Glyphosate-Induced Specific and Widespread Perturbations in the Metabolome of Soil Pseudomonas Species
- 1Department of Biological and Environmental Engineering, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, United States
- 2Soil and Crop Sciences Section, School of Integrative Plant Science, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, United States
- 3Atkinson Center for a Sustainable Future, Cornell University, Ithaca, NY, United States
Previous studies have reported adverse effects of glyphosate on crop-beneficial soil bacterial species, including several soil Pseudomonas species. Of particular interest is the elucidation of the metabolic consequences of glyphosate toxicity in these species. Here we investigated the growth and metabolic responses of soil Pseudomonas species grown on succinate, a common root exudate, and glyphosate at different concentrations. We conducted our experiments with one agricultural soil isolate, P. fluorescens RA12, and three model species, P. putida KT2440, P. putida S12, and P. protegens Pf-5. Our results demonstrated both species- and strain-dependent growth responses to glyphosate. Following exposure to a range of glyphosate concentrations (up to 5 mM), the growth rate of both P. protegens Pf-5 and P. fluorescens RA12 remained unchanged whereas the two P. putida strains exhibited from 0 to 100% growth inhibition. We employed a 13C-assisted metabolomics approach using liquid chromatography-mass spectrometry to monitor disruptions in metabolic homeostasis and fluxes. Profiling of the whole-cell metabolome captured deviations in metabolite levels involved in the tricarboxylic acid cycle, ribonucleotide biosynthesis, and protein biosynthesis. Altered metabolite levels specifically in the biosynthetic pathway of aromatic amino acids (AAs), the target of toxicity for glyphosate in plants, implied the same toxicity target in the soil bacterium. Kinetic flux experiments with 13C-labeled succinate revealed that biosynthetic fluxes of the aromatic AAs were not inhibited in P. fluorescens Pf-5 in the presence of low and high glyphosate doses but these fluxes were inhibited by up to 60% in P. putida KT2440, even at sub-lethal glyphosate exposure. Notably, the greatest inhibition was found for the aromatic AA tryptophan, an important precursor to secondary metabolites. When the growth medium was supplemented with aromatic AAs, P. putida S12 exposed to a lethal dose of glyphosate completely recovered in terms of both growth rate and selected metabolite levels. Collectively, our findings led us to conclude that the glyphosate-induced specific disruption of de novo biosynthesis of aromatic AAs accompanied by widespread metabolic disruptions was responsible for dose-dependent adverse effects of glyphosate on sensitive soil Pseudomonas species.
Introduction
Glyphosate-based herbicides, including notably Roundup®, are the most widely used herbicides in the United States (Woodburn, 2000). This is largely due to the advent of Roundup Ready crops, in which the gene that encodes the enzymatic target of the active ingredient glyphosate is modified (Duke et al., 2012) (Figure 1A). Thus, these genetically-modified crops (e.g., soybean, corn) are protected against glyphosate applications. Glyphosate is lethal to targeted weed plants by inhibiting 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, an essential enzyme in the shikimate pathway that is responsible for the biosynthesis of aromatic amino acids (AAs) (Schulz et al., 1985) (Figure 1B). Although mammalian organisms including humans do not synthesize aromatic AAs and instead obtain these AAs from their diet, microorganisms employ the shikimate pathway similar to plants to synthesize their aromatic AAs. Therefore, of emerging concern are the potential non-targeted effects of glyphosate on soil microorganisms that are important to soil and crop health.
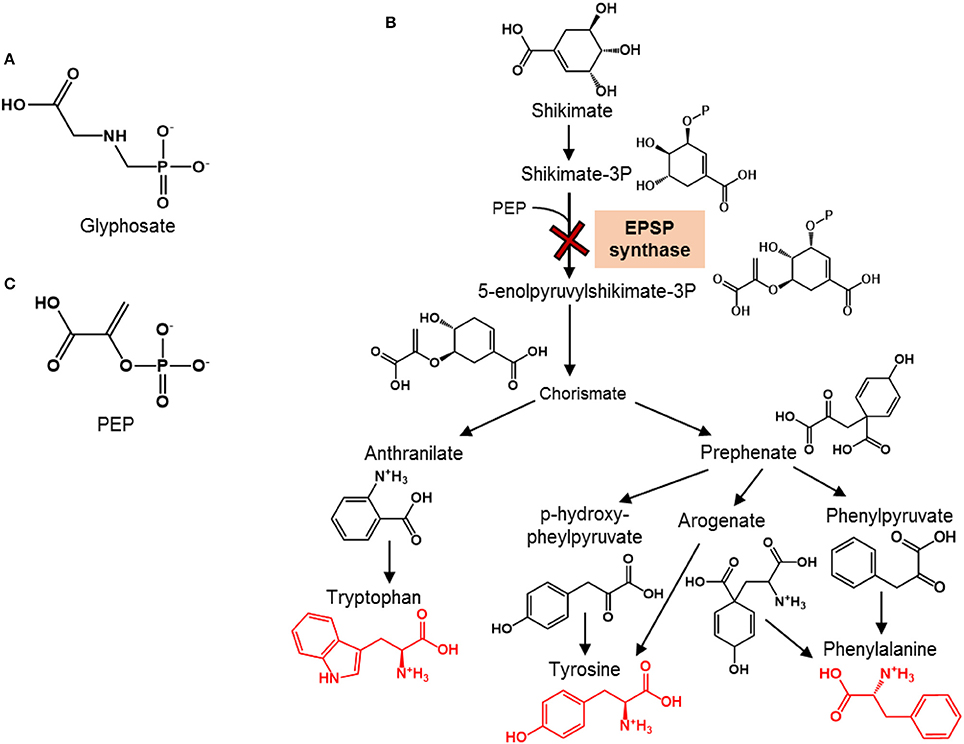
Figure 1. Chemical structures of glyphosate and phosphoenolpyruvate and overview of the shikimate pathway for aromatic amino acid synthesis. (A) Chemical structure of the herbicide glyphosate. (B) Schematic of the shikimate pathway for the biosynthesis of aromatic amino acids (shown in red structures) in plants (adapted from Tzin and Galili, 2010); the toxicity target of glyphosate, which is the enzyme responsible for the synthesis of 5-enolpyruvylshikimate-3-phosphate by combining shikimate-3P and phosphoenolpyruvate (PEP), is shown with the red cross. (C) Chemical structure of PEP, the endogenous substrate at the enzymatic target of glyphosate.
There have been numerous reports of adverse effects of glyphosate treatments on various microbial species and communities (Lévesque et al., 1987; Larson et al., 2006; Kremer and Means, 2009; Zobiole et al., 2011; Lane et al., 2012; Druille et al., 2013, 2015). Here we seek to gain insights into the intracellular targets underlying the response of soil Pseudomonas species, such as P. putida, P. fluorescens, and P. protegens, to glyphosate exposure. Due to their production of a range of antibiotic compounds, iron-scavenging molecules (or siderophores), and plant growth promoters, several Pseudomonas species have been employed as effective biocontrol agents to protect plants against pathogens and promote plant health (Timmis, 2002). A decreased abundance in Pseudomonas species was reported in greenhouse soils wherein glyphosate was applied to soybeans in a manner similar to agricultural applications (Zobiole et al., 2011). Furthermore, a previous study revealed greater inhibition of the EPSP synthase activity in P. fluorescens AFT36 than in P. putida W1616 at the same glyphosate concentration (Schulz et al., 1985). We hypothesize that the sensitivity of glyphosate-exposed Pseudomonas species in glyphosate-containing soils could be due to (1) species-specific growth responses to glyphosate exposure, (2) glyphosate inhibition of the biosynthetic pathway for aromatic AAs, or (3) disruption of other cellular pathways.
Species-specific bacterial growth response to glyphosate have been reported previously (Santos and Flores, 1995; Lancaster et al., 2010; Duke et al., 2012). The glyphosate concentration required for 50% growth inhibition of Escherichia coli, Bacillus subtilis, B. jabonicum, and P. aeruginosa, was estimated to be, respectively, 75 μM, 174 μM, 1.1 mM, and 1.1 mM (Duke et al., 2012). Differences in adverse metabolic effects were also reported for two closely-related nitrogen-fixing bacteria, Azotobacter chroococcum and A. vinelandii, in response to similar glyphosate exposure (Santos and Flores, 1995). Specifically, at 4 kg ha−1 of glyphosate, the nitrogenase activity in A. vinelandii was decreased by 22% but only by 2% in A. chroococcum (Santos and Flores, 1995). At 3 times higher glyphosate application (12 kg ha−1), the reduction in nitrogenase activity was 45 and 13%, respectively, in A. vinelandii and A. chroococcum (Santos and Flores, 1995). Furthermore, a decrease in the cell size of the glyphosate-exposed Azotobacter species was proposed to be a result of a reduction in protein synthesis downstream of EPSP inhibition (Santos and Flores, 1995). Whether the decrease in Pseudomonas species in glyphosate-exposed soils is due to a similar metabolic disruption remains to be determined.
Species-dependent differences in the EPSP synthase are at the basis of the generation of Roundup Ready crops. In these crops, a bacterial gene from Agrobacterium sp. strain CP4 is inserted into the crops in order to produce an EPSP synthase resistant to glyphosate (Funke et al., 2006). Thus, the primary targets of glyphosate-based herbicides are plants with glyphosate-sensitive EPSP synthase in their shikimate pathway. However, in addition to plants, the shikimate pathway is responsible for the de novo biosynthesis of aromatic AAs in microorganisms (Figure 1B). In this pathway, shikimate is eventually converted to chorismate, the common metabolite precursor to the following three aromatic amino AAs: tyrosine, phenylalanine, and tryptophan (Figure 1B). The direct precursor to chorismate, 5-enolpyruvylshikimate-3P, is synthesized by the EPSP enzyme. Therefore, the EPSP synthase represents a gateway to the production of aromatic AAs (Figure 1B). Glyphosate interferes with this gateway by competing with the endogenous substrate, phosphoenolpyruvate (PEP) (Figure 1C), which combines with shikimate-3-phosphate to produce EPSP (Steinrücken and Amrhein, 1980; Schönbrunn et al., 2001). Different extents of inhibition of EPSP synthase were reported previously for different Pseudomonas species (Schulz et al., 1985). Specifically, at about 3.0 μM glyphosate, there was 50% inhibition of the enzyme activity in P. fluorescens AFT36 but no inhibition in P. putida W1616 (Schulz et al., 1985). At one order of magnitude higher concentration (3 mM) of glyphosate, there was 100% inhibition of EPSP synthase in P. fluorescens AFT36 but only 50% inhibition in P. putida W1616 (Schulz et al., 1985). It has not been demonstrated, however, explicitly that the inhibition of the EPSP synthase resulted in the inhibition of the biosynthesis of aromatic AAs in these Pseudomonas species or other crop-beneficial bacterial species susceptible to glyphosate.
A decreased abundance in fungal populations was also reported in soil microbial communities exposed to glyphosate (Johal and Rahe, 1984; Lévesque and Rahe, 1992; Kremer et al., 2005). Stunted root growth of plants in response to increasing glyphosate applications was accompanied by both an increase in the fungal pathogens Fusarium spp. and a decrease in Pseudomonas spp. (Zobiole et al., 2011). Interestingly, certain soil Pseudomonas species are known to produce pyrrolnitrin, an important potent antifungal metabolite, that requires the aromatic AA tryptophan as a metabolite precursor (Kirner et al., 1998). Whether the change in the relative abundance of Fusarium spp. and Pseudomonas spp. is due to disruption in the biosynthetic pathway that feeds the production of pyrrolnitrin was not determined. In addition to fungicides, crop-beneficial Pseudomonas spp. are known to produce auxin-like plant hormones including notably indole-acetic acid (IAA). Similar to pyrrolnitrin, IAA is derived from the aromatic AA tryptophan (Zhao, 2012). In fact, decreased abundance in Pseudomonas spp. was found to be positively correlated with reduced populations of IAA-producing bacteria in a glyphosate-treated soil (Zobiole et al., 2011). Due to the important connection of shikimate pathway to the biosynthesis of both primary and secondary metabolites in Pseudomonas spp., elucidating the effects of glyphosate on both the initiation of this metabolic pathway as well as its associated synthesis of aromatic AAs is warranted.
In the present study, we evaluated our stated hypotheses regarding the factors responsible for the adverse effects of glyphosate exposure on four different soil Pseudomonas species by (1) determining the growth response of these different species to different glyphosate concentrations, (2) evaluating the biosynthesis of aromatic AAs in the shikimate pathway as a specific target of glyphosate, and (3) investigating other cellular pathways susceptible to glyphosate exposure. We achieved our objectives by employing a metabolomics approach involving liquid chromatography-mass spectrometry (LC-MS) and kinetic flux profiling using stable isotope tracers. Our results provide metabolic insights into both specific and widespread metabolic perturbations of glyphosate to a range of soil Pseudomonas species. These findings have broader agricultural relevance in regards to the assessment of potential non-targeted effects of glyphosate to sensitive soil bacterial species.
Experimental Methods
Culturing Conditions
P. putida KT2440, P. putida S12, and P. protegens Pf-5 were obtained from ATCC (American Type Culture Collection, Manassas, VA); the agricultural soil isolate P. fluorescens (strain RA12) was a gift from Dr. Rania Abou-Kandil (Cornell University). Cell cultures (three biological replicates) were grown at 30°C in a G24 environmental incubator shaker (New Brunswick Scientific, Edison, NJ) at 220 rpm (Sasnow et al., 2016). The pH-adjusted (pH 7.0) and filter-sterilized (0.22 μm nylon; Waters Corporation, Massachusetts) growth medium contained both major and minor nutrient salts: 89.4 mM K2HPO4, 56.4 mM NaH2PO4, 0.81 mM MgSO4·7H2O, 18.7 mM NH4Cl, 8.6 mM NaCl, 34 μM CaCl2·2H2O, 30 μM FeSO4·7H2O, 0.86 μM CuSO4·5H2O, 1.9 μM H3BO3, 7.7 μM ZnSO4·7H2O, 0.75 μM MnSO4·5H2O, 0.26 μM NiCl2·6H2O, and 0.31 μM Na2MoO4·5H2O. Additionally, succinate (50 mM) was provided as the source of organic carbon. For glyphosate dosage, the succinate-containing growth medium was prepared with different concentrations of glyphosate (0, 0.5, or 5 mM glyphosate). The choice of glyphosate concentration use here was based on the concentration range used in previous studies on the effects of glyphosate exposure on bacterial species: 0.003–3.0 mM (Schulz et al., 1985); 0.1 mM (Liu et al., 1991); 0.03–10 mM (Forlani et al., 2008); 0.075–1.1 mM (Duke et al., 2012); 0.44–29 mM (Shehata et al., 2013). To determine the influence of aromatic AA supplementation on glyphosate effects on growth, cells were also grown on the same media recipe described above supplemented with a cocktail (0.5 mM each) of the three aromatic AAs (tryptophan, tyrosine, and phenylalanine). All the chemicals listed above were obtained analytical grade from Sigma-Aldrich (St. Louis, MO, USA) and Fisher Scientific (Pittsburg, PA, USA). For all the different growth conditions, the cells were transferred twice into fresh growth medium in order to ensure that cells were fully acclimated in their respective growth medium. Cell growth (three biological replicates) was monitored by measuring the optical density at 600 nm (OD600) using an Agilent Cary UV-visible spectrophotometer (Santa Clara, California).
Sampling Intracellular Metabolite Levels
Intracellular metabolite levels were determined from cell suspensions obtained during exponential growth phase. The suspensions were filtered and the cell-containing filters were immediately quenched by submerging them in a cold (4°C) 2-mL solution of methanol:acetonitrile:water (40:40:20). Solutions with the lysed cells were subsequently filter-centrifuged (Sigma-Aldrich Spin-X 0.22 μm filters). Aliquots of the supernatants were dried under nitrogen gas and re-suspended in LC-MS ultrapure water (Fisher Scientific, Pittsburgh, Pennsylvania) before analysis via LC-MS. Metabolite levels were normalized to biomass quantity at the time of sampling.
Kinetic Intracellular Metabolite Labeling
To capture the influence of glyphosate on in vivo metabolic fluxes, we carried out a kinetic flux experiment. This experimental procedure is described in detail elsewhere (Aristilde, 2017). Briefly, cells were first grown in a succinate-containing growth medium recipe as described above. At early exponential phase after the second transfer, aliquots (3 mL) of the cell suspensions were filtered and the cell-containing filters were transferred to a plate with agar-solidified growth medium with unlabeled succinate. Once the cells growing on the plates had reached mid-exponential phase (OD600 0.5–1.0) as described previously by Aristilde (2017), the cell-containing filters were subsequently transferred to plates that contained labeled [U-13C4]-succinate (50 mM) with 0, 0.5 or 5 mM glyphosate. At specific time points (5, 12, and 30 min) following the isotopic switch, the cell-containing filters were quenched and prepared as described above. Control experiments for the 0-min time point were conducted using cell-containing filters from plates with unlabeled succinate. The labeled substrate was obtained from Cambridge Isotopes (Tewskbury, MA, USA).
Metabolomics Analysis via LC-MS
The metabolite extracts were analyzed by reversed-phase ion-pairing liquid chromatography using ultra-high performance LC (Thermo Scientific DionexUltiMate 3000) coupled with high-resolution/accurate-mass spectrometer (Thermo Scientific Q Exactive quadrupole-Orbitrap hybrid mass spectrometer) with electrospray ionization operated in negative mode. An injection sample of 10 μL was used and the column temperature was set to 25°C. A Waters Acquity UPLC BEH C18 1.7 μm with a column size of 2.1 × 100 mm) (Waters Corporation, Massachusetts) was used. Solvent A contained 97:3 (v:v) LC-MS grade H2O: methanol with acetic acid (15 mM) and tributylamine (10 mM). Solvent B contained 100% methanol. The flow rate was 180 μL min−1 during the entire sample run (25 min). The solvent gradient with respect to solvent A was the following: 0 min, 100%; 2.5 min, 100%; 5 min, 80%; 7.5 min, 80%; 10 min, 45%; 12 min, 45%; 14 min, 5%; 17 min, 5%; 18 min, 0%; 25 min, 0% was run. All metabolite identification and isotopic enrichment were determined using the Metabolomics Analysis and Visualization Engine (MAVEN) software package (Clasquin et al., 2012). Corrections for natural abundance of 13C were conducted on the 13C-labeled fractions.
Statistical Analysis
All experiments were conducted in three independent biological replicates. Unpaired two-tailed t-test analyses were conducted to evaluate statistically-significant differences between two conditions. Statistical significance was determined for the following comparisons: specific growth rate of glyphosate-exposed cells compared to cells grown on succinate alone; changes in metabolite levels in P. putida S12 compared to levels in the other Pseudomonas species; labeled fraction of metabolites after 30-min of incorporated 13C-labeled substrate in glyphosate-exposed KT2440 compared to control; and, specific growth rates of P. putida S12 cells grown on glyphosate-containing growth media with and without aromatic amino acids.
Results
Growth Effects Are Both Strain- and Species-Dependent
Figure 2 illustrates the exponential growth rates determined from monitoring the growth of the four Pseudomonas species in medium containing succinate as the growth substrate in the absence and presence of glyphosate at 0.5 mM and 5 mM. For P. putida S12, the growth rate was reduced by 24.2 ± 2.7% (p = 0.003) during exposure to the low glyphosate dose compared to the control whereas growth was completely compromised with no cell growth obtained at the high glyphosate dose (Figure 2A). For P. putida KT2440, the growth rate at the low glyphosate dose was not statistically-significantly compared to the control (Figure 2B). However, at the high dose of glyphosate, the growth rate of the P. putida KT2440 cells was decreased by 23.4 ± 0.4% (p = 0.02) (Figure 2B). By contrast to the two P. putida strains, both P. protegens Pf-5 and P. fluorescens RA12 did not exhibit any adverse growth effects at both glyphosate exposures (Figures 2C,D). These data thus indicated that the P. putida species investigated here have higher sensitivity to glyphosate than the P. fluorescens and P. protegens species.
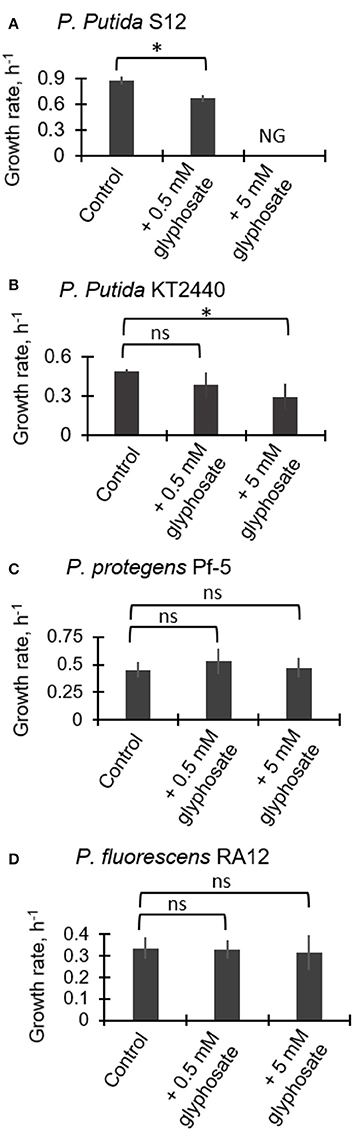
Figure 2. Growth rates of succinate-grown Pseudomonas species in the absence (control) and presence of glyphosate (0.5 mM and 5 mM): (A) P. putida S12; (B) P. putida KT2440; (C) P. protegens Pf-5; (D) P. fluorescens RA12. Abbreviation for (B): NG, no growth. The measured data were from three biological replicates (n = 3). Two-tailed unpaired t-test analysis comparing the specific growth rate of the control experiment to the growth rates of glyphosate-exposed cells: p < 0.05 (*); ns, not statistically significant.
Relevant Glyphosate-Induced Widespread Metabolic Perturbations
To gain insights into the species-specific sensitivity to glyphosate, we profiled the metabolome of glyphosate-exposed cells and compared them to those obtained with cells-grown on succinate alone in the absence of glyphosate (Figure 3). We captured widespread perturbations in the metabolome of all species in response to glyphosate in the growth medium (Figure 3). Given the aforementioned highest sensitivity of P. putida S12 to glyphosate exposure, we used the metabolomic profiling of P. putida S12 as a reference to identify the possible metabolic perturbations responsible for the species-dependent inhibitory effects of glyphosate (Figures 3, 4). We focused on the metabolites that were subjected to the most changes in P. putida S12, at least 10% different from the control in two or more of the three biological replicates in the presence of glyphosate (Figures 3, 4). The relevant metabolites were associated with the shikimate pathway (shikimate-3-phosphate, tyrosine, tryptophan, phenylalanine), precursors to protein biosynthesis (tyrosine, tryptophan, phenylalanine, glutamine, valine), precursors of secondary metabolites (tryptophan, citrulline, ornithine), DNA biosynthesis (thymidine), and the tricarboxylic acid cycle (fumarate) (Figures 3, 4).
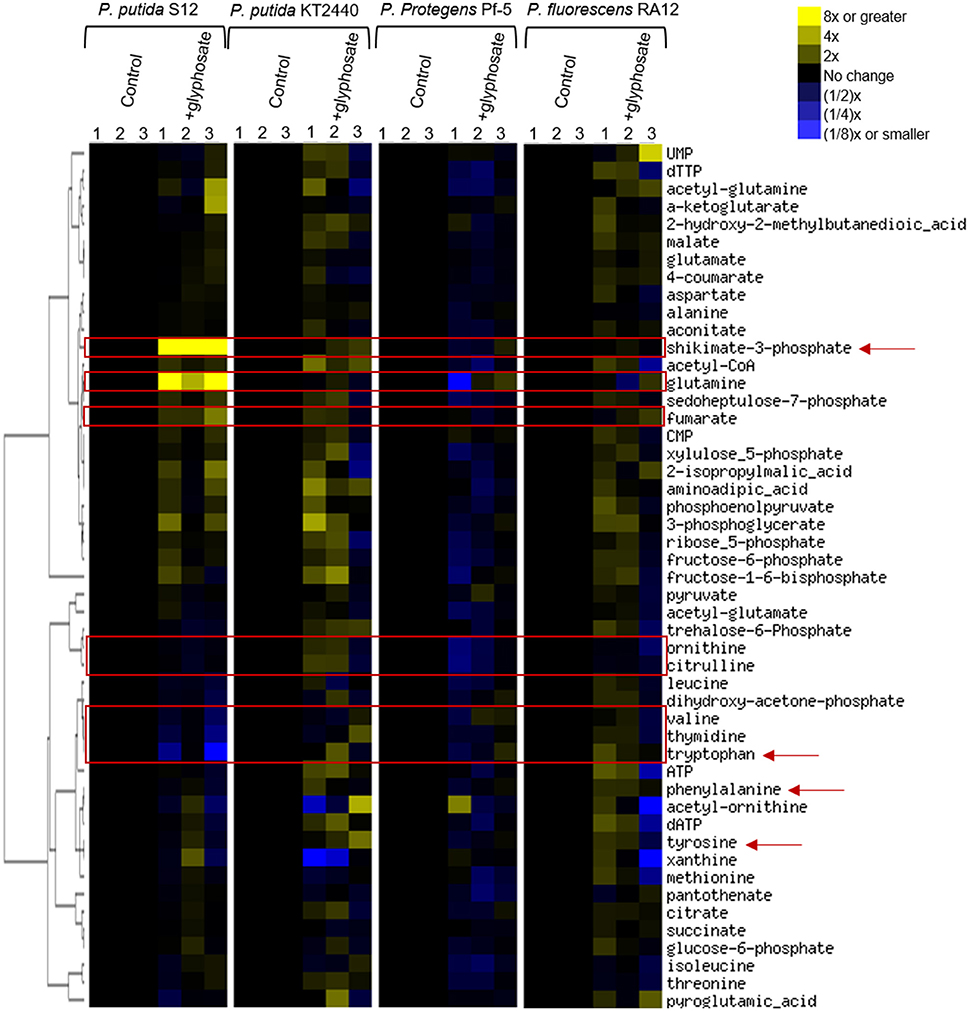
Figure 3. Fingerprinting of the cellular metabolome of P. putida S12, P. putida KT2440, P. protegens Pf-5, P. fluorescens RA12. Unsupervised hierarchical clustering of metabolite levels in succinate-grown cells in the absence (control) and presence of 0.5 mM glyphosate (+glyphosate). Columns 1, 2, and 3 represent data from independent biological replicates. The red rectangular boxes highlight metabolite levels in the metabolome that were disrupted in the presence of glyphosate. The red arrows indicate metabolites that are present in the shikimate pathway.
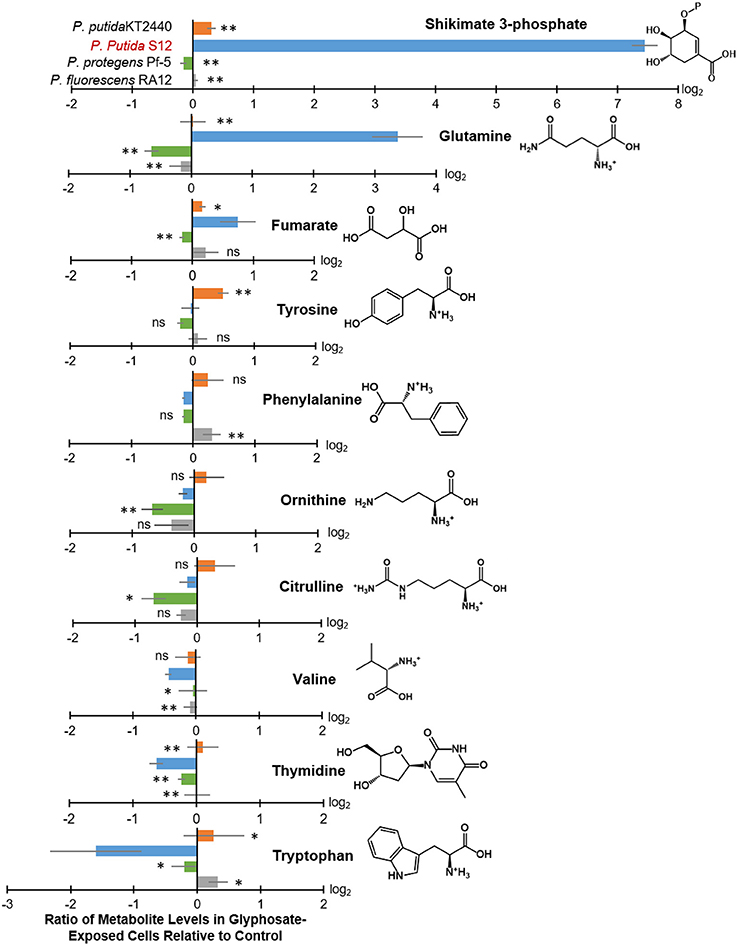
Figure 4. Intracellular levels of selected metabolites in glyphosate-exposed cells relative to control. Extracted from the cellular metabolome shown in Figure 3 are log2-tranformed ratios of metabolite levels in glyphosate-exposed cells (at 0.5 mM) relative to control with statistically-significant differences across the different Pseudomonas species: P. putida KT2440 (orange), P. putida S12 (light blue), P. protegens Pf-5 (green), and P. fluorescens RA12 (gray). Data (average ± standard deviation) were obtained from three independent biological replicates. Two-tailed unpaired t-test analysis comparing the ratios obtained with P. putida S12 (the most sensitive species) to those obtained with each of the other species: p < 0.01 (**), p < 0.05 (*), not statistically significant (ns).
In glyphosate-exposed P. putida S12, we observed depletion in the levels of phenylalanine, ornithine, citrulline, valine, thymidine, and tryptophan (Figure 3). Relative to P. putida S12 cells grown on succinate alone, the levels of phenylalanine, ornithine, and citrulline were only about 10 to 15% less in cells grown on the glyphosate-containing growth medium (Figure 4). There was a more significant depletion of the other metabolites in P. putida S12 in response to glyphosate exposure: valine and thymidine were 35 to 60% less relative to the control and there was an about 5-fold reduction in the tryptophan pool relative to the control (Figure 4). In regards to the other species, there was no statistical difference between the changes in the levels of both ornithine and citrulline recorded in glyphosate-exposed P. putida S12 and the corresponding changes in P. putida KT2440, P. protegens Pf-5, and P. fluorescens RA12 (Figure 4); the change in phenylalanine was either the same as the one recorded for P. putida S12 (P. putida KT2440 and P. protegens Pf-5) or slightly elevated (up to 40% more in P. fluorescens RA12) (Figure 4). Most notably, the changes in the levels of valine, thymidine and tryptophan in the other Pseudomonas species in response to glyphosate were different from the significant depletion obtained with P. putida S12 (p < 0.05, n = 3): valine in the presence of glyphosate remained the same as in the control condition in the other species; there was either no change in thymidine or a small decrease in the presence of glyphosate; and, tryptophan remained the same or was slightly elevated in the other species (Figure 4). These data implied that the severe depletion in these three metabolites (valine, thymidine, and tryptophan) specifically in P. putida S12 may be important in mediating the higher glyphosate toxicity in this bacterium.
In addition to the decrease in the levels of selected metabolites, we obtained a significant buildup of the following metabolites in glyphosate-exposed P. putida S12: shikimate-3-phosphate (up to nearly 200-fold increase), glutamine (up to nearly 16-fold increase), and fumarate (up to 2-fold increase) (Figure 4). The levels of both shikimate-3-phosphate and fumarate were also found to be elevated in P. putida KT2440 (Figure 4). Specifically, shikimate-3-phosphate was about 23% higher and fumarate was about 10% higher than the control in P. putida KT2440 (Figure 4). In contrast, P. protegens Pf-5 and P. fluorescens RA12 grown in the presence of glyphosate did not present any change in the shikimate-3-phosphate level and exhibited no change to a slight increase in the fumarate level (Figure 4). Interestingly, the change in the aromatic AAs was not the same in P. putida S12 (Figure 4). While there was a severe depletion in tryptophan (up to 5-fold decrease) and a modest depletion in phenylalanine (up to 12% decrease), the level of tyrosine remained unchanged in P. putida S12 in response to glyphosate (Figure 4). Whereas, the change in the tyrosine level in both P. protegens Pf-5 and P. putida KT2440 was not statistically different from P. putida S12, P. putida KT2440 had up to 43% increase in tyrosine level in response to glyphosate exposure.
These metabolomics data implied that, in addition to the shikimate pathway, widespread perturbations in the metabolome may exacerbate effects of glyphosate (Figures 3, 4). However, the highest accumulation of shimate-3-phosphate and the greatest depletion of tryptophan pointed specifically to the shikimate pathway as the potential primary toxicity target, as has been proposed previously in other Pseudomonas species (Schulz et al., 1985).
Glyphosate Specifically Targets Aromatic Amino Acid Biosynthesis
It was previously reported that the inhibitory effect of glyphosate on EPSP synthase activity in different Pseudomonas species was species-dependent (Schulz et al., 1985). As a consequence of the inhibition of EPSP synthase in the glyphosate-exposed cells, disruption in downstream metabolic fluxes in the shikimate pathway was expected, specifically in regards to the biosynthesis of the three aromatic AAs (phenylalanine, tyrosine, and tryptophan) downstream this pathway. Focusing on P. protegens Pf-5 and P. putida KT2440, we employed a kinetic flux profiling approach using [U-13C4]-succinate to evaluate explicitly the biosynthesis of aromatic AA as a specific toxicological target of glyphosate (Figure 5). In this approach, evaluation of in vivo flux through a metabolic reaction in the pathway was considered on the basis that the flux would be proportional to the level of the reactant metabolite and the labeling kinetics of the product metabolite (Yuan et al., 2008; Sasnow et al., 2016). Kinetic incorporation of the labeled succinate in both species was monitored at both the low-dose and high-dose glyphosate exposures and we found that the labeling kinetics remained the same in the absence and presence of glyphosate (Figure 5). These results indicated that the uptake of the organic carbon source (i.e., succinate) by the cells was not compromised by the presence of glyphosate. We also found that the labeling kinetics of both PEP, the endogenous analog of glyphosate, and shikimate-3-phosphate remained the same in the glyphosate-exposed cells (Figure 5), thus indicating that the synthesis of these metabolites was not affected by glyphosate exposure.
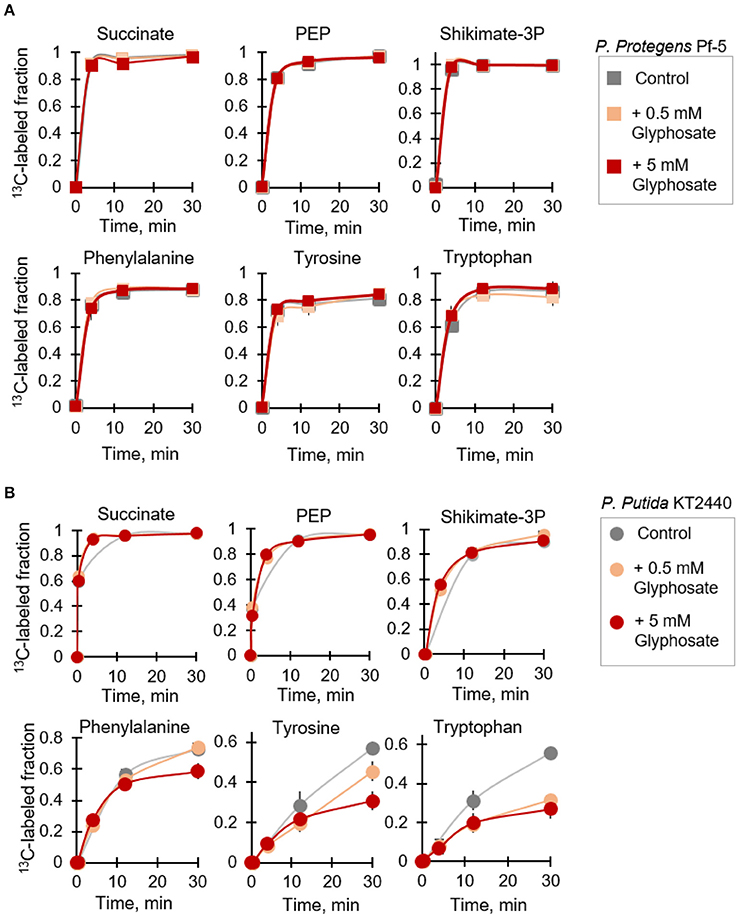
Figure 5. Kinetic isotopic profiling of the shikimate pathway of glyphosate-exposed Pseudomonas species. Dynamic biosynthesis of fully-labeled fractions of metabolites in P. protegens Pf-5 (A) and P. putida KT2440 (B) following isotopic switch from unlabeled succinate to 13C-labeled substrate in the absence (control) or in the presence of glyphosate (at 0.5 mM or 5 mM). The data (average ± standard deviation) are from independent biological replicates (n = 3). Error bars are not visible when they are very small. Compared to the labeling kinetics in the control experiment, the labeling fraction of the aromatic amino acids at 30-min in the P. putida KT2440 cells at the high-glyphosate concentration was statistically-significant (p < 0.05).
On the other hand, we obtained both species-dependent and dose-dependent effects of glyphosate on the biosynthetic fluxes of the aromatic AAs (Figure 5). At the range of glyphosate concentrations investigated here, we did not record any change in the biosynthetic fluxes of all three aromatic AAs in P. protegens Pf-5 (Figure 5A). However, for P. putida KT2440, the 13C isotopic flux in the glyphosate-exposed cells clearly demonstrated an effect of glyphosate exposure compared to the control experiment (Figure 5B). For phenylalanine, the labeling kinetics remained unchanged following exposure of P. putida KT2440 to the low glyphosate dose (Figure 5B). But, when the P. putida KT2440 cells were grown in medium with the high glyphosate dose, the labeling kinetics of phenylalanine remained unchanged initially but there was an approximate 22% decrease in the total fraction of fully labeled phenylalanine (Figure 5B). Compared to P. putida KT2440 cells grown on succinate alone, a slower labeling kinetics was observed in the biosynthetic flux of both tyrosine and tryptophan in the presence of glyphosate (Figure 5B). By 30 min, the total fraction of fully labeled tyrosine was decreased by 21 and 46% at the low-dose and high-dose glyphosate exposures, respectively (Figure 5B). The corresponding decrease for labeled tryptophan was 43 and 52%, respectively (Figure 5B). In accordance with our data, the increasing order of impaired biosynthetic flux with respect to the shikimate-associated aromatic AAs in P. putida KT 2440 in the presence of glyphosate was phenylalanine, tyrosine, and tryptophan (Figure 5B).
Exogenous Supply of Aromatic Amino Acids Overcomes Lethal Glyphosate Dose
Our metabolomics data revealed widespread changes in metabolite levels from different pathways in the cellular metabolome (Figure 4). And, kinetic 13C isotopic flux data with glyphosate-exposed cells demonstrated specific disruption of metabolic fluxes through the shikimate pathway, thereby confirming that the well-known toxicity of glyphosate in plants via the inhibition of the biosynthesis of aromatic AAs was also evident in P. putida KT2440, even at sub-lethal glyphosate exposure (Figure 5B). As previously discussed, amongst the Pseudomonas species investigated here, only the P. putida S12 cells exhibited complete growth inhibition in response to glyphosate exposure, specifically in the high-glyphosate condition (Figure 2A). To determine whether the lethal consequence of glyphosate (at 5 mM) in P. putida S12 was due primarily to a deficiency in the biosynthetic supply of aromatic AAs, we repeated the growth experiments with a growth mixture that contains all three aromatic AAs (tryptophan, tyrosine, phenylalanine) (Figure 6A). Remarkably, the exogenous supply of aromatic AAs in the growth medium led to the complete recovery of the cell growth (Figure 6A). Specifically, the recovered growth rate at 0.61 ± 0.17 h−1 in the glyphosate-containing growth medium supplemented with the aromatic AAs was similar, within an error imprecision of one standard deviation, to the growth rate (0.88 ± 0.11 h−1) of the cells in the absence of glyphosate (Figure 6A). These results implied that P. putida S12 cells lost the biosynthetic ability to produce aromatic AAs in the presence of high glyphosate dose and thus must rely on extracellular supply to survive.
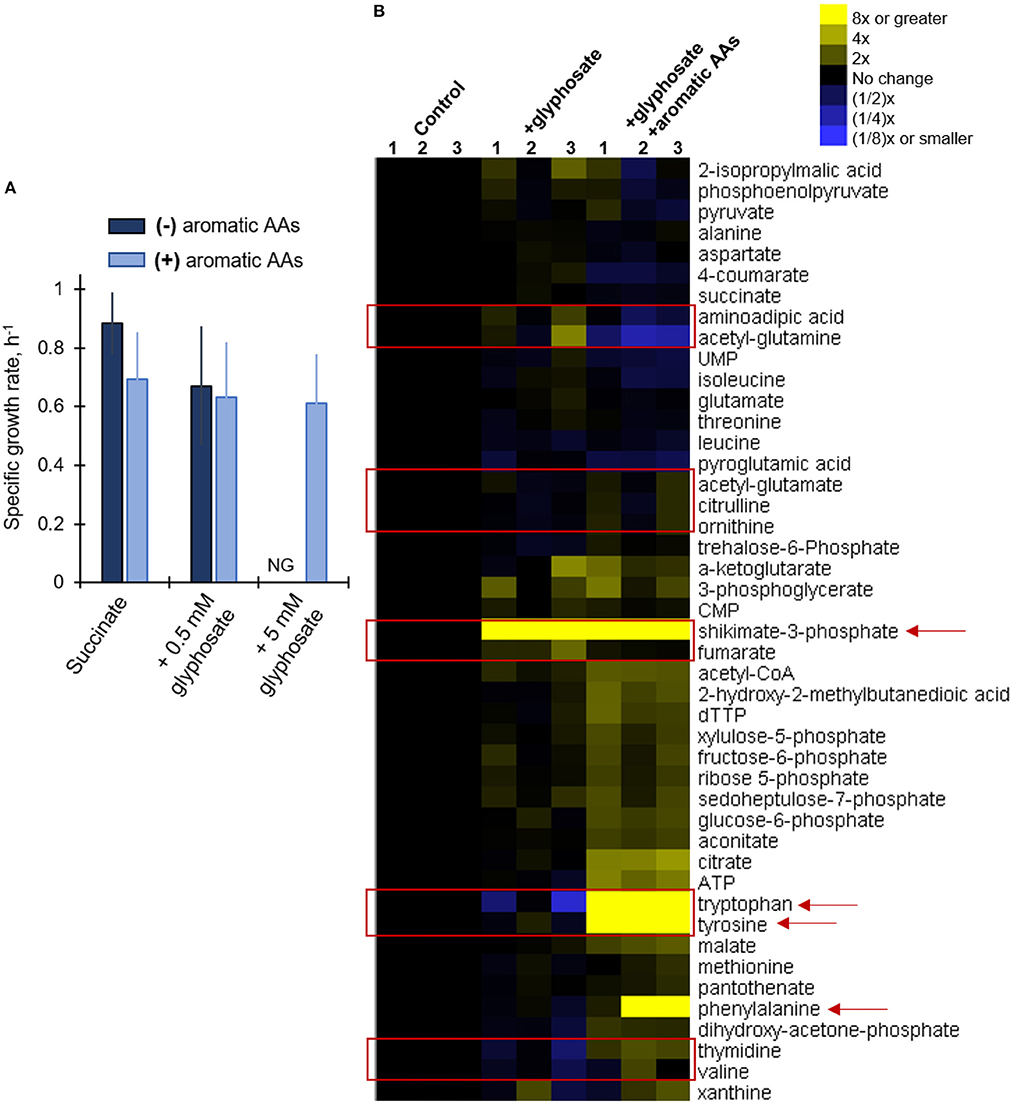
Figure 6. Growth and metabolic phenotypes of glyphosate-exposed P. putida S12 to supplementation of aromatic amino acids in the growth media. (A) Growth rates (average ± standard deviation) of succinate-grown and glyphosate-exposed cells without (dark blue) and with (light blue) added aromatic amino acids (AAs) in the growth medium); see materials and methods for more details. (B) Unsupervised hierarchical clustering of metabolite levels in succinate-grown cells in the absence (control), presence of 0.5 mM glyphosate (+glyphosate) or the lethal glyphosate dose, 5 mM, and aromatic AAs (+glyphosate +aromatic AAs). Columns 1, 2, and 3 represent data from independent biological replicates. The red rectangular boxes highlight metabolite levels in the metabolome that were disrupted in the presence of glyphosate. The red arrows indicate metabolites that are present in the shikimate pathway. Abbreviation for (A): NG, no growth. The measured data in (A) were from three biological replicates (n = 3).
In addition, we also probe whether the exogenous supply aromatic AAs also led to the recovery of metabolic disruptions (Figure 6B). The buildup in shikimate-3-phosphate that was captured in the glyphosate-exposed cells in the absence of the aromatic AAs remained in the cells after the supplement of aromatic AA (Figure 4). This was consistent with continued inhibition of the EPSP synthase by glyphosate (Figure 1B). However, the downstream effect of this inhibition was circumvented by the accumulation of all three aromatic AAs in the cellular metabolome due to the exogenous supply of these AAs in the growth medium (Figure 6B). Our metabolomics profiling of P. putida S12 at the lethal glyphosate level (5 mM) with aromatic AAs revealed that the supplement of aromatic AAs restored the levels of several metabolites beyond those associated with the shikimate pathway (Figure 6B). Specifically, the levels of both the nucleoside thymidine and the AA valine, which were shown to be depleted in P. putida S12 in the presence of glyphosate, restored either to the same level as the control or were slightly elevated in response to the aromatic AA supplement (Figure 6B). We also found that the level of fumarate was restored to the level of the control in cells grown on media containing both glyphosate and the aromatic AAs (Figure 6B). Other major changes in the metabolome of P. putida S12 were also observed when comparing the glyphosate-exposed cells grown in the absence and in the presence of the aromatic AA supplement (Figure 6). Notably, the levels of citrulline, ornithine, acetyl-glutamate increased in the presence of excess aromatic AAs (Figure 6B). On the other hand, the levels of aminoadipic acid and acetyl-glutamine decreased substantially in the presence of excess aromatic AAs (Figure 6B). We are not able to determine the exact relevance of these changes but they suggest that the cellular level of aromatic AAs was connected to the metabolic balance of other amino acids or amino acid-containing metabolites.
Discussion
As stated in the Introduction, we postulated three hypotheses underlying the sensitivity of soil Pseudomonas species to glyphosate exposure: (1) species-specific growth inhibitory effects, (2) inhibition of the shikimate pathway, and (3) disruption of other cellular pathways. In relation to the first hypothesis, we show here that within the same Pseudomonas genus, the growth responses of glyphosate-exposed cells were both species-dependent and strain-dependent (Figure 2). Notably, the growth of both the P. protegens and P. fluorescens species used here was not affected by the range of glyphosate concentration added to the growth medium (Figure 2). By contrast, the P. putida species were more sensitive to the glyphosate-containing growth media whereby, compared to growth in the absence of glyphosate, the strain S12 experienced up to 100% growth inhibition but the strain KT2440 only exhibited up to about 23% growth inhibition (Figure 2). There have been conflicting results on the adverse effects of glyphosate exposure on soil microbial community structure (Weaver et al., 2007; Johal and Huber, 2009; Kremer and Means, 2009; Mijangos et al., 2009; Barriuso et al., 2011; Duke et al., 2012). Our results here support the proposal that species-dependent sensitivities to glyphosate led to the observed change in microbial community structure in soils amended with glyphosate (Ratcliff et al., 2006; Newman et al., 2016). It was found that six different genera of cyanobacteria responded differently to glyphosate concentrations (from 0.03 to 10 mM) and the growth response ranged from no change to lethal effects (Forlani et al., 2008). Along with our findings, these previous reports implied that the wide range of sensitivities of different genera and species of microorganisms will lead to dissimilar effects of glyphosate on different ecosystems.
We evaluated the second and third hypotheses by profiling the cellular metabolome, which revealed species-dependent widespread metabolic disruptions (Figure 3). An accumulation in the levels of metabolites upstream of the EPSP synthase was captured in time-course experiments of glyphosate-exposed pea plant Pisum sativum (Zabalza et al., 2016). We note that the metabolite shikimate-3P is upstream of EPSP synthase, the targeted enzyme in glyphosate-induced toxicity in plants (Figure 1B). Therefore, the buildup of shikimate-3P in P. putida KT2440 and P. putida S12 was indicative of inhibition of EPSP synthase activity by glyphosate in these species (Schönbrunn et al., 2001) (Figures 3, 4). Interestingly, hierarchical clustering of the metabolome indicated that the disruption in non-shikimate pathway metabolites was clustered with several of the shikimate pathway-associated metabolites (Figure 3). Specifically, elevated glutamine and fumarate levels in P. putida S12 were clustered with the significant accumulation of shikimate-3-phosphate (Figure 3). And, the significant depletion in tryptophan level was clustered with depletion of valine and thymidine (Figure 3). In accordance with our results, impairment of pathways involved in energy metabolism (Orcaray et al., 2012) and protein synthesis (Maroli et al., 2016) were found in glyphosate-treated plants. Intracellular accumulation of AAs and depletion in protein synthesis have been reported for the glyphosate-sensitive biotype of the flowering plant Amaranthus palmeri (Maroli et al., 2016). This was proposed to be due to the metabolic response of the plant to the abiotic stressor (Maroli et al., 2016), which may include a decrease in de novo protein synthesis. Thus, we attributed the significant accumulation of glutamine in the glyphosate-sensitive P. putida S12 to either inhibition of protein synthesis or disruption of other glutamine-consuming metabolic pathways by glyphosate (Figure 4). By contrast, in P. protegens Pf-5 and P. fluorescens RA12, there was a depletion in glutamine in the glyphosate-exposed cells, thus indicating that glutamine-consuming pathways were less inhibited in these cells (Figure 4). Furthermore, our results with the two species most sensitive to glyphosate (P. putida S12 and P. putida KT2440) indicated a more severe effect on the level of tryptophan than the other two aromatic AAs (Figure 4). This disproportionate effect on the pools of aromatic AAs in P. putida KT2440 may be due to the differential role of each amino acid in the cellular response to glyphosate exposure. Further metabolic studies are needed to verify this proposal.
Our 13C kinetic flux profiling data revealed a species-dependent inhibition of biosynthetic fluxes of the aromatic AAs that are synthesized downstream of the shikimate pathway (Figure 5). Whereas, these fluxes remained the same in P. protegens Pf-5 upon exposure to glyphosate, we obtained up to 60% decrease in the final isotopic incorporation in the aromatic AAs in P. putida KT2440 (Figure 5). This species-dependent difference in the 13C kinetic flux was consistent with the species-dependent growth responses to glyphosate (Figures 2, 5). Supplementation of growth medium with aromatic AAs led to complete growth recovery of P. putida S12 exposed to lethal dose of glyphosate (Figure 6A). These results implied that the specific impairment of de novo biosynthesis of aromatic AAs was primarily responsible for the growth effects on the different Pseudomonas species. And subsequent metabolomics profiling demonstrated that the aromatic AA supplementation also led to the recovery of metabolic homeostasis as well as changes that deviated from the control condition (Figure 6B). Our findings suggest that rhizospheric bacteria may be less susceptible to glyphosate effects due to the high-carbon environment in the rhizosphere with relatively higher concentration of amino acids and sugars relative to bulk soils.
Four important caveats should be considered in assessing the relevance of our findings. First, we concluded from our data that the inhibition of the biosynthesis of aromatic AAs was a specific target of glyphosate. The different glyphosate-induced growth effects and metabolic phenotypes implied different sensitivities of the metabolome, in addition to the shikimate pathway. In fact, it was found previously that the inhibition of the activity of EPSP synthase, an important enzyme in the initiation of the shikimate pathway, was greater in P. fluorescens AFT36 than in P. putida W1616 (Schulz et al., 1985). We also note that additional factors may be contributing to these differences such as different cell permeability to cellular glyphosate uptake and different ability to break down glyphosate (Kishore and Jacob, 1987; Liu et al., 1991; Zboińska et al., 1992). Second, Pseudomonas species are known to produce auxin-like plant hormones including IAA and antifungal metabolites such as pyrrolnitrin, both of which rely on the aromatic AA tryptophan as an important metabolite precursor derived from the shikimate pathway. It remains to be determined whether the biosynthetic pathway of these plant growth-beneficial molecules is negatively affected by glyphosate treatment due to depletion of aromatic AAs. Third, our study focused on glyphosate, the parental active herbicide ingredient. It is important to note that aminomethylphosphonic acid, the primary degradation product of glyphosate, has been widely detected along with glyphosate in environmental matrices (Reddy et al., 2008; Battaglin et al., 2014). Moreover, glyphosate-based herbicide formulations contain a number of additives including surfactants. An evaluation of synergistic or counteracting effects of glyphosate, its breakdown products, and surfactants is warranted. Fourth, several soil Pseudomonas species are beneficial players in the plant microbiome near the rhizosphere. In addition to these species, we recognize that a diverse number of bacterial and fungal genera exist in this microbiome. A comprehensive understanding of the responses of the entire rhizospheric microbiome is required to assess fully the non-targeted metabolic effects of glyphosate on crop-beneficial microbial communities.
Author Contributions
LA supervised the research, conducted extensive analysis of data, and wrote the manuscript. LA, MLR, RAW, and MAK designed the research. MLR, VK, RAW, and CRSS conducted the different growth experiments. MLR, RAW, and TY conducted the metabolomics experiments. All authors contributed to preliminary data analysis and provided edits to drafts of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Graduate support for RAW, TY, and MAK was provided, respectively, by a graduate fellowship from Cornell University, a scholarship from the Schooner foundation, and an Integrative Graduate Education and Research Traineeship (IGERT) research fellowship from the National Science Foundation. Undergraduate support for MLR, VK, and CRSS was provided by the National Institute of Food and Agriculture (Hatch project 1237419) and an Academic Venture Fund from the Cornell Atkinson Center for a Sustainable Future. This research was supported also in part by a start-up package from Cornell University.
References
Aristilde, L. (2017). Metabolite labeling reveal hierarchies in Clostridium acetobutylicum that selectively channels carbons from sugar mixtures towards biofuel precursors. Microb. Biotechnol. 10, 162–174. doi: 10.1111/1751-7915.12459
Barriuso, J., Marín, S., and Mellado, R. P. (2011). Potential accumulative effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities over a three-year cultivation period. PLoS ONE 6:e27558. doi: 10.1371/journal.pone.0027558
Battaglin, W. A., Meyer, M. T., Kuivila, K. M., and Dietze, J. E. (2014). Glyphosate and its degradation product AMPA occur frequently and widely in U.S. soils, surface water, groundwater, and precipitation. J. Am. Water Resour. Assoc. 50, 275–290. doi: 10.1111/jawr.12159
Clasquin, M., Melamud, E., and Rabinowitz, J. D. (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Prot. Bioinform. 37, 14111–141123.
Druille, M., Cabello, M. N., García Parisi, P. A., Golluscio, R. A., and Omacini, M. (2015). Glyphosate vulnerability explains changes in root-symbionts propagules viability in pampean grasslands. Agric. Ecosyst. Environ. 202, 48–55. doi: 10.1016/j.agee.2014.12.017
Druille, M., Cabello, M. N., Omacini, M., and Golluscio, R. A. (2013). Glyphosate reduces spore viability and root colonization of arbuscular mycorrhizal fungi. Appl. Soil Ecol. 64, 99–103. doi: 10.1016/j.apsoil.2012.10.007
Duke, S. O., Lydon, J., Koskinen, W. C., Moorman, T. B., Chaney, R. L., and Hammerschmidt, R. (2012). Glyphosate effects on plant mineral nutrition, crop rhizosphere microbiota, and plant disease in glyphosate-resistant crops. J. Agric. Food Chem. 60, 10375–10397. doi: 10.1021/jf302436u
Forlani, G., Pavan, M., Gramek, M., Kafarski, P., and Lipok, J. (2008). Biochemical bases for a widespread tolerance of cyanobacteria to the phosphonate herbicide glyphosate. Plant Cell. Physiol. 49, 443–456. doi: 10.1093/pcp/pcn021
Funke, T., Han, H., Healy-Fried, M. L., Fischer, M., and Schönbrunn, E. (2006). Molecular basis for the herbicide resistance of roundup ready crops. Proc. Natl. Acad. Sci. U.S.A. 103, 13010–13015. doi: 10.1073/pnas.0603638103
Johal, G. S., and Huber, D. M. (2009). Glyphosate effects on diseases of plants. Eur. J. Agron. 31, 144–152. doi: 10.1016/j.eja.2009.04.004
Johal, G. S., and Rahe, J. E. (1984). Effect of soilborne plant-pathogenic fungi on the herbicidal action of glyphosate on bean seedlings. Phytopathology 74, 950–955. doi: 10.1094/Phyto-74-950
Kirner, S., Hammer, P. E., Hill, D. S., Altmann, A., Fischer, I., Weislo, L. J., et al. (1998). Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 180, 1939–1943.
Kishore, G. M., and Jacob, G. S. (1987). Degradation of glyphosate by Pseudomonas sp. PG2982 via a sarcosine intermediate. J. Biol. Chem. 262, 12164–12168.
Kremer, R. J., and Means, N. E. (2009). Glyphosate and glyphosate-resistant crop interactions with rhizosphere microorganisms. Eur. J. Agron. 31, 153–161. doi: 10.1016/j.eja.2009.06.004
Kremer, R., Means, N., and Kim, S. (2005). Glyphosate affects soybean root exudation and rhizosphere micro-organisms. Int. J. Environ. Anal. Chem. 85, 1165–1174. doi: 10.1080/03067310500273146
Lancaster, S. H., Hollister, E. B., Senseman, S. A., and Gentry, T. J. (2010). Effects of repeated glyphosate applications on soil microbial community composition and the mineralization of glyphosate. Pest. Manag. Sci. 66, 59–64. doi: 10.1002/ps.1831
Lane, M., Lorenz, N., Saxena, J., Ramsier, C., and Dick, R. P. (2012). The effect of glyphosate on soil microbial activity, microbial community structure, and soil potassium. Pedobiologia 55, 335–342. doi: 10.1016/j.pedobi.2012.08.001
Larson, R. L., Hill, A. L., Fenwick, A., Kniss, A. R., Hanson, L. E., and Miller, S. D. (2006). Influence of glyphosate on rhizoctonia and fusarium root rot in sugar beet. Pest Manag. Sci. 62, 1182–1192. doi: 10.1002/ps.1297
Lévesque, C. A., Rahe, J. E., and Eaves, D. M. (1987). Effects of glyphosate on Fusarium spp.: its influence on root colonization of weeds, propagule density in the soil, and crop emergence. Can. J. Microbiol. 33, 354–360. doi: 10.1139/m87-062
Lévesque, C. A., and Rahe, J. R. (1992). Herbicide Interactions with Fungal root pathogens, with special reference to glyphosate. Annu. Rev. Phytopathol. 30, 579–602. doi: 10.1146/annurev.py.30.090192.003051
Liu, C.-M., McLean, P. A., Sookdeo, C. C., and Cannon, F. C. (1991). Degradation of the herbicide glyphosate by members of the family rhizobiaceae. Appl. Environ. Microbiol. 57, 1799–1804.
Maroli, A., Nandula, V., Duke, S., and Tharayil, N. (2016). Stable isotope resolved metabolomics reveals the role of anabolic and catabolic processes of glyphosate-induced amino acid accumulation in Amaranthus palmeri biotypes. J. Agric. Food Chem. 64, 7040–7048. doi: 10.1021/acs.jafc.6b02196
Mijangos, I., Becerril, J. M., Albizu, I., Epelde, L., and Garbisu, C. (2009). Effects of glyphosate on rhizosphere soil microbial communities under two different plant compositions by cultivation-dependent and -independent methodologies. Soil Biol. Biochem. 41, 505–513. doi: 10.1016/j.soilbio.2008.12.009
Newman, M. M., Hoilett, N., Lorenz, N., Dick, R. P., Liles, M. R., Ramsier, C., et al. (2016). Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci. Tot. Environ. 543, 155–160. doi: 10.1016/j.scitotenv.2015.11.008
Orcaray, L., Zulet, A., Zabalza, A., and Royuela, M. (2012). Impairment of carbon metabolism induced by the herbicide glyphosate. J. Plant Physiol. 169, 27–33. doi: 10.1016/j.jplph.2011.08.009
Ratcliff, A. W., Busse, M. D., and Shestak, C. J. (2006). Changes in microbial community structure following herbicide (glyphosate) additions to forest soils. Appl. Soil. Ecol. 34, 114–124. doi: 10.1016/j.apsoil.2006.03.002
Reddy, K. N., Rimando, A. M., Duke, S. O., and Nandula, V. K. (2008). Aminomethylphosphonic acid accumulation in plant species treated with glyphosate. J. Agric. Food Chem. 56, 2125–2130. doi: 10.1021/jf072954f
Santos, A., and Flores, M. (1995). Effects of glyphosate on nitrogen fixation of free-living heterotrophic bacteria. Lett. Appl. Microbiol. 20, 349–352. doi: 10.1111/j.1472-765X.1995.tb01318.x
Sasnow, S. S., Wei, H., and Aristilde, L. (2016). Bypasses in intracellular glucose metabolism in iron-limited Pseudomonas putida. Microbiologyopen 5, 3–20. doi: 10.1002/mbo3.287
Schönbrunn, E., Eschenburg, S., Shuttleworth, W. A., Schloss, J. V., Amrhein, N., Evans, J. N., et al. (2001). Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380. doi: 10.1073/pnas.98.4.1376
Schulz, A., Krüper, A., and Amrhein, N. (1985). Differential sensitivity of bacterial 5-enolpyruvylshikimate-3-phosphate synthases to the herbicide glyphosate. FEMS Microbiol. Lett. 28, 297–301. doi: 10.1111/j.1574-6968.1985.tb00809.x
Shehata, A. A., Schrödl, W., Aldin, A. A., Hafez, H. M., and Krüger, M. (2013). The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro. Curr. Microbiol. 66, 350–358. doi: 10.1007/s00284-012-0277-2
Steinrücken, H. C., and Amrhein, N. (1980). The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 94, 1207–1212. doi: 10.1016/0006-291X(80)90547-1
Timmis, K. N. (2002). Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ. Microbiol. 4, 779–781. doi: 10.1046/j.1462-2920.2002.00365.x
Tzin, V., and Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways. Mol. Plant 3, 956–972. doi: 10.1093/mp/ssq048
Weaver, M. A., Krutz, L. J., Zablotowicz, R. M., and Reddy, K. N. (2007). Effects of glyphosate on soil microbial communities and its mineralization in a Mississippi soil. Pest Manag. Sci. 63, 388–393. doi: 10.1002/ps.1351
Woodburn, A. T. (2000). Glyphosate: production, pricing and use worldwide. Pest Manag. Sci. 56, 309–312. doi: 10.1002/(SICI)1526-4998(200004)56:4<309::AID-PS143>3.0.CO;2-C
Yuan, J., Bennett, B. D., and Rabinowitz, J. D. (2008). Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat. Protoc. 3, 1328–1340. doi: 10.1038/nprot.2008.131
Zabalza, A., Orcaray, L., Fernández-Escalada, M., Zulet-González, A., and Royuela, M. (2016). The pattern of shikimate pathway and phenylpropanoids after inhibition by glyphosate or quinate feeding in pea roots. Pest. Biochem. Phys. doi: 10.1016/j.pestbp.2016.12.005. [Epub ahead of print].
Zboińska, E., Lejczak, B., and Kafarski, P. (1992). Organophosphonate utilization by the wild-type strain of Pseudomonas fluorescens. Appl. Environ. Microbiol. 58, 2993–2999.
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Keywords: metabolomics of glyphosate effects, soil Pseudomonas species, aromatic amino acid biosynthesis, disruption of cellular metabolome, non-targeted effects
Citation: Aristilde L, Reed ML, Wilkes RA, Youngster T, Kukurugya MA, Katz V and Sasaki CRS (2017) Glyphosate-Induced Specific and Widespread Perturbations in the Metabolome of Soil Pseudomonas Species. Front. Environ. Sci. 5:34. doi: 10.3389/fenvs.2017.00034
Received: 13 March 2017; Accepted: 06 June 2017;
Published: 20 June 2017.
Edited by:
Johann G. Zaller, University of Natural Resources and Life Sciences, Vienna, AustriaReviewed by:
Robert J. Kremer, University of Missouri, United StatesMarcelo Pedrosa Gomes, Universidade Federal de Minas Gerais, Brazil
Copyright © 2017 Aristilde, Reed, Wilkes, Youngster, Kukurugya, Katz and Sasaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludmilla Aristilde, ludmilla@cornell.edu
 Ludmilla Aristilde
Ludmilla Aristilde Michael L. Reed1
Michael L. Reed1  Rebecca A. Wilkes
Rebecca A. Wilkes Tracy Youngster
Tracy Youngster Matthew A. Kukurugya
Matthew A. Kukurugya