- Department of Environmental and Occupational Health Sciences, University of Washington, Seattle, WA, USA
Bacteria can become resistant to antibiotics by mutation, transformation, and/or acquisition of new genes which are normally associated with mobile elements (plasmids, transposons, and integrons). Mobile elements are the main driving force in horizontal gene transfer between strains, species, and genera and are responsible for the rapid spread of particular elements throughout a bacterial community and between ecosystems. Today, antibiotic resistant bacteria are widely distributed throughout the world and have even been isolated from environments that are relatively untouched by human civilization. In this review macrolides, lincosamides, streptogramins, and tetracycline resistance genes and bacteria will be discussed with an emphasis on the resistance genes which are unique to environmental bacteria which are defined for this review as species and genera that are primarily found outside of humans and animals.
Introduction
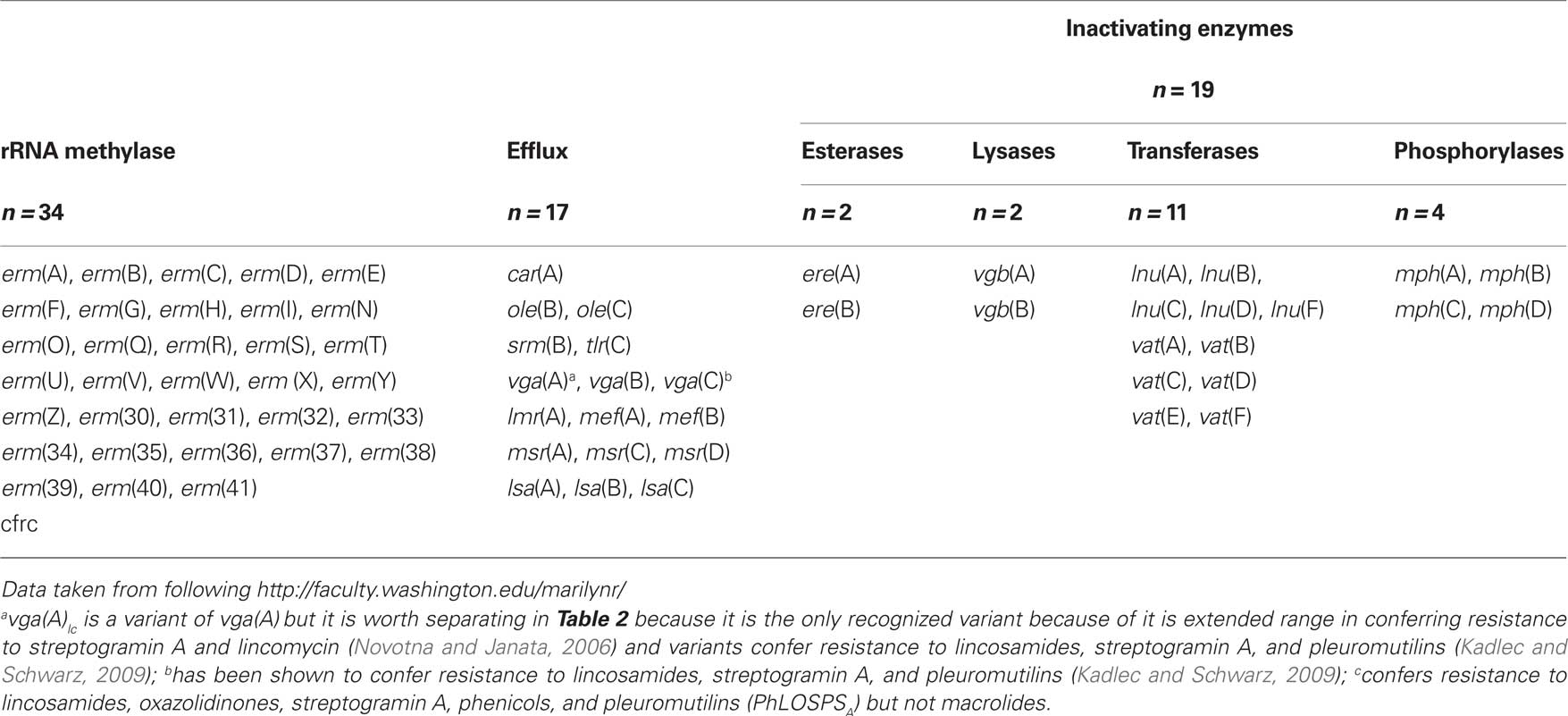
The first macrolide, erythromycin, was discovered in 1952 and since then macrolides have had an important role in treating infectious diseases (Kirst, 2002). Erythromycin had moderate activity against Gram-positive pathogens, while the newer semi-synthetic derivatives azithromycin, clarithromycin, and ketolides have broader antibacterial activity. Macrolides, lincosamides, and streptogramins (MLS), though chemically distinct, are usually considered together because most share overlapping binding sites on the 50S subunit of the ribosome and many bacteria carry acquired resistance genes which confer resistance to more than one drug within this group (Sutcliffe and Leclercq, 2003). These antibiotics inhibit protein synthesis by binding within the exit tunnel, adjacent to the peptidyl transferase center, and inhibit translation by preventing progression of the nascent chain inducing peptidyl-tRNA drop off (Auerbach et al., 2010; Starosta et al., 2010). Different antibiotics within the MLS group interact and bind with different rRNA residues which may account for why a bacterium may be resistant to the macrolide erythromycin but susceptible to semi-synthetic erythromycin telithromycin (Bulkley et al., 2010; Dunkle et al., 2010). Resistance to MLS antibiotics can be due to mutations. However for most bacteria acquisition of new genes coding for; (a) rRNA methylases which generally results in resistance to macrolides, lincosamides, and streptogramin B antibiotics (MLSB); (b) two types of efflux pumps which pump the drug(s) out of the cell; or (c) one of four types of inactivating enzymes which chemically modify the antibiotic preventing it from binding to the ribosome that is the most important way they become MLS resistant (Table 1).
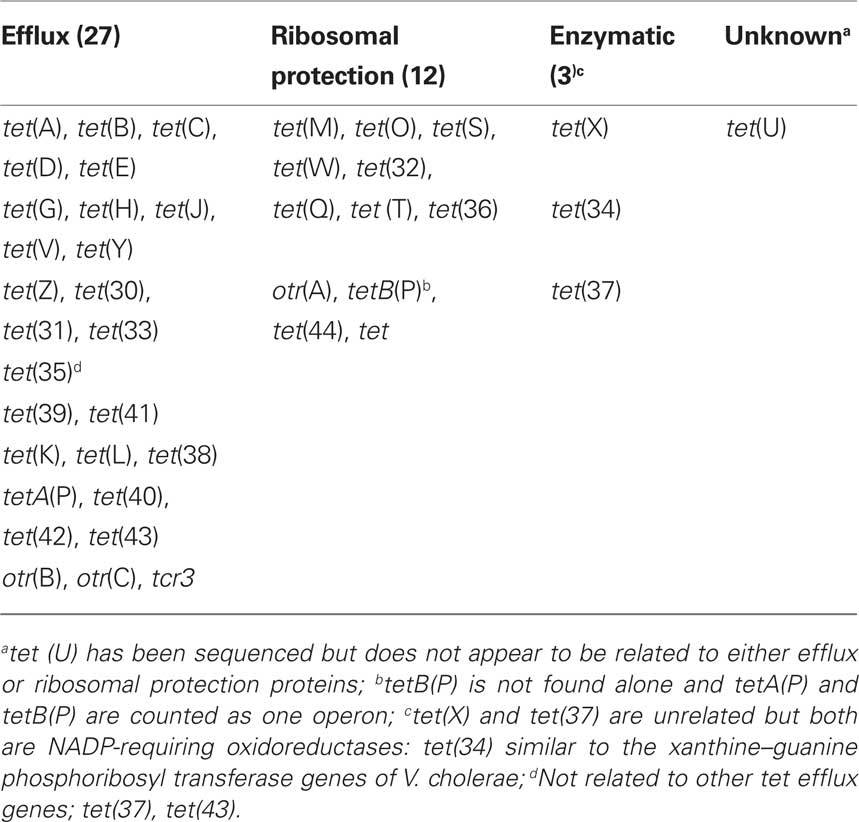
Tetracyclines are one of the oldest classes of antibiotics and the first broad spectrum class of antibiotics. Bacteria may become resistant to tetracyclines by mutation, while the majority of bacteria become tetracycline resistant because they acquire new genes which; (a) pump tetracycline out the cell (efflux); (b) protect the ribosome from the action of tetracyclines; or (c) enzymatically deactivate tetracyclines (Table 2). Tetracyclines interact with the bacterial ribosomes by reversibly attaching to the ribosome which blocks protein synthesis.
Different classes of MLS and tetracycline (tet) resistance genes are defined as having <79% amino acid identity with all previously characterized genes. Two genes are of the same class if they share >80% amino acid sequence identity over the entire length of the protein. For a tet or a MLS resistance gene to be assigned a new gene designation, it has to be completely sequenced and its amino acid composition compared with all currently known classes. This information should be submitted to the MLS resistance nomenclature center (Dr. Roberts, University of Washington) or the tet nomenclature center clearing (Dr. Stuart Levy and Ms. McMurry, Tufts University) are assigned a new resistance gene name which should be used for submission to GenBank and for publication. Demonstration that the gene confers antibiotic resistance is required and data mining genomes looking for DNA and amino acid similarities to known genes is not adequate to be recognized as a new gene. A website1 with the MLS and tetracycline resistance genes provides updated information taken from published papers and abstracts presented at scientific meetings. The majority of both the MLS and tet genes are associated with mobile elements and thus have the capacity to spread through the bacterial ecosystem (Roberts, 1997). Today most characterization of antibiotic resistance genes carried by specific bacteria is done by PCR assays. However only rarely are the PCR products verified by either sequencing or hybridization with a known labeled probe. As a result there is potential for false positive and false negative results to be reported in the literature. Therefore if unusual results occur, such as finding the Gram-negative tet(B) gene in a Gram-positive bacterium, this should be further verified by sequencing of the PCR product and further testing. In the current review only genes that have been given approved MLS or tet genes will be include in the discussion though other genes which confer MLS or tetracycline can be found in the literature and GenBank.
MLS Resistance Genes
There are 70 different MLS genes of which 33 are rRNA methylase genes (erm; Table 1). These genes code for enzymes which add one or two methyl groups to a single adenine (A2058 in Escherichia coli) in 23S rRNA (Roberts, 2005). This modification prevents the MLS antibiotics from attaching to the ribosome and protein synthesis is not impeded. The erm genes confer resistance to MLSB, while the genetically distinct cfr gene confers resistance to lincosamides, oxazolidinones, streptogramin A, phenicols, and pleuromutilins (PhLOSPSA) but not macrolides (Table 1). Sixteen (48%) of the erm genes [erm(H), erm(I), erm(N), erm(O), erm(R), erm(S), erm(W), erm(Z), erm(30), erm(31), erm(32), erm(34), erm(36), erm(39), erm(40), and erm(41)] are unique to environmental bacteria, which for this review are defined as those species and genera which are primarily found outside of humans and animals. The erm(37) gene is found in Mycobacterium tuberculosis and not considered unique to environmental bacteria, while the erm(39), erm(40), and erm(41) genes are found in one or more environmental Mycobacterium species (Nash et al., 2006). There are 17 efflux genes that code for proteins that pump one or more of the MLS antibiotics out of the cell and are either ATP-transporters or major facilitator transporters. Recently variants of the vga(A) gene and the vga(C) genes have been shown to confer resistance not only to lincosamides but to streptogramin A and pleuromutilins (Gentry et al., 2008; Kadlec and Schwarz, 2009). Six (37.5%) of the efflux genes [car(A), lmr(A), ole(B), ole(C), srm(B), tlr(C)] are unique to environmental bacteria and are each found in Streptomyces spp. (Table 3). There are 19 genes which code for inactivating enzymes including 2 esterases, 2 lyases, 11 transferases, and 4 phosphorylases (Table 1). These genes code for proteins which modify the antibiotics by hydrolyzing the lactone ring [ere(A), ere(B)], adenylylating [lnu(A), lnu(B), lnu(F)], acetylating [vat genes], or phosphorylating the antibiotic which disrupts the structure and inactivates the antibiotic. None of the inactivating enzymes are unique to environmental bacteria (Table 3)2.

Table 3. Macrolides, lincosamides, and streptogramins resistance genes unique to environmental bacteria.
Forty-three Gram-negative, 22 Gram-positive genera plus Mycobacterium, Streptomyces, and Ureaplasma spp. have been shown to carry one or more of the MLS resistance gene. This list includes innate genes which are limited to a single genus or in some case, species. For example there are 5 different innate rRNA methylases macrolide resistance genes [erm(37), erm(38), erm(39), erm(40), erm(41)] each found in different intrinsically macrolide resistant Mycobacterium spp., and 10 different rRNA methylases macrolide and 6 different innate efflux resistance genes in antibiotic producing Streptomyces spp. These genes have traditionally been included because they confer MLS resistance and are related to resistance genes found in non-antibiotic producing bacteria and are important for this review because they are unique to environmental bacteria (Table 3).
Among the rRNA methylases classes, the most commonly carried is erm(B) [24 genera], followed by erm(F) [24 genera], erm(C) [24 genera], erm(A) [8 genera], erm(G) and erm(E) [7 genera each], erm(Q) [6 genera], erm(T) and erm(X) [4 genera each], erm(V) [3 genera], erm(D), erm(R), cfr [2 genera each]. The remaining 21 (64%) erm genes are found in a single genus. The erm(A)–erm(G), erm(Q) and erm(V) genes are found in both Gram-positive, Ureaplasma spp., and Gram-negative genera as well as aerobic and anaerobic species, the erm(35) gene is found in Gram-negative genera and the remaining 26 MLS genes are found in Gram-positive, anaerobic Bacteriodes, and/or Streptomyces spp.
The most prevalent efflux gene is mef(A) [27 genera] followed by msr(D) [22 genera] and are found in Gram-positive, Ureaplasma spp., Gram-negative, aerobic, and anaerobic genera. These two genes are normally linked on the same mobile element suggesting that they have the same host range, though msr(D) is often not included in surveillance studies. The msr(A) gene has been identified in eight genera including Gram-positive, Ureaplasma spp., and Gram-negative genera. The recently described mef(B) gene is found in Escherichia spp., while the vga(B) gene is in two genera and the remaining 13 MLS resistance genes (76%) are found in Gram-positive genera or Streptomyces spp. Distributions of the 19 inactivating enzymes are as follows; ere(A) [11 genera] and ere(B) [8 genera] found in both Gram-positive and Gram-negative genera; vgb(A) [2 genera], vgb(B) [1 genus] both Gram-positive genera; lnu(A) [3 genera], lnu(B) [4 genera], ], lnu(D) [1 genus] all Gram-positive genera; lnu(C) [1 Gram-positive, 1 Gram-negative genus] lnu(F) [2 Gram-negative genera]; vat(A), vat(C), vat(D) [1 Gram-positive genus each], vat(B), vat(E) [2 Gram-positive genera each]; vat(F) [1 Gram-negative genus]; mph(A) [11 genera ], mph(B) [4 genera ], mph(D) [6 genera] all Gram-negative genera and mph(C) [1 Gram-positive, 1 Gram-negative genus] (Roberts, 2004)3.
Tetracycline Resistance Genes
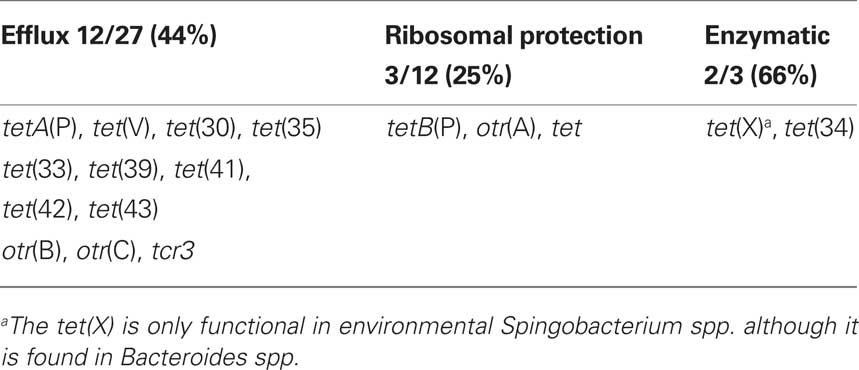
There are 43 different known tet/otr genes which confer resistance to tetracyclines (Table 2) of which 17 (39%) are unique to environmental bacteria (Table 4). The majority of the tet/otr genes are associated with mobile elements which allow for gene exchange and spread throughout different ecosystems (Roberts, 1997). Twenty-seven of the genes encode for energy-dependent efflux proteins, 12 code for ribosomal protection proteins, 3 code for inactivating enzymes and one gene is of unknown mechanism of resistance (Table 2). The efflux proteins are able to reduce intracellular concentrations of tetracycline by exchanging a proton for the tetracycline-cation complex against a concentration gradient and require intact cells to function (Palm et al., 2008). Twenty-six of 27 genetically distinct efflux genes export and confer resistance to tetracycline and doxycycline but not minocycline or tigecycline, while the Gram-negative tet(B) gene exports and confers resistance to tetracycline, doxycycline, and minocycline. Twelve (41%) of the efflux genes [tetA(P), tet(V), tet(Z), tet(30), tet(33), tet(35), tet(39), tet(41), tet(42), otr(B), otr(C), tcr] are unique to environmental bacteria (Table 4).
Seventy-six Gram-negative and 47 Gram-positive genera have been identified that carry tetracycline efflux resistance genes. Many of these genes and genera are found in environmental bacteria but can also be found in bacteria associated with man and/or animals4. The tet(B) gene is the most common efflux gene among Gram-negative bacteria and has been identified in 31 genera, while the tet(A) gene [20 genera], tet(C) gene [10 genera], tet(D) gene [16 genera], tet(E) gene in [10 genera], tet(G) gene [13 genera], the tet(H) gene [8 genera], and the tet(35) [2 genera]. The tet(K) gene is found in 12 Gram-positive genera and the otr(B) gene is found in Mycobacterium and Streptomyces spp. The tet(L) gene is found in 14 Gram-negative and 19 Gram-positive genera, the tet(39) gene is found in 4 Gram-negative and 3 Gram-positive genera, while the tet(42) gene is found in 4 Gram-positive and 2 Gram-negative genera (Table 2). Twelve (44%) of the efflux genes including the tet(J), tetA(P) tet(V), tet(Y), tet(Z), tet(30), tet(31), otr(C), tcr, tet(33), tet(40), and tet(41) are found in a single genera. The tet(43) gene was isolated from a metagenomic DNA library and has yet to be identified in a specific bacterial species or genus.
Tcr C. suis, an obligate intracellular species, contains a13-kb fragment of foreign DNA which includes a truncated repressor gene, tetR(C), and a functional tet(C) gene. The 13-kb region is closely related to the pRAS3.2 plasmid from the environmental water bacterium Aeromonas salmonicida (L’Abbe-Lund and Sorum, 2002). The hypothesis is that C. suis could have acquired the tet(C) gene when other bacteria carrying the 13-kb region with the tet(C) gene co-infected the same eukaryotic cell; this is the first example of horizontal transfer of an antibiotic resistance gene into this group of bacteria. Suchland et al. (2009) have demonstrated that co-infection between Tcr C. suis and susceptible C. trachomatis resulted in Tcr C. trachomatis containing the 13-kb tet(C) region inserted within the ribosomal cluster of the chromosome and is the first example of horizontal transfer between obligate intracellular bacteria. It is likely that other antibiotic resistance genes may be introduced and become established in other obligate intracellular bacteria.
There are 12 ribosomal protection Tcr genes of which three [25%; tetB(P), otr(A), tet] are unique to environmental bacteria (Table 4). The ribosomal protection genes code for cytoplasmic proteins of ∼72.5 kDa which protect the ribosomes from the action of tetracyclines in vitro and in vivo and confer resistance to tetracycline, doxycycline, and minocycline but not tigecycline (Roberts, 2005). These proteins have sequence similarity to the ribosomal elongation factors, EF-G and EF-TU and are grouped in the translation factor superfamily of GTPases (Leipe et al., 2002). The ribosomal protection proteins are hypothesized to interact with the h34 ribosomal protein causing allosteric disruption of the primary tetracycline binding site(s) which releases the bound tetracycline. More details can be found in other publications (Connell et al., 2003a,b). The tet(M) gene has been identified in clinical Enterococcus spp. isolated between 1954 and 1955, approximately the same time as the first Gram-negative tet efflux genes were identified (Watanabe, 1963; Atkinson et al., 1997). Thus both the tet efflux and tet ribosomal protection genes have been in the bacterial population for a considerable time.
Forty-nine Gram-negative genera have been characterized which carry one or more ribosomal protection tet gene(s). Thirty-eight Gram-positive genera carry ribosomal protection genes of which 15 carry a single gene and 23 carry one or more ribosomal protection and/or both ribosomal protection and efflux tet genes5. The tet(M) gene is the most commonly found Tcr gene in bacteria, identified in 36 Gram-negative genera, and 31 Gram-positive genera, Mycoplasma, Mycobacterium, Streptomyces, and Ureaplasma spp. The tet(W) gene has been identified in 25 genera [15 Gram-negative, 10 Gram-positive], the tet(Q) gene has been identified in 22 genera [12 Gram-negative, 10 Gram-positive], the tet(O) gene has been found in 20 genera [8 Gram-negative, 12 Gram-positive], the tet(S) gene has been identified in 7 genera [3 Gram-negative, 4 Gram-positive], while the otr(A) gene has been found in 3 Gram-positive genera, the tet(T), tet(32), and tet(44) genes have been found in 2 Gram-positive genera each, the tet gene has been identified in Streptomyces, and the tetB(P) found in one Gram-positive genera6. The tetB(P) gene is unique among the ribosomal protection genes because all isolates that carry this gene also carry a tetA(P) gene encoding an inducible efflux protein. The two genes are transcribed from a single promoter which is located 529 bp upstream of the tetA(P) start codon; the tetB(P) gene overlaps the tetA(P) gene by 17 nucleotides (Johanesen et al., 2001). The tetA(P) gene alone confers Tcr to the bacterial host, while it is not clear if the tetB(P) gene alone would confer clinically relevant levels of tetracycline resistance.
A few of the ribosomal protection genes have been found to be part of mosaic genes consisting of regions from two or three different known tet genes (Stanton et al., 2005; Patterson et al., 2007; Van Hoek et al., 2008). Identification of mosaic genes requires sequencing of the complete gene which is rarely done in current studies. Hence misnaming the gene present is possible as originally occurred with the tet(32) gene (Patterson et al., 2007). One such gene is a hybrid between the tet(O) at one end and tet(W) at the other end of the gene labeled tet(O/W). Another hybrid between the tet(O) and tet(W) genes has been identified which has a partial tet(O) sequence between the ends of the tet(W) gene [tet(W/O/W)]. Mosaic Tcr genes can only be defined by sequencing the complete gene and at this time, the number of different genera known to have them is very limited (Bifidobacteria, Lactobacillus, Megasphaera). In addition, studies have found mosaic tet genes from metagenomic samples taken from human and animal feces and human saliva samples (Patterson et al., 2007; Kazimierczak et al., 2009). The first mosaic genes sequenced were from Megasphaera elsdenii and a new designation was suggested for hybrid genes which code for proteins made of >50 amino acid residues in a single stretch derived from different genes (Levy et al., 2005; Stanton et al., 2005). Mosaic genes currently identified include tet(O/W), tet(O/W/O), tet(W/32/O), tet(O/32/O), tet(O/W/32/O), tet(O/W/32/W/O), tet(O/W/32/O/W/O) which include genes with fewer then 50 amino acid residues suggesting that a modification of the current recommendation for mosaic genes may need to be considered (Stanton et al., 2005; Patterson et al., 2007; Van Hoek et al., 2008).
The tet(U) gene produces a small protein (105 amino acids) which confers low level tetracycline resistance (Chopra and Roberts, 2001). The TetU protein has 21% similarity over its length to the TetM protein, but it does not include the consensus GTP-binding sequences, which are thought to be important for tetracycline resistance in these proteins. The tet(U) gene has been identified in only one Gram-positive genus.
There are three enzymatic Tcr genes (Table 2). The tet(X) gene encodes for a flavin NADP-dependent monooxygenase that inactivates by region selectively adding a hydroxyl group to the C-11a position of the antibiotic. This action requires oxygen and confers resistance to tetracycline, doxycycline, minocycline, and tigecycline and is found in two genera (Yang et al., 2004; Moore et al., 2005). The tet(37) gene, codes for a NADP-dependent monooxygenase. The tet(37) gene has only been cloned from the oral metagenome and no bacterial isolate has been identified that carrying this gene. The tet(34) gene has been found in three Gram-negative genera [Pseudomonas, Serratia, and Vibrio] and is unique to environmental bacteria (Table 4; Nonaka and Suzuki, 2002).
The Environment
Among the culturable bacteria, some genera are found only in the environment, others are found in the environment and animals and/or man but the distinction between environmental and clinical bacteria has been blurred over the years as more classically environmental bacteria are found as the source of disease in immunocompromised patients. Complication the situation is the fact that some species within a genus are strictly environmental while other species within the same genus are associated with animals and/or man such as M. tuberculosis thus making it difficult to distinguish environmental vs clinical genera. In the past it has been assumed that most environmental bacteria were not well adapted to live in man or animals. However this perception is changing as medicine progresses allowing severely immunocompromised patients to live in the community, others people have productive lives with foreign objects permanently implanted, these and other technological changes in society have provided new opportunities for environmental bacteria to cause disease (Trujillo and Goodfellow, 2003; Rowlinson et al., 2006). Human activity has resulted in only rare remote ecosystems where mankind and civilization’s waste are absent. Thus, today it is more difficult to distinguish environmental from non-environmental bacteria. As a result there is constant mixing of environmental and non-environmental bacteria which provides multiple opportunities for horizontal genetic exchange of antibiotic resistance genes. In addition antibiotic resistance genes associated with mobile elements can be transferred back and forth between environmental and non-environmental bacteria thus reducing the number of resistance genes which are unique to the environment.
Both antibiotics and antibiotic resistant bacteria are moved by water and wind, as well as, by transportation of goods and people around the world. One result of this has been the spread of specific strains around the world such as the recently identified Clostridium difficile NAP1/027/BI (Gould and Limbago, 2010). Originally C. difficile was thought to be a nosocomial disease associated with the hospital setting, but today C. difficile is considered a foodborne and community pathogen. Similarly 25 years ago Acinetobacter baumannii was a rare pathogen and Acinetobacter spp. were thought to be primarily environmental in origin where it is well adapted to grow at different temperatures and pH, using a variety of carbon and energy sources to persist in both moist and dry places for extended time periods. However, today A. baumannii is an opportunistic pathogen which is of major concern to the because it has become multidrug resistant due to the inheritance of a mosaic genomic 86 kb chromosomal antibiotic resistance island which carries integrons and non-integron regions encoding both antibiotic and heavy metal resistance including two copies of the tet(A) gene (Abbo et al., 2005; Fournier et al., 2006). A more recent study from Adams et al. (2010) has demonstrated variability in the composition of the resistance island between various A. baumannii strains suggesting that the resistance in this bacterium is evolving.
Antibiotics are used for both human and agricultural activities for prevention and treatment of infections, as well as food additives and growth promoters in food production in most parts of the world and also used in animal husbandry, aquaculture, fruit crops, and bee-keeping. All of these activities contaminate the environment and provide selective pressure on the resident environmental bacteria to become antibiotic resistant and increases transfer of specific antibiotic resistance genes including tet genes (Facinelli et al., 1993). Antibiotic residues are associated with domestic animal manure which may be transferred to the immediate environment when it is applied to fields or stored in ponds. Antibiotics are sprayed onto crops which contaminate the surrounding soil, sediment, and ground water as well as small wild mammals in the area (Kozak et al., 2009). Antibiotics may be impregnated in food given to farm animals and fish and uneaten food contaminates the surrounding environment. Antibiotics from human therapeutic use, especially from hospital effluents, are a continual source of pollution and are considered part of the “emerging contaminants” in municipal waste with concentrations of tetracycline varying from ng/L to μg/L (Verlicchi et al., 2010). At these levels antibiotics may select for antibiotic resistant environmental bacteria which may persist for extended time periods and become reservoirs of antibiotic resistance genes.
The mobile elements associated with MLS and tet antibiotic resistance genes are often linked to genes which confer resistance to other classes of antibiotic, heavy metal, and detergents (Baker-Austin et al., 2006; Soge et al., 2008). Heavy metals are found naturally in the environment but copper is found in biocides used in agricultural settings, while mercury and silver can be found in antiseptics. Various studies have found a correlation between exposure to heavy metals and co-selection of antibiotic resistance bacteria (Baker-Austin et al., 2006; Stepanauskas et al., 2006; Berg et al., 2010).
Antibiotic resistant bacteria from human activity may contaminate the environment either directly, such as occurs when manure is applied to enrich agricultural fields or indirectly due to sewage contamination. The first description of the tet(M) gene in Bacillus spp. and Tcr Bacillus cereus strains carrying the tet(M) gene, on a functional Tn916 element, were found in animal manure and in the fields where the manure was spread. This suggests that the presence of the tet(M) carrying B. cereus in the fields was a direct result of manure application to the soil. Whether the tet(M) carrying B. cereus would act as a donor and transfer the tet(M) gene to related B. anthracis, and/or B. thuringiensis is unknown, however toxin encoding plasmids are shared between the three species (Agerso et al., 2002).
How human wastes may increase antibiotic resistant bacteria in wild animals was illustrated by a 1980s study which examined three groups of wild baboons in Kenya. Three groups of baboons were examined and the two groups which lived in their natural habitat, with limited or no human contact, had low levels of antibiotic resistant Gram-negative enteric bacteria. The third group of baboons lived close to a tourist lodge and had daily contact with unprocessed human refuse and these animals carried high levels of antibiotic resistant Gram-negative enteric bacteria with >90% Tcr. This study suggests that contact with human refuse greatly increased the carriage of Tcr bacteria in wild animals (Rolland et al., 1985). Unfortunately the surrounding environmental bacteria were not sampled, but one could speculate that the level of Tcr bacteria was most likely higher than in the areas where the other baboon groups lived. Human and animal Tcr bacteria share the majority of the efflux and ribosomal protection genes with environmental bacteria. Recently studies have shown carriage of Tcr bacteria from arctic and sub-arctic seals (Glad et al., 2010); wild boars (Poeta et al., 2009); and wild rabbits (Silva et al., 2010) though the sources of the Tcr E. coli bacteria was not determined.
Tetracyclines have been extensively used in aquaculture and Tcr bacteria including fish pathogens and environmental bacteria associated with fin fish aquaculture have been characterized (DePaola and Roberts, 1995; Furushita et al., 2003; Akinobowale et al., 2007; Jacobs and Chenia, 2007; Nawaz et al., 2008; Seyfried et al., 2010). Tcr bacteria can be found in fish feed, the sediment under the fish pens, as well as the water entering and leaving fresh water ponds (Kerry et al., 1995; Miranda and Zemelman, 2002). Some of the greatest diversity in bacterial Tcr genes has been identified in the aquaculture environment. In one of our studies, we found that 40% of the Tcr bacteria isolated from Chilean salmon fish farms carried previously unidentified Tcr genes suggesting tet resistance gene diversity present in this ecosystem, higher than routinely found in collections from man or food animals (Miranda et al., 2003). We also identified new genera carrying previously characterized tet genes however they were not readily transferred under laboratory conditions raising the question on how some of the genes were being transferred to bacteria across the world and from very different environments (Miranda et al., 2003). The diversity of type and number of Tcr bacteria found in aquaculture suggests that this environment favors rapid evolution of Tcr bacteria which then can be transferred to other environments and ecosystems. More recently, a diverse group of Tcr bacteria, carrying a range of tet genes, isolated from surface water receiving effluent from an oxytetracycline production plant has been reported (Li et al., 2010). Many of the tet gene combinations reported were novel however detection was without verification of the PCR products. This raises the question as to the validity of the study. Nevertheless this study suggests that waste treatment from antibiotic production plants is a potential hot spot for spread of known and uncharacterized antibiotic resistance genes into the environment.
The environments are so physically diverse that many associated bacteria are distinct from those normally found in animals and/or man. Most environmental bacteria have not been characterized and it is estimated that <1% of the total number of bacterial species present in different environments have been identified (Kummerer, 2004). Therefore antibiotic resistance genes associated with unculturable bacteria may only be identified by molecular methods such as those used to isolate the tet(43) gene, which along with other antibiotic resistance genes was isolated from metagenomic analysis of apple orchard soil from an which received repeated treatment with streptomycin (Donato et al., 2010). One can hypothesize that using this approach on other environmental samples will identify additional novel antibiotic resistance genes in the future however the bacteria carrying these genes may never be identified.
Macrolides, lincosamides, and streptogramins resistance genes are found in waste lagoons and groundwater wells at swine farms with a history of tylosin use and erm(A), erm(B), erm(C), and erm(F) genes were detected at high frequency with only a weak correlation with the distribution patterns of tet genes. Many of the RNA methylase genes were associated with bacteria found in animal waste while tylosin resistance genes were rare, this suggests limited levels lack of tylosin-producing Streptomyces genes in either the lagoons or wells and may indicate that lagoon leakage as sources of groundwater contamination (Koike et al., 2010). This study suggests that there could be alternative sources of MLS resistance genes and/or background antibiotic resistant environmental bacteria contributing to the levels of MLS genes, since the most common erm genes, erm(B), and erm(C), identified in the study are also found in environmental bacteria.
Conclusion
Antibiotic resistant bacteria are widely distributed throughout the world and have been isolated from deep subsurface trenches, in waste water, surface and ground water, sediments, and soils. They are present also in places which are relatively untouched by human civilization such as mammals from Antarctica and the Arctic (Kummerer, 2004; Rahman et al., 2008; Donato et al., 2010; Glad et al., 2010). Twenty-two (31%) of the 70 MLS and 17 (40%) of the 43 tet genes are unique to environmental bacteria (Tables 2 and 4). Whether this represents a true separation between MLS and tet genes that are “unique” to environmental bacteria or because the genes are innate to environmental bacteria or that the resistance genes have not been used in surveillance studies of animal/human bacteria is not known. It is possible that over time these “unique tet genes” will move into bacteria associated with animals and/or man as studies with the tet(X) gene suggests (Ghosh et al., 2009). The antibiotic resistance gene complement of environmental bacterial populations should be studied in greater detail as illustrated by the D’Costa et al. (2006) study. This work quantified the density and diversity of antibiotic resistant soil Streptomycetes and even identified genes with novel mechanisms of antibiotic resistance to the recently approved telithromycin and daptomycin. D’Costa et al. (2007) suggest that the number of antibiotic resistance genes in environmental bacteria has been underestimated and given that they found a new mechanisms of drug resistance more studies with environmental bacteria are essential.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abbo, A., Navon-Venezia, S., Hamemer-Muntz, O., Krichali, T., Siegman-IgraY., and Carmeli, Y. (2005). Multidrug-resistant Acinetobacter baumannii. Emerging Infect. Dis. 11, 22–29.
Adams, M. D., Chan, E. R., Neil, D., Molyneaux, N. D., Robert, A., and Bonomo, R. A. (2010). Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 3569–3577.
Agerso, Y., Jensen, L. B., Givskov, M., and Roberts, M. C. (2002). The identification of a tetracycline resistance gene (M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Micobiol Lett. 214, 251–256.
Akinobowale, O. L., Peng, H., and Barton, M. D. (2007). Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J. Appl. Microbiol. 103, 2016–2025.
Atkinson, B. A., Abu-Al-Jaibat, A., and LeBlanc, D. J. (1997). Antibiotic resistance among enterococci isolated from clinical specimens between 1953 and 1954. Antimicrob. Agents Chemother. 41, 1598–1600.
Auerbach, T., Mermershtain, I., Davidovich, C., Bashan, A., Belousoff, M., Wekselman, I., Zimmerman, E., Xiong, L., Klepacki, D., Arakawa, K., Kinashi, H., Mankin, A. S., and Yonat, A. (2010). The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics. Proc. Natl. Acad. Sci. U.S.A. 107, 1983–1988.
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182.
Berg, J., Thorsen, M. K., Holm, P. E., Jensen, J., Nybroe, O., and Brandt, K. K. (2010). Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 44, 8724–8728.
Bulkley, D., Innis, C. A., Blaha, G., and Steitz, T. A. (2010). Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 107, 17158–17163.
Chopra, I., and Roberts, M. C. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260.
Connell, S. R., Tracz, D. M., Nierhaus, K. H., and Taylor, D. E. (2003a). Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47, 3675–3681.
Connell, S. R., Trieber, C. A., Einfeldt, E., Taylor, D. E., and Nierhaus, K. H. (2003b). Mechanism of Tet(O), perturbs the conformation of the ribosomal decoding center. Mol. Microbiol. 45, 1463–1472.
D’Costa, V. M., Griffiths, E., and Wright, G. D. (2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489.
D’Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling antibiotic resistome. Science 311, 374–377.
DePaola, A., and Roberts, M. C. (1995). Class D and E tetracycline resistance determinants in Gram-negative catfish pond bacteria. Mol. Cell. Probes 9, 311–313.
Donato, J. J., Moe, L. A., Converse, B. J., Smart, K. D., Berklein, F. C., McManus, P. S., and Handelsman, J. (2010). Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 76, 4396–4401.
Dunkle, J. A., Xiong, L., Mankin, A. S., and Cate, J. H. D. (2010). Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U.S.A. 107, 17152–17157.
Facinelli, B., Roberts, M. C., Giovanetti, E., Casolari, C., Fabio, U., and Varaldo, P. E. (1993). Genetic basis of tetracycline resistance in food borne isolates of Listeria innocua. Appl. Environ. Microbiol. 59, 614–616.
Fournier, P.-E, Vallenet, D., Barber, V., Audic, S., Ogata, H., Poirel, L., Richet, H., Robert, C., Mangenot, S., Abergel, C., Nordmann, P., Weissenbach, J., Raoult, D., and Claverie, J. M. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2, e7. doi: 10.1371/journal.pgen.0020007
Furushita, M., Shiba, T., Maeda, T., Yahata, M., Kaneoka, A., Takahashi, Y., Torii, K., Hasegawa, T., and Ohta, M. (2003). Similarity of tetracycline resistance genes isolated from fish farm bacteria to those from clinical isolates. Appl. Environ. Microbiol. 69, 5336–5342.
Gentry, D. R., McCloskey, L., Gwynn, M. N., Rittenhouse, S. F., Scangarella, N., Shawar, R., and Holmes, D. J. (2008). Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 4507–4509.
Ghosh, S., Gralnick, J., Roberts, M. C., Sadowsky, M., and LaPara, T. (2009). Sphingobacterium sp. strain PM2-P1-29 harbors a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 106, 1336–1342.
Glad, T., Fristiansen, V. F., Nielsen, K. M., Rusetti, L., Wright, A.-D., and Sundset, M. A. (2010). Ecological characterization of the colonic microbiota in arctic and sub-artic seals. Microb. Ecol. 60, 320–330.
Gould, L. H., and Limbago, B. (2010). Clostridium difficile in food and domestic animals: a new foodborne pathogen? Clin. Infect. Dis. 51, 583–584.
Jacobs, L., and Chenia, H. Y. (2007). Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 114, 295–306.
Johanesen, P. A., Lyras, D., Bannam, T. L., and Rood, J. I. (2001). Transcriptional analysis of the tet(P) operon from Clostridium perfringens. J. Bacteriol. 183, 7110–7119.
Kadlec, K., and Schwarz, S. (2009). Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53, 3589–3591.
Kazimierczak, K. A., Scott, K. P., Kelly, D., and Aminov, R. I. (2009). Tetracycline resistaome of the organic pig gut. Appl. Environ. Microbiol. 75, 1717–1722.
Kerry, J., Hiney, M., Coyne, R., Nicgabhainn, R., Gilroy, D., Cabon, D., and Smith, P. (1995). Fish feed as a source of oxytetracycline-resistant bacteria in the sediments under fish farms. Aquaculture 240, 89–100.
Kirst, H. A. (2002). “Introduction to the macrolide antibiotics,” in Macrolide Antibiotics eds W. Schonfeld and H. A. Kirst (Birkhauser Verlag, Basel), 1–14.
Koike, S., Aminov, R. I., Yannarell, A. C., Gans, H. D., Krapac, I. G., Chee-Sanford, J. C., and Mackie, R. I. (2010). Molecular ecology of macrolide–lincosamide–streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microb. Ecol. 59, 487–498.
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial resistance in Escherichia coli isolated from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566.
L’Abbe-Lund, T. M., and Sorum, H. (2002). A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47, 172–178.
Leipe, D. Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification of and evolution of P-loop GTPase and related ATPases. J. Mol. Biol. 317, 41–72.
Levy, S. B., McMurry, L. M., and Roberts, M. C. (2005). Tet protein hybrids. Antimicrob. Agents Chemother. 49, 3099.
Li, D., Yu, T., Zhang, Y., Yang, M., Li, Z., Liu, M., and Qi, R. (2010). Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and receiving river. Appl. Environ. Microbiol. 76, 3444–3451.
Miranda, C. D., Kehrenberg, C., Ulep, C., Schwarz, S., and Roberts, M. C. (2003). Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 47, 883–888.
Miranda, C. D., and Zemelman, R. (2002). Bacterial resistance to oxytetracycline in Chilean salmon farms. Aquaculture 212, 31–47.
Moore, I. F., Hughes, D. W., and Wright, G. D. (2005). Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44, 11829–11835.
Nash, K. A., Andini, N., Zhang, Y., Brown-Elliott, B. A., and Wallace, R. J. Jr. (2006). Intrinsic macrolide resitance in rapidly growing mycobacteria. Antimicrob. Agents Chemother. 50, 3476–3478.
Nawaz, M., Khan, A. A., Khan, S., Sung, K., and Steele, R. (2008). Isolation and characterization of tetracycline-resistant Citrobacter spp. from catfish. Food Microbiol. 25, 85–91.
Nonaka, L., and Suzuki, S. (2002). New Mg2+ -dependent oxytetracycline resistance determinant Tet 34 in Vibrio isolates from marine fish intestinal contents. Antimicrob. Agents Chemother. 476, 1550–1552.
Novotna, G., and Janata, J. (2006). A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50, 4070–4076.
Palm, G. J., Lederer, T., Orth, P., Saenger, W., Takahashi, M., Hilllen, W., and Hinrichs, W. (2008). Specific binding of divalent metal ions to tetracycline and to the Tet repressor/tetracycline complex. J. Biol. Inorg. Chem. 13, 1097–1110.
Patterson, A. J., Rincon, M. T., Flint, H. J., and Scott, K. P. (2007). Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob. Agents Chemother. 51, 1115–1118.
Poeta, P., Radhouani, H., Pinto, L., Martinho, A., Rego, V., Rodrigues, R., Goncalves, A., Rodrigues, J., Estepa, V., Torres, C., and Igrejas, G. (2009). Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 49, 584–588.
Rahman, M. H., Sakamoto, K. Q., Nonaka, L., and Suzuki, S. (2008). Occurrence and diversity of tetracycline tet(M) in enteric bacteria of Antarctic Adelie penguins. J. Antimicrob. Chemother. 62, 627–628.
Roberts, M. C. (1997). “Genetic mobility and distribution of tetracycline resistance determinants,” in Antibiotic Resistance: Origins, Evolution, Selection and Spread. Ciba Foundation Symposium 207 (Chichester: John Wiley & Sons), 206–218.
Roberts, M. C. (2004). Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol. 28, 47–62.
Roberts, M. C. (2005). Minireview: update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203.
Rolland, R. M., Hausfater, G., Marshall, B., and Levy, S. B. (1985). Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 49, 791–794.
Rowlinson, M.-C., Bruckner, D. A., Hinnebusch, C., Nielsen, K., and Deville, J. G. (2006). Clearance of Cellulosimicrobium cellulans bacteremia in a child without central venous catheter removal. J. Clin. Microbiol. 44, 2650–2654.
Seyfried, E. E., Newton, R. J., Rubert, K. F., Pedersen, J. A., and McMahon, K. D. (2010). Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 59, 799–807.
Silva, N., Igrejas, G., Figueiredo, N., Goncalves, A., Radhouani, H., Rodrigues, H., and Poeta, P. (2010). Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 408, 4871–4876.
Soge, O. O., Beck, N., White, T. M., and Roberts, M. C. (2008). A novel transposon, Tn6009, composed of a Tn916-like element linked to Staphylococcus aureus-like mer operon. J. Antimicrob. Chemother. 62, 674–680.
Stanton, T. B., Humphrey, S. B., Scott, K. P., and Fling, H. J. (2005). Hybrid tet genes and tet gene nomenclature: request for opinions. Antimicrob. Agents Chemother. 49, 1265–1266.
Starosta, A. L., Karpenko, V. V., Shishkina, A. V., Mikolajka, A., Sumbatyan, N. V., Schluenzen, F., Korshunova, G. A., Bogdanov, A. A., and Wilson, D. N. (2010). Interplay between the ribosomal tunnel, nascent chain a, and macrolides influences drug inhibition. Cell 17, 504–514.
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., King, C. J., and McArthur, J. V. (2006). Co-selection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8, 1510–1514.
Suchland, R. J., Sandoz, K. M., Jeffrey, B. M., Stamm, W. E., and Rockey, D. D. (2009). Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob. Agents Chemother. 53, 4604–4611.
Sutcliffe, J. A., and Leclercq, R. (2003). “Mechanisms of resistance to macrolides, lincosamides and ketolides,” in Macrolide Antibiotics, eds W. Schonfeld and H. A. Kirst (Basel: Birkhauser Verlag), 281–317.
Trujillo, M. E., and Goodfellow, M. (2003). Numerical phonetic classification of clinically significant aerobic sporoactinomycetes and related organisms. Antonie Van Leeuwenhoek 84, 39–68.
Van Hoek, A. H. A. M., Mayrhofer, S., Domig, K. J., Florez, A. B., Ammor, M. S., Mayo, B., and Aarts, H. J. M. (2008). Mosaic tetracycline resistance genes and their flanking regions in Bifidobacterium thermophilum and Lactobacillus johnsonii. Antimicrob. Agents Chemother. 52, 248–252.
Verlicchi, P., Galletti, A., and Masotti, L. (2010). Management of hospital wastewaters: the case of the effluent of a large hospital situated in a small town. Water Sci. Technol. 61, 2507–2519.
Watanabe, T. (1963). Infective heredity of multiple drug resistance in bacteria. Bacteriol. Rev. 27, 8–115.
Keywords: macrolide-lincosamide-streptogramin resistance, tetracycline resistance, environmental bacteria, mobile elements, acquisition of new genes
Citation: Roberts MC (2011) Environmental macrolide–lincosamide–streptogramin and tetracycline resistant bacteria. Front. Microbio. 2:40. doi: 10.3389/fmicb.2011.00040
Received: 30 November 2010;
Paper pending published: 22 December 2010;
Accepted: 21 February 2011;
Published online: 02 March 2011.
Edited by:
Julian Davies, The University of British Columbia, CanadaReviewed by:
Julian Davies, The University of British Columbia, CanadaTeresa M. Coque, Hospital Universitario Ramón y Cajal, Spain
Axel Cloeckaert, National Institute of Agronomic Research, France
Copyright: © 2011 Roberts. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Marilyn C. Roberts, Department of Environmental and Occupational Health Sciences, School of Public Health, University of Washington, 1959 NE Pacific Street, Box 357234, Seattle, WA, USA. e-mail: marilynr@u.washington.edu