- Department of Biological Sciences, Oakland University, Rochester, MI, USA
We have developed DNA-based genetic markers for rapid cycling Brassica rapa (RCBr), also known as Fast Plants. Although markers for B. rapa already exist, ours were intentionally designed for use in a teaching laboratory environment. The qualities we selected for were robust amplification in PCR, polymorphism in RCBr strains, and alleles that can be easily resolved in simple agarose slab gels. We have developed two single nucleotide polymorphism (SNP) based markers and 14 variable number tandem repeat (VNTR)-type markers spread over four chromosomes. The DNA sequences of these markers represent variation in a wide range of genomic features. Among the VNTR-type markers, there are examples of variation in a non-genic region, variation within an intron, and variation in the coding sequence of a gene. Among the SNP-based markers there are examples of polymorphism in intronic DNA and synonymous substitution in a coding sequence. Thus these markers can serve laboratory exercises in both transmission genetics and molecular biology.
Introduction
Rapid cycling Brassica rapa (RCBr), also known as Fast Plants, are a widely used model organism in biology education. They were developed by selection of B. rapa for the traits of short time to flowering, rapid seed maturation, lack of seed dormancy, petite growth habit, and high female fertility (Williams and Hill, 1986). The result is a plant with a 7-week generation time that can be cultivated inexpensively by novices. Their fast growth occurs at room temperature under continuous illumination by household fluorescent lights with simple and inexpensive growing materials (Williams, 1997). In addition to the plant strains, the Wisconsin Fast Plants Program1 has developed a large assortment of educational activities and support materials. Topics covered by RCBr activities include the effect of environment on plant growth, plant-herbivore interactions, hormones and growth, and genetics (Musgrave, 2000). Seed stocks and instructional kits are available from Carolina Biological Supply (Burlington, NC, USA). Seeds are also available from the Crucifer Genetics Cooperative (Williams, 1985).
Rapid cycling B. rapa is an excellent organism for teaching genetics. Cross-pollination is easy for students at all levels because like other Brassica, and unlike Arabidopsis, they are self-incompatible for pollination. Lessons in Mendelian inheritance are performed using RCBr stocks that vary in easily scored phenotypes such as stem color (purple versus non-purple) and leaf color (green versus yellow-green; Williams, 1985). There are also traits with complex inheritance such as trichome density which shows additive polygenic inheritance (Lauffer and Fall, 2000) and intensity of anthocyanin pigmentation which is both polygenic and affected by environment (Goldman, 1999). Some molecular genetic markers exist, but have been slower to develop (Wendell and Pickard, 2007).
Although there is a large set of DNA markers for B. rapa in the form of microsatellites and single nucleotide polymorphisms (SNP), they do not lend themselves well to the teaching laboratory where simple agarose slab gels are most common, time and budgets are limited, and the students using them are novices. Microsatellites are highly desirable genetic markers because they tend to have multiple alleles and thus be highly informative (Litt and Luty, 1989; Weber and May, 1989). An extensive list of microsatellite markers for Brassica developed by several groups can be found at the Microsatellite Information Exchange2, and microsatellite markers that have been developed for Brassica crop species are usable and polymorphic in RCBr (Burdzinski and Wendell, 2007; Iniguez-Luy et al., 2009). Instructional use is difficult because the size difference between alleles is usually in the range of 2–20 base pairs which is best resolved in polyacrylamide gels. We have previously reported a set of selected microsatellites and protocols to make them work in a teaching laboratory environment using polyacrylamide mini gels (Wendell and Pickard, 2007). However, the need for polyacrylamide gels still creates a barrier to their use by instructors of undergraduate or advanced high school laboratories who either may not have the needed equipment in a teaching laboratory, or do not wish to work with polyacrylamide. Another type of DNA marker available for B. rapa are SNP (Park et al., 2009). SNPs have grown in significance as genetic markers because they are present at a high density in genomes and SNP genotype data can be collected using automated high throughput methods such as microarrays. Although a single SNP is not as informative as a single polymorphic microsatellite, due to SNPs generally having only two alleles, a string of SNPs can be just as informative as a single microsatellite with multiple alleles (Kruglyak, 1997). However, the methods used to routinely analyze SNPs such as microarrays or automated DNA sequencing cannot be expected to be readily available in an instructional laboratory. Even simpler methods such as TaqMan assays still require more sophisticated equipment (for real time PCR) than most instructional laboratories have on hand.
In order to allow the use of DNA makers with RCBr under the conditions where these plants are most commonly used, in an undergraduate or advanced high school teaching lab, we have developed genetic markers specifically suited to use under simple conditions. The markers we report here have been selected for robust and reliable amplification by PCR, polymorphism in RCBr populations, and alleles that can be readily resolved in small conventional agarose slab gels. For repetitive DNA-based markers we have sought those with longer repeated element like variable number tandem repeats (VNTR) markers, rather than microsatellites, so that the size difference between alleles would be larger. For detection of SNPs, we have identified those that are reliably detected by the technique of PCR-RFLP (Konieczny and Ausubel, 1993).
Materials and methods
Plant Strains
A variety of RCBr strains were used which vary in both Mendelian traits and DNA markers. One of the Mendelian loci is anthocyaninless which has the recessive anl allele for lack of anthocyanin pigment (non-purple stem) and the dominant wild type ANL allele that allows anthocyanin production resulting in purple stems (Williams, 2007). The other Mendelian locus used is yellow-green which has the recessive ygr allele for yellow-green color and the dominant wild type YGR allele for normal green color (Williams, 2007). The Wisconsin Fast Plants strains Standard B. rapa; Purple Stem, Hairy; Non-Purple Stem, Hairless; and Non-Purple Stem, Yellow-Green Leaf were obtained from Carolina Biological Supply Company (Burlington, NC, USA). Strain DWRCBr70 is derived from a purple-stemmed RCBr population by selection for high intensity of purple color. DWRCBr52 was derived from a non-purple stem population by selection for low trichome density. DWRCBr60 was derived from a populations of plants with purple stems and yellow-green leaves.
DNA Purification
DNA was purified from leaf tissue using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) following manufacturer’s instructions with the exception that the tissue was disrupted using a ground glass homogenizer.
PCR
PCR was performed in a 10-μl reaction volume with 50 ng of template DNA and 10 pmol of each primer using either Accuprime Taq DNA polymerase and supplied Buffer I (Invitrogen, Carlsbad, CA, USA) or Syzygy Taq polymerase (Syzygy Biotech, Grand Rapids, MI, USA). The PCR program was an initial incubation at 94°C for 2 min, followed by 25 cycles of 94°C for 30 s, 61°C for 60 s, and 72°C for one 60 s, and a final incubation at 72°C for 4 min.
DNA Sequencing
PCR amplicons were purified for DNA sequencing using a MinElute PCR Cleanup Kit (Qiagen Inc., Valencia, CA, USA) and their purity verified by analytical electrophoresis in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Sequencing reactions were performed using ABI BigDye Terminator v3.1 Cycle Sequencing Kit and analyzed using the Applied Biosystems ABI Prism 3730 DNA Analyzer at the Wayne State University Applied Genomics Technology Center3.
Electrophoresis
PCR products were separated by electrophoresis in 1.2% agarose (Genetic Analysis Grade from Fisher Scientific, Waltham, MA, USA) in 7 cm long slab gels at 150 V for 30–50 min. Bands were detected by ethidium bromide staining.
Identification of Candidate VNTR
In order to identify sequences with potential VNTR polymorphism, bacterial artificial chromosome (BAC) sequences obtained from the B. rapa Genome Project4 were searched on chromosomes 1, 2, 3, and 9. (This work was performed prior to the release of the complete B. rapa genome sequence.) The entire DNA sequence of each BAC was analyzed using the Emboss program eTandem5. eTandem generates a score based on the nature of the putative repeat; for a perfect repeat, the score is equal to the length of the entire repetitive sequence minus the first repeat. From the search results, only those potential VNTR’s with a score greater than 20 and sequences with three or more repeats of 6–100 base pairs were selected, with preference to the longest repeats that were available and/or the highest number of repetitions. The rationale for this choice was that a larger repeat element would produce a larger size difference between alleles and more repeats would increase the probability of polymorphism. Finally only those results with a percent consensus among repeats of 80% or greater were chosen for further analysis.
PCR primers were designed to prime from the sequences flanking the candidate VNTR using Primer-BLAST6. Only primer pairs that were expected to amplify a PCR product size ranging from 200 to approximately 1000 bp were accepted.
Primers designed to amplify potential VNTR markers were tested for suitability by a series of criteria. First they were tested for the ability to robustly and reproducibly amplify a product, i.e., one could always detect a “bright” band on an ethidium bromide stained agarose gel. Those that passed the first test were used to screen for potential polymorphism in a sample of 12 random plants of the strain Standard B. rapa as well as the strains DWRCBr52, DWRCBr60, and DWRCBr70. When more than one band size was detected, the products were tested for evidence that they segregated as alleles of the same locus by genotyping individuals from an F2 generation previously produced by crossing DWRCBr52 and DWRCBr70 strains (Burdzinski and Wendell, 2007). Any primer pair that amplified a product from more than one locus was discarded.
Identification of SNPs in RCBr
To identify SNPs in RCBr, we resequenced sequence-tagged sites (STS) chosen from those reported by Park et al. (2009). For each STS tested, PCR primers were designed using the program Primer-BLAST (see text footnote 6). PCR was performed on three individuals of each strain tested and the amplicons were pooled to provide template for sequencing. Such pools were generated for each of the strains DWRBr52, DWRBr60, and DWRBr70. The resulting sequence data was then aligned using ClustalW27 to identify SNPs between strains.
Development of PCR-RFLP Markers from RCBr SNPs
Single nucleotide polymorphisms were used to develop PCR-RFLP markers using a hierarchical approach. First, the nucleotide sequence surrounding each SNP was screened using NEBcutter V2.08 to identify those SNPs that resided within restriction endonuclease recognition sequences. Next, PCR primers were designed so that the position of the SNP, if cut by the enzyme, would produce restriction fragment lengths on a gel that could be easily resolved from each other and from the uncut band if present.
Genomic Sequence Data
Information on gene sequences and Arabidopsis homologs connected to the markers developed was obtained through the Brassica database BRAD (Cheng et al., 2011).
Genetic Mapping
Markers expected to be on chromosome A09 were genetically mapped relative to the anthocyaninless (ANL) locus in 81 testcross progeny generated by crossing DWRCBr70 (ANL/ANL) with DWRCBr52 (anl/anl) and backcrossing to DWRCBr52. The order of all DNA markers was determined by their position in the B. rapa genome sequence available from BRAD (Cheng et al., 2011) and map distances in Kosambi centimorgans were calculated using MAPMANAGER (Manly et al., 2001). The position of the anthocyaninless locus was determined as that which gave map distances with the highest LOD scores.
Results
VNTR-Type Genetic Markers for RCBr
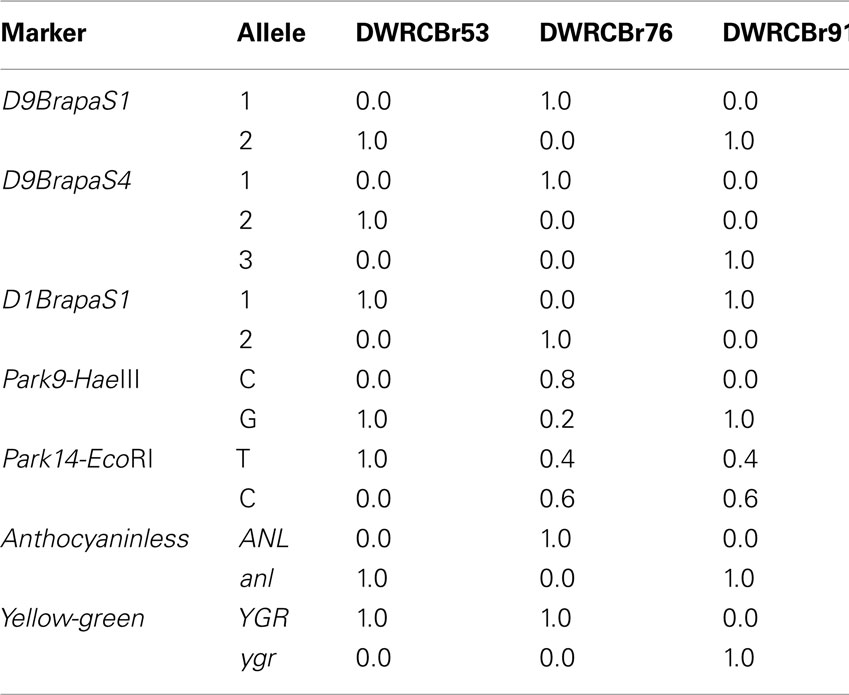
We have developed a total of 14 genetic markers that are based on a VNTR-type repetitive DNA and meet the criteria of robust and reproducible amplification, polymorphism in RCBr strains, and alleles that can be resolved on conventional agarose slab gels (Table 1). Markers are available on chromosomes A01, A02, A03, and A09. With the one exception of D9BrapaS4 which has three alleles, all markers have only two alleles in RCBr populations surveyed. From these 14 VNTR-type markers we chose three to recommend most for use in an educational setting (Table 2) because they are most reliable in producing “bright” bands of alleles that are most readily resolved in small agarose slab gels (Figure 1). We subjected these three markers to further analysis including DNA sequence of their repetitive element to determine the nature of allelic variation.
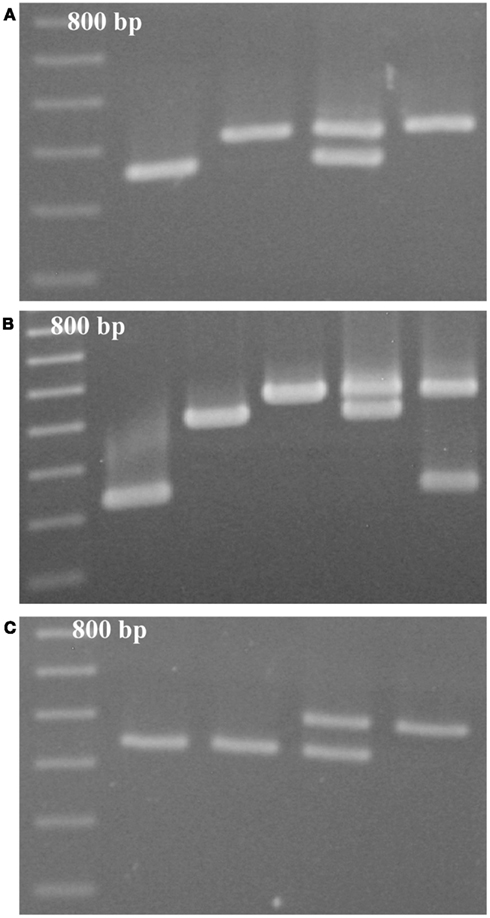
Figure 1. Resolution of alleles of VNTR-type markers (A) D9BrapaS1, (B) D9BrapaS4, and (C) D1BrapaS1 on 1.2% agarose gels. The first lane in each gel is a 100-bp DNA ladder.
D9BrapaS1 resides in a segment of DNA on chromosome A09 that does not contain any annotated genes or other genomic features. Nucleotide sequencing confirms that it contains a VNTR-sized repetitive DNA element (Table 2). However, the two alleles present in RCBr do not differ in the repetitive DNA sequences, but instead vary in a 53-bp insertion/deletion in the single-copy DNA flanking the repetitive element (Figure 2). Alignment of these alleles with the B. rapa genome sequence indicates almost 100% identity except for the 53-bp segment. This indel has been deposited in to the dbSNP9 database (ss490570267).
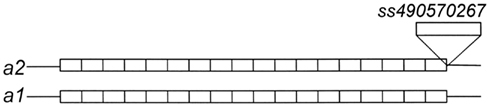
Figure 2. Size variation between the alleles of D9BrapaS1 in RCBr is due to a single insertion ss490570267.
D9BrapaS4 contains a compound repetitive DNA element (Table 2) and the variation between the three alleles we have identified is in these repetitive sequences (Figure 3). A search of the B. rapa genome indicates that it resides within the first intron of the predicted gene Bra007262. Alignment of the comparable portion of the Bra007262 sequence indicates that the largest allele of D9BrapaS4 is nearly identical to the Bra007262 sequence in the BRAD database except for a 20-bp deletion in Bra007262 in the repetitive DNA region of the marker. The indels responsible for the variation between the RCBr alleles have been deposited into the dbSNP database. ss490570286 and ss490570293 are responsible for the size difference between alleles 2 and 3, while the addition of ss490570298 produces allele 1.
Figure 3. Size variation between the alleles of D9BrapaS4 in RCBr is due to a multiple indels designated by 1-1 (ss490570298), 2-1 (ss490570286), and 2-1 (ss490570293).
D1BrapaS1 contains multiple tandem copies of a 16-bp repeat (Table 2), and the variation producing the fragment length difference between alleles is within the repetitive DNA, but the alleles do not vary from each other in numbers of whole repeats. Rather, each allele has several indels (relative to the other alleles) which are mostly smaller than 16 bp. Due to the repetitive DNA sequence, multiple sequence alignments are possible and we cannot presently identify the exact position of the indels.
A search of the B. rapa genome finds that the repetitive DNA element at the core of D1BrapaS1 is in the second exon of the predicted gene Bra011448 and within its predicted open reading frame. Bra011448 is a predicted gene based on similarity to Arabidopsis thaliana gene AT4G33500. Both of these homologous genes are predicted to encode proteins with a protein phosphatase 2C (PP2C) domain near the C-terminus and a ribonuclease E (rne) domain in the second exon. Analysis of the predicted amino acid sequence of Bra011448 using NCBI Conserved Domain Search10 finds that the D1BrapaS1 repetitive DNA lies within the predicted ribonuclease E (rne) domain. Comparison of the nucleotide sequence of the two alleles of D1BrapaS1 with Bra011448, as well as the sequence within AC189637.2 deposited in GenBank, shows that each has a different combination of indels, but all preserve the overall reading frame (Figure 4). They are all in-frame deletions except for one case in allele 2 of D1BrapaS1 where two subsequent deletions preserve the reading frame. In contrast, there is very little variation in the section of the gene predicted to encode a PP2C domain. The nucleotide sequence of predicted exons 6, 7, 8, and 9 of Bra011448 from the RCBr stocks homozygous for either allele of D1BrapaS1 is 97% identical to the sequence in the BRAD database and there are no deletions.

Figure 4. Four different alleles of D1BrapaS1 show four different combinations of in-frame deletions in Bra011448. The sequence shown is the portion of Bra011448 that is predicted to encode an rne domain and lies within the marker D1BrapaS1. AC189637.2 and Bra011448 are Chinese cabbage sequences from public databases and D1BrapaS1a1 and D1BrapaS1a2 are from RCBr.
SNPs Detectable by PCR-RFLP
Among SNPs that we identified within RCBr populations, we found two that were readily assayable by PCR-RFLP under classroom conditions (Table 3). Both of these were detected with primers that amplified robustly in RCBr and when digested with the appropriate enzyme produced bands that were readily resolved within the same gel (Figure 5). The C/G polymorphism identified by Park9-HaeIII lies within the third exon of predicted F-box protein gene Bra026987 on chromosome A09. The substitution is a synonymous polymorphism in the third position of a serine codon. The T/C polymorphism identified by Park14-EcoRI lies within the seventh intron of Bra013780, a predicted transmembrane protein involved in defense or cell death.
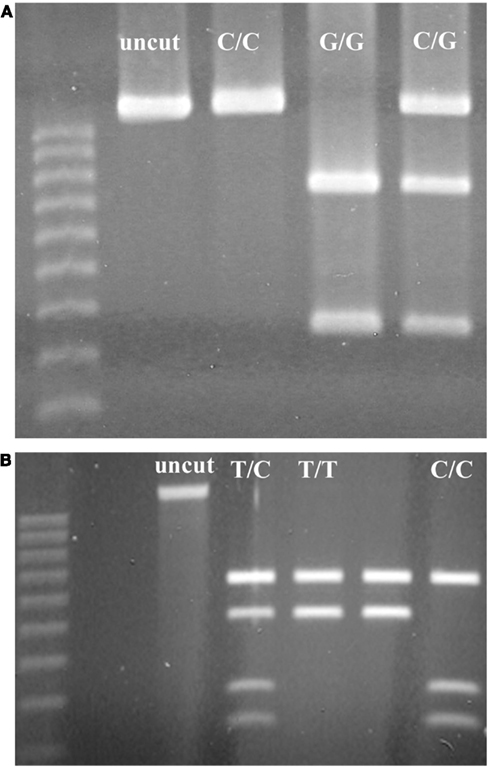
Figure 5. Resolution of alleles of PCR-RFLP markers (A) Park9-HaeIII (ss490571632) and (B) Park14-EcoRI (ss490571635) on 1.2% agarose gels.
Linkage of A09 markers to the Anthocyaninless Locus
Among the markers we have found to be most suitable for teaching laboratory use, two of the VNTR-type and one of the SNPs were expected to be on chromosome A09, the chromosome which we previously reported to hold the anthocyaninless locus (Burdzinski and Wendell, 2007). We genotyped 81 progeny of a testcross between the purple (ANL/ANL) DWRCBR70 and non-purple (anl/anl) DWRCBr53 strains and found that the anthocyaninless locus most likely resides within the 6.3-Mb interval between D9BrapaS4 and Park9-HaeIII (Figure 6).
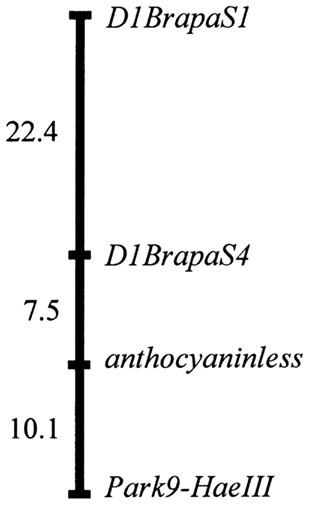
Figure 6. Genetic map of the anthocyaninless locus relative to A09 marker reported in this paper. Map distances are Kosambi centimorgans.
Marker Polymorphism in Fast Plants Strains
To assist instructors who obtain RCBr as Fast Plants seeds from Carolina Biological Supply, we have surveyed the allele frequencies of the markers in four popular Fast Plants strains (Table 4). For each strain, we determined the genotype of 20 randomly chosen plants grown from seeds obtained directly from Carolina Biological Supply. Only the strain “Standard B. rapa” was polymorphic for all five markers tested. It was also the only strain in which we detected all three alleles of D9BrapaS4. For each marker, the strains tested usually had the same major allele. The only two exceptions to this pattern were D9BrapaS1 in the Purple Stem, Hairy strain and Park14-HaeIII in the Non-purple Stem, Hairless strain. The distribution of the genotypes in the plants tested did not deviate from Hardy-Weinberg expectations (not shown).
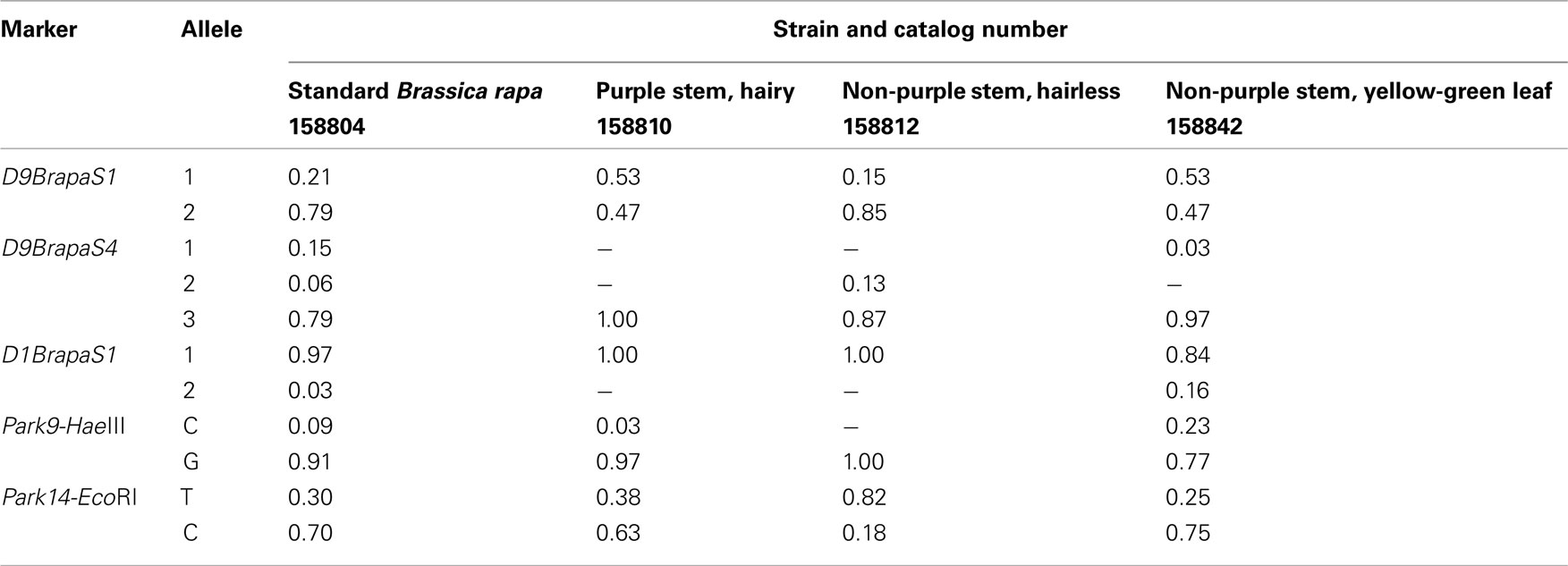
Table 4. Estimated marker allele frequencies in fast plants strains from Carolina Biological Supply.
New RCBr Strains with Defined Marker Genotypes
We have developed three strains of RCBr with genotypes optimized for use of these markers in an instructional lab. The strains vary in both the easy to score Mendelian loci anthocyaninless (purple or non-purple stem color) and yellow-green (green or yellow-green leaf color), and the DNA markers we have developed. For most markers, a strain is fixed for a particular allele so that crosses between strains will be fully informative (Table 5).
Discussion
The DNA-based genetic markers that we have developed were intentionally designed for science education which is the main use of RCBr (also known as Fast Plants). They can be used as both markers for transmission genetics and provide the basis for extensions into molecular biology. The DNA polymorphisms responsible for the observed alleles of D9BrapaS1, D9BrapaS4, Park9-HaeIII, and Park14-EcoRI have been deposited into the dbSNP database (see text footnote 9) so that when students identify alleles of these markers using basic agarose gels as shown in Figure 1, they can then obtain the sequence data underlying these polymorphisms. The sequence data obtained can then be the basis of further exploration of the B. rapa genome through the BRAD database11. The markers turn out to represent a wide variety of genomic features. Of the VNTR-type markers, one is in a non-genic region, one is within the intron of a gene, and one is within the open reading frame of a gene. Of the SNPs, one is in an intron and one is in the open reading from of a gene, although it is a synonymous substitution. Instructors can use the DNA sequence information we present here to develop lessons for students to study the possible impact on gene function of the sequence variation of the alleles.
Three of the markers form a linkage group with the anthocyaninless locus allowing them to be used in laboratory projects in genetic linkage and mapping. They are also excellent tools for projects such as paternity testing. We have previously described a laboratory project using RCBr to perform paternity testing, but the previous design used microsatellite markers (Wendell and Pickard, 2007) which can pose difficulties for lab instructors due to the need for polyacrylamide gels to resolve them. However, the markers we report here can be detected and alleles resolved in the most simple agarose slab gels.
The data we provide on population allele frequencies in RCBr strains will be valuable to instructors using these markers for educational labs. For example, we previously described a simple lab project in which the students perform paternity testing by pollinating one plant (“Mother”) with a mixture of pollen from two other plants (“Alleged Fathers”), but success in this project requires that the Alleged Fathers have different alleles for the markers used (Wendell and Pickard, 2007). An instructor who wishes to perform this project using Fast Plants obtained from Carolina Biological Supply would be best served using the markers D9BrapaS1 and Park14-EcoRI since these are polymorphic in all strains tested (Table 4). To work with a great degree of polymorphism, an instructor should use the strains described in Table 5 because they vary greatly in their genotypes for both our markers and simple Mendelian traits. Seeds for these strains are available by request to Douglas Wendell12.
The VNTR-type markers that we developed are not as polymorphic as expected for repetitive DNA-based markers. Except for D9BrapaS4, we have only found two alleles for each of the markers despite testing numerous RCBr strains, whereas VNTR markers used in mapping and DNA fingerprinting typically have multiple alleles (Nakamura et al., 1987). This could result if the repetitive DNA elements that we have selected are not prone to polymorphism, but could also result if or the RCBr populations have a low rate of polymorphism. The latter explanation is consistent with previous work in which we tested microsatellite markers that had been developed for Brassica crop species for the usefulness in RCBr. Out of 37 primer pairs that amplified a product in RCBr DNA we only found 22 to be polymorphic and only 11 that had more than two alleles in RCBr (Burdzinski and Wendell, 2007), despite the fact that microsatellites usually have multiple alleles.
The reader may wonder why we only report two SNP markers given that SNPs are abundant in organisms, and other groups have reported huge lists of SNPs for B. rapa (Li et al., 2009; Park et al., 2009). We did find several other SNPs (not shown) that lie within restriction sites but we were unable to design a PCR-RFLP around them that gave legible bands. The main source of the problem was the difference in size between “cut” and “uncut” alleles when detected by PCR-RFLP in ethidium bromide stained gels. Because the intensity of staining of DNA in gels by dyes, whether fluorescent or visible, is proportional to the mass of DNA in a band, we encountered a problem of markers where the lower molecular weight bands of the cut allele were too faint for student to reliably detect. Another complication was that in some cases the restriction enzyme that recognized the SNP also had multiple recognition sites close to the SNP.
In addition to developing markers and plant strains, we have developed classroom-tested protocols for their use. We make these publically available at the web site humangeneticsmustard.blogspot.com and will be adding more instructor resources as we develop them.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by an American Recovery and Reinvestment Act grant 5 RC1 RR030293-02 from the National Institutes of Health, USA. We thank Christian Brigolin and Jay Edwards for technical assistance.
- ^www.fastplants.org
- ^http://www.brassica.info/resource/markers/ssr-exchange.php
- ^agtc.wayne.edu
- ^www.brassica-rapa.org
- ^http://emboss.bioinformatics.nl
- ^http://www.ncbi.nlm.nih.gov/tools/primer-blast/
- ^http://www.ebi.ac.uk/Tools/msa/clustalw2/
- ^http://tools.neb.com/NEBcutter2/
- ^http://www.ncbi.nlm.nih.gov/projects/SNP/
- ^http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?
- ^http://brassicadb.org/brad/blastPage.php
- ^wendell@oakland.edu
References
Burdzinski, C., and Wendell, D. L. (2007). Mapping the anthocyaninless (anl) locus in rapid-cycling Brassica rapa (RBr) to linkage group R9. BMC Genet. 8, 64. doi:10.1186/1471-2156-8-64
Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., Li, P., Hua, W., and Wang, X. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11, 136. doi:10.1186/1471-2229-11-136
Goldman, I. L. (1999). Teaching recurrent selection in the classroom with Wisconsin Fast plants. Horttechnology 9, 579–584.
Iniguez-Luy, F. L., Lukens, L., Farnham, M. W., Amasino, R. M., and Osborn, T. C. (2009). Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. TAG. Theor. Appl. Genet. 120, 31–43.
Konieczny, A., and Ausubel, F. M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410.
Kruglyak, L. (1997). The use of a genetic map of biallelic markers in linkage studies. Nat. Genet. 17, 21–24.
Lauffer, D., and Fall, B. (2000). “Evolution by artificial selection and unraveling the mysteries of Hairy’s inheritance,” in 21st Workshop/Conference of the Association for Biology Laboratory Education (ABLE), ed. J. Karcher (Lincoln: Association for Biology Laboratory Education (ABLE)), 147–179.
Li, F., Kitashiba, H., Inaba, K., and Nishio, T. (2009). A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res. 16, 311–323.
Litt, M., and Luty, J. A. (1989). A hypervariable microsatellite revealed by in vitro amplificaation of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 44, 397–401.
Manly, K., Cudmore, R., and Meer, J. (2001). Mapmanager QTX, cross-platform software for genetic mapping. Mamm. Genome 12, 930–932.
Musgrave, M. E. (2000). Realizing the potential of rapid-cycling Brassica as a model system for use in plant biology research. J. Plant Growth Regul. 19, 314–325.
Nakamura, Y., Leppert, M., O’Connell, P., Wolff, R., Holm, T., Culver, M., Martin, C., Fujimoto, E., Hoff, M., Kumlin, E., and White, R. (1987). Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235, 1616–1622.
Park, S., Yu, H.-J., Mun, J.-H., and Lee, S.-C. (2009). Genome-wide discovery of DNA polymorphism in Brassica rapa. Mol. Genet. Genomics 283, 135–145.
Weber, J. L., and May, P. E. (1989). Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet. 44, 388–396.
Wendell, D. L., and Pickard, D. (2007). Teaching human genetics with mustard: rapid cycling Brassica rapa (fast plants type) as a model for human genetics in the classroom laboratory. CBE Life Sci. Educ. 6, 179–185.
Williams, P. H. (2007). The Rapid Cycling Brassica Collection Catalog. Madison, WI: Wisconsin Fast Plants Program.
Keywords: Fast Plants, rapid cycling Brassica rapa, marker, SNP, education, DNA fingerprinting, genetic mapping
Citation: Slankster EE, Chase JM, Jones LA and Wendell DL (2012) DNA-based genetic markers for rapid cycling Brassica rapa (Fast Plants Type) designed for the teaching laboratory. Front. Plant Sci. 3:118. doi: 10.3389/fpls.2012.00118
Received: 28 February 2012; Accepted: 17 May 2012;
Published online: 01 June 2012.
Edited by:
Xiaowu Wang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Dr. Sureshkumar Balasubramanian, Monash University, AustraliaXiyin Wang, Hebei United University, China
Guusje Bonnema, Wageningen University, Netherlands
Copyright: © 2012 Slankster, Chase, Jones and Wendell. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Douglas L. Wendell, Department of Biological Sciences, Oakland University, 2200 N Squirrel Road, Rochester, MI 48309-4401, USA. e-mail: wendell@oakland.edu