Structures and functions in the crowded nucleus: new biophysical insights
- 1Laval University Cancer Research Centre-Centre Hospitalier Universitaire de Québec Oncology, Québec, QC, Canada
- 2Biosystems Group, Institute of Automatic Control, Silesian University of Technology, Gliwice, Poland
Concepts and methods from the physical sciences have catalyzed remarkable progress in understanding the cell nucleus in recent years. To share this excitement with physicists and encourage their interest in this field, this review offers an overview of how the physics which underlies structures and functions in the nucleus is becoming more clear thanks to methods which have been developed to simulate and study macromolecules, polymers, and colloids. The environment in the nucleus is very crowded with macromolecules, making entropic (depletion) forces major determinants of interactions. Simulation and experiments are consistent with their key role in forming membraneless compartments such as nucleoli, PML and Cajal bodies, and discrete “territories” for chromosomes. The chromosomes, giant linear polyelectrolyte polymers, exist in vivo in a state like a polymer melt. Looped conformations are predicted in crowded conditions, and have been confirmed experimentally and are central to the regulation of gene expression. Polymer theory has revealed how the chromosomes are so highly compacted in the nucleus, forming a “crumpled globule” with fractal properties which avoids knots and entanglements in DNA while allowing facile accessibility for its replication and transcription. Entropic repulsion between looped polymers can explain the confinement of each chromosome to a discrete region of the nucleus. Crowding and looping are predicted to facilitate finding the specific targets of factors which modulate activities of DNA. Simulation shows that entropic effects contribute to finding and repairing potentially lethal double-strand breaks in DNA by increasing the mobility of the broken ends, favoring their juxtaposition for repair. Signaling pathways are strongly influenced by crowding, which favors a processive mode of response (consecutive reactions without releasing substrates). This new information contributes to understanding the sometimes counter-intuitive consequences and the evolutionary advantages of a crowded environment in the nucleus.
Introduction
The nucleus can be viewed as a complex colloidal system of proteins, ribonucleoproteins, and giant charged linear polymers (the chromosomes), confined within the lamina of the nuclear envelope. The measured global concentration of macromolecules exceeds 100 mg/ml (reviewed in [1]); the chromosomes occupy ~10% of the nuclear volume [2] and total macromolecules between ~20% [3] and ~40% [4, 5]. As observed 100 years ago, “physical chemists and biochemists have nowadays come to realize that the most fruitful ground of both chemistry and biology lies in the land of colloids” [6] which Ostwald aptly termed “the world of neglected dimensions” [7].
Thinking in this field has been marked by the realization that entropic forces play major roles in interactions between macromolecules in the nucleus, as they do in colloidal systems [8, 9] and, as noted many years ago, in the cell cytoplasm [10], although they are insignificant in the dilute conditions usually used for molecular biological experiments in vitro. Entropic (also termed depletion) forces favor contacts between larger macromolecules or particles in a concentrated mixture, because then the excluded volumes which surround them overlap and more volume is available to smaller molecules [11]. Entropic interactions are highly sensitive to the local shape of macromolecules, conferring a “lock and key” selectivity [12, 13]. A further result of crowding which is likely be important in the nucleus is a significant enhancement of the thermodynamic activity of macromolecules [14] which would allow efficient interactions with fewer members of each species than those required in a dilute medium.
Compartments in the Nucleus
Nuclei contain diverse types of compartments which contain macromolecular complexes with different specialized functions (reviewed in [15]), for example nucleoli where ribosomal RNA is transcribed and other types shown in Figure 1A. These compartments have no external membrane, and with the exception of chromosomes their macromolecules exchange dynamically with the surrounding milieu (for example [16]), they are mobile [17], and they can divide and fuse [17, 18]. RNAs may be essential structural components of compartments [19]. Despite many descriptive studies, the mechanism by which compartments are formed has been unclear.
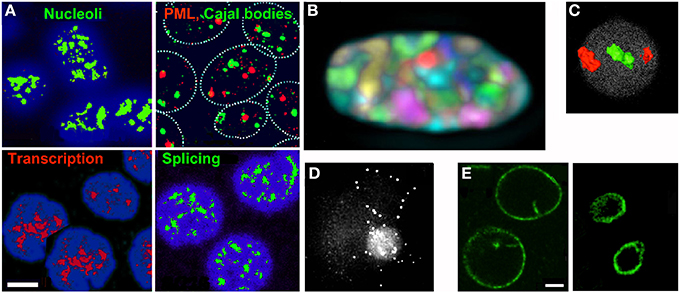
Figure 1. (A–D) Features of the nucleus which can be understood as effects of crowding and entropic forces. (A) Compartments visualized by immunofluorescence (reproduced from [20]). (B) Discrete territories of the 24 chromosomes in a human fibroblast nucleus labeled with different fluorochrome combinations (reproduced from [21]). (C) Gene-poor chromosomes ([18], red) are located more peripherally than gene-rich chromosomes ([19], green) in the nucleus of human lymphocytes (reproduced from [22] by permission from Macmillan Publishers Ltd, 2001). (D) A loop of DNA containing ~220 kb of the human dystrophin gene visualized by fluorescence in situ hybridization (FISH) (reproduced from [23] by permission of Oxford University Press). (E) The nuclear lamina confines and compresses the contents of the nucleus. The nuclear lamina of K562 cells was visualized by immunofluorescence before (left) or after (right) DNA was digested with restriction enzymes and chromatin was removed by electroelution (R. Hancock, unpublished). Scale bars 5 μm.
Experimental and simulation studies are consistent with a key role for crowding in the association of macromolecules to form compartments. Simulations show that model particles form clusters in crowded conditions (Figure 2A) [24]. Cluster formation is observed quite commonly in colloidal systems [25], and the concentration of protein in clusters formed in a crowded solution may reach up to ~700 mg/ml [26] (Figure 2B). The formation of compartments can also be regarded as phase separation, where entropic attractions in a mixture of macromolecules result in expulsion of one component as a separate phase [27]. Since entropic effects favor the positioning of particles on a surface [28] they may contribute to the frequently-observed localization of Cajal and PML bodies in contact with chromosomes (for example [29, 30]).
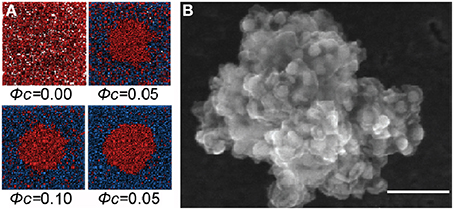
Figure 2. Formation of protein clusters in a crowded solution. (A) Molecular Dynamics simulation of Lennard-Jones particles (red) shows clustering induced by crowding particles (blue) which is more pronounced at higher volume fractions (ϕc) of crowder (reproduced from [24] with permission from Elsevier, © 2012). (B) A cluster of monoclonal antibody molecules in a solution with trehalose as crowder (scanning electron microscopy); similar clusters form using polyethylene glycol as crowder. Scale bar 100 μm (reproduced from [26] with permission from the American Chemical Society, © 2012).
Structure and Packing of Chromosomes
Chromatin Fibers
DNA in eukaryotic cells is associated with spherical protein subunits (nucleosomes) as a giant linear polyelectrolyte polymer, and until recently thinking was dominated by the model that this fiber has a regular helical conformation with a diameter of ~30 nm. Nevertheless, irregular conformations were seen quite commonly in vivo in studies by electron and optical microscopy (for example [31, 32]) (Figure 3B) and were also predicted by simulation of the response to crowding of linear polyelectrolyte polymers [9, 33] (Figure 3A) and by considering that chromosomes resemble block copolymers [34, 35] with interspersed regions of repeated DNA sequences [36], methylated cytosine-containing DNA which has particular conformational properties [37, 38], and nucleosomes containing variant or modified histones which influence fiber interactions [39, 40]. These discrepancies have been resolved by recent cryo-electron microscopy and X-ray scattering studies, which show conclusively that chromatin fibers exist in vivo in a disordered, interdigitated state resembling a polymer melt [41].
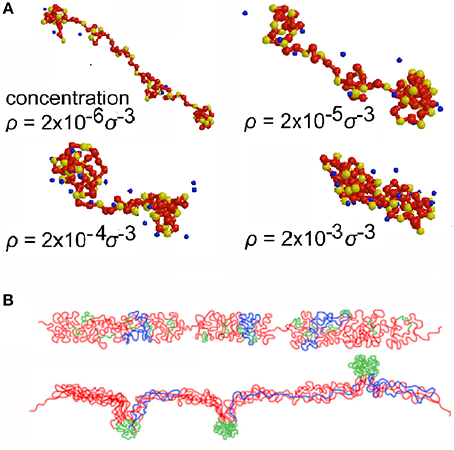
Figure 3. Conformations of chromatin fibers. (A) Simulation by Molecular Dynamics of a linear polyelectrolyte polymer resembling chromatin at increasing concentration (ρ) (self-crowding) (reproduced from [34] with permission of ACS Publications, © 1999). (B) Conformations in vivo of the DNA of a transgene deduced from studies using FISH; the transgene DNA is red, bacterial artificial chromosome DNA is blue, and lac operator DNA is green (reproduced from [32] by permission of Rockefeller University Press, © 2010).
Loops in Chromatin Fibers In vivo
The existence of loops in DNA in vivo has been a common theme in optical and electron microscopy studies of lysed nuclei (for example [42, 43]), and is now realized to be central to understanding chromatin fiber conformations in vivo [44]. Looping must be invoked in order to reconcile the spatial distance between two points on a chromosome in vivo, measured by 3D fluorescence in situ hybridization (FISH), with their linear distance along the chromosome [45, 46]. Simulation shows that spontaneous looping of a polymer is favored by crowding (Figure 4A) and is more frequent and persistent in longer chains [47]. These predictions have been confirmed experimentally by mapping the contact points at the base of chromatin loops after crosslinking them in vivo (chromosome conformation capture, 3C) which reveals loops a few kb to tens of Mb in length [48, 49]. Looping is at least to some extent a stochastic process, and in the context of gene regulation is clearly an attractive model for bringing regulatory sequences in DNA into proximity to the genes which they control [50, 51] (Figure 4B). Nucleoli [52] and transcription factories [53] are proposed to be formed by the assembly of numerous loops.
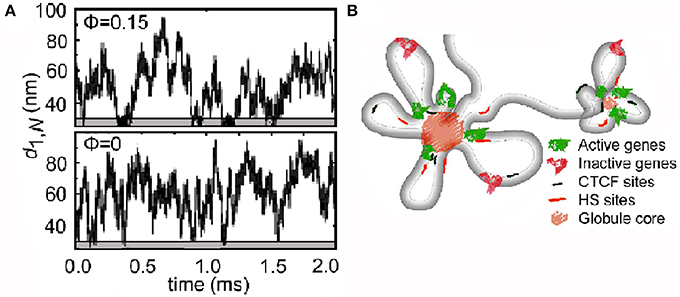
Figure 4. Loops in chromatin. (A) Simulation by Langevin dynamics of the spontaneous formation of loops in a linear polymer. In the presence of crowding particles (upper panel), the minima of the end-to-end distance (d1,N) which reflect looping are more frequent and persistent (lower gray areas) (reproduced from [47] with permission from the American Physical Society, © 2006). (B) Chromatin loops in the region of the human α-globin gene in vivo deduced from chromosome conformation capture (3C) experiments (reproduced from [48] by permission from Macmillan Publishers Ltd, © 2014).
Chromosome Territories
Each chromosome is confined to a discrete territory in the nucleus, with little or no intermingling (reviewed in [21, 22]) (Figure 1B). Simulations show that this segregation can be understood by the entropic repulsion which occurs between polymers containing loops [44, 54]. The preferential positioning of gene-rich and transcriptionally active chromosomes in central regions of the nucleus while inactive chromosomes are more peripheral [55] (Figure 1C) can be explained by entropic effects resulting from a higher frequency of loops in more compact inactive chromatin [52].
Packing the Genome into the Nucleus
The ~2 m of DNA in human cells are packed as chromatin fibers into a nucleus ~10 μm in diameter, a formidable level of compaction. How this is achieved has been revealed by the seminal simulation studies of the collapse of a linear polymer by Grosberg et al. [57], which show how a compact “crumpled globule” with fractal properties is formed (a notable example of the value of non-translational research). Experiments and simulations strongly support this fractal manner of packing chromatin in the nucleus [58–61] (with the exception of yeast), which was suggested earlier by neutron diffraction studies of nuclei [62]. Data from 3C studies of human chromosomes are consistent with a fractal organization, but not with the alternative “equilibrium globule” conformation [58]. On could speculate that the fractal globule conformation has been selected during evolution because the chromatin fiber does not contain knots or entanglements, DNA is easily accessible, and chromosomes are localized in a territorial pattern without intermingling (Figure 5) [58, 59]. A more evolved model which is consistent with the fractal globule allows dynamic variations of chromatin folding and switch-like changes of genome architecture to be captured [63]. These models will replace the common textbook depictions of chromatin packed through a hierarchical series of largely speculative coiled intermediates.
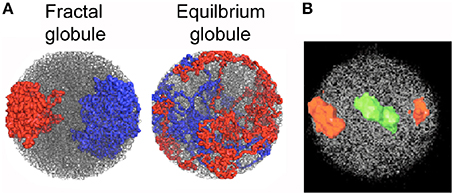
Figure 5. Chromosome territories. (A) Simulation of two polymer chains compacted into a fractal globule shows a territorial organization like that of chromosomes 18 and 19 in a human lymphocyte nucleus (B), whereas they are mixed in an equilibrium globule. (A) reproduced from [58], (B) reproduced from [22] by permission from Macmillan Publishers Ltd, © 2001.
Diffusion and Signaling
Diffusion of molecules is central to all cellular activities, from biochemical reactions to metabolic networks, signaling pathways, and control of gene expression. Diffusion of macromolecules is slowed in the nucleus ([64, 65], reviewed in [66]) probably due to collosions with chromatin and other large obstacles or to viscoelasticity, but nevertheless most macromolecules and multiprotein complexes can explore the entire nuclear volume [67]. Large particles and macromolecules show anomalous diffusion in the nucleus, a lesss-than-linear increase of mean-square displacement with time like that seen in crowded solutions [68]. Remarkably, subdiffusion can increase the probability of finding a nearby target compared to normal diffusion [68–71].
Signaling pathways depend on diffusion and are therefore influenced by crowding, as illustrated by fascinating recent studies of a step in the Mitogen-activated Protein Kinase (MAPK) pathway which transmits signals from the cell surface to DNA in the nucleus. This has the typical structure of a cascade of kinases in which each kinase phosphorylates the next and activates it; phosphorylation can be reversed by a phosphatase and must occur at two sites for complete activation. In dilute conditions, after phosphorylating its substrate the kinase dissociates leading to a significant probability that a different kinase molecule will phosphorylate the second site, a distributive mode. In contrast, in crowded conditions when diffusion is slower it is more probable that the first kinase molecule will remains bound or close to its substrate while regaining activity by binding ATP and will then phosphorylate the second site, a processive mode (Figure 6A) [72]. This prediction has been confirmed experimentally using purified kinases and a crowding agent in vitro [73] (Figure 6B). Thus responses of pathways of this type appear to be distributive in in vitro experiments, but in conditions in vivo are actually processive with different downstream responses to signals [74].
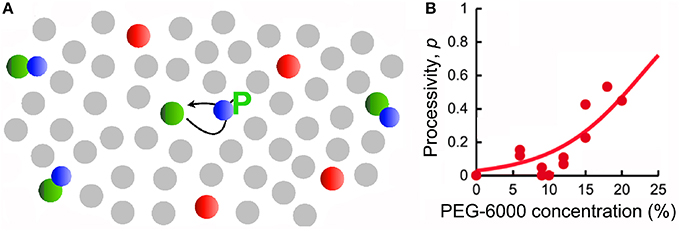
Figure 6. Stimulation by crowding of the processivity of a step in a signaling pathway. (A) In the MAPK pathway MEK (Mitogen/Extracellular signal-regulated Kinase) (green) phosphorylates ERK (Extracellular signal-Regulated Kinase) (blue) at two positions, on tyrosine and threonine. The system also contains phosphatases (red). In the presence of crowders (gray) the reactants diffuse more slowly, and therefore the probability of rebinding and a second phosphorylation of ERK on threonine is increased (reproduced from [72] with permission from Elsevier, © 2014). (B) Experimental data using purified kinases in vitro showing how the frequency of a second phosphorylation of ERK by the same molecule of MEK (processivity) increases as a function of the concentration of the crowder PEG-6000 (reproduced from [74] by permission from Macmillan Publishers Ltd, © 2013).
Finding Targets in the Genome
Proteins which regulate activities of DNA are believed to find their target in chromatin by facilitated diffusion, a combination of 3-dimensional diffusion in the medium and 1-dimensional sliding on chromatin [75]. Target finding is predicted to be accelerated by crowding [76, 77], and also by DNA looping which facilitates the bypassing of factors which could block sliding [76]. The fractal organization of chromatin [67] also has implications for the kinetics of target finding; chromatin-binding proteins have a long residence time in compact (hetero-) chromatin, suggesting that they bind to all available sites, while on the other hand exploration is faster and less redundant in less compact (eu-) chromatin which offers more exposed DNA, presumably facilitating the detection of less frequent regulatory elements [60].
Target finding is crucial for the survival of a cell when potentially lethal double-strand breaks (DSBs) in DNA must be repaired by rejoining one extremity of the broken DNA correctly to the other extremity. A DSB causes local changes in the mobility of chromatin; modeling predicts an increased mobility due to entropic effects [78], but motion in vivo is subdiffusional [79] which reduces the probability of long-range movements. The broken ends adopt more peripheral positions in chromosomes [78], favoring their meeting and rejoining.
Future Challenges and Directions
The new insights discussed here suggest that many features of the nucleus which are apparently complex can be understood by the operation of relatively simple physicochemical principles. Many aspects of the biophysical implications of crowding in the nucleus remain to be explored. Experimentally-accessible questions include:
- the effects of crowding on the structure in vivo of chromatin which contains DNA in conformations other than the classical B-form double helix, such as the DNA in telomeres whose conformation in vitro is strongly influenced by crowding [80];
- the consequences of crowding for the structures of RNAs and ribonucleoproteins in vivo; the folding and stability of RNA in vitro are enhanced significantly by crowding [81–83];
- loops in chromatin fibers in vivo are usually thought to be stabilized by proteins such as cohesin (for example [84]). Reports that nucleosomes which contain identical DNA sequences can self-associate preferentially [85] raise the possibility that similar interactions could contribute to the formation and stabilization of loops [86]. Could this be one of the still obscure functions of “junk” DNA [87]?
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to acknowledge the hospitality of Drs. Joanna Rzeszowska and Andrzej Swierniak during the preparation of this review.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fphy.2014.00053/abstract
References
1. Hancock R. The crowded nucleus. Int Rev Cell Mol Biol. (2014) 307:15–26. doi: 10.1016/B978-0-12-800046-5.00002-3
2. Weidemann T, Wachsmuth M, Knoch TA, Müller G, Waldeck W, Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J Mol Biol. (2003) 334:229–40. doi: 10.1016/j.jmb.2003.08.063
3. Finan JD, Chalut KJ, Wax A, Guilak F. Nonlinear osmotic properties of the cell nucleus. Ann Biomed Eng. (2009) 37:477–91. doi: 10.1007/s10439-008-9v618-5
4. Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J. (2006) 91:4649–64. doi: 10.1529/biophysj.106.086454
5. Fritsch CC, Langowski J. Anomalous diffusion in the interphase cell nucleus: the effect of spatial correlations of chromatin. J Chem Phys. (2010) 133:025101. doi: 10.1063/1.3435345
7. Ostwald W. An Introduction to Theoretical and Applied Colloid Chemistry. The world of Neglected Dimensions. New York, NY: John Wiley & Sons (1917). p. 232.
8. Yodh AG, Lin K, Crocker JC, Dinsmore AD, Kaplan PD. Entropically driven self-assembly and interaction in suspension. Phil Trans R Soc Lond A (2001) 359:921–37. doi: 10.1098/rsta.2000.0810
9. Lekkerkerker HNW, Tuinier R. Colloids and the Depletion Interaction. Lecture Notes in Physics, vol. 833. Berlin, Heidelberg: Springer (2011). p. 233. doi: 10.1007/978-94-007-1223-2
10. Walter H, Brooks DE. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. (1995) 361:135–9. doi: 10.1016/0014-5793(95)00159-7
11. Asakura S, Oosawa F. On interaction between two bodies immersed in a solution of macromolecules. J Chem Phys. (1954) 22:1255–6. doi: 10.1063/1.1740347
12. Kinoshita M, Oguni T. Depletion effects on the lock and key steric interactions between macromolecules. Chem Phys Lett. (2002) 351:79–84. doi: 10.1016/S0009-2614(01)01346-X
13. Sacanna S, Irvine WTM, Chaikin PM, Pine DJ. Lock and key colloids. Nature (2008) 464:575–8. doi: 10.1038/nature08906
14. Schnell S, Hancock R. The intranuclear environment. Methods Mol Biol. (2008) 463:3–19. doi: 10.1007/978-1-59745-406-3_1
15. Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol. (2014) 28C:76–83. doi: 10.1016/j.ceb.2014.03.004
16. Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, et al. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. (2008) 121:2731–43. doi: 10.1242/jcs.031922
17. Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. (2000) 151:1561–74. doi: 10.1083/jcb.151.7.1561
18. Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA (2011) 108:4334–9. doi: 10.1073/pnas.1017150108
19. Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. (2012) 22:179–87. doi: 10.1016/j.gde.2011.12.005
20. Hancock R, Hadj-Sahraoui Y. Isolation of cell nuclei using inert macromolecules to mimic the crowded Cytoplasm. PLoS ONE (2009) 4:e7560. doi: 10.1371/journal.pone.0007560
21. Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and Prometaphase rosettes. PLoS Biol. (2005) 3:e157. doi: 10.1371/journal.pbio.0030157
22. Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. (2001) 2:292–301. doi: 10.1038/35066075
23. Iarovaia OV, Bystritskiy A, Ravcheev D, Hancock R, Razin SV. Visualization of individual DNA loops and a map of loop domains in the human dystrophin gene. Nucleic Acids Res. (2004) 32:2079–86. doi: 10.1093/nar/gkh532
24. Cho EJ, Kim JS. Crowding effects on the formation and maintenance of nuclear bodies: insights from molecular-dynamics simulations of simple spherical model particles. Biophys J. (2012) 103:424–33. doi: 10.1016/j.bpj.2012.07.007
25. Kovalchuk N, Starov V, Langston P, Hilal N. Formation of stable clusters in colloidal suspensions, Adv Colloid Interface Sci. (2009) 147–148:144–54. doi: 10.1016/j.cis.2008.11.001
26. Johnston KP, Maynard JA, Truskett TM, Borwankar AU, Miller MA, Wilson BK, et al. Concentrated dispersions of equilibrium protein nanoclusters that reversibly dissociate into active monomers. ACS Nano (2012) 6:1357–69. doi: 10.1021/nn204166z
27. Aumiller WM Jr., Davis BW, Keating CD. Phase separation as a possible means of nuclear compartmentalization. Int Rev Cell Mol Biol. (2014) 307:109–49. doi: 10.1016/B978-0-12-800046-5.00005-9
28. Dinsmore AD, Wong DT, Nelson P, Yodh AG. Hard spheres in vesicles: curvature-induced forces and particle-induced curvature. Phys Rev Lett. (1998) 80:409–12. doi: 10.1103/PhysRevLett.80.409
29. Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell (2001) 12:565–76. doi: 10.1091/mbc.12.3.565
30. Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, et al. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol. (2004) 164:515–26. doi: 10.1083/jcb.200305142
31. Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol (1994) 125:1–10. doi: 10.1083/jcb.125.1.1
32. Sinclair P, Bian Q, Plutz M, Heard E, Belmont AS. Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J Cell Biol. (2010) 190:761–76. doi: 10.1083/jcb.200912167
33. Micka U, Holm C, Kremer K. Strongly charged, flexible polyelectrolytes in poor solvents: molecular dynamics simulations. Langmuir (1999) 15:4033–44. doi: 10.1021/la981191a
34. Woloszczuk S, Banaszak M, Knychala P, Lewandowski K, Radosz M. Alternating multiblock copolymers exhibiting protein-like transitions in selective solvents: a Monte Carlo study. J Non-Cryst Solids (2008) 354:4138–42. doi: 10.1016/j.jnoncrysol.2008.06.022
35. Neratova IV, Komarov PV, Pavlov AS, Ivanov VA. Collapse of an AB copolymer single chain with alternating blocks of different stiffness. Russ Chem B (2011) 60:229–37. doi: 10.1007/s11172-011-0038-6
36. Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. (2008) 72:686–727. doi: 10.1128/MMBR.00011-08
37. Severin PM, Zou X, Gaub HE, Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res (2011) 39:8740–51. doi: 10.1093/nar/gkr578
38. Shimooka Y, Nishikawa J, Ohyama T. Most Methylation-Susceptible DNA sequences in human embryonic stem cells undergo a change in conformation or flexibility upon methylation. Biochemistry (2013) 52:1344–53. doi: 10.1021/bi301319y
39. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z. et al. High-resolution profiling of histone methylations in the human genome. Cell (2007) 129:823–37. doi: 10.1016/j.cell.2007.05.009
40. Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science (2006) 311:844–7. doi: 10.1126/science.1124000
41. Nozaki T, Kaizu K, Pack CG, Tamura S, Tani T, Hihara S, et al. Flexible and dynamic nucleosome fiber in living mammalian cells. Nucleus (2013) 4:349–56. doi: 10.4161/nucl.26053
42. Benyajati C, Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell (1976) 9:393–407. doi: 10.1016/0092-8674(76)90084-2
43. Hancock R, Hughes ME. Organization of DNA in the interphase nucleus. Biol Cell (1982) 44:201–12.
44. Heermann DW. Physical nuclear organization: loops and entropy. Curr Opin Cell Biol. (2011) 23:332–7. doi: 10.1016/j.ceb.2011.03.010
45. Tark-Dame M, van Driel R, Heermann DW. Chromatin folding - from biology to polymer models and back. J Cell Sci. (2011) 124:839–45. doi: 10.1242/jcs.077628
46. Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci USA (2009) 106:3812–7. doi: 10.1073/pnas.0809501106
47. Toan NM, Marenduzzo D, Cook PR, Micheletti C. Depletion effects and loop formation in self-avoiding polymers. Phys Rev Lett. (2006) 97:178302. doi: 10.1103/PhysRevLett.97.178302
48. Baù D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. (2011) 18:107–14. doi: 10.1038/nsmb.1936
49. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (2009) 326:289–93. doi: 10.1126/science.1181369
50. Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. (2013) 14:390–403. doi: 10.1038/nrg3454
51. Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta (2009) 1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002
52. Canals-Hamann AZ, das Neves RP, Reittie JE, Iñiguez C, Soneji S, Enver T, et al. A biophysical model for transcription factories. BMC Biophys (2013) 6:2. doi: 10.1186/2046-1682-6-2
53. Doyle B, Fudenberg G, Imakaev M, Mirny LA. Chromatin loops as modulators of enhancer-promoter interactions in their vicinity. BioRxiv (2014) arXiv:1403.1236 (q-bio.GN). doi: 10.1101/003087
54. de Nooijer S, Wellink J, Mulder BM, Bisseling T. Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei. Nucleic Acids Res. (2009) 37:3558–68. doi: 10.1093/nar/gkp219
55. Dietzel S, Zolghadr K, Hepperger C, Belmont AS. Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J Cell Sci. (2004) 117:4603–14. doi: 10.1242/jcs.01330
56. Marenduzzo D, Cook PR. Entropic organization of interphase chromosomes. J Cell Biol. (2009) 186:825–34. doi: 10.1083/jcb.200903083
57. Grosberg AY, Nechaev SK, Shakhnovich EI. The role of topological constraints in the kinetics of collapse of macromolecules. J Phys France (1988) 49:2095–100. doi: 10.1051/jphys:0198800490120209500
58. Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. (2011) 19:37–51. doi: 10.1007/s10577-010-9177-0
59. Grosberg AY. How two meters of DNA fit into a cell nucleus: polymer models with topological constraints and experimental data. Polymer Science Ser C (2012) 54:1–10. doi: 10.1134/S1811238212070028
60. Bancaud A, Lavelle C, Huet S, Ellenberg J. A fractal model for nuclear organization: current evidence and biological implications. Nucl Acids Res. (2012) 40:8783–92. doi: 10.1093/nar/gks586
61. Rosa A, Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput Biol. (2008) 4:e1000153. doi: 10.1371/journal.pcbi.1000153
62. Lebedev DV, Filatov MV, Kuklin AI, Islamov AK, Kentzinger E, Pantina R, et al. Fractal nature of chromatin organization in interphase chicken erythrocyte nuclei: DNA structure exhibits biphasic fractal properties. FEBS Lett. (2005) 579:1465–68. doi: 10.1016/j.febslet.2005.01.052
63. Barbieri M, Chotalia M, Fraser J, Lavitas LM, Dostie J, Pombo A, et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci USA (2012) 109:16173–8. doi: 10.1073/pnas.1204799109
64. Seksek O, Biwersi J, Verkman AS. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J Cell Biol. (1997) 138:131–42. doi: 10.1083/jcb.138.1.131
65. Ritland Politz JC, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell (2003) 14:4805–12. doi: 10.1091/mbc.E03-06-0395
66. Weiss M. Crowding, diffusion, and biochemical reactions. Int Rev Cell Mol Biol. (2014) 307:383–417. doi: 10.1016/B978-0-12-800046-5.00011-4
67. Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. (2009) 28:3785–98. doi: 10.1038/emboj.2009.340
68. Banks DS, Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys J. (2005) 89:2960–71. doi: 10.1529/biophysj.104.051078
69. Guigas G, Weiss M. Sampling the cell with anomalous diffusion - the discovery of slowness. Biophys J. (2008) 94:90–4. doi: 10.1529/biophysj.107.117044
70. Leijnse N, Jeon JH, Loft S, Metzler R, Oddershede LB. Diffusion inside living human cells. Eur Phys J Special Topics (2012) 204:75–84. doi: 10.1140/epjst/e2012-01553-y
71. Bauer M, Metzler R. In vivo facilitated diffusion model. PLoS ONE (2013) 8:e53956. doi: 10.1371/journal.pone.0053956
72. ten Wolde PR, Mugler A. Importance of crowding in signaling, genetic, and metabolic networks. Int Rev Cell Mol Biol. (2014) 307:419–42. doi: 10.1016/B978-0-12-800046-5.00012-6
73. Takahashi K, Tanase-Nicola S, ten Wolde PR. Spatio-temporal correlations can drastically change the response of a MAPK pathway. Proc Natl Acad Sci USA (2010) 107:2473–8. doi: 10.1073/pnas.0906885107
74. Aoki K, Takahashi K, Kaizu K, Matsuda M. A quantitative model of ERK MAP kinase phosphorylation in crowded media. Sci Rep. (2013) 3:1541. doi: 10.1038/srep01541
75. Mirny L, Slutsky M, Wunderlich Z, Tafvizi A, Leith J, Kosmrlj A. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J Phys A (2009) 42:1751–8121. doi: 10.1088/1751-8113/42/43/434013
76. Li G, Berg OG, Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nat Phys. (2009) 5:294–7. doi: 10.1038/nphys1222
77. Brackley CA, Cates ME, Marenduzzo D. Intracellular facilitated diffusion: searchers, crowders, and blockers. Phys Rev Lett. (2013) 111:108101. doi: 10.1103/PhysRevLett.111.108101
78. Zhang Y, Heermann DW. DNA double-strand breaks: linking gene expression to chromosome morphology and mobility. Chromosoma (2014) 123:103–15. doi: 10.1007/s00412-013-0432-y
79. Girst S, Hable V, Drexler GA, Greubel C, Siebenwirth C, Haum M, et al. Subdiffusion supports joining of correct ends during repair of DNA double-strand breaks. Sci Rep. (2013) 3:2511. doi: 10.1038/srep02511
80. Sugimoto N. Noncanonical structures and their thermodynamics of DNA and RNA under molecular crowding: beyond the Watson-Crick double helix. Int Rev Cell Mol Biol. (2014) 307:205–73. doi: 10.1016/B978-0-12-800046-5.00008-4
81. Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc. (2010) 132:8690–6. doi: 10.1021/ja101500g
82. Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA (2014) 20:331–47. doi: 10.1261/rna.042747.113
83. Dupuis NF, Holmstrom ED, Nesbitt DJ. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc Natl Acad Sci USA (2014) 111:8464–9. doi: 10.1073/pnas.1316039111
84. Cuylen S, Haering CH. A new cohesive team to mediate DNA looping. Cell Stem Cell (2010) 7:424–6. doi: 10.1016/j.stem.2010.09.006
85. Nishikawa J, Ohyama T. Selective association between nucleosomes with identical DNA sequences. Nucl Acids Res. (2013) 41:1544–54. doi: 10.1093/nar/gks1269
86. Cherstvy AG, Teif VB. Structure-driven homology pairing of chromatin fibers: the role of electrostatics and protein-induced bridging. J Biol Phys. (2013) 39:363–85. doi: 10.1007/s10867-012-9294-4
Keywords: nucleus, crowding, entropic forces, nuclear compartments, chromatin loops, fractal globule, signaling, target finding
Citation: Hancock R (2014) Structures and functions in the crowded nucleus: new biophysical insights. Front. Phys. 2:53. doi: 10.3389/fphy.2014.00053
Received: 26 June 2014; Paper pending published: 30 July 2014;
Accepted: 22 August 2014; Published online: 12 September 2014.
Edited by:
Francesco Piazza, University of Orléans, FranceReviewed by:
Haroldo Valentin Ribeiro, Universidade Estadual de Maringa, BrazilDavide Marenduzzo, University of Edinburgh, UK
Copyright © 2014 Hancock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronald Hancock, Laval University Cancer Research Centre-CHUQ, 9 rue MacMahon, Québec, QC G1R 2J6, Canada e-mail: ronald.hancock@crhdq.ulaval.ca
 Ronald Hancock
Ronald Hancock