Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni
- 1 LUNAM Université, Oniris, University of Nantes, Nantes, France
- 2 UMR1014 Sécurité des Aliments et Microbiologie, INRA, Nantes, France
- 3 Unité Hygiène et Qualité des Produits Aviaires et Porcins, Administración Nacional de la Seguridad Social, Ploufragan, France
- 4 UMR1319 MICALIS, Plateforme d’Analyse Protéomique de Paris Sud-Ouest, INRA, Jouy-en-Josas, France
- 5 Department of Microbiology, Immunology, and Cancer Biology, University of Virginia, Charlottesville, VA, USA
Polynucleotide phosphorylase (PNPase), encoded by the pnp gene, is known to degrade mRNA, mediating post-transcriptional regulation and may affect cellular functions. The role of PNPase is pleiotropic. As orthologs of the two major ribonucleases (RNase E and RNase II) of Escherichia coli are missing in the Campylobacter jejuni genome, in the current study the focus has been on the C. jejuni ortholog of PNPase. The effect of PNPase mutation on C. jejuni phenotypes and proteome was investigated. The inactivation of the pnp gene reduced significantly the ability of C. jejuni to adhere and to invade Ht-29 cells. Moreover, the pnp mutant strain exhibited a decrease in C. jejuni swimming ability and chick colonization. To explain effects of PNPase on C. jejuni 81-176 phenotype, the proteome of the pnp mutant and parental strains were compared. Overall, little variation in protein production was observed. Despite the predicted role of PNPase in mRNA regulation, the pnp mutation did not induce profound proteomic changes suggesting that other ribonucleases in C. jejuni might ensure this biological function in the absence of PNPase. Nevertheless, synthesis of proteins which are involved in virulence (LuxS, PEB3), motility (N-acetylneuraminic acid synthetase), stress-response (KatA, DnaK, Hsp90), and translation system (EF-Tu, EF-G) were modified in the pnp mutant strain suggesting a more specific role of PNPase in C. jejuni. In conclusion, PNPase deficiency induces limited but important consequences on C. jejuni biology that could explain swimming limitation, chick colonization delay, and the decrease of cell adhesion/invasion ability.
Introduction
Campylobacter jejuni is considered as the leading cause of human bacterial gastroenteritis in the world (Skirrow, 1994; Altekruse et al., 1999). The main reservoir of C. jejuni is the guts of avian species with up to 109 CFU/g of feces (Newell and Fearnley, 2003). Although gut colonization is asymptomatic in mammals and birds, this bacterium causes diarrhea, fever, and abdominal pain in humans. Moreover, in rare but significant cases, C. jejuni triggers autoimmune disease such as Guillain–Barré syndrome (Nachamkin et al., 1998). Consumption and handling poultry meat products are the major sources of campylobacteriosis in developed countries (Cohn et al., 2007).
To analyze gene regulation in C. jejuni and its impact on cellular functions, several regulators have been identified and characterized. They are involved in oxidative stress (PerR; Palyada et al., 2009), in iron homeostasis (Fur; Holmes et al., 2005), in host cells interaction (Andersen et al., 2005; Fields and Thompson, 2008), in colonization (Guo et al., 2008; Palyada et al., 2009), and other have pleiotropic functions such as HspR (Andersen et al., 2005). Besides these regulators, RNA degradation mechanisms have shown to be determinant for the post-transcriptional control of gene expression and are also known to be involved in cellular functions. RNases mediate the processing, decay and quality control of RNA, and could be endo- or exoribonucleases (Arraiano et al., 2010). In bacteria, many times mRNAs are first degraded into smaller fragments by endonucleases including RNase E, RNase G, RNase III, and RNase P (Mackie, 1998; Linton et al., 2000; Deutscher, 2006) and subsequently they are degraded in the 3′-5′ direction by exonucleases such as RNase II, polynucleotide phosphorylase (PNPase), and RNase R, (Donovan and Kushner, 1986; Deutscher, 2006; Andrade et al., 2009). Complexes of RNases have been also described as RNA-degrading machines namely degradosomes, involved in RNA degradation control of bacteria including Escherichia coli (Carpousis, 2007; Arraiano et al., 2010). Such a protein complex has not been yet described in C. jejuni and little is known about post-transcriptional regulation mediated by ribonucleases. In C. jejuni, RNase E and RNase II responsible for the major RNA degradation in Gram-negative bacteria are absent, however a pnp gene has been identified in all sequenced genomes of the species. PNPase encoded by pnp gene is a 3′-5′ exoribonuclease identified in a large number of both eukaryotic and prokaryotic organisms. It has both 3′-5′ exoribonuclease activity and 3′-terminal oligonucleotide polymerase activity (Mohanty and Kushner, 2000) and plays important roles in mRNA degradation, tRNA processing, and small RNA (sRNA) turnover. In most of bacterial sequenced genomes, orthologous genes to pnp have been identified (Leszczyniecka et al., 2004).
Polynucleotide phosphorylase does not seem to be essential at optimal temperature in E. coli, unless other exoribonucleases (RNase II or RNase R) are also missing. PNPase is however essential for E. coli growth at low temperatures (Luttinger et al., 1996; Zangrossi et al., 2000): It is required to grow after a cold shock in E. coli (García-Mena et al., 1999; Beran and Simons, 2001; Briani et al., 2007; Matus-Ortega et al., 2007) and it is involved in cold adaptation of Bacillus subtilis (Wang and Bechhofer, 1996), Salmonella enterica (Clements et al., 2002), and the psychrotrophic bacterium Yersinia enterocolitica (Goverde et al, 1998). C. jejuni can not grow below 30°C, but can survive several weeks at 4°C. In this context, we have previously shown that PNPase was also involved in the survival at refrigerated temperatures (under 10°C; Haddad et al., 2009). The role of PNPase in the other cellular functions in diverse bacterial species has been also investigated. It was reported to be implicated in antibiotic resistance and competence of B. subtilis (Luttinger et al., 1996; Bechhofer and Stasinopoulos, 1998; Bechhofer and Wang, 1998), and in resistance to oxidative stress in E. coli (Wu et al., 2009). This exoribonuclease enzyme also regulates virulence determinants such as host cell invasion, type III secretion systems, and infection in mouse in S. enterica, Y. pestis, and Y pseudotuberculosis pathogens (Clements et al., 2002; Rosenzweig et al., 2005, 2007; Ygberg et al., 2006).
Considering the wide potential cellular functions of PNPase in the bacterial world and the absence of gene encoding the major exoribonuclease RNase II in C. jejuni, the aim of this study was to characterize the phenotypic effects of a pnp mutation in C. jejuni strain on the proteome, cell motility, in vitro cell adhesion/invasion and chick in vivo colonization.
Materials and Methods
Bacterial Strains and Growth Conditions
Campylobacter jejuni 81-176 and F38011 strains and its derivative mutants defective in PNPase (C. jejuni 81Δpnp and F38 Δpnp) were described in our previous paper (Haddad et al., 2009). The pnp mutant was obtained by double crossover homologous recombination. C. jejuni was routinely cultured on Karmali agar plates (Oxoid, Basingstoke, Hampshire, England), or in brain and heart infusion (BHI) broth (Merck, Darmstadt, Germany) under 110 rpm shaking. C. jejuni was grown under microaerobic conditions (10% carbon dioxide, 5% oxygen, and 85% nitrogen) at 37° or 42°C C. jejuni enumeration was performed on Karmali agar plates incubated 48 h at 42° C under microaerobic conditions. Kanamycin (Km) 50 μg/ml was used for C. jejuni 81 Δ pnp strain growth. Strains and plasmids used in this study are listed in Table 1.
To determine the doubling time of the parental and the mutant strains, we first determined the growth rate in exponential growth phase. C. jejuni cultures were grown in BHI and incubated at different temperatures (32, 34, 37, and 42°C). Experiments were repeated three times independently.
Complementation
As described in Haddad et al. (2009), trans-complementation of the derivative strains was done using the entire C. jejuni pnp gene and its natural promoter cloned into pRY111 vector. The recombinant vector was introduced in the C. jejuni derivative strain (Δpnp) by electroporation, and transformants resistant to both kanamycin at 50 μg/ml and chloramphenicol at 15 μg/ml were isolated.
Motility Assay
The parental strains (C. jejuni 81-176 and F38011), the mutant strains (C. jejuni 81 Δpnp and F38Δpnp) and the F38Δpnp complemented strain were grown overnight on Karmali plates at 42° C under microaerobic conditions. They were harvested using BHI broth to obtain a cell suspension (OD600 = 0.1). One microliter of the cell suspension was deposited in the center of BHI soft agar plates (0.25 or 0.4% agar). After 48 h incubation in microaerobic conditions at 37 or 42°C, the ability of the strain to move in the soft agar was evaluated by measuring the diameter of the spread colonies. The assays were done four times independently.
Ht-29 Cell Infection
Bacterial adhesion and invasion into Ht-29 cells was studied using the gentamicin protection assay as described previously (Haddad et al., 2010). Briefly, microplate wells were seeded with 2 × 105 Ht-29 cells then incubated for 5 days at 37°C in a humidified, 5% CO2 incubator. After washing, the Ht-29 monolayers were infected with a suspension of approximately 2 to 5 × 107 CFU Campylobacter. To measure cell adhesion, the infected monolayers incubated for 1 h at 37°C in a humidified 5% CO2 incubator were washed, then Ht-29 adherent Campylobacter were enumerated on Karmali agar. To evaluate bacterial invasion, the infected monolayers were incubated during 3 h and consecutively 2 h with gentamicin (250 μg/ml). After washes and Ht-29 cell lyses, intracellular Campylobacter was enumerated on Karmali agar. Each experiment was done in duplicate and performed at least three times independently. The significance of differences in adhesion and invasion efficiency was determined using Student’s t-test. A P-value < 0.05 was defined as significant.
Chick Colonization
The chick model was used to determine colonization potential of C. jejuni. Specific pathogen free (SPF) 2 days old chickens were obtained from the high-security livestock at the French Agency of Food, Environmental and Occupational Health Safety (ANSES). Chicks were housed and treated in accordance with the regulation of the local veterinary office (Direction des Services Vétérinaires des Côtes d’Armor, France). All the animals were reared in isolators with controlled airflow. Sixty SPF chickens were distributed into three groups of 20 animals and maintained in separate isolators with unlimited food and water. One group of 20 animals was kept as negative controls and placed in a separate isolator. At 2 days old, chicks were each dosed orally, by gavage, with 107 CFU of C. jejuni in 0.1 ml of 0.1 M PBS, pH 7.2. During 4 weeks, every 7 days, chicks were sacrificed by cervical dislocation. Ceca were aseptically collected then C. jejuni enumerated on Karmali agar plate. The non-parametric Mann–Whitney test was used to assess the statistical significance of differences in colonization levels (P < 0.01).
A second independent experiment was performed using the same protocol but with 5 days old chicks.
Bacterial RNA Isolation and Purification
To study expression of pnp gene versus growth phase and temperature incubation, C. jejuni 81-176 was grown in BHI broth at 37 or 42°C until the mid-log phase (5 h) or the stationary phase (16 h). C. jejuni cells from 10 ml of mid-log phase and from 1 ml of stationary phase cultures were harvested (3,300 g, 6 min, 4°C) then resuspended in 1 ml of Extract All (Eurobio, Courtaboeuf, France) and 0.2 volumes of chloroform. After centrifugation at 12,000 g for 15 min at 4°C, the aqueous phase was removed. Total RNA was precipitated in isopropanol, rinsed in cold ethanol 75% then solubilized in 50 μl of sterile water. Isolation of total RNA was performed in triplicate from three independent cultures. DNA was degraded by treatment with DNase I Amplification Grade (Sigma-Aldrich, Saint Quentin Fallavier, France) then DNA removal was checked by PCR using MO001 primers (Table 2). Quality and quantity of RNA were checked using a NanoDrop 2000 (2000c Spectrophotometer, Thermo Scientific). The integrity of RNA samples was checked on a 2% agarose gel; electrophoresis was carried out in Tris–acetate–EDTA buffer for 45 min at 100 V. RNA concentrations were then standardized to 20 ng/μl for each sample prior to reverse transcription.
Quantitative Real-Time Reverse Transcription PCR
cDNA synthesis was performed using M-MLV (Moloney murine leukemia virus) Reverse Transcriptase (Promega, Charbonnieres, France) with random hexamer primers according to the manufacturer’s instructions. Quantitative real-time PCR assay was performed using an Applied Biosystems 7300 Real-time PCR System and MO002 to MO007 primers (Table 2). The composition of the PCR mix was as follows: 5 μl of sample, reverse primer (1 μM), forward primer (1 μM), and qsp 25 μl of SYBR Green I Master Mix (Applied Biosystems, Foster City, CA, USA). Amplification program included an initial denaturing step at 95° C (10 min), followed by 40 cycles of 95°C (15 s) and 60°C (1 min). A negative control (without cDNA) was included to each run. A melting curve was obtained from a first step starting from 60 to 95°C, to control specificities of quantitative PCR reaction for each primer pair. Efficiency of amplifications was determined by running a standard curve with serial dilutions of cDNA. The efficiency (E) was calculated with the formula E = [10(1/—s)] × 100, where “s” is the slope of the standard curve. The CT (critical threshold) stood for the first PCR cycle during which the fluorescence value was significantly distinct from the background and corresponded to a signal. Results were analyzed using the comparative critical threshold (ΔΔ CT) method in which the amount of target RNA was adjusted to a reference [internal target RNA, rpoA (51)]. Relative expression level was obtained by the following formula: 2ΔΔCT (37). RT-qPCR was performed in triplicate with, at least, three templates of RNA extracted from independent cultures.
Protein Extraction
To avoid possible misinterpretation of results, proteins were extracted from C. jejuni 81-176 or 81 Δpnp cultures at the late exponential phase showing similar optical density (OD) at 600 nm and cell counts on agar. Thus, the two strains were assumed to be in the same physiological state. Prior to each experiment, one colony grown on Karmali agar for 48 h at 42° C was resuspended in 50 ml of BHI broth then incubated at 42° C for 24 h in microaerobic conditions with shaking at 110 rpm. Two milliliters of the resulting culture were used to inoculate 200 ml of BHI broth incubated for 7 h in the same conditions (until OD600 reached 0.2). Cells were harvested by centrifugation at 7,000 g for 20 min, washed consecutively with 200 mM glycine solution (Sigma-Aldrich) and 100 mM Tris-HCl pH 7.0 solution (Sigma-Aldrich) then resuspended in 10 ml of a 10 mM Tris–HCl pH 7.0 buffer. Cells were disrupted by series of 6 × 30 s sonications with 6 min intervals on ice (Vibracell 72434, Bioblock Scientific, Illkirch, France). To eliminate cell debris, samples were centrifuged twice at 10,000 g for 20 min at 4°C Then, cytoplasmic proteins were separated from membrane fractions by ultracentrifugation at 188,000 g for 1 h at 4°C The cytoplasmic protein fraction in the supernatant was treated with protease inhibitor cocktail tablets Complete (Roche Diagnostics, Mannheim, Germany) and nuclease solution containing RNase and DNase (Sigma-Aldrich). Protein samples were dialyzed using cellulose membrane tubing with a cut-off at 12,000 Da (Sigma-Aldrich) against Milli-Q water at 4°C in shaking conditions during 3 days, refreshing the dialyze bath each day. Total protein concentration was determined using the Micro BCATM Protein Assay Kit (Perbio-Science, Brebieres, France).
Two-Dimensional Gel Electrophoresis of Bacterial Proteins
A quantity of 100 μg of protein was concentrated using Concentrator 5301 (Eppendorf, Le Pecq, France), at room temperature. Samples were diluted with 275 μl of a solution of 6 M urea, 2 M thiourea, 4% CHAPS, 0.4% DTT, bromophenol blue (BB; Sigma-Aldrich) and 2% Biolyte 3/10 (Bio-Rad, Marnes-la-Coquette, France). Proteins in the rehydratation solution were absorbed overnight by 17 cm pH 4–7 or 6–11 IPG strips (Bio-Rad, GE Healthcare, Orsay, France). Then, the iso-electro-focalization (IEF) was performed using the Bio-Rad IEF program as follows: from 50 to 250 V for 3 h, from 250 to 6,000 V for 3 h, and at 6,000 V until reaching 54,000 V/h. Finally, each strip was soaked for 20 min in equilibration buffer [1.5 ml of a 6-M urea, 2% SDS, 50 mM Tris–HCl (pH 8.8), 30% glycerol and BB] supplemented with 2% DTT and subsequently for 20 min in equilibration buffer with 4% iodoacetamide (Bio-Rad). The second dimension was performed in 12% acrylamide gels covered with 1% low-melting point agarose (Bio-Rad) and run at 40 mA/gel at 14°C using a Protean II xi cell (Bio-Rad) until the migration reached the base of the gels. Proteins in gels were finally silver stained and scanned with a GS-800 densitometer (Bio-Rad) operated with the Quantity One® software (Bio-Rad) at the resolution of 42.3 μm.
Image, Statistical Analysis, and Protein Identifications
The image analysis was performed using the Progenesis Samespots® software (NonLinear Dynamics, Newcastle upon Tyne, UK). The statistical analysis of the results was performed with two technical replicates of three independent experiments resulting in six gels for each strain. Differences between the parental and mutant strain proteome (2 × 6 gels) were validated by principal component analysis (PCA) and differences among matched spot intensities were statistically validated by ANOVA (at a 5% significance level) and by using a Power superior to 0.8. The PCA analysis ensured the reproducibility of the experiments and has shown a significant effect of the pnp disruption on C. jejuni proteome. Over- and down-expressed proteins were taken into account only if the mean difference in spot intensities passed the threshold of 1.8. When spots of interest were located, 2D-electrophoresis gels were performed again using 700 μg of proteins and were stained with BioSafe colloidal Coomassie blue (Bio-Rad). Spots of interest were excised from preparative gels and protein identification was performed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry using PAPPSO platform facilities1. Gel plugs were first washed twice with 50% (v/v) acetonitrile, 25 mM ammonium carbonate in water, and then dried in a vacuum speed concentrator. Aliquot (10 μl) of a trypsin solution (Promega, 12.5 ng/μl in 50 mM NH4HCO3) was added to each sample and digestion was performed for 6 h at 37°C A 1-μl aliquot of each supernatant was spotted directly onto the MALDI plate then dried at room temperature before adding 1 ill of the matrix solution [α-cyano-4-hydroxycinnamic acid, 3 mg/ml in 50% (v/v) acetonitrile 0.1% (v/v) trifluoroacetic acid]. Mass spectra were acquired on a Voyager-DE-STR (Applied Biosystems, Framingham, MA, USA), equipped with a nitrogen laser (Laser Science, Franklin, MA, USA). Tryptic autodigestion ion peaks, (M + H)+ = 2211.104 and 842.509 Da, were used for internal calibration of spectra. The peptide mass lists were used for protein identification in a C. jejuni, strain 81-176, amino acid sequence database (UniProtKB, 09.09.2010), using the Protein Prospector v 3.2.1 software set with the following parameters: mass tolerance < 20 ppm, four peptides required to match, one missed cleavage, oxidized methionine and carbamidomethylated cysteine respectively as possible and fixed modifications.
Poly(A) Degrading Activity in C. jejuni Cell Lysates
Campylobacter jejuni was grown in Mueller-Hinton medium to late log phase, lysates prepared using EasyLyse (Epicenter), and assayed for ribonuclease activity as described (Deutscher and Reuven, 1991), except that the reactions were done in a buffer optimized for measurement of RNase R (Cheng et al., 1998). Degradation of [3H]poly(A) (Amersham Product TRK.480) was quantified by scintillation counting [3H]nucleotide released into acid soluble form. Dependence of the reaction on lysates of C. jejuni parental and pnp mutant strains was measured in the presence and absence of 10 mM KPO4 to distinguish hydrolytic from phosphorolytic activities. Less than 10% of substrate was consumed in the reactions.
In Silico Analysis
DNA and protein homology searches were performed by BLAST analysis 2. Oligonucleotide primers were designed using Primer 3 program 3.
Results
Bacterial Growth Curves and pnp Gene Expression in C. jejuni 81-176
To determine the expression profile of pnp gene, quantitative RT-PCR experiments were assayed in four different growth conditions. C. jejuni 81-176 was grown in BHI broth at 37 or 42°C until the mid-log phase (5 h) or the stationary phase (16 h). CT values were determined in four replicates from cDNA synthesized from RNA extracted from three independent cultures for each growth condition. The specificity of each primer pair (MO002 toMO007) was controlled by analyzing melting curves. PCR efficiencies of the primer pair used were homogenous and all over 90%. The rpoA gene was used as an internal control (Ritz et al., 2009) and the expressions at 37°C in mid-log phase were used as calibrators to calculate relative expression levels. No significant difference (P < 0.05) in expression of pnp gene was observed for the four growth conditions tested (Figure 1).
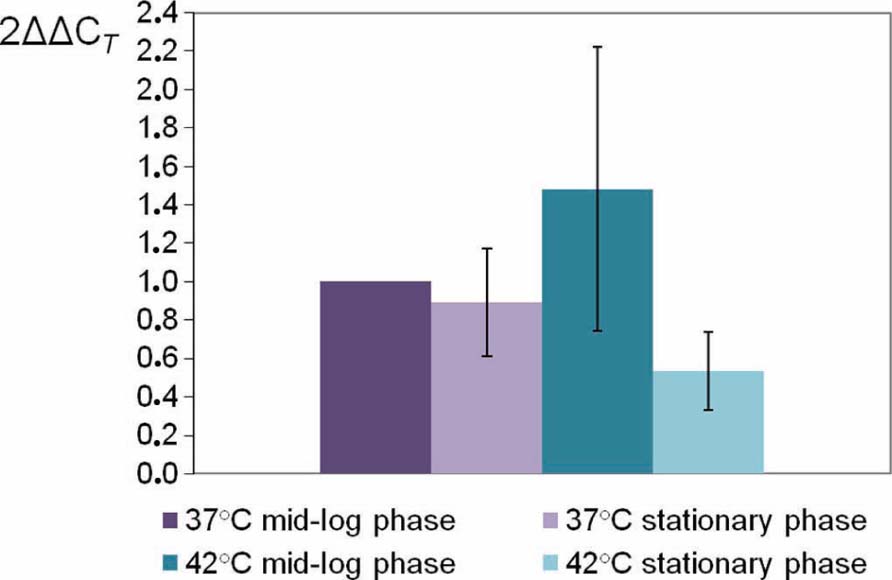
FIGURE 1. Relative expression levels of C. jejuni 81-176 pnp gene at different conditions. Gene expression has been estimated using RT-qPCR and the comparative critical threshold (ΔΔCT) method. The rpoA gene was used as the internal control, and the expression at 37°C in mid-log phase as the calibrator. Error bars represent the standard deviations from the mean of three independent experiments.
There is a difference between the growth of the parental and the mutant strain, however this difference is only significant below 35°C as shown in Figure 2.
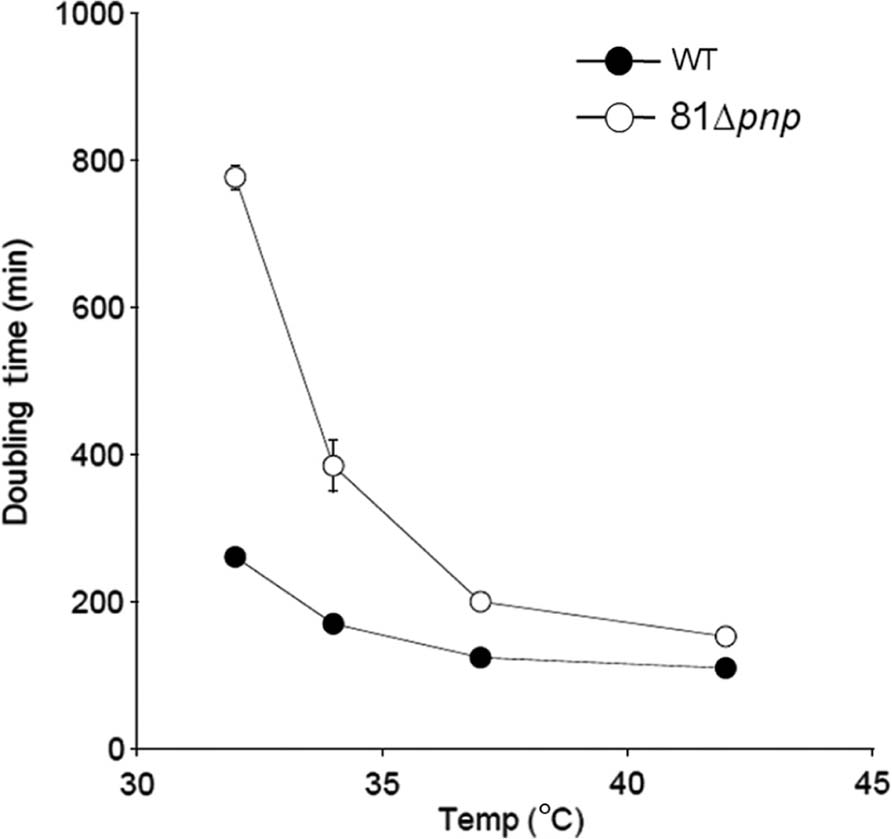
FIGURE 2. Mean generation time of the parental strain (solid circles) versus the mutant strain (open circles). C. jejuni was cultivated in BHI broth and incubated in microaerobic condition at 32, 34, 37, and 42°C. Experiments were performed three times independently.
To check that phenotypic changes in the pnp mutant strain were specifically due to PNPase absence, we attempted genetic complementation of pnp in C. jejuni 81-176. Despite several attempts, we never obtained such a complemented strain in C. jejuni 81-176, but were able to complement the pnp mutation in C. jejuni strain F38011 (Haddad et al., 2009).
Effect of pnp Mutation on C. jejuni 81-176 Motility
Motility is a common bacterial virulence factor involved in colonization and invasion of host cells by pathogens. Swimming abilities of C. jejuni 81-176 parental and C. jejuni 81 Δpnp mutant strains are presented Figure 3 on soft agar plate 0.4%. Results indicated that inactivation of pnp gene caused a 5.3-fold reduction in motility on agar plates compared to the parental strain, at both 37 and 42° C. Because motility is phase variable in C. jejuni, measurements were achieved using independent cultures on different days. The results of four experiments were essentially identical, suggesting that reduced motility of the mutant strain is unlikely due to phase variation. To test if the motility defect observed is specifically linked to the inactivation of the pnp gene, we also tested the effect of genetic complementation of the pnp mutant with the wild-type pnp gene. As we were unable to complement the pnp mutation in C. jejuni 81-176, we used the complemented mutant of strain F38011 (Haddad et al., 2009). As shown in Figure 3C, complementation of the pnp mutation in trans restored partially wild-type motility, confirming that absence of PNPase results in reduced motility and associated virulence phenotypes.
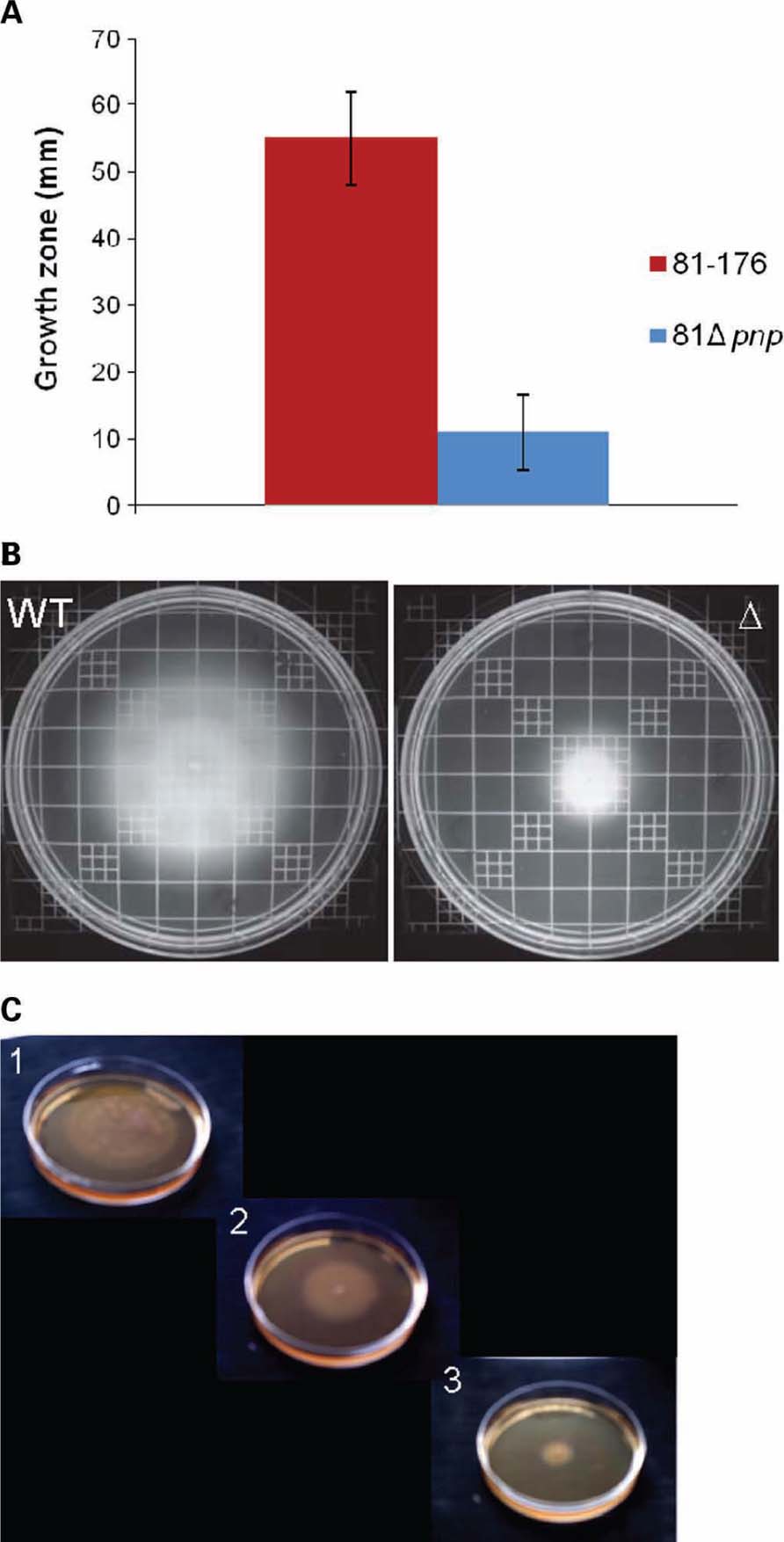
FIGURE 3. Motility phenotypes of C. jejuni strains. Swimming ability was assessed on BHI agar containing 0.4% agar. Strains were inoculated into BHI motility agar and incubated for 48 h at 37°C under microaerobic conditions. (A) Mean diameters of four independent experiments were represented. (B) Motility agar obtained after an incubation at 37°C during 48 h. (C) Motility assay of the parental C. jejuni F38011 strain (1), the F38Δpnp complemented strain (2), and the mutant strain F38Δpnp (3), on BHI plates containing 0.4% agar.
Moreover, impaired mutant strain motility was also observed on soft agar plate 0.25% (data not shown).
Effect of pnp Mutation on Adhesion to and Invasion in Ht-29 Cells
In C. jejuni, motility is critical both for invasion of gastrointestinal epithelial cells and for colonization of the mucus lining of the gastrointestinal tract (Yao et al., 1994). To examine the role of the pnp gene in C. jejuni infection, the in vitro adhesion and invasion abilities of the pnp mutant and parental C. jejuni 81-176 strains were compared. The C. jejuni 81 Δpnp mutant exhibited a significant (P ≤ 0.02) decrease in adhesion ability compared to the parental strain (Figure 4A), and a 3 log-fold decrease in invasion of Ht-29 cells (P < 0.02; Figure 4B). No difference in susceptibility to gentamicin for both strains was observed, indicating that the invasion phenotypes of the strains were not due to differences in gentamicin susceptibilities (data not shown). Results of invasion assays are consistent with the lowered adherence of the mutant strain and indicate that inactivation of the pnp locus indirectly affects the interaction of C. jejuni with epithelial cells, most likely via the reduced motility resulting from the absence of PNPase.
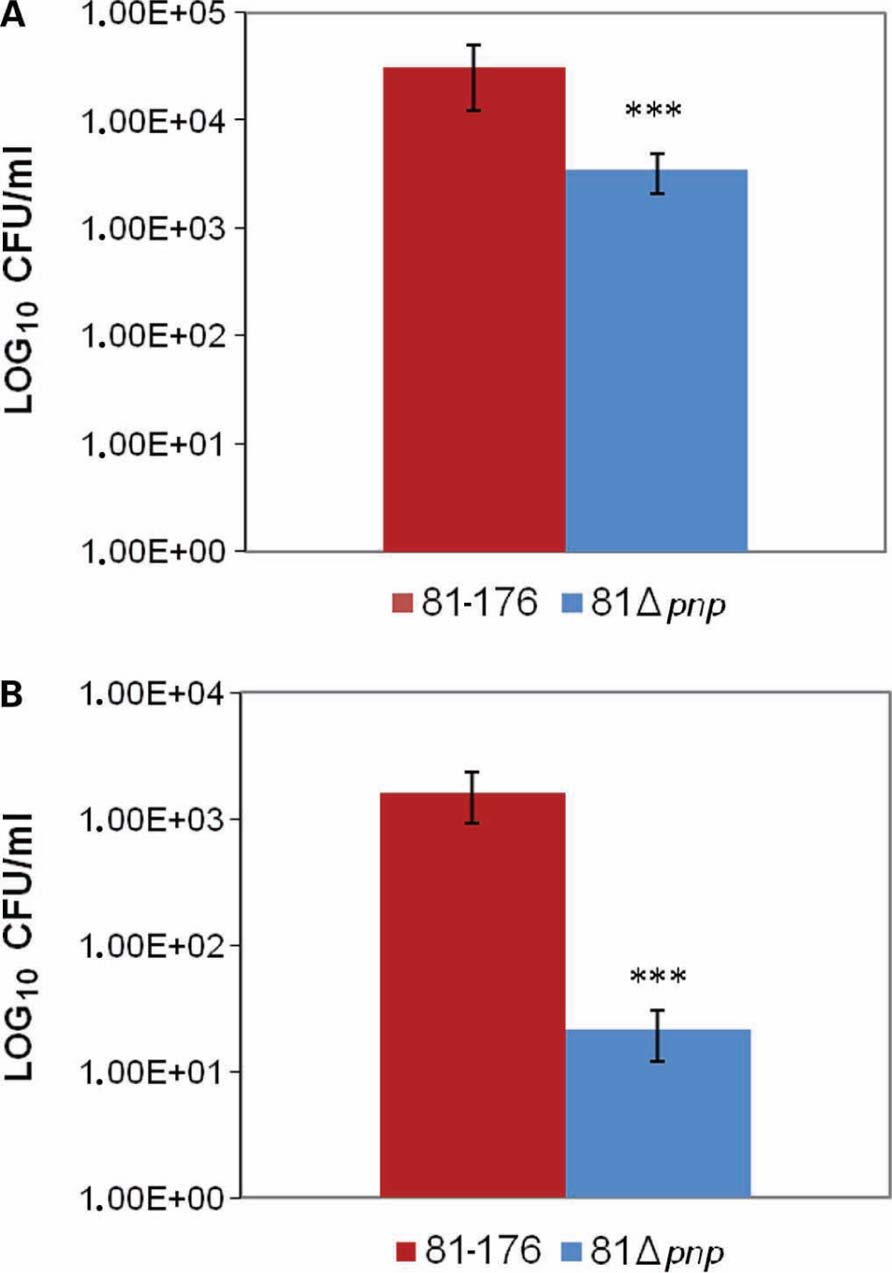
FIGURE 4. Adhesion and invasion of C. jejuni in Ht-29 cells. (A) Effect of mutation of the pnp locus on C. jejuni adhesion to Ht-29 human intestinal epithelial cells. (B) Ht-29 cell invasion assay. Cells were infected with the parental and mutant strains for 3 h, and invasion values were calculated from the number of bacteria that survived 2 h of incubation in the presence of gentamicin. The experiment was performed in duplicate from at least three independent cultures. The values plotted represent means plusmn; ± standard deviations (error bars). Statistical significance was assessed with an unpaired Student’s t-test. ***P < 0.02.
Effect of PNPase Deficiency on Colonization of Chick Gut by C. jejuni
According to the motility and adhesion results, colonization experiments were also conducted. To determine the relative importance of PNPase in chicken colonization, we inoculated 2 days old chicks with the parental and PNPase mutant strains. Three groups of five birds were infected with 107 CFU of parental or mutant strains. On day 7, viable bacteria were found in all five chickens fed with the parental strain, whereas viable mutant bacteria were recovered from only two. In addition, the mean colonization concentration of the chicken inoculated with the parental strain was 1.5 × 107 CFU/g on day 7 and was significantly lower (2.4 × 105 CFU/g; P < 0.01) in the chickens inoculated with the mutant strain. On days 14, 21, and 28, viable mutant bacteria were detected in all the chickens. However, throughout the experiment, the mean colonization level of the chicken inoculated with the mutant strain was significantly reduced compared to those inoculated with the parental C. jejuni (Figure 5; P < 0.01). The defect in colonization of the pnp mutant strain was confirmed by a second experiment using SPF 5 days old chicken in which, the mutant strain was unable to colonize chicken gut, while the parental strain reached a colonization level of 9.3 × 107 CFU/g at 29 days (data not shown).
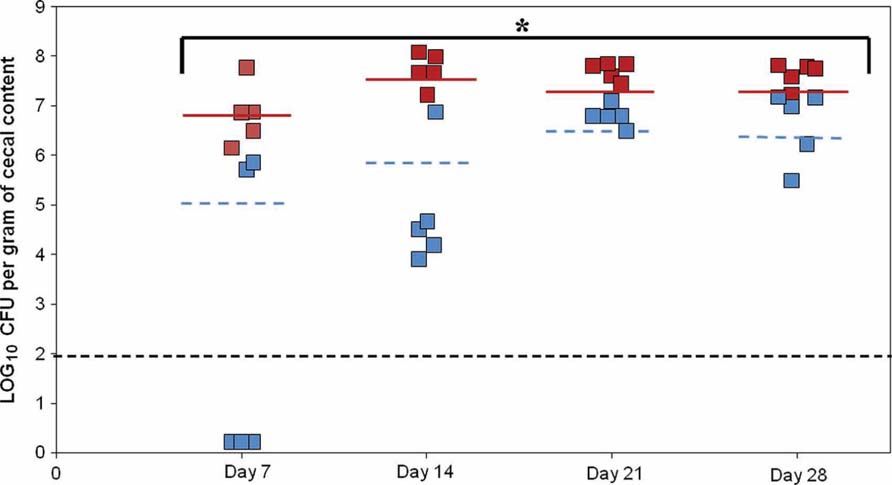
FIGURE 5. Effect of the PNPase mutation on chick gut colonization by C. jejuni. The experiment included three groups of birds. Two days old chicks were orally gavaged with 1.4 to 1.5 × 107 CFU of C. jejuni 81-176 (red squares) or 81-176 derivative strain lacking production of PNPase (blue squares). Colonization levels were measured by enumeration of bacteria present in ceca on days 7, 14, 21, and 28 post-inoculation (LOG10 CFU/g of ceca). Each dot represents the load of C. jejuni in the cecum of an individual chick. The geometric mean of the bacterial loads from each set of chicks is denoted by the horizontal bar. The line designates the detection limit of 2 × 102 CFU/g of ceca. Statistical analysis was performed using the Mann–Whitney test (mean and P-value < 0.01). *Significant difference between the parental and pnp mutant strains.
Comparative Proteomic Analysis
In order to pinpoint the effect of pnp mutation at a protein level, proteomic analyses were performed. Proteomes of the pnp mutant and its parental strains were compared using Progenesis SameSpots® software. Examples of digital images from 2-DE gels achieved in the 4-7 and 6-11 pI ranges are given in Figures 6A,B. This comparative analysis resulted in nine preponderant proteins showing a significant difference in expression, amongst which three were over-expressed and six were repressed in the pnp mutant strain compared to the parental strain (Table 3). As expected, spot occurrences corresponding to PNPase were absent in the mutant strain (Table 3). Among the down-expressed proteins, the heat-shock protein Hsp90, catalase KatA, the elongation factor EF-Tu, the N-acetylneuraminic acid (NANA) synthetase and, an antigenic peptide, PEB3 were identified. Among the over-expressed proteins, the elongation factor EF-G, the chaperone DnaK and S-ribosylhomocysteinase, LuxS were found (Table 3).
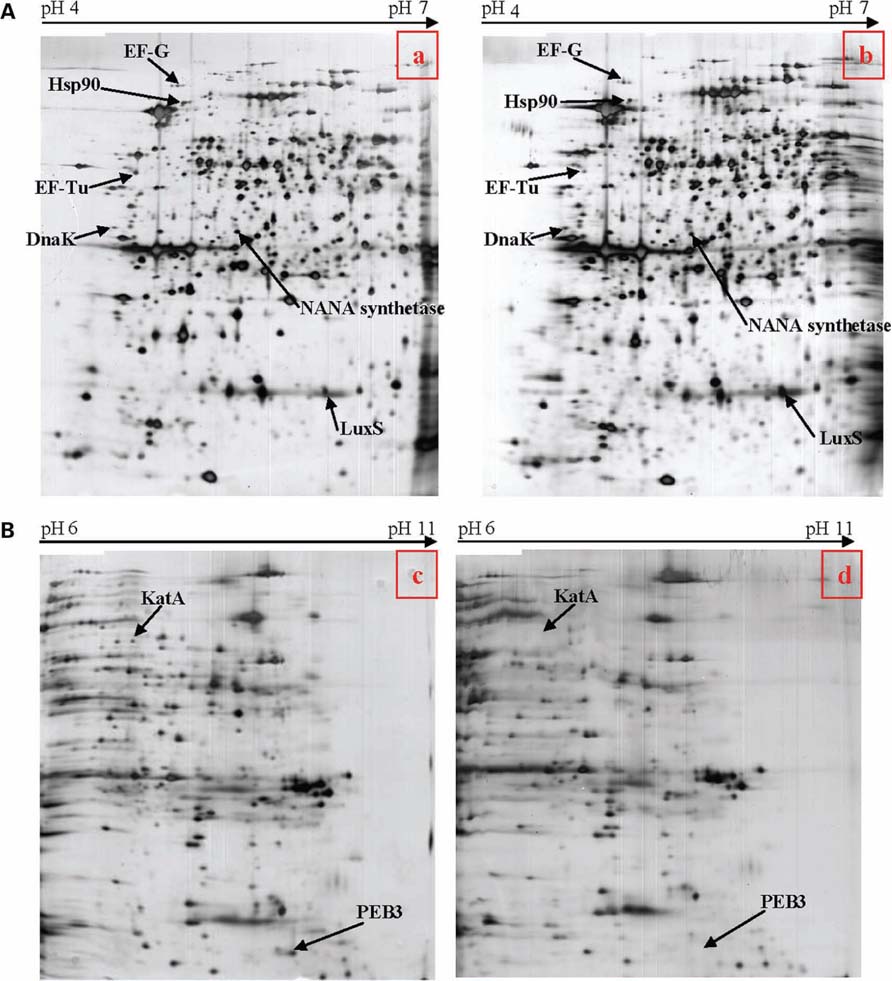
FIGURE 6. (A) Two-dimensional electrophoresis gels of acidic proteins extracted from C. jejuni strain 81-176 (a) and the pnp mutant (b) derived from this strain. Proteins were extracted from bacteria cultivated in BHI at 42°C collected when OD600 reached 0.2. (B) Two-dimensional protein gel analysis of basic proteins extracted from C. jejuni strain 81-176 (c) and the pnp mutant (d) derived from this strain. Proteins were extracted from bacteria cultivated in BHI at 42°C collected when OD600 reached 0.2.
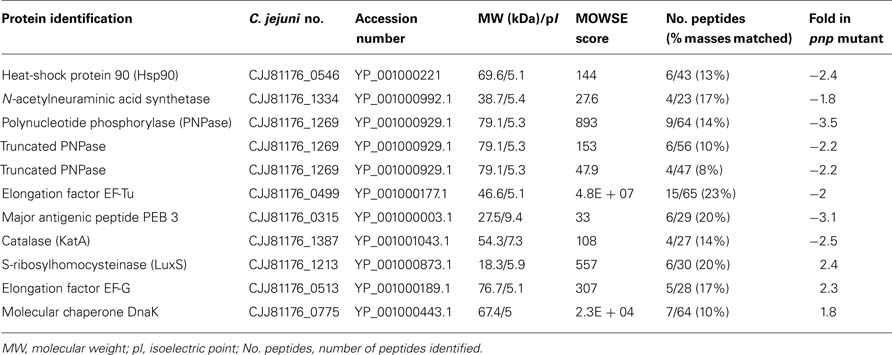
Table 3. Campylobacter jejuni over- and down-expressed proteins in the pnp mutant as compared to the parental strain.
Effect of pnp Mutation on luxS, peb3, katA, and hsp90 Genes Expression
To analyze if our proteomic data are correlated to gene expression, a quantitative RT-PCR analysis was performed on four genes known to be involved in adhesion, invasion, or colonization ability of C. jejuni. Therefore, gene expression of luxS, peb3, katA, and hsp90 was measured (Figure 7). Levels of peb3 and katA mRNA was significantly decreased in the pnp mutant strain as compared to the parental strain, showing the same trend of protein expression, while gene expression of luxS and hsp90 remained unaffected by pnp mutant, but not protein expression.
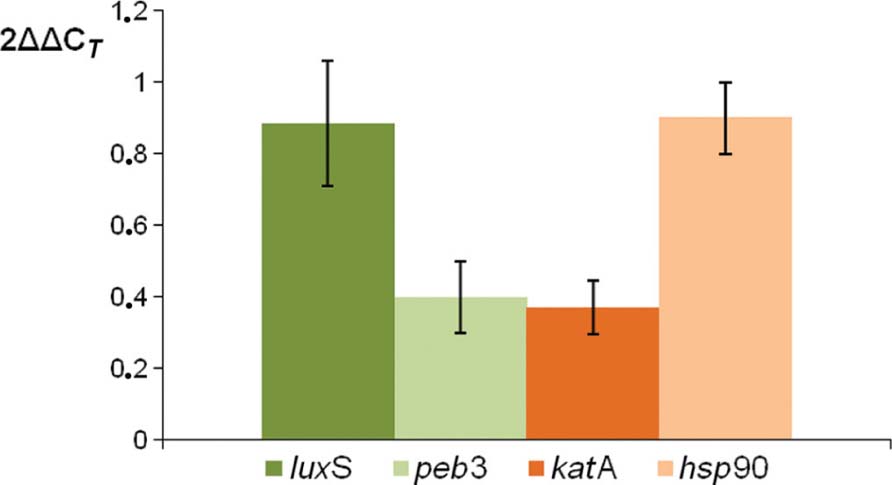
FIGURE 7. Comparison of relative transcript levels of luxS, peb3, katA, and hsp90 genes between C. jejuni pnp mutant and the parental strain, assessed by quantitative RT-PCR. Each bar represents relative expression level of a gene in the mutant strain according to the 2AACj method described by Livak and Schmittgen (2001). The internal reference gene used was rpoA and the calibrator condition was the expression in the wild-type strain (equivalent to 1). Each value is the mean of at least three independent RNA extractions and error bars indicate the standard deviations.
RNA Decay in C. jejuni
An established method to assess the relative contributions of phosphorolytic and hydrolytic degradation in bacterial lysates was employed to characterize behavior of C. jejuni. Reactions were done to confirm measurements were made in the linear range of the assay. Figure 8 shows that poly(A) can be degraded in the parental C. jejuni cell lysates by a hydrolytic mechanism, revealing the existence of hydrolytic ribonucleases. However, phosphorolytic RNA degradation predominates as phosphate addition greatly increased activity. Moreover, poly(A) degradation by lysates of the pnp mutant strain was almost totally non-responsive to phosphate addition (Figure 8), indicating that the majority of phosphorolytic activity, and therefore the majority of poly(A) degrading activity in the parental cells, depends on the pnp gene. Comparison of hydrolytic poly(A) degrading activity in parental and mutant strains lysates revealed that pnp deletion increases this activity by almost fivefold.
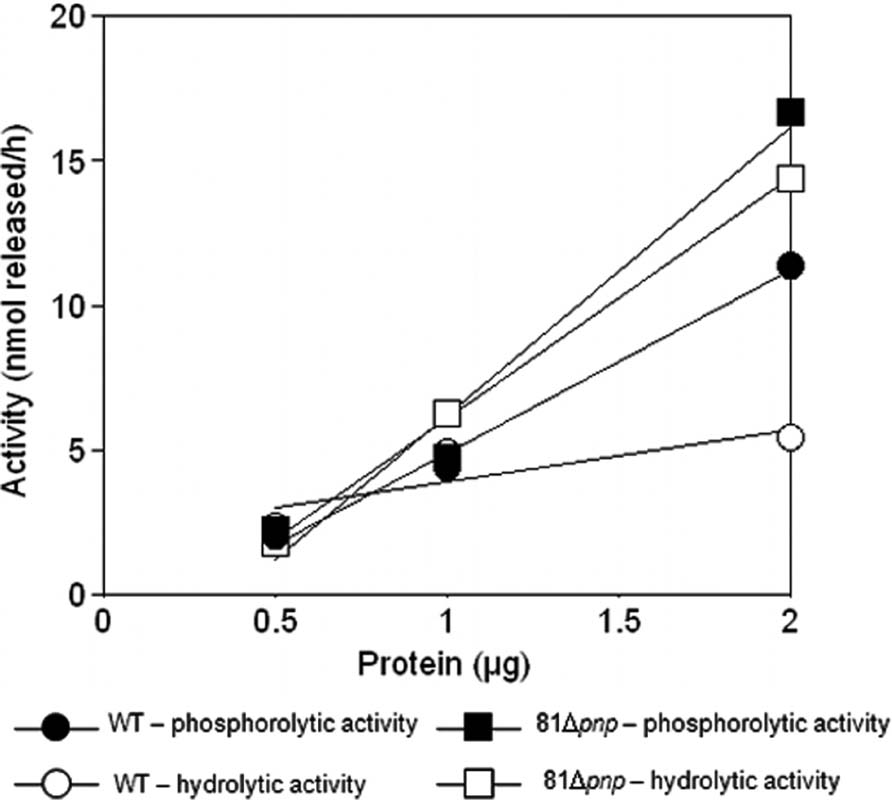
FIGURE 8. Poly(A) degrading activity in C. jejuni cell lysates. Degradation of [3H]poly(A) was quantified by scintillation counting [3H]nucleotide released into acid soluble form. Dependence of the reaction on lysates of the parental strain (circle) and the pnp mutant (square) was measured in the presence (closed) and absence (open) of 10 mM KPO4 to distinguish hydrolytic from phosphorolytic activities.
Discussion
Ribonucleases are enzymes involved in post-transcriptional regulation of genes by mediating RNA degradation. In some Gram-negative bacteria, the endoribonuclease RNase E is essential for cellular growth and inactivation of rne gene (encoding RNase E) impedes processing and prolongs the lifetime of bulk mRNA (Cam et al., 1996; Ow and Kushner, 2002). For the exoribonuclease activity, RNase II is responsible for the majority of RNA phosphodiester bond cleavage in E. coli as most RNA (90%) is degraded via a hydrolytic mechanism (Deutscher and Reuven, 1991). Genes encoding RNase E and RNase II enzymes are absent in the C. jejuni genome, suggesting a major role is played by other ribonucleases like PNPase. Because of the pleiotropic functions of PNPase described in various bacteria (Luttinger et al., 1996; Wang and Bechhofer, 1996; Bechhofer and Stasinopoulos, 1998; Goverde et al., 1998; Yamanaka et al., 1999; Clements et al., 2002; Rosenzweig et al., 2005, 2007; Ygberg et al., 2006; Wu et al., 2009), the present study broadly analyzed the role of PNPase in C. jejuni, beyond its role in long-term survival to cold (Haddad et al., 2009).
In the current study, we have examined the role of the putative post-transcriptional regulator PNPase and its consequences on C. jejuni proteome, which may then affect some biological functions of the pathogen. Growth curve data show that, inactivation of the pnp gene has no significant effect on C. jejuni 81-176 growth rate at optimal temperature. Generation time increases with the decrease of the temperature of incubation, a finding that confirms the hypothesis that PNPase is involved in C. jejuni survival at temperatures below the optimal temperature of growth.
Transcript analyses revealed that expression of C. jejuni 81-176 pnp gene is not influenced by growth phase or by optimal temperatures of growth. In order to define the role of PNPase in the cellular biology of C. jejuni, proteomic profiles of parental and PNPase mutant strains were compared. The pnp gene mutation affects synthesis of proteins in C. jejuni 81-176; however, only few proteins vary significantly between the parental and the pnp-deleted strains Although comparison of proteome of these two strains verified the non-essential role of pnp gene in C. jejuni 81-176 strain, the majority of phosphorolytic ribonuclease activity seems to depend on PNPase. Moreover, deletion of pnp gene increases hydrolytic ribonuclease activity. These results suggest that PNPase either down regulates, or antagonizes function, of a hydrolytic ribonuclease in C. jejuni. The enzyme responsible for hydrolytic poly(A) degrading activity is unknown, but comparative sequence analysis predicts a single hydrolytic mRNA degrading exoribonuclease encoded by the C. jejuni genome, RNase R.
The swarming ability of the C. jejuni pnp mutant strain is reduced. As motility is directly linked to C. jejuni pathogenic features, we confirmed that the motility defect observed with the pnp mutant strain is due to the pnp mutation using the F38011 complemented strain. The motility of C. jejuni is conferred by polar flagella, and approximately 40 genes are predicted to be involved in flagella biosynthesis or function (Yao et al., 1994; Parkhill et al., 2000). Flagella-mediated motility is responsible for colonization of the mucous lining of the mammalian and avian gastrointestinal tracts as well as invasion of intestinal epithelial cells (Wassenaar et al., 1991; Grant et al., 1993; Rosenzweig et al., 2007). In proteomic analysis, the abundance of protein related to flagella was not affected by the mutation however a decrease in NANA synthetase production was observed in the pnp mutant strain, a factor supposed to be involved in C. jejuni motility. NANA synthetase, encoded by the neuB gene, catalyzes formation of NANA, the main component of C. jejuni lipo-oligosaccharide (LOS). Linton et al. (2000) hypothesized that neuB3 gene is involved in the post translational modification of the flagellin subunit and in motility, as inactivation of neuB3 leads to non-motile and aflagellate cells (Linton et al., 2000). This information is particularly interesting in that for the C. jejuni 81-176 genome, neuB is located in a gene locus encoding proteins involved in the formation of flagella or mobility. Because motility is a prerequisite to eukaryotic cells invasion, we expected that the pnp mutant would be less invasive than its parental strain. Studies using Ht-29 cells confirmed that mutation of pnp decreased the invasiveness of C. jejuni and also a decrease in adhesion ability. The putative adhesion PEB3 was 3.1-fold decreased in the pnp mutant and quantitative RT-PCR also revealed a diminution in peb3 mRNA synthesis. PEB3 protein shares 51.8% sequence identity to Paa, an E. coli adhesion (Batisson et al., 2003) and 49% identity to AcfC, an accessory colonization factor from Vibrio cholerae (Peterson and Mekalanos, 1988). PEB3 of C. jejuni is considered as an adhesion based on the work performed with Hep-2 cells by Wren et al. (Drs. B. W. Wren and R. Langdon, personal communication; Rangarajan et al., 2007).
Interestingly, PNPase was described as a factor affecting S. enterica invasion and establishment of chronic infection in a mouse model (Clements et al., 2002). In contrast with results obtained in C. jejuni, S. enterica pathogenicity islands are over-expressed in PNPase mutants, resulting in increased invasion into epithelial MDCK cells (Clements et al., 2002). However, the cell line used in Clements et al. (2002) study has been isolated from canine kidney and is largely different from those utilized in the current study. Moreover, studies on S. enterica PNPase use the typhoid strain of the bacteria while the current study showed interaction of C. jejuni with intestinal cells. Other ribonucleases have been shown to be involved in bacterial virulence. RNase R of Shigella flexneri has been reported to be required for the expression of the invasion factors IpaB, IpaC, and IpaD (Cheng et al., 1998).
As chickens are the principal origin of campylobacteriosis in humans, the potential colonization of the parental and mutant strains were also investigated in the avian intestine. Initial colonization was markedly different between the variants, indicating that PNPase is required to establish colonization. Hanel et al. (2004) have used a chick model to study the colonization ability of different strains of C. jejuni (Hanel et al., 2004). Three groups of strains have been determined. The first group corresponds to strains which could not be re-isolated after chicken inoculation; the second one contains weak or delayed colonizers; the last group includes strong colonizers that could be re-isolated from all animals 7 days after inoculation. In the current study, the wild-type strain C. jejuni 81-176 could be considered as a “strong colonizer” because this strain was present in all animals up to 7 days after inoculation. However, pnp disruption results in a delayed colonization of the C. jejuni mutant strain, which can be linked to the group 2 of Hanel et al. (2004). This defect in colonization could be due to the defect in motility and adhesion abilities of the mutant strain. Moreover, the observed lower expression of katA gene and KatA protein in the pnp mutant strain could be linked to the deficiency in colonization. C. jejuni expresses a single heme-containing catalase, KatA, which is part of a defense system against oxidative stress (Haas and Goebel, 1992). In addition, KatA contributes to intramacrophage survival of C. jejuni and chick gut colonization (Day et al., 2000; Palyada et al., 2009). A ΔkatA mutant is unable to colonize the chick ceca after 4 days of incubation (Palyada et al., 2009).
Proteomic experiments showed that LuxS, another protein involved in colonization, is 2.4-fold overproduced in the PNPase mutant strain, but no variation was observed in luxS mRNA level. Being the last enzyme in the AutoInducer-2 (AI-2) biosynthesis pathway, LuxS controls production of AI-2. While Holmes et al. (2009) have shown that AI-2 does not function as a quorum sensing molecule in C. jejuni during exponential growth in vitro (Holmes et al., 2009), LuxS seems to be involved in C. jejuni motility by modifying transcription rate of flagellin flaA mRNA without change the level of total flagellin protein in the cells (Jeon et al., 2003). Quiñones et al. (2009) have also demonstrated the implication of LuxS in chick gastrointestinal tract colonization and adherence to avian cells LMH, which could be correlated with phenotypic results obtained in this work (Jeon et al., 2005; Quiñones et al., 2009).
Proteomic results indicated that synthesis of two heat-shock proteins were modified by inactivation of the pnp gene suggesting a probable stress-like response in the mutant strain. Hsp90 was down-expressed, whereas DnaK chaperone was up-regulated in the pnp mutant. In the C. jejuni mutant strain, Hsp90 was 2.4-fold less abundant than in parental strain; however, transcript analysis did not reveal such variation. Transcript hsp90 could be more stable in the absence of PNPase enzyme and accumulation of this mRNA would lead to a greater production of the protein. In E. coli, Hsp90 participates in de novo protein folding during a heat-shock condition by facilitating the ability of the DnaK-DnaJ-GrpE complex to interact with newly synthesized polypeptides (Thomas and Baneyx, 2000); however, Homuth et al. (2000) suggested that the probable role of this protein is in response to acid or oxidative stress, because Hsp90 is not over-expressed in Helicobacter pylori following a heat-shock (Homuth et al., 2000). Moreover, Hsp90 has been implicated in Salmonella pathogenesis by contributing to bacterial survival within host cells (Osman et al., 2009).
In the case of DnaK, this protein is known to be involved in DNA replication, protein folding, and in stress-response. DnaK was up-regulated in the C. jejuni pnp mutant compared to the parental strain, suggesting accumulation of denatured proteins in the mutant cells. Interestingly in E. coli, DnaK is preferentially associated to PNPase in the degradosome, a multiprotein complex involved in RNA degradation (Regonesi et al., 2006); however, no degradosome has been yet described in C. jejuni particularly because the major component of this complex, RNase E, is not detected in the genome of C. jejuni. Production of two elongation factors (EF-Tu and EF-G) was also modified in the mutant strain, suggesting a role of PNPase in the translation mechanism or a rearrangement of protein production in absence of this exoribonuclease.
In conclusion, deficiency in PNPase production in C. jejuni results in a defect in its ability to swim, to adhere and invade intestinal cells, and to colonize chick gut. All these phenotypic features could be linked to the motility or export function of the flagella. Further analyses should focus on a possible link existing between PNPase and C. jejuni flagella. Moreover, mutation of the pnp gene induces a stress-like state of the bacterial cell, observed with overproduction of proteins such as Hsp90, DnaK, KatA, suggesting that PNPase could be involved in C. jejuni resistance to stress. Proteomic experiments showed that PNPase inactivation causes little variation in C. jejuni protein production, but, specific proteins such as PEB3 and KatA are nonetheless affected. These data suggest that in C. jejuni, PNPase is involved in gene expression in multiple ways, in addition to directly degrading the primary transcript; however, PNPase is responsible for adding nucleotides to an existing poly(A) tail of mRNA through a poly(A) polymerase activity (Li and Deutscher, 1994; Mohanty and Kushner, 2000; Oussenko et al., 2005; Viegas et al., 2007). The heteropolymeric tails synthesized by PNPase may further enhance the degradation of mRNA intermediates targets. Further molecular studies using mRNA stability assays will be conducted to verify if the regulation of the genes identified in this study are potential targets of PNPase, either directly or through an alternative mechanism. Indeed, RNase R can be substituting PNPase since the pnp mutant strain has the hydrolytic activity increased. This could explain the little variation observed in proteomic results. In E. coli, RNase II antagonizes PNPase action on polyadenylated transcripts. In C. jejuni, PNPase may antagonize RNase R action. However, direct measurement of hydrolytic ribonuclease gene expression or combinatorial in vitro RNA degradation assays may also be required.
Acknowledgments
Legal authorization for the use of animals in experiments was kindly provided by Catherine Magras. We thank the supervisors of the Molecular Biology Platform of Oniris, in which some experiments were done. The work was supported by the GENICAMP project founded by Région Pays de la Loire, France.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Altekruse, S. F., Stern, N. J., Fields, P. I., and Swerdlow, D. L. (1999). Campylobacter jejuni: an emerging foodborne pathogen. Emerg. Infect Dis. 5, 28–35.
Andersen, M. T., Brøndsted, L., Pearson, B. M., Mulholland, F., Parker, M., Pin, C, Wells, J. M., and Ingmer, H. (2005). Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151, 905–915.
Andrade, J. M., Pobre, V., Silva, I. J., Domingues, S., and Arraiano, C. M. (2009). The role of 3′–5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 85,187–229.
Arraiano, C. M., Andrade, J. M., Domingues, S., Guinote, I. B., Malecki, M., Matos, R. G., Moreira, R. N., Pobre, V., Reis, F. P., Saramago, M., Silva, I. J., and Viegas, S. C. (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 34, 883–923.
Batisson, I., Guimond, M. P., Girard, F., An, H., Zhu, C., Oswald, E., Fairbrother, J. M., Jacques, M., and Harel, J. (2003). Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 71,4516–4525.
Bechhofer, D. H., and Stasinopoulos, S. J. (1998). tetA(L) mutants of a tetracycline-sensitive strain of Bacillus subtilis with the polynucleotide phosphorylase gene deleted. J. Bacteriol. 180, 3470–3473.
Bechhofer, D. H., and Wang, W. (1998). Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J. Bacteriol. 180, 5968–5977.
Beran, R. K., and Simons, R. W. (2001). Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 39, 112–125.
Briani, F., Del Favero, M., Capizzuto, R., Consonni, C., Zangrossi, S., Greco, C., De Gioia, L., Tortora, P., and Deho, G. (2007). Genetic analysis of polynucleotide phosphorylase structure and functions. Biochimie 89, 145–157.
Cam, K., Rome, G., Krisch, H. M., and Bouché, J. P. (1996). RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 24, 3065–3070.
Carpousis, A. J. (2007). The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87.
Cheng, Z. F., Zuo, Y. H., Li, Z. W., Rudd, K. E., and Deutscher, M. P. (1998). The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273, 14077–14080.
Clements, M. O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J. C. D., Normark, S., and Rhen, M. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 99, 8784–8789.
Cohn, M. T., Ingmer, H., Mulholland, F., Jorgensen, K., Wells, J. M., and Brondsted, L. (2007). Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 73, 7803–7813.
Day, W. A., Sajecki, J. L., Pitts, T. M., and Joens, L. A. (2000). Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68, 6337–6345.
Deutscher, M. P. (2006). Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34, 659–666.
Deutscher, M. P., and Reuven, N. B. (1991). Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 88, 3277–3280.
Donovan, W. P., and Kushner, S. R. (1986). Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 83, 120–124.
Fields, J. A., and Thompson, S. A. (2008). Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190, 3411–3416.
García-Mena, J., Das, A., Sánchez-Trujillo, A., Portier, C., and Montañez, C. (1999). A novel mutation in the KH domain of polynucleotide phosphorylase affects autoregulation and mRNA decay in Escherichia coli. Mol. Microbiol. 33, 235–248.
Goverde, R. L. J., Veld, J., Kusters, J. G., and Mooi, F. R. (1998). The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 degrees C). Mol. Microbiol. 28, 555–569.
Grant, C. C. R., Konkel, M. E., Cieplak, W., and Tompkins, L. S. (1993). Role of flagella in adherence, internalisation, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial-cell cultures. Infect. Immun. 61, 1764–1771.
Guo, B. Q., Wang, Y., Shi, F., Barton, Y. W., Plummer, P., Reynolds, D. L., Nettleton, D., Grinnage-Pulley, T., Lin, J., and Zhang, Q.J. (2008). CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J. Bacteriol. 190, 1879–1890.
Haas, A., and Goebel, W. (1992). Microbial strategies to prevent oxygen dependent killing. Free Radic. Res. Commun. 16, 137–157.
Haddad, N., Burns, C. M., Bolla, J. M., Prévost, H., Fédérighi, M., Drider, D., and Cappelier, J. M. (2009). Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl. Environ. Microbiol. 75, 7310–7318.
Haddad, N., Maillart, G., Garenaux, A., Jugiau, F., Federighi, M., and Cappelier, J. M. (2010). Adhesion ability of Campylobacter jejuni to Ht-29 cells increases with the augmentation of oxidant agent concentration. Curr. Microbiol. 61,500–505.
Hanel, I., Muller, J., Muller, W., and Schulze, E. (2004). Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 101, 75–82.
Holmes, K., Mulholland, F., Pearson, B. M., Pin, C, Mcnicholl-Kennedy, J., Ketley, J. M., and Wells, J. M. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151, 243–257.
Holmes, K., Tavender, T. J., Winzer, K., Wells, J. M., and Hardie, K. R. (2009). AI-2 does not function as a quorum sensing molecule in Campylobacter jejuni during exponential growth in vitro. BMC Microbiol. 9, 214. doi: 10.1186/1471-2180-9-214
Homuth, G., Domm, S., Kleiner, D., and Schumann, W. (2000). Transcriptional analysis of major heat shock genes of Helicobacter pylori. J. Bacteriol 182,4257–4263.
Jeon, B., Itoh, K., Misawa, N., and Ryu, S. (2003). Effects of quorum sensing on flaA transcription and autoagglutination in Campylobacter jejuni. Microbiol. Immunol. 47, 833–839.
Jeon, B., Itoh, K., and Ryu, S. (2005). Promoter analysis of cytolethal distending toxin genes (cdtA, B, and C) and effect of a luxS mutation on CDT production in Campylobacter jejuni. Microbiol. Immunol. 49, 599–603.
Korlath, J. A., Osterholm, M. T., Judy, L. A., Forfang, J. C., and Robinson, R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596.
Leszczyniecka, M., Desalle, R., Kang, D. C, and Fisher, P. B. (2004). The origin of polynucleotide phosphorylase domains. Mol Phylogenet. Evol 31, 123–130.
Li, Z., and Deutscher, M. P. (1994). The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem. 269, 6064–6071.
Linton, D., Gilbert, M., Hitchen, P. G., Dell, A., Morris, H. R., Wakarchuk, W. W., Gregson, N. A., and Wren, B. W (2000). Phase variation of a beta-1,3 galactosyltransferase involved in generation of the ganglioside GM(1)-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37, 501–514.
Luttinger, A., Hahn, J., and Dubnau, D. (1996). Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19, 343–356.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25,402–408.
Matus-Ortega, M. E., Regonesi, M. E., Piña-Escobedo, A., Tortora, P., Dehò, G., and García-Mena, J. (2007). The KH and S1 domains of Escherichia coli polynucleotide phosphorylase are necessary for autoregulation and growth at low temperature. Biochim. Biophys. Acta 1769, 194–203.
Mohanty B. K., and Kushner, S. R. (2000). Polynucleotide phosphorylase functions both as a 3′ → 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 11966–11971.
Nachamkin, I., Allos, B. M., and Ho, T. (1998). Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11,555–567.
Newell, D. G., and Fearnley, C. (2003). Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69, 4343–4351.
Osman, K. M., Ali, M. M., Radwan, M. I., Kim, H. K., and Han, J. (2009). Comparative proteomic analysis on Salmonella gallinarum and Salmonella enteritidis exploring proteins that may incorporate host adaptation in poultry. J. Proteomics 72, 815–821.
Oussenko, I. A., Abe T., Ujiie H., Muto A., and Bechhofer D. H. (2005). Participation of 3/-to-5/ exoribonucleases in the turnover of Bacillus subtilis mRNA.J. Bacteriol. 187,2758–2767.
Ow, M. C, and Kushner, S. R. (2002). Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 16, 1102–1115.
Palyada, K., Yi-Qian, S., Flint, A., Butcher, J., Naikare, H., and Stintzi, A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10, 481. doi: 10.1186/1471-2164-10-481
Parkhill, J., Wren, B. W, Mungall, K., Ketley, J. M., Churcher, C, Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., Jagels, K., Karlyshev, A. V., Moule, S., Pallen, M. J., Penn, C. W, Quail, M. A., Rajandream, M. A., Rutherford, K. M., Van Vliet, A. H. M., Whitehead, S., and Barrell, B. G. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668.
Peterson, K. M., and Mekalanos, J. J. (1988). Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56, 2822–2829.
Quiñones, B., Miller, W. G., Bates, A. H., and Mandrell, R. E. (2009). Autoinducer-2 production in Campylobacter jejuni contributes to chicken colonization. Appl. Environ. Microbiol. 75, 281–285.
Rangarajan, E. S., Bhatia, S., Watson, D. C., Munger, C., Cygler, M., Matte, A., and Young, N. M. (2007). Structural context for protein N-glycosylation in bacteria: the structure of PEB3, an adhesin from Campylobacter jejuni. Protein Sci. 16, 990–995.
Regonesi, M. E., Del Favero, M., Basilico, F., Briani, F., Benazzi, L., Tortora, P., Mauri, P., and Deho, G. (2006). Analysis of the Escherichia coli RNA degradosome composition by a proteomic approach. Biochimie 88, 151–161.
Ritz, M., Garenaux, A., Berge, M., and Federighi, M. (2009). Determination of rpoA as the most suitable internal control to study stress response in C. jejuni by RT-qPCR and application to oxidative stress. J. Microbiol. Methods 76, 196–200.
Rosenzweig, J. A., Chromy, B., Echeverry, A., Yang, J., Adkins, B., Plano, G. V., Mccutchen-Maloney, S., and Schesser, K. (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270, 255–264.
Rosenzweig, J. A., Weltman, G., Plano, G. V., and Schesser, K. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280, 156–163.
Skirrow, M. B. (1994). Diseases due to Campylobacter, Helicobacter, and related bacteria. J. Comp. Pathol. 111, 113–149.
Thomas, J. G., and Baneyx, F. (2000). ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 36, 1360–1370.
Viegas, S. C., Pfeiffer V., Sittka A., Silva I. J., Vogel J., and Arraiano C. M. (2007). Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35, 7651–7664.
Wang, W., and Bechhofer, D. H. (1996). Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178, 2375–2382.
Wassenaar, T. M., Bleumink-Pluym, N. M. C, and Van Der Zeijst, B. A. M. (1991). Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10, 2055–2061.
Wu, J. H., Jiang, Z., Liu, M., Gong, X., Wu, S. H., Burns, C. M., and Li, Z. W. (2009). Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 48, 2012–2020.
Yamanaka, K., Mitta, M., and Inouye, M. (1999). Mutation analysis of the 5; untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181, 6284–6291.
Yao, R. J., Burr, D. H., Doig, P., Trust, T. J., Niu, H.Y, and Guerry, P. (1994). Isolation of motile and nonmotile insertional mutants of Campylobacter jejuni- the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14, 883–893.
Ygberg, S. E., Clements, M. O., Rytkonen, A., Thompson, A., Holden, D. W., Hinton, J. C. D., and Rhen, M. (2006). Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74, 1243–1254.
Keywords: Campylobacter jejuni, polynucleotide phosphorylase, in vitro virulence tests, chick colonization, 2D-electrophoresis
Citation: Haddad N, Tresse O, Rivoal K, Chevret D, Nonglaton Q, Burns CM, Prévost H and Cappelier JM (2012) Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni. Front. Cell. Inf. Microbio. 2:30. doi: 10.3389/fcimb.2012.00030
Received: 02 December 2011; Accepted: 29 February 2012;
Published online: 14 March 2012.
Edited by:
Alain Stintzi, Ottawa Institute of Systems Biology, CanadaReviewed by:
Arnoud H. M. Van Vliet, Institute of Food Research, UKCecilia M. Arraiano, Instituto de Tecnologia Química e Biológica/Universidade Nova de Lisboa, Portugal
Copyright: © 2012 Haddad, Tresse, Rivoal, Chevret, Nonglaton, Burns, Prévost and Cappelier. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Nabila Haddad, ONIRIS, UMR1014 Sécurité des Aliments et Microbiologie, INRA, rue de la Géraudière, 44322 Nantes, France. e-mail: nabila.haddad@oniris-nantes.fr

