- Department of Biological Sciences, Graduate School of Science, Osaka University, Toyonaka, Osaka, Japan
Land plants have evolved a single layer of epidermal cells, which are characterized by mostly anticlinal cell division patterns, formation of a waterproof coat called cuticle, and unique cell types such as stomatal guard cells and trichomes. The shoot epidermis plays important roles not only to protect plants from dehydration and pathogens but also to ensure their proper organogenesis and growth control. Extensive molecular genetic studies in Arabidopsis and maize have identified a number of genes that are required for epidermal cell differentiation. However, the mechanism that specifies shoot epidermal cell fate during plant organogenesis remains largely unknown. Particularly, little is known regarding positional information that should restrict epidermal cell fate to the outermost cell layer of the developing organs. Recent studies suggested that certain members of the HD-ZIP class IV homeobox genes are possible master regulators of shoot epidermal cell fate. Here, we summarize the roles of the regulatory genes that are involved in epidermal cell fate specification and discuss the possible mechanisms that limit the expression and/or activity of the master transcriptional regulators to the outermost cell layer in plant shoots.
Introduction
The shoot epidermis is a single layer of surface cells that are morphologically characterized by anticlinal cell division patterns. The outer surface of the shoot epidermis is covered with a hydrophobic structure called a cuticle, which prevents water loss, pathogen attacks, and post-genital fusion of organs (Yeats and Rose, 2013). Besides basic pavement cells, leaf epidermis contains specialized cell types such as hair cells (trichomes) and stomatal guard cells, which function to cope with dehydration and pathogen attacks. In addition to its protective function, the epidermis plays roles in the regulation of organ growth and shoot stem cell maintenance (Savaldi-Goldstein et al., 2007; Knauer et al., 2013; Nobusawa et al., 2013). In addition, protodermal cells in the shoot meristem and the embryo are necessary for the production and transport of the phytohormone auxin, which drives embryonic axis formation and lateral organ primordia initiation (Reinhardt et al., 2003; Kierzkowski et al., 2013; Robert et al., 2013; Wabnik et al., 2013).
Determination of shoot epidermal fate relies on a “position” rather than a cell “lineage,” as clonal analyses has shown that there is no strict lineage restriction in developing leaves; cells can flexibly change their fates, and only the cells finally located at the surface of the organ develop into epidermis (Stewart and Dermen, 1975). However, positional cues that determine shoot epidermal cell fate remain largely unknown. This review describes recent advances in the studies of epidermal cell specification in the shoots, focusing mainly on the regulation of key transcription factors.
HD-ZIP Class IV Transcription Factors are Key Regulators for Epidermal Cell Specification
ARABIDOPSIS THALIANA MERISTEM L1 LAYER (ATML1) was identified as an epidermis-specific homeobox gene that belongs to the HD-ZIP class IV family (Lu et al., 1996). ATML1 expression is first detected in the embryos as early as the one-cell stage and its expression is restricted to the outermost cells around the 16-cell stage (dermatogen stage) after the embryos have undergone tangential cell divisions to generate outer protodermal cells and inner cells (Lu et al., 1996; Sessions et al., 1999; Takada and Jürgens, 2007; Figures 1A–E).
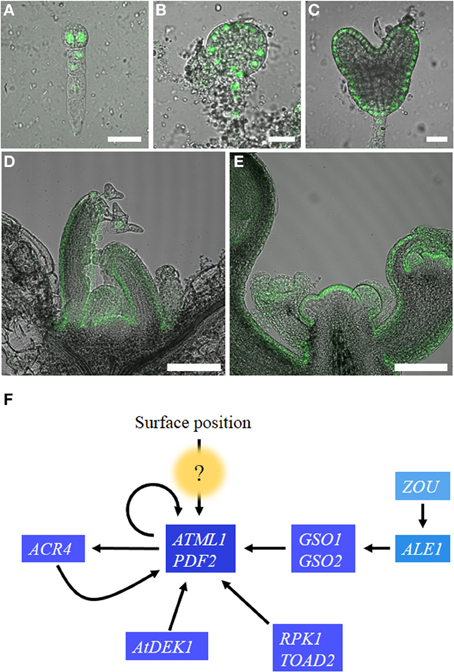
Figure 1. Regulation of ATML1 expression in the embryos and shoot apices. ATML1 promoter activity visualized using a nuclear-localized GFP reporter in the developing embryos (A–C) and the shoot apices (D,E). Two-cell stage (A), early globular stage (B), and heart stage (C) embryos are shown. GFP expression is restricted to the outermost cell layer after the 16-cell stage. Epidermis-specific expression is observed in vegetative (D) and inflorescence (E) shoot apices. (F) Genetic interactions among genes involved in epidermal cell fate specification during Arabidopsis thaliana embryogenesis. Arrows indicate positive interactions. The question mark indicates currently unknown components. For details, please see the main text. Scale bars, 20 μm in (A–C); 100 μm in (D,E).
Mutations in ATML1 and its closest homolog PROTODERMAL FACTOR2 (PDF2) caused severe phenotypes associated with defects in epidermal cell specification (Abe et al., 2003). Strong mutant alleles of atml1;pdf2 showed embryo-lethal phenotypes with irregular division patterns of the protoderm, whereas weak mutant alleles of atml1;pdf2 produced a few leaves that lack an epidermis (Abe et al., 2003; San-Bento et al., 2014; Supplementary Table 1). ATML1 homologs have been isolated in several species and most of them are preferentially expressed in the epidermis (Ito et al., 2003; Nakamura et al., 2006; Javelle et al., 2011). Several of these genes are implicated in epidermis-related functions, not only for the initial specification of surface cell fate but also for the generation of distinct cell types within the epidermis (Rerie et al., 1994; Roeder et al., 2012; Peterson et al., 2013; Takada, 2013).
HD-ZIP class IV transcription factors may function as transcriptional activators or repressors (Ohashi et al., 2003; Yu et al., 2008; Javelle et al., 2010; Depège-Fargeix et al., 2011; Peterson et al., 2013). ATML1 and PDF2 were shown to bind in vitro to an 8-bp sequence called the L1 box (Abe et al., 2001, 2003). Considering that an L1 box is often found in the promoters of epidermis-specific genes including ATML1 and PDF2, ATML1 and PDF2 have been proposed to positively regulate the expression of epidermis-specific genes (Abe et al., 2001, 2003). In fact, expression of epidermis-related genes was decreased in atml1;pdf2 (Abe et al., 2003; Takada et al., 2013). Moreover, gain-of-function experiments suggest that ATML1 activates expression of several epidermis specific genes during the initiation of new epidermal cell fate, but may also function as a negative regulator in maintaining expression levels (Takada et al., 2013; San-Bento et al., 2014). Notably, overexpression of ATML1 was sufficient to induce differentiation of epidermal cells such as stomata and trichomes in the inner tissues of leaves (Takada et al., 2013; Supplementary Table 1). These results are consistent with the idea that ATML1 is a master transcriptional regulator for epidermal cell specification in shoots.
Importantly, expression of ATML1 and its putative orthologs depends on a “surface” position, irrespectively of epidermal cell identity or cell lineage, as indicated by the presence of ATML1 promoter activity at the surface mesophyll cells of atml1;pdf2 leaves (Takada et al., 2013). In addition, expression of RICE OUTERMOST CELL-SPECIFIC GENE1 (ROC1), a rice HD-ZIP class IV gene, was induced on the cut surface during callus regeneration (Ito et al., 2002). In the maize extra cell layers1 (xcl1) mutant, which develops multilayered epidermis by amplification of differentiated epidermal cells, expression of an HD-ZIP class IV gene was detected only in the outermost epidermal layer (Kessler et al., 2002). These reports suggest that identification of upstream regulators that determine the outermost cell-specific expression of ATML1 homologs would be an effective strategy for identifying positional signals that specify shoot epidermal cell fate.
Positional Signals that Specify Epidermal Cell Fate
Deletion and mutational analyses of an ATML1 promoter revealed the involvement of several positive regulators in the protoderm-specific activation of ATML1 (Takada and Jürgens, 2007). MicroRNAs and phytohormone auxin, two major components that are known for morphogen-like activity, appeared not to be involved in the outermost cell-specific expression of ATML1 (Takada and Jürgens, 2007; Nodine and Bartel, 2010). Below, we discuss the candidate molecules or genes that may provide positional cues for shoot epidermal cell specification (Figure 1F).
Cuticle
Cuticle is a hydrophobic lipid layer formed on the surface of the shoot epidermis. In cuticle-deficient plants, trichome numbers, stomatal density, and regular anticlinal cell division of the epidermis are impaired, suggesting that cuticular components and/or their precursors are required for the patterning of epidermis (Yephremov et al., 1999; Gray et al., 2000; Sieber et al., 2000). Cuticle can be observed in the zygote and is maintained only in the outermost cells of the embryo even before a layer of protoderm is visible (Bruck and Walker, 1985). Therefore, the presence of cuticle or cuticle biogenesis may be instructive for epidermal cell specification and/or maintenance. In fact, expression of ROC1 was reduced in a rice mutant defective in the biosynthesis of very-long-chain fatty acids (VLCFAs), which serve as precursor of cuticular wax (Tsuda et al., 2013). Although it has been shown that ATML1 and other HD-ZIP class IV genes positively regulate expression of cuticle biosynthesis genes and facilitate cuticle deposition, these results suggest that cuticle also functions to maintain epidermal cell identity as a positive feedback mechanism (Javelle et al., 2010; Wu et al., 2011; Takada, 2013; Takada et al., 2013). VLCFAs or its derivatives produced in the epidermis have been recently suggested to function as non-cell autonomous signals that promote cell proliferation in internal tissues (Nobusawa et al., 2013). Therefore, some intermediates or byproducts formed during cuticle biosynthesis may play roles in pattern formation in plants.
Signaling from the Endosperm
In the angiosperms, the embryo is surrounded by endosperm tissues, which provide nutrients to the developing embryo. Recent studies show that signaling from the endosperm is necessary for epidermal cell differentiation during embryogenesis. abnormal leaf shape1 (ale1) and zhoupi (zou) mutants are defective in cuticle formation in organs generated during embryogenesis (Tanaka et al., 2001; Yang et al., 2008; Supplementary Table 1). ZOU encodes an endosperm-specific transcriptional regulator that promotes the expression of ALE1 and other genes required for the breakdown of the endosperm (Yang et al., 2008; Supplementary Table 1; Figure 1F). ALE1 encodes a subtilisin-like serine protease, and its expression in the endosperm is sufficient to rescue cuticle-deficient phenotypes of ale1 and zou, suggesting that ALE1 non-cell-autonomously promotes epidermal cell differentiation (Tanaka et al., 2001; Xing et al., 2013; Supplementary Table 1; Figure 1F). Considering that subtilisin-like proteases are involved in the processing of peptide hormone precursors in animals, the simplest scenario would be that ALE1 produces ligands promoting epidermal cell specification in the endosperm and the outermost cells of the embryos that receive those ligands differentiate into the epidermis (Steiner, 1998; Tanaka et al., 2001). GASSHO1 (GSO1) and GSO2 are candidate receptor-like kinases that receive signals produced by ALE1. GSO1 and GSO2 are preferentially expressed in the embryo, and gso1;gso2 shows severe cuticle-deficient phenotypes epistatic to those of ale1 (Tsuwamoto et al., 2008; Xing et al., 2013; Supplementary Table 1; Figure 1F).
It is not certain whether ALE1 and GSO1/GSO2 are generally required for the initiation of epidermal cell fate or specifically required for the cuticle formation on the surface of the embryo. In fact, epidermal cell specification can occur in the absence of the endosperm (such as during organ regeneration from calli, somatic embryogenesis, and aerial organ initiation in post-embryonic development). Plant embryos may require ALE1 and GSO1/GSO2-mediated signaling for efficient deposition of cuticle on the surface of the protodermal cells that develop in close physical contact with surrounding endosperm cells.
CRINKLY4
crinkly4 (cr4) is a maize mutant with defects in the development of leaf epidermis and aleurone layer (Becraft et al., 1996). CR4 encodes a plant-specific receptor-like kinase, and mutations in a homolog of CR4 in Arabidopsis [ARABIDOPSIS THALIANA HOMOLOGUE OF CRINKLY4 (ACR4)] cause phenotypes defective in epidermal cell differentiation, lateral root initiation, and root initial cell maintenance (Gifford et al., 2003; Watanabe et al., 2004; De Smet et al., 2008; Stahl et al., 2009, 2013; Supplementary Table 1). Although acr4 shows a mild effect on epidermal cell differentiation, acr4;ale1 double mutants show severe phenotypes with reduced ATML1 expression in the embryo, suggesting that ACR4 acts in parallel with the “endosperm pathway” to positively regulate epidermal cell differentiation upstream of ATML1 (Tanaka et al., 2007; Supplementary Table 1; Figure 1F).
However, expression of ACR4 is restricted to the epidermis of the embryos and shoots, and its epidermis-specific expression depends on an L1 box in the promoter, suggesting that ACR4 is a downstream target of HD-ZIP class IV transcription factors (Tanaka et al., 2002; Gifford et al., 2003; San-Bento et al., 2014; Supplementary Table 1; Figure 1F). ACR4 expression consistently began later than ATML1 expression during embryogenesis (Tanaka et al., 2002; Gifford et al., 2003). Moreover, ATML1 and PDF2 were associated with an ACR4 promoter in planta, and ACR4 expression was reduced in atml1;pdf2 (Abe et al., 2003; San-Bento et al., 2014).
ACR4 is localized to the basal and lateral membranes of the epidermis, suggesting that it is involved in intercellular communication between the same and different layers (Gifford et al., 2003, 2005; Watanabe et al., 2004). ACR4 and CR4 were shown to localize to plasmodesmata (PD), pores connecting plant cells, in the aleurone cells and the cotyledon epidermis (Tian et al., 2007; Stahl et al., 2013). Pore sizes of the PD connecting aleurone cells were wider than those connecting aleurone and underlying starchy endosperm cells (Tian et al., 2007). These observations may suggest that CR4 is involved in the modulation of the size of PD pores and facilitates intercellular communication between the same layer, maintaining epidermal/aleurone cell fate. Considering together, these results suggest that ACR4 is possibly more involved in the maintenance of epidermal cell fate downstream of ATML1 than in the perception of positional signals for epidermal cell specification (Figure 1F).
Leucine-Rich Repeat Receptor-Like Kinase in the Embryo
RECEPTOR-LIKE PROTEIN KINASE1 (RPK1) and TOADSTOOL2 (TOAD2) are leucine-rich repeat receptor-like kinases redundantly required for epidermal cell differentiation in the embryo (Nodine et al., 2007). rpk1;toad2 shows embryo-lethal phenotypes with disorganized cell division patterns particularly in the basal half of the embryo proper (Nodine et al., 2007; Supplementary Table 1). ATML1 mRNA was detected in the outer cell layer of rpk1;toad2 at the dermatogen stage but disappeared after the early globular stage, suggesting that these receptor-like kinases are necessary for the maintenance but not for the initial specification of epidermal cell fate (Nodine et al., 2007; Supplementary Table 1; Figure 1F). Because of the embryonic lethality, the roles of RPK1 and TOAD2 in shoot epidermal cell differentiation in the post-embryonic development are unknown.
Defective Kernel 1: A Calpain-Like Protease
DEFECTIVE KERNEL 1 (DEK1) is a calpain like cysteine protease that is conserved among land plants (Lid et al., 2002; Liang et al., 2013). Downregulation of DEK1 expression in several species causes phenotypes associated with defects in epidermal differentiation such as reduced deposition of cuticle, disorganization of cell division planes, and ectopic differentiation of mesophyll plastids (chloroplasts) in the surface cells (Becraft et al., 2002; Ahn et al., 2004; Johnson et al., 2005; Tian et al., 2007; Supplementary Table 1). Strong mutant alleles of dek1 cause embryo-lethal phenotypes in Arabidopsis, maize, and rice, and expression of ATML1 homologs disappears in these embryos, implying that DEK1 is necessary for the initiation of epidermal fate in the early embryo (Lid et al., 2002, 2005; Johnson et al., 2005; Hibara et al., 2009; Supplementary Table 1; Figure 1F).
DEK1 mRNA is expressed ubiquitously, suggesting that its activity is regulated post-translationally (Wang et al., 2003; Johnson et al., 2005; Lid et al., 2005; Hibara et al., 2009). DEK1 is composed of an N-terminal membrane-spanning region and a C-terminal cytosolic region that includes the calpain cysteine protease (Lid et al., 2002; Tian et al., 2007). It has been hypothesized that upon binding of epidermis-promoting ligands to the N-terminal region, DEK1 is cleaved by its autocatalytic activity to release the C-terminal region, an active form of the calpain (Wang et al., 2003; Tian et al., 2007; Johnson et al., 2008). In animals, calpain-like proteases are involved in the activation/inactivation of several signaling molecules, suggesting that DEK1 transduces signals for epidermal specification (Storr et al., 2011). However, overexpression of active truncated forms of DEK1 in Arabidopsis was not sufficient to upregulate the expression of ATML1 (Johnson et al., 2008; Supplementary Table 1). Moreover, modulation of DEK1 activity affected cell division and growth also in internal tissues, suggesting that the action of DEK1 is not epidermis-specific (Johnson et al., 2008; Supplementary Table 1). Johnson et al. (2008) proposed that DEK1 controls mainly cell division in developing leaves and that epidermal cells respond more sensitively to the amount of DEK1; reduction in cell division rate in dek1 may cause discontinuity and abortion of the epidermis (Johnson et al., 2008). In this scenario, DEK1 is possibly more involved in the maintenance rather than the initiation of the epidermal cell layer.
Post-Transcriptional Regulation of HD-ZIP Class IV Activity
Localization of some HD-ZIP class IV transcription factors was not limited to nuclei of heterologous cells (Zhang et al., 2010; Yang et al., 2011). The HD-ZIP class IV transcription factor GLABRA2 (GL2), a positive regulator of trichome formation, was restricted to the nuclei only in trichome cells and not in internal cells, suggesting a cell-type-specific regulation of nuclear transport (Szymanski et al., 1998). HD-ZIP class IV transcription factors contain a putative lipid/sterol binding domain (START) and a dimerization motif (ZLZ), implying a regulation of their activities by dimerization and binding of lipid/sterol ligands (Schrick et al., 2004). In fact, sterol and VLCFA biosynthesis-deficient mutants are defective in proper distribution of stomata and trichomes, respectively, (Yephremov et al., 1999; Qian et al., 2013). However, to date the roles of START domains have not been investigated, except for an observation showing that an N-terminal part of a START domain can function as a transcriptional activation domain in yeast and maize suspension cells (Depège-Fargeix et al., 2011).
ATML1 was shown to heterodimerize with PDF2 in planta and it is possible that dimerization with other HD-ZIP class IV proteins changes the activity of ATML1 in a cell-type dependent manner, although ectopic expression of ATML1 alone was sufficient to induce epidermal cell fate in inner tissues (Takada et al., 2013; San-Bento et al., 2014). In related transcription factors, DNA-binding was inhibited by oxidation of Cys residues in the ZLZ motif, suggesting that redox signals are also involved in the regulation of ATML1 activity (Tron et al., 2002).
Repression of Inner Cell Fate
Several lines of evidence show that acquisition of epidermal cell fate is associated with a loss of mesophyll or internal cell fate. First, epidermis-deficient atml1;pdf2 and DEK1 knockdown lines showed ectopic differentiation of mesophyll cells on the surfaces of leaves and cotyledons, respectively, (Abe et al., 2003; Johnson et al., 2005). Second, rpk1;toad2 embryos exhibited ectopic subepidermal marker expression in the outermost cell layer (Nodine et al., 2007; Supplementary Table 1). Third, overexpression of ATML1 decreased differentiation of green mesophyll cells in leaves (Takada et al., 2013). Moreover, cr4 and dek1, which are defective in “surface” aleurone layer differentiation in the maize endosperm, cause ectopic differentiation of “inner” starchy endosperm cells on the surface of the endosperm (Becraft et al., 1996; Becraft and Asuncion-Crabb, 2000). Therefore, repression of internal or “default” cell fate may be a general requirement for surface cell differentiation in plants. It is not possible, however, to test whether or not mesophyll cell differentiation represses epidermal cell fate because no positive regulators of inner cell fate are available at present. Chloroplast development itself appears not to exert a negative effect on epidermal cell differentiation, considering that stomatal guard cells possess chloroplasts and no ectopic epidermal cell differentiation has been reported in the mesophyll tissues of albino plants (Stewart and Dermen, 1975).
Mesophyll cells possibly represent a primitive state of leaf cells, considering that ancestral aquatic algae are composed mainly of mesophyll-like cells. Land plants may have repressed mesophyll cell differentiation to evolve an epidermis on the surface. Evolutionary studies, including comparative genomics, may be useful for identifying molecular components that promote epidermal cell formation (Zalewski et al., 2013).
Future Perspectives
Despite the extensive molecular genetic studies in model plants, positional signals that specify shoot epidermal cell fate remain unknown (Figure 1F). Most of the receptor-like kinases, characterized by their roles in shoot epidermal cell differentiation, are possibly involved in the maintenance than in the specification of epidermal cell fate. This appearance may be because of the difficulties in distinguishing between phenotypes associated with “specification” and those related to “maintenance” of epidermal cell fate in forward genetic screens.
The cuticle-bearing outermost cells should have distinct mechanical properties compared with inner cells. Moreover, cells located at the surface are unique, in that they are in constant contact with the environment. These unique properties could influence the differentiation of epidermal cells. Attempts to directly isolate epidermis-promoting biomolecules and to identify physical/environmental constraints influencing epidermal cell fate may shed new light on the issue.
Author Contributions
Shinobu Takada wrote the main manuscript text and Hiroyuki Iida prepared Figure 1 and Supplementary Table 1. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the grants from the Japan Society for the Promotion of Science [20657012, 22687003, and 23657036 to Shinobu Takada].
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00049/abstract
References
Abe, M., Katsumata, H., Komeda, Y., and Takahashi, T. (2003). Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–643. doi: 10.1242/dev.00292
Abe, M., Takahashi, T., and Komeda, Y. (2001). Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494. doi: 10.1046/j.1365-313x.2001.01047.x
Ahn, J. W., Kim, M., Lim, J. H., Kim, G. T., and Pai, H. S. (2004). Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 38, 969–981. doi: 10.1111/j.1365-313X.2004.02102.x
Becraft, P. W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127, 4039–4048.
Becraft, P. W., Kang, S. H., and Suh, S. G. (2001). The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiol. 127, 486–496. doi: 10.1104/pp.010299
Becraft, P. W., Li, K., Dey, N., and Asuncion-Crabb, Y. (2002). The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129, 5217–5225.
Becraft, P. W., Stinard, P. S., and McCarty, D. R. (1996). CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409. doi: 10.1126/science.273.5280.1406
Bruck, D. K., and Walker, D. B. (1985). Cell determination during embryogenesis in Citrus jambhiri. I. Ontogeny of the epidermis. Bot. Gaz. 146, 188–195. doi: 10.1086/337514
Depège-Fargeix, N., Javelle, M., Chambrier, P., Frangne, N., Gerentes, D., Perez, P., et al. (2011). Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J. Exp. Bot. 62, 293–305. doi: 10.1093/jxb/erq267
De Smet, I., Vassileva, V., De Rybel, B., Levesque, M. P., Grunewald, W., Van Damme, D., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. doi: 10.1126/science.1160158
Gifford, M. L., Dean, S., and Ingram, G. C. (2003). The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130, 4249–4258. doi: 10.1242/dev.00634
Gifford, M. L., Robertson, F. C., Soares, D. C., and Ingram, G. C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17, 1154–1166. doi: 10.1105/tpc.104.029975
Gray, J. E., Holroyd, G. H., van der Lee, F. M., Bahrami, A. R., Sijmons, P. C., Woodward, F. I., et al. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408, 713–716. doi: 10.1038/35042663
Hibara, K., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 15, 345–354. doi: 10.1016/j.ydbio.2009.07.042
Ito, M., Sentoku, N., Nishimura, A., Hong, S. K., Sato, Y., and Matsuoka, M. (2002). Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 29, 497–507. doi: 10.1046/j.1365-313x.2002.01234.x
Ito, M., Sentoku, N., Nishimura, A., Hong, S.-K., Sato, Y., and Matsuoka, M. (2003). Roles of rice GL2 type homeobox genes in epidermis differentiation. Breed. Sci. 53, 245–253. doi: 10.1270/jsbbs.53.245
Javelle, M., Klein-Cosson, C., Vernoud, V., Boltz, V., Maher, C., Timmermans, M., et al. (2011). Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 157, 790–803. doi: 10.1104/pp.111.182147
Javelle, M., Vernoud, V., Depège-Fargeix, N., Arnould, C., Oursel, D., Domergue, F., et al. (2010). Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor OUTER CELL LAYER1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 154, 273–286. doi: 10.1104/pp.109.150540
Johnson, K. L., Degnan, K. A., Ross Walker, J., and Ingram, G. C. (2005). AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44, 114–127. doi: 10.1111/j.1365-313X.2005.02514.x
Johnson, K. L., Faulkner, C., Jeffree, C. E., and Ingram, G. C. (2008). The phytocalpain DEFECTIVE KERNEL 1 is a novel Arabidopsis growth regulator whose activity is regulated by proteolytic processing. Plant Cell 20, 2619–2630. doi: 10.1105/tpc.108.059964
Kessler, S., Seiki, S., and Sinha, N. (2002). Xcl1 causes delayed oblique periclinal cell divisions in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development 129, 1859–1869.
Kierzkowski, D., Lenhard, M., Smith, R., and Kuhlemeier, C. (2013). Interaction between meristem tissue layers controls phyllotaxis. Dev. Cell 26, 616–628. doi: 10.1016/j.devcel.2013.08.017
Knauer, S., Holt, A. L., Rubio-Somoza, I., Tucker, E. J., Hinze, A., Pisch, M., et al. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132. doi: 10.1016/j.devcel.2012.12.009
Liang, Z., Demko, V., Wilson, R. C., Johnson, K. A., Ahmad, R., Perroud, P. F., et al. (2013). The catalytic domain CysPc of the DEK1 calpain is functionally conserved in land plants. Plant J. 75, 742–754. doi: 10.1111/tpj.12235
Lid, S. E., Gruis, D., Jung, R., Lorentzen, J. A., Ananiev, E., Chamberlin, M., et al. (2002). The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. U.S.A. 99, 5460–5465. doi: 10.1073/pnas.042098799
Lid, S. E., Olsen, L., Nestestog, R., Aukerman, M., Brown, R. C., Lemmon, B., et al. (2005). Mutation in the Arabidopisis thaliana DEK1 calpain gene perturbs endosperm and embryo development while over-expression affects organ development globally. Planta 221, 339–351. doi: 10.1007/s00425-004-1448-6
Lu, P., Porat, R., Nadeau, J. A., and O'Neill, S. D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168.
Nakamura, M., Katsumata, H., Abe, M., Yabe, N., Komeda, Y., Yamamoto, K. T., et al. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141, 1363–1375. doi: 10.1104/pp.106.077388
Nobusawa, T., Okushima, Y., Nagata, N., Kojima, M., Sakakibara, H., and Umeda, M. (2013). Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 11:e1001531. doi: 10.1371/journal.pbio.1001531
Nodine, M. D., and Bartel, D. P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24, 2678–2692. doi: 10.1101/gad.1986710
Nodine, M. D., Yadegari, R., and Tax, F. E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12, 943–956. doi: 10.1016/j.devcel.2007.04.003
Ohashi, Y., Oka, A., Rodrigues-Pousada, R., Possenti, M., Ruberti, I., Morelli, G., et al. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430. doi: 10.1126/science.1083695
Peterson, K. M., Shyu, C., Burr, C. A., Horst, R. J., Kanaoka, M. M., Omae, M., et al. (2013). Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140, 1924–1935. doi: 10.1242/dev.090209
Qian, P., Han, B., Forestier, E., Hu, Z., Gao, N., Lu, W., et al. (2013). Sterols are required for cell-fate commitment and maintenance of the stomatal lineage in Arabidopsis. Plant J. 74, 1029–1044. doi: 10.1111/tpj.12190
Racolta, A., Bryan, A. C., and Tax, F. E. (2014). The receptor-like kinases GSO1 and GSO2 together regulate root growth in Arabidopsis through control of cell division and cell fate specification. Dev. Dyn. 243, 257–278. doi: 10.1002/dvdy.24066
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081
Rerie, W. G., Feldmann, K. A., and Marks, M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. doi: 10.1101/gad.8.12.1388
Robert, H. S., Grones, P., Stepanova, A. N., Robles, L. M., Lokerse, A. S., Alonso, J. M., et al. (2013). Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23, 2506–2512. doi: 10.1016/j.cub.2013.09.039
Roeder, A. H., Cunha, A., Ohno, C. K., and Meyerowitz, E. M. (2012). Cell cycle regulates cell type in the Arabidopsis sepal. Development 139, 4416–4427. doi: 10.1242/dev.082925
San-Bento, R., Farcot, E., Galletti, R., Creff, A., and Ingram, G. (2014). Epidermal identity is maintained by cell–cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J. 77, 46–58. doi: 10.1111/tpj.12360
Savaldi-Goldstein, S., Peto, C., and Chory, J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446, 199–202. doi: 10.1038/nature05618
Schrick, K., Nguyen, D., Karlowski, W. M., and Mayer, K. F. (2004). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5:R41. doi: 10.1186/gb-2004-5-6-r41
Sessions, A., Weigel, D., and Yanofsky, M. F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. doi: 10.1046/j.1365-313x.1999.00594.x
Shen, B., Li, C., Min, Z., Meeley, R. B., Tarczynski, M. C., and Olsen, O. A. (2003). sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc. Natl. Acad. Sci. U.S.A. 100, 6552–6557. doi: 10.1073/pnas.0732023100
Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J. P., et al. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–738. doi: 10.2307/3870997
Stahl, Y., Grabowski, S., Bleckmann, A., Kühnemuth, R., Weidtkamp-Peters, S., Pinto, K. G., et al. (2013). Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23, 362–371. doi: 10.1016/j.cub.2013.01.045
Stahl, Y., Wink, R. H., Ingram, G. C., and Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909–914. doi: 10.1016/j.cub.2009.03.060
Steiner, D. F. (1998). The proprotein convertases. Curr. Opin. Chem. Biol. 2, 31–39. doi: 10.1016/S1367-5931(98)80033-1
Stewart, R. N., and Dermen, H. (1975). Flexibility in ontogeny as shown by the contribution of the shoot apical layers to leaves of periclinal chimeras. Am. J. Bot. 62, 935–947. doi: 10.2307/2441638
Storr, S. J., Carragher, N. O., Frame, M. C., Parr, T., and Martin, S. G. (2011). The calpain system and cancer. Nat. Rev. Cancer 11, 364–374. doi: 10.1038/nrc3050
Szymanski, D. B., Jilk, R. A., Pollock, S. M., and Marks, M. D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171.
Takada, S. (2013). Post-embryonic induction of ATML1-SRDX alters the morphology of seedlings. PLoS ONE 8:e79312. doi: 10.1371/journal.pone.0079312
Takada, S., and Jürgens, G. (2007). Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141–1150. doi: 10.1242/dev.02803
Takada, S., Takada, N., and Yoshida, A. (2013). ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140, 1919–1923. doi: 10.1242/dev.094417
Tanaka, H., Onouchi, H., Kondo, M., Hara-Nishimura, I., Nishimura, M., Machida, C., et al. (2001). A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development 128, 4681–4689.
Tanaka, H., Watanabe, M., Sasabe, M., Hiroe, T., Tanaka, T., Tsukaya, H., et al. (2007). Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development 134, 1643–1652. doi: 10.1242/dev.003533
Tanaka, H., Watanabe, M., Watanabe, D., Tanaka, T., Machida, C., and Machida, Y. (2002). ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 43, 419–428. doi: 10.1093/pcp/pcf052
Tian, Q., Olsen, L., Sun, B., Lid, S. E., Brown, R. C., Lemmon, B. E., et al. (2007). Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 19, 3127–3145. doi: 10.1105/tpc.106.048868
Tron, A. E., Bertoncini, C. W., Chan, R. L., and Gonzalez, D. H. (2002). Redox regulation of plant homeodomain transcription factors. J. Biol. Chem. 277, 34800–34807. doi: 10.1074/jbc.M203297200
Tsuda, K., Akiba, T., Kimura, F., Ishibashi, M., Moriya, C., Nakagawa, K., et al. (2013). ONION2 fatty acid elongase is required for shoot development in rice. Plant Cell Physiol. 54, 209–217. doi: 10.1093/pcp/pcs169
Tsuwamoto, R., Fukuoka, H., and Takahata, Y. (2008). GASSHO1 and GASSHO2 encoding a putative leucine-rich repeat transmembrane-type receptor kinase are essential for the normal development of the epidermal surface in Arabidopsis embryos. Plant J. 54, 30–42. doi: 10.1111/j.1365-313X.2007.03395.x
Wabnik, K., Robert, H. S., Smith, R. S., and Friml, J. (2013). Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 23, 2513–2518. doi: 10.1016/j.cub.2013.10.038
Wang, C., Barry, J. K., Min, Z., Tordsen, G., Rao, A. G., and Olsen, O. A. (2003). The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J. Biol. Chem. 278, 34467–34474. doi: 10.1074/jbc.M300745200
Watanabe, M., Tanaka, H., Watanabe, D., Machida, C., and Machida, Y. (2004). The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39, 298–308. doi: 10.1111/j.1365-313X.2004.02132.x
Wu, R., Li, S., He, S., Wassmann, F., Yu, C., Qin, G., et al. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411. doi: 10.1105/tpc.111.088625
Xing, Q., Creff, A., Waters, A., Tanaka, H., Goodrich, J., and Ingram, G. C. (2013). ZHOUPI controls embryonic cuticle formation via a signalling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 140, 770–779. doi: 10.1242/dev.088898
Yang, C., Li, H., Zhang, J., Luo, Z., Gong, P., Zhang, C., et al. (2011). A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. U.S.A. 108, 11836–11841. doi: 10.1073/pnas.1100532108
Yang, S., Johnston, N., Talideh, E., Mitchell, S., Jeffree, C., Goodrich. J., et al. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135, 3501–3509. doi: 10.1242/dev.026708
Yeats, T. H., and Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.1104/pp.113.222737
Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11, 2187–2201.
Yu, H., Chen, X., Hong, Y. Y., Wang, Y., Xu, P., Ke, S. D., et al. (2008). Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20, 1134–1151. doi: 10.1105/tpc.108.058263
Zalewski, C. S., Floyd, S. K., Furumizu, C., Sakakibara, K., Stevenson, D. W., and Bowman, J. L. (2013). Evolution of the class IV HD-zip gene family in streptophytes. Mol. Biol. Evol. 30, 2347–2365. doi: 10.1093/molbev/mst132
Keywords: epidermal cell differentiation, positional signal, HD-ZIP class IV transcription factor, cuticle, endosperm, receptor-like kinase, calpain-like cysteine protease, Arabidopsis thaliana
Citation: Takada S and Iida H (2014) Specification of epidermal cell fate in plant shoots. Front. Plant Sci. 5:49. doi: 10.3389/fpls.2014.00049
Received: 30 December 2013; Paper pending published: 19 January 2014;
Accepted: 01 February 2014; Published online: 25 February 2014.
Edited by:
Shucai Wang, Northeast Normal University, ChinaReviewed by:
Sergey Morozov, Moscow State University, RussiaRumi Tominaga-Wada, Hiroshima University, Japan
Copyright © 2014 Takada and Iida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinobu Takada, Department of Biological Sciences, Graduate School of Science, Osaka University, 1-1 Machikaneyama-cho, Toyonaka, Osaka 560-0043, Japan e-mail: shinobu_takada@bio.sci.osaka-u.ac.jp
 Shinobu Takada
Shinobu Takada Hiroyuki Iida
Hiroyuki Iida