- 1Department of Molecular Microbiology and Immunology, Oregon Health and Science University, Portland, OR, USA
- 2Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA, USA
- 3Department of Infectious Diseases, J. Craig Venter Institute, Rockville, MD, USA
The extracytoplasmic functioning sigma factor σE is known to play an essential role for Salmonella enterica serovar Typhimurium to survive and proliferate in macrophages and mice. However, its regulatory network is not well-characterized, especially during infection. Here we used microarray to identify genes regulated by σE in Salmonella grown in three conditions: a nutrient-rich condition and two others that mimic early and late intracellular infection. We found that in each condition σE regulated different sets of genes, and notably, several global regulators. When comparing nutrient-rich and infection-like conditions, large changes were observed in the expression of genes involved in Salmonella pathogenesis island (SPI)-1 type-three secretion system (TTSS), SPI-2 TTSS, protein synthesis, and stress responses. In total, the expression of 58% of Salmonella genes was affected by σE in at least one of the three conditions. An important finding is that σE up-regulates SPI-2 genes, which are essential for Salmonella intracellular survival, by up-regulating SPI-2 activator ssrB expression at the early stage of infection and down-regulating SPI-2 repressor hns expression at a later stage. Moreover, σE is capable of countering the silencing of H-NS, releasing the expression of SPI-2 genes. This connection between σE and SPI-2 genes, combined with the global regulatory effect of σE, may account for the lethality of rpoE-deficient Salmonella in murine infection.
Introduction
Salmonella enterica enterica serovar Typhimurium (strain 14028s; referred to as Salmonella hereafter) is an invasive enteric pathogen with remarkable adaptability to diverse environments. The host as well as the residential niche of Salmonella varies, requiring the pathogen to sense its location within the host and to adjust its gene expression accordingly. Salmonella has evolved a number of strategies to sense the environment and to modulate its production of virulence factors appropriately (Alpuche Aranda et al., 1992; Raffatellu et al., 2005; Yoon et al., 2009). The bacterial surface is the frontline of the host-pathogen interaction, making it both a major target of the host immune response and a primary location for the pathogen to activate its own defensive strategies (Rowley et al., 2006).
In Gram-negative bacteria, stresses that affect components of the cell envelope, such as periplasmic and outer-membrane proteins, illicit a variety of responses in the cell which are collectively known as extracytoplasmic stress responses (ESRs). There are at least four signal transduction systems in Gram-negative bacteria that govern the ESRs: the alternative sigma factor σE (encoded by rpoE), the two-component regulatory systems CpxR/CpxA and BaeS/BaeR, and the phage shock protein (Psp) system (4). Of these systems only the absence of σE causes a strong virulence defect, although there is overlap and crosstalk between the systems when reacting to different extracytoplasmic stresses (Connolly et al., 1997; Jones et al., 1997; Humphreys et al., 1999, 2004; Kenyon et al., 2002; Becker et al., 2005; Karlinsey et al., 2010). The availability of σE is controlled by antisigma factor, which sequesters σE to the membrane in an inactive state when membrane stress is absent. However, in the presence of stresses that lead to accumulation of misfolded proteins in the periplasm, a proteolytic cascade is initiated, releasing σE from antisigma factor. Free σE recognizes specific promoters and initiates transcription with core RNA polymerase (Rowley et al., 2006; Osterberg et al., 2011).
Using molecular genetic approaches and DNA microarray analyses, the σE regulon has been extensively studied in E. coli (Dartigalongue et al., 2001; Rezuchova et al., 2003; Kabir et al., 2005; Rhodius et al., 2006). However, the σE regulon obtained from E. coli is not directly applicable to Salmonella because σE plays distinct roles in these two bacteria. For instance, σE is required for viability in E. coli, while in Salmonella, it is essential for resisting reactive oxygen species, antimicrobial peptides, acid stress, and likely additional virulence related functions (Bang et al., 2005; Crouch et al., 2005; Muller et al., 2009). One of the rpoE promoters, rpoEp3, is more strongly induced by cold shock than by heat shock in Salmonella, but not in E. coli, reflecting the functional variation of σE in combating different stresses in these two organisms (Miticka et al., 2003). The Salmonella σE regulon has been studied using an E. coli two-plasmid screening system and by microarrays, however, the growth conditions exploited by previous studies did not discriminate different stages of Salmonella infection (Skovierova et al., 2006; Yoon et al., 2009).
Compared to other regulators important for Salmonella virulence (e.g., fruR, ssrAB, slyA, crp, rpoS, etc.), an rpoE mutant is the most sensitive to the intracellular environment. Specifically, the LD50 of an rpoE mutant in BALB/c mice by intraperitoneal infection is greater than 106 CFU, whereas that of the parent strain is 1–2 CFU. Only a few, if any, viable rpoE mutants are recovered from primary macrophages after 30 min of infection (Yoon et al., 2009). It is unclear why the absence of rpoE in Salmonella has such an extreme phenotype during intracellular growth. Previous studies have shown that hundreds of genes were regulated by σE, including genes in the Salmonella Pathogenicity Island 2 (SPI-2) (Osborne and Coombes, 2009; Yoon et al., 2009). SPI-2 genes encode components of a type III secretion system (TTSS) and its associated effectors, which are required for intracellular survival. The expression of SPI-2 genes is tightly regulated temporally and spatially. Without induction (e.g., carbon limitation, low concentrations of Mg2+ or Ca2+, and acidic pH), SPI-2 is bound by nucleoid-associated protein H-NS to silence transcription and avoid detrimental consequences of inappropriate expression. Inside professional phagocytic cells, SPI-2 expression is induced by the two-component systems PhoP/PhoQ, OmpR/EnvZ, and SsrA/SsrB (Lee et al., 2000; Garmendia et al., 2003; Bijlsma and Groisman, 2005).
To obtain a more complete picture of the σE regulatory network in Salmonella, in this paper we analyzed the transcriptional profile of σE by microarray on wild-type (WT) and ΔrpoE mutant cultured under three conditions: in nutrient-rich Luria-Bertani (LB) broth to log phase, in acidic minimal medium (LPM) for 4 h to mimic early intracellular infection, and in LPM for 20 h to mimic late intracellular infection. We established that a large number of Salmonella genes involving various functional categories are regulated by σE, both directly and indirectly. Notably, we found that σE up-regulates SPI-2 gene expression through different mechanisms at different stages of infection: by increasing the transcription of SPI-2 activator ssrB in early stage, and by decreasing the transcription of SPI-2 repressor hns in late stage. σE can also counter the silencing of H-NS on SPI-2 genes.
Materials and Methods
Bacterial Strains and Growth Conditions
Salmonella STM ATCC 14028s was used as the parent strain in this study. The rpoE-deletion strain (ΔrpoE) was constructed using λ red recombination system as described (Yoon et al., 2009). Bacteria were grown under 3 different conditions to cover expression of a large number of Salmonella genes. They were: in Luria-Bertani (LB) medium to log phase (OD600 = 0.5), in pH 5.8, low phosphate, low magnesium-containing medium (LPM) for 4 h (OD600 ~ 0.5) or 20 h (OD600 ~ 1.0) (for the LPM culture, bacteria were grown in LB to stationary phase, washed twice in LPM, and resuspended in LPM at 1:10 dilution for an additional 4 h or 20 h) (Niemann et al., 2011). The latter two conditions partially mimic the intracellular environment of the Salmonella-containing vacuole and represent the early and late stage Salmonella infection. All the bacterial cultures were grown in triplicate. For microarray analysis 3 ml of culture was centrifuged, pellet was collected and treated with RNAlater (Ambion), then stored at -20°C prior to processing. The plasmid (pASK- H-NS -3xFLAG) expressing H-NS was constructed by cloning a DNA fragment containing coding sequence of hns on pASK-3xFLAG (constructed on pASK-IBA33plus by Hyunjin Yoon) via EcoRI and AvrII. Primers used in the PCR amplification of hns are shown in Table S1.
Expression and Purification of Recombinant RpoE and H-NS
S. Typhimurium 14028s rpoE and hns genes were cloned into the plasmid pET200/D-TOPO (Invitrogen) using directional TOPO cloning method following the vendor's instructions. The genes were inserted downstream of the hexahistidine tag coding sequence under an isopropyl-1-thio-3/4-D- galactopyranoside (IPTG)-inducible T7 promoter. The bacterial expression constructs were confirmed by sequencing and transformed into BL21 (DE3) E. coli strain (Invitrogen). Transformed bacteria were grown in LB broth containing 60 μg/ml kanamycin to mid-log phase. IPTG was added to a final concentration of 1 mM and the incubation was allowed to proceed for 3 h at 37°C. Bacteria were harvested by centrifugation, stored at -80°C, and lysed in lysis buffer (10 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA) supplemented with fresh protease inhibitor cocktail (Sigma) and 1 mg/ml lysozyme for 30 min on ice. Subsequently, the lysate was sonicated and clarified by centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was incubated with HisPur™ Cobalt resin (Pierce) in batch and the beads were washed extensively with Wash buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole; pH 7.4). Bound proteins were eluted with Elution buffer (50 mM sodium phosphate, 300 mM NaCl, 150 mM imidazole; pH 7.4). Elution buffer was replaced with exchange buffer (50 mM Tris-HCl pH 8.0, 1% SDS, 1 mM EDTA, 10% glycerol) using an Amicon Ultra-4 Centrifugal filter unit (millipore) with a molecular weight cutoff of 3000 Da. A small portion of the eluted protein was withdrawn and subjected to immunoblot to confirm that RpoE and H-NS were expressed at the correct size. The remaining eluted protein sample was separated on SDS-PAGE, and stained with Coomassie blue. The single gel bands at the size of recombinant RpoE and H-NS (His tagged) were excised and sent to Pacific Immunology Corp. (Ramona, CA) for polyclonal antisera production.
Purification of RpoE and H-NS Antisera
The RpoE and H-NS antisera generated in rabbits were purified by affinity chromatography. Briefly, the purified recombinant RpoE or H-NS in coupling buffer (500 mM NaCl, 100 mM NaHCO3, pH 8.0) was mixed with activated CH Sepharose 4B (GE Healthcare) at 1:1 ratio and rotated for 4 h at 4°C. After washing away excess antigens, the remaining active groups on sepharose was blocked with 0.1 M Tris-HCl, then washed with buffers of alternating pH for 5 cycles. Each cycle consisted of a wash with 100 mM acetic acid/sodium acetate, pH 4.0 containing 500 mM NaCl followed by a wash with 100 mM Tris-HCl buffer pH 8.0 containing 500 mM NaCl. The RpoE or H-NS antisera were loaded onto antigen-coupled CH Sepharose 4B in a chromatography column (Bio-Rad). After sequential washes with PBS and lithium chloride solution (1 M LiCl, 150 mM NaCl, 0.5% Nonidet P-40, 10 mM Tris-HCl, pH 8) the column was eluted with 200 mM glycine pH 2 in 10 ml fractions by the addition of 10× PBS and 1% BSA. The pH of the eluates was adjusted to 7 with 5 N NaOH. Fractions containing the purified anti-RpoE and anti-H-NS antibodies as judged by SDS-PAGE were frozen at −20°C. Protein concentration determination was performed according to modified Lowry method using bovine serum albumin (BSA) as reference protein (Sandermann and Strominger, 1972).
Microarray Analysis
For each of the three experimental conditions (LB, LPM 4 h and LPM 20 h), we identified genes that were differentially expressed between the WT and ΔrpoE strains of Salmonella. The samples were assayed to the Salmonella Typhimurium/Typhi microarray (version 8), a two-channel spotted array (70-mer probes) designed by the Pathogen Functional Genomics Resource Center at the J. Craig Venter Institute (JCVI). The analysis consisted of quantifying spot intensities, handling low quality probes, correcting for background intensities, imputing missing values, summarizing replicate probes, normalizing the summarized intensities, and finally, finding differentially expressed genes.
First, we calculated a single, background-corrected intensity for the probes (spots) on each of our arrays. Using the scanned array image, we quantified the probe intensities using the Spotfinder tool from the TM4 Microarray Software Suite (Saeed et al., 2003, 2006), giving us an MEV file for each array. To load and manipulate the intensity data in the MEV files, we used Bioconductor's limma package (Gentleman et al., 2004; Smyth, 2005) We subtracted the background intensities from the foreground, giving us a background-corrected intensity for the probes on each array. If, after this step a probe had a negative intensity, we ignored it treating it as a missing value. We then identified and removed any replicate samples that did not have at least a 0.7 correlation with other replicates. For WT strain there were 13, 12, and 7 replicates that passed this array-level QC step in LB, LPM 4 h, and LPM 20 h conditions, respectively. For the rpoE-deletion strain, the corresponding numbers were 2, 4, and 2.
Next, we summarized replicate probe intensities into a single, normalized expression value for each gene. Before summarization, we imputed missing values using a k- nearest neighbors approach, as implemented in Bioconductor's impute package (Gentleman et al., 2004; Hastie et al.1). We then summarized the replicate intensities (there were two identical probes per gene) by calculating their mean. We finally normalized all of the mutant and WT expression values using quantile normalization, as implemented in the normalize.quantiles function of the preprocessCore R package (Bolstad et al., 2003; Bolstad2). We performed a separate normalization for each of the three experimental conditions.
Using the normalized expression values, we next identified differentially regulated genes between the ΔσE and WT strains. Since our sample size for the knockouts was small, we decided to use the methodology described by Smyth et al., which involves using a moderated t-statistic that is more reliable for a small number of arrays (Smyth, 2004). The differential expression analysis was performed using functions available in the limma package. All microarray data is deposited on the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI), accession numbers GSE25441 and GSE26755.
Chromatin Immunoprecipitation
Salmonellla 14028s grown in LB to log phase, or in LPM for 4 h or 20 h were cross-linked by 1% formaldehyde at room temperature for 25 min, then quenched using 125 mM glycine for an additional 5 min of incubation at room temperature. After cell lysis and sonication, cell debris was removed by centrifugation at 13,000 rpm for 10 min at 4°C, and the supernatant was collected for the immunoprecipitation. The supernatant was split into two samples. One was mixed with affinity purified rabbit anti-RpoE antibody to immunoprecipitate σE–DNA complex, and the other sample was mixed with rabbit monoclonal antibody to GFP as the control (normal rabbit serum contains anti-Salmonella antibody so that was not used as control). They were next incubated at 4°C overnight, and 50 μl of the Dynabead M-280 sheep anti-rabbit IgG (Invitrogen) was added into the mixture. After 6 h of incubation at 4°C with rotating, the beads were washed with a serials of stringent buffers (Cho et al., 2008). Beads were resuspended in 200 μl of elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, and 1% SDS) and incubated at 65°C overnight to reverse the cross-linking. The supernatant was incubated with 1 μl of RNaseA (QIAGEN) for 2 h at 37°C to remove RNAs, followed by incubation with 4 μl of proteinase K solution (Invitrogen) for 2 h at 55°C to remove proteins. The sample was then purified with a PCR purification kit (QIAGEN). Gene-specific quantitative PCR was carried out using the DNA samples.
Quantitative RT-PCR Analysis
Total RNA was isolated using RNAlater Solution (Ambion), RNeasy mini kit (Qiagen), and DNase to remove residual chromosomal DNA (Qiagen) according to manufacturer's instructions. RNA concentration was measured by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.). cDNA was synthesized using the iScript cDNA synthesis kit (Bio Rad) and cDNA corresponding to 10 ng of input RNA was used as template for real-time reaction containing Power SYBR green (Applied Biosystems) and gene-specific primers. The primers were designed with Primer Express 3.0 software and tested for their amplification efficiencies (Table S2). The gyrB gene, encoding for the B subunit of the DNA gyrase, was utilized as endogenous control. The RT-PCR reactions were carried out at 95°C for 10 min, 95°C for 15 s and 60°C for 1 min for 40 cycles (ABI 7700, Applied Biosystems). The expression ratio of each gene was the average from three independent RNA samples and was normalized to the level of gyrB.
Immunoblot Analysis
The WT strain containing pASK-H-NS-3FLAG plasmid was grown in LPM for 20 h in the presence or absence of anhydrotetracycline (AHT) for 4 h. The cells were washed and ~5 × 107 colony-forming units were pelleted and re-suspended in Laemmli sample buffer, boiled for 5 min, and then separated on SDS-PAGE. Proteins on the gel were then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). After blocking in Tris-buffered saline (TBS) plus 5% non-fat dry milk for 1 h, membranes were probed with anti-FLAG monoclonal antibody (Sigma). Membranes were washed and probed with secondary antibody (anti-mouse IgG conjugated with peroxidase) (Sigma). The immune complexes were detected via chemiluminescence using Western Lightning™ (PerkinElmer), and then exposed to XAR Biofilm (Kodak). For immunoblot on H-NS, WT and ΔrpoE strains were grown in LB to log phase, or in LPM for 4 h or 20 h. Affinity purified rabbit anti-H-NS antibody was used as primary antibody, and goat anti-rabbit (IgG) antibody conjugated to horseradish peroxidase (Cell Signaling Technology) was used as the secondary antibody. In both cases, DnaK was probed at the same time as loading control.
Results
Identification of σE-Dependent Genes by Transcriptional Profiling
To study the global regulatory effect of σE on Salmonella gene expression, WT (14028s) and ΔrpoE strains were grown in nutrient-rich LB broth to log phase, or in LPM for 4 h and 20 h to mimic early and late intracellular infection, respectively. Salmonella Typhimurium/Typhi microarray designed by the Pathogen Functional Genomics Resource Center at the J. Craig Venter Institute (JCVI) was used to analyze the transcriptional changes under each condition. Gene expression values were calculated as the log2 ratio of fold change of rpoE mutant to WT (Table S2), and based on positive or negative value of the ratio, the genes affected by σE are described as “activated (up-regulated)” vs. “repressed (down-regulated),” which does not necessarily imply a direct regulatory activity. In LB log phase, a total of 1334 genes were differentially expressed (p ≤ 0.05 and fold change = 1.5); in the LPM 4 h condition, 1295 genes were differentially expressed; in the LPM 20 h condition, 911 genes were differentially expressed. In total, 58% (2533 genes) of all 4355 Salmonella genes measured by the microarray were differentially expressed in at least one of the three conditions (i.e., their expression was σE-dependent) (Figure 1A and Table S2). In each of the three conditions, the number of genes that σE activated and repressed was very similar, especially within the LPM 4 h and 20 h conditions (Figures 1B,C). Sigma factors are generally associated with positive regulation of gene expression, but because microarrays cannot discriminate between direct and indirect regulation, we expected and observed both positive and negative regulation. There were 81 genes, belonging to miscellaneous functional categories, which exhibited the same regulatory trend across all three conditions. Sixteen of these genes were up-regulated by σE in all 3 in vitro conditions, while 65 were down-regulated (Figures 1B,C and Table S3). To explain why σE has such a widespread effect on gene expression, we investigated the influence of σE on other Salmonella regulators. Approximately 40% of the known/predicted regulators (168 out of 411) were transcriptionally regulated by σE in at least one condition (Table S4). This group included most of the global regulators and many were differentially affected under the different growth conditions including Crp, Fis, ArcA, SlyA, Fur, and H-NS (Figures 1D–F). The regulation of these regulators by σE either directly or indirectly contributes to the complex regulatory network at different stages of Salmonella infection.
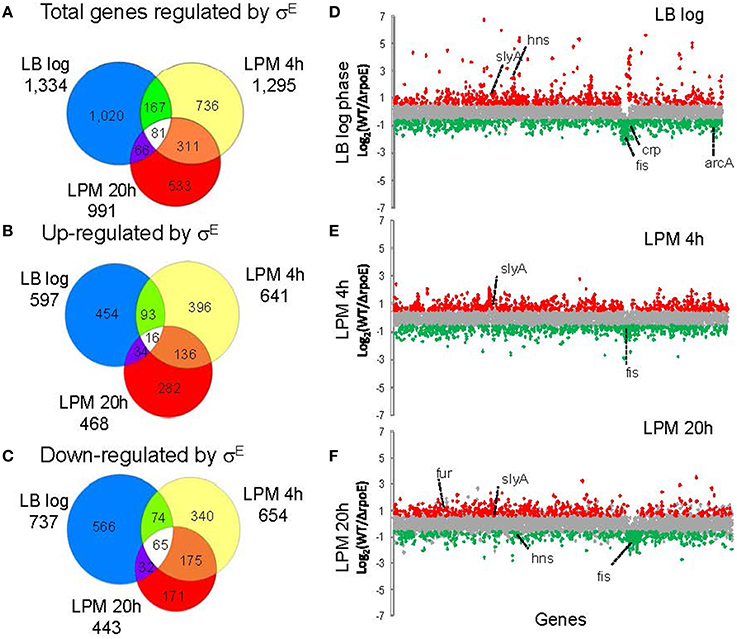
Figure 1. Overview of gene expression regulated by σE in Salmonella Typhimurium cultured under three growth conditions. Salmonella WT and rpoE-deletion strains were grown in LB medium to log phase or in acidic minimal medium (LPM) for 4 h or 20 h in triplicate. Total RNA was isolated and analyzed by the Salmonella Typhimurium/Typhi microarray (version 8). The Venn diagrams show overlaps of total genes regulated by σE (A), genes up-regulated by σE (B) and genes down-regulated by σE (C) in the three growth conditions. The charts show differential gene transcription regulated by σE in LB log phase (D), LPM 4 h condition (E) and LPM 20 h condition (F). Each dot represents one gene of the Salmonella 14028s genome with the x-axis showing gene order in relation to the gene location on chromosome, and the y-axis showing log2-based fold changes of transcript of WT vs. rpoE-deletion strains. Genes activated by σE are red, while genes repressed by σE are green. Major global regulators regulated by σE are labeled.
To further validate the microarray results, we conducted qRT-PCR on 13 genes that the microarrays identified as up- or down-regulated; in each case, qRT-PCR confirmed the microarray results (Figure 2). Moreover, out of the 62 σE-dependent Salmonella genes previously identified by Skovierova et al., 44 (71%) of them were in agreement with our findings, including rseA, rpoH, fusA, htrA, recB, eno, tolA, apl, yggT (Skovierova et al., 2006).
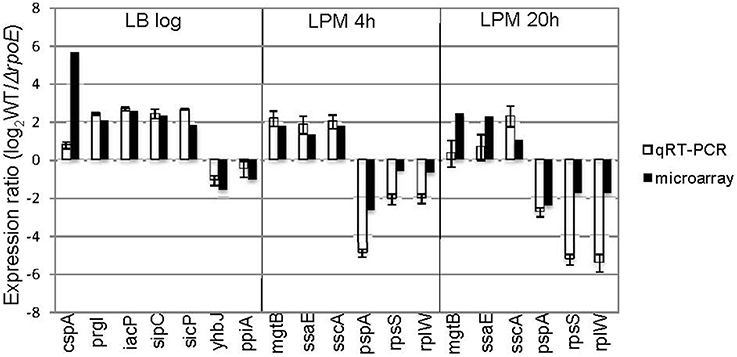
Figure 2. Validation of microarray results by qRT-PCR. Groups of genes both up- and down-regulated by σE were selected from three growth conditions (LB log, LPM 4 h, and LPM 20 h) based on microarray results, and validated by qRT-PCR using primers designed inside those genes. The results are plotted on a log2-scale comparing WT strain to rpoE-deletion strains. Values are normalized with gyrB mRNA levels and represent the average of RNA prepared from independent biological triplicates.
Functional Categories and Groups of Genes Regulated by σE
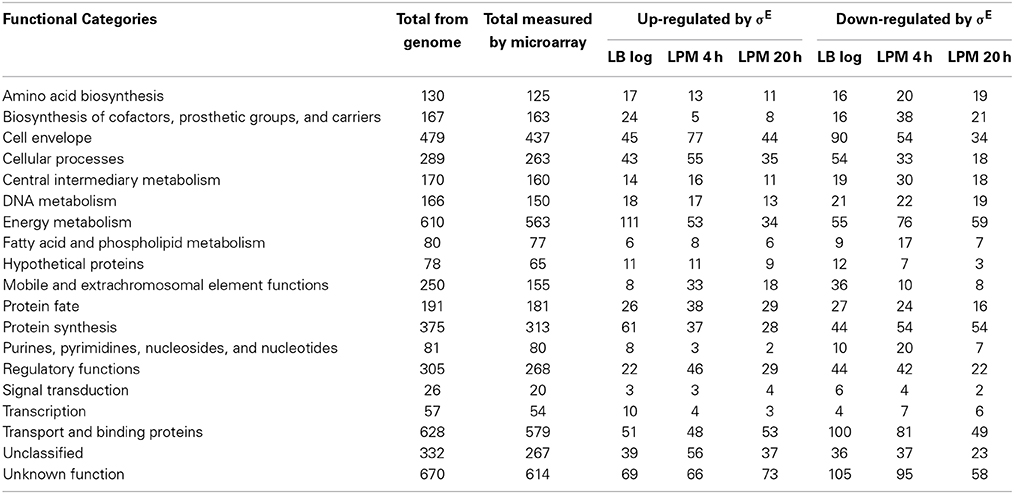
The genes regulated by σE were involved in a broad spectrum of cellular functions, and classified into 19 categories according to JCVI annotation (Table 1). Comparing the genome annotation to the genes measured by microarray, our experiments covered 92–99% of each category. For the LB log phase, when cells are actively dividing, energy metabolism and protein synthesis were the two most abundant functional categories associated with up-regulated genes, whereas transport and binding and cell envelope functional categories were the most represented among down-regulated genes. For the LPM 20 h condition, where cells were exposed to significant stress, we observed the opposite scenario. Here, genes involved in energy metabolism and protein synthesis were mostly down-regulated by σE, while genes encoding transport and binding and cell envelope proteins were up-regulated. For the LPM 4 h condition, the cell envelope category was most represented for up-regulated genes (the second most representative category of up-regulated genes at LPM 20 h), while transport and binding and energy metabolism was most represented for down-regulated genes. Across the three growth conditions, cell envelope, energy metabolism, and transport and binding functions were the top three categories of σE-regulated genes.
Notably, using the three growth conditions, we observed that σE differentially regulated four important groups of genes: (1) SPI-1; (2) SPI-2; (3) protein synthesis; and (4) stress response. SPI-1 genes were up-regulated by σE in LB log phase and in LPM 4 h condition, whereas in LPM 20 h condition, most of the SPI-1 genes were unaffected (Figure 3A). In contrast, SPI-2 genes were up-regulated by σE in LPM 4 h and 20 h conditions, but were unaffected by σE in LB log phase (Figure 3B). Our observations are consistent with previous report that σE activates the expression of most SPI-2 genes only when Salmonella are grown in minimal acidic media (Yoon et al., 2009). Genes associated with protein synthesis (e.g., translational initiation factors, protein translocation, and elongation) were up-regulated by σE in LB log phase, which contrasted with samples from the LPM 20 h condition, where they were down-regulated (Figure 3C). Consistent with previous findings that the phage shock protein (Psp) system and two-component regulatory system CpxR/CpxA compensate for the loss of σE function, we found that genes in the psp operon, including pspA, pspB, pspD, and pspE, were up-regulated in ΔrpoE under LPM 20 h condition when compared to WT (Figure 3D) (Connolly et al., 1997; Becker et al., 2005). The expression of cpxP, an indicator of the activation status of the CpxR/CpxA system, was also found to be elevated in ΔrpoE when compared to WT in LPM 4 h and 20 h samples (Figure 3D) (Kato et al., 2012). Moreover, σE up-regulated the cold-shock protein genes cspA, cspC, and cspE; the universal stress genes uspA and ynaF; the oxidative stress-response genes sodA, sodB, and sodC_2; the hyperosmotic stress genes osmC, osmE, and osmY; and the heat shock protein genes ibpA and ibpB under LB log phase condition. These results suggest that σE coordinates multiple stress response systems to achieve appropriate gene regulation.
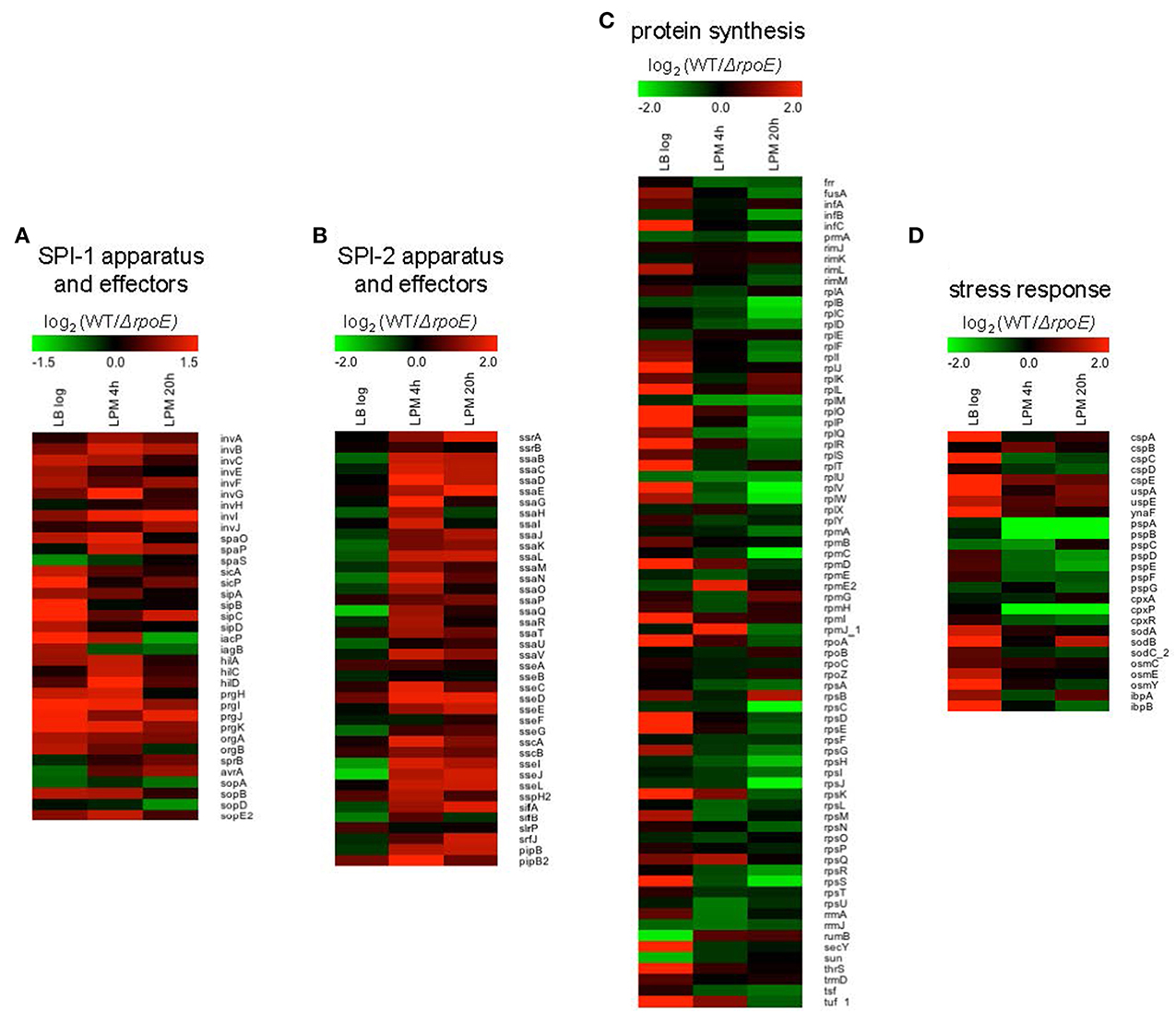
Figure 3. Heat maps of four groups of genes that are differentially regulated by σE in Salmonella grown in LB to log phase, or in LPM for 4 h or 20 h. Shown are genes involved in SPI-1 apparatus and effectors (A), SPI-2 apparatus and effectors (B), protein synthesis (C), and stress response (D). Red represents up-regulation of genes by σE while green represents down-regulation.
σE Up-Regulates SPI-2 Gene Expression by Increasing ssrB Transcription in the Condition that Mimics Early Infection
Although σE has been found to regulate SPI-2 gene expression during infection-like conditions in previous studies, the mechanism of how it accomplishes this is still not clear (Osborne and Coombes, 2009; Yoon et al., 2009). To investigate how σE manipulates SPI-2 gene expression, we selected four genes within SPI-2 in different operons (ssrB, ssaE, sscA, and ssaJ) and two genes outside of SPI-2 (sseI and pipB) that encode the effectors secreted by the SPI-2 TSSS, and then compared their transcription in WT vs. ΔrpoE strain in LB log phase, LPM 4 h and LPM 20 h conditions (Figure 4A). We observed that σE up-regulated the expression of all the above genes in the condition that mimics early infection (LPM 4 h) (Figure 4A, patterned bars). Since ssrB encodes a general activator of SPI-2 genes, we investigated whether or not σE up-regulated SPI-2 gene expression in LPM 4 h condition by increasing ssrB transcription (Coombes et al., 2007; Yoon et al., 2009). We confirmed that the presence of SsrB increased the expression of the selected SPI-2 genes studied here using qRT-PCR, by comparing the expression ratio of these SPI-2 genes in WT vs. ΔssrB strains (Figure 4B, black bars). To eliminate the effects of SsrB on σE–mediated SPI-2 gene expression, we constructed an rpoE/ssrB double mutant strain and compared ssaE, sscA, sseI, ssaJ, and pipB expression in ΔssrB vs. (ΔrpoE, ΔssrB). The expression ratios of the above 5 genes showed no significant difference between these two strains (Figure 4B, white bars), which means deletion of ssrB abolished the effects of σE on SPI-2 genes in LPM 4 h condition. Altogether, our results suggest that σE up-regulates ssrB transcription which results in increased SPI-2 gene expression in the condition that mimics early infection.
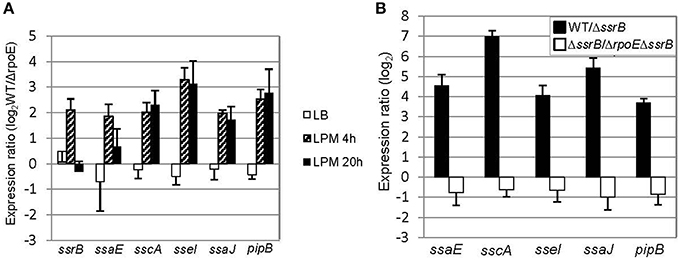
Figure 4. σE up-regulates SPI-2 gene expression by increasing ssrB transcription under LPM 4 h condition. (A) σE regulates SPI-2 gene expression differently in three growth conditions. The WT and ΔrpoE strains were grown in LB to log phase, or in LPM for 4 h or 20 h. SPI-2 gene expression was measured by qRT-PCR using gyrB as an internal control. Shown are expression ratios comparing SPI-2 gene level in WT vs. ΔrpoE strains prepared from independent biological triplicates. (B) The effects of SsrB and σE on SPI-2 gene expression in LPM 4 h condition. The WT, ΔssrB, and (ΔssrB, ΔrpoE) strains were grown in LPM for 4 h. The transcription of SPI-2 genes was measured by qRT-PCR. Expression ratio comparing WT to ΔssrB indicates that SsrB activates SPI-2 gene expression; Expression ratio comparing ΔssrB to (ΔssrB, ΔrpoE) indicates that the up-regulating effects of σE on SPI-2 gene expression are through SsrB in LPM 4 h condition.
σE Up-Regulates SPI-2 Gene Expression by Repressing hns Transcription in the Condition that Mimics Late Infection
In the condition that mimics late infection (LPM 20 h), the expression of ssrB was unaffected by the presence of σE (Figure 4A), suggesting that the general activator SsrB was not causing the up-regulation of SPI-2 genes at this time point. We next considered whether a change to the transcription of general negative regulators might indirectly induce the expression of SPI-2 genes. We focused on the nucleoid-associated protein H-NS because it has been shown to be a general repressor of SPI-2 genes (Navarre et al., 2006). Under LPM 20 h condition, microarray data showed that σE repressed the expression of hns (Table S2), which was further confirmed with qRT-PCR (Figure 5A). Although other nucleoid-associated proteins, such as YdgT, Hha, and StpA, also recognize and selectively silence the expression of foreign DNA, none of them were down-regulated by σE (Table S2), and therefore were not further investigated. Western blot analysis indicated that σE–mediated hns down-regulation was also reflected at the protein level in LPM 20 h condition (Figure 5B). The effects of H-NS on SPI-2 gene expression in LPM 20 h condition was examined by over-producing H-NS instead of deleting hns, as it is required for viability in Salmonella. Construction of a Δhns strain requires deletion of an extra gene (rpoS or phoP), which would complicate the analysis (Navarre et al., 2006). The WT strain was transformed with a plasmid containing a FLAG-tagged H-NS under a tetracycline-regulated promoter. As expected, H-NS was strongly induced by the addition of anhydrotetracycline (AHT) as visualized by western blot using anti-FLAG antibody (Figure 5C), and was further verified using H-NS specific rabbit polyclonal antibodies (data not shown). We measured the expression ratios of SPI-2 genes when H-NS was induced vs. non-induced and found that induction of H-NS resulted in 2–16 fold decrease of SPI-2 gene expression (Figure 5D). Therefore, under LPM 20 h condition, H-NS functions to repress SPI-2 gene expression. Since σE represses hns transcription under the same condition, collectively our results suggest that under the condition that mimics late infection, σE up-regulates SPI-2 expression by repressing hns transcription.
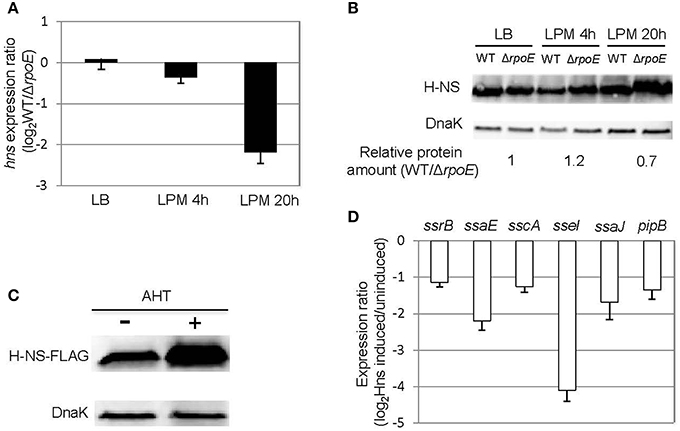
Figure 5. σE up-regulates SPI-2 gene expression by repressing hns transcription under LPM 20 h condition. (A) σE down-regulates hns expression in LPM 20 h condition. The WT and ΔrpoE strains were grown in LB to log phase, or in LPM for 4 h or 20 h in biological triplicates. The transcription of hns was measured by qRT-PCR using gyrB as an internal control. The expression ratio compares the level of hns in WT vs. ΔrpoE in each condition. (B) The effects of σE on H-NS expression in LB log phase, LPM 4 h, and LPM 20 h conditions. Western blots of H-NS in protein lysates from WT and ΔrpoE strains were generated using affinity purified rabbit anti-H-NS antibody. DnaK was used as loading control. For quantification, H-NS level in each strain under each condition was normalized to DnaK level, then relative protein amount (WT/ΔrpoE) for each condition was calculated. The ratios of protein amount are shown at the bottom. (C) Overexpression of H-NS through AHT induction. Salmonella 14028s strain was transformed with a plasmid containing FLAG-tagged H-NS (pASK-H-NS-3xFLAG) under AHT-inducible promoter. The bacteria were grown in LPM for 20 h in the presence or absence of AHT. The level of H-NS was detected by Western blot using monoclonal antibody to FLAG. (D) Overexpression of H-NS represses SPI-2 gene expression. The WT strain containing pASK-H-NS-3xFLAG plasmid was grown in LPM for 20 h with or without AHT induction. SPI-2 gene expression was measured by qRT-PCR. The results are presented as expression ratio comparing the strain in which H-NS is induced vs. un-induced on a logarithmic scale.
σE Counters the Silencing of H-NS on SPI-2 Gene Expression
Several regulators (e.g., σS, SsrB and SlyA) have been shown to be capable of relieving H-NS silencing of transcription (Mujacic and Baneyx, 2006; Perez et al., 2008; Walthers et al., 2011; Galagan et al., 2013). To investigate if σE can counter the H-NS silencing of SPI-2 gene expression, we transformed both WT and ΔrpoE strains with the AHT-inducible plasmid (pASK-H-NS-3xFLAG), which allowed us to change the expression of both hns and rpoE independently. The expression of SPI-2 apparatus (ssaE, sscA) and effector (sseI) genes were measured using qRT-PCR with samples generated from LPM 20 h conditions with and without AHT induction (Figure 6). Whether hns was induced or not, the expression of ssaE, sscA, and sseI were higher in WT than in ΔrpoE strain (comparing “WT hns uninduced” to “ΔrpoE hns uninduced,” or “WT hns induced” to “ΔrpoE hns induced”), which is consistent with results in Figure 4A. The overexpression of H-NS repressed SPI-2 gene expression (comparing “ΔrpoE hns uninduced” to “ΔrpoE hns induced”), however when σE was present, the repression of H-NS on SPI-2 was relieved (compare “WT hns induced” to “ΔrpoE hns induced”). These results suggest that σE is capable of countering the silencing of H-NS on SPI-2 genes.
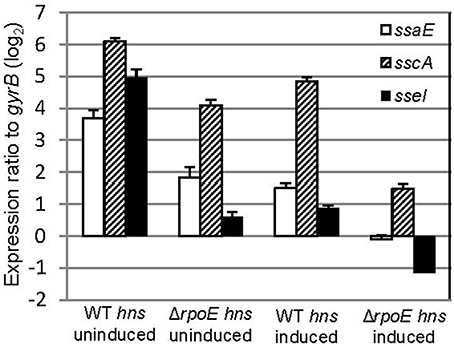
Figure 6. σE counters the silencing of H-NS on SPI-2 gene expression. The WT and ΔrpoE strain were transformed with plasmid pASK-H-NS-3xFLAG, and grown in LPM for 20 h in the presence or absence of AHT. SPI2 gene (ssaE, sscA, and sseI) expression were measured by qRT-PCR in each condition (triplicate biological samples) and standardized to gyrB in a logarithmic scale.
σE Regulates ssrB and hns Expression Indirectly
σE activates transcription by recognizing a canonical binding motif at the promoter region (Skovierova et al., 2006; Osterberg et al., 2011). To investigate if ssrB and hns are directly regulated by σE, we searched the -35 and -10 elements of ssrB and hns but failed to identify σE–dependent promoter (data not shown). Since not all of the σE-binding sites in Salmonella contain the specific motif (in vivo ChIP-seq, unpublished data), which is also true for other regulators (Galagan et al., 2013), we performed a chromatin immunoprecipitation combined with quantitative PCR assay (ChIP-qPCR) that compared the enrichment of target regions in pulldowns using anti-σE or control (anti-GFP) antibody (Figure 7). Compared to rpoE promoter 3, which is a known binding site of σE, primers designed around the promoter region of ssrB and hns yielded no significant enrichment. Another general regulator of SPI-2 gene, SlyA, was also studied, and similarly did not exhibit elevated binding to σE than control. Our results suggest that there is no in vivo occupancy of ssrB and hns promoters by σE, therefore, it is likely that σE indirectly regulates ssrB and hns, which up-regulates SPI-2 gene expression.
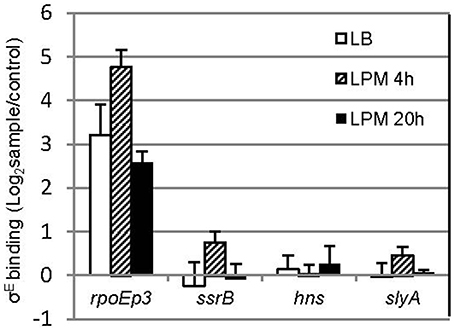
Figure 7. σE does not bind to the promoter of ssrB, hns, and slyA in vivo. Salmonellla 14028s was grown in LB to log phase, or in LPM for 4 h or 20 h, crosslinked, sonicated, and immunoprecipitated with affinity purified rabbit anti-RpoE antibody (sample) or rabbit monoclonal antibody to GFP (control) to pull down σE interactors, after removal of proteins and RNAs, purified DNA was used as template for quantitative PCR with primers designed around the promoter region of each gene. The promoter 3 region of rpoE (rpoEp3) was used as a positive control. Shown are the binding ratios comparing σE-specific vs. non-specific binding displayed in a logarithmic scale. The mean and S.D. values were obtained from independent biological triplicates.
Discussion
As an alternative sigma factor, σE functions to coordinate gene expression in response to extracytoplasmic stresses. It recognizes a “canonical sequence” at the promoter region of target genes and initiates transcription with core RNA polymerase (Skovierova et al., 2006; Osterberg et al., 2011). Compared to the genes reported to be regulated by σE in Salmonella, our study expanded the σE regulon enormously, partially because multiple growth conditions that mimic different stages of Salmonella infection were exploited (Skovierova et al., 2006; Yoon et al., 2009). A large number of genes regulated by σE are involved in various biological processes including cell envelope biosynthesis and degradation, energy metabolism, protein synthesis, transport and binding, as well as functions that are not known. Since the activation of SPI-2 genes is essential for Salmonella intracellular survival and our current and previous studies indicated that σE up-regulates SPI-2 gene expression under infection-like conditions, we further investigated how σE manipulates SPI-2 gene expression (Yoon et al., 2009). We found that σE indirectly regulates ssrB and hns transcription, also counters the silencing of H-NS on SPI-2 genes. Together with more than a 100 other Salmonella regulators transcriptionally regulated by σE, it is likely the strong regulatory effects of σE may account for the extremely attenuated phenotype exhibited in rpoE null mutant.
Many regulators of Salmonella act on SPI-2 by regulating the two-component regulator SsrB and the MarR-type regulator, SlyA (Yoon et al., 2009). It is likely the effects of σE on SPI-2 transcription is mediated by SsrB because overexpression of SsrB, but not SlyA, can complement the decrease of SPI-2 expression as a result of rpoE deletion (Yoon et al., 2009). Therefore, in this study, we focused on the coordinated function of ssrB and σE on SPI-2 regulation without examining the involvement of SlyA (Figure 4A). We found that σE up-regulated SPI-2 gene expression through ssrB in early stages of infection, and through hns during late stages of infection. It is not known why Salmonella utilizes different mechanisms to meet its need in activating SPI-2 genes, yet in late stages of infection overall protein synthesis was down-regulated by σE (Figure 3C), consistent with its effect on hns. Moreover, neither ssrB nor hns was regulated by σE directly (Figure 7), on the contrary, both ssrB and hns contain σD-recognizable promoters, consistent with previous findings (Kroger et al., 2012). Since σD has been shown to be directly regulated by σE, we speculate that σE might manipulate ssrB and hns transcription through a σE−σD-ssrB/hns cascade (Skovierova et al., 2006). Alternatively, the cascade could be represented as σE−σD-ompR/slyA/phoP- ssrB/hns as OmpR, SlyA, and PhoP have been shown to directly regulate ssrB and all of their transcriptional start sites are associated with σD (Lee et al., 2000; Feng et al., 2003; Bijlsma and Groisman, 2005; Okada et al., 2007; Kroger et al., 2012). Further investigations are needed to verify the above hypotheses.
This is the first report that σE is capable of countering the silence of H-NS on SPI-2 genes (Figure 6). The SPI-2 genes of Salmonella are normally bound by H-NS, which is a barrier to transiting RNA polymerase. However, this transcriptional barrier is relatively weak (~7pN) and is easily overcome in conditions that induce SPI-2 expression (Fang and Rimsky, 2008). It has been reported that SsrB counters the silencing of H-NS on SPI-2 genes by displacing H-NS bound in its polymerization mode, and subsequently activates SPI-2 transcription (Walthers et al., 2007, 2011). In contrast, SlyA does not displace H-NS from the DNA, but remodels the H-NS-DNA nucleoprotein complex to recruit RNA polymerase and promotes PhoP-mediated gene transcription (Perez et al., 2008). σS counters the silencing of H-NS on selective genes that can be transcribed by both σD and σS, since H-NS assembles nucleoprotein complexes with σD but not σS RNA polymerase holoenzyme, σS is able to escape H-NS trapping and the genes can be transcribed through σS–dependent promoters (Shin et al., 2005; Typas et al., 2007). Whether σE exploits similar mechanisms as SsrB, SlyA, or σS or uses another mechanism to relieve H-NS silencing on SPI-2 genes is under investigation.
While entering into stationary phase is known to induce σE, the actual signal that activates σE is the accumulation of misfolded outer membrane proteins within the periplasm, which occurs under a variety of conditions (Mecsas et al., 1993; Missiakas et al., 1996; Raivio and Silhavy, 1999). Here, we reported that a large amount of genes are regulated by σE in LB log phase (Figure 1), consistent with previous results (Kabir et al., 2005). Although the house-keeping sigma factor σD is considered to play the major role to maintain metabolism during exponential phase and is required for viability as expected, σE is also required for growth in both normal and stress conditions (De Las Penas et al., 1997). σE and σD recognize different binding motifs, however, both of them can activate multiple general regulators, through which the regulation effect is magnified (Hook-Barnard et al., 2006; Rhodius et al., 2006). We also found that alternative sigma factors function together to regulate stress response genes. For instance, the heat shock protein genes ibpA and ibpB are directly regulated by σH, and indirectly, by the action of σE on σH (Figure 3D) (Nonaka et al., 2006; Skovierova et al., 2006). σS is the master regulator of stress response genes, and plays a dominant role in regulating hyperosmotic stress in Salmonella (Hengge-Aronis, 2002; McMeechan et al., 2007). We observed that the osmolarity stress genes osmB, osmC, and osmY are also regulated by σE (Figure 3D) (Bang et al., 2005). The co-regulation of gene expression by sigma factors benefits the pathogen by simultaneously inducing general and specific stress responses to many environmental factors, even if that resistance is not immediately required (Battesti et al., 2011).
In addition to up-regulating gene expression, σE was found to down-regulate a large number of genes in our study (Table S2). This phenomenon has been observed before in both Salmonella and E. coli, however at a much smaller scale (Bang et al., 2005; Kabir et al., 2005). The down-regulation of gene expression may not be a direct effect of σE, rather an indirect repression through its downstream transcriptional regulators, or by binding to small non-coding RNAs (sRNAs), such as RybB and MicA, both of which have been found to function as global regulators (Papenfort et al., 2006; Gogol et al., 2011). Some of the genes repressed by MicA and RybB were also found to be down-regulated by σE in our study, such as pal and gloA (Table S2). Additionally, we found that σE regulates hfq expression in both nutrient-rich and infection-like conditions (Table S2). Hfq is a sRNA-binding protein that bridges sRNA to act on trans-encoded target mRNAs, therefore modulates the stability and translation of the target mRNAs (Vogel and Luisi, 2011). Both RybB and MicA are regulated by Hfq in repressing outer membrane protein expression. For instance, Hfq significantly enhances RybB binding to ompC and ompD mRNAs, facilitating the decay of omp mRNAs when the cell experiences extracytoplamic stress (Udekwu et al., 2005; Papenfort et al., 2006). Hfq controls the expression of at least 20% of all Salmonella genes directly or indirectly, and has a pleiotropic effect on Salmonella virulence through regulation of motility, outer membrane protein expression, invasion, and intracellular growth (Sittka et al., 2007, 2008; Ansong et al., 2009). As the list of sRNAs in Salmonella grows, these newly identified sRNAs may help to explain the global impact of σE on gene expression (Kroger et al., 2012).
By comparing the global transcription profiles of σE across three growth conditions, we established that σE coordinated the gene expression of Salmonella for proliferation and/or survival in response to various environments. The role of σE in global gene regulation and SPI-2 gene activation observed in this study explains why σE is required for systemic mouse infection, and shows that σE is the conductor of the Salmonella gene regulation orchestra, whether under stress or not.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (5R01 AI022933 and IAA Y1-AI-8401) and the National Institute of General Medical Sciences (GM094623).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00027/abstract
Table S1. List of primers used in this study.
Table S2. List of genes regulated by σE under three growth conditions.
Table S3. List of genes up- and down-regulated by σE in all three growth conditions.
Table S4. List of regulators regulated by σE in at least one of the three growth conditions.
Footnotes
1. ^Hastie, T., Tibshirani, R., Narasimhan, B., and Chu, G. Impute: Impute: Imputation for Microarray Data. R package version 1.40.40.
2. ^Bolstad, B. M. preprocessCore: A Collection of Pre-Processing Functions. R Package Version 1.28.20.
References
Alpuche Aranda, C. M., Swanson, J. A., Loomis, W. P., and Miller, S. I. (1992). Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. U.S.A. 89, 10079–10083. doi: 10.1073/pnas.89.21.10079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ansong, C., Yoon, H., Porwollik, S., Mottaz-Brewer, H., Petritis, B. O., Jaitly, N., et al. (2009). Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS ONE 4:e4809. doi: 10.1371/journal.pone.0004809
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bang, I. S., Frye, J. G., McClelland, M., Velayudhan, J., and Fang, F. C. (2005). Alternative sigma factor interactions in Salmonella: σE and σH promote antioxidant defences by enhancing σS levels. Mol. Microbiol. 56, 811–823. doi: 10.1111/j.1365-2958.2005.04580.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Battesti, A., Majdalani, N., and Gottesman, S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi: 10.1146/annurev-micro-090110-102946
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becker, L. A., Bang, I. S., Crouch, M. L., and Fang, F. C. (2005). Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol. Microbiol. 56, 1004–1016. doi: 10.1111/j.1365-2958.2005.04604.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bijlsma, J. J., and Groisman, E. A. (2005). The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57, 85–96. doi: 10.1111/j.1365-2958.2005.04668.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bolstad, B. M., Irizarry, R. A., Astrand, M., and Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. doi: 10.1093/bioinformatics/19.2.185
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cho, B. K., Knight, E. M., Barrett, C. L., and Palsson, B. O. (2008). Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910. doi: 10.1101/gr.070276.107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Connolly, L., De Las Penas, A., Alba, B. M., and Gross, C. A. (1997). The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 11, 2012–2021. doi: 10.1101/gad.11.15.2012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Coombes, B. K., Lowden, M. J., Bishop, J. L., Wickham, M. E., Brown, N. F., Duong, N., et al. (2007). SseL is a Salmonella-specific translocated effector integrated into the SsrB-controlled Salmonella pathogenicity island 2 type III secretion system. Infect. Immun. 75, 574–580. doi: 10.1128/IAI.00985-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Crouch, M. L., Becker, L. A., Bang, I. S., Tanabe, H., Ouellette, A. J., and Fang, F. C. (2005). The alternative sigma factor σE is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol. Microbiol. 56, 789–799. doi: 10.1111/j.1365-2958.2005.04578.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dartigalongue, C., Missiakas, D., and Raina, S. (2001). Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 276, 20866–20875. doi: 10.1074/jbc.M100464200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Las Penas, A., Connolly, L., and Gross, C. A. (1997). σE is an essential sigma factor in Escherichia coli. J. Bacteriol. 179, 6862–6864.
Fang, F. C., and Rimsky, S. (2008). New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11, 113–120. doi: 10.1016/j.mib.2008.02.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feng, X., Oropeza, R., and Kenney, L. J. (2003). Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48, 1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Galagan, J., Lyubetskaya, A., and Gomes, A. (2013). ChIP-Seq and the complexity of bacterial transcriptional regulation. Curr. Top. Microbiol. Immunol. 363, 43–68. doi: 10.1007/82_2012_257
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garmendia, J., Beuzon, C. R., Ruiz-Albert, J., and Holden, D. W. (2003). The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149, 2385–2396. doi: 10.1099/mic.0.26397-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. doi: 10.1186/gb-2004-5-10-r80
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gogol, E. B., Rhodius, V. A., Papenfort, K., Vogel, J., and Gross, C. A. (2011). Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. U.S.A. 108, 12875–12880. doi: 10.1073/pnas.1109379108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hengge-Aronis, R. (2002). Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/MMBR.66.3.373-395.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hook-Barnard, I., Johnson, X. B., and Hinton, D. M. (2006). Escherichia coli RNA polymerase recognition of a sigma70-dependent promoter requiring a -35 DNA element and an extended -10 TGn motif. J. Bacteriol. 188, 8352–8359. doi: 10.1128/JB.00853-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humphreys, S., Rowley, G., Stevenson, A., Anjum, M. F., Woodward, M. J., Gilbert, S., et al. (2004). Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72, 4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humphreys, S., Stevenson, A., Bacon, A., Weinhardt, A. B., and Roberts, M. (1999). The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67, 1560–1568.
Jones, C. H., Danese, P. N., Pinkner, J. S., Silhavy, T. J., and Hultgren, S. J. (1997). The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16, 6394–6406. doi: 10.1093/emboj/16.21.6394
Kabir, M. S., Yamashita, D., Koyama, S., Oshima, T., Kurokawa, K., Maeda, M., et al. (2005). Cell lysis directed by σE in early stationary phase and effect of induction of the rpoE gene on global gene expression in Escherichia coli. Microbiology 151, 2721–2735. doi: 10.1099/mic.0.28004-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karlinsey, J. E., Maguire, M. E., Becker, L. A., Crouch, M. L., and Fang, F. C. (2010). The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium. Mol. Microbiol. 78, 669–685. doi: 10.1111/j.1365-2958.2010.07357.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kato, A., Hayashi, H., Nomura, W., Emori, H., Hagihara, K., and Utsumi, R. (2012). A connecter-like factor, CacA, links RssB/RpoS and the CpxR/CpxA two-component system in Salmonella. BMC Microbiol. 12:224. doi: 10.1186/1471-2180-12-224
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kenyon, W. J., Sayers, D. G., Humphreys, S., Roberts, M., and Spector, M. P. (2002). The starvation-stress response of Salmonella enterica serovar Typhimurium requires σE, but not CpxR-regulated extracytoplasmic functions. Microbiology 148, 113–122.
Kroger, C., Dillon, S. C., Cameron, A. D., Papenfort, K., Sivasankaran, S. K., Hokamp, K., et al. (2012). The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 109, E1277–E1286. doi: 10.1073/pnas.1201061109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, A. K., Detweiler, C. S., and Falkow, S. (2000). OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182, 771–781. doi: 10.1128/JB.182.3.771-781.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McMeechan, A., Roberts, M., Cogan, T. A., Jorgensen, F., Stevenson, A., Lewis, C., et al. (2007). Role of the alternative sigma factors σE and σS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 153, 263–269. doi: 10.1099/mic.0.29235-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mecsas, J., Rouviere, P. E., Erickson, J. W., Donohue, T. J., and Gross, C. A. (1993). The activity of σE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628. doi: 10.1101/gad.7.12b.2618
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Missiakas, D., Betton, J. M., and Raina, S. (1996). New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21, 871–884. doi: 10.1046/j.1365-2958.1996.561412.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miticka, H., Rowley, G., Rezuchova, B., Homerova, D., Humphreys, S., Farn, J., et al. (2003). Transcriptional analysis of the rpoE gene encoding extracytoplasmic stress response sigma factor σE in Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 226, 307–314. doi: 10.1016/S0378-1097(03)00600-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mujacic, M., and Baneyx, F. (2006). Regulation of Escherichia coli hchA, a stress-inducible gene encoding molecular chaperone Hsp31. Mol. Microbiol. 60, 1576–1589. doi: 10.1111/j.1365-2958.2006.05207.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muller, C., Bang, I. S., Velayudhan, J., Karlinsey, J., Papenfort, K., Vogel, J., et al. (2009). Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 71, 1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niemann, G. S., Brown, R. N., Gustin, J. K., Stufkens, A., Shaikh-Kidwai, A. S., Li, J., et al. (2011). Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43. doi: 10.1128/IAI.00771-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nonaka, G., Blankschien, M., Herman, C., Gross, C. A., and Rhodius, V. A. (2006). Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20, 1776–1789. doi: 10.1101/gad.1428206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okada, N., Oi, Y., Takeda-Shitaka, M., Kanou, K., Umeyama, H., Haneda, T., et al. (2007). Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology 153, 548–560. doi: 10.1099/mic.0.29259-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osborne, S. E., and Coombes, B. K. (2009). RpoE fine tunes expression of a subset of SsrB-regulated virulence factors in Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:45. doi: 10.1186/1471-2180-9-45
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Osterberg, S., Del Peso-Santos, T., and Shingler, V. (2011). Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65, 37–55. doi: 10.1146/annurev.micro.112408.134219
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Papenfort, K., Pfeiffer, V., Mika, F., Lucchini, S., Hinton, J. C., and Vogel, J. (2006). σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perez, J. C., Latifi, T., and Groisman, E. A. (2008). Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J. Biol. Chem. 283, 10773–10783. doi: 10.1074/jbc.M709843200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect. Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raivio, T. L., and Silhavy, T. J. (1999). The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2, 159–165. doi: 10.1016/S1369-5274(99)80028-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rezuchova, B., Miticka, H., Homerova, D., Roberts, M., and Kormanec, J. (2003). New members of the Escherichia coli σE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225, 1–7. doi: 10.1016/S0378-1097(03)00480-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rhodius, V. A., Suh, W. C., Nonaka, G., West, J., and Gross, C. A. (2006). Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. doi: 10.1371/journal.pbio.0040002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rowley, G., Spector, M., Kormanec, J., and Roberts, M. (2006). Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4, 383–394. doi: 10.1038/nrmicro1394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Liang, W., Sharov, V., Howe, E. A., et al. (2006). TM4 microarray software suite. Meth. Enzymol. 411, 134–193. doi: 10.1016/S0076-6879(06)11009-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378.
Sandermann, H. Jr., and Strominger, J. L. (1972). Purification and properties of C 55 -isoprenoid alcohol phosphokinase from Staphylococcus aureus. J. Biol. Chem. 247, 5123–5131.
Shin, M., Song, M., Rhee, J. H., Hong, Y., Kim, Y. J., Seok, Y. J., et al. (2005). DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 19, 2388–2398. doi: 10.1101/gad.1316305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sittka, A., Lucchini, S., Papenfort, K., Sharma, C. M., Rolle, K., Binnewies, T. T., et al. (2008). Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. doi: 10.1371/journal.pgen.1000163
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sittka, A., Pfeiffer, V., Tedin, K., and Vogel, J. (2007). The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63, 193–217. doi: 10.1111/j.1365-2958.2006.05489.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Skovierova, H., Rowley, G., Rezuchova, B., Homerova, D., Lewis, C., Roberts, M., et al. (2006). Identification of the σE regulon of Salmonella enterica serovar Typhimurium. Microbiology 152, 1347–1359. doi: 10.1099/mic.0.28744-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. doi: 10.2202/1544-6115.1027
Smyth, G. K. (2005). “Limma: linear models for microarray data,” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, eds R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (New York, NY: Springer), 397–420.
Typas, A., Becker, G., and Hengge, R. (2007). The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 63, 1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Udekwu, K. I., Darfeuille, F., Vogel, J., Reimegard, J., Holmqvist, E., and Wagner, E. G. (2005). Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19, 2355–2366. doi: 10.1101/gad.354405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walthers, D., Carroll, R. K., Navarre, W. W., Libby, S. J., Fang, F. C., and Kenney, L. J. (2007). The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol. Microbiol. 65, 477–493. doi: 10.1111/j.1365-2958.2007.05800.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walthers, D., Li, Y., Liu, Y., Anand, G., Yan, J., and Kenney, L. J. (2011). Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 286, 1895–1902. doi: 10.1074/jbc.M110.164962
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yoon, H., McDermott, J. E., Porwollik, S., McClelland, M., and Heffron, F. (2009). Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306. doi: 10.1371/journal.ppat.1000306
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Salmonella, RpoE, microarray, SPI-2, H-NS, regulation, ChIP-seq
Citation: Li J, Overall CC, Nakayasu ES, Kidwai AS, Jones MB, Johnson RC, Nguyen NT, McDermott JE, Ansong C, Heffron F, Cambronne ED and Adkins JN (2015) Analysis of the Salmonella regulatory network suggests involvement of SsrB and H-NS in σE-regulated SPI-2 gene expression. Front. Microbiol. 6:27. doi: 10.3389/fmicb.2015.00027
Received: 11 November 2014; Paper pending published: 25 November 2014;
Accepted: 08 January 2015; Published online: 10 February 2015.
Edited by:
Beiyan Nan, University of California, Berkeley, USAReviewed by:
Martin Wiedmann, Cornell University, USAQiaobin Xiao, University of Notre Dame, USA
Jinlei Zhao, University of Pennsylvania, USA
Copyright © 2015 Li, Overall, Nakayasu, Kidwai, Jones, Johnson, Nguyen, McDermott, Ansong, Heffron, Cambronne and Adkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joshua N. Adkins, Biological Sciences Division, Pacific Northwest National Laboratory, 902 Battelle Boulevard, PO Box 999, MSIN K8-98, Richland, WA 99352, USA e-mail: joshua.adkins@pnnl.gov
†Present address: Ernesto S. Nakayasu, Bindley Bioscience Center, Purdue University, West Lafayette, IN, USA
‡These authors have contributed equally to this work.
 Jie Li
Jie Li Christopher C. Overall
Christopher C. Overall Ernesto S. Nakayasu
Ernesto S. Nakayasu Afshan S. Kidwai1
Afshan S. Kidwai1 Nhu T. Nguyen
Nhu T. Nguyen Jason E. McDermott
Jason E. McDermott Eric D. Cambronne
Eric D. Cambronne Joshua N. Adkins
Joshua N. Adkins