Epigenetic Regulation of Cell-Fate Changes That Determine Adult Liver Regeneration After Injury
- MRC Laboratory for Molecular Cell Biology, University College London, London, United Kingdom
The adult liver has excellent regenerative potential following injury. In contrast to other organs of the body that have high cellular turnover during homeostasis (e.g., intestine, stomach, and skin), the adult liver is a slowly self-renewing organ and does not contain a defined stem-cell compartment that maintains homeostasis. However, tissue damage induces significant proliferation across the liver and can trigger cell-fate changes, such as trans-differentiation and de-differentiation into liver progenitors, which contribute to efficient tissue regeneration and restoration of liver functions. Epigenetic mechanisms have been shown to regulate cell-fate decisions in both embryonic and adult tissues in response to environmental cues. Underlying their relevance in liver biology, expression levels and epigenetic activity of chromatin modifiers are often altered in chronic liver disease and liver cancer. In this review, I examine the role of several chromatin modifiers in the regulation of cell-fate changes that determine efficient adult liver epithelial regeneration in response to tissue injury in mouse models. Specifically, I focus on epigenetic mechanisms such as chromatin remodelling, DNA methylation and hydroxymethylation, and histone methylation and deacetylation. Finally, I address how altered epigenetic mechanisms and the interplay between epigenetics and metabolism may contribute to the initiation and progression of liver disease and cancer.
Introduction
The adult liver has extraordinary regenerative potential. This knowledge dates back to the myth of Prometheus, who was condemned to an eternal punishment caused by an eagle eating his liver, which would replenish itself every day. Indeed, after partial hepatectomy, which leads to removal of up to 70% of the liver, liver mass and functions become significantly restored within a few days. Mechanisms underlying restoration of liver mass and function in response to hepatectomy have been elegantly reviewed elsewhere (Michalopoulos and DeFrances, 1997; Michalopoulos, 2007). In this review, I focus on adult liver regeneration in response to tissue damage that can normally challenge the liver, in line with its central role in nutrient metabolism and detoxification in the adult body.
The adult liver is a slowly self-renewing organ and does not exhibit a defined stem-cell compartment that maintains homeostasis, opposite to other organs that have excellent regenerative capacity, including intestine, stomach, and skin. A great number of experimental models indicate that cellular plasticity, intended as the capacity of liver cells to adopt alternative fates and functions in response to environmental changes, determines efficient adult liver regeneration following damage (Tetteh et al., 2015; Aloia et al., 2016; Kopp et al., 2016; Gadd et al., 2020). A key question is: what molecular mechanisms regulate cellular plasticity and cell-fate changes to achieve efficient liver regeneration upon injury? In 1957, Conrad Hal Waddington postulated that the epigenetic landscape contributes to determine cell-fate decisions and acquisition of cell-identity during embryo development (Waddington, 1957). Increasing evidence indicates that the epigenetic landscape is dynamically regulated in the adulthood and allows cell-fate changes and cellular plasticity in adult tissues in response to environmental cues and tissue injury (Takahashi and Yamanaka, 2015; Rajagopal and Stanger, 2016; Flavahan et al., 2017; Paksa and Rajagopal, 2017). Of note, here I refer to epigenetic mechanisms as modifications of the chromatin (the ensemble of DNA and histones), which are involved in the regulation of gene transcription. Importantly, epigenetic mechanisms are inheritable by the cell progeny and reversible in response to environmental changes. One intriguing hypothesis is that dynamic regulation of epigenetic mechanisms might represent an efficient way for liver cells to respond to injury, repair the damaged tissue, and return to the homeostatic state once the injury is resolved. Supporting this hypothesis, our data indicated that transient epigenetic remodelling, mediated by the methylcytosine dioxygenase TET1, triggers liver cellular plasticity and contributes to liver regeneration following injury (Aloia et al., 2019). Here, I review the role of several chromatin modifiers in the regulation of cell-fate changes required for liver epithelial regeneration after injury in mouse models (Table 1). In addition, I address the interplay between epigenetics and metabolism in liver regeneration and chronic liver disease and discuss how epigenetic alterations may contribute to liver disease and cancer.
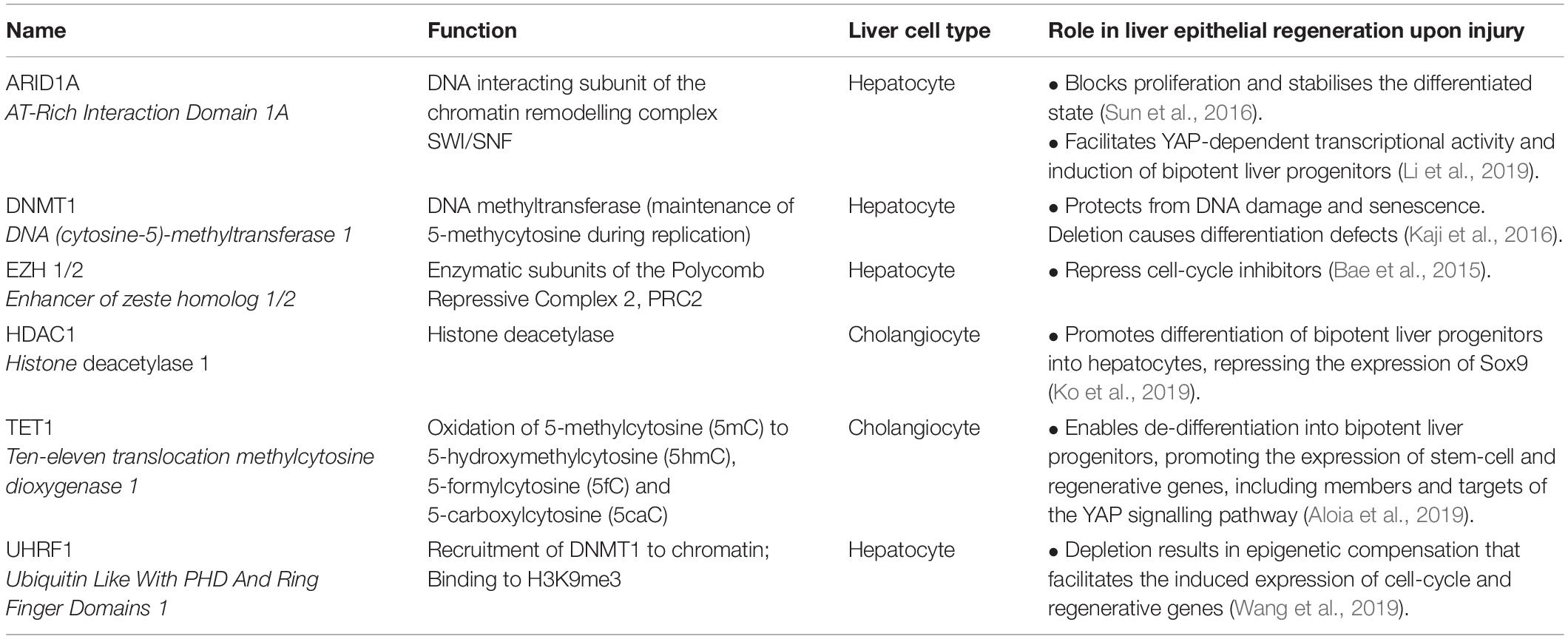
Table 1. Role of selected chromatin modifiers in liver epithelial regeneration following tissue injury.
Hepatocytes and Cholangiocytes Exhibit Remarkable Plasticity in Response to Liver Injury
Two epithelial cell types are present in the adult liver: (i) hepatocytes, which represent >60% of total liver cells and perform most of the metabolic and detoxification functions of the organ and (ii) cholangiocytes, which represent 3–5% of total liver cells and form biliary ducts that collect the bile produced by the hepatocytes and export it to the intestine for digestive purposes (Ober and Lemaigre, 2018). Both hepatocytes and cholangiocytes exhibit remarkable plasticity in response to injury. The role of epithelial plasticity in liver regeneration has been elegantly reviewed by Stuart Forbes and colleagues (Gadd et al., 2020).
Increasing evidence indicates that the vast majority of the hepatocytes, if not all, can acquire proliferation to ensure efficient restoration of liver mass and functions (Chen et al., 2020; Matsumoto et al., 2020; Monga, 2020; Sun et al., 2020). Upon liver injury induced by chemicals or toxins, hepatocytes can trans-differentiate into cholangiocytes (Yanger et al., 2013) or de-differentiate into bipotent liver progenitors, capable of generating both liver epithelial cell types (Tarlow et al., 2014; Yimlamai et al., 2014; Han et al., 2019). Liver injury can also trigger cholangiocyte proliferation and expansion in the liver parenchyma, a process known as ductular reaction (Roskams et al., 2004). Importantly, cholangiocytes can de-differentiate in vivo into bipotent liver progenitors that give rise to both hepatocytes and cholangiocytes (Itoh and Miyajima, 2014; Ko et al., 2020; So et al., 2020). The role of cholangiocytes in liver regeneration has been extensively debated. Several experimental models indicated that hepatocytes are solely responsible for restoration of the epithelial compartment after liver injury and the contribution of cholangiocytes is neglectable (Malato et al., 2011; Schaub et al., 2014; Yanger et al., 2014; Font-Burgada et al., 2015; Lin et al., 2018). However, different mouse models demonstrated that cholangiocytes significantly contribute to liver regeneration when hepatocytes are compromised. Data from Lu et al. (2015) indicated that cholangiocytes have liver repopulation capacity upon transplantation into adult mouse liver that shows extensive hepatocyte senescence and necrosis induced by genetic deletion of the E3 ubiquitin ligase Mdm2. Moreover, lineage-tracing experiments demonstrated that cholangiocytes can restore from 10 to 70% of total liver hepatocytes following diet-induced liver injury in mouse models exhibiting genetic deletion of β1-integrin (Raven et al., 2017) or β-catenin (Russell et al., 2018) or over-expression of p21 (Raven et al., 2017) in hepatocytes or after prolonged liver injury in the absence of genetic alterations that might compromise hepatocyte functions (Deng et al., 2018; Manco et al., 2019).
The capacity of both hepatocytes and cholangiocytes to acquire a bipotent progenitor state relies on their origin from a common embryonic progenitor, the hepatoblast (Zaret and Grompe, 2008; Ober and Lemaigre, 2018). Of note, chromatin modifications such as trimethylation of lysine 9 and 27 of histone H3 (H3K9me3 and H3K27me3) play a crucial role in the establishment of hepatoblast identity (Nicetto et al., 2019), supporting the relevance of epigenetic mechanisms in liver cell-identity and cell-fate decisions. In the following sections, I examine the role of several epigenetic modifiers in liver regeneration mediated by adult hepatocytes and cholangiocytes following tissue damage.
Epigenetic Mechanisms Regulating Hepatocyte-Mediated Liver Regeneration
The hepatocytes play a crucial role in adult liver regeneration after injury (Malato et al., 2011; Schaub et al., 2014; Yanger et al., 2014; Font-Burgada et al., 2015; Lin et al., 2018). Arid1a, a DNA interacting subunit of the SWI/SNF chromatin remodelling complex, has been shown to regulate hepatocyte plasticity in mouse models carrying liver-specific deletion of Arid1a (by using an Albumin-Cre/Arid1a flx/flx mouse line) or conditional deletion of Arid1a in hepatocytes (mediated by infection with an AAV-Cre in Arid1a flx/flx mice) (Sun et al., 2016; Li et al., 2019). Sun et al. (2016) found that Arid1a deletion increased hepatocyte proliferation and reduced necrosis, inflammation, and fibrosis in response to liver damage induced by carbon tetrachloride, CCl4. Moreover, Arid1a deletion resulted in accelerated weight recovery and extended survival following treatment with the toxic diet 3,5-diethoxycarbonyl-1,4-dihydrocollidine, DDC (6 weeks). Depletion of Arid1a following damage, in the recovering liver after a 12-week treatment with CCl4 or a 4-week treatment with DDC, reduced fibrosis and accelerated regain of normal weight. Thus, these data indicated that Arid1a deletion facilitated liver recovery from damage. At the molecular level, Sun et al. (2016) showed that Arid1a genomic binding might facilitate chromatin accessibility to key transcription factors that repress proliferation (e.g., E2F4) and maintain the hepatocyte differentiated cell state, such as C/EBPa and Hnf4a (Sun et al., 2016). Together, these findings suggested that Arid1a might impair liver regeneration by stabilising the hepatocyte differentiated state and reducing hepatocyte plasticity (Sun et al., 2016). This hypothesis is challenged by the findings of Li et al. (2019), who showed that conditional Arid1a deletion resulted in a significant reduction of Hnf4a+/Sox9+ bipotent liver progenitors in response to DDC treatment (2 weeks). At the molecular level, they found that Arid1a determined increased chromatin accessibility in the homeostatic liver, which facilitates YAP genomic binding after liver injury. YAP transcriptional activity was previously shown to promote hepatocyte de-differentiation into liver progenitors (Yimlamai et al., 2014). Consistently, Li et al. (2019) found that the number of YAP+/Sox9+ liver progenitors was significantly reduced in Arid1a-depleted mice upon over-expression of a constitutively active YAPS127A. Of note, they found that Arid1a deletion resulted in hepatomegaly, hepatocyte hypertrophy, and impaired liver functions upon DDC-mediated liver damage, whereas its depletion following DDC-induced damage did not affect liver recovery (Li et al., 2019). The different role of Arid1a in liver regeneration and recovery after injury reported by Sun et al. (2016) and Li et al. (2019) could be explained by the different length of DDC treatment and recovery, which might result in different levels of tissue injury at the time of the analyses performed (4–6 weeks DCC followed by 0–10 days recovery vs 2 weeks DDC followed by 8 weeks recovery, respectively). The different length of injury/recovery might result in a different timing of Arid1a binding to DNA and SWI/SNF-mediated chromatin remodelling activity. This, in turn, might influence the induction of hepatocyte de-differentiation into progenitors and their return to the homeostatic state. Of note, in their experimental conditions, Sun et al. (2016) reported increased hepatocyte proliferation following Arid1a depletion, whereas Li et al. (2019) did not observe a significant difference. This has important consequences on hepatocyte plasticity, since increased proliferation of mature hepatocytes is likely to determine faster regeneration, thus reducing the requirement of hepatocyte de-differentiation into progenitors.
To achieve efficient liver regeneration, it is crucial that adult hepatocytes maintain their functions during proliferation and cell division. Bae et al. (2015) found that EZH1 and EZH2 regulated hepatocyte proliferation and promoted increased mouse survival following CCl4. EZH1/2 are the catalytic subunits of the Polycomb repressive complex 2 (PRC2), which result in trimethylation of lysine 27 of histone 3 (H3K27me3), an epigenetic mark mainly associated to gene repression. Bae et al. (2015) found that key target genes of EZH1/2 are the cell-cycle inhibitors Cdkn2a and Cdkn2b, which became up-regulated in Ezh1/2 knock-out (KO) livers. Consistent with the importance of hepatocyte proliferation and division in liver regeneration, epigenetic regulation mediated by the maintenance DNA methyltransferase DNMT1 and its regulator UHRF1 has been shown to play an important role in hepatocyte viability and regenerative capacity (Kaji et al., 2016; Wang et al., 2019). Kaji et al. (2016) and Wang et al. (2019) performed liver-specific deletion of Dnmt1 and Uhrf1, respectively, crossing Albumin-Cre with specific flx/flx mouse lines. Wang et al. (2019) showed that Uhrf1 deletion resulted in global hypomethylation, which induced redistribution of repressive H3K27me3 from promoters to transposable elements to block their aberrant expression. This has implications in liver regeneration, since reduced H3K27me3 levels at promoter facilitated the induced expression of cell-cyle and regenerative genes (Wang et al., 2019). Kaji et al. (2016) showed that Dnmt1 deletion induced liver fibrosis and inflammation and resulted in hepatocyte senescence. Intriguingly, Dnmt1 deletion resulted in maturation defects and lack of expression of the cytochrome Cyp2e1, which is responsible for the bioactivation of CCl4, thus making Dnmt1 KO mice resistant to liver injury induced by CCl4 (Kaji et al., 2016). Of note, Bae et al. (2015) and Kaji et al. (2016) showed that impaired hepatocyte regenerative capacity or tissue damage caused by hepatocyte senescence and necrosis triggered ductular reaction.
These findings highlight the relevance of epigenetic mechanisms in the regulation of hepatocyte viability, proliferation, and plasticity. Further work will determine how dynamic epigenetic regulation influences the balance between proliferation of mature hepatocytes and hepatocyte de-differentiation into progenitors and how those two processes act to coordinate liver response to injury to achieve efficient regeneration.
Epigenetic Mechanisms Regulating Cholangiocyte-Mediated Liver Regeneration
Cholangiocyte expansion and de-differentiation into liver progenitors have been extensively reported in a variety of injury models in mammals (Okabe et al., 2009; Sackett et al., 2009; Dorrell et al., 2011; Shin et al., 2011; Espanol-Suner et al., 2012; Huch et al., 2013). Lineage-tracing experiments in mouse models demonstrated a crucial role for cholangiocytes in adult liver regeneration when hepatocyte response to injury is compromised by genetic alterations or following severe and prolonged liver damage (Raven et al., 2017; Deng et al., 2018; Russell et al., 2018; Manco et al., 2019).
What mechanisms regulate cholangiocyte plasticity? Our data demonstrated that upon liver injury, cholangiocytes undergo epigenetic remodelling mediated by the methylcytosine dioxygenase TET1 (Aloia et al., 2019). TET1/2/3 oxidise the repressive DNA mark 5-methylcytosine into 5-hydroxymethylcytosine (5hmC). 5hmC can act as a stable epigenetic mark associated with gene activation or represent an intermediate of complete de-methylation after further oxidation catalysed by TET proteins (Rasmussen and Helin, 2016). We found that TET1 was lowly expressed in homeostatic adult cholangiocytes and was up-regulated following DDC-mediated liver injury in vivo vs healthy liver, opposite to Tet2/3, which became down-regulated. By using a Tet1 hypomorphic mouse, we found that TET1 reduced levels impaired cholangiocyte proliferation after acute DDC treatment (5 days) and resulted in liver fibrosis after prolonged DDC-mediated liver injury (∼8 weeks including off-treatment intervals) (Aloia et al., 2019). Lineage-tracing experiments demonstrated that TET1 was required for the formation of cholangiocyte-derived hepatocyte regenerative clusters following DDC-mediated liver injury (Aloia et al., 2019), using a liver injury model developed by Raven et al. (2017). Together, our findings indicated that TET1 triggers cholangiocyte plasticity in response to liver injury and is required for efficient liver epithelial regeneration. At the molecular level, we found that TET1 regulates the expression of stem-cell genes that identify hepatoblasts (e.g., Lgr5; Prior et al., 2019) and adult human and mouse bipotent liver progenitors (e.g., Trop2; Okabe et al., 2009; Aizarani et al., 2019). Consistently, TET1 was required for the establishment of mouse intrahepatic cholangiocyte organoids, which exhibit induced expression of stem-cell genes (e.g., Lgr5 and Trop2) and can differentiate into hepatocytes (Huch et al., 2013). Moreover, genome-wide TET1 DamID-sequencing (van den Ameele et al., 2019) in intrahepatic cholangiocyte organoids revealed that TET1 regulates the expression of components and targets of signalling pathways required for liver regeneration, including YAP (Yimlamai et al., 2014; Pepe-Mooney et al., 2019; Planas-Paz et al., 2019). Together, our data suggested that TET1 enables de-differentiation of cholangiocytes into bipotent liver progenitors both in vitro and in vivo to ensure efficient liver regeneration and organoid formation (Aloia et al., 2019).
Of note, the increase in Tet1 levels after liver injury in vivo (day 3 following DDC treatment) was only transient, since Tet1 expression decreased at the peak of DDC-mediated liver damage (day 5). In line with this, we found that 5hmC levels were transiently increased in cholangiocytes in vivo at day 3 at the transcriptional start site of >3000 genes, which are mainly involved in biological processes such as development, chromatin modifications, and cell-cycle (Aloia et al., 2019). Therefore, this suggested that dynamic epigenetic remodelling in vivo allows cholangiocytes to both respond to liver injury and return to the homeostatic state to avoid the detrimental effects of an aberrant progenitor response. Supporting this hypothesis, Lgr5+ cells have been identified as tumour initiating cells in mouse liver cancer (Cao et al., 2020) and YAP can induce liver cancer (Dong et al., 2007).
What epigenetic mechanisms regulate cholangiocyte differentiation into hepatocytes? Following experiments in zebrafish that led to the identification of a regenerative role for Hdac1 by repressing the expression of Sox9 and modulating the Notch signalling, Ko et al. (2019) showed that the HDAC inhibitor MS-275 impaired cholangiocyte-mediated hepatocyte regeneration in a mouse model developed by Russell et al. (2018). Of note, MS-275 treatment did not affect cholangiocyte proliferation and ductular reaction (Ko et al., 2019), thus suggesting that Hdac1 specifically regulates cholangiocyte differentiation into hepatocytes without affecting cholangiocyte response to liver injury.
These findings highlight the relevance of dynamic epigenetic regulation of cholangiocyte plasticity for liver regeneration in response to severe or persistent liver injury that compromises the hepatocytes. Of note, ductular reaction is often observed in human chronic liver disease, concomitantly with massive hepatocyte alterations and necrosis (Ko et al., 2020). Therefore, understanding the epigenetic mechanisms that regulate cholangiocyte plasticity might indicate novel therapeutic strategies aimed at stimulating cholangiocyte regenerative capacity to ameliorate human chronic liver disease.
The Interplay Between Epigenetics and Metabolism in Liver Regeneration and Disease
Epigenetic mechanisms are often regulated by metabolic inputs (Etchegaray and Mostoslavsky, 2016). For example, TET1 epigenetic activity is dependent on 2-oxoglutarate, which is an intermediate of the Krebs cycle (Rasmussen and Helin, 2016). The adult liver is the central organ of the body for nutrient metabolism and presents metabolic zonation with specialised metabolic functions. Liver zonation follows decreasing oxygen and nutrient gradient across the liver lobule from the portal triad (formed by the hepatic artery, portal vein, and biliary ducts), to the central vein, where the blood meets the systemic circulation (Kietzmann, 2017). This suggests that different types and durations of liver injuries may affect different metabolic zones and trigger specific epigenetic mechanisms, which, in turn, promote distinct cellular response to damage. Importantly, metabolic alterations might cause chronic liver disease. For example, metabolic disorders are associated with the most common form of human chronic liver disease, non-alcoholic fatty liver disease (NAFLD). Patients with NAFLD exhibit increased fat accumulation and can present significant inflammation, named as non-alcoholic steatohepatitis (NASH) (Benedict and Zhang, 2017). At late stages, chronic liver disease is characterised by cirrhosis, which accounts for >1 million deaths per year worldwide and can predispose to liver cancer (Asrani et al., 2019). Therefore, it is reasonable to speculate that metabolic dysfunctions induce epigenetic alterations that, in turn, might impair liver regeneration capacity and exacerbate liver disease and favour its progression towards liver cirrhosis. The interplay between epigenetics and metabolism is bidirectional, since altered epigenetic mechanisms may also cause metabolic dysfunctions and trigger liver disease. A circadian rhythm was shown to determine chromatin recruitment of the histone deacetylase Hdac3, which in turn controls the expression of genes related to lipid metabolism, and determines the balance between gluconeogenesis and lipid synthesis (Feng et al., 2011; Sun et al., 2012). Consistently, Hdac3 deletion in mouse resulted in hepatic steatosis (Feng et al., 2011). In addition, Arid1a was found to regulate lipogenesis and fatty acid oxidation and its deletion was implicated in NASH in mouse (Fang et al., 2015; Moore et al., 2019).
Together, this suggests that investigating the complex interplay between epigenetics and metabolism will represent an important step to understand how the liver achieves efficient regeneration and identify the molecular mechanisms that influence initiation and progression of chronic liver disease.
Discussion
In this review, I have examined the role of several chromatin modifiers that regulate epithelial cell-fate changes to achieve efficient liver regeneration after injury in mouse models (Figure 1). It is worth mentioning that whether mouse models allow recapitulating several aspects of human liver regeneration and disease, species-specific difference should be taken in account, and the epigenetic mechanisms identified in mouse models should be validated also in human models such as cell lines, organoid systems, or primary specimens.
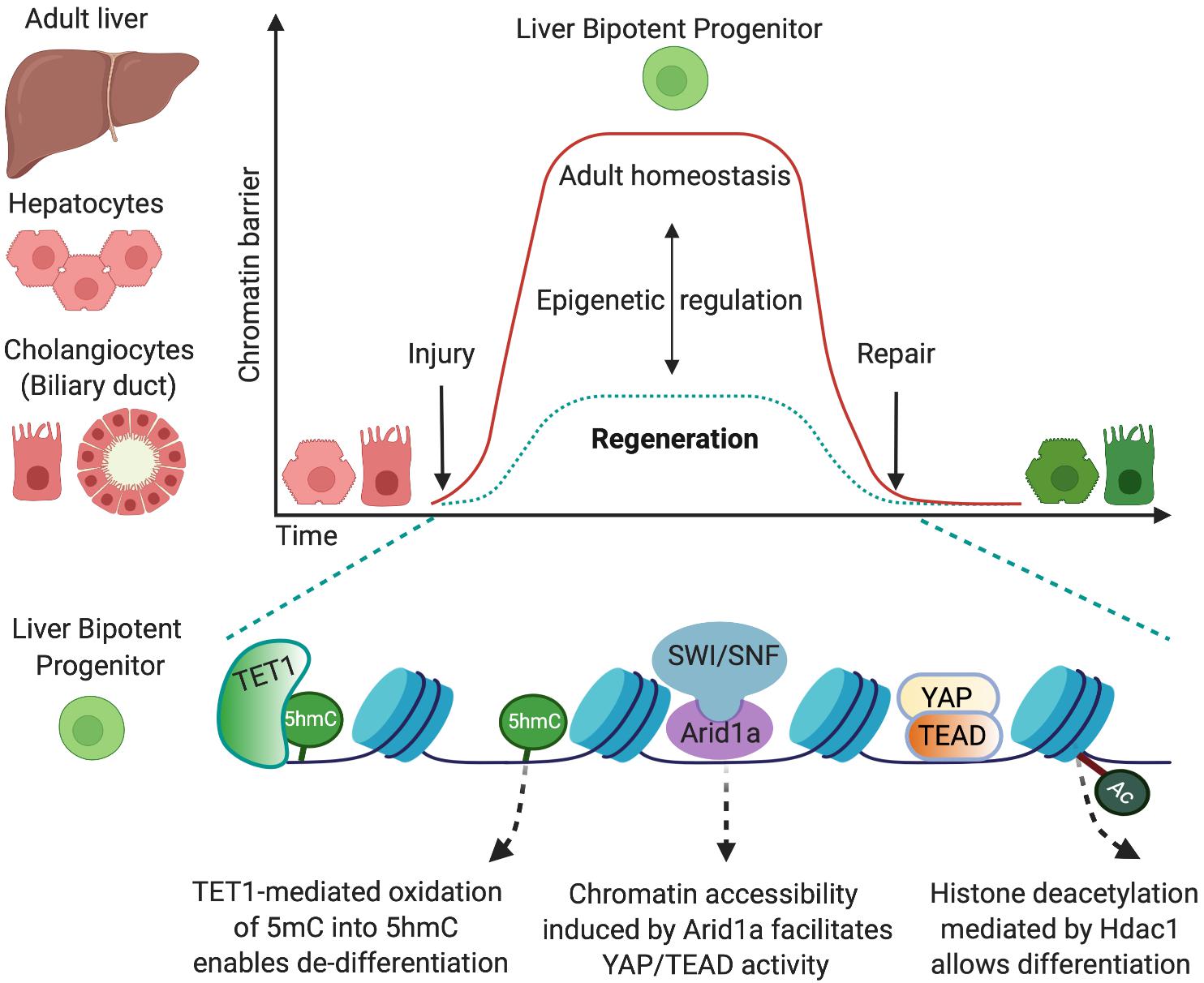
Figure 1. Epigenetic mechanisms that allow cell-fate changes into bipotent liver progenitors. The adult liver is formed by two epithelial cell types, hepatocytes and cholangiocytes, which derive from a common bipotent embryonic progenitor, the hepatoblast. In the adulthood, in homeostatic conditions, the epigenetic landscape preserves cell-identity by maintaining chromatin conformations that stabilise the differentiated state (red). Upon injury, both adult hepatocytes and cholangiocytes can de-differentiate into bipotent liver progenitors that give rise to both liver epithelial cell types (green) and restore the liver epithelial compartment. This cell-fate change into progenitors is enabled by epigenetic mechanisms that determine permissive chromatin states, including increased chromatin accessibility mediated by Arid1a and the chromatin remodelling complex SWI/SNF (Li et al., 2019) and oxidation of 5-methycytosine (5mC) into 5-hydromethylcytosyne (5hmC) mediated by the methylcytosine dioxygenase TET1 (Aloia et al., 2019). Such permissive states facilitate the chromatin binding of the transcriptional machinery (e.g., YAP/TEAD) that promotes the establishment of liver progenitor identity. Importantly, dynamic epigenetic regulation allows liver epithelial cells to return to the homeostatic state once the injury is resolved and the tissue is repaired. In this regard, TET1 and 5hmC levels are only transiently enriched in cholangiocytes at early stages upon liver injury (Aloia et al., 2019), and histone deacetylation mediated by Hdac1 is required for differentiation of cholangiocyte progenitors into hepatocytes (Ko et al., 2019).
The expression levels of chromatin modifiers and genomic distribution of their epigenetic marks are often altered in human chronic liver disease and liver cancer (Hardy and Mann, 2016). For example, loss of function mutations in ARID1A have been reported in >16% of human hepatocellular carcinoma (Guichard et al., 2012). Arid1a was shown to have a dual role as both oncogene and tumour suppressor in mouse, supporting the hypothesis that epigenetic mechanisms determine specific cellular phenotypes according to the context of the surrounding tissue. Specifically, Arid1a triggered liver cancer initiation by increasing reactive oxygen species, whereas its loss in pre-existing tumours promoted cancer growth and metastasis (Sun et al., 2017). UHRF1 is often over-expressed in human liver cancer and high UHRF1 levels are associated with a poor prognosis. Interestingly, UHRF1 over-expression was shown to induce hypomethylation in human liver cancer (Mudbhary et al., 2014), as observed upon its depletion in mouse, where it induced epigenetic compensation (Wang et al., 2019). Further studies will determine the relevance of dynamic epigenetic remodelling in human liver cancer.
Pharmacological interventions based on drugs targeting epigenetic activity have a huge potential for the development of therapeutic strategies in human disease and cancer (Cheng et al., 2019). Therefore, advances in our knowledge of the epigenetic mechanisms that regulate liver plasticity might open new avenues for the treatment of chronic liver disease and liver cancer. In this regard, 3D liver organoid cultures derived from adult liver biopsies or induced pluripotent stem cells have proven reliable models of homeostatic and regenerative cholangiocytes (Huch et al., 2013, 2015; Sampaziotis et al., 2017; Aloia et al., 2019; Rimland et al., 2020) and hepatocytes (Hu et al., 2018; Peng et al., 2018), steatohepatitis (Ouchi et al., 2019; Ramli et al., 2020), and primary liver cancer (Broutier et al., 2017; Nuciforo et al., 2018), thus representing promising platforms to uncover the molecular mechanisms underlying liver regeneration, disease, and cancer.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by MRC core funding to the MRC Laboratory for Molecular Cell Biology at University College London, award code (MC_U12266B).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
LA thanks Laura Broutier and Serena Mirra for critical reading of the manuscript. The figure was created with BioRender.com.
References
Aizarani, N., Saviano, A., Sagar, Mailly, L., Durand, S., Herman, J. S., et al. (2019). A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204. doi: 10.1038/s41586-019-1373-2
Aloia, L., McKie, M. A., and Huch, M. (2016). Cellular plasticity in the adult liver and stomach. J. Physiol. 594, 4815–4825. doi: 10.1113/jp271769
Aloia, L., McKie, M. A., Vernaz, G., Cordero-Espinoza, L., Aleksieva, N., van den Ameele, J., et al. (2019). Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 21, 1321–1333. doi: 10.1038/s41556-019-0402-6
Asrani, S. K., Devarbhavi, H., Eaton, J., and Kamath, P. S. (2019). Burden of liver diseases in the world. J. Hepatol. 70, 151–171. doi: 10.1016/j.jhep.2018.09.014
Bae, W. K., Kang, K., Yu, J. H., Yoo, K. H., Factor, V. M., Kaji, K., et al. (2015). The methyltransferases enhancer of zeste homolog (EZH) 1 and EZH2 control hepatocyte homeostasis and regeneration. FASEB J. 29, 1653–1662. doi: 10.1096/fj.14-261537
Benedict, M., and Zhang, X. (2017). Non-alcoholic fatty liver disease: an expanded review. World J. Hepatol. 9, 715–732. doi: 10.4254/wjh.v9.i16.715
Broutier, L., Mastrogiovanni, G., Verstegen, M. M., Francies, H. E., Gavarro, L. M., Bradshaw, C. R., et al. (2017). Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435. doi: 10.1038/nm.4438
Cao, W., Li, M., Liu, J., Zhang, S., Noordam, L., Verstegen, M. M. A., et al. (2020). LGR5 marks targetable tumor-initiating cells in mouse liver cancer. Nat. Commun. 11:1961.
Chen, F., Jimenez, R. J., Sharma, K., Luu, H. Y., Hsu, B. Y., Ravindranathan, A., et al. (2020). Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell 26, 27–33.e4.
Cheng, Y., He, C., Wang, M., Ma, X., Mo, F., Yang, S., et al. (2019). Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 4:62.
Deng, X., Zhang, X., Li, W., Feng, R. X., Li, L., Yi, G. R., et al. (2018). Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23, 114–122.e3.
Dong, J., Feldmann, G., Huang, J., Wu, S., Zhang, N., Comerford, S. A., et al. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133. doi: 10.1016/j.cell.2007.07.019
Dorrell, C., Erker, L., Schug, J., Kopp, J. L., Canaday, P. S., Fox, A. J., et al. (2011). Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 25, 1193–1203. doi: 10.1101/gad.2029411
Espanol-Suner, R., Carpentier, R., Van Hul, N., Legry, V., Achouri, Y., Cordi, S., et al. (2012). Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology 143, 1564–1575.e7.
Etchegaray, J. P., and Mostoslavsky, R. (2016). Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol. Cell 62, 695–711. doi: 10.1016/j.molcel.2016.05.029
Fang, J. Z., Li, C., Liu, X. Y., Hu, T. T., Fan, Z. S., and Han, Z. G. (2015). Hepatocyte-specific arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS One. 10:e0143042. doi: 10.1371/journal.pone.0143042
Feng, D., Liu, T., Sun, Z., Bugge, A., Mullican, S. E., Alenghat, T., et al. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319. doi: 10.1126/science.1198125
Flavahan, W. A., Gaskell, E., and Bernstein, B. E. (2017). Epigenetic plasticity and the hallmarks of cancer. Science 357:eaal2380. doi: 10.1126/science.aal2380
Font-Burgada, J., Shalapour, S., Ramaswamy, S., Hsueh, B., Rossell, D., Umemura, A., et al. (2015). Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779. doi: 10.1016/j.cell.2015.07.026
Gadd, V. L., Aleksieva, N., and Forbes, S. J. (2020). Epithelial Plasticity during liver injury and regeneration. Cell Stem Cell 27, 557–573. doi: 10.1016/j.stem.2020.08.016
Guichard, C., Amaddeo, G., Imbeaud, S., Ladeiro, Y., Pelletier, L., Maad, I. B., et al. (2012). Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698. doi: 10.1038/ng.2256
Han, X., Wang, Y., Pu, W., Huang, X., Qiu, L., Li, Y., et al. (2019). Lineage tracing reveals the bipotency of SOX9(+) hepatocytes during liver regeneration. Stem Cell Rep. 12, 624–638. doi: 10.1016/j.stemcr.2019.01.010
Hardy, T., and Mann, D. A. (2016). Epigenetics in liver disease: from biology to therapeutics. Gut 65, 1895–1905. doi: 10.1136/gutjnl-2015-311292
Hu, H., Gehart, H., Artegiani, B., LÖpez-Iglesias, C., Dekkers, F., Basak, O., et al. (2018). Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19.
Huch, M., Dorrell, C., Boj, S. F., van Es, J. H., Li, V. S., van de Wetering, M., et al. (2013). In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250. doi: 10.1038/nature11826
Huch, M., Gehart, H., van Boxtel, R., Hamer, K., Blokzijl, F., Verstegen, M. M., et al. (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312. doi: 10.1016/j.cell.2014.11.050
Itoh, T., and Miyajima, A. (2014). Liver regeneration by stem/progenitor cells. Hepatology 59, 1617–1626.
Kaji, K., Factor, V. M., Andersen, J. B., Durkin, M. E., Tomokuni, A., Marquardt, J. U., et al. (2016). DNMT1 is a required genomic regulator for murine liver histogenesis and regeneration. Hepatology 64, 582–598. doi: 10.1002/hep.28563
Kietzmann, T. (2017). Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 11, 622–630. doi: 10.1016/j.redox.2017.01.012
Ko, S., Russell, J. O., Molina, L. M., and Monga, S. P. (2020). Liver progenitors and adult cell plasticity in hepatic injury and repair: knowns and unknowns. Annu. Rev. Pathol. 15, 23–50. doi: 10.1146/annurev-pathmechdis-012419-032824
Ko, S., Russell, J. O., Tian, J., Gao, C., Kobayashi, M., Feng, R., et al. (2019). Hdac1 regulates differentiation of bipotent liver progenitor cells during regeneration via Sox9b and Cdk8. Gastroenterology 156, 187–202.e14.
Kopp, J. L., Grompe, M., and Sander, M. (2016). Stem cells versus plasticity in liver and pancreas regeneration. Nat. cell Biol. 18, 238–245. doi: 10.1038/ncb3309
Li, W., Yang, L., He, Q., Hu, C., Zhu, L., Ma, X., et al. (2019). A homeostatic arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated YAP signaling. Cell Stem Cell 25, 54–68.e5.
Lin, S., Nascimento, E. M., Gajera, C. R., Chen, L., Neuhofer, P., Garbuzov, A., et al. (2018). Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248. doi: 10.1038/s41586-018-0004-7
Lu, W. Y., Bird, T. G., Boulter, L., Tsuchiya, A., Cole, A. M., Hay, T., et al. (2015). Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 17, 971–983. doi: 10.1038/ncb3203
Malato, Y., Naqvi, S., Schurmann, N., Ng, R., Wang, B., Zape, J., et al. (2011). Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 121, 4850–4860. doi: 10.1172/jci59261
Manco, R., Clerbaux, L. A., Verhulst, S., Bou Nader, M., Sempoux, C., Ambroise, J., et al. (2019). Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J. Hepatol. 70, 1180–1191. doi: 10.1016/j.jhep.2019.02.003
Matsumoto, T., Wakefield, L., Tarlow, B. D., and Grompe, M. (2020). In Vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell 26, 34–47.e3.
Monga, S. P. (2020). No zones left behind: democratic hepatocytes contribute to liver homeostasis and repair. Cell Stem Cell 26, 2–3. doi: 10.1016/j.stem.2019.12.002
Moore, A., Wu, L., Chuang, J. C., Sun, X., Luo, X., Gopal, P., et al. (2019). Arid1a loss drives nonalcoholic steatohepatitis in mice through epigenetic dysregulation of hepatic lipogenesis and fatty acid oxidation. Hepatology 69, 1931–1945. doi: 10.1002/hep.30487
Mudbhary, R., Hoshida, Y., Chernyavskaya, Y., Jacob, V., Villanueva, A., Fiel, M. I., et al. (2014). UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209. doi: 10.1016/j.ccr.2014.01.003
Nicetto, D., Donahue, G., Jain, T., Peng, T., Sidoli, S., Sheng, L., et al. (2019). H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363, 294–297. doi: 10.1126/science.aau0583
Nuciforo, S., Fofana, I., Matter, M. S., Blumer, T., Calabrese, D., Boldanova, T., et al. (2018). Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. 24, 1363–1376. doi: 10.1016/j.celrep.2018.07.001
Ober, E. A., and Lemaigre, F. P. (2018). Development of the liver: insights into organ and tissue morphogenesis. J. Hepatol. 68, 1049–1062. doi: 10.1016/j.jhep.2018.01.005
Okabe, M., Tsukahara, Y., Tanaka, M., Suzuki, K., Saito, S., Kamiya, Y., et al. (2009). Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 136, 1951–1960. doi: 10.1242/dev.031369
Ouchi, R., Togo, S., Kimura, M., Shinozawa, T., Koido, M., Koike, H., et al. (2019). Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e6.
Paksa, A., and Rajagopal, J. (2017). The epigenetic basis of cellular plasticity. Curr. Opin. Cell Biol. 49, 116–122. doi: 10.1016/j.ceb.2018.01.003
Peng, W. C., Logan, C. Y., Fish, M., Anbarchian, T., Aguisanda, F., Alvarez-Varela, A., et al. (2018). Inflammatory cytokine TNFalpha promotes the long-term expansion of primary hepatocytes in 3D culture. Cell 175, 1607–1619.e15.
Pepe-Mooney, B. J., Dill, M. T., Alemany, A., Ordovas-Montanes, J., Matsushita, Y., Rao, A., et al. (2019). Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and regeneration. Cell Stem Cell 25, 23–38.e8.
Planas-Paz, L., Sun, T., Pikiolek, M., Cochran, N. R., Bergling, S., Orsini, V., et al. (2019). YAP, but Not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell 25, 39–53.e10.
Prior, N., Hindley, C. J., Rost, F., Melendez, E., Lau, W. W. Y., Gottgens, B., et al. (2019). Lgr5(+) stem and progenitor cells reside at the apex of a heterogeneous embryonic hepatoblast pool. Development 146:dev174557. doi: 10.1242/dev.174557
Rajagopal, J., and Stanger, B. Z. (2016). Plasticity in the adult: how should the waddington diagram be applied to regenerating tissues? Dev. Cell 36, 133–137. doi: 10.1016/j.devcel.2015.12.021
Ramli, M. N. B., Lim, Y. S., Koe, C. T., Demircioglu, D., Tng, W., Gonzales, K. A. U., et al. (2020). Human pluripotent stem cell-derived organoids as models of liver disease. Gastroenterology 159, 1471–1486.e12.
Rasmussen, K. D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. doi: 10.1101/gad.276568.115
Raven, A., Lu, W. Y., Man, T. Y., Ferreira-Gonzalez, S., O’Duibhir, E., Dwyer, B. J., et al. (2017). Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354. doi: 10.1038/nature23015
Rimland, C. A., Tilson, S. G., Morell, C. M., Tomaz, R. A., Lu, W. Y., Adams, S. E., et al. (2020). Regional differences in human biliary tissues and corresponding in vitro derived organoids. Hepatology doi: 10.1002/hep.31252 [Epub ahead of print].
Roskams, T. A., Theise, N. D., Balabaud, C., Bhagat, G., Bhathal, P. S., Bioulac-Sage, P., et al. (2004). Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology 39, 1739–1745. doi: 10.1002/hep.20130
Russell, J. O., Lu, W. Y., Okabe, H., Abrams, M., Oertel, M., Poddar, M., et al. (2018). Hepatocyte-specific beta-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 69, 742–759. doi: 10.1002/hep.30270
Sackett, S. D., Li, Z., Hurtt, R., Gao, Y., Wells, R. G., Brondell, K., et al. (2009). Foxl1 is a marker of bipotential hepatic progenitor cells in mice. Hepatology 49, 920–929. doi: 10.1002/hep.22705
Sampaziotis, F., Justin, A. W., Tysoe, O. C., Sawiak, S., Godfrey, E. M., Upponi, S. S., et al. (2017). Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 23, 954–963.
Schaub, J. R., Malato, Y., Gormond, C., and Willenbring, H. (2014). Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 8, 933–939. doi: 10.1016/j.celrep.2014.07.003
Shin, S., Walton, G., Aoki, R., Brondell, K., Schug, J., Fox, A., et al. (2011). Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 25, 1185–1192. doi: 10.1101/gad.2027811
So, J., Kim, A., Lee, S. H., and Shin, D. (2020). Liver progenitor cell-driven liver regeneration. Exp. Mol. Med. 52, 1230–1238. doi: 10.1038/s12276-020-0483-0
Sun, T., Pikiolek, M., Orsini, V., Bergling, S., Holwerda, S., Morelli, L., et al. (2020). AXIN2(+) pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell 26, 97–107.e6.
Sun, X., Chuang, J. C., Kanchwala, M., Wu, L., Celen, C., Li, L., et al. (2016). Suppression of the SWI/SNF component arid1a promotes mammalian regeneration. Cell Stem Cell 18, 456–466. doi: 10.1016/j.stem.2016.03.001
Sun, X., Wang, S. C., Wei, Y., Luo, X., Jia, Y., Li, L., et al. (2017). Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 32, 574–589.e6.
Sun, Z., Miller, R. A., Patel, R. T., Chen, J., Dhir, R., Wang, H., et al. (2012). Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 18, 934–942. doi: 10.1038/nm.2744
Takahashi, K., and Yamanaka, S. (2015). A developmental framework for induced pluripotency. Development 142, 3274–3285. doi: 10.1242/dev.114249
Tarlow, B. D., Pelz, C., Naugler, W. E., Wakefield, L., Wilson, E. M., Finegold, M. J., et al. (2014). Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618. doi: 10.1016/j.stem.2014.09.008
Tetteh, P. W., Farin, H. F., and Clevers, H. (2015). Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 25, 100–108. doi: 10.1016/j.tcb.2014.09.003
van den Ameele, J., Krautz, R., and Brand, A. H. (2019). TaDa! analysing cell type-specific chromatin in vivo with Targeted DamID. Curr. Opin. Neurobiol. 56, 160–166. doi: 10.1016/j.conb.2019.01.021
Waddington, C. (1957). “A discussion of some aspects of theoretical biology,” in The Strategy of the Genes, ed. C. H. Waddington (London: London George Allen and Unwin).
Wang, S., Zhang, C., Hasson, D., Desai, A., SenBanerjee, S., Magnani, E., et al. (2019). Epigenetic compensation promotes liver regeneration. Dev. Cell 50, 43–56.e6.
Yanger, K., Knigin, D., Zong, Y., Maggs, L., Gu, G., Akiyama, H., et al. (2014). Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 15, 340–349. doi: 10.1016/j.stem.2014.06.003
Yanger, K., Zong, Y., Maggs, L. R., Shapira, S. N., Maddipati, R., Aiello, N. M., et al. (2013). Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 27, 719–724. doi: 10.1101/gad.207803.112
Yimlamai, D., Christodoulou, C., Galli, G. G., Yanger, K., Pepe-Mooney, B., Gurung, B., et al. (2014). Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338. doi: 10.1016/j.cell.2014.03.060
Keywords: plasticity, cell-fate change, epigenetics, liver cancer, chronic liver disease, liver regeneration, liver progenitor, metabolism
Citation: Aloia L (2021) Epigenetic Regulation of Cell-Fate Changes That Determine Adult Liver Regeneration After Injury. Front. Cell Dev. Biol. 9:643055. doi: 10.3389/fcell.2021.643055
Received: 17 December 2020; Accepted: 05 February 2021;
Published: 01 March 2021.
Edited by:
Justin Brumbaugh, University of Colorado Boulder, United StatesReviewed by:
Kirsten Sadler, New York University Abu Dhabi, United Arab EmiratesWei-Yu Lu, University of Birmingham, United Kingdom
Copyright © 2021 Aloia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luigi Aloia, l.aloia@ucl.ac.uk
 Luigi Aloia
Luigi Aloia