CEACAM1 Activation by CbpF-Expressing E. coli
- 1The Institute of Dental Sciences, The Hebrew University-Hadassah School of Dental Medicine, Jerusalem, Israel
- 2The Concern Foundation Laboratories at the Lautenberg Center for General and Tumor Immunology, Department of Immunology and Cancer Research, Institute for Medical Research Israel Canada (IMRIC), Faculty of Medicine, The Hebrew University Medical School, Jerusalem, Israel
- 3I. Department of Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
- 4Department of Biochemistry and Molecular Biology, Institute for Medical Research Israel Canada (IMRIC), Faculty of Medicine, The Hebrew University Medical School, Jerusalem, Israel
- 5Department of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
Recent studies on the oral, anaerobic, gram-negative bacterium Fusobacterium nucleatum revealed its presence and involvement in colorectal, esophageal and breast cancer. We previously demonstrated that F. nucleatum binds and activates the human inhibitory receptors TIGIT and CEACAM1 leading to inhibition of T and NK cell anti-tumor immunity. CEACAM1 was found to be bound and activated by the fusobacterial trimeric autotransporter adhesin CbpF. Here we report the generation of a recombinant E. coli expressing full-length CbpF that efficiently binds and activates CEACAM1.
Introduction
Fusobacterium nucleatum is an oral, gram-negative, anaerobic bacterium and one of the most abundant species found in the oral cavity (Socransky et al., 1998; Nozawa et al., 2020). It is one of the phathobionts (Hajishengallis and Lamont, 2016; Lamont et al., 2018) that outgrow during dysbiosis preceding periodontal disease (Socransky et al., 1998; Nozawa et al., 2020). F. nucleatum is also found in colon adenocarcinoma (Castellarin et al., 2012; Kostic et al., 2012), esophageal cancer (Yamamura et al., 2016), pancreatic cancer (Mitsuhashi et al., 2015) and breast cancer (Parhi et al., 2020). The presence of F. nucleatum in pancreatic colon and esophageal cancer has been associated with poor prognosis (Mitsuhashi et al., 2015; Mima et al., 2016; Yamamura et al., 2016; Yamaoka et al., 2018). Generation of a pro-tumorigenic immune microenvironment (Kostic et al., 2013) and inhibition of the recruitment of tumor infiltrating lymphocytes (TILs) (Mima et al., 2015; Parhi et al., 2020) are among the mechanisms manipulated by F. nucleatum to promote cancer development. In addition to inhibition of the recruitment of TILs to the tumor, F. nucleatum activates the T and natural killer (NK) cells inhibitory receptors TIGIT and CEACAM1, leading to a reduction in their ability to kill tumor cells (Gur et al., 2015; Brewer et al., 2019; Gur et al., 2019b; Galaski et al., 2021).
CEACAM1 is a member of the human carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) that mediate cell-cell interactions and cellular signaling events. On various immune cell subsets, CEACAM1 acts as an inhibitory receptor (Gray-Owen and Blumberg, 2006; Yamin et al., 2016; Gur et al., 2019a; Gur et al., 2019b). The checkpoint-inhibitory activity of CEACAM1 can be activated by several ligands: CEACAM1 (CEACAM1-CEACAM1 interactions); Opacity-associated (Opa) proteins of pathogenic Neisseria (Gray-Owen and Blumberg, 2006); ubiquitous surface proteins A1 (UspA1) and A2V (UspA2V) of Moraxella catarrhalis (Hill and Virji, 2003; Hill et al., 2012); HopQ of Helicobacter pylori (Javaheri et al., 2016; Koniger et al., 2016; Gur et al., 2019a; Tegtmeyer et al., 2019) and by the CEACAM binding protein of Fusobacterium (CbpF) that facilitates fusobacterial colonization in the CEACAM1 displaying dentogingival junction (Brewer et al., 2019), and activates CEACAM1 signaling in immune cells (Galaski et al., 2021).
CEACAM1 is expressed on cells of the myeloid lineage, epithelial cells of human mucosa and some endothelial cells (Prall et al., 1996; Obrink, 1997; Hammarstrom, 1999). In addition, it is found on a wide variety of tumor cells, where it is considered to be a biomarker correlated with tumor progression, metastasis and poor prognosis (Wang et al., 2014; Dankner et al., 2017).
Tumor infiltrating lymphocytes (TILs) found in the colorectal cancer microenvironment are characterized by high levels of CEACAM1, along with reduced levels of cytotoxic activity when compared to paraneoplastic T cells (Zhang et al., 2017). These findings suggest a role of CEACAM1 in mediating T cell exhaustion. The homophilic interactions of CEACAM1, occurring between CEACAM1+ TILs, and CEACAM1+ tumor cells, as well as the interaction of CbpF with CEACAM1 expressed on NK and T cells, may protect tumors from killing by these immune cells in a synergistic mechanism.
Analysis of the CbpF amino acid sequence indicated that it belongs to the autotransporter proteins superfamily of the type Vc secretion pathway (Brewer et al., 2019). All autotransporters contain a three-domain structure which include an N-terminal leader peptide, a passenger domain and a C-terminal β-barrel domain. The C-terminal β-barrel autotransporter domain creates a β-barrel channel in the bacterial outer membrane that allows the passage of the passenger domain through the outer membrane to be presented on the bacterial outer surface (Henderson et al., 2004). Unlike other autotransporter subfamilies, the members of the type Vc secretion systems, often referred to as trimeric autotransporter adhesins (TAAs), form trimeric autotransporter proteins that consist of three identical polypeptide chains (Leo et al., 2012).
The C-terminus hydrophobic β-barrel domain of autotransporters can lead to solubility issues during the production of full-length recombinant autotransporter proteins. To the best of our knowledge, TAAs have most often been studied using truncated recombinant proteins expressed in E. coli (Nordstrom et al., 2004; Riess et al., 2004; Tan et al., 2005; Leo et al., 2008; Szczesny et al., 2008; Leo and Goldman, 2009; Hallstrom et al., 2011; Leo et al., 2011; Bentancor et al., 2012; Brewer et al., 2019). The features of truncated recombinant proteins might not fully match those of the native full-length proteins. Therefore, studies on a full-length recombinant TAA might expand our knowledge about the function and molecular properties of these protein family members.
Materials and Methods
Bacterial Strains and Cell Lines Growth Conditions
Escherichia coli strains were grown in LB broth (Difco) or on LB agar plates (Difco), at 37°C under aerobic conditions.
E. coli strain C43(DE3) was used to express the CbpF variants, and strain Top10 was used to amplify the CbpF variants plasmids. All plasmids-transformed E. coli strains were grown in the presence of 100 µg/ml ampicillin (Sigma).
721.221 cells and BW cells were grown in RPMI supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% each of non-essential amino acids, L-glutamine, sodium pyruvate and with penicillin-streptomycin (all from Biological Industries). Cells were grown at 37°C in a humidified 5% CO2 incubator.
Plasmid Transformation and Purification
100 ng of recombinant plasmid was gently mixed with 100 μl chemically competent bacteria. After 30 min incubation on ice, the mixture was heat shocked at 42°C for 60 seconds and then placed on ice for 2 min. Next, 900 μl of LB medium was added to the cell mixture and tubes incubated for 1 h at 37°C with shaking at 220 rpm. Next, 100 μl of the transformation mixture were spread on LB agar plates supplemented with 100 μg/ml ampicillin.
For plasmid purification, single colonies of transformed-E. coli Top10 were inoculated into 5ml selective LB medium and cultivated overnight at 37°C with shaking at 220 rpm. Plasmid was extracted using the QIAprep Spin Miniprep Kit (Qiagen), according to the manufacturer’s instructions.
Protein Expression and Purification
For expression, E. coli C43(DE3) was transformed with either the rCbpF-1 or rCbpF-2 encoding plasmids pSA-OCbpF1 and pSA-OCbpF2 respectively. A single colony was then inoculated in LB broth and grown overnight. The starter culture was then diluted x 100 in 2xYT medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl) containing, 100 µg/ml ampicillin (Sigma), 1 x NPS buffer (3.3 g/L (NH4)2SO4, 6.8 g/L KH2PO4, 7.1 g/L Na2HPO4) and 0.1% glucose, and grown at 37 °C until reaching an OD 600nm of 0.6–0.8. Cultures were then induced with 0.4 mM isopropyl-β-d-thio-galactoside (IPTG, Sigma) at 22°C overnight while shaking at 220 rpm. The cells were harvested by centrifuging at 5000 x g for 15 min, growth medium and part of the cell pellets were stored at -20°C until further use. The retained pellets were then resuspended with 1ml Lysis Buffer (50mM Tris pH 8.0, 10% glycerol, 1% Triton X-1 and 100 µg/ml lysozyme) and incubated 30 minutes in room temperature. Samples were then rapidly frozen by immersion in liquid nitrogen (2 min) and thawed in 42°C for 5 min. This freeze-thaw procedure was repeated three times, and the samples were centrifuged at 8,000 x g for 5 minutes at 4°C. The supernatant was transferred to a new test tube and both the pellet and the supernatant were kept also at -20°C for further analysis.
For rCbpF-1 purification, the bacterial growth medium was filtered with a 0.2 µm vacuum filter (Merck Millipore) and then concentrated x80 using a 50 kDa centrifugal filter (Amicon Ultra – 15, Merck Millipore). The concentrated growth medium was then loaded on to the 5 ml His-Trap columns (GE Healthcare), washed with washing buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM imidazole, pH 8.0), and eluted with a linear gradient of imidazole (15–300 mM). Fractions containing pure proteins were pooled and dialyzed against a buffer containing 20 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol.
Gel Electrophoresis
Samples were dissolved in a denaturing sample buffer (192 mM Tris-HCl [pH 6.8], 30% glycerol, 9% SDS, 0.01% bromophenol blue, 2% β-mercaptoethanol), or in a non-reducing sample buffer (lacking β-mercaptoethanol). The denaturing samples were also boiled at 95°C for 10 minutes, and samples were subjected to SDS-PAGE gels. Following electrophoresis, the gels were stained with Coomassie brilliant blue R-250 (Bio-Rad).
Western Blot and Far Western
Following electrophoresis, the proteins were transferred to a nitrocellulose membrane (110 V, 1 h, 4°C) using the Mini Trans-Blot cell apparatus (Bio-Rad). Nonspecific binding sites were blocked using PBS-Tween 20 (0.05% Tween 20, J.T. Baker) containing 5% non-fat dry milk for 1 h at room temperature. The membrane was then overlaid with mouse anti-His antibody (Bio-Rad), diluted 1:1000 in the blocking solution and incubated overnight at 4°C. Next day, the membrane was washed three times with PBS-Tween 20 and overlaid with peroxidase-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch Laboratories) diluted 1:5000 in blocking solution, for 1 h at room temperature. The membrane was then washed three times with PBS-Tween 20. Before imaging (using the ChemiDoc MP imaging system, Bio-Rad), the membrane was incubated in EZ-ECL (Biological Industries) solution for 5 min.
For far western blotting, the experiment was performed similar to the western blot experiment, but CEACAM1-Ig (2 µg/ml) (Gur et al., 2019b) was used instead of the first antibody and the HRP-conjugated α - human IgG (Jackson ImmunoResearch Laboratories, diluted 1:5,000) was used as a secondary antibody.
FITC Labeling of Bacteria and Flow Cytometry
For FITC-labeling, bacteria were washed twice in PBS, and incubated with 0.1 mg/ml FITC (Sigma) in PBS at room temperature in the dark for 30 minutes. Subsequently, bacteria were washed three times in PBS to remove unbound FITC. For flow cytometry, 721.221 cells were used as carrier cells to facilitate gating. To this end, bacteria were divided into 96-well plates and incubated with 721.221 cells for 30 minutes on ice to allow for bacterial adhesion to the cells (6 x 107 bacteria were placed together with 1 x 105 721.221 cells per well). Next, cells were washed and incubated with 3 µg of CEACAM1-Ig on ice for 1 hour followed by a 30-minute incubation with Alexa Fluor 647-conjugated donkey anti-human IgG (Jackson ImmunoResearch Laboratories). Histograms of cell-bound bacteria stained with CEACAM1-Ig were gated on FITC-positive cells.
BW Assay
The generation of BW cells expressing chimeric CEACAM1 (composed of the extracellular portion of human CEACAM1 fused to the mouse CD3ζ chain) was previously described (Markel et al., 2002).
E. coli were inactivated at 60°C for 40 minutes, divided into 96-well plates (3x107 bacteria per well) and incubated for one hour at 37°C in complete RPMI. Subsequently, BW CEACAM1 cells were added at 5x104 per well and incubated with the bacteria for 48 hours at 37°C. Next, supernatants were collected and mouse IL-2 levels were quantified by a sandwich ELISA.
Results
Production of a Recombinant Trimeric CbpF Autotransporter Adhesin
In an attempt to obtain a functional recombinant CbpF, we synthesized genes for two new CbpF variants (rCbpF-1 and rCbpF-2). As cloning and expression of AT-rich DNA in E. coli is difficult (Coppenhagen-Glazer et al., 2015), and the fusobacterial genome possesses a high (~ 70%) AT content (Kapatral et al., 2002), both genes were optimized for E. coli by performing codon optimization (Genscript). In addition to codon optimization, in cbpF-1, the fusobacterial CbpF signal peptide was replaced with that of the E. coli OmpA. In cbpF-2 the last four amino acids of the fusobacterial CbpF signal peptide were kept, followed by the CbpF passenger domain. Thus the CbpF autotransporter β-barrel domain was omitted as described previously (Brewer et al., 2019). The optimized genes (shown in Supplementary Figure 1) were cloned into pET-11a to generate pSA-OCbpF1 and pSA-OCbpF2, respectively (Supplementary Figure 2).
Expression of cbpF-1 in E. coli C43 resulted in the secretion of a full-length recombinant trimeric CbpF-1(rCbpF-1) to the growth medium, while cbpF-2 expression resulted in an intracellular monomeric truncated variant which required bacterial lysis for purification (Figures 1A, B). Next, rCbpF-1 was purified from cbpF-1 -expression growth medium concentrated 80 - fold (Growth med. conc. X80) using Ni-NTA (Figures 1C, D).
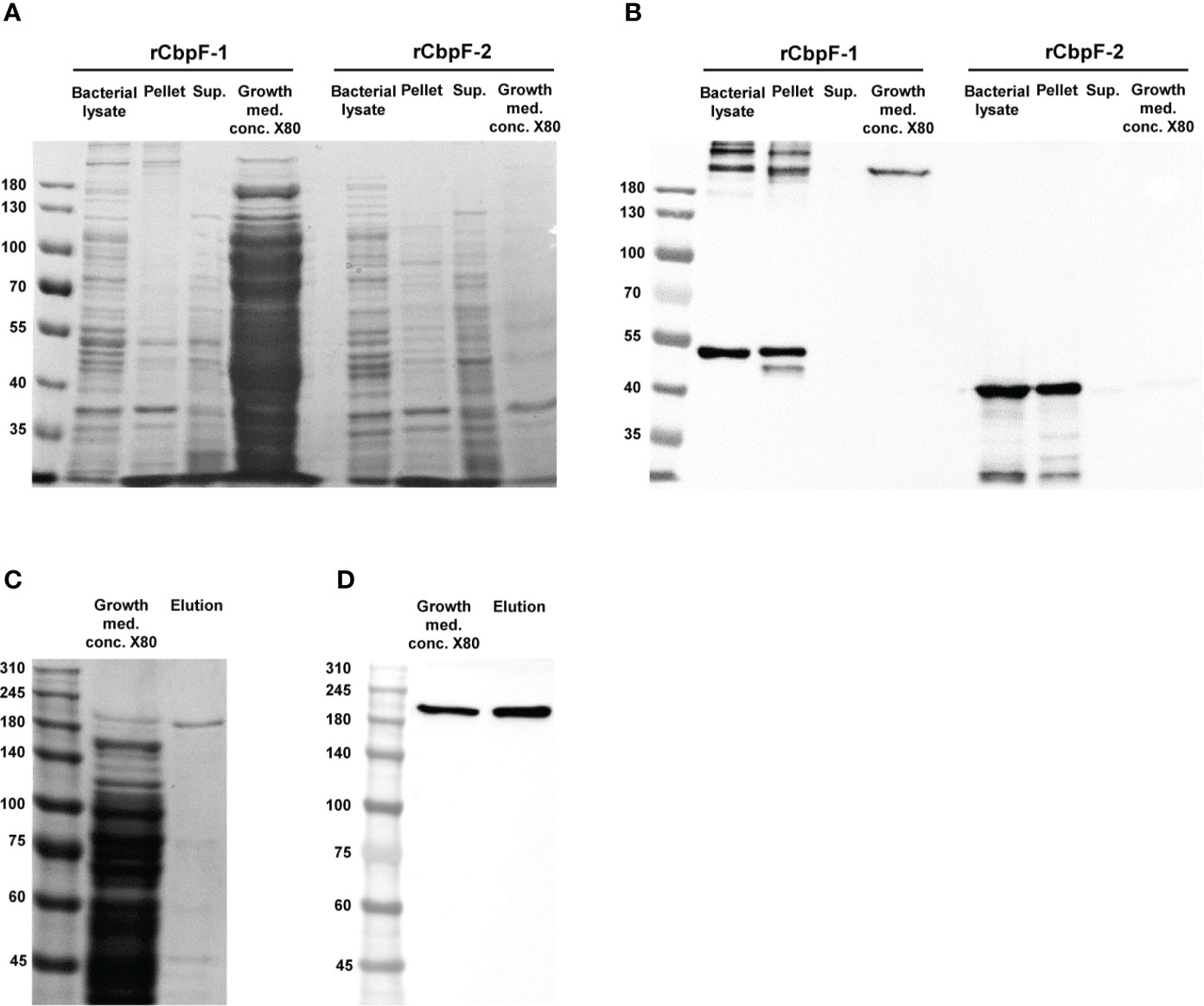
Figure 1 Expression of cbpF-1 and cbpF-2, and purification of rCbpF-1. cbpF-1 and cbpF-2 were expressed in E. coli C43 transformed with pSA-OCbpF1 and pSA-OCbpF2 respectively. Whole bacterial lysate, Pellet and Supernatant (Sup.) fractions after bacterial lysis, and growth medium concentrated X80 (Growth med. conc. X80) of E. coli C43 expressing either cbpF-1 or cbpF-2 were subjected to 7.5% gels SDS-PAGE (A), and Western immunodetection using anti-His monoclonal antibody (B). Recombinant CbpF-1 (rCbpF-1) was purified using Ni-NTA from growth medium concentrated X80 and subjected to 6% SDS-PAGE (C), and Western immunodetection using anti-His monoclonal antibody (D). Molecular weights are indicated on the left.
Similar to native CbpF expressed by F. nucleatum (Brewer et al., 2019), rCbpF-1 is a stable trimer that migrates in SDS-PAGE predominantly in a trimeric form following heating in reducing conditions.
CbpF-1-Expressing Recombinant E. coli Binds and Activates CEACAM1
Far western analysis under denaturing (Figure 2A) and non-reducing (Figure 2B) conditions was performed in order to test if the recombinant CbpF variants can bind CEACAM1. A CEACAM1-Ig fusion protein in which the extracellular domain of human CEACAM1 is fused to the Fc portion of human IgG1 (CEACAM1-Ig) was used as a probe. While the rCbpF-2 monomer failed to bind CEACAM1-Ig, the E. coli-bound CbpF-1 trimer, and the Ni-NTA -purified, secreted rCbpF-1 trimer were able to bind CEACAM1-Ig in both denaturing and native far western assays.
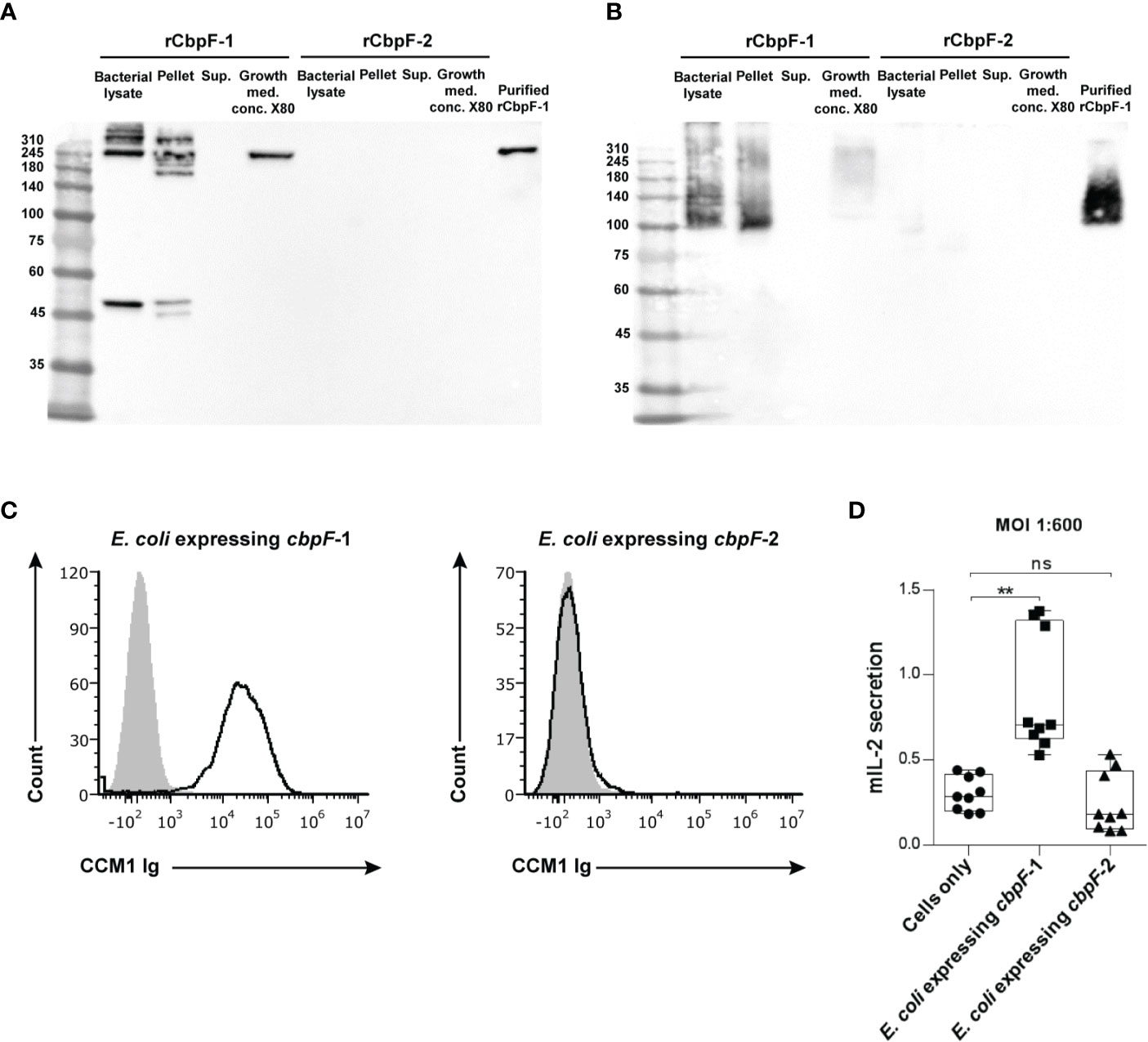
Figure 2 Recombinant E. coli expressing CbpF-1 binds CEACAM1. Whole bacterial lysate; Pellet and Supernatant (Sup.) fractions after lysis; growth medium concentrated X80 (Growth med. conc. X80) of E. coli C43 expressing cbpF-1 or cbpF-2; and Ni-NTA -purified rCbpF1 were subjected to denaturing (A), and native (B) 7.5% SDS-PAGE followed by far western analysis using CEACAM1-Ig (see materials and methods). Molecular weights are indicated on the left. (C) FITC-labeled E. coli expressing cbpF-1 or E. coli expressing cbpF-2 were stained with 2 μg of human CEACAM1-Ig. Human B lymphoblastoid cell line 721.221 cells were used as carrier cells. Filled grey histograms represent staining with secondary antibody only. Presented results represents one of two independent repeats. (D) CEACAM1-reporter BW cells were incubated with E. coli expressing cbpF-1 or cbpF-2 at a ratio of 1:600. Mouse IL-2 (mIL-2) in the supernatants 48 hours later was determined by ELISA. Boxplot whiskers represent extrema, box bounds represent upper and lower quartiles, and center-line represents the median value of nine observations. n.s., non-significant, **p = 0.0033 two-tail, as determined by the Dunn post-hoc multiple-comparisons procedure following the Kruskal-Wallis test (GraphPad Prism 6).
Flow cytometry using CEACAM1-Ig was used next to confirm that rCpbF-1 is displayed on the surface of cbpF-1 -expressing E. coli. As can be seen in Figure 2C, binding of human CEACAM1-Ig was observed only to cbpF-1-expressing E. coli but not to cbpF-2-expressing ones.
We next wanted to test whether E. coli that displays rCbpF-1 can activate CEACAM1. For this aim, we used a reporter system that consists of murine thymoma BW cells that were transfected with chimeric proteins composed of the extracellular domain of CEACAM1 fused to a mouse zeta chain. When the CEACAM1 is bound and activated by specific ligands in this system, mouse IL-2 is secreted to the medium. CEACAM1-reporter BW cells were incubated with E. coli induced to express cbpF-1 or cbpF-2. Activation of CEACAM1-expressing BW cells was observed only when incubated with cbpF-1-expressing E. coli (Figure 2D).
Discussion
CEACAM1 plays an important immunomodulatory role (Gray-Owen and Blumberg, 2006; Kim et al., 2019). Its checkpoint activity is programed to prevent self-harming autoimmune responses. Selection often guides tumors to overexpress CEACAM1 in order to bind and activate CEACAM1 on immune cells and escape anti-tumor activity (Kim et al., 2019). Bacteria have also evolved to express CEACAM1 ligands that assist in attachment and colonization of host tissues (Brewer et al., 2019), but more importantly, impair anti-microbial immunity (Dankner et al., 2017; Gur et al., 2019a; Gur et al., 2019b). As F. nucleatum is found in several cancer types (including Colon, esophageal, pancreatic and breast) it is plausible to hypothesize that CEACAM1 activation by tumor-colonized fusobacteria might suppress anti-tumor immunity and affect disease outcome. Therefore, creating tools for analyzing checkpoints activators, that might eventually lead to the generation of novel checkpoint inhibitors is important. Using fusobacterial mutagenesis we recently confirmed that CbpF indeed activates CAECAM1 (Galaski et al., 2021). Here we aimed to generate the first recombinant bacterial immune checkpoint activator. We chose the fusobacterial CbpF, and by optimizing the codon-preference of its gene to that of E. coli, and replacing the fusobacterial leader peptide with that of the E. coli OmpA, we managed to express a functional full-length trimeric recombinant CbpF-1 on the outer-surface of E. coli.
The predicted molecular weight of the rCbpF monomer is 50.5 kDa. SDS-PAGE indicated that rCbpF-1 is in a multimer form (Figures 1, 2). The elution profile of purified rCbpF-1 in gel filtration indicated a high molecular weight, confirming that purified rCbpF-1 is not in a monomeric state (Supplementary Figure 3). rCbpF appeared in gels with a higher molecular mass than the predicted molecular weight of the trimer (Figures 1, 2). Such size differences were detectable when rCbpF-1 was subjected to SDS-PAGE gels with different polyacrylamide percentage. In a 6% gel, rCbpF-1 migrated as ~200 kDa (Figures 1C, D), while in a 7.5% gel, it migrated as 250 kDa (Figure 2A). This observation might point to slight aggregation that occurs during electrophoresis. In native gels, rCbpF-1 migrated at a molecular mass close to the predicted one (Figure 2B), suggesting that pre-heating promotes such aggregation due to the exposure of hydrophobic regions (Fink, 1998).
Interestingly, while the rCbpF-2 monomer failed to bind CEACAM1-Ig in the far-western assay, the E. coli -bound rCbpF-1 trimer, and the rCbpF1 trimer purified using Ni-NTA from the growth medium, were able to bind CEACAM1-Ig in both denaturing and native far western assays. This might point to a role of the C-terminal autotransporter domain in the proper folding of CbpF and that proper folding of the CEACAM-binding domain of CbpF is required for CEACAM1 binding.
Our results demonstrate the production of the first functional full length CbpF-expressing recombinant E. coli, and one of the few full-length recombinant TAAs studied (Grosskinsky et al., 2007; Atack et al., 2020). To the best of our knowledge, it is the only full-length recombinant TAA CEACAM-activating ligand produced (Nordstrom et al., 2004; Tan et al., 2005; Hallstrom et al., 2011; Brewer et al., 2019; Mikula et al., 2019). Recombinant bacterial checkpoint activators might pave the way for future generation of a new class of bacterial-based checkpoints inhibitors.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AS and JG designed and carried out experiments, analyzed the data and participated in writing the manuscript. TA-M carried out experiments. JF and RW performed gel-filtration. DS analyzed the data. GB and OM supervised the project and participated in writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Israel Cancer Research Fund Project grant (GB), the Israel Science Foundation Moked grant and the Israel Ministry of Science and Technology Personalized Medicine grant (GB and OM). AS is supported by the Planning and Budgeting Committee of the Israeli Council for Higher Education. JG is supported by the German Research Foundation (DFG) with a postdoctoral research fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.699015/full#supplementary-material
References
Atack, J. M., Day, C. J., Poole, J., Brockman, K. L., Timms, J. R. L., Winter, L. E., et al. (2020). The Nontypeable Haemophilus influenzae Major Adhesin Hia Is a Dual-Function Lectin That Binds to Human-Specific Respiratory Tract Sialic Acid Glycan Receptors. mBio 11, e02714–20. doi: 10.1128/mBio.02714-20
Bentancor, L. V., Camacho-Peiro, A., Bozkurt-Guzel, C., Pier, G. B., Maira-Litran, T. (2012). Identification of Ata, a Multifunctional Trimeric Autotransporter of Acinetobacter Baumannii. J. Bacteriol. 194, 3950–3960. doi: 10.1128/JB.06769-11
Brewer, M. L., Dymock, D., Brady, R. L., Singer, B. B., Virji, M., Hill, D. J. (2019). Fusobacterium Spp. Target Human CEACAM1 via the Trimeric Autotransporter Adhesin CbpF. J. Oral. Microbiol. 11, 1565043. doi: 10.1080/20002297.2018.1565043
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum Infection Is Prevalent in Human Colorectal Carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Coppenhagen-Glazer, S., Sol, A., Abed, J., Naor, R., Zhang, X., Han, Y. W., et al. (2015). Fap2 of Fusobacterium nucleatum Is a Galactose-Inhibitable Adhesin Involved in Coaggregation, Cell Adhesion, and Preterm Birth. Infect. Immun. 83, 1104–1113. doi: 10.1128/IAI.02838-14
Dankner, M., Gray-Owen, S. D., Huang, Y. H., Blumberg, R. S., Beauchemin, N. (2017). CEACAM1 as a Multi-Purpose Target for Cancer Immunotherapy. Oncoimmunology 6, e1328336. doi: 10.1080/2162402X.2017.1328336
Fink, A. L. (1998). Protein Aggregation: Folding Aggregates, Inclusion Bodies and Amyloid. Fold. Des. 3, R9–R23. doi: 10.1016/S1359-0278(98)00002-9
Galaski, A., Shhadeh, A., Umaña, A., Yoo, C. C., Arpinati, L., Isaacson, B., et al (2021). Fusobacterium Nucleatum CbpF Mediates Inhibition of T Cell Function Through CEACAM1 Activation. Front. Cell Infect. Microbiol. 11, 692544. doi: 10.3389/fcimb.2021.692544
Gray-Owen, S. D., Blumberg, R. S. (2006). CEACAM1: Contact-Dependent Control of Immunity. Nat. Rev. Immunol. 6, 433–446. doi: 10.1038/nri1864
Grosskinsky, U., Schutz, M., Fritz, M., Schmid, Y., Lamparter, M. C., Szczesny, P., et al. (2007). A Conserved Glycine Residue of Trimeric Autotransporter Domains Plays a Key Role in Yersinia Adhesin A Autotransport. J. Bacteriol. 189, 9011–9019. doi: 10.1128/JB.00985-07
Gur, C., Ibrahim, Y., Isaacson, B., Yamin, R., Abed, J., Gamliel, M., et al. (2015). Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors From Immune Cell Attack. Immunity 42, 344–355. doi: 10.1016/j.immuni.2015.01.010
Gur, C., Maalouf, N., Gerhard, M., Singer, B. B., Emgard, J., Temper, V., et al. (2019a). The Helicobacter pylori HopQ Outermembrane Protein Inhibits Immune Cell Activities. Oncoimmunology 8, e1553487. doi: 10.1080/2162402X.2018.1553487
Gur, C., Maalouf, N., Shhadeh, A., Berhani, O., Singer, B. B., Bachrach, G., et al. (2019b). Fusobacterium nucleatum Supresses Anti-Tumor Immunity by Activating CEACAM1. Oncoimmunology 8, e1581531. doi: 10.1080/2162402X.2019.1581531
Hajishengallis, G., Lamont, R. J. (2016). Dancing With the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 24, 477–489. doi: 10.1016/j.tim.2016.02.010
Hallstrom, T., Nordstrom, T., Tan, T. T., Manolov, T., Lambris, J. D., Isenman, D. E., et al. (2011). Immune Evasion of Moraxella catarrhalis Involves Ubiquitous Surface Protein A-Dependent C3d Binding. J. Immunol. 186, 3120–3129. doi: 10.4049/jimmunol.1002621
Hammarstrom, S. (1999). The Carcinoembryonic Antigen (CEA) Family: Structures, Suggested Functions and Expression in Normal and Malignant Tissues. Semin. Cancer Biol. 9, 67–81. doi: 10.1006/scbi.1998.0119
Henderson, I. R., Navarro-Garcia, F., Desvaux, M., Fernandez, R. C., Ala’aldeen, D. (2004). Type V Protein Secretion Pathway: The Autotransporter Story. Microbiol. Mol. Biol. Rev. 68, 692–744. doi: 10.1128/MMBR.68.4.692-744.2004
Hill, D. J., Virji, M. (2003). A Novel Cell-Binding Mechanism of Moraxella catarrhalis Ubiquitous Surface Protein UspA: Specific Targeting of the N-Domain of Carcinoembryonic Antigen-Related Cell Adhesion Molecules by Uspa1. Mol. Microbiol. 48, 117–129. doi: 10.1046/j.1365-2958.2003.03433.x
Hill, D. J., Whittles, C., Virji, M. (2012). A Novel Group of Moraxella Catarrhalis UspA Proteins Mediates Cellular Adhesion via CEACAMs and Vitronectin. PloS One 7, e45452. doi: 10.1371/journal.pone.0045452
Javaheri, A., Kruse, T., Moonens, K., Mejias-Luque, R., Debraekeleer, A., Asche, C. I., et al. (2016). Helicobacter pylori Adhesin HopQ Engages in a Virulence-Enhancing Interaction With Human CEACAMs. Nat. Microbiol. 2, 16189. doi: 10.1038/nmicrobiol.2016.189
Kapatral, V., Anderson, I., Ivanova, N., Reznik, G., Los, T., Lykidis, A., et al. (2002). Genome Sequence and Analysis of the Oral Bacterium Fusobacterium nucleatum Strain ATCC 25586. J. Bacteriol. 184, 2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002
Kim, W. M., Huang, Y. H., Gandhi, A., Blumberg, R. S. (2019). CEACAM1 Structure and Function in Immunity and Its Therapeutic Implications. Semin. Immunol. 42, 101296. doi: 10.1016/j.smim.2019.101296
Koniger, V., Holsten, L., Harrison, U., Busch, B., Loell, E., Zhao, Q., et al. (2016). Helicobacter pylori Exploits Human CEACAMs via HopQ for Adherence and Translocation of CagA. Nat. Microbiol. 2, 16188. doi: 10.1038/nmicrobiol.2016.188
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic Analysis Identifies Association of Fusobacterium With Colorectal Carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Lamont, R. J., Koo, H., Hajishengallis, G. (2018). The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 16, 745–759. doi: 10.1038/s41579-018-0089-x
Leo, J. C., Elovaara, H., Brodsky, B., Skurnik, M., Goldman, A. (2008). The Yersinia Adhesin YadA Binds to a Collagenous Triple-Helical Conformation But Without Sequence Specificity. Protein Eng. Des. Sel. 21, 475–484. doi: 10.1093/protein/gzn025
Leo, J. C., Goldman, A. (2009). The Immunoglobulin-Binding Eib Proteins From Escherichia coli Are Receptors for IgG Fc. Mol. Immunol. 46, 1860–1866. doi: 10.1016/j.molimm.2009.02.024
Leo, J. C., Grin, I., Linke, D. (2012). Type V Secretion: Mechanism(s) of Autotransport Through the Bacterial Outer Membrane. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 1088–1101. doi: 10.1098/rstb.2011.0208
Leo, J. C., Lyskowski, A., Hattula, K., Hartmann, M. D., Schwarz, H., Butcher, S. J., et al. (2011). The Structure of E. Coli IgG-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins. Structure 19, 1021–1030. doi: 10.1016/j.str.2011.03.021
Markel, G., Wolf, D., Hanna, J., Gazit, R., Goldman-Wohl, D., Lavy, Y., et al. (2002). Pivotal Role of CEACAM1 Protein in the Inhibition of Activated Decidual Lymphocyte Functions. J. Clin. Invest. 110, 943–953. doi: 10.1172/JCI0215643
Mikula, K. M., Kolodziejczyk, R., Goldman, A. (2019). Structure of the UspA1 Protein Fragment From Moraxella catarrhalis Responsible for C3d Binding. J. Struct. Biol. 208, 77–85. doi: 10.1016/j.jsb.2019.08.002
Mima, K., Nishihara, R., Qian, Z. R., Cao, Y., Sukawa, Y., Nowak, J. A., et al. (2016). Fusobacterium nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut 65, 1973–1980. doi: 10.1136/gutjnl-2015-310101
Mima, K., Sukawa, Y., Nishihara, R., Qian, Z. R., Yamauchi, M., Inamura, K., et al. (2015). Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 1 (5), 653–661. doi: 10.1001/jamaoncol.2015.1377
Mitsuhashi, K., Nosho, K., Sukawa, Y., Matsunaga, Y., Ito, M., Kurihara, H., et al. (2015). Association of Fusobacterium Species in Pancreatic Cancer Tissues With Molecular Features and Prognosis. Oncotarget 6, 7209–7220. doi: 10.18632/oncotarget.3109
Nordstrom, T., Blom, A. M., Forsgren, A., Riesbeck, K. (2004). The Emerging Pathogen Moraxella catarrhalis Interacts With Complement Inhibitor C4b Binding Protein Through Ubiquitous Surface Proteins A1 and A2. J. Immunol. 173, 4598–4606. doi: 10.4049/jimmunol.173.7.4598
Nozawa, A., Oshima, H., Togawa, N., Nozaki, T., Murakami, S. (2020). Development of Oral Care Chip, a Novel Device for Quantitative Detection of the Oral Microbiota Associated With Periodontal Disease. PloS One 15, e0229485. doi: 10.1371/journal.pone.0229485
Obrink, B. (1997). CEA Adhesion Molecules: Multifunctional Proteins With Signal-Regulatory Properties. Curr. Opin. Cell Biol. 9, 616–626. doi: 10.1016/S0955-0674(97)80114-7
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast Cancer Colonization by Fusobacterium nucleatum Accelerates Tumor Growth and Metastatic Progression. Nat. Commun. 11, 3259. doi: 10.1038/s41467-020-16967-2
Prall, F., Nollau, P., Neumaier, M., Haubeck, H. D., Drzeniek, Z., Helmchen, U., et al. (1996). CD66a (BGP), an Adhesion Molecule of the Carcinoembryonic Antigen Family, Is Expressed in Epithelium, Endothelium, and Myeloid Cells in a Wide Range of Normal Human Tissues. J. Histochem. Cytochem. 44, 35–41. doi: 10.1177/44.1.8543780
Riess, T., Andersson, S. G., Lupas, A., Schaller, M., Schafer, A., Kyme, P., et al. (2004). Bartonella Adhesin a Mediates a Proangiogenic Host Cell Response. J. Exp. Med. 200, 1267–1278. doi: 10.1084/jem.20040500
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., Kent, R. L., Jr. (1998). Microbial Complexes in Subgingival Plaque. J. Clin. Periodontol. 25, 134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x
Szczesny, P., Linke, D., Ursinus, A., Bar, K., Schwarz, H., Riess, T. M., et al. (2008). Structure of the Head of the Bartonella Adhesin BadA. PloS Pathog. 4, e1000119. doi: 10.1371/journal.ppat.1000119
Tan, T. T., Nordstrom, T., Forsgren, A., Riesbeck, K. (2005). The Respiratory Pathogen Moraxella catarrhalis Adheres to Epithelial Cells by Interacting With Fibronectin Through Ubiquitous Surface Proteins A1 and A2. J. Infect. Dis. 192, 1029–1038. doi: 10.1086/432759
Tegtmeyer, N., Harrer, A., Schmitt, V., Singer, B. B., Backert, S. (2019). Expression of CEACAM1 or CEACAM5 in AZ-521 Cells Restores the Type IV Secretion Deficiency for Translocation of CagA by Helicobacter Pylori. Cell Microbiol. 21, e12965. doi: 10.1111/cmi.12965
Wang, N., Feng, Y., Wang, Q., Liu, S., Xiang, L., Sun, M., et al. (2014). Neutrophils Infiltration in the Tongue Squamous Cell Carcinoma and Its Correlation With CEACAM1 Expression on Tumor Cells. PloS One 9, e89991. doi: 10.1371/journal.pone.0089991
Yamamura, K., Baba, Y., Nakagawa, S., Mima, K., Miyake, K., Nakamura, K., et al. (2016). Human Microbiome Fusobacterium nucleatum in Esophageal Cancer Tissue Is Associated With Prognosis. Clin. Cancer Res. 22, 5574–5581. doi: 10.1158/1078-0432.CCR-16-1786
Yamaoka, Y., Suehiro, Y., Hashimoto, S., Hoshida, T., Fujimoto, M., Watanabe, M., et al. (2018). Fusobacterium nucleatum as a Prognostic Marker of Colorectal Cancer in a Japanese Population. J. Gastroenterol. 53, 517–524. doi: 10.1007/s00535-017-1382-6
Yamin, R., Lecker, L. S. M., Weisblum, Y., Vitenshtein, A., Le-Trilling, V. T. K., Wolf, D. G., et al. (2016). HCMV Vcxcl1 Binds Several Chemokine Receptors and Preferentially Attracts Neutrophils Over NK Cells by Interacting With CXCR2. Cell Rep. 15, 1542–1553. doi: 10.1016/j.celrep.2016.04.042
Keywords: F. nucleatum, CbpF, trimeric autotransporter adhesin, CEACAM1, NK cells
Citation: Shhadeh A, Galaski J, Alon-Maimon T, Fahoum J, Wiener R, Slade DJ, Mandelboim O and Bachrach G (2021) CEACAM1 Activation by CbpF-Expressing E. coli. Front. Cell. Infect. Microbiol. 11:699015. doi: 10.3389/fcimb.2021.699015
Received: 22 April 2021; Accepted: 13 July 2021;
Published: 29 July 2021.
Edited by:
Ozlem Yilmaz, Medical University of South Carolina, United StatesReviewed by:
Ashu Sharma, University at Buffalo, United StatesAndreas Zanzoni, Aix-Marseille Université, France
Copyright © 2021 Shhadeh, Galaski, Alon-Maimon, Fahoum, Wiener, Slade, Mandelboim and Bachrach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilad Bachrach, giladba@ekmd.huji.ac.il
†These authors have contributed equally to this work
 Amjad Shhadeh
Amjad Shhadeh Johanna Galaski
Johanna Galaski Tamar Alon-Maimon1
Tamar Alon-Maimon1  Daniel J. Slade
Daniel J. Slade Ofer Mandelboim
Ofer Mandelboim Gilad Bachrach
Gilad Bachrach