, the TUDOR protein interacts with PIWI proteins, and consequently with piRNAs, in coordinating the regulation of LINE-1 retrotransposon activity (Chen et al., 2009; Reuter et al., 2009; Siomi et al., 2010; Yabuta et al., 2011). Together with the fact that LINE-1 influences experience-dependent neuronal plasticity and somatic mosaicism within the hippocampus (Coufal et al., 2009; Muotri et al., 2009; Muotri et al., 2010), one can envision how the activity of piRNAs might be integral to these processes. It is remarkable that the current focus on the role of piRNAs is limited to the silencing of transposable elements in early development when in fact they may be potent regulators of neural plasticity and cognitive function.
Long ncRNAs
Long ncRNAs are transcripts over 200 nt long that can be found in both the cytoplasm and the nucleus where they exhibit tightly regulated spatiotemporal patterns of expression (Kapranov et al., 2007; Guttman et al., 2009; Khalil et al., 2009). Initially, long ncRNAs were believed to be the least conserved of all ncRNAs and to mainly serve as precursors for small RNAs (Kapranov et al., 2007). Their low level of expression also fueled the debate that they might merely be the result of transcriptional noise or artifacts of high-throughput sequencing (Struhl, 2007; Van Bakel et al., 2010). However, it has been shown that some long intergenic RNAs are under purifying selection in mammals (Ponjavic et al., 2007; Guttman et al., 2009) and exhibit cross-species conservation as well as function-specific stability (Marques and Ponting, 2009; Ulitsky et al., 2011; Clark et al., 2012). Interestingly, nuclear long ncRNAs undergo a high rate of turnover and are relatively unstable, which is concordant with the notion that they provide a mechanism for rapid response to external stimuli (Mercer et al., 2009; Clark et al., 2012). Due to these characteristics, long ncRNAs appear to be in a state of constant and rapid evolutionary development. However, short sequences nested within long ncRNAs, particularly those close to the promoter region, are highly conserved (Ponjavic et al., 2007; Guttman et al., 2010; Ulitsky et al., 2011), which suggests that they might represent core motifs for RNA binding proteins responsible for the extensive chromatin remodeling functions attributed to a variety of long ncRNAs. This would support the idea that it is the function of long ncRNAs in relation to protein-coding genes, and not their primary sequence, that is critical for their influence on gene expression and positions long ncRNAs as flexible modular scaffolds (Guttman and Rinn, 2012).
Although, the precise biogenic pathway of long ncRNAs is not yet fully understood, it involves RNAPol II-mediated transcription, polyadenylation, and end-capping (Guttman et al., 2009). Long ncRNAs are derived from sense and antisense strands, intergenic and intronic regions, and overlapping regions of protein-coding genes. Furthermore, novel subsets of long ncRNAs called macroRNAs are believed to serve as precursors for other small and long non-coding transcripts (Furuno et al., 2006). These might originate via de novo transcription of precursor non-coding transcripts, by duplication of another ncRNA or by loss of function of an ancestral protein-coding gene (Ulitsky et al., 2011). An interesting example of a long ncRNA derived from a former protein-coding sequence is Xist, which has arisen from the mutated protein-coding gene, Lnx3 (Duret et al., 2006). Moreover, Tsx is a long ncRNA derived from the loss of protein-coding function within the same locus (Anguera et al., 2011). It has also been proposed that long ncRNAs originate from genomic sequences from exonic regions, from chromosome rearrangement, or by duplication of the non-coding gene through retrotransposition activity and insertion of transposable elements (Ponting et al., 2009).
Long ncRNAs may function as enhancers as well as repressors through coordinated regulation of epigenetic processes, both in cis and in trans, as illustrated in Figure 2 (Khalil et al., 2009; Orom et al., 2010a,b; Guttman et al., 2011). Considering that they account for the majority of the non-coding transcriptome (Mattick and Makunin, 2006; Jia et al., 2010), that 20% of long ncRNAs found in the human genome are bound to the polycomb group protein repressive complex (PRC), and that more than 50% of all known long ncRNAs interact with other chromatin modifying complexes such as CoREST and SMCX (Khalil et al., 2009), they are likely to be heavily involved in the epigenetic regulation of neural plasticity. Prototypical examples of epigenetic regulation directed by long ncRNAs include chromatin remodeling by Xist, acting in cis, to drive X chromosome inactivation (Duret et al., 2006), and HOTAIR, operating in trans, to silence the HOXD locus by recruitment of PRC2 (Rinn et al., 2007). Their role in imprinting is also evident in the activity of the paternally expressed lncRNA, Air, which directs silencing of IGF2R, Slc22a2, and Slc22a3, through the recruitment of G9a histone methyltransferase and methylation of H3K9 domains (Nagano et al., 2008). Another example of a long ncRNA that induces chromatin remodeling involves Kcnq1ot1, which interacts with Dnmt1 to regulate methylation of the Kcnq1 gene through the recruitment of the histone methyltransferases Ezh2 and G9a (Mohammad et al., 2010, 2008; Pandey et al., 2008). Each of the examples further emphasizes the important role of long ncRNAs in directing epigenetic processes. Given the emerging appreciation for epigenetic mechanisms in learning and memory (Day and Sweatt, 2010), an understanding of how long ncRNAs direct the epigenome within the context of neural plasticity, cognition, and neuropsychiatric disorders is on the horizon.
Long ncRNAs have already been associated with several neurodegenerative disorders characterized by impaired cognitive function. For instance, along ncRNA isoform of the overlapping SOX2 gene transcript called SOX2OT regulates CNS vertebrate development, as well as neurogenesis in adult mice (Amaral et al., 2009), and thus represents a candidate biomarker for Alzheimer’s disease (Arisi et al., 2011). Furthermore, the novel long ncRNA17A directs the splicing of GPR51, thereby producing an alternative GABAB receptor isoform, which results in a significant impairment in GABAB receptor signaling in Alzheimer’s disease (Massone et al., 2011). An additional example of the versatility of long ncRNAs in driving gene expression within the human brain is the BACE1 antisense transcript, which prevents miR-485-5p induced expression of BCAE1 gene by competing for the same binding site on its mRNA. Indeed, dysregulation of this long ncRNA has also been reported in Alzheimer’s disease (Faghihi et al., 2010). Disruption of key long ncRNAs may also play a role in the development of psychiatric disorders. For example, the long ncRNA disrupted in schizophrenia 2 (DISC2), is antisense to, and overlaps with, its protein-coding transcript DISC1. This long ncRNA has been implicated in schizophrenia, bipolar depression, and autism spectrum disorder (Millar et al., 2000; Chubb et al., 2008; Williams et al., 2009).
It is becoming clear that long ncRNAs have a direct role in the regulation of genes involved in neural plasticity and cognitive function. For example, BDNF supports neuronal survival and synaptic plasticity and is critically involved in learning and memory (Hall et al., 2000; Rattiner et al., 2004a,b; Bredy et al., 2007). A human antisense transcript called BDNFOS or antiBDNF was previously shown to interact with its sense mRNA, BDNF (Pruunsild et al., 2007). Interestingly, very recently a conserved antisense transcript to BDNF has been discovered in mice, which inhibits BDNF transcription by recruitment of EZH2, a key component of the epigenetic silencing complex, PRC2. Additionally, administration of a sequence-specific inhibitor called an antagoNAT or siRNA-mediated knockdown of BDNF-AS both lead toup-regulation of BDNF mRNA and protein expression in vitro and in vivo, resulting in increased neurite outgrowth and maturation (Modarresi et al., 2012).
The potential importance of long ncRNA activity related to higher order cognitive processing is also highlighted by the discovery of human accelerated region 1 (HAR1F), which is expressed in Cajal–Retzius neurons and was one of the first long ncRNAs shown to be involved in human neocortical development (Pollard et al., 2006). Recently it was found that repression of HAR1F by REST leads to its decreased expression in the striatum of Huntington’s disease (Johnson et al., 2010), and that REST also interacts with DGCR5 and TUG1 lncRNAs leading to DiGeorge syndrome and Huntington’s disease (Johnson et al., 2009; Johnson, 2011). Johnson (2012) extended these investigations and confirmed that both TUG1 and NEAT1 are significantly up-regulated in Huntington’s disease brains, whereas MEG3 and DGCR5 showed decreased expression.
Among other functions, the organization of the nuclear architecture also seems to require the transcription of the polyadenylated long ncRNA NEAT1, which is associated with nuclear paraspeckles (Clemson et al., 2009; Souquere et al., 2010), and NEAT2 (also known as MALAT1 – metastasis-associated lung adenocarcinoma transcript 1), which regulates synapse formation (Bernard et al., 2010). Furthermore, the long ncRNA Cycling D1 (CCND1) interacting with a protein called translocated in liposarcoma (TLS; as indicated in Figure 3), acts as a transcriptional co-repressor and as a sensor of DNA damage by binding and repressing both CREB and p300 acetyltransferase leading to the silencing of their target gene (Wang et al., 2008a). Given the important role of CREB and p300 in regulating neural plasticity and cognitive function, it would be interesting to determine whether a disruption of the long ncRNA CCND1 can cause impairments in cognitive function similar to those observed when either CREB or p300 is inhibited.
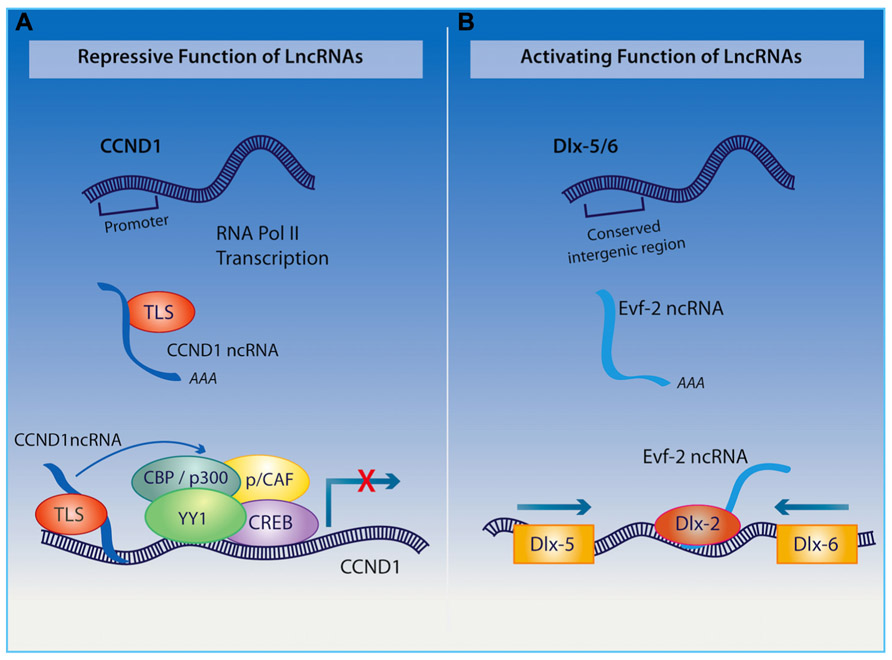
FIGURE 3.(A) Upon receiving a signal for DNA damage, the long ncRNA CCND1 binds to the TLS protein to direct a protein complex to the CCND1 target gene where allosteric modification of CREB and the histone acetyltranserases (HATs) CBP and p300 are produced, which results in gene silencing. (B) Long ncRNA Evf2 is transcribed from an ultra conserved intergenic region within the Dlx-5/6 locus. Binding of this long ncRNA to the Dlx-2 protein triggers transcriptional activation of the Dlx-5/6 enhancer region in a homeodomain-specific manner.
The ability of long ncRNAs to interact with transcription factors also endows them with a remarkable capacity to orchestrate mammalian neurogenesis, which will undoubtedly have an influence on cognition. For example, the long non-coding transcript Evf2, derived from the Dlx5/6 gene locus, binds to the homeodomain of Dlx-2 forming an enhancer-like complex that activates Dlx5/6 expression (as described in Figure 3; Feng et al., 2006). The relevance of this long ncRNA in the hippocampus and cognition is exemplified through studies on Evf2 knockout mice, which exhibit impaired synaptic plasticity and spatial memory as a result of aberrant interneuron development (Bond et al., 2009). Extending these discoveries, Ng et al. (2012) have demonstrated that long ncRNAs interact with transcription factor SOX2 as well as PRC2, with the authors suggesting that they are required for pluripotency of neural progenitor cells. Additionally, long ncRNAs have been shown to direct that activity of other ncRNAs within the CNS. Indeed, the cytoplasmic long ncRNA, lncRNA_N2, harboring miRNA miR-125B, regulates the transcription of this small RNA and indirectly promotes neurogenesis.
More excitingly, long ncRNAs have also been associated with drug addiction and, potentially, fear-related anxiety disorder. For instance, the long ncRNAs MIAT, MEG3, and NEAT1/2 are up-regulated within the nucleus accumbens of heroin abusers (Michelhaugh et al., 2011). Accordingly, a previous genome-wide association study has implicated MEG3 with potential predisposition to heroin addiction (Nielsen et al., 2008). Interestingly, MIAT is associated with the nuclear matrix and is involved in oligodendrocyte development (Mercer et al., 2010), whereas MEG3 is a cAMP-responsive, maternally imprinted, long ncRNA that is expressed in distinct subpopulations of neurons (Zhao et al., 2006; Michelhaugh et al., 2011) and contributes to early neurogenesis (Mercer et al., 2010). More recently, the lncRNA RMST, also known as dorsomedial telencephalon gene 2 (DMT2), was revealed to interact with nuclear and chromatin factors during the development of dopaminergic neurons (Ng et al., 2012). Additionally, the long ncRNA BC1 has been shown to direct the activity of the dopamine 2 receptor (Drd2) as well as metabotropic glutamate receptors (mGluR), which are required for balancing neuronal excitability at the synapse (Zhong et al., 2009), and that BC1-/- mice develop increased anxiety (Centonze et al., 2007). Considering the important role of Drd2 and mGluRs in regulating the extinction of conditioned fear, it is possible that a dysregulation of the long ncRNA BC1 contributes to the development of fear related anxiety disorders.
Through their interaction with various epigenetic regulatory proteins, long ncRNAs might serve as molecular brakes on transcriptional activity, as originally proposed by McQuown and Wood (2011) within the context of HDAC activity and memory formation, which in our opinion would also contribute greatly to rapid, experience-dependent, variations in genomic function. Consequently, it is anticipated that long ncRNAs will have profound effects on neural plasticity and cognition associated with neuropsychiatric disorders. In summary, cell type-specific patterns of long ncRNA expression may serve to coordinate the fidelity of synaptic plasticity and neural connectivity by dynamically monitoring and integrating multiple transcriptional and post-transcriptional events (Mercer et al., 2008; Qureshi and Mehler, 2011).
Finally, as described in Table 1, there are also many other classes of ncRNAs not discussed in this review, the understanding of which is only starting to emerge.
Although the biogenesis of most of these non-coding transcripts remains unclear, their tissue-specificity and developmental stage-dependent expression provides strong evidence for their biological relevance justifying the need for further research into the complex regulatory potential hidden within the eukaryotic transcriptome.
Outlook and Conclusion
The huge repertoire of ncRNAs, that we are only beginning to recognize, suggests a sophisticated RNA-mediated layer of control over genomic function that has evolved to coordinate tissue- and developmental stage-specific regulation of gene expression. The redundancy in consensus sequences typical of most ncRNAs allows for a highly efficient transcriptional regulation of protein-coding genes, whereas the potential stability provided by the methylation status of most small ncRNAs supports their well established role in regulating gene expression. In the case of some long ncRNAs, their low level of conservation may contribute directly to their function by enabling the cell to respond to external stimuli with greater flexibility, thereby conferring a rapidly adaptive plasticity that is distinct from, and more complex than, the much slower acting protein-coding genes (Mattick, 2011). Consequently and given that ncRNAs are highly expressed in the brain, it is anticipated that they will play a significant role in driving neural connectivity within learning and behavioral adaptation of higher eukaryotes. This firmly positions ncRNAs at the forefront of cell-to-cell communication within the context of cognitive function and potentially in the development of neuropsychiatric disorders. Given that a significant proportion of known long ncRNAs interact with repressive complexes, future investigations will expand on these observations through the application of innovative techniques including high-throughput sequencing together with UV-crosslinking and immunoprecipitation (HITS-CLIP; Zhang and Darnell, 2011), chromatin isolation by RNA purification (ChIRP; Chu et al., 2011), capture hybridization analysis of RNA combined with RNA sequencing (CHART; Simon et al., 2011), and RNA-capture-Seq (Mercer et al., 2011). Such studies will aid in the determination of novel ncRNAs and their genome-wide binding sites, which will be important to elucidate the essential role of RNA-directed epigenetic regulation of gene function. Most studies on ncRNAs have been based on stem cell and embryonic development, with the function of miRNAs in the adult brain only recently becoming appreciated. Moreover, in vitro studies examining long ncRNAs and piRNAs, and their influence on neural plasticity, are emerging. It will be exciting to see how these in vitro findings translate in vivo and to elucidate whether the biological significance of these ncRNAs will extend to memory and cognition. Furthermore, RNAi will be a useful tool to facilitate the knockdown of ncRNAs that are associated to neuropsychiatric disorders, such as TUG1 and NEAT1. Consequently, if lncRNAs truly represent modular scaffolds, then it will be imperative to determine the stability of the epigenetic processes directed by all classes of ncRNAs, particularly those resulting from rapidly changing behavioral responses. Given the fact that endogenous siRNAs are mostly represented in oocytes, whereas piRNAs are strongly expressed in the male germ line, each capable of exerting their own genomic function, also makes them attractive candidates to investigate their potential influence on genomic imprinting and transgenerational epigenetic inheritance. Finally, since many ncRNAs remain transcriptionally quiescent until a specific developmental stage is met or an appropriate environmental signal is received (Kretz et al., 2012), it will be important to decipher the mechanisms coordinating such precise and rapid genomic responsiveness. The central dogma of protein-coding genes controlling the eukaryotic genome represents an archaic constraint on our journey to truly expand the understanding of gene–epigenome–environmental interactions. It is undeniable that this layer of gene regulation has many fundamental biological and functional roles yet to be explained and it might be within neural plasticity, cognition, and neuropsychiatric disorders where the lasting signature of ncRNAs could be at their most significant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge grant support from the National Health and Medical Research Council (APP1023127) and the Australian Research Council (DP1096148) to Timothy W. Bredy. Paola A. Spadaro is supported by an Australian Postgraduate Award from the University of Queensland. The authors give special thanks to Rowan Tweedale for helpful editing and Dee McGrath for graphics support.
References
Amaral, P. P., Neyt, C., Wilkins, S. J., Askarian-Amiri, M. E., Sunkin, S. M., Perkins, A. C., and Mattick, J. S. (2009). Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15, 2013–2027.
Anguera, M. C., Ma, W., Clift, D., Namekawa, S., Kelleher, R. J., III, and Lee, J. T. (2011). Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 7:e1002248. doi: 10.1371/journal.pgen.1002248
Aravin, A. A., and Bourc’his, D. (2008). Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 22, 970–975.
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., Morris, P., Brownstein, M. J., Kuramochi-Miyagawa, S., Nakano, T., Chien, M., Russo, J. J., Ju, J., Sheridan, R., Sander, C., Zavolan, M., and Tuschl, T. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207.
Aravin, A. A., Hannon, G. J., and Brennecke, J. (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764.
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027.
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Toth, K. F., Bestor, T., and Hannon, G. J. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799.
Arisi, I., D’onofrio, M., Brandi, R., Felsani, A., Capsoni, S., Drovandi, G., Felici, G., Weitschek, E., Bertolazzi, P., and Cattaneo, A. (2011). Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 24, 721–738.
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801.
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M. Q., Sedel, F., Jourdren, L., Coulpier, F., Triller, A., Spector, D. L., and Bessis, A. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093.
Beyret, E., and Lin, H. (2011). Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 355, 215–226.
Blackwell, B., Lopez, M. F., Wang, J., Krastins B., Sarracino, D., Tollervey, J. R., Dobke, M., Jordan, I. K., and Lunyak, V. V. (2012). Protin interaction indicates putative participation of retroRNA in the cell, DNA repair and chromatin assembly. Mob. Genet. Elements 2, 1–10.
Bond, A. M., Vangompel, M. J., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., Kohtz, J. D. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027.
Borchert, G. M., Lanier, W., and Davidson, B. L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13, 1097–1101.
Bredy, T. W., Lin, Q., Wei, W., Baker-Andresen, D., and Mattick, J. S. (2011). MicroRNA regulation of neural plasticity and memory. Neurobiol. Learn. Mem. 96, 89–94.
Bredy, T. W., Wu, H., Crego, C., Zellhoefer, J., Sun, Y. E., and Barad, M. (2007). Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem. 14, 268–276.
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., and Hannon, G. J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103.
Burkhart, K. B., Guang, S., Buckley, B. A., Wong, L., Bochner, A. F., and Kennedy, S. (2011). A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7:e1002249. doi: 10.1371/journal.pgen.1002249
Cao, F., Li, X., Hiew, S., Brady, H., Liu, Y., and Dou, Y. (2009). Dicer independent small RNAs associate with telomeric heterochromatin. RNA 15, 1274–1281.
Carmell, M. A., Girard, A., Van De Kant, H. J., Bourc’his, D., Bestor, T. H., De Rooij, D. G., and Hannon, G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514.
Carninci, P., Yasuda, J., and Hayashizaki, Y. (2008). Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 20, 274–280.
Cech, T. R. (2011). The RNA worlds in context. Cold Spring Harb Perspect Biol. pii, cshperspect.a006742v1.
Cech, T. R., Zaug, A. J., and Grabowski, P. J. (1981). In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27, 487–496.
Centonze, D., Rossi, S., Napoli, I., Mercaldo, V., Lacoux, C., Ferrari, F., Ciotti, M. T., De Chiara, V., Prosperetti, C., Maccarrone, M., Fezza, F., Calabresi, P., Bernardi, G., and Bagni, C. (2007). The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 27, 8885–8892.
Chandrasekar, V., and Dreyer, J. L. (2009). microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42, 350–362.
Chandrasekar, V., and Dreyer, J. L. (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36, 1149–1164.
Chen, C., Jin, J., James, D. A., Adams-Cioaba, M. A., Park, J. G., Guo, Y., Tenaglia, E., Xu, C., Gish, G., Min, J., and Pawson, T. (2009). Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. U.S.A. 106, 20336–20341.
Chendrimada, T. P., Gregory, R. I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744.
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E., and Chang, H. Y. (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678.
Chubb, J. E., Bradshaw, N. J., Soares, D. C., Porteous, D. J., and Millar, J. K. (2008). The DISC locus in psychiatric illness. Mol. Psychiatry 13, 36–64.
Chung, W. J., Okamura, K., Martin, R., and Lai, E. C. (2008). Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802.
Clark, M. B., Johnston, R. L., Inostroza-Ponta, M., Fox, A. H., Fortini, E., Moscato, P., Dinger, M. E., and Mattick, J. S. (2012). Genome-wide analysis of long noncoding RNA stability. Genome Res. 22, 885–898.
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., and Lawrence, J. B. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726.
Corthals, S. L., Sun, S. M., Kuiper, R., De Knegt, Y., Broyl, A., Van Der Holt, B., Beverloo, H. B., Peeters, J. K., El Jarari, L., Lokhorst, H. M., Zweegman, S., Jongen-Lavrencic, M., and Sonneveld, P. (2011). MicroRNA signatures characterize multiple myeloma patients. Leukemia 25, 1784–1789.
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., Morell, M., O’Shea, K. S., Moran, J. V., and Gage, F. H. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131.
Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727.
Das, S. R., Jensen, R., Kelsay, R., Shumaker, M., Bochart, R., Brim, B., Zamzow, D., and Magnusson, K. R. (2012). Reducing expression of GluN1(0XX) subunit splice variants of the NMDA receptor interferes with spatial reference memory. Behav. Brain Res. 230, 317–324.
Davis, T. H., Cuellar, T. L., Koch, S. M., Barker, A. J., Harfe, B. D., Mcmanus, M. T., and Ullian, E. M. (2008). Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330.
Day, J. J., and Sweatt, J. D. (2010). DNA methylation and memory formation. Nat. Neurosci. 13, 1319–1323.
De Pietri Tonelli, D., Pulvers, J. N., Haffner, C., Murchison, E. P., Hannon, G. J., and Huttner, W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921.
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H., and Lieberman, P. M. (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413.
Derrien, T., Guigo, R., and Johnson, R. (2011). The long non-coding RNAs: a new (p)layer in the “dark matter.” Front. Genet. 2:107. doi: 10.3389/fgene.2011.00107
Dharap, A., Nakka, V. P., and Vemuganti, R. (2011). Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke 42, 1105–1109.
Difiglia, M., Sena-Esteves, M., Chase, K., Sapp, E., Pfister, E., Sass, M., Yoder, J., Reeves, P., Pandey, R. K., Rajeev, K. G., Manoharan, M., Sah, D. W., Zamore, P. D., and Aronin, N. (2007). Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 104, 17204–17209.
Dominska, M., and Dykxhoorn, D. M. (2010). Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 123, 1183–1189.
Duret, L., Chureau, C., Samain, S., Weissenbach, J., and Avner, P. (2006). The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312, 1653–1655.
Eckstein, A., Grossl, T., Geisler, A., Wang, X., Pinkert, S., Pozzuto, T., Schwer, C., Kurreck, J., Weger, S., Vetter, R., Poller, W., and Fechner, H. (2010). Inhibition of adenovirus infections by siRNA-mediated silencing of early and late adenoviral gene functions. Antiviral Res. 88, 86–94.
Emara, M. M., Ivanov, P., Hickman, T., Dawra, N., Tisdale, S., Kedersha, N., Hu, G. F., and Anderson, P. (2010). Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285, 10959–10968.
ENCODE Transcriptome Project. (2009). Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457, 1028–1032.
Fagegaltier, D., Bouge, A. L., Berry, B., Poisot, E., Sismeiro, O., Coppee, J. Y., Theodore, L., Voinnet, O., and Antoniewski, C. (2009). The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 21258–21263.
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van Der Brug, M. P., Nalls, M. A., Cookson, M. R., St-Laurent, G. III, and Wahlestedt, C. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56.
Feng, J., Bi, C., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811.
Flavell, S. W., Cowan, C. W., Kim, T. K., Greer, P. L., Lin, Y., Paradis, S., Griffith, E. C., Hu, L. S., Chen, C., and Greenberg, M. E. (2006). Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012.
Foshay, K. M., and Gallicano, G. I. (2009). miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 326, 431–443.
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105.
Furuno, M., Pang, K. C., Ninomiya, N., Fukuda, S., Frith, M. C., Bult, C., Kai, C., Kawai, J., Carninci, P., Hayashizaki, Y., Mattick, J. S., and Suzuki, H. (2006). Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2:e37. doi: 10.1371/journal.pgen.0020037
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202.
Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S. R., and Bateman, A. (2005). Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33, D121–D124.
Grimson, A., Srivastava, M., Fahey, B., Woodcroft, B. J., Chiang, H. R., King, N., Degnan, B. M., Rokhsar, D. S., and Bartel, D. P. (2008). Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197.
Grivna, S. T., Pyhtila, B., and Lin, H. (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 13415–13420.
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. (1983). The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857.
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., Huarte, M., Zuk, O., Carey, B. W., Cassady, J. P., Cabili, M. N., Jaenisch, R., Mikkelsen, T. S., Jacks, T., Hacohen, N., Bernstein, B. E., Kellis, M., Regev, A., Rinn, J. L., and Lander, E. S. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227.
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., Young, G., Lucas, A. B., Ach, R., Bruhn, L., Yang, X., Amit, I., Meissner, A., Regev, A., Rinn, J. L., Root, D. E., and Lander, E. S. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300.
Guttman, M., Garber, M., Levin, J. Z., Donaghey, J., Robinson, J., Adiconis, X., Fan, L., Koziol, M. J., Gnirke, A., Nusbaum, C., Rinn, J. L., Lander, E. S., and Regev, A. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510.
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346.
Hamdan, F. F., Gauthier, J., Spiegelman, D., Noreau, A., Yang, Y., Pellerin, S., Dobrzeniecka, S., Cote, M., Perreau-Linck, E., Carmant, L., D’Anjou, G., Fombonne, E., Addington, A. M., Rapoport, J. L., Delisi, L. E., Krebs, M. O., Mouaffak, F., Joober, R., Mottron, L., Drapeau, P., Marineau, C., Lafreniere, R. G., Lacaille, J. C., Rouleau, G. A., and Michaud, J. L. (2009). Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N. Engl. J. Med. 360, 599–605.
Haramati, S., Navon, I., Issler, O., Ezra-Nevo, G., Gil, S., Zwang, R., Hornstein, E., and Chen, A. (2011). MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J. Neurosci. 31, 14191–14203.
Hatfield, S. D., Shcherbata, H. R., Fischer, K. A., Nakahara, K., Carthew, R. W., and Ruohola-Baker, H. (2005). Stem cell division is regulated by the microRNA pathway. Nature 435, 974–978.
Hebert, S. S., Papadopoulou, A. S., Smith, P., Galas, M. C., Planel, E., Silahtaroglu, A. N., Sergeant, N., Buee, L., and De Strooper, B. (2010). Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19, 3959–3969.
Heimberg, A. M., Sempere, L. F., Moy, V. N., Donoghue, P. C., and Peterson, K. J. (2008). MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl. Acad. Sci. U.S.A. 105, 2946–2950.
Hollander, J. A., Im, H. I., Amelio, A. L., Kocerha, J., Bali, P., Lu, Q., Willoughby, D., Wahlestedt, C., Conkright, M. D., and Kenny, P. J. (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature 466, 197–202.
Hommel, J. D., Sears, R. M., Georgescu, D., Simmons, D. L., and Dileone, R. J. (2003). Local gene knockdown in the brain using viral-mediated RNA interference. Nat. Med. 9, 1539–1544.
Huang, T., Liu, Y., Huang, M., Zhao, X., and Cheng, L. (2010). Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J. Mol. Cell Biol. 2, 152–163.
Hall, J., Thomas, K. L., and Everitt, B. J. (2000). Rapid and selective induction of BDNF expression in the hippocampus during contextual learning. Nat. Neurosci. 3, 533–535.
Im, H. I., Hollander, J. A., Bali, P., and Kenny, P. J. (2010). MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13, 1120–1127.
Ito, H. (2011). Small RNAs and transposon silencing in plants. Dev. Growth Differ. doi: 10.1111/j.1440-169X.2011.01309.x [Epub ahead of print].
Jia, H., Osak, M., Bogu, G. K., Stanton, L. W., Johnson, R., and Lipovich, L. (2010). Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16, 1478–1487.
Ji, L., and Chen, X. (2012). Regulation of small RNA stability: methylation and beyond. Cell Res. 22, 624–636.
Johnson, J. M., Edwards, S., Shoemaker, D., and Schadt, E. E. (2005). Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21, 93–102.
Johnson, R. (2011). Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 46, 245–254.
Johnson, R., Richter, N., Jauch, R., Gaughwin, P. M., Zuccato, C., Cattaneo, E., and Stanton, L. W. (2010). The human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol. Genomics 41, 269–274.
Johnson, R., Teh, C. H., Jia, H., Vanisri, R. R., Pandey, T., Lu, Z. H., Buckley, N. J., Stanton, L. W., and Lipovich, L. (2009). Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15, 85–96.
Johnson, R. (2012). Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 46, 245–254.
Kaplan, J., Kaplan, F. S., and Shore, E. M. (2011). Restoration of normal BMP signaling levels and osteogenic differentiation in FOP mesenchymal progenitor cells by mutant allele-specific targeting. Gene Ther. doi: 10.1038/gt.2011.152 [Epub ahead of print].
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., Stadler, P. F., Hertel, J., Hackermuller, J., Hofacker, I. L., Bell, I., Cheung, E., Drenkow, J., Dumais, E., Patel, S., Helt, G., Ganesh, M., Ghosh, S., Piccolboni, A., Sementchenko, V., Tammana, H., and Gingeras, T. R. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488.
Kapranov, P., Ozsolak, F., Kim, S. W., Foissac, S., Lipson, D., Hart, C., Roels, S., Borel, C., Antonarakis, S. E., Monaghan, A. P., John, B., and Milos, P. M. (2010). New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature 466, 642–646.
Kawaoka, S., Mitsutake, H., Kiuchi, T., Kobayashi, M., Yoshikawa, M., Suzuki, Y., Sugano, S., Shimada, T., Kobayashi, J., Tomari, Y., and Katsuma, S. (2012). A role for transcription from a piRNA cluster in de novo piRNA production. RNA 18, 265–273.
Kawase-Koga, Y., Low, R., Otaegi, G., Pollock, A., Deng, H., Eisenhaber, F., Maurer-Stroh, S., and Sun, T. (2010). RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell Sci. 123, 586–594.
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., Thomas, K., Presser, A., Bernstein, B. E., Van Oudenaarden, A., Regev, A., Lander, E. S., and Rinn, J. L. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672.
Kim, M., Patel, B., Schroeder, K. E., Raza, A., and Dejong, J. (2008). Organization and transcriptional output of a novel mRNA-like piRNA gene (mpiR) located on mouse chromosome 10. RNA 14, 1005–1011.
Kim, T. K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., Harmin, D. A., Laptewicz, M., Barbara-Haley, K., Kuersten, S., Markenscoff-Papadimitriou, E., Kuhl, D., Bito, H., Worley, P. F., Kreiman, G., and Greenberg, M. E. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187.
Kim, V. N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385.
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139.
Klattenhoff, C., and Theurkauf, W. (2008). Biogenesis and germline functions of piRNAs. Development 135, 3–9.
Kocerha, J., Faghihi, M. A., Lopez-Toledano, M. A., Huang, J., Ramsey, A. J., Caron, M. G., Sales, N., Willoughby, D., Elmen, J., Hansen, H. F., Orum, H., Kauppinen, S., Kenny, P. J., and Wahlestedt, C. (2009). MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. U.S.A. 106, 3507–3512.
Konopka, W., Kiryk, A., Novak, M., Herwerth, M., Parkitna, J. R., Wawrzyniak, M., Kowarsch, A., Michaluk, P., Dzwonek, J., Arnsperger, T., Wilczynski, G., Merkenschlager, M., Theis, F. J., Kohr, G., Kaczmarek, L., and Schutz, G. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842.
Kretz, M., Webster, D. E., Flockhart, R. J., Lee, C. S., Zehnder, A., Lopez-Pajares, V., Qu, K., Zheng, G. X., Chow, J., Kim, G. E., Rinn, J. L., Chang, H. Y., Siprashvili, Z., and Khavari, P. A. (2012). Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 26, 338–343.
Krichevsky, A. M., and Kosik, K. S. (2002). RNAi functions in cultured mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 99, 11926–11929.
Kruger, K., Grabowski, P. J., Zaug, A. J., Sands, J., Gottschling, D. E., and Cech, T. R. (1982). Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157.
Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., Asada, N., Kojima, K., Yamaguchi, Y., Ijiri, T. W., Hata, K., Li, E., Matsuda, Y., Kimura, T., Okabe, M., Sakaki, Y., Sasaki, H., and Nakano, T. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917.
Langenberger, D., Bermudez-Santana, C., Hertel, J., Hoffmann, S., Khaitovich, P., and Stadler, P. F. (2009). Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics 25, 2298–2301.
Lau, N. C., Seto, A. G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D. P., and Kingston, R. E. (2006). Characterization of the piRNA complex from rat testes. Science 313, 363–367.
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., and Kosik, K. S. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099.
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., and Kim, V. N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060.
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20.
Li, P., Liu, D., Sun, X., Liu, C., Liu, Y., and Zhang, N. (2011). A novel cationic liposome formulation for efficient gene delivery via a pulmonary route. Nanotechnology 22, 245104.
Lin, Q., Wei, W., Coelho, C. M., Li, X., Baker-Andresen, D., Dudley, K., Ratnu, V. S., Boskovic, Z., Kobor, M. S., Sun, Y. E., and Bredy, T. W. (2011). The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat. Neurosci. 14, 1115–1117.
Liu, H., and Kohane, I. S. (2009). Tissue and process specific microRNA–mRNA co-expression in mammalian development and malignancy. PLoS ONE 4:e5436. doi:10.1371/journal.pone.0005436
Liu, J., Yang, A. R., Kelly, T., Puche, A., Esoga, C., June, H. L. Jr., Elnabawi, A., Merchenthaler, I., Sieghart, W., June, H. L., Sr., and Aurelian, L. (2011). Binge alcohol drinking is associated with GABAA alpha2-regulated Toll-like receptor 4 (TLR4) expression in the central amygdala. Proc. Natl. Acad. Sci. U.S.A. 108, 4465–4470.
Liu, W. M., Pang, R. T., Chiu, P. C., Wong, B. P., Lao, K., Lee, K. F., and Yeung, W. S. (2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. U.S.A. 109, 490–494.
Lombardi, M. S., Jaspers, L., Spronkmans, C., Gellera, C., Taroni, F., Di Maria, E., Donato, S. D., and Kaemmerer, W. F. (2009). A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 217, 312–319.
Malone, C. D., Brennecke, J., Dus, M., Stark, A., Mccombie, W. R., Sachidanandam, R., and Hannon, G. J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535.
Malone, C. D., and Hannon, G. J. (2009). Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 74, 225–234.
Marques, A. C., and Ponting, C. P. (2009). Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 10, R124.
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., Tabaton, M., Robello, M., Gatta, E., Russo, C., Florio, T., Dieci, G., Cancedda, R., and Pagano, A. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317.
McBride, J. L., Pitzer, M. R., Boudreau, R. L., Dufour, B., Hobbs, T., Ojeda, S. R., and Davidson, B. L. (2011). Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 19, 2152–2162.
McQuown, S. C., and Wood, M. A. (2011). HDAC3 and the molecular brake pad hypothesis. Neurobiol. Learn. Mem. 96, 27–34.
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159.
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721.
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., and Mehler, M. F. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Mercer, T. R., Wilhelm, D., Dinger, M. E., Solda, G., Korbie, D. J., Glazov, E. A., Truong, V., Schwenke, M., Simons, C., Matthaei, K. I., Saint, R., Koopman, P., and Mattick, J. S. (2011). Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res. 39, 2393–2403.
Michelhaugh, S. K., Lipovich, L., Blythe, J., Jia, H., Kapatos, G., and Bannon, M. J. (2011). Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 116, 459–466.
Millar, J. K., Wilson-Annan, J. C., Anderson, S., Christie, S., Taylor, M. S., Semple, C. A., Devon, R. S., St Clair, D. M., Muir, W. J., Blackwood, D. H., and Porteous, D. J. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423.
Miller, B. H., Zeier, Z., Xi, L., Lanz, T. A., Deng, S., Strathmann, J., Willoughby, D., Kenny, P. J., Elsworth, J. D., Lawrence, M. S., Roth, R. H., Edbauer, D., Kleiman, R. J., and Wahlestedt, C. (2012). MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. U.S.A. 109, 3125–3130.
Mohammad, F., Mondal, T., Guseva, N., Pandey, G. K., and Kanduri, C. (2010). Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137, 2493–2499.
Mohammad, F., Pandey, R. R., Nagano, T., Chakalova, L., Mondal, T., Fraser, P., and Kanduri, C. (2008). Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell. Biol. 28, 3713–3728.
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., Van Der Brug, M. P., and Wahlestedt, C. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459
Moreau, M. P., Bruse, S. E., David-Rus, R., Buyske, S., and Brzustowicz, L. M. (2011). Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 69, 188–193.
Morrissey, D. V., Lockridge, J. A., Shaw, L., Blanchard, K., Jensen, K., Breen, W., Hartsough, K., Machemer, L., Radka, S., Jadhav, V., Vaish, N., Zinnen, S., Vargeese, C., Bowman, K., Shaffer, C. S., Jeffs, L. B., Judge, A., Maclachlan, I., and Polisky, B. (2005). Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23, 1002–1007.
Muerdter, F., Olovnikov, I., Molaro, A., Rozhkov, N. V., Czech, B., Gordon, A., Hannon, G. J., and Aravin, A. A. (2012). Production of artificial piRNAs in flies and mice. RNA 18, 42–52.
Muotri, A. R., Marchetto, M. C., Coufal, N. G., Oefner, R., Yeo, G., Nakashima, K., and Gage, F. H. (2010). L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446.
Muotri, A. R., Zhao, C., Marchetto, M. C., and Gage, F. H. (2009). Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19, 1002–1007.
Nagano, T., Mitchell, J. A., Sanz, L. A., Pauler, F. M., Ferguson-Smith, A. C., Feil, R., and Fraser, P. (2008). The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720.
Nejepinska, J., Malik, R., Filkowski, J., Flemr, M., Filipowicz, W., and Svoboda, P. (2012). dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 40, 399–413.
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533.
Nie, J., Liu, L., Zheng, W., Chen, L., Wu, X., Xu, Y., Du, X., and Han, W. (2012). microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis 33, 220–225.
Nielsen, D. A., Ji, F., Yuferov, V., Ho, A., Chen, A., Levran, O., Ott, J., and Kreek, M. J. (2008). Genotype patterns that contribute to increased risk for or protection from developing heroin addiction. Mol. Psychiatry 13, 417–428.
O’Connor, R. M., Dinan, T. G., and Cryan, J. F. (2011). Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol. Psychiatry. 17, 359–376.
O’Donnell, K. A., and Boeke, J. D. (2007). Mighty Piwis defend the germline against genome intruders. Cell 129, 37–44.
Orom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G., Lai, F., Zytnicki, M., Notredame, C., Huang, Q., Guigo, R., and Shiekhattar, R. (2010a). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58.
Orom, U. A., Derrien, T., Guigo, R., and Shiekhattar, R. (2010b). Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb. Symp. Quant. Biol. 75, 325–331.
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., Nagano, T., Mancini-Dinardo, D., and Kanduri, C. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246.
Panossian, A., Wikman, G., Kaur, P., and Asea, A. (2012). Adaptogens stimulate neuropeptide y and hsp72 expression and release in neuroglia cells. Front. Neurosci. 6:6. doi: 10.3389/fnins.2012.00006
Parker, J. S., Roe, S. M., and Barford, D. (2005). Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666.
Peer, D., and Lieberman, J. (2011). Special delivery: targeted therapy with small RNAs. Gene Ther. 18, 1127–1133.
Pfister, E. L., Kennington, L., Straubhaar, J., Wagh, S., Liu, W., Difiglia, M., Landwehrmeyer, B., Vonsattel, J. P., Zamore, P. D., and Aronin, N. (2009). Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 19, 774–778.
Pillai, R. S., Bhattacharyya, S. N., Artus, C. G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309, 1573–1576.
Pillai, R. S., and Chuma, S. (2012). piRNAs and their involvement in male germline development in mice. Dev. Growth Differ. 54, 78–92.
Podesta, J. E., Al-Jamal, K. T., Herrero, M. A., Tian, B., Ali-Boucetta, H., Hegde, V., Bianco, A., Prato, M., and Kostarelos, K. (2009). Antitumor activity and prolonged survival by carbon-nanotube-mediated therapeutic siRNA silencing in a human lung xenograft model. Small 5, 1176–1185.
Pollard, K. S., Salama, S. R., Lambert, N., Lambot, M. A., Coppens, S., Pedersen, J. S., Katzman, S., King, B., Onodera, C., Siepel, A., Kern, A. D., Dehay, C., Igel, H., Ares, M. Jr., Vanderhaeghen, P., and Haussler, D. (2006). An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172.
Ponjavic, J., Ponting, C. P., and Lunter, G. (2007). Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17, 556–565.
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641.
Preker, P., Almvig, K., Christensen, M. S., Valen, E., Mapendano, C. K., Sandelin, A., and Jensen, T. H. (2011). PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 39, 7179–7193.
Preker, P., Nielsen, J., Kammler, S., Lykke-Andersen, S., Christensen, M. S., Mapendano, C. K., Schierup, M. H., and Jensen, T. H. (2008). RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854.
Pruunsild, P., Kazantseva, A., Aid, T., Palm, K., and Timmusk, T. (2007). Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90, 397–406.
Pulipparacharuvil, S., Renthal, W., Hale, C. F., Taniguchi, M., Xiao, G., Kumar, A., Russo, S. J., Sikder, D., Dewey, C. M., Davis, M. M., Greengard, P., Nairn, A. C., Nestler, E. J., and Cowan, C. W. (2008). Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron 59, 621–633.
Qi, J., Yu, J. Y., Shcherbata, H. R., Mathieu, J., Wang, A. J., Seal, S., Zhou, W., Stadler, B. M., Bourgin, D., Wang, L., Nelson, A., Ware, C., Raymond, C., Lim, L. P., Magnus, J., Ivanovska, I., Diaz, R., Ball, A., Cleary, M. A., and Ruohola-Baker, H. (2009). microRNAs regulate human embryonic stem cell division. Cell Cycle 8, 3729–3741.
Qureshi, I. A., and Mehler, M. F. (2011). Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol. Med. 17, 337–346.
Rajasethupathy, P., Antonov, I., Sheridan, R., Frey, S., Sander, C., Tuschl, T., and Kandel, E. R. (2012). A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149, 693–707.
Rattiner, L. M., Davis, M., French, C. T., and Ressler, K. J. (2004a). Brain-derived neurotrophic factor and tyrosine kinase receptor B involvement in amygdala-dependent fear conditioning. J. Neurosci. 24, 4796–4806.
Rattiner, L. M., Davis, M., and Ressler, K. J. (2004b). Differential regulation of brain-derived neurotrophic factor transcripts during the consolidation of fear learning. Learn. Mem. 11, 727–731.
Redon, S., Reichenbach, P., and Lingner, J. (2010). The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38, 5797–5806.
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906.
Renthal, W., Kumar, A., Xiao, G., Wilkinson, M., Covington, H. E. III, Maze, I., Sikder, D., Robison, A. J., Laplant, Q., Dietz, D. M., Russo, S. J., Vialou, V., Chakravarty, S., Kodadek, T. J., Stack, A., Kabbaj, M., and Nestler, E. J. (2009). Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62, 335–348.
Reuter, M., Chuma, S., Tanaka, T., Franz, T., Stark, A., and Pillai, R. S. (2009). Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 16, 639–646.
Rexach, J. E., Clark, P. M., Mason, D. E., Neve, R. L., Peters, E. C., and Hsieh-Wilson, L. C. (2012). Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 8, 253–261.
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., Goodnough, L. H., Helms, J. A., Farnham, P. J., Segal, E., and Chang, H. Y. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323.
Rissland, O. S., Hong, S. J., and Bartel, D. P. (2011). MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell 43, 993–1004.
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86.
Saito, K., Nishida, K. M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., and Siomi, M. C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222.
Sakurai, K., Amarzguioui, M., Kim, D. H., Alluin, J., Heale, B., Song, M. S., Gatignol, A., Behlke, M. A., and Rossi, J. J. (2011). A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 39, 1510–1525.
Sapru, M. K., Yates, J. W., Hogan, S., Jiang, L., Halter, J., and Bohn, M. C. (2006). Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp. Neurol. 198, 382–390.
Saulnier, A., Pelletier, I., Labadie, K., and Colbere-Garapin, F. (2006). Complete cure of persistent virus infections by antiviral siRNAs. Mol. Ther. 13, 142–150.
Schaefer, A., Im, H. I., Veno, M. T., Fowler, C. D., Min, A., Intrator, A., Kjems, J., Kenny, P. J., O’Carroll, D., and Greengard, P. (2010). Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med. 207, 1843–1851.
Schoeftner, S., and Blasco, M. A. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236.
Shcherbata, H. R., Hatfield, S., Ward, E. J., Reynolds, S., Fischer, K. A., and Ruohola-Baker, H. (2006). The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle 5, 172–175.
Simon, M. D., Wang, C. I., Kharchenko, P. V., West, J. A., Chapman, B. A., Alekseyenko, A. A., Borowsky, M. L., Kuroda, M. I., and Kingston, R. E. (2011). The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20497–20502.
Siomi, M. C., Mannen, T., and Siomi, H. (2010). How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24, 636–646.
Siomi, M. C., Sato, K., Pezic, D., and Aravin, A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258.
Smalheiser, N. R., Lugli, G., Lenon, A. L., Davis, J. M., Torvik, V. I., and Larson, J. (2010). Olfactory discrimination training up-regulates and reorganizes expression of microRNAs in adult mouse hippocampus. ASN Neurol. 2, e00028.
Smalheiser, N. R., Lugli, G., Thimmapuram, J., Cook, E. H., and Larson, J. (2011). Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA 17, 166–181.
Song, R., Hennig, G. W., Wu, Q., Jose, C., Zheng, H., and Yan, W. (2011). Male germ cells express abundant endogenous siRNAs. Proc. Natl. Acad. Sci. U.S.A. 108, 13159–13164.
Song, E., Lee, S. K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J., Shankar, P., and Lieberman, J. (2003). RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9, 347–351.
Souquere, S., Beauclair, G., Harper, F., Fox, A., and Pierron, G. (2010). Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell 21, 4020–4027.
Sreekumar, R., Sayan, B. S., Mirnezami, A. H., and Sayan, A. E. (2011). MicroRNA control of invasion and metastasis pathways. Front. Genet. 2:58. doi: 10.3389/fgene.2011.00058
Stiles, D. K., Zhang, Z., Ge, P., Nelson, B., Grondin, R., Ai, Y., Hardy, P., Nelson, P. T., Guzaev, A. P., Butt, M. T., Charisse, K., Kosovrasti, V., Tchangov, L., Meys, M., Maier, M., Nechev, L., Manoharan, M., Kaemmerer, W. F., Gwost, D., Stewart, G. R., Gash, D. M., and Sah, D. W. (2012). Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp. Neurol. 233, 463–471.
Struhl, K. (2007). Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14, 103–105.
Tam, O. H., Aravin, A. A., Stein, P., Girard, A., Murchison, E. P., Cheloufi, S., Hodges, E., Anger, M., Sachidanandam, R., Schultz, R. M., and Hannon, G. J. (2008). Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538.
Taft, R. J., Glazov, E. A., Cloonan, N., Simons, C., Stephen, S., Faulkner, G. J., Lassmann, T., Forrest, A. R., Grimmond, S. M., Schroder, K., Irvine, K., Arakawa, T., Nakamura, M., Kubosaki, A., Hayashida, K., Kawazu, C., Murata, M., Nishiyori, H., Fukuda, S., Kawai, J., Daub, C. O., Hume, D. A., Suzuki, H., Orlando, V., Carninci, P., Hayashizaki, Y., and Mattick, J. S. (2009). Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578.
Taft, R. J., Pheasant, M., and Mattick, J. S. (2007). The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299.
Taft, R. J., Simons, C., Nahkuri, S., Oey, H., Korbie, D. J., Mercer, T. R., Holst, J., Ritchie, W., Wong, J. J., Rasko, J. E., Rokhsar, D. S., Degnan, B. M., and Mattick, J. S. (2010). Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 17, 1030–1034.
Takahashi, M., Katagiri, T., Furuya, H., and Hohjoh, H. (2011). Disease-causing allele-specific silencing against the ALK2 mutants, R206H and G356D, in fibrodysplasia ossificans progressiva. Gene Ther. doi: 10.1038/gt.2011.193 [Epub ahead of print].
Tan, E. L., Tan, T. M., Tak Kwong Chow, V., and Poh, C. L. (2007). Inhibition of enterovirus 71 in virus-infected mice by RNA interference. Mol. Ther. 15, 1931–1938.
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H., and Bartel, D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550.
Umbach, J. L., and Cullen, B. R. (2009). The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23, 1151–1164.
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324.
Valencia-Sanchez, M. A., Liu, J., Hannon, G. J., and Parker, R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524.
Van Bakel, H., Nislow, C., Blencowe, B. J., and Hughes, T. R. (2010). Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8:e1000371. doi: 10.1371/journal.pbio.1000371
Verdel, A., Vavasseur, A., Le Gorrec, M., and Touat-Todeschini, L. (2009). Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 53, 245–257 .
Volk, N., and Shomron, N. (2011). Versatility of MicroRNA biogenesis. PLoS ONE 6:e19391. doi: 10.1371/journal.pone.0019391
Wang, P., Zou, F., Zhang, X., Li, H., Dulak, A., Tomko, R. J. Jr., Lazo, J. S., Wang, Z., Zhang, L., and Yu, J. (2009). microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 69, 8157–8165.
Wang, X., Arai, S., Song, X., Reichart, D., Du, K., Pascual, G., Tempst, P., Rosenfeld, M. G., Glass, C. K., and Kurokawa, R. (2008a). Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130.
Wang, Y., Baskerville, S., Shenoy, A., Babiarz, J. E., Baehner, L., and Blelloch, R. (2008b). Embryonic stem cell-specific microRNAs regulate the G1–S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483.
Watanabe, T., Takeda, A., Tsukiyama, T., Mise, K., Okuno, T., Sasaki, H., Minami, N., and Imai, H. (2006). Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743 .
Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., Chiba, H., Kohara, Y., Kono, T., Nakano, T., Surani, M. A., Sakaki, Y., and Sasaki, H. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543.
Watanabe, T., Tomizawa, S., Mitsuya, K., Totoki, Y., Yamamoto, Y., Kuramochi-Miyagawa, S., Iida, N., Hoki, Y., Murphy, P. J., Toyoda, A., Gotoh, K., Hiura, H., Arima, T., Fujiyama, A., Sado, T., Shibata, T., Nakano, T., Lin, H., Ichiyanagi, K., Soloway, P. D., and Sasaki, H. (2011). Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852.
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862.
Wilkins, C., Dishongh, R., Moore, S. C., Whitt, M. A., Chow, M., and Machaca, K. (2005). RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436, 1044–1047.
Williams, J. M., Beck, T. F., Pearson, D. M., Proud, M.B., Cheung, S. W., and Scott, D. A. (2009). A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am. J. Med. Genet. A. 149A, 1758–1762.
Woese, C. R. (1967). The Genetic Code; the Molecular Basis for Genetic Expression. New York: Harper & Row.
Xu, N., Papagiannakopoulos, T., Pan, G., Thomson, J. A., and Kosik, K. S. (2009). MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658.
Yabuta, Y., Ohta, H., Abe, T., Kurimoto, K., Chuma, S., and Saitou, M. (2011). TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 192, 781–795.
Yan, Z., Hu, H. Y., Jiang, X., Maierhofer, V., Neb, E., He, L., Hu, Y., Hu, H., Li, N., Chen, W., and Khaitovich, P. (2011). Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39, 6596–6607.
Yang, J. S., and Lai, E. C. (2011). Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 43, 892–903.
Yehezkel, S., Rebibo-Sabbah, A., Segev, Y., Tzukerman, M., Shaked, R., Huber, I., Gepstein, L., Skorecki, K., and Selig, S. (2011). Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics 6, 63–75.
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016.
Yi, C., Wang, Q., Wang, L., Huang, Y., Li, L., Liu, L., Zhou, X., Xie, G., Kang, T., Wang, H., Zeng, M., Ma, J., Zeng, Y., and Yun, J. P. (2012). MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene doi: 10.1038/onc.2011.629 [Epub ahead of print].
Zhang, C., and Darnell, R. B. (2011). Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 29, 607–614.
Zhang, R., Zhang, L., and Yu, W. (2012a). Genome-wide expression of non-coding RNA and global chromatin modification. Acta Biochim. Biophys. Sin. (Shanghai) 44, 40–47.
Zhang, Y., Engelman, J., and Friedlander, R. M. (2009). Allele-specific silencing of mutant Huntington’s disease gene. J. Neurochem. 108, 82–90.
Zhang, Z. J., Zhang, H., Kang, Y., Sheng, P. Y., Ma, Y. C., Yang, Z. B., Zhang, Z. Q., Fu, M., He, A. S., and Liao, W. M. (2012b). miRNA expression profile during osteogenic differentiation of human adipose-derived stem cells. J. Cell. Biochem. 113, 888–898.
Zhang, W., Zhou, X., Xia, J., and Zhou, X. (2012c). Identification of microRNAs and natural antisense transcript-originated endogenous siRNAs from small-RNA deep sequencing data. Methods Mol. Biol. 883, 221–227.
Zhao, J., Zhang, X., Zhou, Y., Ansell, P. J., and Klibanski, A. (2006). Cyclic AMP stimulates MEG3 gene expression in cells through a cAMP-response element (CRE) in the MEG3 proximal promoter region. Int. J. Biochem. Cell Biol. 38, 1808–1820.
Zhao, Y., Yu, Y., Zhai, J., Ramachandran, V., Dinh, T. T., Meyers, B. C., Mo, B., and Chen, X. (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694.
Zhong, J., Chuang, S. C., Bianchi, R., Zhao, W., Lee, H., Fenton, A. A., Wong, R. K., and Tiedge, H. (2009). BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 29, 9977–9986.
Zhong, Y., Xia, Z., Liu, J., Lin, Y., and Zan, H. (2012). The effects of cyclooxygenase-2 gene silencing by siRNA on cell proliferation, cell apoptosis, cell cycle and tumorigenicity of Capan-2 human pancreatic cancer cells. Oncol. Rep. 27, 1003–1010.
Keywords: non-coding RNA, neuropsychiatric disorder, addiction, anxiety
Citation: Spadaro PA and Bredy TW (2012) Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuropsychiatric disorders. Front. Genet. 3:132. doi: 10.3389/fgene.2012.00132
Received: 10 April 2012; Accepted: 28 June 2012;
Published online: 13 July 2012.
Edited by:
Peng Jin, Emory University School of Medicine, USAReviewed by:
Eran Hornstein, Weizmann Institute of Science, IsraelDaniel Wai Hung Ho, The Polytechnic University of Hong Kong, Hong Kong
Yujing Li, Emory University, USA
Copyright: © 2012 Spadaro and Bredy. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Timothy W. Bredy, ARC Australian Research Fellow, Queensland Brain Institute, Psychiatric Epigenomics Laboratory, The University of Queensland, Brisbane, QLD 4072, Australia. e-mail: t.bredy@uq.edu.au