- 1Department of Environmental Biology, University of Rome “Sapienza”, Rome, Italy
- 2Istituto di Entomologia e Patologia Vegetale, Università Cattolica del Sacro Cuore, Piacenza, Italy
- 3Food Chemistry and Natural Substances Unit, Department of Organic and Industrial Chemistry, University of Parma, Parma, Italy
- 4Department of Agricultural, Food and Forestry Science, University of Turin, Torino, Italy
Oxylipins are fatty acid-derived signaling compounds produced by all eukaryotes so far investigated; in mycotoxigenic fungi, they modulate toxin production and interactions with the host plants. Among the many enzymes responsible for oxylipin generation, Linoleate Diol Synthase 1 (LDS1) produces mainly 8-hydroperoxyoctadecenoic acid and subsequently different di-hydroxyoctadecenoic acids. In this study, we inactivated a copy of the putative LDS1 ortholog (acc. N. FVEG_09294.3) of Fusarium verticillioides, with the aim to investigate its influence on the oxylipin profile of the fungus, on its development, secondary metabolism and virulence. LC-MS/MS oxylipin profiling carried out on the selected mutant strain revealed significant quali-quantitative differences for several oxylipins when compared to the WT strain. The Fvlds1-deleted mutant grew better, produced more conidia, synthesized more fumonisins and infected maize cobs faster than the WT strain. We hypothesize that oxylipins may act as regulators of gene expression in the toxigenic plant pathogen F. verticillioides, in turn causing notable changes in its phenotype. These changes could relate to the ability of oxylipins to re-shape the transcriptional profile of F. verticillioides by inducing chromatin modifications and exerting a direct control on the transcription of secondary metabolism in fungi.
Introduction
Fusarium verticillioides causes the ear and stalk rot of maize (Zea mays) by entering through roots, stalks and ears at different growth stages. Tissue invasion is often asymptomatic even in the presence of massive growth inside the kernels (Estrada et al., 2012), where F. verticillioides produces fumonisins. This is a family of mycotoxins that, especially in their B series (FB), are hazardous for human and animal health, while apparently not directly related to fungal virulence and aggressiveness. In particular, fumonisin B1 (FB1) has been included in the 2B class by the International Agency for Research on Cancer because of its possible carcinogenic effect in humans. Moreover, the European Union has enforced the legislation to fix a threshold of FB1 + FB2 content in raw maize and derived products intended for human consumption (EU Commission Regulation No. 1126/2007); recommendations have also been given for animal feeding (EU Commission Regulation No. 576/2006).
Fatty acids (FAs) and FA-metabolites are major structural and metabolic constituents of the cell, functioning also as modulators of signal transduction pathways or transcription factors induced by several stimuli (Duplus et al., 2000). For instance, the enzymatic and non-enzymatic peroxidation of FAs is one of the processes switched on during early plant defense signaling related to pathogen perception (Shea and Del Poeta, 2006; Walley et al., 2013). The products of FA peroxidation, named oxylipins, constitute a large family of oxidized FAs, and their by-products are present in almost every living kingdom (Koo and Howe, 2009; Brodhun and Feussner, 2011). The non-enzymatic pathway of oxylipin synthesis derives from spontaneous formation of reactive oxygen species (ROS) such as hydroxyl radical, which may initiate lipid peroxidation (Brodhun and Feussner, 2011). Besides this route, oxylipins are mainly produced through three enzyme-dependent pathways, the ones initiated by lipoxygenases (LOX) or dioxygenases, and pathways involving cytochromes (Brodhun and Feussner, 2011). Oxylipins possess pivotal functions as signal molecules (Christensen and Kolomiets, 2011).
In fungi, the first described oxylipins (Precocious sexual inducers, Psi-factors) owe the name to their involvement in the regulation of the sexual and asexual phases in Aspergillus (Tsitsigiannis et al., 2005; Brown et al., 2008). Psi factors are also involved in host-pathogen communication and promote fungal infection of plants (Tsitsigiannis and Keller, 2006; Brodhun and Feussner, 2011; Christensen and Kolomiets, 2011). The enzymes responsible for synthesizing the Psi-factors were initially identified in Aspergillus nidulans as orthologs of the 7,8-linoleate diol synthase (7,8-LDS) from Gaeumannomyces graminis, and were named Psi factor-producing oxygenases (Ppo) (Tsitsigiannis et al., 2005; Garscha et al., 2007). Over recent years, 7,8-LDS has been considered functionally and structurally similar to the mammalian prostaglandin H2-synthase. 7,8-LDS-encoding genes have been detected in several pathogenic and non-pathogenic fungi such as: Cercospora zeae-maydis (Shim and Dunkle, 2002), Magnaporthe grisea (Cristea et al., 2003), Magnaporthe oryzae (Jerneren et al., 2010), Ustilago maydis (Huber et al., 2002), and A. nidulans. In the latter fungus, three different genes with significant similarity to the gene encoding 7,8-LDS were identified (PpoA, PpoB, and PpoC) (Tsitsigiannis et al., 2005) and a fourth gene (PpoD) was characterized in Aspergillus flavus (Brown et al., 2009). Information on LDS enzymatic activity is scanty: it is known that PpoA is responsible for the formation of 5S,8R-dihydroxyoctadecenoic acid (diHODE), whilst PpoC could code for a 10R-dioxygenase, producing 10-hydroperoxyoctadecenoic acid (HPODE) (Garscha et al., 2007; Brodhun et al., 2009).
Plant and fungal lipids and their oxidized derivatives may drive the fate (compatible/incompatible) of plant-pathogen interactions and mycotoxin production (Tsitsigiannis and Keller, 2007; Christensen et al., 2014; Scarpari et al., 2014). During its interaction with the host, F. verticillioides induces the expression of the maize LOX, ZmLOX3 as well as of its close homolog ZmLOX2 (Gao et al., 2007). Infections by Aspergillus spp. increase 9S-HPODE levels in peanut seeds while suppressing 13S-HPODE synthesis (Brodhagen et al., 2008). Mycotoxigenic fungi may hijack the host 9-LOX pathway that, in turn, induces mycotoxin biosynthesis; thus, some 9-oxylipins function as susceptibility factors in the producing host plants (Gao et al., 2007; Tsitsigiannis and Keller, 2007; Brodhagen et al., 2008; Christensen and Kolomiets, 2011; Scarpari et al., 2014). Coherently, the pathogen-derived oxylipins may induce the expression of the host LOX genes. Brodhagen et al. (2008) indeed showed that whilst wild-type (WT) A. nidulans triggers the expression of a peanut LOX gene, its oxylipin-deficient ppo mutants fail to do so. Thus, an additional function for the oxylipins produced by pathogenic fungi might be in manipulating the expression of the host genes involved in lipid and oxylipin metabolism (Reverberi et al., 2010).
In this study, we investigated the functions of endogenous LDS1-derived oxylipins in affecting the biology of the maize pathogen F. verticillioides and its interaction with the host maize plant. To this purpose, we created a ΔFvlds1 strain (LDS1-deleted) and we studied its phenotype, secondary metabolism and virulence. Moreover, we explored the possibility that oxylipins—acting as chromatin modifiers—may control the expression of fumonisin biosynthetic genes (FUM) and, in turn, fumonisin synthesis. Results indicate that LDS1 deletion changes the whole oxylipin and FB-production profile of F. verticillioides. We here suggest that oxylipins may induce these alterations by controlling gene transcription also through a direct influence on the chromatin status.
Materials and Methods
Fungal Strains and Media
The WT strain deposited as ITEM 10027 in the collection of the Institute of Sciences of Food Production (ISPA-CNR, Bari, Italy; http://server.ispa.cnr.it/ITEM/Collection) was isolated from maize in Northern Italy and maintained at −80°C. The V8 juice agar with 100 ppm of hygromycin (200 mL of V8 juice, 3 g of CaCO3 and 20 g agar per L) was used for the first step of mutant selection. Monoconidial cultures of all strains were obtained and cultivated in liquid media in the dark and at 25°C on a rotary shaker (150 rpm) for 7 days unless noted otherwise.
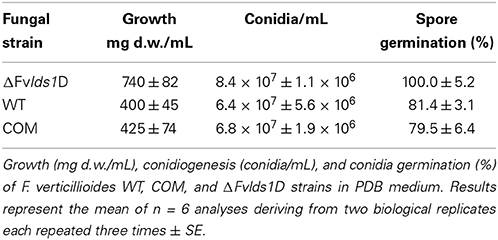
Fungal growth, conidiogenesis and spore production were determined by inoculating all the tested strains into Potato Dextrose Broth (PDB) liquid medium at 25°C. Since PDB is not conducive for FB production, we also used Czapek medium amended with cracked maize 2 g/L (abbreviated CDYM) which, as reported (Scala et al., 2013), induces FB production. CDYM (50 mL) was inoculated with 1 × 106 conidia/mL. Cultures were incubated up to 15 days after inoculation (DAI) in the dark at 25°C.
Molecular Biology Techniques
Gene deletion, fungal transformation and selection of transformants
To generate the construct for disrupting the F. verticillioides FVEG_09294.3 gene (putative LDS1) by targeted homologous recombination, specific oligonucleotides were designed to amplify the upstream (primers Lds_for_PciI and Lds_rev_StuI, presenting a PciI and StuI site respectively) and downstream (primers Lds_for_NaeI and Lds_rev_SbfI, presenting a NaeI and SfbI site respectively) flanking region of LDS1 (Supplementary Table 1). The amplicons (about 1.0 Kb each) were initially T/A cloned in the pGEM-T Easy vector and then sub-cloned, alongside with the hygromycin cassette (Hph box) derived from the hygromycin B resistance gene, into the vector pAN7.1 (6.7 Kb) previously digested with PciI–StuI at 5′ and NaeI–SbfI at 3′. WT protoplasts were transformed with the resulting disruption cassette. Fifty hygromycin-resistant colonies were collected and transferred onto Potato Dextrose Agar (PDA) plates supplemented with 100 μg/mL of hygromycin B (Duchefa Biochemie, Haarlem, NL). Monoconidial cultures of all resistant mutants were obtained and preliminarily screened by PCR using the primer pair panlds1F-panldsR that amplified in the vector used for the deletion (Supplementary Table 1). Finally, 20 colonies were selected and further sub-cultured. The stability of these transformants was tested by two additional single-spore transfers on non-selective, and then again on selective, media. The deletion of the gene was tested by Southern blot hybridization (see dedicated paragraph).
The ΔFvlds1 complementation strains (COM) were generated by transformation of the Fvlds1-deleted mutant with the WT LDS1 allele using the geneticin resistance gene, GenR, as a selectable marker as described elsewhere (Gruber et al., 2012). The selection and purification of the putative LDS1-complemented strains was performed as described above concerning the generation of deleted mutants, i.e., by transferring the transformants individually onto PDA plates amended with 300 μg/mL geneticin (G418, Sigma-Aldrich, St Louis, USA). Cassette integration was verified by PCR using primers nptII_for and nptII_rev designed in the GenR box (nptII) (Supplementary Table 1).
Southern blot hybridization
Genomic DNA (10 μ g), extracted as previously described (Scala et al., 2013) from WT F. verticillioides ITEM 10027 and ΔFvlds1 strains, was digested with EcoRI (10 U) at 37°C for 4 h (Fermentas). EcoRI-restricted DNA fragments were separated by electrophoresis and then blotted onto Hybond-N+ nylon membrane (Roche). Fluorescent DNA probes [FvLDS1 (0.8 Kb) and Hph (1.0 Kb)] were prepared according to the PCR digoxigenin (DIG)-labeling method (Roche) by using primers Lds_1 F and Lds_1 R (Supplementary Table 1; EcoRI did not restrict the 800 bp-probe fragment). The membranes were hybridized for 12–16 h in DIG-easy hybridization buffer (Roche) containing 250 ng of digoxigenin (DIG)-labeled FvLDS1 or Hph probes at 65°C.
Transcript quantification assay
For the in vitro experiments, total RNA from the mycelia of F. verticillioides WT, ΔFvlds1D, and COM strains was extracted as reported (Scala et al., 2013) at 0, 2, 5, 7, 10, and 15 DAI in CDYM cultures and used to develop reverse-transcriptase quantitative PCR (RT-qPCR) assays for FvLDS1 (FVEG_09294.3), FvLDS2 (FVEG_12540.3), FvLDS3 (FVEG_11670.3), and FvLOX (FVEG_09897.3). For the in vivo experiments, total RNA from maize cobs infected and non-infected by F. verticillioides WT and ΔFvlds1D was extracted as reported (Scarpari et al., 2014) at 0, 2, 5, 7, 10, and 15 DAI. Related cDNA was used to develop RT-qPCR experiments for FvLDS1, FvLDS2, FvLDS3, FvLOX, ZmLOX3 (AF329371), and ZmPR4 (AJ969166). The sequence of the primers used for the RT-qPCR assays are reported in Scala et al. (2013), Brodhagen et al. (2008) and in Supplementary Table 1. Reverse transcriptase reactions were performed as previously described (Scala et al., 2013). Gene expression in the WT, ΔFvlds1D, and COM strains, and in maize cobs, was calculated by using the 2−ΔΔCt method, i.e., by normalizing transcript levels of the gene of interest onto the transcript of a housekeeping gene [β-Tubulin for F. verticillioides (FVEG_05512.3) (Scala et al., 2013) and α-Actin for Zea mays (DQ492681.1) (Supplementary Table 1)] and onto their value at the time of inoculation (time zero). The software for relative expression quantification provided with the Line GeneK thermocycler (Bioer, PRC) was used.
Chromatin immunoprecipitation (ChIP)
The ChIP procedure was performed as described by the manufacturer of the EZ ChIP chromatin immunoprecipitation kit (Upstate, Charlottesville, VA). F. verticillioides WT and ΔFvlds1D strains were cultured in CDYM up to 15 DAI to induce FB production. Protein/DNA cross-linking was performed at harvest by incubating the cultures for 10 min in 1% formaldehyde (final concentration in the growth medium). The cross-linking reaction was stopped by addition of 0.125 M Glycine (final concentration). The mycelium was rinsed in PBS (20 mM Na2HPO4/KH2PO4 and 150 mM NaCl, pH 7.5) and then frozen in liquid nitrogen. Frozen mycelium (0.5 g) was ground in liquid nitrogen with a mortar and pestle, and the powder was resuspended in 2 mL of ice-cold DNA extraction buffer (Upstate) according to the manufacturer's instructions. Chromatin was sheared by sonication in a Hewlett-Packard V14 with 5 pulses of 60 s each at full power. Cross-linked DNA and proteins were immunoprecipitated with antibodies against hyper-acetylated histone H4 (Penta; Upstate) on the cross-linked DNA. Chromatin was then washed and eluted, and cross-links were reversed according to the manufacturer's instructions. Quantification was performed by qPCR under conditions that mirrored those for RT-qPCR but with immunoprecipitated DNA as a template (samples normalized to fresh weight). The primers used here are designed on the FUM1 promoter region (Visentin et al., 2012) (Supplementary Table 1). We quantified the abundance of target DNA by the relative standard curve method with β-TUB as the endogenous reference for normalization. Two biological and three analytical repeats were performed and standard errors were calculated. Calibration curves were calculated by linear regression.
Sequencing analysis
Paired-end libraries with an average insert size of 500 bp were produced and sequenced with the Illumina HiSeq 1500 technology at Genomix4life (Baronissi, Italy). On average, 15 × 106 reads were obtained for each F. verticillioides strain with a read length of 100 bp. A homemade pipeline called SUPERW (Simply Unified Pair-End Read Workflow) was developed to create a dynamic and fast tool to analyze the variation data produced from the re-sequencing experiments. The SUPERW pipeline is divided in three steps: the filtering and mapping, the variation calling and the output of the data. The first step automatically recognizes the quality of the samples redirecting them either to filtering or directly to mapping. Filtering of the reads creates a new high-quality subset of reads suitable for mapping analyses. After the filtering step, all the reads are mapped against a reference genome using the bwa mem (Li et al., 2009) algorithm and allowing the user to select all the possible bwa options. The mapped files are filtered for PCR duplicates, compressed in bam files, sorted and indexed (Li et al., 2009) creating as output a bam file from each mapping and a text file with the statistics about the trimming and the mapping steps. The mapped bam files and the reference genome are the input files for the variation-calling step of the pipeline.
Calling large structural variations
A depth of coverage approach was used to extract the large structural variations (CNVs) using the CNVnator tool (Abyzov et al., 2011), a method based on combining the established mean-shift approach to broaden the range of discovered CNVs. All CNVs were filtered for read-depth (RD) of ± 0.5 and p-value of 0.001. Finally the CNVs falling in the LDS1 region were extracted and analyzed with a window approach of 100 bp to better study the “gene deletion” event.
Phenotypic Characterization of F. verticillioides Strains
Fungal growth in vitro and spore germination
Fungal growth was measured as dry weight (d.w.) by weighing the mycelium after filtration (Millipore filters, 0.45 μm) and drying for 48 h at 80°C. To evaluate spore germination rates, 1 × 105 spores/mL of F. verticillioides were inoculated in PDB and the percentage of germinated spores was estimated at 25°C 24 h after inoculation.
Mating-type identification
The mating types (MAT-1 and MAT-2) of F. verticillioides WT and mutant strains were identified by PCR using the primers fusALPHArev—fusALPHAfor for MAT-1, and fusHMGfor—fusHMGrev for MAT-2 as described (Kerenyi et al., 2004). The annealing temperature for hybridization of both primers was adjusted to 56°C. The amplicons of the MAT-1 and MAT-2 idiomorphs are approximately 200 and 260 bp, respectively. For the definition of inter-strain fertility and ability to produce perithecia, the general crossing protocol was applied as described by Leslie and Summerell (2006). Reference strains obtained from the official collection of ISPA-CNR, Bari, Italy, were used: ITEM 15575 (KSU A-149) for MAT-1 and ITEM 15574 (KSU A-999) for MAT-2 respectively. Strains of the female parent were inoculated onto Petri dishes (Ø 9 cm) containing carrot agar, and the strain serving as the male was inoculated on Petri dishes (Ø 5 cm) containing water agar on the same day. The strains were incubated at 25°C in the dark. After 7 days, conidia from the male parent were suspended in 5 mL of 0.25% Tween®60 (Sigma-Aldrich, St. Louis, MO, USA) solution and 1 mL of the conidial suspension was spread on the surface of the female culture and worked into the mycelia with a L-shape rod. Fertilized plates were then incubated at 25°C in darkness and weekly checked for perithecia formation up to 8 weeks after the crossing. Crossings were repeated 5 times for each fungal strain and considered positive when perithecia were found in at least 2 out of 5 crossing trials and as negative when perithecia were absent on at least 3 out of 5 crossing trials (Leslie and Summerell, 2006).
Virulence assays on artificially inoculated maize cobs
WT and mutant F. verticillioides strains were inoculated on Potato Dextrose Agar (PDA) plates and incubated at 25°C in darkness for 1 week; then, conidial suspensions were prepared by adding 5 mL of sterile water on each colony. Suspensions were adjusted to a concentration of 105 conidia/mL and used to infect cobs of a commercial maize hybrid. These were collected at dough stage and infected with the pin bar technique. Wounds were made all around the middle area of the cob, without leaf removal, obtaining three portions with visible holes. After inoculation with 104 conidia per wound, cobs were placed in plastic bottles containing 50 mL of Hoagland's solution at the bottom (1 mL KH2PO4 1 M, 5 mL KNO3 1 M, 5 mL Ca(NO3)2 × 4H2O 1 M, 2 mL MgSO4 × 7H2O 1 M and sterile water to 1 L) as nutritional source (Hoagland and Arnon, 1950) and incubated at RT. Non-inoculated cobs were included. We performed our experiments under environmental conditions conducive for F. verticillioides infection. In our lab, room temperature during summer is stable around 23–25°C and there is natural light with natural day/night duration. Even if these may not be the most conducive conditions possible, they are largely permissive, as demonstrated in previous works (Marin et al., 2010). Different incubation times were considered (0, 2, 4, 7, and 15 DAI) and each thesis was managed in triplicate. After incubation, cobs were stored at −18°C until analysis. After hand de-husking, the severity of the ear attack by WT and ΔFvlds1D strains of F. verticillioides was evaluated using a visual rating scale including seven classes based on the percentage of visibly infected kernels (Disease Severity Rating-DSR: 1 = 0%-no infection; 2 = 1–3%; 3 = 4–10%; 4 = 11–25%; 5 = 26–50%, 6 = 51–75%; 7 = 76–100%) as reported (Reid et al., 2002). Visual rating is a common practice for in-field evaluation of infection severity. The use of this kind of scales has been reported on detached maize cobs in several papers (Reid et al., 2002 and reference therein) as they are quite effective in representing maize infection severity. In our case, also because infection was the outcome of artificial inoculation under controlled conditions, cob symptoms were only due to WT or ΔFvlds1D-mutant F. verticillioides strains, given that control cobs were clear of infection symptoms.
In order to monitor fungal growth into the maize cobs, a specific SYBR green qPCR method was set by using FUM1 primers (Supplementary Table 1). Real-time PCR was prepared in a 20 μ L reaction mixture containing SYBR green JumpStart Taq Ready as described in Reverberi et al. (2013). Standard calibration was performed plotting the Real-time PCR signals obtained for F. verticillioides genomic DNA in the concentration range 1 pg–100 ng. The equation describing the increase of DNA concentration was calculated (y = −0,9754x + 28,591, R2 = 0.991) and used afterwards as a reference standard for the extrapolation of quantitative information of DNA targets of unknown concentrations. The final amount of fungal DNA (ng) was referred to 1 μ g of total DNA. The efficiency of the PCR reaction (101%) was obtained from the calibration curve slope (E = 10−1 / slope − 1).
Metabolite Analyses
Oxylipins
Oxylipins were extracted as described by Scala et al. (2013) with slight modifications. Samples were analyzed by liquid chromatography (HPLC 1200 series rapid resolution, Agilent Technologies, Santa Clara, CA, USA) coupled to triple quadrupole (G6410A series triple quadrupole, QqQ, Agilent Technologies, CA, USA) equipped with an electrospray ionization (ESI). Experiments in MRM in negative ion mode [M-H]− were performed and data processed as reported by Ludovici et al. (2014).
Free and total fumonisins
Free and total fumonisins were determined according to our previous work (Dall'Asta et al., 2010). Fumonisins obtained after sample hydrolysis were measured as the sum of hydrolyzed FB1, FB2, and FB3. All of the results are expressed as the sum of FB1, FB2, and FB3 equivalents, considering a correction factor due to the different molecular weight of parent and hydrolyzed compounds, and referred to as “total fumonisins” (FBtot).
Statistical analysis
Data are presented as the mean value (±SE) of three independent measurements from two separate experiments. In each experiment, data sets were pooled and compared using Mann-Whitney's test and the differences were considered significant when the p-value was <0.05.
Results
Molecular Profile of Fvlds1-Deleted F. verticillioides
Our previous study (Scala et al., 2013) identified the bona fide LDS1 gene homolog as one of the putative oxylipin-forming genes most highly expressed during the saprophytic growth of WT F. verticillioides (ITEM 10027) on a FB-inducing medium containing cracked maize. To study its function in F. verticillioides, the FvLDS1 gene (acc. N. FVEG_09294.3 as annotated on the reference, sequenced strain 7600, see below) was targeted for deletion to create ΔFvlds1 mutants. Putative deleted strains were PCR-screened and those, which demonstrated clear molecular differences in comparison with the WT were selected (data not shown). One out of 20 PCR-selected mutants was further characterized and the genomic organization of FvLDS1 was studied by Southern blot hybridization using fragments of LDS1 and Hph as molecular probes. The results indicated the presence of two hybridization signals with the LDS1 probe whilst at least one putative mutant (thereafter named ΔFvlds1D) presented a recognizable deletion pattern (Supplementary Image 1). The same mutant strain carried the Hph box in its genome (Supplementary Image 1).
At least three LDS-encoding genes—namely, LDS1, LDS2, and LDS3—are present in the genome of F. verticillioides strain 7600 released by the Broad institute (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html). LDS2 and LDS3 are quite dissimilar in their nucleotide sequence from the LDS1 hybridization probe (32 and 29% identity, respectively). Thus, it may be assumed that the hybridization pattern in Supplementary Image 1 is indicative of the presence of two LDS1 paralogs, LDS1a, and LDS1b in our WT strain. Since only one FvLDS1 gene is reported in the genome of F. verticillioides reference strain 7600, we sequenced our WT strain (http://www.ncbi.nlm.nih.gov/bioproject/263314) as well as its mutant ΔFvlds1D for pinpointing any difference with the reference strain. A depth-of-coverage approach was used to search for copy number variation (CNV) of genes. Mapped files were used to call CNV regions. Two thousand and twenty-six CNV regions were identified; among them, only those with a minimum read depth (RD) of 1.5 and a p-value of 0.05 were considered. In this way, 1407 total CNVs (695 duplications and 713 deletions) were identified. By comparing the ΔFvlds1D and WT strains against the genome of the reference strain, we established a significant CNV in the LDS1 locus (genomic region Supercontig_3.12 coord. 1,190,484 to 1,187,862 reverse strand). In particular, the comparison between our WT and the FV7600 reference strain showed the presence of a massive duplication (~24 Kb; RD 1.8) in the LDS1 locus of our WT genotype (Supplementary Image 2A). The comparison of ΔFvlds1D genome against the genome of the reference strain (FV7600) did not show differences in the depth of coverage (Supplementary Image 2B). These results can be explained with the presence of two LDS1 genes in the WT and a consequent deletion of one copy (thereafter named LSD1b) in the mutant strain ΔFvlds1D, confirming the Southern blot results.
LDS1 function was recovered in the deleted genetic background of ΔFvlds1D by ectopic expression of the WT LDS1b genomic sequence under the control of the endogenous promoter. Insertion of the complementation cassette was verified by PCR of the GenR box (nptII). The expected 447-bp fragment was amplified in 2 out of 5 putative complemented strains (COM) (Supplementary Image 3).
In vitro Phenotypic Analysis of Fvlds1-Deleted F. verticillioides
The morphological features and the spore differentiation ability of the ΔFvlds1D strain and of WT strain were further characterized (Figure 1). The most obvious differences with the WT were related to the mycelium network consistency (fluffy in the WT, thin and leathery in the ΔFvlds1D strain), in the color (pink in the WT and deep pink in the ΔFvlds1D strain) and in the growth mode on solid medium (aerial in the WT vs. submerged in the ΔFvlds1D strain). The COM strain recovered the morphological features of the WT strain (Figure 1).
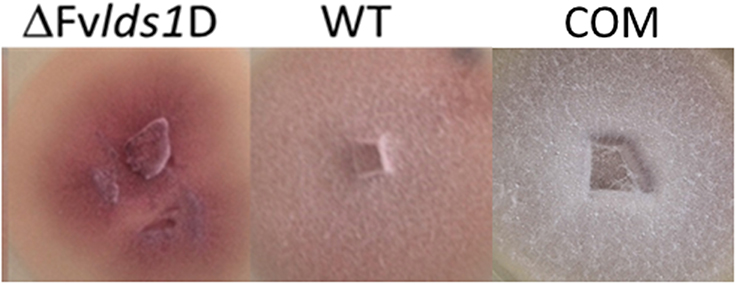
Figure 1. Phenotype characterization. Phenotype of F. verticillioides WT, ΔFvlds1D, and COM strains grown in PDA medium 7 DAI.
In liquid media, the deleted strain grew faster than the WT and COM at 15 DAI (Table 1) whereas in solid media its growth rate was similar (data not shown). ΔFvlds1D produced more conidia—which germinated significantly faster—than the WT and COM, starting 24 h after inoculation at 25°C (Table 1). These results demonstrate that the deletion of LDS1b significantly affected colony morphology, growth rate on liquid medium (p < 0.001), conidia production (p < 0.01) and their germination speed (p < 0.05) in F. verticillioides.
Preliminary but suggestive results were obtained in sexual crossing experiments. We firstly identified the mating type of our strains (MAT-1; Supplementary Image 4) and then we crossed them with the reference strain for MAT-2, as suggested by Leslie and Summerell (2006). Both the WT and ΔFvlds1D were able to produce perithecia in at least two independent experiments, even if with different efficiencies (number of fertile crosses/number of total crosses: WT-100%; ΔFvlds1D-83%, Supplementary Image 4).
Oxylipin Biosynthesis in vitro: Gene Expression and Mass Spectrometry Analysis
As stated previously, at least four LDS genes (LDS1a and b, LDS2, and LDS3) and one gene encoding a putative 13S-LOX (based on its high similarity with LOX2 in F. oxysporum, Brodhun et al., 2013) are present in our F. verticillioides WT strain. We tested their transcript level in the WT, ΔFvlds1D and COM strains grown on CDYM by RT-qPCR. All LDS genes were strongly down-regulated in ΔFvlds1D (Figures 2A–C; LDS1a and b were not discriminated in this analysis). It is also worth mentioning that the expression of the putative 13S-LOX gene, instead, was significantly up-regulated early after inoculation (2 and 5 DAI) when compared to the WT (Figure 2D). Transcript levels of oxylipin genes in the COM strain were similar to the WT (Figures 2A–D).
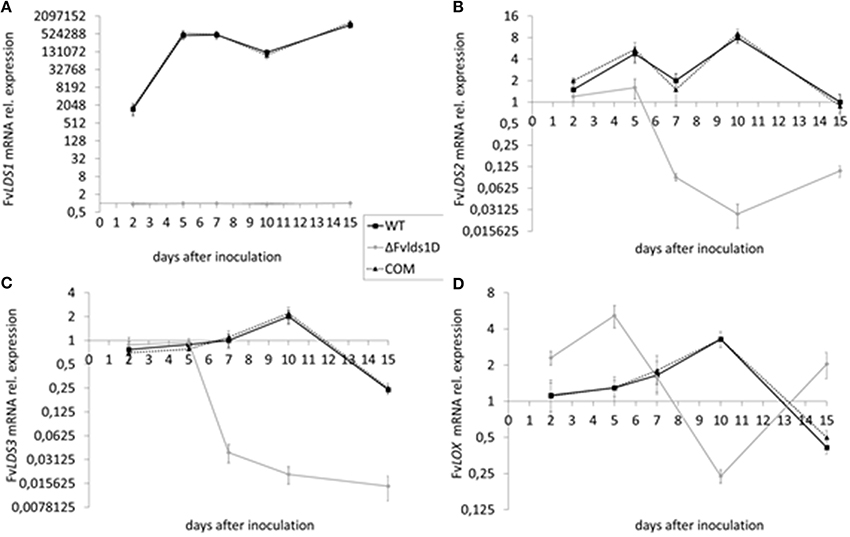
Figure 2. mRNA levels of LDS1-3 (A–C) and LOX (D) at 2-15 DAI. WT, mutant (ΔFvlds1D) and COM strains of F. verticillioides were grown in FB-inducing CDYM. Relative mRNA expression is calculated by using the 2−ΔΔCt method, i.e., by normalizing gene of interest expression onto housekeeping gene expression and to their value at the time of inoculation (time zero). LDS1a and b were not discriminated in this analysis. Results are the mean (± SE) of six replicates from two independent experiments.
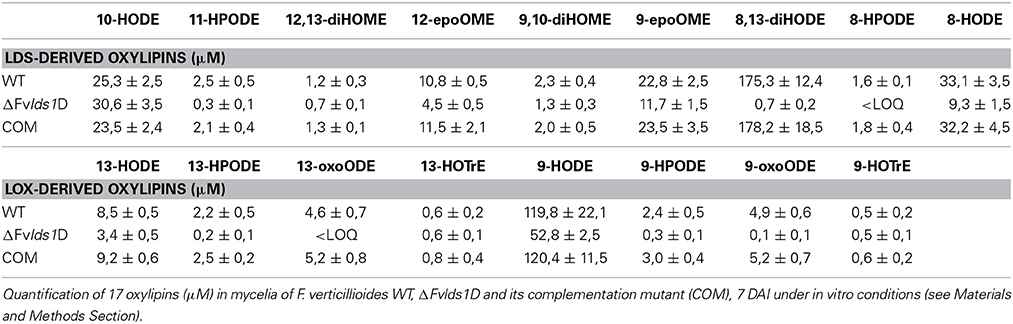
In order to quantify the effect of this general re-modulation of the expression of oxylipin-related genes in the ΔFvlds1D strain, a set of 17 oxylipins derived from both linoleic and linolenic acid was analyzed (Table 2; Supplementary Data Sheet 1). The oxylipin set was divided accordingly to its putative enzymatic origin (specifically, if LDS- or LOX-derived) following a published protocol (Strassburg et al., 2012). The oxylipin profile originated by the comparison between ΔFvlds1D and the WT strains partly confirmed RT-qPCR results reported in Figures 2A–D. The COM strain recovered the oxylipin profile of the WT strain (Table 2).
The accumulation trend for the whole set of oxylipins was followed from 1 up to 15 DAI (Supplementary Data Sheet 1). Table 2 summarizes the results of 7 DAI, i.e., when the difference between the strains is more marked. Putative LDS1 products, specifically 8-HPODE, 8,13-diHODE and 8-HODE, were severely down-represented in the ΔFvlds1D mutant as compared to the WT, while 10-HODE was up-regulated in the mutant strain. Oxylipins in the 9- and 13-LOX pathways (i.e., HPODE, HODE, and oxoODE) were also affected in ΔFvlds1D (Table 2).
Fumonisin B Production and Gene Expression
The main B-series fumonisins (FBtot = FB1, FB2, and FB3) were quantified in WT, COM and ΔFvlds1D strains at 2–15 DAI, i.e., at the peak of toxin production under our experimental conditions. ΔFvlds1D produced significantly (p < 0.001) more FB than the WT and COM strains starting from 7 DAI onwards (Figure 3A), with a comparable effect on FB1, FB2, and FB3 (data not shown). The fumonisin B increase was apparently modulated at transcriptional level. In fact, FUM1 was more expressed at 7 and 10 DAI (p < 0.001) in the ΔFvlds1D strain compared to the WT as well as to the COM strain (Figure 3B).
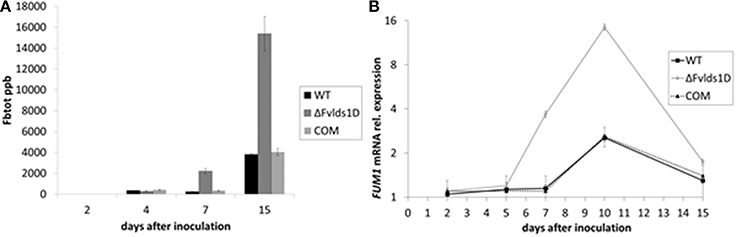
Figure 3. Fumonisin production and transcript of the key biosynthetic gene FUM1 in vitro. (A) Quantification of the main B-series fumonisins (FBtot = FB1 + FB2 + FB3, expressed as ppb) and (B) FUM1 mRNA quantification in the WT, mutant (ΔFvlds1D) and COM strains of F. verticillioides relative to inoculation time 0. Fungi were grown for 2–15 DAI in FB-inducing CDYM; results are the mean (± SE) of six replications deriving from two independent experiments.
Effects of LDS1b Deletion on Growth and Virulence of F. verticillioides on Maize Cobs
Oxylipins play a significant role in fungal virulence and in the cross talk with the host (Tsitsigiannis and Keller, 2007; Brodhun et al., 2009; Christensen and Kolomiets, 2011; Scarpari et al., 2014). To assess the virulence phenotype of the ΔFvlds1D strain, we inoculated maize cobs with the mutant or the WT strain. Rapid mold development was noticeable for the ΔFvlds1D mutant, slightly slower for the WT. Cobs wounded but not inoculated did not show any sign of fungal growth (Figures 4A,B). The severity of ear rot disease symptoms was rated by a published index (Reid et al., 2002). As shown in Figure 4C, the mutant strain caused more damage than the WT at 15 DAI. qPCR aimed at quantifying fungal growth into plant tissues confirmed this result. The mutant indeed colonized the host tissue significantly faster (p < 0.01) than the WT strain at 7 DAI, whereas this difference was lost at 15 DAI (Figure 4D).
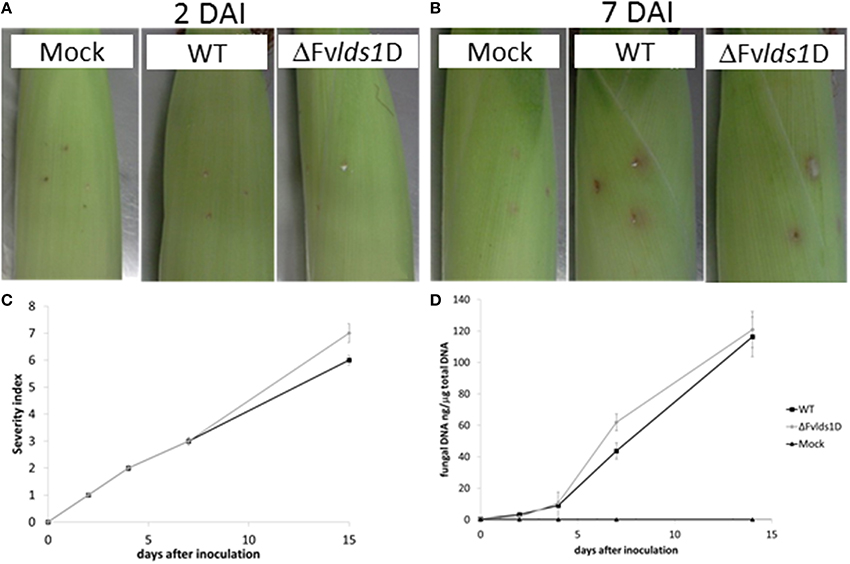
Figure 4. Virulence analysis of the two strains of F. verticillioides (WT and ΔFvlds1D mutant). Visual appearance of cobs artificially wounded, non-inoculated (mock) and inoculated with the two strains of F. verticillioides (WT and ΔFvlds1D) (A) 2 and (B) 7 DAI at 25°C in bottles containing Hoagland's solution. (C) Severity of F. verticillioides ear attack evaluated by a visual rating scale based on the SCIA (wounding and non-wounding) method (Marin et al., 2010). (D) Abundance of fungal DNA in maize ears at different DAI, as a measure of tissue colonization. This method adopts 7 classes based on the percentage of visibly infected kernels (Disease Severity Rating: 1 = 0%-no infection; 2 = 1–3%; 3 = 4–10%; 4 = 11–25%; 5 = 26–50%, 6 = 51–75%; 7 = 76–100%).
Oxylipin Formation in vivo: Gene Expression and Mass Spectrometry Analysis
Fungal pathogens as well as host plants produce oxylipins during the interaction. The shared chemical structure is at the basis of their mutual ability to affect defense onset (in the host) and virulence (in the fungus) (Christensen and Kolomiets, 2011). We analyzed the expression of some fungal oxylipin-related genes and one of the most studied maize genes (ZmLOX3) coding for LOX enzymes and known to be important in the interaction of maize with other mycotoxin-producing fungi (Gao et al., 2009). In particular, we monitored the expression of fungal LDS1-3 and LOX as well as of maize ZmLOX3 (Figures 5A–E) in maize cobs artificially inoculated. As reported in Figure 5A, total FvLDS1 transcript (LDS1a plus b) is conspicuously less abundant, but not completely absent, in the ΔFvlds1D strain as compared to the WT. Since we deleted only one out of the two LDS1 copies present in the genome of our parental strain, the results suggest that the non-deleted copy LDS1a may be expressed in vivo (Figure 5A) while it is not in vitro (see Figure 2A for comparison). The transcript abundance of other oxylipin-related genes was similar (LDS2, Figure 5B) or higher (LDS3, LOX, Figures 5C,D) in the ΔFvlds1D strain when compared to the WT. It is noteworthy that while LDS2 and LDS3 were down-regulated under in vitro conditions at all time-points (p < 0.01), both are up-regulated in vivo, likely upon the influence of the host tissues. Conversely, FvLOX expression was consistently up-regulated in the ΔFvlds1D strain as compared to the WT (p < 0.001), even more conspicuously than under in vitro conditions (see Figure 2D for comparison).
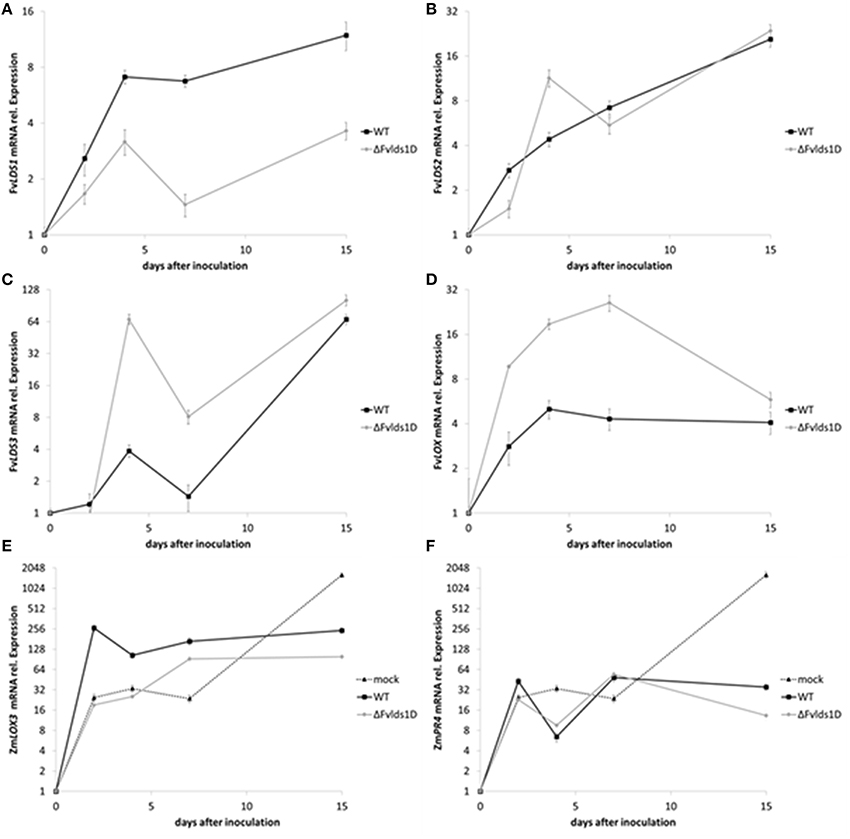
Figure 5. (A–F) mRNA levels of fungal and plant genes in infected cobs. F. verticillioides LDS1 (A), LDS2 (B), LDS3 (C), and LOX (D), Zea mays LOX3 (E), and PR4 (F) were quantified at 2–15 DAI in maize cobs non-inoculated (mock), or inoculated with the WT or mutant strain (ΔFvlds1D). Relative mRNA expression is calculated by using the 2−ΔΔCt method, i.e., by normalizing gene of interest expression onto housekeeping gene expression and to their value at the time of inoculation (time zero). Results are the mean (± SE) of six replications deriving from two independent experiments.
Maize up-regulated ZmLOX3 in the early stage of fungal infection (up to 7 DAI; p < 0.01). Then, at 15 DAI, i.e. when the pathogen achieved its maximum development in the host tissues, ZmLOX3 was significantly (p < 0.001) repressed both upon WT as well as ΔFvlds1D infection (Figure 5E) as compared to uninfected maize. Despite this shared trend, significant differences emerged between the two F. verticillioides strains. Namely, the ZmLOX3 transcript was more abundant in the WT- than the ΔFvlds1D-infected maize tissues (p < 0.001).
Maize defense responses include activation of class 4 pathogenesis-related (PR) proteins. PR4 proteins are chitinases contributing to the degradation of the fungal cell wall (Wang et al., 2011); ethylene- and jasmonic acid-related pathways (Van Loon et al., 2006) control PR4 expression in Arabidopsis. As jasmonic acid is an oxylipin, and given the broad-spectrum alteration of the oxylipin profile in pathogen-challenged maize cobs observed in this and in recent works (Christensen et al., 2014; Scarpari et al., 2014), we monitored ZmPR4 expression as a marker of plant defense responses triggered by oxylipins. The infection by both F. verticillioides strains (WT and ΔFvlds1D) modulated ZmPR4 expression in a trend intriguingly similar to that of ZmLOX3 (Figures 5E,F in comparison). Both ZmPR4 and ZmLOX3 expression were sharply up-regulated (to a level comparable to the infected cobs) in the mock-inoculated cobs at early time-points, indicating that wounding itself induces their expression as reported in other studies (Wang et al., 2011). However, the transcript levels of both genes were seemingly kept at bay by the invading fungus at later time-points, and more efficiently by the ΔFvlds1D mutant than by the WT. Specifically, ZmPR4 expression was up-regulated at 15 DAI even if significantly (p < 0.01) less in the mutant- than in the WT-infected tissues (Figure 5F). Therefore, as whole, in vivo experiments suggest that ZmLOX3 as well as ZmPR4 were affected consistently and similarly by pathogen invasion, and that the ΔFvlds1D strain triggers plant defense responses less vigorously than the WT.
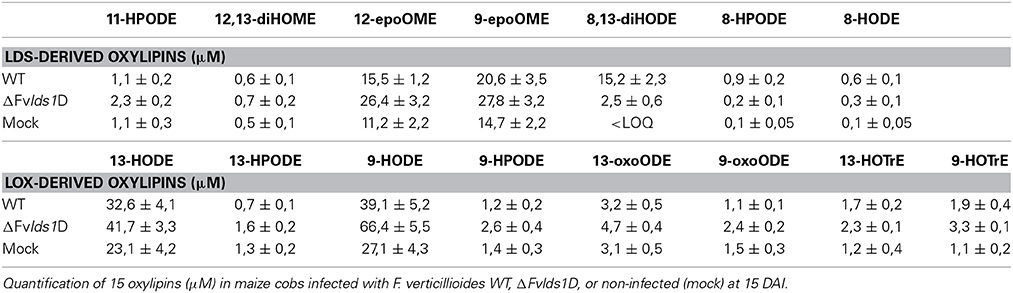
To validate the RT-qPCR results on the expression of the oxylipin-related genes at the metabolic level, we quantified 15 different oxylipins in the WT- and ΔFvlds1D-infected and non-infected cobs at 2–15 DAI (Table 3; Supplementary Data Sheet 2). Almost every oxylipin monitored achieved its maximum level at 15 DAI (Supplementary Data Sheet 2). In Table 3, their amount is reported at 15 DAI in maize cobs non-infected (mock), infected with the WT or ΔFvlds1D strain. Firstly, it can be pinpointed that the only oxylipins consistently down-modulated in the ΔFvlds1D compared to the WT-infected samples (both in vitro—see Table 2—and in vivo, Table 3) were 8-HPODE, 8,13-diHODE and 8-HODE. This may be because they are the main putative products of fungal LDS, so their synthesis would be directly affected by LDS1b deletion. Intriguingly, the other oxylipins (LDS- as well as LOX-derived ones) are up regulated in the maize cobs infected with ΔFvlds1D strain compared to those infected with the WT strain (Table 3). This result confirmed the hypothesis that oxylipin biosynthesis is tightly coordinated in F. verticillioides even during maize infection, although with different trends in vitro and in vivo.
The ΔFvlds1D mutant strain produced significantly more FBtot (p < 0.001) than the WT at 15 DAI in maize ears (5828 ± 458 vs. 867 ± 96 ppb). As shown in Table 3, 9-H(P)ODE is produced at higher levels in ΔFvlds1D-challenged maize ears than in WT-infected cobs (e.g., 66.42 vs. 39.11 μM for 9-HODE). 9-H(P)ODE is known to promote mycotoxin synthesis, specifically during the interaction between F. verticillioides and maize (Christensen et al., 2014). It can thus be suggested that two factors may contribute to the enhanced FB production in the mutant strain: the down-modulation of LDS products such as 8-H(P)ODE, and the enhancement of 9-H(P)ODE synthesis, possibly by ZmLOX3.
How do Oxylipins Affect Fumonisin Synthesis?
We previously showed that FB levels are enhanced in oxylipin-defective mutants of F. verticillioides because of transcription activation (see FBtot and FUM1 transcript quantification in Figures 3A,B, for comparison). We speculated that this may be ascribed to altered acetylation levels of histone proteins at PFUM1, as reported under FB-inducing conditions for another WT strain of F. verticillioides (Visentin et al., 2012). To test this hypothesis, we performed a chromatin immunoprecipitation (ChIP) assay with a commercial antibody targeted to the hyper-acetylated form of histone H4 and focused our analysis on PFUM1 of the WT and ΔFvlds1D strains grown in FB-inducing medium (CDYM). The quantities of immunoprecipitated DNA fragments containing PFUM1 were significantly (p < 0.001) greater in the ΔFvlds1D than in the WT strain (Figure 6). This result suggests a role for oxylipins in modulating the expression of gene(s) in the FUM cluster at the chromatin level.
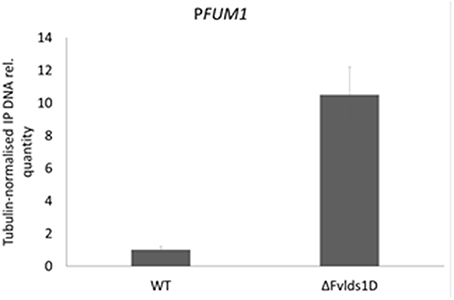
Figure 6. Relative acetylation levels of the FUM1 gene promoter (PFUM1) in F. verticillioides WT and ΔFvlds1D strains. Acetylation levels were analyzed by ChIP with antibodies specific to hyper-acetylated histone H4 on DNA from fungi cultured in FB-inducing CDYM. The abundance of target DNA was quantified by the comparative CT method with β-TUB as the endogenous reference for normalization. Error bars indicate the standard errors of two biological and three technical repetitions.
Discussion
F. verticillioides is a ubiquitous colonizer of maize kernels even if its presence is often asymptomatic. The major concern related to maize ear rot is not the disease per se but the production of the neurotoxic and carcinogenic fumonisins (Brown et al., 2006). Moreover, the presence of conjugated fumonisins, which alone may equalize the content of free fumonisins, worsens the safety issues, being the plant matrix-complexes released in free form upon animal digestion, virtually doubling the content of mycotoxins in feed and food (Dall'Asta et al., 2010). The description of the biology of the fungus, its invasion procedures and regulation of mycotoxin biosynthesis is still under way and yet absolutely needed for an efficient control of this highly invasive and often “silent” pathogen. This study explores some molecular mechanisms linking several features of its biology. Specifically, we describe how one of the most—inter-kingdom—conserved classes of signaling molecules, the oxylipins, may control F. verticillioides biology. Over recent years, several studies illustrated the role of oxylipins in fungal pathogens and in plant-pathogen interactions (Tsitsigiannis and Keller, 2007; Gao et al., 2009; Reverberi et al., 2010; Christensen et al., 2014; Scarpari et al., 2014). Oxylipins may represent almost stable and yet reactive molecular species able to transduce environmental stimuli efficiently into transcriptional reprogramming. They control multiple aspects of fungal as well as of plant biology (Tsitsigiannis and Keller, 2007; De Geyter et al., 2012). Most recently, Kolomiets group showed that a unique monocot-specific 9-LOX of maize (ZmLOX12) is crucial in driving defense responses against F. verticillioides, possibly by indirectly affecting the synthesis of jasmonic acid (Christensen et al., 2014). Moreover, Dall'Asta et al. (2014) demonstrated that in natural field conditions, the LOX-derived 9-HODE marks the maize hybrids containing the highest amount of fumonisins. In particular, the authors demonstrated that along the different growing stages, 9-HODE became a discriminant entity for maize hybrids only at the harvest stage.
The structure of oxylipins, conserved in both the pathogen and its host, allows inter-kingdom communication and makes them play a demonstrated role in plant-pathogen interactions (Christensen and Kolomiets, 2011). In some instances, oxylipins produced by the host as part of its defensive molecular apparatus (such as 9-HPODE) may end up inducing mycotoxin synthesis rather than controlling cell death processes in the plant and pathogen (Rusterucci et al., 1999; Shlezinger et al., 2011).
Very little information is available on the role of fungal oxylipins in the plant cell. A previous study on the A. ochraceus/Triticum durum pathosystem indicated that the deletion of a fungal LOX gene (AoLOX1) hides the fungus to the host. In fact, the wheat defense system does not react upon mutant strain infection (Reverberi et al., 2010). In the A. flavus/Z. mays interaction, the scarce production of HPODEs by the fungus enhances the capacity of the host to produce them (Scarpari et al., 2014). This means that other fungal oxylipins, such as LDS-derived products, may act as DAMPs or MAMPs and be perceived by the host surveillance system to activate defense responses (Savchenko et al., 2010). The leading hypothesis therefore is that the fungus produces its own oxylipins to drive its attack/reproductive machinery, but these compounds may end up alerting the plant defense system.
In this study, we demonstrated that LDS-derived oxylipins are involved in controlling growth, sporulation and mycotoxin production by the fungus, as well as in shaping the oxylipin profile in the producing microbe, also during its interaction with the host. In fact, even if it is almost impossible to discriminate between oxylipins produced by the host and by the pathogen, a change in the oxylipin profile can be described during the interaction—the most relevant trend being an up-regulation of the oxylipin machinery, likely in both organisms. Analogously to what recently described for the interaction between A. flavus and the kernels of Z. mays (Scarpari et al., 2014), also in our pathosystem the over-production of 9-H(P)ODE and the decrease in LDS products [such as 8H(P)ODE] may enhance fumonisin synthesis compared to kernels infected by the WT strain. The in vivo results confirmed the role of LDS-derived oxylipins in the pathogenic behavior of F. verticillioides on Z. mays. In spite of the lower tissue colonization rate by the WT fungus as established by visual analysis of infection and qPCR results, maize reacted more promptly to WT than to ΔFvlds1D invasion. The absence of LDS1b influenced the two-way interaction and made the pathogen trigger less the host-defense responses, confirming a role for fungal oxylipins as elicitors of plant defenses. Schenk and colleagues demonstrated indeed that, in Arabidopsis thaliana as well as in our pathosystem, oxylipins influence host defense and fortify resistance against pathogens (Schenk et al., 2014).
The pleiotropic effect, whereby a perturbation of oxylipin metabolism that leads to a deregulation of mycotoxin production, is similar to what reported previously in the IRT4 strain of A. flavus (Brown et al., 2009). This strain is silenced in all four Ppo genes and has dramatically increased aflatoxin production when compared to the WT. Thus, we suggest that also in F. verticillioides some products of Ppo (LDS) genes repress or at least down-modulate mycotoxin production. This hypothesis is supported by the significant up-regulation of the key biosynthetic gene FUM1 in the ΔFvlds1D strain compared to the WT (Figure 3B). Our results would suggest that LDS1-related products might coordinate the synthesis of the other oxylipins and, in turn, re-shape F. verticillioides lifestyle, namely growth, aggressiveness and toxin biosynthesis.
How may oxylipins affect gene expression and cell metabolism in fungi? Recently, different hypotheses on how the cell perceive and transduce oxylipins to reprogram the cell transcriptome have been proposed. Keller's group identified G protein-coupled receptors (GPCRs) as main components in exogenous oxylipin perceiving and signaling (Affeldt et al., 2012). However, as for endogenous oxylipin perception, a nuclear receptor such as the mammalian PPARγ through which oxylipins modulate the transcription in a hormone-like fashion (Itoh et al., 2008) does not seem to exist in fungi. A recent paper proposed peroxisomal proteins such as PEX11 as potential targets for oxylipins, even if the relation with the transcriptional reprogramming of secondary metabolism has still to be demonstrated (Reverberi et al., 2012). In the present study, we explored the possibility that oxylipins may indirectly modify chromatin in order to contribute to transcriptional re-programming. Animal histone deacetylases (HDACs) and histone acetyl transferases (HATs) can be post-translationally modified by carbonylation through direct reaction with phlogogenous oxylipins such as prostaglandins (Ravindra et al., 2012). Carbonylation inhibits HDAC and HAT enzymatic activity, and is thereby thought to mediate the genome-wide changes in histone acetylation patterns typical of the inflammatory responses. Chromatin modification represents, also in filamentous fungi, a rapid and efficient way to switch gene expression on/off (Strauss and Reyes-Dominguez, 2011), and this control level applies to toxin biosynthetic genes, too (Li et al., 2009). Previous observations of oxylipin increases in plant and fungal cells during host-pathogen interactions led us to hypothesize that in a cascade effect, this would affect histone acetylation, thus modulating the global chromatin landscape and contributing to reprogramming the plant and fungal transcriptome. This, in turn, could affect also the expression of genes for secondary metabolite synthesis (mycotoxins included), and trigger fungal toxigenicity. Histone H4 proteins at the promoter of FUM1 (PFUM1) are hyper-acetylated under FB-inducing conditions (Visentin et al., 2012). Our results indicate that oxylipins may control the transcription of FUM genes via local histone hyper-acetylation. Whether this happens through direct carbonylation and modulation of histone-modifying enzymes is still to be proven.
This study reports the LDS1 gene duplication of our WT compared to the reference strain pinpointing the importance of oxylipin-related genes in these pathogenic fungi. It may be worth emphasizing here that gene duplication is thought to have a crucial role in evolution (Ohno, 1970). Gene duplication may result as a form of adaption when ecological stress occurs, leading to either dosage benefits or neo-functionalization of duplicated copies (Chang and Duda, 2012). In a recently proven, complementary theory, diversification of gene function predates duplication and stabilizes it. Subsequent specialization of the paralogs would allow an efficiency increase that outweighs fitness costs directly linked to gene duplication itself (Näsvall et al., 2012). In this perspective, it may be interesting to assess the transcription pattern of the only LDS1 gene of the reference WT strain (7600) compared to LDS1a and b in our WT strain.
Experiments, aimed to reproduce the in vivo interaction among oxylipins and their potential target, may demonstrate how the lipid-derived signal can shape expression of the fungal genome, opening new perspectives in the understanding of how exogenous cues are perceived, transduced into reactive molecules and used for adapting the fungus to an ever-changing environment.
Author Contributions
Valeria Scala conceived of the study, participated in its design and coordination and helped to draft the manuscript. She performed the in vitro assays, generated the mutants, characterized them, and performed all the RT-qPCR assays. Paola Giorni performed the sexual reproduction and the pathogenicity assays. Martina Cirlini performed the fumonisin analysis. Matteo Ludovici performed the oxylipin LC-MS/MS analyses. Ivan Visentin performed the ChiP assay. Francesca Cardinale participated in the design of the study and coordination and helped to draft the manuscript. Anna A. Fabbri conceived of the study, participated in its design and coordination and helped to draft the manuscript. Corrado Fanelli conceived of the study, participated in its design and coordination and helped to draft the manuscript. Massimo Reverberi conceived of the study, participated in its design and coordination and helped to draft the manuscript. Paola Battilani conceived of the study, and participated in its design and coordination and helped to draft the manuscript. Gianni Galaverna participated to the design of the study. Chiara Dall'Asta conceived of the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
The present study was funded by the Ministry of Research and Education through the project FIRB2008 “Futuro in Ricerca,” grant N. FIRB-RBFR08JKHI.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors dedicate special thanks to Prof. Claudio Scazzocchio for his invaluable help in ameliorating this paper through his suggestions and criticisms and to Walter Sanseverino, Sequentia-Biotech, for his important contribute in bioinformatic analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00669/abstract
Supplementary Image 1. Characterization of the genomic organization of FvLDS1 in WT F. verticillioides and putative Fvlds1-deleted D strain (ΔFvlds1D). Southern blot hybridization of EcoRI-restricted genomic DNA was carried out using PCR digoxigenin (DIG)-labeled fragments of LDS1 and Hph as molecular probes.
Supplementary Image 2. (A) Duplicated region inferred with a depth of coverage approach. The X axis shows the Supercontig coordinates and the Y axis the depth of coverage. The black line shows for each 100 bp bin the depth of coverage and the green one the normalized coverage value. The blue lines highlight the LDS1 locus. (B) Non-duplicated region inferred with a depth-of-coverage approach. The X axis shows the Supercontig coordinates and the Y axis the depth of coverage. The black line shows for each 100 bp bin the depth of coverage and the green one the normalized coverage value. The blue lines highlight the LDS1 locus.
Supplementary Image 3. Fvlds1 complementation. Complemented strains were generated by transformation of the ΔFvlds1D mutant with the WT LDS1b allele using the geneticin resistance gene, GenR, as a selectable marker. The figure shows the results of PCR-screening of five putative complementation strains using primers designed on the GenR box.
Supplementary Image 4. Sexual fertility assays. Images of sexual crossings and summary of the presence of perithecia (reported also as mean percentage of 5 repetitions) after 8 weeks of incubation between F. verticillioides WT and ΔFvlds1D with the opposite MAT-type reference strain of F. verticillioides (ITEM 15574). Agarose gel of PCR products amplified from DNA isolated from F. verticillioides ITEM 10027 (WT) and its mutant ΔFvlds1D. The F. verticillioides-specific fusALPHA primers (MAT-1) were capable of amplifying; not so fusHMG primers (MAT-2).
Supplementary Data Sheet 1. Oxylipin production in vitro. MRM analysis of 17 different oxylipins in F. verticillioides WT, ΔFvlds1D and COM strains at different days after inoculation (2–15 DAI) under in vitro conditions (see Materials and Methods section for growing conditions). Results are the mean (± SE) of six replications from two independent experiments and are expressed as μM.
Supplementary Data Sheet 2. Oxylipin production in vivo. MRM analysis of 15 different oxylipins in maize infected with F. verticillioides WT and ΔFvlds1D strains or non-infected (mock) at different days after inoculation (2–15 DAI) under in vivo conditions (see Materials and Methods section for inoculation conditions into maize cobs). Results are the mean (± SE) of six replications from two independent experiments and are expressed as μM.
Supplementary Table 1. Primers used in this study.
Abbreviations
CDYM, Basal medium with the addition of 0.2% (w/v) cracked maize; ChIP, chromatin immunoprecipitation; CNV, copy number variation; DAI, days after inoculation; diHODE, dihydroxyoctadecenoic acid; FAs, Fatty acids; FB1, fumonisin B1; HATs, histone acetyl transferases; HDACs, histone deacetylases; HPODE, hydroperoxyoctadecenoic acid; LDS1, Linoleate Diol Synthase 1; LOX, lipoxygenases; MRM, multiple reaction monitoring; PR, pathogenesis-related; PFUM1, promoter of FUM1; RD, read depth; RT-qPCR, reverse transcription quantitative PCR; SVs, structural variations; FBtot, total fumonisins.
References
Abyzov, A., Urban, A. E., Snyder, M., and Gerstein, M. (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21, 974–984. doi: 10.1101/gr.114876.110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Affeldt, K. J., Brodhagen, M., and Keller, N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins 4, 695–717. doi: 10.3390/toxins4090695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brodhagen, M., Tsitsigiannis, D. I., Hornung, E., Göbel, C., Feussner, I., and Keller, N. P. (2008). Reciprocal oxylipin mediated cross-talk in the Aspergillus-seed pathosystem. Mol. Microbiol. 67, 378–391. doi: 10.1111/j.1365-2958.2007.06045.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brodhun, F., Cristobal-Sarramian, A., Zabel, S., Newie, J. M. H., Hamberg, M., and Feussner, I. (2013). An iron 13S-lipoxygenase with an α-linolenic acid-specific hydroperoxidase activity from Fusarium oxysporum. PLoS ONE 8:e64919. doi: 10.1371/journal.pone.0064919
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brodhun, F., and Feussner, I. (2011). Oxylipins in fungi. FEBS J. 278, 1047–1063. doi: 10.1111/j.1742-4658.2011.08027.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brodhun, F., Göbel, C., Hornung, E., and Feussner, I. (2009). Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/peroxidase and a cytochrome P450. J. Biol. Chem. 284, 11792–11805. doi: 10.1074/jbc.M809152200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, D. W., Butchko, R. A. E., and Proctor, R. H. (2006). Fusarium genomic resources: tools to limit crop diseases and mycotoxin contamination. Mycopathologia 162, 191–199. doi: 10.1007/s11046-006-0053-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, S., Scott, J. B., Bhaheetharan, J., Sharpee, W. C., Milde, L., Wilson, R. A., et al. (2009). Oxygenase coordination is required for morphological transition and the host-fungus interaction of Aspergillus flavus. Mol. Plant-Microbe Interact. 22, 882–894. doi: 10.1094/MPMI-22-7-0882
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, S., Zarnowski, R., Sharpee, W. C., and Keller, N. P. (2008). Morphological transitions governed by density dependence and lipoxygenase activity in Aspergillus flavus. Appl. Environ. Microbiol. 74, 5674–5685. doi: 10.1128/AEM.00565-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, D., and Duda, T. F. J. (2012). Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 29, 2019–2029. doi: 10.1093/molbev/mss068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christensen, A. A., Nemchenko, A., Park, Y.-S., Borrego, E., Huang, P.-C., Schmelz, E. A., et al. (2014). The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against fusarium verticillioides in Maize. Mol. Plant-Microbe Interact. 27, 1263–1276. doi: 10.1094/MPMI-06-13-0184-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christensen, S. A., and Kolomiets, M. V. (2011). The lipid language of plant-fungal interactions. Fungal Genet. Biol. 48, 4–14. doi: 10.1016/j.fgb.2010.05.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cristea, M., Osbourn, A., and Oliw, E. (2003). Linoleate diol synthase of the rice blast fungus Magnaporthe grisea. Lipids 38, 1275–1280. doi: 10.1007/s11745-003-1189-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dall'Asta, C., Giorni, P., Cirlini, M., Reverberi, M., Gregori, R., Ludovici, M., et al. (2014). “Maize lipids play a pivotal role in the fumonisin accumulation,” in Role of The Plant-Pathogen Cross Talking in Fusarium Mycotoxin Production and Masking in Maize, 1, 50–76. doi: 10.3920/WMJ2014.1754
Dall'Asta, C., Falavigna, C., Galaverna, G., Dossena, A., and Marchelli, R. (2010). In vitro digestion assay for determination of hidden fumonisins in maize. J. Agric. Food Chem. 58, 12042–12047. doi: 10.1021/jf103799q
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Geyter, N., Gholami, A., Goormachtig, S., and Goossens, A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17, 349–359. doi: 10.1016/j.tplants.2012.03.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duplus, E., Glorian, M., and Forest, C. (2000). Fatty acid regulation of gene transcription. J. Biol. Chem. 275, 30749–30752. doi: 10.1074/jbc.R000015200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Estrada, A. E. R., Jonkers, W., Kistler, H. C., and May, G. (2012). Interactions between Fusarium verticillioides, Ustilago maydis, and Zea mays: an endophyte, a pathogen, and their shared plant host. Fungal Genet. Biol. 49, 578–587. doi: 10.1016/j.fgb.2012.05.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, X., Brodhagen, M., Isakeit, T., Brown, S. H., Göbel, C., Betran, J., et al. (2009). Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant-Microbe Interact. 22, 222–231. doi: 10.1094/MPMI-22-2-0222
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, X., Shim, W. B., Gobel, C., Kunze, S., Feussner, I., Meeley, R., et al. (2007). Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant-Microbe Interact. 20, 922–933. doi: 10.1094/MPMI-20-8-0922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garscha, U., Jerneren, F., Chung, D., Keller, N. P., Hamberg, M., and Oliw, E. H. (2007). Identification of dioxygenases required for Aspergillus development: studies of products, stereochemistry, and the reaction mechanism. J. Biol. Chem. 282, 34707–34718. doi: 10.1074/jbc.M705366200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gruber, S., Omann, M., Escobar Rodrìguez, C., Radebner, T., and Zeilinger, S. (2012). Generation of Trichoderma atroviride mutants with constitutively activated G protein signaling by using geneticin resistance as selection marker. BMC Res. Notes 5:641. doi: 10.1186/1756-0500-5-641
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoagland, D. R., and Arnon, D. I. (1950). The Water-Culture Method for Growing Plants without Soil. Berkley, CA: University of California Agricultural Experiment Station.
Huber, S. F., Lottspeich, F., and Kämper, J. (2002). A gene that encodes a product with similarity to dioxygenases is highly expressed in teliospores of Ustilago maydis. Mol. Genet. Genomics 267, 757–771. doi: 10.1007/s00438-002-0717-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Itoh, T., Fairall, L., Amin, K., Inaba, Y., Szanto, A., Balint, B. L., et al. (2008). Structural basis for the activation of PPARγ by oxidized fatty acids. Nat. Struct. Mol. Biol. 15, 924–931. doi: 10.1038/nsmb.1474
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jerneren, F., Sesma, A., Francheschetti, M., Hamberg, M., and Oliw, E. H. (2010). Gene deletion of 7,8-linoleate diol synthase of the rice blast fungus. J. Biol. Chem. 285, 5308–5316. doi: 10.1074/jbc.M109.062810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kerenyi, Z., Moretti, A., Waalwijk, C., Olah, B., and Hornok, L. (2004). Mating type sequences in asexually reproducing Fusarium species. Appl. Environ. Microbiol. 70, 4419–4423. doi: 10.1128/AEM.70.8.4419-4423.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koo, A. J. K., and Howe, G. A. (2009). The wound hormone jasmonate. Phytochem 70, 1571–1580. doi: 10.1016/j.phytochem.2009.07.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leslie, J. F., and Summerell, B. A. (2006). The Fusarium Laboratory Manual. Lowa, IA: Blackwell Publishing.
Li, H., Handsaker, B., Wysoker, A., Fennel, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ludovici, M., Ialongo, C., Reverberi, M., Beccaccioli, M., Scarpari, M., and Scala, V. (2014). Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis of Fusarium verticillioides and maize kernels. Food Addit. Contam. 31, 2026–2033. doi: 10.1080/19440049.2014.968810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marin, P., Magan, N., Vázquez, C., and González-Jaén, M. T. (2010). Differential effect of environmental conditions on the growth and regulation of the fumonisin biosynthetic gene FUM1 in the maize pathogens and fumonisin producers Fusarium verticillioides and Fusarium proliferatum. FEMS Microbiol. Ecol. 73, 303–311. doi: 10.1111/j.1574-6941.2010.00894.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Näsvall, J., Sun, L., Roth, J. R., and Andersson, D. I. (2012). Real-time evolution of new genes by innovation, amplification, and divergence. Science 338, 384–387. doi: 10.1126/science.1226521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ravindra, K. C., Narayan, V., Lushington, G. H., Peterson, B. R., and Prabhu, K. S. (2012). Targeting of histone acetyltransferase p300 by cyclopentenone prostaglandin Δ (12)-PGJ(2) through covalent binding to Cys(1438). Chem. Res. Toxicol. 25, 337–347. doi: 10.1021/tx200383c
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reid, L. M., Woldemariam, T., Zhu, X., Stewart, D. W., and Schaafsma, A. W. (2002). Effect of inoculation time and point of entry on disease severity in Fusarium graminearum, Fusarium verticillioides, or Fusarium subglutinans inoculated maize ears. Can. J. Plant Pathol. 24, 162–167. doi: 10.1080/07060660309506991
Reverberi, M., Punelli, F., Scarpari, M., Camera, E., Zjalic, S., Ricelli, A., et al. (2010). Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 85, 1935–1946. doi: 10.1007/s00253-009-2220-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reverberi, M., Punelli, M., Scala, V., Scarpari, M., Uva, P., Mentzen, W., et al. (2013). Genotypic and Phenotypic Versatility of Aspergillus flavus during Maize Exploitation. PLoS ONE 8:e68735. doi: 10.1371/journal.pone.0068735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reverberi, M., Punelli, M., Smith, C. A., Zjalic, S., Scarpari, M., Scala, V., et al. (2012). How peroxisomes affect aflatoxin biosynthesis in Aspergillus flavus. PLoS ONE 7:e48097. doi: 10.1371/journal.pone.0048097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rusterucci, C., Montillet, J. L., Agnel, J. P., Battesti, C., Alonso, B., Knoll, A., et al. (1999). Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 274, 36446–36455. doi: 10.1074/jbc.274.51.36446
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Savchenko, T., Walley, J. W., Chehab, E. W., Xiao, Y., Kaspi, R., Pye, M. F., et al. (2010). Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22, 3193–3205. doi: 10.1105/tpc.110.073858
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scala, V., Camera, E., Ludovici, M., Dall'Asta, C., Cirlini, M., Giorni, P., et al. (2013). Fusarium verticillioides and maize interaction in vitro: relation between oxylipin cross-talk and fumonisin synthesis. World Mycotoxin J. 6, 343–351. doi: 10.3920/WMJ2012.1527
Scarpari, M., Punelli, M., Scala, V., Zaccaria, M., Nobili, C., Ludovici, M., et al. (2014). Lipids in Aspergillus flavus-maize interaction. Front. Microbiol. 5:74. doi: 10.3389/fmicb.2014.00074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schenk, S. T., Hernández-Reyes, C., Samans, B., Stein, E., Neumann, C., Schikora, M., Bigeard, J., et al. (2014). N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26, 2708–2723. doi: 10.1105/tpc.114.126763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shea, J. M., and Del Poeta, M. (2006). Lipid signaling in pathogenic fungi. Curr. Opin. Microbiol. 9, 352–358. doi: 10.1016/j.mib.2006.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shim, W. B., and Dunkle, L. D. (2002). Identification of genes expressed during cercosporin biosynthesis in Cercospora zeae-maydis. Physiol. Mol. Plant Pathol. 61, 237–248. doi: 10.1006/pmpp.2002.0437
Shlezinger, N., Minz, A., Gur, Y., Hatam, I., Dagdas, Y. F., Talbot, N. J., et al. (2011). Anti-apoptotic machinery protects the necrotrophic fungus botrytis cinerea from host-induced apoptotic-like cell death during plant infection. PLoS Pathog. 7:e1002185. doi: 10.1371/journal.ppat.1002185
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Strassburg, K., Huijbrechts, A. M., Kortekaas, K. A., Lindeman, J. H., Pedersen, T. L., Dane, A., et al. (2012). Quantitative profiling of oxylipins through comprehensive LC-MS/MS analysis: application in cardiac surgery. Anal. Bioanal. Chem. 404, 1413–1426. doi: 10.1007/s00216-012-6226-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Strauss, J., and Reyes-Dominguez, Y. (2011). Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 48, 62–69. doi: 10.1016/j.fgb.2010.07.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsitsigiannis, D. I., and Keller, N. P. (2006). Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol. Microbiol. 59, 882–892. doi: 10.1111/j.1365-2958.2005.05000.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsitsigiannis, D. I., and Keller, N. P. (2007). Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15, 109–118. doi: 10.1016/j.tim.2007.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsitsigiannis, D. I., Kowieski, R., Zarnowski, R., and Keller, N. P. (2005). Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology 151, 1809–1821. doi: 10.1099/mic.0.27880-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Loon, L. C., Rep, M., and Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Visentin, I., Montis, V., Doll, K., Alabouvette, C., Tamietti, G., Karlovsky, P., et al. (2012). Transcription of genes in the biosynthetic pathway for Fumonisin mycotoxins is epigenetically and differentially regulated in the fungal maize pathogen Fusarium verticillioides. Eukaryot. Cell 11, 252–259. doi: 10.1128/EC.05159-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walley, J. W., Kliebenstein, D. J., Bostock, R. M., and Dehesh, K (2013). Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 16, 520–526. doi: 10.1016/j.pbi.2013.06.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, N., Xiao, B., and Xiong, L. (2011). Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224. doi: 10.1016/j.jplph.2011.07.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Fusarium verticillioides, Zea mays, lipidomic, genome sequencing, chromatin immunoprecipitation
Citation: Scala V, Giorni P, Cirlini M, Ludovici M, Visentin I, Cardinale F, Fabbri AA, Fanelli C, Reverberi M, Battilani P, Galaverna G and Dall'Asta C (2014) LDS1-produced oxylipins are negative regulators of growth, conidiation and fumonisin synthesis in the fungal maize pathogen Fusarium verticillioides. Front. Microbiol. 5:669. doi: 10.3389/fmicb.2014.00669
Received: 05 September 2014; Accepted: 18 November 2014;
Published online: 11 December 2014.
Edited by:
Mehdi Razzaghi-Abyaneh, Pasteur Institute of Iran, IranReviewed by:
Eva-Guadalupe Lizárraga-Paulín, Instituto Tecnológico y de Estudios Superiores de Monterrey, MexicoPerng-Kuang Chang, Southern Regional Research Center, USA
Charles P. Woloshuk, Purdue University, USA
Copyright © 2014 Scala, Giorni, Cirlini, Ludovici, Visentin, Cardinale, Fabbri, Fanelli, Reverberi, Battilani, Galaverna and Dall'Asta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimo Reverberi, Dipartimento di Biologia Ambientale, Università Sapienza, Largo Cristina di Svezia 24, 00165 Roma, Italy e-mail: massimo.reverberi@uniroma1.it
 Valeria Scala
Valeria Scala Paola Giorni2
Paola Giorni2 Martina Cirlini
Martina Cirlini Ivan Visentin
Ivan Visentin Francesca Cardinale
Francesca Cardinale Anna A. Fabbri
Anna A. Fabbri Corrado Fanelli
Corrado Fanelli Massimo Reverberi
Massimo Reverberi Paola Battilani
Paola Battilani Gianni Galaverna
Gianni Galaverna Chiara Dall'Asta
Chiara Dall'Asta