- Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC, USA
The phosphoinositide pathway and inositol-1,4,5-triphosphate (InsP3) have been implicated in plant responses to many abiotic stresses; however, their role in response to biotic stress is not well characterized. In the current study, we show that both basal defense and systemic acquired resistance responses are affected in transgenic plants constitutively expressing the human type I inositol polyphosphate 5-phosphatase (InsP 5-ptase) which have greatly reduced InsP3 levels. Flagellin induced Ca2+-release as well as the expressions of some flg22 responsive genes were attenuated in the InsP 5-ptase plants. Furthermore, the InsP 5-ptase plants were more susceptible to virulent and avirulent strains of Pseudomonas syringae pv. tomato (Pst) DC3000. The InsP 5-ptase plants had lower basal salicylic acid (SA) levels and the induction of SAR in systemic leaves was reduced and delayed. Reciprocal exudate experiments showed that although the InsP 5-ptase plants produced equally effective molecules that could trigger PR-1 gene expression in wild type plants, exudates collected from either wild type or InsP 5-ptase plants triggered less PR-1 gene expression in InsP 5-ptase plants. Additionally, expression profiles indicated that several defense genes including PR-1, PR-2, PR-5, and AIG1 were basally down regulated in the InsP 5-ptase plants compared with wild type. Upon pathogen attack, expression of these genes was either not induced or showed delayed induction in systemic leaves. Our study shows that phosphoinositide signaling is one component of the plant defense network and is involved in both basal and systemic responses. The dampening of InsP3-mediated signaling affects Ca2+ release, modulates defense gene expression and compromises plant defense responses.
Introduction
Plants have developed a multilayered strategy to effectively control and combat pathogen invasion (Jones and Dangl, 2006). The first line of defense is triggered by the recognition of microbial- or pathogen-associated molecular patterns (MAMP or PAMP) by the membrane associated pattern recognition receptors (PRRs) (Nürnberger et al., 2004; Delledonne, 2005). Pathogen triggered immunity (PTI) also known as basal resistance (Jones and Dangl, 2006), is non-specific and against a range of virulent pathogens. A series of rapid responses are initiated within seconds to minutes of encountering MAMPs including ionic fluxes, production of reactive oxygen species (ROS) and activation of kinase cascades followed shortly by transcriptional reprogramming (reviewed in Postel and Kemmerling, 2009; Gimenez-Ibanez and Rathjen, 2010; Tsuda and Katagiri, 2010).
The second line of defense known as effector-triggered immunity (ETI) involves recognition of specific pathogen avirulent proteins by their counterpart plant disease resistance (R) proteins located within the cell (reviewed in Jones and Dangl, 2006). The local infected sites undergo programmed cell death to halt pathogen growth as part of the hypersensitive response (HR). HR also involves rapid ion fluxes and production of ROS (reviewed in Torres et al., 2006) as well as induction of pathogenesis-related (PR) proteins. In addition to the local response, distal parts of the plant develop immunity against a broad range of pathogens, known as systemic acquired resistance (SAR). A hallmark of SAR is an increase in salicylic acid (SA). SA accumulates in both distal and infected leaves in response to pathogen attack and treatment with SA or SA analogs can induce the PR genes such as PR-1, PR-2, and PR-5 (reviewed in Durrant and Dong, 2004; Fu and Dong, 2013). More recently several other metabolites have been implicated as mobile signals in the development of SAR (Park et al., 2007; reviewed in Kachroo and Kachroo, 2009; Gao et al., 2014).
A transient increase in cytosolic Ca2+ ([Ca2+]in) is one of the early events associated with both PTI and ETI. The initial increase in Ca2+ upon MAMP recognition is accepted to be via an influx through plasma membrane localized Ca2+ channels, such as cyclic nucleotide gated channels (CNGC) and/or glutamate receptor (GluRs) channels. In addition, the involvement of intracellular Ca2+ stores in the propagation of the Ca2+ signal is very likely (reviewed in Ma and Berkowitz, 2007; McAinsh and Pittman, 2009; Manzoor et al., 2012). Several downstream defense related events are regulated by Ca2+ including activation of mitogen-activated protein kinase (MAPK) and calcium-dependent protein kinase (CDPK) cascades and modulation of Ca2+ binding proteins (reviewed in Ma and Berkowitz, 2011; Wurzinger et al., 2011; Boudsocq and Sheen, 2013; Romeis and Herde, 2014). The MAMP-triggered oxidative burst also appears to dependent on the initial cytosolic Ca2+ elevation. The NADP oxidase RbohD is directly and indirectly regulated by Ca2+ (Sagi and Fluhr, 2006; Dubiella et al., 2013). In response to elicitation with flg22, early membrane depolarization events are unaffected in the rbohD mutant (Jeworutzki et al., 2010) and the cytosolic Ca2+ elevation is similar to wild type plants (Ranf et al., 2011). A rise in cytosolic Ca2+ is also important for ETI responses and may precede the oxidative burst in response to avirulent pathogens (Grant et al., 2000). Furthermore, several Ca2+ or calmodulin (CaM) binding transcription factors TGA, CPB60g and CAMTA3, (reviewed in Reddy et al., 2011) act as both positive and negative regulators of SA accumulation and SA-regulated PR gene expression.
Many lipids and lipid related molecules are thought to play roles in plant defense signaling and responses to both biotrophic and necrotrophic pathogens (reviewed in Shah, 2005; Walley et al., 2013). However, there is less information on the potential involvement of the phosphoinositide signaling pathway in plant defense. The membrane-associated phospholipids along with the soluble inositol phosphates (collectively known as phosphoinositides) are present in all eukaryotic cells and are implicated in plant responses to many environmental stimuli (see reviews in Stevenson et al., 2000; Meijer and Munnik, 2003; Krinke et al., 2007a; Im et al., 2010, 2011). In the canonical pathway, phosphatidylinositol is sequentially phosphorylated to phosphatidylinositol 4-phosphate (PtdIns4P) and phosphatidylinositol 4,5-bisphosphate (PtdInsP2) by the enzymes phosphatidylinositol 4-kinase (PI4K) and phosphatidylinositol 4-phosphate 5-kinase (PIP5K), respectively. Activation of phospholipase C (PLC) in response to a stimulus or stress leads to the hydrolysis of PtdInsP2 to generate the soluble second messenger inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG). While many aspects of the phosphoinositide pathway are conserved in plants there are several differences between plants and animals (reviewed in Munnik and Nielsen, 2011; Delage et al., 2013; Pokotylo et al., 2014). In plants, DAG is converted to phosphatidic acid (PtdOH), while InsP3 maybe further phosphorylated to form inositol hexakisphosphate (InsP6). Both InsP3 and InsP6 are thought to release Ca2+ from intracellular stores (reviewed in Krinke et al., 2007a; Im et al., 2010; Gillaspy, 2011). Additionally, InsP5 and InsP6 are implicated in hormone signaling via their interaction with the jasmonic acid receptor COI (Sheard et al., 2010; Mosblech et al., 2011) and the auxin receptor TIR1 (Tan et al., 2007), respectively. PtdOH may also be generated by hydrolysis of structural phospholipids such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the phospholipase D (PLD) enzymes. Recent reports implicate PtdOH as an important signaling molecule in plants in both abiotic and biotic stresses (reviewed in Testerink and Munnik, 2011). Using a transgenic inducible system to express type III effectors (AvrRpm1 or AvrRpt2) in planta, a biphasic accumulation of PtdOH was reported involving first PLC and then PLD activation (Andersson et al., 2006). Furthermore, activation of both PI4K and PLD has been shown to be an early response of suspension cells to treatment with SA (Krinke et al., 2007b, 2009). PtdOH-binding targets include the NAD oxidase RbohD and MPK6 protein kinase and PtdOH is implicated in ROS and HR responses (reviewed in Canonne et al., 2011; Testerink and Munnik, 2011). Specific PLC isoforms may also regulate defense responses (Vossen et al., 2010; Canonne et al., 2011). Arabidopsis mutants which have constitutively low InsP6 levels exhibited an increased susceptibility to microbial infections (Murphy et al., 2008). Collectively these results support the involvement of the phosphoinositides in plant defense; however there are many missing links in our understanding.
In this study, our goal was to further investigate the function of phosphoinositide-mediated signaling in the plant defense network. In previous work, we generated transgenic Arabidopsis plants expressing the human type I inositol polyphosphate 5-phosphatase (InsP 5-ptase), which specifically hydrolyzes soluble inositol phosphates and terminates InsP3-mediated signals. These InsP 5-ptase transgenic plants have normal growth and morphology under optimal growth conditions. However, their basal InsP3 levels are greatly reduced, to only ~5% of wild type level (Perera et al., 2006). The InsP 5-ptase plants exhibit delayed and reduced responses to gravity and impaired Ca2+ signaling in response to salt and cold stimuli (Perera et al., 2006, 2008). The InsP 5-ptase plants were also shown to be less resistant to wounding and herbivory (Mosblech et al., 2008). Interestingly, a global comparison of basal transcript profiles between wild type and InsP 5-ptase plants revealed that 16 out of 62 basally downregulated genes in transgenic plants were defense related (Perera et al., 2008), including PR-1, PR-2, PR-5, and AIG1(avrRpt2- induced gene1). In the current study, we demonstrate that InsP 5-ptase plants show impaired Ca2+ elevation in response to flg22, decreased expression of several defense related genes and delayed and reduced SAR. Our results support the role of InsP3 as a mediator of the intracellular Ca2+ cascade and highlight the involvement of phosphoinositide-mediated signaling in the development of a robust defense response in plants.
Materials and Methods
Growth Conditions for Plants and Bacteria
Transgenic Arabidopsis plants expressing the human type I inositol polyphosphate 5-phosphatase generated via Agrobacterium mediated transformation were generated previously (Perera et al., 2006). Plants were grown in a growth chamber under 8 h light/16 h dark, light intensity of ~150 μmol m−2 s−1, 35–50% humidity and temperature of 21°C. All experiments were carried out using 6–8 week old healthy well watered plants with fully expanded leaves (prior to bolting). For most experiments, wild type (Wt) and two independent transgenic lines (T6 and T8), were tested along with the empty vector control (C2). Bacterial strains Pst DC3000 and isogenic lines carrying avrRpt2 or avrRpm1genes were cultured at room temperature in King's medium B (40 g L−1 peptone, 2% (v/v) glycerol, 1.5 g L−1 K2HPO4 and 1.5 g L−1 MgSO4, pH 7) containing 50 μg ml−1 kanamycin, and 100 μg ml−1 rifampacin. The two non-pathogenic strains, PstDC3000 with mutated effector protein hrcC− and P. syringae pv. Phaseolicola 6, were also cultured at room temperature on King's medium B plates containing 100 ug ml− rifampacin, and an additional 34 μg ml−1 chloramphenicol for growing Pst DC3000 hrcC−.
Pathogen Inoculation
For preparing bacteria used in pathogen inoculation experiments, cells from an overnight culture were first resuspended in 10 mM MgCl2, and the concentration of bacteria was adjusted based on the absorbance reading at OD600. The actual titer was also determined by counting serial dilutions on a selection plate after 48 h at room temperature. To examine the growth of bacteria in planta, leaves were inoculated either by hand using a 1 ml needle-less syringe or by spraying with bacteria solution containing an additional 0.02% (v/v) silwet L-77 (Lehle Seeds, Round Rock, TX). The bacterial titer was determined from 4 leaf discs (total area is 1 cm2) collected from 4 separate leaves, (one leaf per plant). Pooled leaf discs were ground in 10 mM MgCl2 in a final volume of 1 ml. The colony-forming units (cfu) were obtained by spotting 2 μl of a serial 10-fold titrated bacteria onto King's medium B plates containing appropriate antibiotics and additional 50 μg ml−1 cyclohexamine for preventing fungus contamination. In each experiment, a total of 4 plants per line were used. For statistical analysis either Student's t-test or analysis of variance (ANOVA) were performed.
Flagellin Treatment
The elicitor flg22 was synthesized by EZBiolab custom peptide service (Westfield, IN) based on the peptide sequence reported in Felix et al. (1999). For root growth inhibition experiments, 0.1 μM of flg22 was used which caused the half-maximal growth inhibition in Arabidopsis seedlings reported in Gomez-Gomez et al. (1999). For induction of ROS production in leaf discs, the procedure was adapted from Gomez-Gomez et al. (1999) with a flg22 concentration of 1 μM. For induction of Ca2+ release and qRT-PCR analysis of flg22 responsive gene expression, 10 μM flg22 was used according to Navarro et al. (2004) and Zipfel et al. (2004).
SAR Assay
To induce SAR, two fully expanded leaves of wild type and InsP 5-ptase plants were hand inoculated with avirulent strain Pst DC3000+avrRpt2 (OD600 = 0.001; 7.5 × 105 cfu/ml). The un-induced plants were mock inoculated with 10 mM MgCl2, (two leaves per plant). Both induced and un-induced plants were then separated into 4 groups for challenging with the virulent strain PstDC3000 (OD600 = 0.004; 2 × 106 cfu/ml) at day 0, 1, 2, or 3 post inoculation by spraying the bacteria onto the whole plants. To examine the development of SAR, systemic leaves sprayed with PstDC3000 were collected. Leaf extracts were prepared and the cfu was counted as described previously. The growth of virulent bacteria in induced and un-induced plants was compared for determining the level of SAR.
Inositol (1,4,5) Trisphosphate Assay
For measuring the InsP3 level of inoculated leaves, 4–5 plants per experiment for each bacterial strain were grouped together and sprayed with bacterial solution prepared as before. The treated leaves were harvested and immediately frozen in liquid nitrogen. Frozen tissues (around 0.08 g) were ground into powder in liquid nitrogen and then incubated with 160 μl of 20% perchloric acid on ice for 15 min. Samples were centrifuged to remove the debris and the supernatant was transferred to a new tube followed by pH adjustment to 7.5 using 1.5 M KOH/3 mM HEPES. InsP3 assays were carried out using the TRK1000 InsP3 assay kit (Amersham-Pharmacia Biotech) as described previously (Perera et al., 2006).
SA Measurement and Application
For analysis of total SA at the local inoculated leaves, plants were syringe-inoculated with PstDC3000+avrRpt2, and inoculated leaves were harvested at day 2 post inoculation. To extract total SA (free SA + SA conjugates), the protocol described in Nandi et al. (2004) was used. In brief, leaves were first ground into powder in liquid nitrogen, and then ~0.2 g of ground tissue was extracted first with 90% methanol and again with 100% methanol. The combined extracts were subjected to evaporation under N2 gas. After evaporation, the residue was first resuspended in 5% trichloroacetic acid and then submitted to acid hydrolysis by HCl at 100°C for 30 min. SA was extracted with cyclohexane:ethylacetate:isopropanol (50:50:1). The organic phase was evaporated and resuspended in HPLC eluent. Chromatography was performed on a 3.9 × 300-mm C18 reverse-phase μBondapak column (Waters, Milford, MA, USA).
For exogenous SA application, SA (Sigma-Aldrich, St. Louis, MO) was first dissolved in ethanol, then diluted to 300 μM in water with additional 0.02% (v/v) silwet L-77 (Lehle Seeds, Round Rock, TX), and the pH was adjusted to 7 with 0.1 M KOH. For control solution, equal amount of ethanol without SA was diluted in the same way. Solutions were gently and evenly sprayed on plants. Treated plants were then kept in a growth chamber with plastic covers until harvest.
Calcium-Dependent Aequorin Bioluminescence
The wild type and two independent InsP 5-ptase plants (T6 and T8) expressing cytosolic aequorin (Knight et al., 1996) were created previously (Perera et al., 2008). For measuring Ca2+ changes after treatment with flg22, 5-day old seedlings were first incubated with 2 μM coelenterazine (Molecular Probe®, Eugene, OR) solution in the dark for 16 h. Seedlings were transferred into tubes containing water and placed in the luminometer and a baseline reading was obtained. After 1 min, seedlings were treated with 10 μM flg22 which was injected automatically to avoid any handling. The luminescence signals were recorded at 10 s intervals for 35 min. The Ca2+ concentration were calculated after measuring the discharged Ca2+ as described previously (Knight et al., 1996; Perera et al., 2008). For monitoring the Ca2+ increase after pathogen treatment, fully expanded leaves were first infiltrated with 2 μM coelenterazine (Molecular Probe®, Eugene, OR) solution and kept in the dark for 16 h, then the reconstituted leaves were inoculated with Pst DC3000+avrRpm1 (OD600 = 0.5; 5 × 108 cfu/ml) or 10 mM MgCl2 mock solution. Immediately, detached leaves were placed into a tube containing 100 μl water to prevent drying out. The luminescence signals were recorded every 5 s for 3 h. The timing of the peak luminescence were taken for calculation. Luminescence measurements were made using a Sirius luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany).
Chemiluminscent and in situ Detection of ROS
For chemiluminescent detection of ROS, uniform leaf discs were first incubated overnight in sterile water in the dark. Two leaf discs were placed in a test tube containing 105 μL sterile water, with 1 μg horseradish peroxidase (HRP), 20 μM luminol, and 1μM flg22. Luminescence resulting from the reaction between hydrogen peroxide, HRP, and luminol was measured for 35 min in Sirius luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany). For in situ detection of ROS, plants were either sprayed or hand-inoculated with PstDC3000+avrRpt2. Infected leaves were carefully removed at 6 or 48 h, and vacuum-infiltrated with fresh 0.1% (w/v) of 3,3′-diaminobenzidine (DAB) (Sigma-Aldrich, St. Lious, MO) dissolved in 0.01 N HCl. Infiltrated leaves were kept in the dark for 3 h, then de-stained in a destaining solution of 95% EtOH:85% acetic acids: glycerol (3:1:1 in volume) and photographed.
RNA Isolation and qRT-PCR
Total RNA was isolated by RNeasy plant mini kits (Qiagen, Germantown, MD). For RT-PCR, the first strand cDNA was synthesized from total RNA using Ominiscript reverse transcriptase (Qiagen) and random primers, and PCR of specific genes was carried out using Hot Start Taq DNA polymerase (Qiagen). For qRT-PCR, the first strand cDNA was synthesized using StrataScript QPCR cDNA synthesis kits (Agilent Technologies Inc., Santa Clara, CA) and random primers, and the PCR reaction was performed using Full Velocity SYBR-Green QPCR Master mix (Agilent Technologies Inc.) on the Mx3000p thermocycler (Agilent Technologies Inc.). Genes that were analyzed from the Primary Library for Arabidopsis Pathogen-Inducible Genes (cat# PR0100, Sigma-Aldrich, St. Lious, MO) are listed in Supplementary Table 1. Primer sequences of other examined genes are listed in Supplementary Table 2. The fold change in relative gene expression was calculated based on the 2−ΔΔCt method (Schmittgen and Livak, 2008) using wild type as the normalizer and ACT2 or PP2A as the reference genes.
Petiole Exudate Experiment
For collecting the petiole exudates, we followed the method described in Maldonado et al. (2002) and Chaturvedi et al. (2008). Leaves were hand inoculated with PstDC3000+avrRpt2 (OD600 = 0.01; 5.5 × 106 cfu/ml) or mock inoculation with 10 mM MgCl2. Treated plants were placed in the growth chamber and covered with plastic lids. After 6–7 h, inoculated leaves were cut from their bases and immediately dipped in 50% ethanol. They were rinsed once briefly with 0.0005% Clorox bleach and then placed in 1 mM EDTA solution. A group of 10 leaves was placed in a collection tube containing 2.5 ml of 1mM EDTA and 50 μg ml−1 ampicillin, pH 8. They were kept in a covered tray with wet paper towels at the base of the tray to maintain the humidity in the growth chamber under continuous light. After 16 h, collected exudates were diluted 2 fold with water, and filter-sterilized before use.
Results
The Response of InsP 5-ptase Plants to flg22
The bacterial elicitor flagellin is a well characterized MAMP used to investigate plant innate immunity responses. We first treated wild type and InsP 5-ptase seedlings with the active elicitor peptide flg22 and measured root growth. A greater inhibition of root growth was observed in the InsP 5-ptase seedlings compared to wild type (Figure 1A), indicating that the transgenic plants are more sensitive to flg22. We next monitored Ca2+ induction, ROS production and flg22-associated pathogen-responsive gene expression; which are all known to be triggered by flg22 treatment in Arabidopsis (Gomez-Gomez et al., 1999; Asai et al., 2002; Navarro et al., 2004; Zipfel et al., 2004; Boudsocq et al., 2010; Ma et al., 2012). Treatment with flg22 elicits a rapid rise in cytosolic Ca2+ which can be monitored using aequorin-mediated bioluminescence (Jeworutzki et al., 2010; Ranf et al., 2011). InsP 5-ptase seedlings expressing aequorin (Perera et al., 2008) were treated with flg22 and the intracellular Ca2+ changes were monitored. Upon treatment with flg22, a rapid rise in [Ca2+]cyt with a peak at ~2 min was observed in both wild type and InsP 5-ptase plants; however, the induction level was greatly reduced in InsP 5-ptase plants (Figure 1B) with an average reduction in the Ca2+ signal of ~44% in the transgenic plants (Figure 1C). As shown by the root growth experiment (Figure 1A), the InsP 5-ptase plants are responsive to flg22; the increased root growth inhibition correlates with the attenuation of the Ca2+ signal. We also monitored the oxidative burst (by measuring H2O2 generation) after elicitation with flg22. An increase in ROS was detected at the first 1–2 min post treatment, which peaked ~5 min, and tapered off returning to the baseline within 25 min (Figure 1D), which was different from the sustained Ca2+ elevation (Figure 1B). Surprisingly, there was no significant difference in ROS generation between wild type and InsP 5-ptase plants. These results demonstrate that although the Ca2+ release after flg22 treatment was dampened, the oxidative burst was not affected.
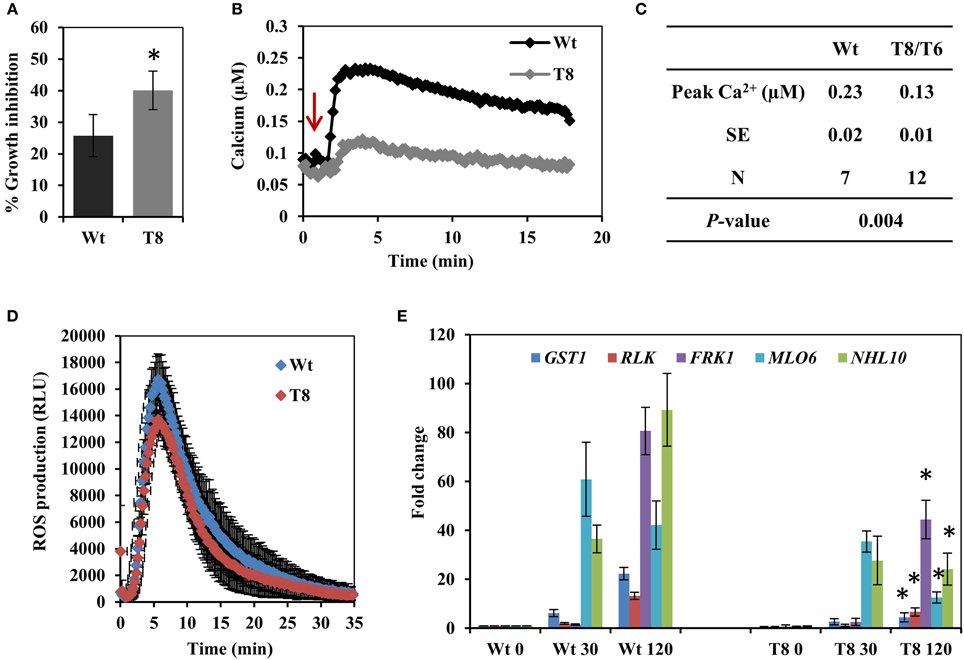
Figure 1. The response of the InsP 5-ptase plants to flagellin. (A) One week old wild type (Wt) and InsP 5-ptase (T8) seedlings were incubated in media containing either 0 or 0.1 μM flg22. Root length was measured after 5 days. Growth inhibition was calculated as the percentage of the non-treated control. Data is the average of four independent experiments (n = 10 seedlings per experiment) ± SE. (B) Five-day old seedlings expressing aequorin were reconstituted with coelenterazine overnight and placed in the luminometer. Seedlings were treated with 10 μM flg22 after 1 min (indicated by red arrow). Ca2+ concentrations were calculated after measuring the discharged Ca2+. A representative trace is shown. (C) The table lists the average peak Ca2+ concentration. (N = independent biological replicates). The average baseline concentrations were 0.097 and 0.098 for wild type and transgenic respectively. (D) ROS production was measured in leaf discs treated with 1 μM flg22. Luminescence was measured at 14 s intervals. Data plotted is the average of five biological replicates ± SD. (E) qRT-PCR analysis of flg22 responsive gene expression at 30 and 120 min post treatment. Data plotted is the average ± SE normalized to Wt control from three independent experiments. *Indicates significant difference between Wt and T8 expression at the same time point (P < 0.05).
Our data suggested that the InsP 5-ptase plants could be a tool to potentially distinguish between parallel downstream pathways of MAMP-triggered immunity. In order to further investigate the downstream responses, a group of genes known to be induced by flg22-FLS2 interaction (Asai et al., 2002; Qutob et al., 2006; Gust et al., 2007; Boudsocq et al., 2010) were selected and their expression levels were quantified by qRT-PCR. The five selected genes were GST1, RLK, FRK1, NHL10, and MLO6. The peak expression of MLO6 was at 30 min after flg22 treatment, whereas the other four genes showed a peak expression at 120 min post flg22 treatment (Figure 1E). Both wild type and InsP 5-ptase plants, showed similar kinetics of induction; however, expression levels at 120 min were significantly reduced in the InsP 5-ptase plants compared to the wild type. The reduction in expression suggests that the InsP 5-ptase plants are unable to sustain or maintain maximal gene induction. Interestingly, FRK1 and NHL10 are both considered to be regulated via MAPK dependent pathways (Boudsocq et al., 2010) and these results suggest a role for phosphoinositide signaling interacting with the MAPK cascade.
The Response of InsP 5-ptase Plants to Virulent and Avirulent Strains of P. syringae
We next examined the interaction between the InsP 5-ptase transgenic plants and the plant pathogenic bacteria, P. syringae pv. tomato (Pst) DC3000 which is widely used to study plant disease resistance (Nishimura and Dangl, 2010). In Arabidopsis, PstDC3000 bacteria multiply rapidly and disease symptoms can be observed ~2 day post-infection with characteristic tissue necrosis and chlorosis (Katagiri et al., 2002). Both virulent and avirulent strains of P. syringae were used to evaluate the susceptibility of InsP 5-ptase transgenic plants compared to wild type.
Figure 2A shows that transgenic plants exhibited more severe symptoms after infection with the virulent strain of P. syringae (Pst DC3000). Moreover, when those infected leaves were harvested and bacterial concentration was quantified, the InsP 5-ptase transgenic plants harbored more bacteria at OD600 = 0.001 and 0.0005 compared to the wild type, indicating they are more susceptible to Pst DC3000 (Figure 2B). Similarly, the transgenic plants also showed a slight elevated bacterial growth of the avirulent strain PstDC3000+avrRpt2 at the low concentration of inoculants OD600 = 0.0005 (Figure 2C). Compared to the wild type the InsP 5-ptase plants are more susceptible to the avirulent strain; however the response to the avirulent strain was not as intense as the response to the virulent strain, suggesting that the gene-for-gene defense mechanism is still functional.
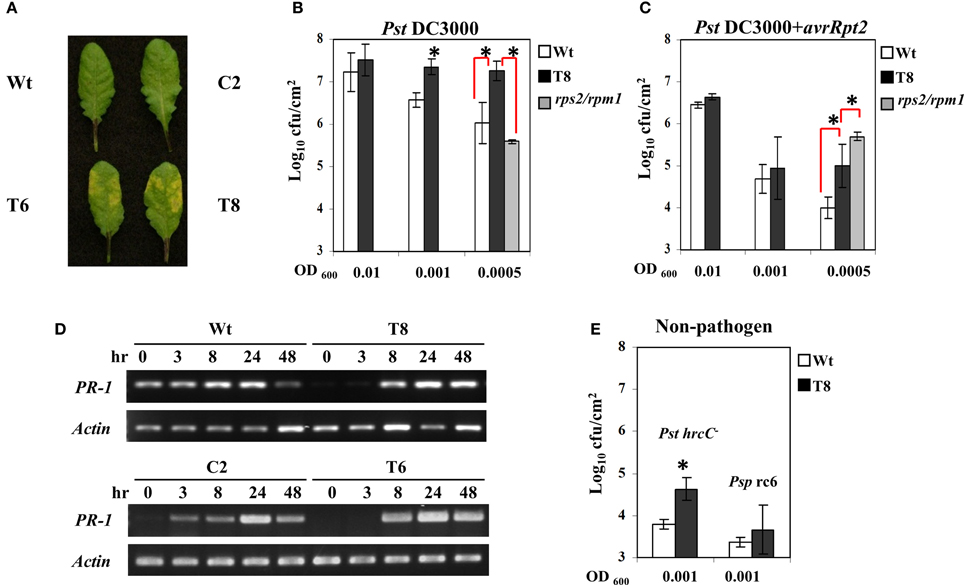
Figure 2. The response of InsP 5-ptase plants to Pseudomonas syringae. Leaves of Arabidopsis plants inoculated with Pst DC3000 at OD600 = 0.001 (7 × 105 cfu/ml) were photographed at day 2 after inoculation (A). Bacterial growth measured from plants at day 2 after inoculation with Pst DC3000 (B) or PstDC3000+avrRpt2 (C). Starting bacterial cultures are OD600 = 0.01 (8 × 106 cfu/ml), 0.001 (7 × 105 cfu/ml) or 0.0005 (3.5 × 105 cfu/ml). (D) Infected leaves used for RT-PCR were inoculated with PstDC3000+avrRpt2 (OD600 = 0.001, 7 × 105 cfu/ml). RT-PCR was carried out with gene-specific primers for PR-1 and Actin. (E) For bacterial growth counts taken at day 4 after inoculation with the two non-pathogens, PstDC3000+hrcC- (Pst hrcC-) and P. syringae pv. Phaseolicola race 6 (Psp rc6), the concentration of inoculants were OD600 = 0.001 (7 × 105 cfu/ml). Plant lines used were Wt (white), T8 (black), and the rps2/rpm1 double knock-out mutant (gray). *P < 0.05.
We also compared the disease response of InsP 5-ptase plants to that of rps2/rpm1 double mutants (lacking both of the Rps2 and Rpm1 R genes) which are equally susceptible to both virulent and avirulent strains of PstDC3000 even at low concentrations of inoculant. We found that transgenic plants were less affected than the rps2/rpm1 mutants when challenged with avirulent strains PstDC3000+avrRpt2; and did not have the same high level of bacterial growth (Figure 2C). Therefore, the response of InsP 5-ptase plants to PstDC3000+avrRpt2 is not similar to rps2/rpm1 double mutant which lacks the R gene mediated resistance. The enhanced susceptibility pattern observed in InsP 5-ptase plants is more similar to previously reported mutants with a defect in SA-mediated disease resistance, such as nahG and eds16/sid2 mutants (Gaffney et al., 1993; Delaney et al., 1994; Dewdney et al., 2000; Wildermuth et al., 2001). These plants with a defect in SA-mediated resistance are more susceptible to avirulent bacterial strains, but not so severely as to virulent bacterial strains, indicating that the R genes in these mutant plants are not affected (Dewdney et al., 2000). It is possible that the enhanced susceptibility toward avirulent strains might be the result of dampened signal transduction leading to disease resistance. To test this hypothesis, we examined PR-1 gene induction as a downstream response during the course of infection with PstDC3000+avrRpt2 at 3, 8, 24, and 48 h, and found that the two independent InsP 5-ptase lines T6 and T8 show a delayed induction of gene expression (the response was observed at 8 h instead of 3 h) compared to the wild type and vector control plants C2 (Figure 2D).
To further demonstrate that basal defense is compromised, we challenged InsP 5-ptase plants with a TTSS-defective hrcC mutant of PstDC3000 (in which pathogen effector proteins are not delivered into host plants such that only a few bacteria multiply in wild type plants). We found a slight elevation in bacterial growth in InsP 5-ptase plants (Figure 2E), suggesting that the basal defense in InsP 5-ptase plants was weakened. However, we did not observe any bacterial growth or symptoms when challenged with the non-host pathogen strain P. syringae pv. Phaseolicola strain race 6 in both wild type and transgenic plants, even after 12 days (Figure 2E). These results suggest that the non-host resistance mechanism in transgenic plants is not affected.
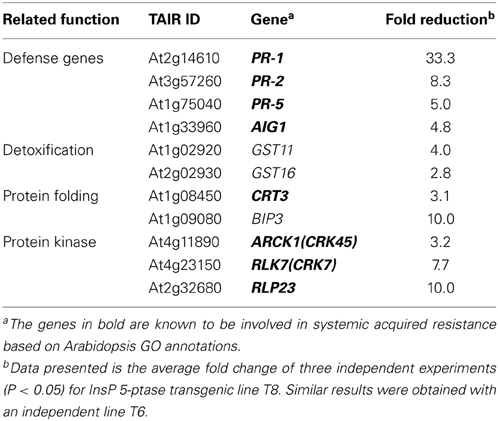
Previous microarray data indicated that a group of disease resistance-related genes are basally downregulated under normal growth conditions in two independent InsP 5-ptase plants compared to wild type and vector control plants. These include PR-1, PR-5, and AIG1(avrRpt2- induced gene1), as well as genes encoding proteins involved in Ca2+ storage in the endoplasmic reticulum (ER) such as CRT3 and BiP3 (Perera et al., 2008). In this study we used qRT-PCR to analyze a set of selected genes. We determined that, in addition to previously identified defense-related genes, PR-2, two putative glutathione-S-transferase genes (GST11 and GST16) and three protein kinases (CRK7, CRK45, and RLP23) were also basally downregulated in InsP 5-ptase plants (Table 1). Among the eleven genes tested, eight of them are also required for SAR according to the GO annotation in TAIR. Although BiP3 has not been annotated as the other eight genes, other BiP isoforms have been shown to play a role in promoting plant immunity (Wang et al., 2005; Carvalho et al., 2014). The data suggest that reduced expression of these genes might contribute to the enhanced susceptibility of the transgenic plants to P. syringae.
InsP3 Changes and Calcium Release During ETI
In order to determine whether InsP3 is induced in wild type plants when infected with P. syringae, we examined InsP3 levels during a 6 h period of post spray with P. syringae in wild type Arabidopsis leaves. When spray-infected with a virulent strain Pst DC3000, InsP3 levels increased ~two-fold at 4 h post spray (Figure 3A). When infected with avirulent strains PstDC3000+avrRpm1 or avrRpt2, InsP3 levels increased 2–3 fold at a shorter time, 20 min and 1 h post spray, respectively. No change in InsP3 levels was observed with mock spray (Figure 3A). These results demonstrate that the induction of InsP3 is one of the signals generated during plant-pathogen interaction. The difference in the induction times of InsP3 between virulent and avirulent pathogens is in good agreement with the rapid induction of HR by avirulent strains compared to the slower development of disease symptoms by virulent strains (Katagiri et al., 2002).
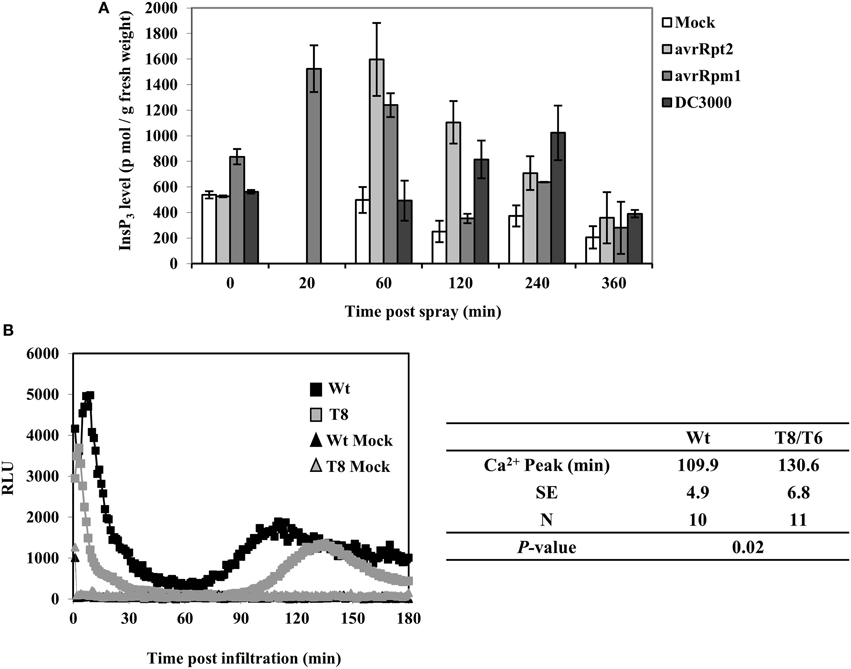
Figure 3. InsP3 changes and Ca2+ release in response to avirulent Pseudomonas syringae pv tomato DC3000. (A) Wild type Arabidopsis plants were sprayed with Pst DC3000 or Pst DC3000+avrRpt2 or + avrRpm1 at a concentration of OD600 = 0.004 (2 × 106 cfu/ml). Mock spray was 10 mM MgCl2. Treated leaves were harvested at different time points and InsP3 was quantified. Data shown is the average of 5 independent experiments ± SE. (B) Wild type (Wt) or InsP 5-ptase seedlings carrying aequorin were reconstituted with coelenterazine and inoculation with Pst DC3000+avrRpm1 (OD600 = 0.5, 5 × 108 cfu/ml) or mock solution (10 mM MgCl2). A representative experiment is shown with luminescence counts taken in every 5 s. The table lists the average time of the second sustained Ca2+ peak (N = independent biological replicates).
In a previous study, a biphasic increase in cytosolic Ca2+ was measured by aequorin-mediated bioluminescence in Arabidopsis leaves infected with avirulent strains of PstDC3000+avrB or avrRpm1 (Grant et al., 2000). We tested aequorin lines of wild type and InsP 5-ptase plants infected with PstDC3000+avrRpm1. We found that the early transient rise in [Ca2+]in was similar in both wild type and InsP 5-ptase plants but there were differences in the second sustained increase in [Ca2+]in. The wild type showed a peak of Ca2+ at 110 min post inoculation which is similar to the previous report (Grant et al., 2000). In InsP 5-ptase plants, there was a ~20 min delay in the timing of the Ca2+ release (Figure 3B) suggesting that InsP3-mediated signaling contributes to the cytosolic Ca2+ increase observed in avirulent pathogen infected leaves.
ROS Production and SA-mediated Signaling During ETI
The oxidative burst in avirulent pathogen infected leaves is part of the disease resistance mechanism (see review in Lamb and Dixon, 1997). It leads to the generation of ROS, which presumably could confine bacterial growth (see review in Torres et al., 2006). To examine the ROS production in avirulent pathogen infected leaves, we used 3,3′-diaminobenzidine (DAB) to detect ROS production in situ. Similar to the result observed with flagellin treatment (Figure 1A), no difference in ROS production between wild type and InsP 5-ptase plants was observed (Supplementary Figure 1). In both lines staining was visible at 6 h and disappeared at 48 h either after inoculation or spraying with PstDC3000+avrRpt2. The results demonstrate that the HR-induced oxidative burst and ROS production are unaffected in InsP 5-ptase plants and suggest that ROS generation in plant ETI does not involve InsP3-mediated pathways.
Because SA is a key systemic signal generated during ETI, we compared the SA levels in wild type and InsP 5-ptase plants (Figure 4A). We measured total SA including the conjugated form. The basal SA level in transgenic plants was ~50% of the amount in wild type. After inoculating with PstDC3000+avrRpt2, the SA levels increased in local inoculated wild type leaves and to a lesser extent in the InsP 5-ptase leaves. Although the SA reduction post inoculation in the transgenic plants is not highly significant (P = 0.1), the results do show that the total basal SA in InsP 5-ptase plants was significantly lower compared to wild type. It was not impaired as much as in nahG plants and sid2 mutants, but this low efficiency might be enough to reduce the SA-mediated signaling and cause delay in downstream gene expression, such as PR-1.
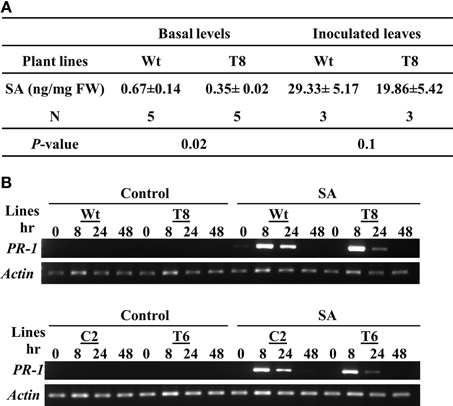
Figure 4. InsP 5-ptase plants have a low basal level of SA; however the local response to exogenous SA is normal. (A) SA levels were measured in untreated leaves (basal) or in leaves harvested from plants inoculated with Pst DC3000+avrRpt2 at a concentration of OD600 = 0.1 (1 × 108 cfu/ml) at day 2 post-inoculation. Data is the average of three to five experiments ± SE. Each experiment consisted of a pool of leaves from four individual plants/line. (B) Wild type (Wt), two independent InsP 5-ptase lines (T6 and T8), and vector control (C2) plants were sprayed with 300 μM of SA or control solution (0.05% ethanol). Leaves were harvested before (0) and at 3, 8, 24, and 48 h after treatment. RT-PCR was carried out with gene-specific primers for PR-1 and Actin.
To further investigate whether the delayed PR-1 gene induction observed in InsP 5-ptase plants (Figure 2D) was due to a delayed response to SA, we applied exogenous SA and monitored PR-1 gene induction over a two day period. The wild type and vector control plants (C2) as well as two independent InsP 5-ptase lines were treated with 300 μM SA by mist spray, treated leaves were harvested at 0, 8, 24, and 48 h and PR-1 transcript levels was monitored by RT-PCR (Figure 4B). Both InsP 5-ptase lines have similar induction of PR-1 as wild type and C2 in response to exogenous SA, suggesting that the InsP 5-ptase plants can respond to SA similar to the wild type. The data suggest that the delay in PR-1 gene induction is probably due to the delayed synthesis of SA. This result excludes the possibility of InsP3 being an intermediate between SA and PR-1 gene expression.
The SAR Response in InsP 5-ptase Plants
In order to examine whether the reduced SA basal levels and delayed PR-1 induction would compromise SAR in InsP 5-ptase plants, we carried out an SAR assay (Zhang et al., 2010). Plants were first inoculated with PstDC3000+avrRpt2, (two leaves per plant), and then challenged with PstDC3000 by spraying whole plants at day 0, 1, 2, and 3 post first inoculation. To monitor the progression of bacterial growth in systemic leaves, bacterial counts were measured in leaves at day 4 post spray. We found that wild type plants acquired resistance to PstDC3000 around day 3 post first inoculation showing a reduced bacterial growth compared to that at day 0, 1, and 2 (Figure 5A). Although the bacterial growth in InsP 5-ptase plants also declined slightly at day 3, the reduced growth is not statistically significant compared to that at day 0, 1, and 2. The assay result indicates that the SAR response was affected in InsP 5-ptase plants. It could be that the acquired resistance was not induced as much as in the wild type at day 3, (i.e., a delayed response), or it could be that the level of resistance was substantially reduced. No acquired resistance was detected when plants were first mock inoculated with 10 mM MgCl2 (Figure 5B).
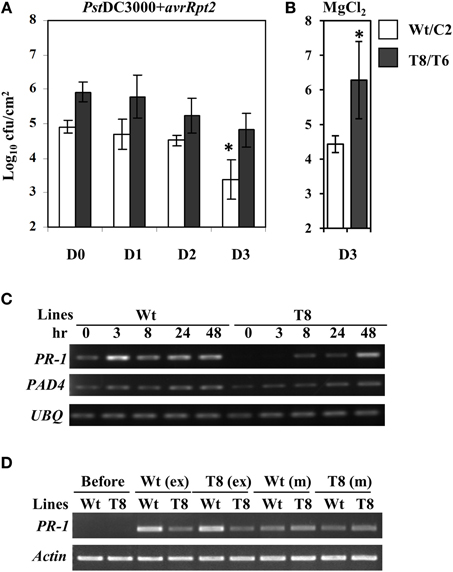
Figure 5. The SAR response is delayed in the InsP 5-ptase plants. The SAR assay was carried out using wild type (Wt), vector control (C2) and two independent InsP 5-ptase lines (T6 and T8). Plants were either first inoculated with Pst DC3000+avrRpt2 (OD600 = 0.001, 7 × 105 cfu/ml) (A), or 10 mM MgCl2 mock solution (B), then sprayed with PstDC3000 (OD600 = 0.004, 2 × 106 cfu/ml) at day 0, 1, 2, or 3. Systemic leaves were harvested and the bacterial growth was quantified at day 4 post spray. Data is the average of three experiments ± SD. Each experiment has three plants per line. Results from Wt and C2 (Wt/C2), and T6 and T8 (T8/T6) were pooled for analysis. *P < 0.05 (C) Systemic leaves were also harvested before (0) and after initial inoculation at 3, 8, 24, and 48 h for RT-PCR carried out with gene-specific primers for PR-1, PAD4, and UBQ10. (D) For the exudate experiment, only Wt and T8 and their reciprocal treatments are shown. Leaves were harvested before or at 48 h post exudate (ex) or mock (m) infiltration for RT-PCR carried out with gene-specific primers for PR-1 and Actin.
To investigate the expression levels of defense-related genes during SAR induction in systemic leaves, we performed qRT-PCR analysis over a time course. We first inoculated plants with PstDC3000+avrRpt2 (two leaves per plant), then the systemic leaves were harvested at 3 h, 8 h, day 1, and day 2 post-inoculation. We monitored expression of a total of 47 genes (for a complete gene list see Supplementary Table 1). We found 14 genes that were differentially expressed in the wild type and two transgenic plant lines that were either only induced in the wild type or showed reduced and delayed induction in the transgenic plants (Table 2, calculated data are in Supplementary Table 3). Among these 14 genes, eight of them had low basal expression in the transgenic plants. The remaining six genes, which were not basally low, exhibited a reduced and delayed induction. These genes included NHL10, RLK1, PAD4, and NIMIN-2, which are implicated in SAR and SA-mediated signaling (Zhou et al., 1998; Li et al., 2004; Navarro et al., 2004; Weigel et al., 2005). To further confirm the qRT-PCR result, we also performed an independent RT-PCR analysis on systemic leaves (Figure 5C) using the marker genes PR-1 and PAD4. The RT-PCR results were consistent with the qRT-PCR data. The results indicate that the expression of a subset of genes involved in SAR were altered, both in timing and extent in the InsP 5-ptase plants.
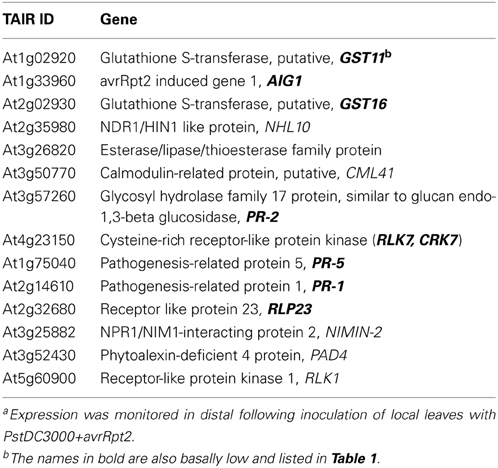
Table 2. Genes which were either not induced or showed delayed expression in distal leaves of InsP 5-ptase plants compared to Wta.
Signal Molecules Generated in InsP 5-ptase Plants
The onset of SAR requires an array of mobile signals which are generated from the primary infected leaves (reviewed in Gao et al., 2014). These molecules are presumed to be translocated from local inoculated leaves to distal leaves through the phloem tissue. Phloem exudate collected from local leaves provides a means to study the effects of the mobile signals. We performed exudate experiments to examine whether the signal molecules generated from the local infected leaves in wild type can trigger the same response in InsP 5-ptase plants and vice versa. The experimental design is to mimic the process of delivering signal molecules from primary infected leaves to systemic leaves by bypassing the plant delivery system. The exudate from infected plants containing putative signal molecules was collected and injected into healthy uninfected plants. In these experiments, PR-1 gene expression is expected to increase in the new leaves (similar to systemic leaves) upon injection in response to the presence of the signal molecules. We found that exudates collected from InsP 5-ptase plants were equally effective in inducing PR-1 gene expression in wild type plants as exudates from wild type plants (Figure 5D). However, InsP 5-ptase plants showed diminished PR-1 gene expression in response to exudates from either wild type or transgenic plants. These results indicate that the generation of mobile signals at the primary infected site is not impaired in the InsP 5-ptase plants and the exudates were as effective as those produced in wild type. Since the response to exogenous application of SA was also normal in the InsP 5-ptase plants (Figure 4B), these results suggest that InsP 5-ptase plants are affected in either sensing the mobile signal and/or in the synthesis of SA at the systemic site.
Discussion
A summary of our major results is illustrated in Figure 6. In the InsP 5-ptase plants, PTI responses including MAMP-triggered Ca2+ increase and gene expression were affected, although ROS generation was normal. Additionally, ETI responses occurring at the local infected leaves were compromised in InsP 5-ptase plants leading to a reduced and delayed induction of the systemic response.
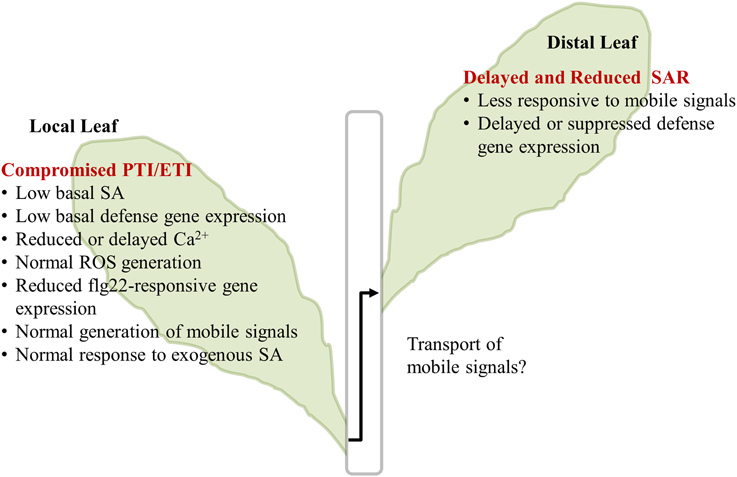
Figure 6. Summary of InsP 5-ptase plants responses to flg22 and Pseudomonas syringae. The major results described in the paper are listed. The transport of mobile signals from local to distal leaves was not investigated and is indicated by the “?.” PTI, PAMP triggered immunity; ETI, Effector triggered immunity; SA, salicylic acid; ROS, reactive oxygen species; SAR, systemic acquired resistance.
A reduction in basal InsP3 levels is the most dramatic and direct consequence of the constitutive expression of InsP 5-ptase in the InsP 5-ptase transgenic Arabidopsis plants (Perera et al., 2006, 2008). It is reasonable to speculate therefore, that the inability to propagate an InsP3 signal is the primary basis for the altered defense responses exhibited by these plants. However, we cannot rule out potential up and downstream effects of the increased turnover of InsP3, since InsP3 is an intermediate in the phosphoinositide pathway and is linked to both the phospholipids and the inositol phosphates. In previous work, we showed that PIP5K activity and PtdInsP2 synthesis was upregulated in transgenic tobacco cells in suspension culture expressing InsP 5-ptase (Perera et al., 2002). Under normal growth conditions, in our hands, we have not detected a up regulation of PtdInsP2 synthesis in transgenic Arabidopsis seedlings or plants. It is possible that the InsP 5-ptase plants have altered localized perturbations in membrane phospholipids under specific conditions (such as hydroponic growth, König et al., 2007) or in response to stimuli such as pathogens.
InsP3 signaling in plants remains a controversial topic primarily due to the fact that at a molecular level, an InsP3 responsive Ca2+ channel (analogous to the animal InsP3 receptor) has not been identified in plant genomes (discussed in Krinke et al., 2007a; Munnik and Nielsen, 2011). Nevertheless, many studies have demonstrated that InsP3 is a physiological ligand (reviewed in Krinke et al., 2007a; Dodd et al., 2010; Pokotylo et al., 2014) and InsP3 changes occur rapidly and transiently in response to wide variety of both abiotic and biotic signals/stimuli (reviewed in Krinke et al., 2007a; Im et al., 2010). It has also been suggested that InsP6 rather than InsP3 is the ligand responsible for Ca2+ release from intracellular plant stores (Lemtiri-Chlieh et al., 2003). Since InsP3 is an intermediate in the lipid-dependent route of InsP6 biosynthesis (reviewed in Gillaspy, 2011, 2013), increased InsP3 turnover could be expected to affect InsP6 levels and indeed the InsP 5-ptase plants have lower InsP6 levels compared to wild type (Perera et al., 2008). Increased susceptibility to several microbial pathogens was reported in low InsP6 mutants (Murphy et al., 2008). However, (unlike the InsP 5-ptase plants), these mutants had normal basal levels of SA and normal SA induction upon infection. Therefore we cannot attribute all of the attenuated defense responses of the InsP 5-ptase plants to a decrease in InsP6. Other inositol phosphate intermediates, including InsP4 and InsP5 are implicated in stress responses (reviewed in Pokotylo et al., 2014). It is conceivable that InsP6 (and/or other inositol phosphates) act synergistically with InsP3 to mediate signaling leading to defense responses.
Attenuation of Ca2+ Signals
It is clear that both PTI and ETI share similar signal transduction components and downstream targets (discussed in Ma and Berkowitz, 2007; Tsuda and Katagiri, 2010) and may represent an overlapping and interconnected network. The complexity of this signaling network is illustrated by the convergence and crosstalk between parallel branches (Boudsocq and Sheen, 2013). A rise in cytosolic Ca2+ is one common signaling element in response to both PTI and ETI and Ca2+ may be a primary signal essential for initiating many of the early events; however there are differences in the timing and duration of the PTI and ETI associated Ca2+ signals. Additionally there are feedback loops that modulate the propagation of the Ca2+ signal (Ma and Berkowitz, 2007, 2011).
We showed that the InsP 5-ptase seedlings have a greatly attenuated Ca2+ signal in response to flg22 compared to wild type (Figure 1B). A similar result was reported using intact leaves exposed to flg22 (Ma et al., 2012). Intriguingly, the InsP 5-ptase plants exhibited a normal Ca2+ response to the plant derived peptide elicitor Pep3 (Ma et al., 2012). The Pep receptor PEPR is linked to the cyclic nucleotide gated cation channel CNGC2 (Qi et al., 2010; Ma et al., 2012). Interestingly, the cngc2 mutant showed an opposite Ca2+ response to the InsP 5-ptase plants; an attenuated Ca2+ rise in response to Pep3 but a normal Ca2+ response to flg22 (Ma et al., 2012). This result supports the involvement of different pathways of Ca2+ influx in response to Pep and flg22. It was suggested that Pep primarily targets extracellular stores (via influx by CNGC2) while flg22 may involve an additional contribution from intracellular stores. Detailed studies of the amplitude and kinetics of flg22 dose-dependent Ca2+ changes also suggested that two processes or two different stores may be involved in the flg22 induced Ca2+ response (Jeworutzki et al., 2010; Ranf et al., 2011).
A biphasic Ca2+ response was previously reported in response to avirulent pathogens (Grant et al., 2000). Ma and Berkowitz (2007) proposed that the initial pathogen-associated Ca2+ rise is primarily via influx from the apoplast while the subsequent Ca2+ rise is from intracellular stores. We observed that while the initial Ca2+ peak was similar to wild type, the timing of the second Ca2+ peak was delayed in the InsP 5-ptase plants (Figure 3B) which supports a role for InsP3 in sustaining and propagating the pathogen-associated cytosolic Ca2+ signal.
Our results are consistent with a model in which InsP3 (and or its derivatives) may act in an intracellular Ca2+ relay which would be downstream of the initial Ca2+ influx from the apoplast. Because MAMP-triggered ROS was unaffected in the InsP 5-ptase plants we propose that the initial rise in cytosolic Ca2+ in response to MAMP elicitation may activate some downstream targets (such as CDPKs) as well as trigger the early oxidative burst. Alternatively, the ROS response may be “primed” by BIK1 in a parallel Ca2+ independent manner (Kadota et al., 2014; Li et al., 2014).
In plants several intracellular compartments serve as Ca2+ stores (reviewed in Dodd et al., 2010). The InsP3 responsive Ca2+ store is likely to be the ER or vacuole; however we cannot rule out the involvement of the chloroplast (Manzoor et al., 2012). A plant specific Ca2+ sensor protein CAS, which is located in the chloroplasts, was recently shown to play a role in both PTI and ETI (Nomura et al., 2012). There are shared similarities in the response of the cas-1 mutant and the InsP 5-ptase transgenic plants in defense signaling, such as increased susceptibility to both virulent and avirulent bacterial strains, basally reduced PR-1 gene expression and normal ROS generation in response to flg22. However at present, we have no information on whether InsP3 (and/or InsP6) may affect Ca2+ stores in the chloroplast. The basal down regulation of PR genes and the reduced activation of PR gene expression in the InsP 5-ptase plants may also reflect alterations in ER Ca2+ homeostasis as discussed further below.
SA Biosynthesis and SA Mediated Signaling
InsP 5-ptase plants have low basal SA (under normal control conditions) and when infected with avirulent pathogens, SA levels were lower than wild type plants at day 2 post inoculation (Figure 4), suggesting that SA biosynthesis maybe reduced in the transgenic plants. The reduced levels of SA in InsP 5-ptase plants suggest a link between PI-metabolism and SA accumulation/pathway through a yet unknown mechanism that ultimately affects plant defense, either directly or indirectly. At a transcriptional level, based on previous microarray results (Perera et al., 2008), we have not detected lower expression of the two key genes involved in SA biosynthesis, namely phenylalanine ammonia lyase (PAL) and isochorismate synthase (ICS1), (Wildermuth et al., 2001). ABA has been implicated as a negative regulator of SA due to their overlapping biosynthetic pathways (de Torres Zabala et al., 2009; reviewed in Cao et al., 2011). However, we did not detect increased basal ABA levels in InsP 5-ptase plants (Perera et al., 2008). Another possibility is that expression of genes encoding positive regulators of SA accumulation (reviewed in Lu, 2009; Ng et al., 2011), could be impaired in InsP 5-ptase plants, resulting in lowered SA levels. One of these genes is PAD4, which showed a delayed induction in systemic leaves (Table 2). Similar to InsP 5-ptase plants, mutants of this group of genes have reduced SA accumulation and enhanced susceptibility to pathogens which could be rescued by exogenous SA treatment (Zhou et al., 1998). Recent studies also point to the involvement of Ca2+/calmodulin (CaM) in modulating SA biosynthesis. SARD1 and CBP60g, are two related transcription factors which control SA biosynthesis by regulating ICS1 expression. Interestingly, CBP60g is a calmodulin-binding protein and its activity is Ca2+-dependent (Zhang et al., 2010; Truman and Glazebrook, 2012). It is possible that the reduced Ca2+ response in InsP 5-ptase plants might affect induction and accumulation of SA via CBP60g activity.
Attenuated Gene Expression
We have reported that select early flg22 responsive genes showed reduced expression at 120 min in the InsP 5-ptase plants (Figure 1E). We suspect that some downstream transcriptional responses may require the continued propagation of the Ca2+ signal.
We have also shown that the InsP 5-ptase plants have basally reduced levels of some defense-related genes (Table 1). Additionally, induction of PR-1 was delayed in local leaves and several SAR related genes were either not induced or delayed in induction in systemic leaves (Figure 2D, Table 2). We suggest that these genes are in part under the control of SA and Ca2+/CaM which can regulate transcription by either directly or indirectly interacting with transcription factors (review Dong, 2004; Kim et al., 2009; Reddy et al., 2011). The DNA-binding activity of the transcription factor CAMTA (or SR) is enhanced by Ca2+/CaM binding (review, Finkler et al., 2007). Arabidopsis CAMTA3 acts as a negative regulator of biotic stress responses. The loss-of-function mutants have elevated levels of SA and exhibit enhanced disease resistance. Additionally, several defense-associated genes (including PR genes) are found to be up-regulated in camta3 mutants (Galon et al., 2008; Du et al., 2009).
The cyclic nucleotide-gated channel (CNGC) mutant cngc2 also shows constitutively high PR-1 gene expression and high SA production (Chan et al., 2008). The phenotypes of both the camta3 and cngc2 mutants are opposite to what we observed in the InsP 5-ptase plants, which have low PR-1 gene expression and low SA levels. We have noted a striking inverse correlation between genes that are basally down regulated in the InsP 5-ptase plants and genes that are upregulated in camta3 and cngc2 (Supplementary Table 4). Twelve of the basally upregulated genes in cngc2, and thirteen of the upregulated genes in camta3 were found downregulated in InsP 5-ptase plants (Perera et al., 2008) including PR-1, PR-2, and PR-5 (Supplementary Table 4). Furthermore, there was strong overlap between this subset of genes and the top 50 candidates in the PR-1 coexpression network as well as with genes that upregulated in wild type seedlings grown under high Ca2+ conditions (Chan et al., 2008). These results are further support for Ca2+ mediated regulation of gene expression although at present, the exact mechanism of activation of these over lapping genes is not known.
SA is also implicated in the regulation of PR gene expression via the activation of the TGA (TGACG Motif-Binding Factor) transcription factors (review, Dong, 2004; Fu and Dong, 2013). Accumulation of SA affects the cellular redox and controls the translocation of the cofactor NPR1 into the nucleus where it binds and activates TGAs. Most TGAs act as positive regulators to induce PR genes as well as the expression of genes encoding ER resident proteins involved in protein folding and Ca2+ storage. The induction of the protein secretory pathway is required for SAR (Wang et al., 2005). InsP 5-ptase plants have low basal levels of PR-1, PR-5, CRT3, and BiP3 (Table 1), which may be in part due to low basal SA and/or low ER Ca2+.
In conclusion, we show that the constitutive expression of InsP 5-ptase affects Ca2+ release, expression of a subset of defense related genes and both basal and SAR responses but does not affect ROS production. We favor the model postulated by Ranf et al. (2011) in which Ca2+ acts as an “on/off switch” and a threshold level of Ca2+ is required for full activation of the downstream pathways. The InsP 5-ptase plants are impaired in their ability to propagate and maintain the Ca2+ signal and therefore are unable to mount a full and robust defense response.
Author Contributions
Imara Y. Perera and Chiu-Yueh Hung designed and supervised the experiments and constructed the final manuscript. Chiu-Yueh Hung, Peter Aspesi Jr, and Aaron W. Lomax carried out the pathogen infection and SAR experiments; Chiu-Yueh Hung and Melissa R. Hunter designed and carried out qRT-PCR and analysis. Aaron W. Lomax also prepared samples for SA analysis. Imara Y. Perera supervised and carried out the flg22 treatment, ROS, and Ca2+ assays.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service, Grant number 2004-35100-14892 to Imara Y. Perera and in part by the summer NSF-REU program in Integrative Plant Molecular Systems at North Carolina State University. The bacterial strains and Arabidopsis rps2/rpm1 double mutant were kindly provided by Dr. Jeff Dangl (University of North Carolina at Chapel Hill, USA). We thank Dr. Kirk Overton for suggestions with experimental design. We thank Dr. Brian Phillippy for the SA analysis by HPLC. We acknowledge former NSF-REU students Christian Estes and Michael Rydberg for their help with the flg22 experiments. We also thank Dr. Wendy Boss (North Carolina State University) for support and helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00267/abstract
References
Andersson, M. X., Kourtchenko, O., Dangl, J. L., Mackey, D., and Ellerström, M. (2006). Phospholipase−dependent signalling during the AvrRpm1−and AvrRpt2−induced disease resistance responses in Arabidopsis thaliana. Plant J. 47, 947–959. doi: 10.1111/j.1365-313X.2006.02844.x
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Canonne, J., Froidure-Nicolas, S., and Rivas, S. (2011). Phospholipases in action during plant defense signaling. Plant Signal. Behav. 6, 13–18. doi: 10.4161/psb.6.1.14037
Cao, F. Y., Yoshioka, K., and Desveaux, D. (2011). The roles of ABA in plant-pathogen interactions. J. Plant Res. 124, 489–499. doi: 10.1007/s10265-011-0409-y
Carvalho, H. H., Silva, P. A., Mendes, G. C., Brustolini, O. J., Pimenta, M. R., Gouveia, B. C., et al. (2014). The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 164, 654–670. doi: 10.1104/pp.113.231928
Chan, C. W., Wohlbach, D. J., Rodesch, M. J., and Sussman, M. R. (2008). Transcriptional changes in response to growth of Arabidopsis in high external calcium. FEBS Lett. 582, 967–976. doi: 10.1016/j.febslet.2008.02.043
Chaturvedi, R., Krothapalli, K., Makandar, R., Nandi, A., Sparks, A. A., Roth, M. R., et al. (2008). Plastid ω3−fatty acid desaturase−dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54, 106–117. doi: 10.1111/j.1365-313X.2007.03400.x
Delage, E., Puyaubert, J., Zachowski, A., and Ruelland, E. (2013). Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4, 5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Prog. Lipid Res. 52, 1–14. doi: 10.1016/j.plipres.2012.08.003
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
Delledonne, M. (2005). NO news is good news for plants. Curr. Opin. Plant Biol. 8, 390–396. doi: 10.1016/j.pbi.2005.05.002
de Torres Zabala, M., Bennett, M. H., Truman, W. H., and Grant, M. R. (2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. doi: 10.1111/j.1365-313X.2009.03875.x
Dewdney, J., Reuber, T. L., Wildermuth, M. C., Devoto, A., Cui, J., Stutius, L. M., et al. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. doi: 10.1046/j.1365-313x.2000.00870.x
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S. N., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. doi: 10.1073/pnas.1221294110
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313X.1999.00265.x
Finkler, A., Ashery-Padan, R., and Fromm, H. (2007). CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. doi: 10.1016/j.febslet.2007.07.051
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Chemical inducers of systemic immunity in plants. J. Exp. Bot. 65, 849–1855. doi: 10.1093/jxb/eru010
Gillaspy, G. E. (2011). The cellular language of myo−inositol signaling. New Phytol. 192, 823–839. doi: 10.1111/j.1469-8137.2011.03939.x
Gillaspy, G. E. (2013). “The role of phosphoinositides and inositol phosphates in plant cell signaling,” in Lipid-Mediated Protein Signaling, ed D. G. S. Capelluto (Dordrecht: Springer), 141–157. doi: 10.1007/978-94-007-6331-9_8
Gimenez-Ibanez, S., and Rathjen, J. P. (2010). The case for the defense: plants versus Pseudomonas syringae. Microbes Infect. 12, 428–437. doi: 10.1016/j.micinf.2010.03.002
Gomez-Gomez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18, 277–284. doi: 10.1046/j.1365-313X.1999.00451.x
Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. doi: 10.1046/j.1365-313x.2000.00804.x
Gust, A. A., Biswas, R., Lenz, H. D., Rauhut, T., Ranf, S., Kemmerling, B., et al. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282, 32338–32348. doi: 10.1074/jbc.M704886200
Im, Y. J., Heilmann, I., and Perera, I. Y. (2011). “The hull of fame: lipid signaling in the plasma membrane,” in The Plant Plasma Membrane, Plant Cell Monographs, Vol. 19, eds A. S. Murphy, B. Schulz, and W. Peer (Berlin; Heidelberg: Springer), 437–455.
Im, Y. J., Phillippy, B. Q., and Perera, I. Y. (2010). “InsP3 in plant cells,” in Lipid Signaling in Plants, ed T. Munnik (Berlin; Heidelberg: Springer), 145–160.
Jeworutzki, E., Roelfsema, M. R. G., Anschütz, U., Krol, E., Elzenga, J. T. M., Felix, G., et al. (2010). Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels. Plant J. 62, 367–378. doi: 10.1111/j.1365-313X.2010.04155.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kachroo, A., and Kachroo, P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 54, 43–55. doi: 10.1016/j.molcel.2014.02.021
Katagiri, F., Thilmony, R., and He, S. Y. (2002). “The Arabidopsis thaliana- Pseudomonas syringae interaction,” in The Arabidopsis Book, eds C. R. Somerville and E. M. Meyerowitz (Rockville, MD: American Society of Plant Biologists), 1:e0039. Available online at: http://www.aspb.org/publications/arabidopsis
Kim, M. C., Chung, W. S., Yun, D. J., and Cho, M. J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2, 13–21. doi: 10.1093/mp/ssn091
Knight, H., Trewavas, A. J., and Knight, M. R. (1996). Cold calcium signalling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8, 489–503.
König, S., Mosblech, A., and Heilmann, I. (2007). Stress-inducible and constitutive phosphoinositide pools have distinctive fatty acid patterns in Arabidopsis thaliana. FASEB J. 21, 1958–1967. doi: 10.1096/fj.06-7887com
Krinke, O., Flemr, M., Vergnolle, C., Collin, S., Renou, J. P., Taconnat, L., et al. (2009). Phospholipase D activation is an early component of the salicylic acid signaling pathway in Arabidopsis cell suspensions. Plant Physiol. 150, 424–436. doi: 10.1104/pp.108.133595
Krinke, O., Novotná, Z., Valentová, O., and Martinec, J. (2007a). Inositol trisphosphate receptor in higher plants: is it real?. J. Exp. Bot. 58, 361–376. doi: 10.1093/jxb/erl220
Krinke, O., Ruelland, E., Valentová, O., Vergnolle, C., Renou, J. P., Taconnat, L., et al. (2007b). Phosphatidylinositol 4-kinase activation is an early response to salicylic acid in Arabidopsis suspension cells. Plant Physiol. 144, 1347–1359. doi: 10.1104/pp.107.100842
Lamb, C., and Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Lemtiri-Chlieh, F., MacRobbie, E. A., Webb, A. A., Manison, N. F., Brownlee, C., and Skepper, J. N. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. U.S.A. 100, 10091–10095. doi: 10.1073/pnas.1133289100
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Lu, H. (2009). Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 4. 713–717. doi: 10.4161/psb.4.8.9173
Ma, W., and Berkowitz, G. A. (2007). The grateful dead: calcium and cell death in plant innate immunity. Cell. Microbial. 9, 2571–2585. doi: 10.1111/j.1462-5822.2007.01031.x
Ma, W., and Berkowitz, G. A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190, 566–572. doi: 10.1111/j.1469-8137.2010.03577.x
Ma, Y., Walker, R. K., Zhao, Y., and Berkowitz, G. A. (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 19852–19857. doi: 10.1073/pnas.1205448109
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., and Cameron, R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. doi: 10.1038/nature00962
Manzoor, H., Chiltz, A., Madani, S., Vatsa, P., Schoefs, B., Pugin, A., et al. (2012). Calcium signatures and signaling in cytosol and organelles of tobacco cells induced by plant defense elicitors. Cell Calcium 51, 434–444. doi: 10.1016/j.ceca.2012.02.006
McAinsh, M. R., and Pittman, J. K. (2009). Shaping the calcium signature. New Phytol. 181, 275–294. doi: 10.1111/j.1469-8137.2008.02682.x
Meijer, H. J., and Munnik, T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306. doi: 10.1146/annurev.arplant.54.031902.134748
Mosblech, A., König, S., Stenzel, I., Grzeganek, P., Feussner, I., and Heilmann, I. (2008). Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol. Plant 1, 249–261. doi: 10.1093/mp/ssm028
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957. doi: 10.1111/j.1365-313X.2011.04480.x
Munnik, T., and Nielsen, E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497. doi: 10.1016/j.pbi.2011.06.007
Murphy, A. M., Otto, B., Brearley, C. A., Carr, J. P., and Hanke, D. E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56, 638–652. doi: 10.1111/j.1365-313X.2008.03629.x
Nandi, A., Welti, R., and Shah, J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16, 465–477. doi: 10.1105/tpc.016907
Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., et al. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128. doi: 10.1104/pp.103.036749
Ng, G., Seabolt, S., Zhang, C., Salimian, S., Watkins, T. A., and Lu, H. (2011). Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189, 851–859. doi: 10.1534/genetics.111.132332
Nishimura, M. T., and Dangl, J. L. (2010). Arabidopsis and the plant immune system. Plant J. 61, 1053–1066. doi: 10.1111/j.1365-313X.2010.04131.x
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926. doi: 10.1038/ncomms1926
Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H., and Kinoshita, K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991. doi: 10.1093/nar/gkn807
Park, S. W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Perera, I. Y., Hung, C.-Y., Brady, S., Muday, G. K., and Boss, W. F. (2006). A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiol. 140, 746–760. doi: 10.1104/pp.105.075119
Perera, I. Y., Hung, C.-Y., Moore, C. D., Stevenson-Paulik, J., and Boss, W. F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20, 2876–2893. doi: 10.1105/tpc.108.061374
Perera, I. Y., Love, J., Heilmann, I., Thompson, W. F., and Boss, W. F. (2002). Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant Physiol. 129, 1795–1806. doi: 10.1104/pp.003426
Pokotylo, I., Kolesnikov, Y., Kravets, V., Zachowski, A., and Ruelland, E. (2014). Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 96, 144–157. doi: 10.1016/j.biochi.2013.07.004
Postel, S., and Kemmerling, B. (2009). Plant systems for recognition of pathogen-associated molecular patterns. Semin. Cell Dev. Biol. 20, 1025–1031. doi: 10.1016/j.semcdb.2009.06.002
Qi, Z., Verma, R., Gehring, C., Yamaguchi, Y., Zhao, Y., Ryan, C. A., et al. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. U.S.A. 107, 21193–21198. doi: 10.1073/pnas.1000191107
Qutob, D., Kemmerling, B., Brunner, F., Küfner, I., Engelhardt, S., Gust, A. A., et al. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18, 3721–3744. doi: 10.1105/tpc.106.044180
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signaling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68, 100–113. doi: 10.1111/j.1365-313X.2011.04671.x
Reddy, A. S., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Romeis, T., and Herde, M. (2014). From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 20, 1–10. doi: 10.1016/j.pbi.2014.03.002
Sagi, M., and Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. doi: 10.1104/pp.106.078089
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shah, J. (2005). Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytol. 43, 229–260. doi: 10.1146/annurev.phyto.43.040204.135951
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Stevenson, J. M., Perera, I. Y., Heilmann, I. I., Persson, S., and Boss, W. F. (2000). Inositol signaling and plant growth. Trends Plant Sci. 5, 252–258. doi: 10.1016/S1360-1385(00)01652-6
Tan, X., Calderon-Villalobos, L. I. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Testerink, C., and Munnik, T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361. doi: 10.1093/jxb/err079
Torres, M. A., Jones, J. D. G., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Truman, W., and Glazebrook, J. (2012). Co-expression analysis identifies putative targets for CBP60g and SARD1 regulation. BMC Plant Biol. 12:216. doi: 10.1186/1471-2229-12-216
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Vossen, J. H., Abd-El-Haliem, A., Fradin, E. F., van den Berg, G., Ekengren, S. K., Meijer, H. J., et al. (2010). Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 62, 224–239. doi: 10.1111/j.1365-313X.2010.04136.x
Walley, J. W., Kliebenstein, D. J., Bostock, R. M., and Dehesh, K. (2013). Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 16, 520–526. doi: 10.1016/j.pbi.2013.06.011
Wang, D., Weaver, N. D., Kesarwani, M., and Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. doi: 10.1126/science.1108791
Weigel, R. R., Pfitzner, U. M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17, 1279–1291. doi: 10.1105/tpc.104.027441
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wurzinger, B., Mair, A., Pfister, B., and Teige, M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6, 8–12. doi: 10.4161/psb.6.1.14012
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18220–18225. doi: 10.1073/pnas.1005225107
Zhou, N., Tootle, T. L., Tsui, F., Klessig, D. F., and Glazebrook, J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10, 1021–1030. doi: 10.1105/tpc.10.6.1021
Keywords: Arabidopsis, Ca2+, salicylic acid, phosphoinositides, InsP3, plant defense signaling, SAR
Citation: Hung C-Y, Aspesi P Jr, Hunter MR, Lomax AW and Perera IY (2014) Phosphoinositide-signaling is one component of a robust plant defense response. Front. Plant Sci. 5:267. doi: 10.3389/fpls.2014.00267
Received: 23 April 2014; Accepted: 22 May 2014;
Published online: 11 June 2014.
Edited by:
Vera Bonardi, University of North Carolina at Chapel Hill, USAReviewed by:
Ingo Heilmann, Martin-Luther-University Halle-Wittenberg, GermanyMats X. Andersson, University of Gothenburg, Sweden
Yijian He, NC State University, USA
Copyright © 2014 Hung, Aspesi, Hunter, Lomax and Perera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Imara Y. Perera, Department of Plant and Microbial Biology, North Carolina State University, Box 7612, Raleigh, NC 27695, USA e-mail: imara_perera@ncsu.edu
†Present address: Chiu-Yueh Hung, Department of Pharmaceutical Sciences, Biomanufacturing Research Institute and Technology Enterprise, North Carolina Central University, Durham, USA;
Peter Aspesi Jr, Novartis Institutes for BioMedical Research, Cambridge, USA;
Melissa R. Hunter, Department of Pharmacy Services, Virginia Commonwealth University Medical Center, Richmond, USA;
Aaron W. Lomax, Department of Biochemistry, University of Wisconsin, Madison, USA
 Chiu-Yueh Hung
Chiu-Yueh Hung Peter Aspesi Jr
Peter Aspesi Jr Melissa R. Hunter
Melissa R. Hunter Aaron W. Lomax†
Aaron W. Lomax† Imara Y. Perera
Imara Y. Perera