Escherichia coli isolated from urinary tract infections of Lebanese patients between 2005 and 2012: epidemiology and profiles of resistance
- 1Faculty of Medicine and Medical Sciences, University of Balamand, Tripoli, Lebanon
- 2“Centre Hospitalier du Nord” Hospital, Zgharta, Lebanon
- 3Faculty of Medicine, American University of Beirut, Beirut, Lebanon
- 4Division of Geographic and Infectious Diseases, Tufts Medical Center, Boston, MA, USA
The early treatment of urinary tract infections (UTIs) is directly related to decrease in morbidity, which makes the empirical treatment of great importance. Recently, beta lactamases of several types have emerged as significant mechanisms of resistance in Gram-negative bacilli, especially Escherichia coli. Our aim was to study the urinary E. coli isolated from Lebanese patients and to characterize their mechanisms of resistance. The study analyzed data between 2005 and 2012 of UTIs caused by E. coli. The mechanisms of resistance were characterized by phenotypic and genotypic methods and the pulsed-field gel electrophoresis (PFGE) was used to determine the different bacterial clusters. As expected, the highest incidence was observed with E. coli (60.53–73.98%) followed by K. pneumoniae (5.32–8.33%). ICU isolates were constantly associated with the lowest rates of susceptibility to extended-spectrum cephalosporins, ciprofloxacin, as well as most of the tested antibiotics. A 100% occurrence of CTX-M in extended-spectrum β-lactamase (ESBL)-producing isolates was recorded, followed by TEM, SHV, and OXA. In addition, 15.9% harbored 4 different ESBL enzymes and only 13 isolates (14.8%) harbored only one enzyme (CTX-M). Over the years, the simultaneous susceptibility of E. coli to ceftazidime and ciprofloxacin decreased from 62.5% in 2006 to 48.7% in 2012. PFGE results demonstrated that 10 clusters were 32 generated, denoting diversity among detected isolates. Understanding the epidemiology of resistance is 33 instrumental for the implementation of recommendations for the management of antimicrobials, infection 34 control measures, as well as active surveillance and antimicrobial stewardship.
Introduction
The antimicrobials use and misuse have led to bacterial resistance. Urinary tract infections (UTIs) are among the most common infectious diseases encountered in the community and in the hospital; they result in high rates of morbidity and high economic costs associated with treatment (1–3). In recent decades, the extended-spectrum β-lactamases (ESBLs) of the TEM, SHV, CTX-M, and OXA type, as well as the CTX-M have emerged as significant mechanisms of resistance in Gram-negative bacilli (4). ESBLs are enzymes able to efficiently hydrolyze extended-spectrum cephalosporins and monobactams and have been associated with therapeutic failures. These enzymes have widely spread to geographic regions, and this is due, in part, to the fact that many resistance genes are often carried on self-transmissible or mobile plasmids that are capable of spreading horizontally between and within species (4, 5). In addition, carbapenem resistance, a worrisome public health threat, is being reported throughout the world.
Early treatment of UTI is directly related to decrease in morbidity, which makes the selection of empirical therapy of great importance (6). The correct choice of empirical antimicrobial requires a good understanding of the typical bacteriology involved in UTIs (4–6), local resistance patterns, as well as the specific patient’s antimicrobial and microbiologic history, all of which affect the relative likelihoods of various organisms. Appropriate selection of antimicrobial therapy is an important strategy in the prevention of the spread of resistance, since use of overly broad therapy can lead to development of resistance. Escherichia coli, a major component of the intestinal flora, has been described as the most frequent uropathogen involved in community as well as hospital-acquired UTI (6–9). According to a study previously performed in Beirut (10), E. coli was the most frequent isolate (60.64% of the total isolates) and an increase in the production of ESBL was observed between the years 2000 and 2009 (2.3–16.8%). Unfortunately, very limited data concerning UTIs is available from other regions of the country. The north of Lebanon represents an important site for the investigation of bacterial infection and resistance in view of the many socio-cultural, economic, and medical practice differences. The purpose of this study was to investigate the bacterial etiology of UTIs in one of the busiest hospitals of the north of Lebanon (Centre Hospitalier du Nord) and to examine the epidemiologic and microbiologic properties, including phenotypic and genotypic characterization of the mechanisms of resistance of E. coli isolated from UTIs of Lebanese patients over an 8-year period.
Materials and Methods
Study Design
This study includes a retrospective part analyzing the data generated between January 2005 and January 2011, and a prospective part dealing with the occurrence of beta-lactamases genes in clinical isolates of E. coli between January 2011 and January 2013 and the related epidemiology of infection. Isolates of E. coli that showed intermediate or full resistance to carbapenems were stored at −80°C until they were processed for analysis. Over the 8-year-study, the population included all in and outpatients with positive urine cultures for a Gram-negative bacillus (6284) regardless of the type of UTI (complicated UTI, non-complicated UTI, acute pyelonephritis, etc.). Histological and microbiological criteria for positivity are cited below. In both the retrospective and prospective parts of the study, all urinary isolates data were collected and entered into a database using WHONET software. These data were classified based on frequency, department of origin, antimicrobial susceptibility profiles, as well as etiology. Among the UTIs caused by Gram-negative bacilli, there were 4,097 (65.2%) samples from female patients and 2,187 (34.8%) from males. Adult patients were sampled by clean catch midstream urine, and children aged <3 years were sampled using sterile urine bags. Only a single positive culture per patient was included in the analysis within the period of 3 months.
Isolation and Identification of Organisms
Samples for urine culture were tested within half an hour of sampling. All samples were inoculated on Uriselect agar and MacConkey agar and incubated at 37°C for 24 h, and for 48 h in case no growth was observed after 24 h. A specimen was considered positive for UTI in the light of the number of yielded colonies (≥105 cfu/mL) and the cytology of the urine through microscopic detection of bacteriuria and PMNs (≥8 leukocytes/mm3). However, lower colony counts associated with significant pyuria or low PMN count associated with significant colony counts was considered and analyzed in the light of the clinical picture and the patient’s immunological status. Bacterial identification was based on standard culture and biochemical characteristics of isolates. Gram-negative bacteria were identified by standard biochemical tests (11, 12).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of Enterobacteriaceae was tested by the disk diffusion method according to CLSI recommendations, using Mueller–Hinton agar (13). Antimicrobial agents tested were ampicillin, amoxicillin–clavulanic acid, aztreonam, cefoxitin, cefuroxime, cefotaxime, ceftriaxone, ceftazidime, cefepime, piperacillin, piperacillin–tazobactam, imipenem, meropenem, gentamicin, tobramycin, norfloxacin, ciprofloxacin, trimethoprim–sulfamethoxazole, and tigecycline (all disk charges were chosen as per the CLSI recommendations)
Statistical Analysis
Variables were expressed as percentages.
Phenotypic Detection of Resistance
The phenotypic detection of ESBL was done by the double disk synergy test (DDST). In brief, ceftazidime (30 mg), cefepime (30 mg), aztreonem (30 mg), and cefotaxime (30 mg) disks (Oxoid) were placed 25 mm (center to center) from the amoxicillin/clavulanic acid (20/10 mg) disk on Mueller–Hinton agar plate inoculated with the test organism; the presence of a keyhole effect was recorded 24 h after incubation. Resistance to carbapenems was detected as described by Birgy et al. (14). In this method, ertapenem, imipenem, and meropenem were tested on Mueller–Hinton agar plates (MHA) impregnated with 5 mM of EDTA, 10 mg/mL of phenyl boronic acid (PBA), and 250 μg/mL cloxacillin (embedded) were used, respectively, for the detection of metallo-β-lactamases (MBLs), Klebsiella pneumoniae carbapenemases (KPCs), and overproduction of AmpC. Additionally, disks of amoxicillin–clavulanic acid, ceftazidime, cefotaxime, and cefepime were added and arranged in a manner where a keyhole effect could be observed for the detection of ESBL production, and this provided an additional step for ESBL detection in view of the enhanced keyhole effect observed in presence of EDTA and/or PBA. Resistance to temocillin was used for the phenotypic detection of Oxa-48 carbapenemase. E. coli ATCC 25922 was used as ESBL-negative and K. pneumoniae 700603 was used as ESBL-positive reference strains.
Genotypic Detection of Resistance Genes
Multiplex PCR (four plex) was used to determine the presence blaTEM, blaSHV, blaCTX-M, and blaOXA (15). Primers used for these genes were: blaSHV – 237bp (F5′-CTTTATCGGCCCTCACTCAA-3′, R5′-AGGTGCTCATCATGGGAAAG-3′), blaTEM – 445bp (F5′-CGCCGCATACACTATTCTCAGAATGA-3 ′, R5′-ACGCTCACCGGCTCCAGATTTAT-3′), blaCTX-M – 593 bp(5′-ATGTGCAGYACCAGTAARGTKATGGC-3′, R5′-TGGGTRAARTARGTSACCAGAAYCAGCGG-3′), blaOXA – 813 bp (F5′-ACACAATACATATCAACTTCGC-3′, R5′-AGTGTGTTTAGAATGGTGATC-3′). PCR was performed as follows: 15 min at 95°C, 30 s at 94°C (30 cycles), 90 s at 62°C (30 cycles), 60 s at 72°C (30 cycles), 10 min at 72°C. The amplicons were tested by gel electrophoresis. The PCR mix of this reaction included 10 μl of master mix, 4 μl of primers mix, 1 μl of DNA, 5 μl of water.
For the detection of carbapenemases, 2 μl of total DNA was subjected to multiplex PCR in a 50-μL reaction mixture. The mix for the detection of blaIMP, blaVIM, and blaSPM genes contained 1× PCR buffer [10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl], 1.5 mmol/L of MgCl2, 0.125 mmol/L of each deoxynucleotide triphosphate, 10 μmol/L of each primer, and 2 U of AmpliTaq Gold Polymerase (Roche, Meylan, France). The mix for the detection of blaKPC, blaNDM, blaBIC, and blaOXA-48 was the same concentration. The mix for the detection of blaAIM, blaDIM, blaGIM, and blaSIM contained 1× PCR buffer [10 mmol/L Tris–HCl (pH 8.3), 50 mmol/L KCl], 3 mmol/L of MgCl2, 0.125 mmol/L of each deoxynucleotide triphosphate, 10 μmol/L of each primer, 3 μL of dimethyl sulfoxide, and 2 U of AmpliTaq Gold Polymerase. Amplification was carried out with the following thermal cycling conditions: 10 min at 94°C and 36 cycles of amplification consisting of 30 s at 94°C, 40 s at 52°C, and 50 s at 72°C, with 5 min at 72°C for the final extension. DNA fragments were analyzed by electrophoresis in a 2% agarose gel at 100 V for 1 h in 1× TAE [40 mmol/L Tris–HCl (pH 8.3), 2 mmol/L acetate, 1 mmol/L EDTA] containing 0.05 mg/L ethidium bromide. The primers were as follows (5’–3’): blaIMP – 232 bp (F GGAATAGAGTGGCTTAAYTCTC, R GGTTTAAYAAAACAACCACC), blaSPM – 271 (F AAAATCTGGGTACGCAAACG, R ACATTATCCGCTGGAACAGG), blaAIM – 322 (F CTGAAGGTGTACGGAAACAC, R GTTCGGCCACCTCGAATTG), blaVIM – 390 (F GATGGTGTTTGGTCGCATA, R CGAATGCGCAGCACCAG), blaOXA-48 – 438 (F GCGTGGTTAAGGATGAACAC, R CATCAAGTTCAACCCAACCG), blaGIM – 477 (F TCGACACACCTTGGTCTGAA, R AACTTCCAACTTTGCCATGC), blaBIC – 537 (FTATGCAGCTCCTTTAAGGGC, R TCATTGGCGGTGCCGTACAC), blaSIM – 570 (F TACAAGGGATTCGGCATCG, R TAATGGCCTGTTCCCATGTG), blaNDM – 621 (F GGTTTGGCGATCTGGTTTTC, R CGGAATGGCTCATCACGATC), blaDIM – 699 (F GCTTGTCTTCGCTTGCTAACG, R CGTTCGGCTGGATTGATTTG), blaKPC – 798 (F CGTCTAGTTCTGCTGTCTTG, R CTTGTCATCCTTGTTAGGCG).
Genotyping
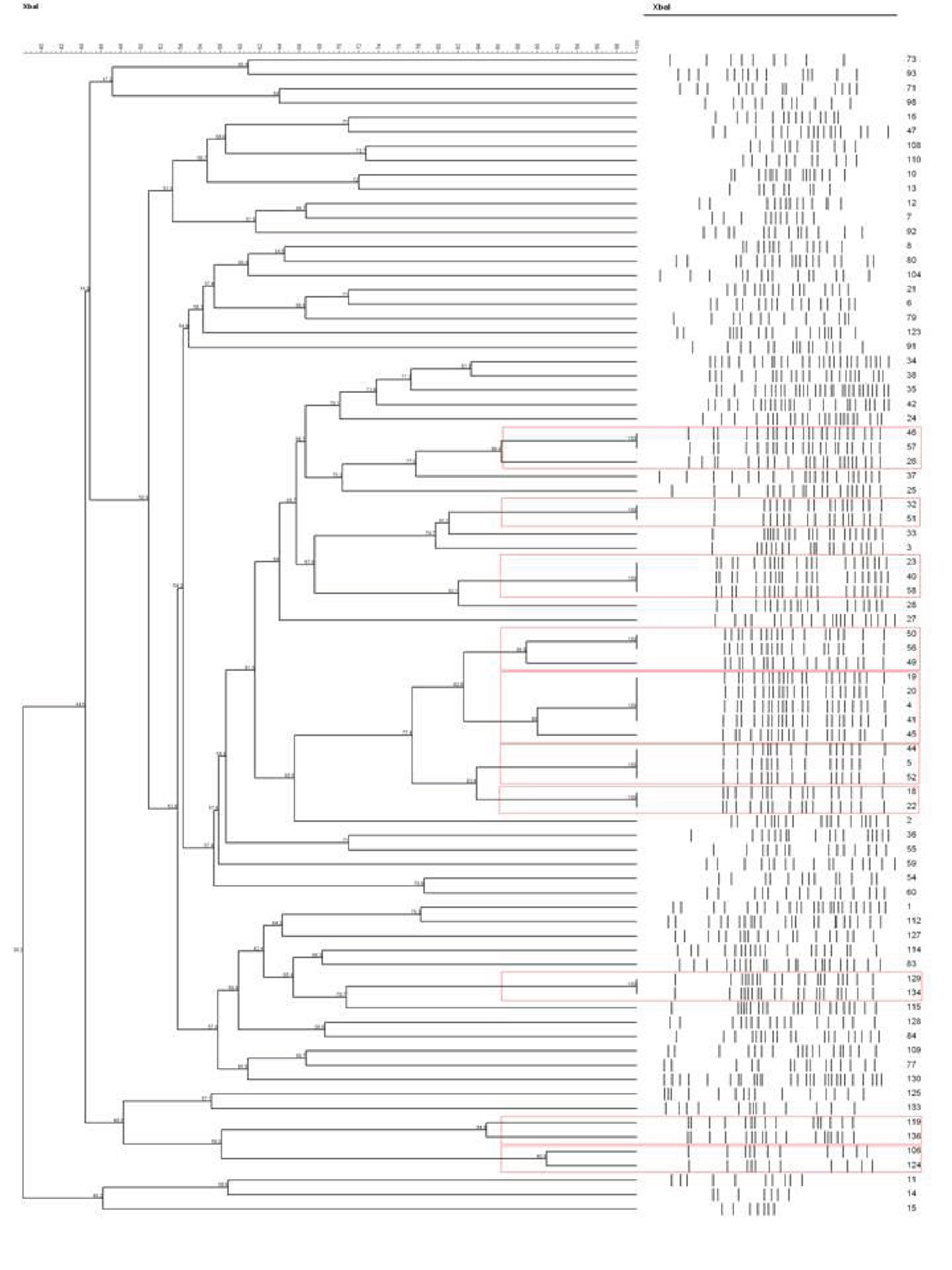
Pulsed-field gel electrophoresis (PFGE) was carried out using restriction enzyme XbaI (Invitrogen) and the PFGE CHEF MAPPER (Bio-Rad). The comparison for E. coli was made using the Dice coefficient. The dendrograms were generated adopting the UPGMA method (unweighted pair group method using arithmetic averages) using Bionumerics software. DNA ratio was calculated on the Dice coefficient, and isolates were considered genetically related if the Dice coefficient correlation was 85% or higher. Isolates with 85% and above relatedness were clustered using a red frame.
Results
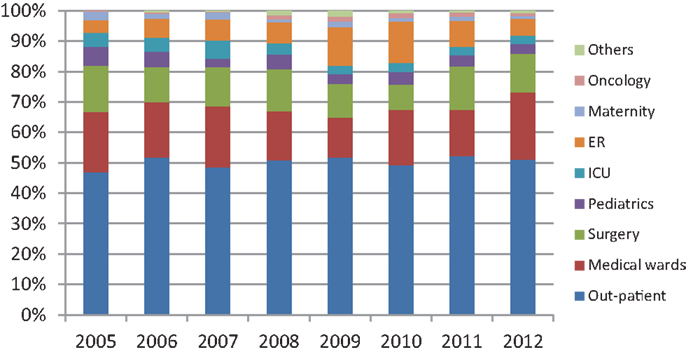
Over the 8-year period, a total of 6284 Gram-negative bacterial isolates was recovered from documented UTIs. Table 1 shows the Gram-negative bacilli most frequently isolated from these positive urine cultures. As expected, the highest frequency was observed with E. coli (60.53–73.98%) followed by K. pneumoniae (5.32–8.33%), Proteus mirabilis (3.36–6.17%), Pseudomonas aeruginosa (2.3–3.9%), and Enterobacter cloacae (0.76–2.23%). Figure 1 shows the distribution of urinary isolates of E. coli by location. Out-patient isolates predominate over the years with a more or less stable percent of occurrence.
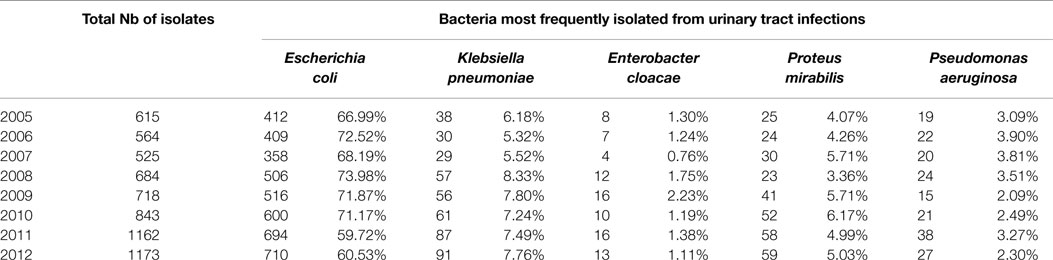
Table 1. Frequency of Gram-negative Bacilli-documented UTI isolates at the CHN hospital classified by year.
Figure 2 is plotted based on scatterplot analysis of co-susceptibility/non-susceptibility to ceftazidime and ciprofloxacin. Over the years, the isolation of E. coli that are susceptible to both antibiotics is decreasing (62.5% in 2006 and 48.7% in 2012), while strains that are non-sensitive to both antibiotics are increasing (13.5% in 2006 and 23.2% in 2012).
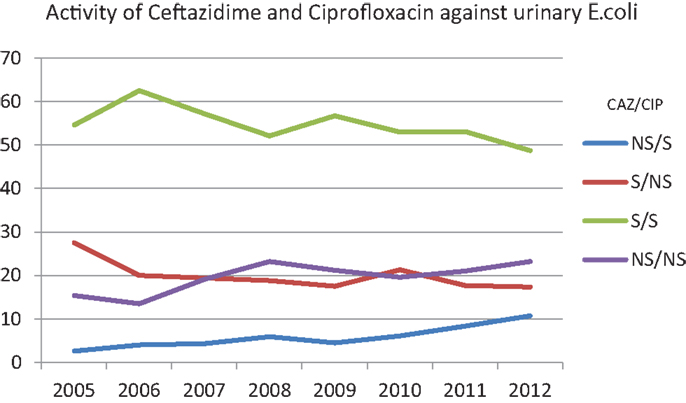
Figure 2. Percentages of susceptibility of urinary E. coli to ceftazidime and ciprofloxacin over the years. NS/S: trends of the isolates non-susceptible to ceftazidime/susceptible to ciprofloxacin. S/NS: trends of the isolates susceptible to ceftazidime/non-susceptible to ciprofloxacin. S/S: trends of the isolates susceptible to ceftazidime/susceptible to ciprofloxacin. NS/NS: trends of the isolates non-susceptible to ceftazidime/non-susceptible to ciprofloxacin.
Table 2 shows the susceptibility profiles of E. coli to the different antimicrobial agents. No major difference in susceptibility is observed between the overall isolates and the urinary isolates. The lowest percentage of susceptibility was manifested against ampicillin reaching 26% in 2007. Over successive years, the susceptibility to third and fourth generations’ cephalosporins (represented by Ceftazidime in Figure 3F) and aztreonam (Figure 3G) shows an obvious and constant decrease; this is coupled by an increase in the production of ESBL as shown in Table 2 (12.6% in 2005 and 29.4% in 2012 for all E. coli isolates). A similar pattern of ESBL production is noted for the urinary isolates (11.6% in 2005 and 25.3% in 2012). Since 2008, a minimum of 10% susceptibility decrease to third and fourth generation cephalosporin between in and out-patients isolates was observed. Such a difference is most likely due to the higher occurrence of ESBL and AmpC in the hospital isolates. Figure 3K shows a considerably higher susceptibility of isolates from outpatients to ciprofloxacin, although the general patterns of in- and out-patients’ isolates are similar. The susceptibility to trimethoprim–sulfamethoxazole is essentially the same for in and outpatients; however, it is decreasing with time (Figure 3J). Susceptibility to nitrofurantoin manifests an irregular pattern where the high rates in 2007 are significantly decreased in in-patients, affecting therefore the overall percentage of susceptibility; however, this antibiotic seems to keep its high activity against E. coli. Two imipenem-resistant E. coli were isolated from UTIs in 2011 and 2012; of course, this does not affect the pattern of susceptibility to this antibiotic (Figure 3H). Isolates with decreased susceptibility to fosfomycin were observed in 2011 and 2012. Figures 3A and 3E show the patterns of susceptibility to amikacin and cefoxitin respectively.
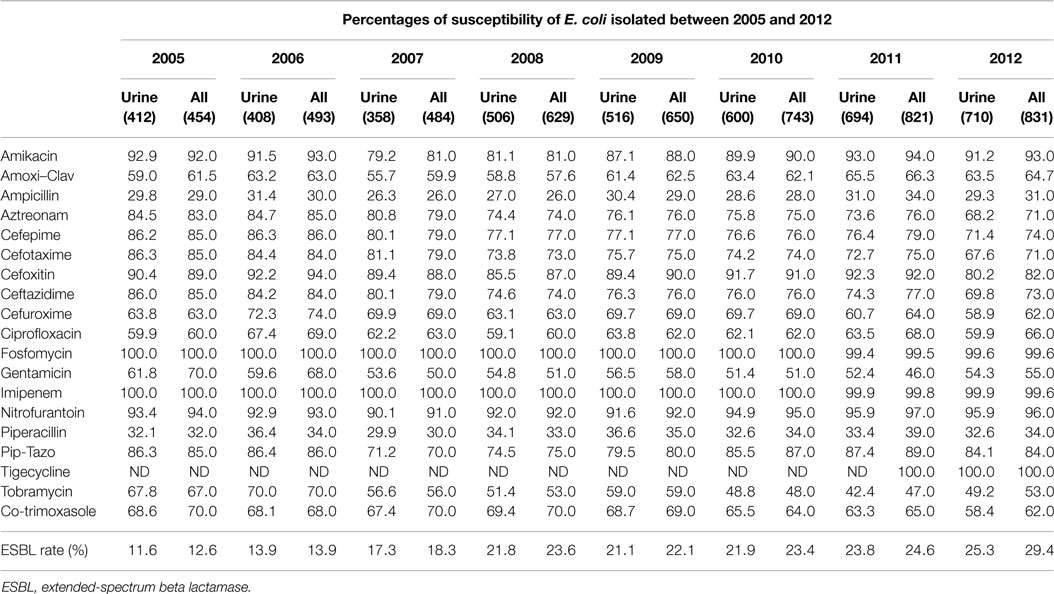
Table 2. The rates of susceptibility of urinary and all isolates of E. coli between 2005 and 2012 is shown.
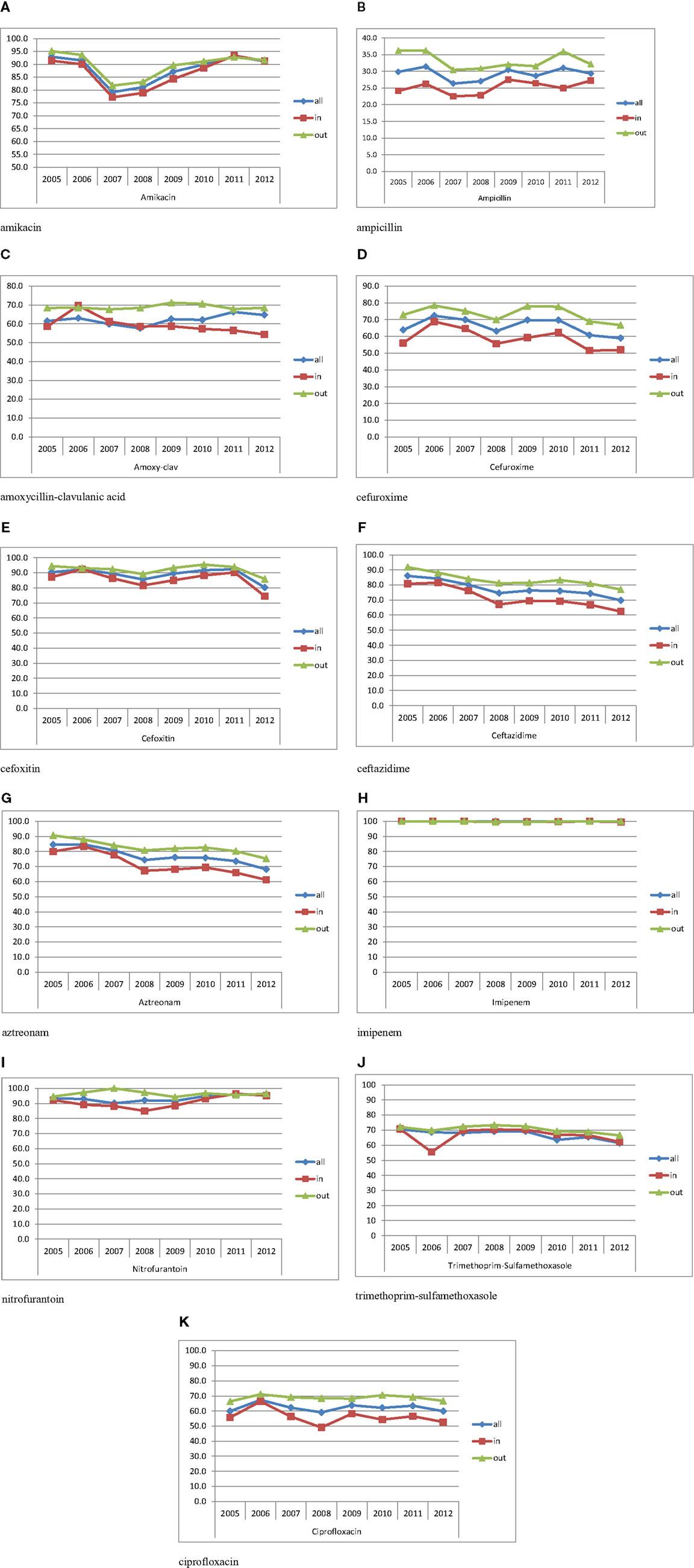
Figure 3. Percentages of susceptibility of urinary isolates of E. coli in all/in/and out-patients. (A) Amikacin. (B) Ampicillin. (C) Amoxicillin–clavulanic acid. (D) Cefuroxime. (E) Cefoxitin. (F) Ceftazidime. (G) Aztreonam. (H) Imipenem. (I) Nitrofurantoin. (J) Trimethoprim–sulfamethoxazole. (K) Ciprofloxacin.
Tables 3 and 4 report the susceptibility profiles of the urinary isolates of E. coli by location. ICU isolates were constantly associated with the lowest rates of susceptibility to extended-spectrum cephalosporins, ciprofloxacin, as well as most of the tested antibiotics. Isolates coming from ER patients showed rates of susceptibility similar to out-patients’ isolates.
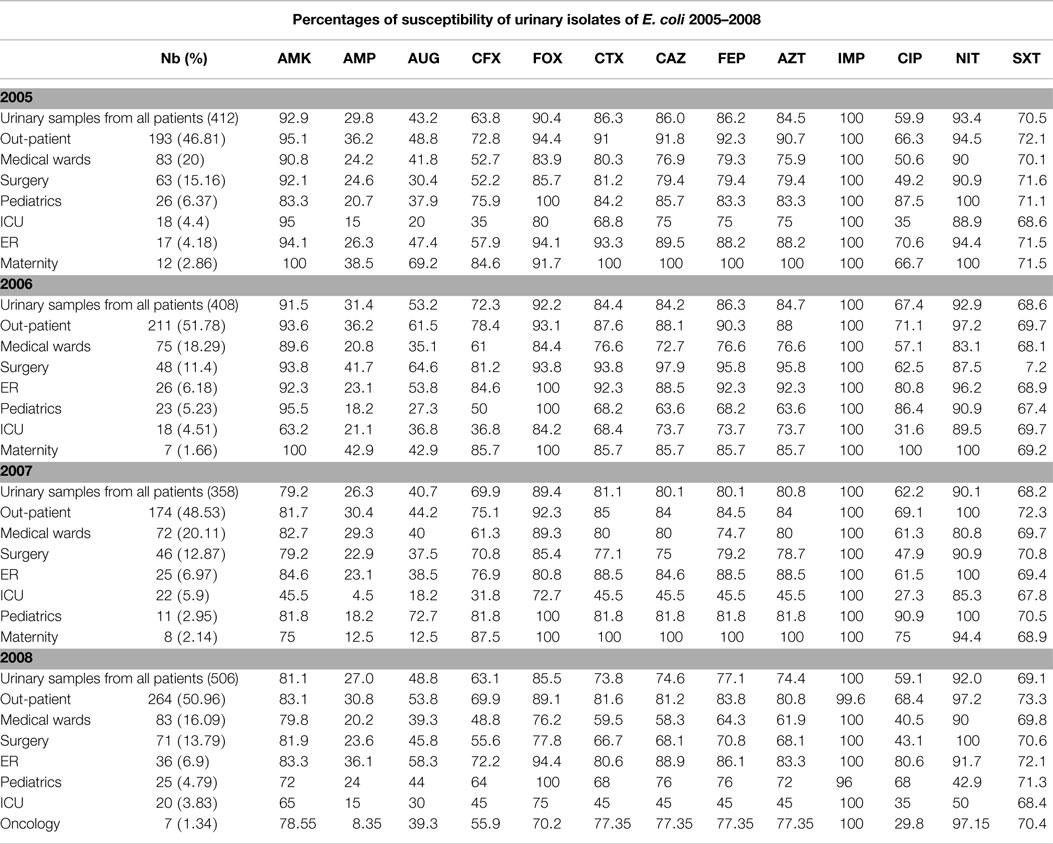
Table 3. Profiles of susceptibility of urinary E. coli isolated between 2005 and 2008 grouped by location.
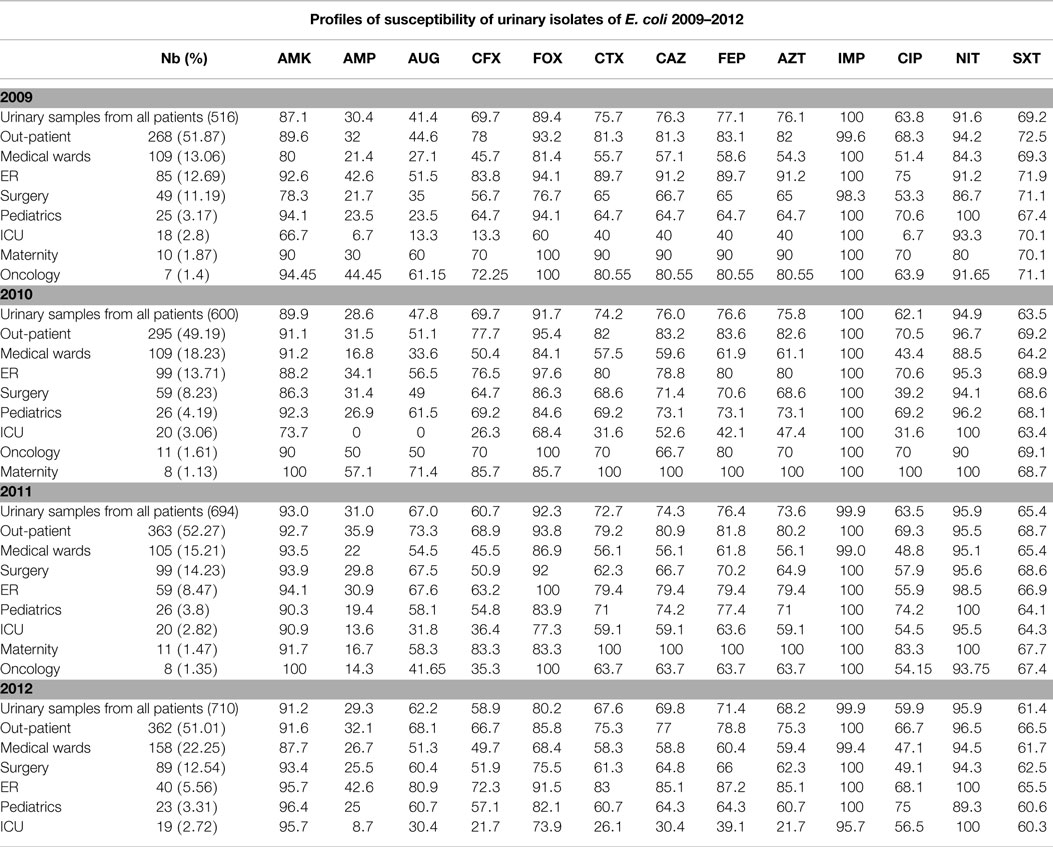
Table 4. Profiles of susceptibility of urinary E. coli isolated between 2009 and 2012 grouped by location.
A total of 88 isolates isolated in 2012 were subjected to molecular testing for ESBL enzymes. Our results suggest a high correlation between phenotypic and genotypic testing. A 100% occurrence of CTX-M in ESBL-producing isolates is recorded (Table 5), followed by TEM, SHV, and OXA. In addition, 15.9% harbored 4 different ESBL enzymes and only 13 isolates (14.8%) harbored only 1 enzyme (CTX-M).
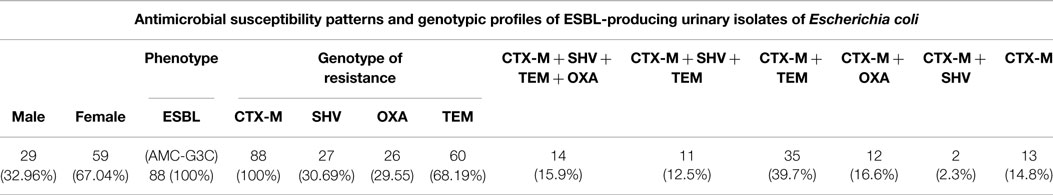
Table 5. Phenotypic and genotypic detection of the mechanisms of resistance in 88 urinary E. coli isolated in 2012.
On the other hand, concerning the five carbapenem-resistant isolates of E. coli (two were urinary isolates), both phenotypic and genotypic testing showed that OXA-48 was the most common mechanism of carbapenem resistance (Table 6). Two of these isolates came from pus, two from urine, and one from sputum cultures. In both urinary isolates, OXA-48 was responsible for resistance to imipenem.
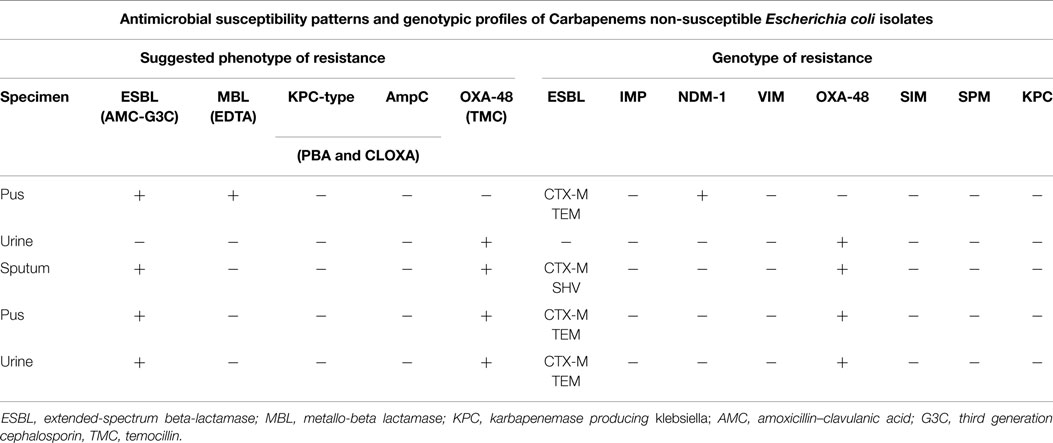
Table 6. Phenotypic and genotypic characterization of the mechanisms of resistance in 5 E. coli isolated in 2012.
On the other hand, the PFGE results of E. coli isolated in 2013 (Figure 4) demonstrated that 10 clusters were generated, denoting diversity among detected isolates. However, those isolates falling within the same cluster have a relatively high degree of relatedness among them.
Discussion
Lebanon is currently witnessing an increase in the number of bacterial infections that are associated with a wide spectrum of resistance to common antimicrobial agents (16). The presence of ESBL-producing organisms has been reported in hospitals as well as in the community (17). In 1997, the percentage of ESBL-producing E. coli and K. pneumoniae was 1.3 and 7.5%, respectively, as compared to non-ESBL-producing strains of the same species. This percentage, however, has increased over the years [in 2001, the percentage of ESBL-producing E. coli reached 4% (16, 18)]. Overall, there is a scarcity of data relating to antimicrobial resistance in the Middle East (16). Molecular analysis revealed that CTX-M-15 is the most widespread ESBL since it was present in 83% of the resistant strains. The reason behind this increase of the frequency of occurrence of ESBL-producing organisms is likely due to the misuse, and abuse of antibiotics (19).
Our results show that the northern part of Lebanon is not different from the rest of the country. The rate of ESBLs increased considerably over the course of the study period from 12.6 to 29.4 in all E. coli isolates and from 11.6 to 25.3% in urinary isolates. This result correlates with the general trend in the country. In addition to containing CTX-M-15, 30.7% of isolates tested positive for carriage of SHV, 29.6% for OXA, and 68.2% for TEM. These results indicate that ESBL-producing organisms frequently harbor more than one β-lactamase gene. In fact, 15.9% of E. coli harbored all four ESBL enzymes and 14.8% harbored only one gene of resistance (Table 5). Though this does not impact the treatment strategy, harboring more than one gene of ESBL enzymes makes the diversity of resistance gene carriage considerably bigger.
It is known that the occurrence of ESBL-producing E. coli in high-risk areas of the hospital, such as ICUs, has increased significantly (20, 21). In this context, bigger concern has resulted from reports of the increasing frequency of ESBL-producing organisms causing infections in outpatients. In our study, most UTIs were isolated from out-patients. This suggests an important spread of these enzymes in the community, even if these outpatients’ infections are not entirely community acquired and might correlate with hospital or health care facilities visits.
Appropriate antibiotic treatment for infections caused by ESBL-producing E. coli is significantly affected by the cross-resistance to other antibiotics such as quinolones (22). Resistance to these drugs has been shown to be related to the presence of a conjugative plasmid associated with the ESBL phenotype. In this context, carbapenems represent a good choice when therapy is needed for serious infections.
In our study, co-resistance has been observed with fluoroquinolones and ceftazidime (Figure 2). While susceptible isolates to both antibiotics remain the majority, the trend of occurrence of these isolates is clearly decreasing over the years and is coupled by an increase in resistance to either antibiotics. This implies that fluoroquinolone resistance could be driven by cephalosporin use at both hospital and community levels. Another implication of this observation is that ciprofloxacin resistance may be associated with limited treatment options for other classes of agents, as observed in our study. The isolation of two carbapenem-resistant E. coli from UTIs should be carefully considered; their occurrence foreshadows of a possible spread of carbapenemases into the community.
Although the PFGE results show 10 clusters among the urinary isolates of E. coli collected in 2013 with high level of similarity (>95%), these results indicate genetic diversity yet all encoded the CTX-M-15 type enzyme.
Limitations of the study include that some of the isolates included in the analysis may have been associated with asymptomatic bacteriuria rather than true UTI. While we used standard surveillance criteria to define UTI, we allowed for some isolates to be included without meeting those criteria if their clinical picture was highly consistent with UTI. This had the potential to introduce inter-observer variability in the determination of which isolates were included. Furthermore, asymptomatic bacteriuria is frequently associated with both high colony counts of bacteria and significant pyuria, especially in the presence of a bladder catheter. While the epidemiology of isolates obtained from patients with asymptomatic bacteriuria should be similar to those from patients with true infection, the public health implications are different, since asymptomatic bacteriuria does not require treatment except in rare circumstances.
In conclusion, ESBL-producing E. coli in urinary isolates is a growing problem and is spreading over the whole country. This study is among the first to provide information on the prevalence and distribution of isolated E. coli with ESBL producer, possible types of enzymes produced, clonal relationship, and susceptibility to potentially active drugs in the north (2005–2012). This information contributes to the understanding of the epidemiology of resistance in the whole country as well as the implementation of recommendations for the management of antimicrobials, infection control measures, and active surveillance and antimicrobial stewardship is highly needed.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Arjunan M, Al-Salamah AA, Amuthan M. Prevalence and antibiotics susceptibility of uropathogens in patients from a rural environment, Tamilnadu. Am J Infect Dis (2010) 6:29–33. doi: 10.3844/ajidsp.2010.29.33
2. Rahman F, Chowdhury S, Rahman MM, Ahmed D, Hossain A. Antimicrobial resistance pattern of Gram-negative bacteria causing urinary tract infection. Stamford J Pharm Sci (2009) 2:44–55. doi:10.3329/sjps.v2i1.5815
3. Hryniewicz K, Szczypa K, Sulikowska A, Jankowski K, Betltjewska K, Hryniewicz W. Antibiotic susceptibility of bacterial strains isolated from urinary tract. J Antimicrob Chemother (2001) 47:773–80. doi:10.1093/jac/47.6.773
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Apisarnthanarak A, Kiratisin P, Mundy LM. Clinical and molecular epidemiology of healthcare-associated infections due to extended-spectrum beta-lactamase (ESBL)-producing strains of Escherichia coli and Klebsiella pneumoniae that harbor multiple ESBL genes. Infect Control Hosp Epidemiol (2008) 29:1026–34. doi:10.1086/591864
5. Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother (2004) 48:1–14. doi:10.1128/AAC.48.1.1-14.2004
6. Francesco MA, Ravizzola G, Peroni L, Negrini R, Manca N. Urinary tract infections in Brescia, Italy: etiology of uropathogens and antimicrobial resistance of common uropathogens. Med Sci Monit (2007) 6:136–44.
7. Neto JAD, Martins ACP, Silva LDM, Tiraboshi RB, Domingos ALA, Cologna AJ, et al. Community acquired urinary tract infection etiology and bacterial susceptibility. Acta Cir Bras (2003) 18:33–6.
8. Wagenlehner F, Naber K. Antibiotics and resistance of uropathogens. EAU Update Ser (2004) 2:125–35. doi:10.1016/j.euus.2004.06.003
9. Drekonja DM, Johnson JR. Urinary tract infections. Prim Care (2008) 35:345–67. doi:10.1016/j.pop.2008.01.001
10. Daoud Z, Afif C. Escherichia coli isolated from urinary tract infections of Lebanese patients between 2000 and 2009: epidemiology and profiles of resistance. Chemother Res Pract (2011) 2011:1–6. doi:10.1155/2011/218431
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Pezzlo M. Detection of urinary tract infections by rapid methods. Clin Microbiol Rev (1988) 1:268–80.
12. Bonadio M, Meini M, Spitaleri P, Gigli C. Current microbiological and clinical aspects of urinary tract infections. Eur Urol (2001) 40:439–45. doi:10.1159/000049813
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; 15th Informational Supplement. CLSI Document M100-S15. Wayne, PA: Clinical and Laboratory Standards Institute (2005).
14. Birgy A, Bidet P, Genel N, Doit C, Decré D, Arlet G, et al. Phenotypic screening of carbapenemases and associated β-lactamases in carbapenem-resistant Enterobacteriaceae. J Clin Microbiol (2012) 50:1295–302. doi:10.1128/JCM.06131-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum beta lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J Clin Microbiol (2008) 46:707–12. doi:10.1128/JCM.01943-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Moubareck C, Daoud Z, Hakimé NI, Hamze M, Mangeney N, Matta H, et al. Countrywide spread of community- and hospital-acquired extended-spectrum β-lactamase (CTX-M-15)-producing enterobacteriaceae in lebanon. J Clin Microbiol (2005) 43:3309–13. doi:10.1128/JCM.43.7.3309-3313.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Daoud Z, Moubareck C, Hakime N, Doucet-Populaire F. Extended spectrum β-lactamase producing enterobacteriaceae in Lebanese ICU patients: epidemiology and patterns of resistance. J Gen Appl Microbiol (2006) 52:169–78. doi:10.2323/jgam.52.169
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Daoud Z, Hakime N. Prevalence and susceptibility patterns of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a general university hospital in Beirut, Lebanon. Rev Esp Quimioter (2003) 16:233–8.
19. Kanafani ZA, Mehio-Sibai A, Araj GF, Kanaan M, Kanj SS. Epidemiology and risk factors for extended-spectrum beta-lactamase-producing organisms: a case control study at a tertiary care center in Lebanon. Am J Infect Control (2005) 33:326–32. doi:10.1016/j.ajic.2005.03.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Winokur PL, Canton R, Casellas JM, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum β-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific Region. Clin Infect Dis (2001) 32:S94–103. doi:10.1086/320182
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Itokazu GS, Quinn JP, Bell-Dixon C, Kahan FM, Weinstein RA. Antimicrobial resistance rates among aerobic Gram-negative bacilli recovered from patients in intensive care units: evaluation of a national postmarketing surveillance program. Clin Infect Dis (1996) 23:779–84. doi:10.1093/clinids/23.4.779
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Yagüe A, Cebrian L, Rodriguez-Diaz JC, Gonzalo-Jiménez N, Royo G, Campillos P, et al. Expanded-spectrum beta-lactamase-producing strains of E. coli: origin, characteristics and incidence in southern Alicante (Spain) in the period 1999–2003. Enferm Infecc Microbiol Clin (2005) 23:76–9. doi:10.1157/1307161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: ESBL, carbapenemases, E. coli, urinary infections, bacterial resistance
Citation: Daoud Z, Salem Sokhn E, Masri K, Cheaito K, Haidar-Ahmad N, Matar GM and Doron S (2015) Escherichia coli isolated from urinary tract infections of Lebanese patients between 2005 and 2012: epidemiology and profiles of resistance. Front. Med. 2:26. doi: 10.3389/fmed.2015.00026
Received: 27 February 2015; Accepted: 09 April 2015;
Published: 28 April 2015
Edited by:
Evangelos Giamarellos-Bourboulis, University of Athens, GreeceReviewed by:
Thomas Tsaganos, Athens Medical School, GreeceGeorgia Vrioni, University of Athens, Greece
Copyright: © 2015 Daoud, Salem Sokhn, Masri, Cheaito, Haidar-Ahmad, Matar and Doron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziad Daoud, Beirut, Lebanon, ziad.daoud@balamand.edu.lb
 Ziad Daoud
Ziad Daoud Elie Salem Sokhn
Elie Salem Sokhn Khalil Masri2
Khalil Masri2
 Nathaline Haidar-Ahmad
Nathaline Haidar-Ahmad Ghassan M. Matar
Ghassan M. Matar