Clonal Relationship and Resistance Profiles Among ESBL-Producing Escherichia coli
- 1Department of Microbiology, Faculty of Advanced Sciences and Technology, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
- 2Department of Microbiology, Research Center of Reference Health Laboratory, Ministry of Health and Medical Education, Tehran, Iran
- 3Division of Microbiology, Department of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
AmpC β-lactamases hydrolyze all β-lactams except cefepime and carbapenems. The study of AmpC-producing E. coli has high priority for the infection control committee. This research is aimed to investigate the resistant urinary AmpC-generating E. coli isolates and identify their genetic variety. Some 230 E. coli isolates from patients suffering urinary tract infection symptoms were studied in 2017–2018 to assess their susceptibility toward antimicrobial agents. AmpC gene was evaluated by PCR and molecular typing using the 10-loci MLVA method. MLVA images were examined by BioNumerics 6.6 software through the use of the UPGMA algorithms. Thirty-eight AmpC-generating E. coli isolates were detected. The most abundant determinant was blaCIT and blaEBC, blaFOX, and blaDHA had the next ranks, respectively. Six major clusters and a singleton were identified by MLVA. AmpC beta-lactamases in urinary isolates of E. coli in the hospital under study and high rate of additional resistance to gentamicin, cotrimoxazole and ciprofloxacin. The most frequent gene determinant of AmpC beta-lactamase was blaCIT and vary depending on time and geographical location.
Background
Escherichia coli is the first infectious agent in the development of urinary tract infection (UTI) (Connie et al., 2014) accounting for more than 85% UTI cases (Queenan and Bush, 2007). One hundred and fifty million cases of urinary tract infection are annually reported throughout the globe (Dallenne et al., 2010). It is 30 times more prevalent among females and almost 60% of women have the experience of UTI at least once in their lives (Dallenne et al., 2010). Antimicrobial agents can greatly contribute to the clinical control of UTI. Antimicrobial resistance, in particular, multidrug-resistant Gram-negative bacilli, has posed a remarkable challenge for the clinical settings (Saffar et al., 2016). Diverse drug resistance mechanisms have been introduced in Gram-negative bacteria among which, extended-spectrum β-lactamases (ESBL) production, AmpC β-lactamase generation, efflux, and porin deficiency can be mentioned. AmpC β-lactamases and ESBLs can be identified in the clinical laboratories. AmpC β-lactamases are capable of causing resistance to both narrow- and broad-spectrum cephalosporins, β-lactam/β-lactamase inhibitor combinations, and aztreonam (Xu et al., 2015). They can be chromosomally modified or plasmid-mediated. The plasmid-mediated AmpC β-lactamases can hydrolyze the entire β-lactam antibiotics, except Cefepime and carbapenems. The plasmid-mediated AmpC genes are originated from inducible chromosomal genes mobilized in different organisms. blaACC, blaFOX, blaMOX, blaDHA, blabla, blaCIT, and blaEBC are among the well-known gene determinants (Turnidge, 1998; Nikaido and Pagès, 2012; Nakano et al., 2017). AmpC detection plays a crucial role in the clinical management of infections as it can offer epidemiological data. Despite specific tests to detect beta-lactamases such as using inhibitor based test with phenylboronic acid (PBA) or cloxacillin, there is no mandatory guideline for their diagnosis in clinical laboratories (Hopkins et al., 2006). Considering the scarce epidemiological data, this study is aimed to assess the frequency of blaACC, blaFOX, blaMOX, blaDHA, blaCIT, and blaEBC genes in the urinary AmpC-generating E. coli strains collected from the patients with UTI. The second goal is the determination of genetic diversity in the AmpC-generating strains.
Material and Methods
During 2017–2018, we collected 230 isolates of AmpC and ESBL-producing E. coli from patients with UTI symptoms who were hospitalized at Milad hospital. The Milad hospital is a 1,000-bed tertiary care hospital, affiliated with the social assurance organization. The isolates were identified using standard biochemical tests. Antimicrobial susceptibility testing was performed by the E-test (MIC method). All isolates were stored at −80°C in tryptic soy broth containing 15% glycerol for future molecular studies.
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility tests were conducted on E. coli isolates according to the E-test method (Liofilchem® MIC Test Strips). The MICs (minimum inhibitory concentration) of eight antibiotics (ceftazidime, ceftriaxone, gentamicin, meropenem, ciprofloxacin, piperacillin/tazobactam, and trimethoprim/sulfamethoxazole) against the E. coli isolates were investigated. The results were recorded based on M100-CLSI 2019 guideline and categorized into three groups: resistant, intermediate, and susceptible.
Detection of AmpC-Positive Isolates
The screening tests were carried out on isolated E. coli strains to detect AmpC-Positive Isolates by the use of cefoxitin (30 µg) disk. Subsequently, the suspected strains were further verified by an AmpC detection set (MAST DISCS™ID, UK). AmpC detection set has been validated for identification of AmpC β-lactamase. E. coli ATCC 25922 and Enterobacter cloacae ATCC 13047 served as negative and positive controls, respectively, for the production of AmpC β-lactamase. The findings of Antimicrobial susceptibility testing (AST) and AmpC detection tests were entered in WHONET software.
Detection of ESBL-Positive Isolates
The ESBL-resistance E. coli isolates were confirmed using the combination disc based on CLSI guidelines (CLSI 2019). In a typical procedure, a ceftazidime disc (CAZ) (30 μg) alone and one in combination with clavulanic acid (CAC) (30/10 μg) were applied to confirm the isolates. Both discs were placed on a Mueller Hinton Agar (MHA) plate followed by overnight incubation at 37°C. The distance between the centers of the discs was set to 20 mm. ESBL-positive involved A ≥5 mm increase in zone diameter of either antimicrobial agents combined with clavulanic acid as compared to their zone upon the use of antimicrobial agents alone. K. pneumoniae ATCC 700603 and E. coli ATCC 25922 were utilized as ESBL-positive and negative controls, respectively.
PCR Reactions for AmpC-Producing Genes
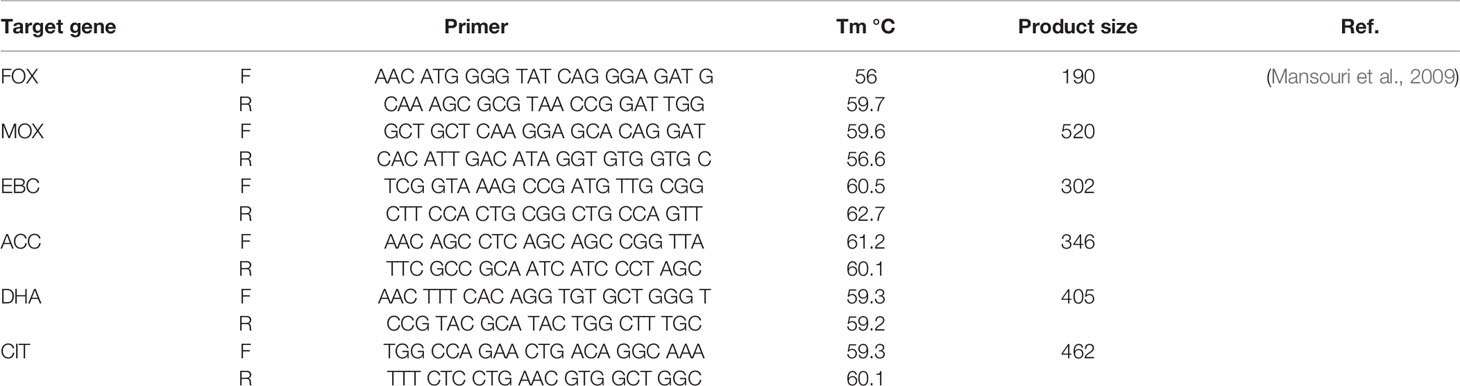
Genomic DNA extraction was carried out by High Pure PCR Template Preparation kit (Roche, Germany). The detection of AmpC- β-lactamase and ESBL genes in urinary E. coli isolates were conducted by PCR employing the specific oligonucleotide primers as presented in Table 1. PCR amplification was achieved in a Peqlab PCR thermal cycler with the PCR Master Mix (Ampliqon Inc., Denmark) following the manufacturer’s guidelines. The PCR reactions were carried out on 25 µl volume. The master mix included 12 µl Master Mix Red (Ampliqon Inc., Denmark) and 1 µl target DNA. Forward and reverse primers were added as mentioned in Table 1. The reaction volume was increased to 25 μl by adding sterile distilled water. The addition of forward and reverse primers and temperature profile was performed by the Queenan’s protocol (Mansouri et al., 2009).
Multi-Locus Variable Number of Tandem Repeats Analysis (MLVA)
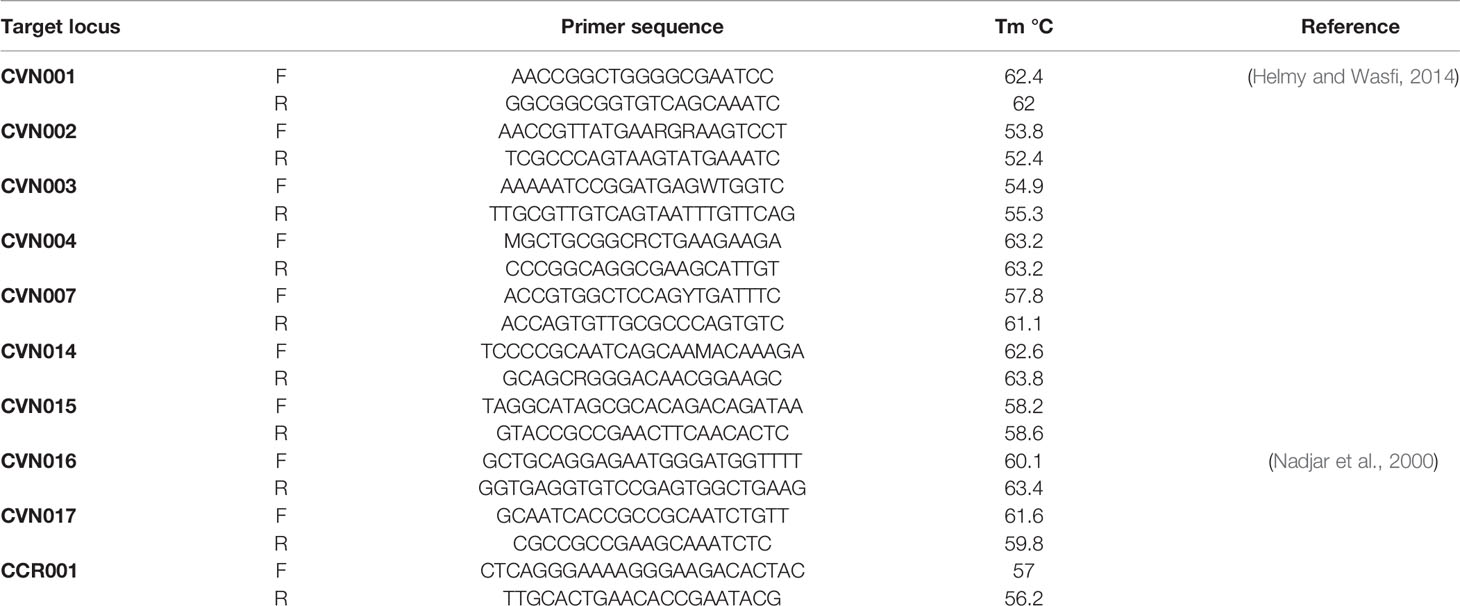
The complete E. coli DNA was provided from overnight cultured samples utilizing High Pure PCR Template Preparation Kit (Roche, Germany). E. coli MLVA was conducted by the seven tandem sequence repeats (CVN001, CVN002, CVN003, CVN004, CVN007, CVN014, CVN015) according to Lindstedt et al. (Helmy and Wasfi, 2014). Moreover, CVN016, CVN017, and a regularly-interspersed short palindromic repeat (CCR001), which have been reported to enhance the discriminatory power of the MLVA, were included with 10-loci E. coli MLVA (Nadjar et al., 2000). Table 2 lists the applied primers. Repeats were amplified by PCR and assessed on 3% agarose. Here, the size of PCR products was simply assayed on agarose gel with no complication using 100 bp and 20 bp size markers (Bio-Rad Laboratories Inc.).
The following formula was employed to determine the VNTR (Variable-Number Tandem Repeat) repeat numbers of each locus; ((NPS − OF)/RL, in which PS = product size, OF = offset region (region of repeat-free sequence) and RL = length of one repeat unit (Tan et al., 2009).
Results
Among 230 urinary E. coli isolates, 87 samples were from the male cases, while the remaining 142 were sampled from female subjects. The patients’ age varied one to 93 years (average: 58.81 ± 25.07).
Phenotypic Detection of AmpC- and ESBL-Producing Isolates
Thirty-eight AmpC-producing E. coli isolates were detected by cefoxitin (30 µg) among which, 35 isolates were confirmed by the AmpC detecting test (MASTDISCS™ID, UK). Among these 35 AmpC-producing E. coli isolates, nineteen cases were AmpC and ESBL coproducer; while sixteen isolates were AmpC-positive and non-ESBL.
About 40% (14/35) of AmpC-producing isolates showed resistance toward gentamicin. Some 65.7% (23/35) and 74.2% (26/35) of the studied isolates were resistant against trimethoprim-sulfamethoxazole and ciprofloxacin, respectively.
As suggested by Figure 1, no blaMOX and blaECC genes were detected in uropathogenic E. coli isolates, while, 73.6% of the tested samples contained blaCIT. blaFOX (blaFOX-1 to blaFOX-5), blaDHA (blaDH7 and blaDHA-2) and blaEBC were detected in 10.5, 10.5 and 15.8% of AmpC-producing E. coli isolates, respectively. Four isolates had both blaCIT and blaEBC. blaCIT and blaFOX were simultaneously present in four isolates. Two isolates also possessed both blaCIT and blaDHA. The blaCIT group of AmpC β-lactamases prevalently encompassed blaLAT-1 to blabla, blaBIL-1, blaCMY-2 to blaCMY-7, blaCMY-12 to blaCMY-18, and blaCMY-21 to blaCMY-23 (Dallenne et al., 2010). AmpC and ESBL-coproducing isolates had blaCIT, and four isolates having blaCIT and blaEBC, were AmpC and ESBL-coproducer.
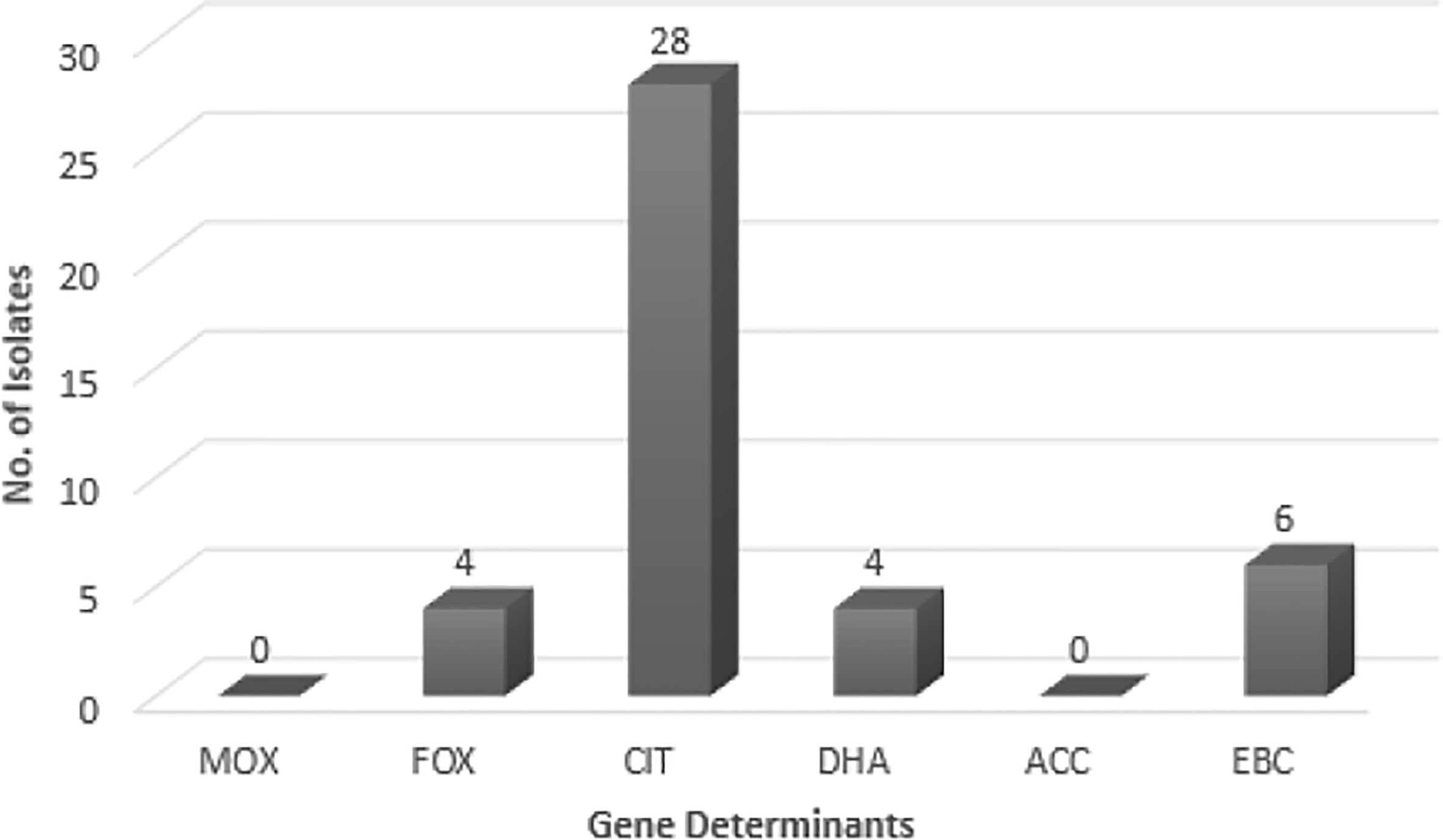
Figure 1 The frequency of AmpC genes in E. coli isolates. MOX, Active on Moxalactam; FOX, Active on Cefoxitin; CIT, Firstly isolated from Citrobacter freundii; DHA, Firstly isolated at Dhahran Hospitals in Saudi Arabia; ACC, Ambler class C; EBC, Isolated from Enterobacter cloacae.
Typing of AmpC-Producing E. coli Isolates by MLVA
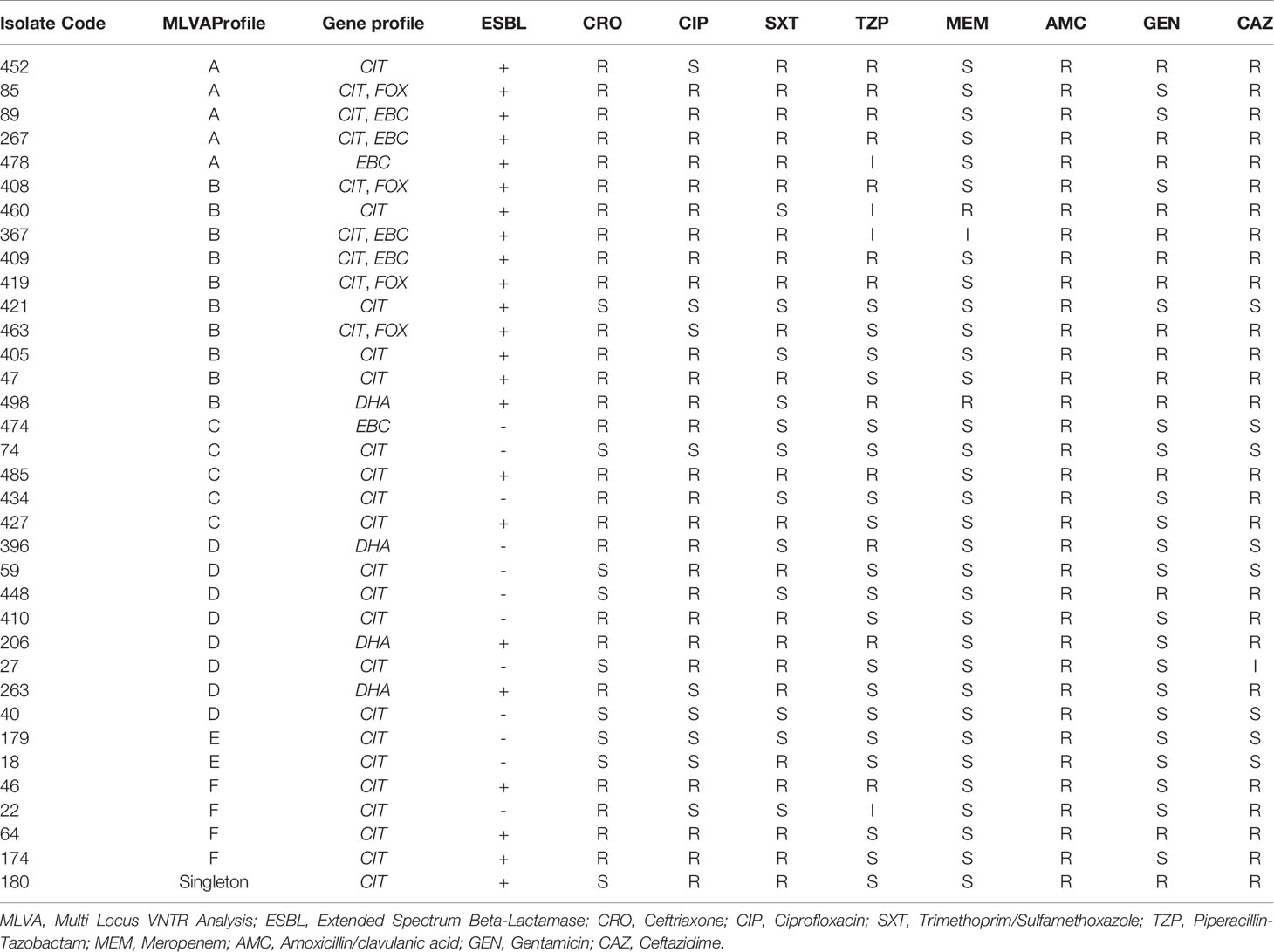
A dendrogram was formed based on 10-loci using BioNumerics software ver.6.6 and the 31 MLVA types created six major clusters (labeled A to F) by 10-loci based according to their genetic similarity (cutoff of 80%). Moreover, a singleton was detected in AmpC-producing E. coli isolate sampled from UTI patients. In the present research, 30 distinct genotypes were categorized by 10-loci MLVA typing of 35 AmpC-producing E. coli isolates (Figure 2).
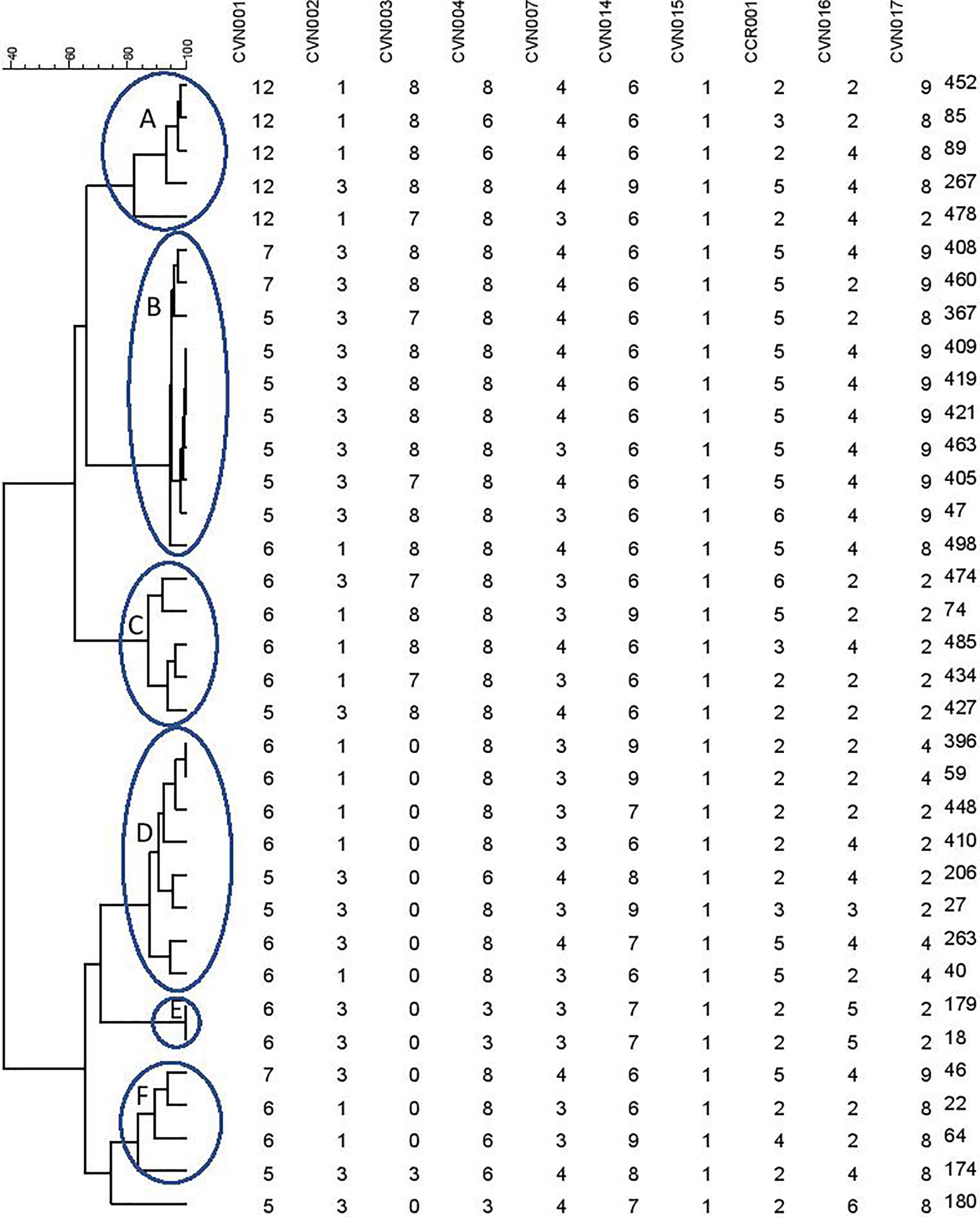
Figure 2 Clustering results of b-lactamase-producing E. coli using 10-loci MLVA (A–F clusters) differentiated UPGMA with the categorical coefficient of similarity.
As mentioned in Table 3, the resistance profile exhibited no significant relationship with MLVA pattern and the AmpC or ESBL production ability of E. coli.
Discussion
Plasmid-mediated AmpC β-lactamases have clinical significance due to causing serious challenges in the treatment procedure, drug resistance surveillance, epidemiological process, and infection management programs. In this context, the determination of the AmpC-producing isolates can greatly contribute to both surveillance and infection control for preventing nosocomial outbreaks and treatment failure (Saffar et al., 2016). β-Lactamase resistance is more prevalent in Enterobacterales compared to other gram-negative bacilli (Xu et al., 2015). Some factors can explain this condition: the extensive use of β-lactam antibiotics, in particular, broad-spectrum cephalosporins (Turnidge, 1998). No strategy is currently available to detect and confirm AmpC β-lactamases in clinical microbiology labs (Nikaido and Pagès, 2012). Nonetheless, the molecular evaluation of these isolates highlights its clinical detection. Thus, the molecular study of β-lactamases, in particular, AmpC type, is of crucial significance in terms of gene determinants and molecular epidemiology. Here, more than 54% (19.35) of E. coli isolates coproduced AmpC and ESBL and the remaining 46% of isolated E. coli showed only AmpC-producing features. It was previously reported that AmpCs β-lactamase is capable of hiding the second resistance phenotypes like ESBL (Nakano et al., 2017). As a result, the production of plasmid-mediated AmpC in an isolate may lead to false-negative results concerning ESBLs. These results can be helpful in epidemiological studies or infection management. The ESBL-suspected E. coli isolates must be tested for AmpC β-lactamases. In this research, screening tests on 16 E. coli isolates showed positive AmpC β-lactamase findings, meanwhile negative-ESBL results based on confirmation test (CAZ-CLA versus CAZ). About 40% (14.35) of AmpC-producing isolates exhibited resistance upon exposure to gentamicin. Moreover, 65.7% (23.35) and 74.2% (26.35) isolates were resistant against trimethoprim-sulfamethoxazole and ciprofloxacin, respectively.
Observing plasmids possessing AmpC-encoding genes next to other resistance genes to other antibiotics can be alarming for treatment setting (Hopkins et al., 2006). Based on antimicrobial susceptibility assays, AmpC-positive isolates exhibited a great resistance to cephalosporins when compared with their AmpC-negative counterparts, some of them showed resistance to aminoglycosides and quinolones (Table 3). This highlights the significance of detecting AmpC-producing isolates. This important issue should be considered by clinicians when using cephalosporins. The detection of the AmpC-producing isolates could influence antimicrobial therapy.
The AmpC-encoding genes such as blaFOX, blaCIT, and blaEBC have been found in previous works (Mansouri and Hoseini), while blaACC class was not reported in any of them (Mansouri et al., 2009; Nahid Hoseini et al., 2017). blaFOX family showed the highest prevalence in these works. While blaMOX class was not detected in the Mansouri’s research, Hoseini reported the presence of blaFOX and blaCIT. Other studies in different countries (e.g. Egypt) indicated 57.7% Gram-negative AmpC-producing bacilli among which, 22 isolates carried blaMOX, blaFOX, and blaCIT families (Mansouri et al., 2009; Helmy and Wasfi, 2014; Nahid Hoseini et al., 2017). In Egypt, Helmy (Helmy and Wasfi, 2014) reported the blaCMY homologs as the most dominant gene (86.9%); blaDHA (21.7%), blaFOX (17.3%), blaEBC (13%), and blaMOX (13%) had the next ranks. Helmy’s study did not observe the blaACC class. In this research, however, blaCIT had the highest frequency (73.7%) among the AmpC-encoding families; blaEBC (15.8%), blaFOX (10.5%), and blaDHA (10.5%) allocated the subsequent ranks, respectively. No members of blaACC class were found. This enzyme could not be fully inhibited by cefoxitin (Nadjar et al., 2000; Helmy and Wasfi, 2014). Thean et al. (Tan et al., 2009) found that screening to find cefoxitin resistance has a lower sensitivity toward blaACC family. Therefore, cefoxitin resistance could be exploited as a screening approach for the differentiation of blaACC enzymes.
The geographical region and the duration of the research also affected the prevalence and type of plasmid-mediated AmpCs (Ruppé et al., 2006; Adler et al., 2007). Numerous clinical microbiological laboratories fail in diagnosing the resistance mechanisms (Kashef et al., 2012). In the cases of simultaneous various antibiotics-resistance mechanisms, the phenotypic detection of AmpC enzymes could be very difficult; thus, molecular methods are recommended (Moland et al., 2008). According to our findings, AmpC β-lactamases can be related to false results of antimicrobial susceptibility tests for cephalosporins, therefore, the detection of AmpC-producing isolates is important for ESBL-negative isolates (Moland et al., 2008; Gupta et al., 2014).
The MLVA method was also employed to determine the genotypes of AmpC-producing E. coli. Three isolates were lost; hence, the remaining thirty-five AmpC-producing E. coli isolates were evaluated to indicate their clonal association by MLVA approach. MLVA showed thirty-one distinct patterns of AmpC-producing E. coli isolates which can be classified into six clusters and one singleton (Cutoff 80%). The singleton and MLVA type E isolates may be transmitted to the hospital by the patients, visitors, or medical staff as they had not been established in the hospital. MLVA Type B and MLVA Type D showed the highest prevalence and MLVA Type A, MLVA Type C, and MLVA Type F had the subsequent ranks, respectively. The mentioned MLVA types could be nosocomial infections; highlighting the necessity of evaluating MLVA types in the strains isolated from the patients and hospital staff. The two most prevalent MLVA types (Types A and B) exhibited two determinant genes of AmpC enzymes (blaCIT-blaEBC and blaCIT-blaFOX), whereas the other MLVA types only held one AmpC-encoding gene (Table 1). These results suggest the existence of the plasmids containing two genes in MLVA types A and B which can be transmitted by conjugating with other isolates. Thus, it is essential to identify these isolates and determine their transmission routes to establish a proper infection control strategy to prevent the release of resistance genes.
The determination of the frequency of AmpC-producing urinary E. coli isolates plays an important role in the antibiotic resistance and infection management programs in hospitals. The appearance of plasmid-mediated AmpC-producing E. coli can result in the spread of antibiotics resistance in the clinical settings. This dissemination can occur because of plasmid-mediated genes that may serve as the reservoir for antibiotic resistance. Therefore, the study of common gene determinants of AmpC resistance and its distribution in the bacterial species is of urgent necessity.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Islamic Azad University in IRAN (registration number IR.IAU.PS.REC.1397.306). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AD: study design and implementation and funding. SH, MR, MF and MD: scientific support. MF: supplier of laboratory site and equipment. MR: study management. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the kind collaboration of all the staff in Milad hospital and Iranian reference health laboratory for technical supports and data collection.
References
Adler, H., Fenner, L., Walter, P., Hohler, D., Schultheiss, E., Oezcan, S., et al. (2007). Plasmid-Mediated AmpC β-Lactamases in Enterobacteriaceae Lacking Inducible Chromosomal AmpC Genes: Prevalence At a Swiss University Hospital and Occurrence of the Different Molecular Types in Switzerland. J. Antimicrob. Chemother. 61 (2), 457–458. doi: 10.1093/jac/dkm472
Connie, R., Mahon, D. C. L., Manuselis, G. (2014). Textbook of Diagnostic Microbiology-E-Book (Netherlands: Elsevier Health Sciences).
Dallenne, C., Da Costa, A., Decré, D., Favier, C., Arlet, G. (2010). Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65 (3), 490–495. doi: 10.1093/jac/dkp498
Gupta, G., Tak, V., Mathur, P. (2014). Detection of AmpC β Lactamases in Gram-Negative Bacteria. J. Lab. Physicians 6 (1), 1–6. doi: 10.4103/0974-2727.129082
Helmy, M. M., Wasfi, R. (2014). Phenotypic and Molecular Characterization of Plasmid Mediated AmpC β-Lactamases Among Escherichia Coli, Klebsiella Spp., and Proteus Mirabilis Isolated From Urinary Tract Infections in Egyptian Hospitals. BioMed. Res. Int. 2014, 171548. doi: 10.1155/2014/171548
Hopkins, K. L., Deheer-Graham, A., Karisik, E., Batchelor, M. J., Liebana, E., Threlfall, E. J. (2006). New Plasmid-Mediated AmpC Beta-Lactamase (CMY-21) in Escherichia Coli Isolated in the UK. Int. J. Antimicrob. Agents 28 (1), 80–82. doi: 10.1016/j.ijantimicag.2006.03.020
Kashef, M., Hosny, A. E.-D. M., Hosny, M. S. (2012). A Study on Occurrence of Plasmid Mediated AmpC ß-Lactamases Among Gram Negative Clinical Isolates and Evaluation of Different Methods Used for Their Detection. J. Appl. Sci. Res. 8 (4), 2280–2285.
Mansouri, S., Chitsaz, M., Haji Hosseini, R., Mirzaei, M., Gheyni, M. H. (2009). Determination of Resistance Pattern of Plasmid-Mediated Ampc β–Lactamases Producing Isolate of Escherichia Coli. Daneshvar Med. 16 (80), 61–70.
Moland, E. S., Kim, S.-Y., Hong, S. G., Thomson, K. S. (2008). Newer β-Lactamases: Clinical and Laboratory Implications, Part II* *Editor’s Note: Part I of This Article Was Published in the May 15, 2008 Issue of CMN (Vol. 30, No. 10). Clin. Microbiol. Newslett. 30 (11), 79–85. doi: 10.1016/j.clinmicnews.2008.05.001
Nadjar, D., Rouveau, M., Verdet, C., Donay, J.-L., Herrmann, J.-L., Lagrange, P. H., et al. (2000). Outbreak of Klebsiella Pneumoniae Producing Transferable AmpC-Type β-Lactamase (ACC-1) Originating From Hafnia Alvei. FEMS Microbiol. Lett. 187 (1), 35–40. doi: 10.1016/S0378-1097(00)00166-X
Nahid Hoseini, I. S., Nejad, A. S. M., Alikhani, M. Y. (2017). Phenotypic and Genotypic Detection of AmpC Enzymes in Clinical Isolates of Escherichia Coli and Klebsiella Pneumoniae. J. Krishna Inst. Med. Sci. Univ. 6 (3), 10–18.
Nakano, R., Nakano, A., Yano, H., Okamoto, R. (2017). Role of AmpR in the High Expression of the Plasmid-Encoded Ampc β-Lactamase CFE-1. mSphere 2 (4), e00192–17. doi: 10.1128/mSphere.00192-17
Nikaido, H., Pagès, J. M. (2012). Broad-Specificity Efflux Pumps and Their Role in Multidrug Resistance of Gram-Negative Bacteria. FEMS Microbiol. Rev. 36 (2), 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Queenan, A. M., Bush, K. (2007). Carbapenemases: The Versatile Beta-Lactamases. Clin. Microbiol. Rev. 20 (3), 440–458, table of contents. doi: 10.1128/CMR.00001-07
Ruppé, E., Bidet, P., Verdet, C., Arlet, G., Bingen, E. (2006). First Detection of the Ambler Class C 1 AmpC Beta-Lactamase in Citrobacter Freundii by a New, Simple Double-Disk Synergy Test. J. Clin. Microbiol. 44 (11), 4204–4207. doi: 10.1128/JCM.00918-06
Saffar, H., Asgari Niaraki, N., Ghahroudi Tali, A., Baseri, Z., Abdollahi, A., Yalfani, R. (2016). Prevalence of AmpC β-Lactamase in Clinical Isolates of Escherichia Coli, Klebsiella Spp., and Proteus Mirabilis in a Tertiary Hospital in Tehran, Iran. Jundishapur J. Microbiol. 9 (12), e39121. doi: 10.5812/jjm.39121
Tan, T. Y., Ng, L. S., He, J., Koh, T. H., Hsu, L. Y. (2009). Evaluation of Screening Methods to Detect Plasmid-Mediated AmpC in Escherichia Coli, Klebsiella Pneumoniae, and Proteus Mirabilis. Antimicrob. Agents Chemother. 53 (1), 146–149. doi: 10.1128/AAC.00862-08
Turnidge, J. D. (1998). The Pharmacodynamics of Beta-Lactams. Clin. Infect. Dis. 27 (1), 10–22. doi: 10.1086/514622
Keywords: E. coli, AmpC, molecular typing, resistance, gene
Citation: Dolatyar Dehkharghani A, Haghighat S, Rahnamaye Farzami M, Rahbar M and Douraghi M (2021) Clonal Relationship and Resistance Profiles Among ESBL-Producing Escherichia coli. Front. Cell. Infect. Microbiol. 11:560622. doi: 10.3389/fcimb.2021.560622
Received: 09 May 2020; Accepted: 19 May 2021;
Published: 23 June 2021.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Krisztina M. Papp-Wallace, Louis Stokes Cleveland VA Medical Center, United StatesBranka Bedenić, University of Zagreb, Croatia
Copyright © 2021 Dolatyar Dehkharghani, Haghighat, Rahnamaye Farzami, Rahbar and Douraghi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Rahbar, rahbar.reflab@gmail.com
 Alireza Dolatyar Dehkharghani
Alireza Dolatyar Dehkharghani Setareh Haghighat
Setareh Haghighat Marjan Rahnamaye Farzami2
Marjan Rahnamaye Farzami2