- Laboratory of Virology, Department of Plant Sciences, Wageningen University, Wageningen, Netherlands
To establish a successful infection plant viruses have to overcome a defense system composed of several layers. This review will overview the various strategies plants employ to combat viral infections with main emphasis on the current status of single dominant resistance (R) genes identified against plant viruses and the corresponding avirulence (Avr) genes identified so far. The most common models to explain the mode of action of dominant R genes will be presented. Finally, in brief the hypersensitive response (HR) and extreme resistance (ER), and the functional and structural similarity of R genes to sensors of innate immunity in mammalian cell systems will be described.
Introduction
During the past decades it has become clear that plants have a unique and complex defense system that consists of several layers, which enables them to avoid, suppress, or actively defend against pathogens from all kingdoms like fungi, bacteria, nematodes, and viruses. Of all plant viruses known, only a few cause serious diseases and, if so, mostly limited to a very small number of crops. In general, most viruses have a limited (natural) host range and the number of so-called non-hosts exceeds those of hosts. In those plants that are hosts, viruses encounter different mechanisms of defense. Some act general against all viruses and this response is part of the innate immune system, while others are virus-specific and involve resistance genes. Triggering of the latter simultaneously mediates a rapid necrosis at the site of virus entry and prevents further spread of the virus throughout the host. In several cases resistance genes do not confer absolute resistance and low levels of virus replication can still be observed. In those cases the genes are referred to as partial resistance genes or tolerance genes.
While throughout the years reviews on resistance genes have appeared with regular intervals, these mostly had their main focus on fungal and bacterial resistance genes, primarily due to the large amount of data available. This review aims to present an overview on the current status of resistance genes against plant viruses, with emphasis on single dominant resistance genes. The very basis of plant pathogens (among others plant viruses) not being able to infect all plants is due to a mechanism called non-host resistance (NHR) (For an extensive review on this, see Uma et al., 2011). NHR holds for all plant pathogens and is a generic, nonspecific resistance that can be divided into two main types, distinguished by the mechanism and mode of recognition (Mysore and Ryu, 2004). Type 1 is the most pre-dominant type of NHR and presents a basic defense mechanism that prevents pathogen invasion, e.g., thickening of the cell-wall, secondary metabolite production, etc. This type of resistance usually is symptomless. In contrast, type 2 NHR is associated with induction of necrosis at the site of infection, and is induced when pathogens overcome type 1 resistance. Here, the pathogen is recognized through specific structures or proteins that are associated with the pathogen. The recognition of these structures/proteins, so called microbe associated molecular patterns (MAMPs) or PAMPS (Pathogen), takes place by pattern recognition receptors (PRRs) on plant plasma membranes. These PRRs recognize conserved structures of pathogens, like flaggelin from the flagella of bacteria or chitin from the cell wall of fungi, and induce a so called PAMP triggered immunity (PTI) response (Jones and Dangl, 2006). Since plant viruses need to overcome the physical barrier of a cell wall, they enter their host cells either via mechanical inoculation or the infection is mediated by vectors like insects, nematodes, or even fungi. Direct recognition of viruses probably does not occur in the apoplast. However, a study recently reported on the possible involvement of (intracellular) receptor-like kinases (RLKs), of the like that are involved in PAMP recognition by PRRs, in plant-virus interactions (Kørner et al., 2013).
One of the first innate immune responses all plant viruses encounter when invading a host consists of antiviral RNA silencing [also called RNA interference (RNAi) and in the very early days post-transcriptional gene silencing (PTSG)]. RNA silencing is a host response triggered by double stranded (ds)RNA. These molecules thus act as a MAMP/PAMP and in which RNAi can be regarded as PTI. The main difference with pathogens such as fungi and bacteria is that recognition of viral MAMPs/PAMPs occurs intracellularly (Ding and Voinnet, 2007).
RNA silencing consists of two major “branches”; the first one is that of small-interfering (si)RNAs, and one of the hallmarks for antiviral RNAi, and the second one is that of (host-gene encoded) micro (mi)RNAs involved in gene regulation. The antiviral RNAi response is induced by viral double stranded (ds)RNA molecules that arise from replicative intermediates or secondary RNA folding structures. These structures are sensed by a host RNase type III-like enzyme called Dicer-like (DCL) protein and cleaved into short interfering (si)RNA of 21–24 nucleotides (nt) in size (Sharma et al., 2013). The siRNAs generated are unwound and only one strand, the so-called guide-strand, is uploaded into a functional protein complex termed RNA-induced silencing complex (RISC). This activated complex next surveils and subsequently degrades (viral) RNA target molecules with sequence complementarity to the guide-strand. Degradation of the target RNA is mediated by slicer, the core component of RISC, which is represented by a member of the Argonaut (AGO) family of proteins (Vaucheret, 2008; Sharma et al., 2013). After primary siRNAs have been generated, in plants an amplification of siRNAs follows, which is required to mount an RNAi response to effectively combat virus infections locally and systemically. This amplification involves host RNA dependent RNA polymerases (RDRs) that are able to convert (aberrant) viral (m)RNAs into dsRNA in a siRNA-dependent and -independent manner (Csorba et al., 2009). Their subsequent processing by DCL leads to the generation of secondary siRNAs that correspond to sequences outside the primary target sequence, a process also called transitive silencing (Sijen et al., 2001). The antiviral RNAi response acts against all RNA and DNA viruses (Incarbone and Dunoyer, 2013), but in general is a relatively slow process that does not lead to complete clearance of viral infections. For an extensive description of RNAi readers are referred to nice reviews from e.g., Ding (2010) and Sharma et al. (2013).
Besides RNAi, viruses may also run into another, second layer of defense that involves resistance genes. While most of these are triggered by and confer resistance to a specific virus only, some act against several (related) viruses. The major class of these genes represent single dominant resistance genes (and of which the biggest group consists of the NB-LRR type), while others are recessive, tolerance, or partial resistance genes. A very nice example of a dominant resistance gene of the latter case has recently been described with the cloning and characterization of the Ty-1 resistance gene from tomato against Tomato yellow leaf curl geminivirus (TYLCV) This gene encodes an RNA-dependent RNA polymerase (RdRp) and is proposed to confer resistance to TYLCV by amplifying the RNAi signal (Verlaan et al., 2013). Tomato plants containing Ty-1 do not show symptoms upon a challenge with TYLCV, but low levels of virus can still be detected.
Recessive resistance (Truniger and Aranda, 2009) acting against viruses, relies on the observation that viruses require host factors (also called susceptibility factors) to enable an infection. The inability of interaction between such host factor and the virus leads to resistance. Since susceptibility factors are dominant, a resistance based on these requires all gene copies to be in the (resistant) recessive state. This explains why such resistance is generally termed recessive resistance. The majority of the recessive resistance genes known against plant viruses have been reported for potyviruses (Kang et al., 2005) and encode translation initiation factors of the 4E or 4G family (eIF4E/eIF4G) (Truniger and Aranda, 2009). The latter proteins need to interact with the cap-structure on (viral) transcripts, to allow for translation. Potyviral transcripts do not contain a cap structure, but provide a VPg (Virus-protein genome linked) to render their transcripts translatable in a cap-independent manner. Potyvirus infection leads to host shut off of cap-dependent transcripts, but only allow the cap-independent transcripts to be translated mediated by a subgroup of translation initiation factors; eIF(iso)4E/G. Viruses that encode their own cap-like structure (like potyviruses: VPg) require interaction with the translation initiation factors eIF4E/eIF4G for translation, this in turn induces a selection pressure on the host to escape the interaction between VPg and eIF4e, leading to recessive resistance. Recessive resistance genes toward other pathogens, such as fungi and bacteria have only been described to a limited extent and their encoded susceptibility factors (S genes) are proposed to provide a more durable resistance than dominant R genes. However due to their functions they may cause pleiotropic effects when knocked out from the host genome (Gawehns et al., 2013).
Dominant Resistance
Effector-Mediated Triggering of Single Dominant Resistance Genes
Plant pathogens need to evade or suppress the PTI response in plants and achieve the latter by encoding effector proteins that can interfere with the recognition by PRRs, usually by binding to the substrate that PRRs would otherwise recognize. This process allows the pathogen to establish a successful infection, and is referred to as Effector Triggered Susceptibility (ETS) (Figure 1): a strategy that also applies to antiviral RNAi. One of the most common strategies plant viruses use to counteract RNAi is to encode RNA silencing suppressors (RSS), viral proteins that interfere with a specific part of the RNAi pathway and thereby reduce its effectiveness against plant viruses (Burgyan and Havelda, 2011). The majority of plant virus RSS proteins exert this activity through binding of small interfering (si)RNAs, or sometimes (also) long dsRNA, and thereby prevent their uploading into RISC and Dicer-cleavage, respectively (Lakatos et al., 2006). In recent years some RSS have also been discovered to inhibit the RNAi pathway in other ways, e.g., by binding directly to key-enzyme proteins like AGO1, the core component of RISC during the antiviral RNAi response (Zhang et al., 2006; Giner et al., 2010). Viral suppression of RNAi leads to a stage of ETS during which viruses are able to establish a successful infection.
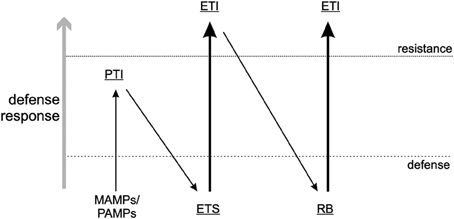
Figure 1. Zig-zag-model. A visual presentation of the arms race between pathogen and host according to Jones and Dangl (2006). Here, a slightly modified version of that model is presented and as described in this review. MAMPs/PAMPs, Microbe/Pathogen associated molecular patterns; PTI, PAMP triggered immunity; ETS, Effector triggered susceptibility; ETI, Effector-triggered immunity; RB, resistance breaker.
Single dominant resistance (R) gene products (in)directly sense the presence of a specific pathogen by their effector, termed avirulence factors (Avr), as a counter defense against ETS, leading to a stage called Effector-Triggered Immunity (ETI) (Figure 1). Triggering of R genes is generally associated with a (concomitant) induction of a programmed cell death response, as visualized by the rapid appearance of necrotic lesions (a hypersensitive response, HR) or in rare occasions extreme resistance (ER) during which no necrosis is observed at all. However, more and more evidence is presented, that there is an uncoupling of the resistance response from the programmed cell death response, although both can work in concert. Due to these responses, viruses (and other pathogens) are confined to the site of entry/invasion where infections are prevented. In contrast to the slower onset of antiviral RNAi, the R gene response generally is rapid and within ~3/4 days lead to containment of the virus.
Dominant R genes basically can be grouped into two classes, namely those that encode NB-LRRs and all others. The major class of R genes consists of the NB-LRR type and encode proteins that, irrespective of the pathogen they recognize, consist of three domains; (1) the Nucleotide Binding Site (NBS) in the center of the protein, (2) a Leucine Rich Repeat (LRR) at the C-terminal end, and (3) a Coiled-coil (CC) or Toll and Interleukin-1 Receptor (TIR) domain at the N-terminal end of the resistance gene product (Moffett, 2009). The LRR determines the specificity of the target protein and is the most variable part of the protein, therefore considered to be under selection pressure to evolve for recognition of (new) target proteins. The NBS is composed of a conserved part that contains the Nucleotide Binding site (NB) and an ARC-domain, both required to bind and hydrolyze ATP. R genes that contain an N-terminal TIR domain are only found in dicots from the angiosperms (Collier et al., 2011), and through this domain share homology to Toll-like receptor (TLR) proteins, that act as PRRs in the innate immunity response in animal systems. Those with no predicted structure at its N-terminus, are grouped with the CC-domain (Maekawa et al., 2011a; Hao et al., 2013) in the non-TIR group. All three domains are involved in an interaction with each other and change conformationally upon activation to subsequently induce the resistance response (Lukasik and Takken, 2009; Slootweg et al., 2010).
Only a few cases have been described in which the dominant R gene product recognizes an Avr protein through direct interaction (Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Krasileva et al., 2010; Chen et al., 2012; Cesari et al., 2013), of which one is the TMV-p50 helicase domain (Ueda et al., 2006). In the majority of known R genes recognition of the pathogen occurs indirectly and involves host proteins, which are considered guardees, decoys, or baits, depending on the model, as further discussed below (Model of R Gene Recognition) (Van Der Biezen and Jones, 1998; Jones and Dangl, 2006; Van Der Hoorn and Kamoun, 2008; Collier and Moffett, 2009).
Cloned R Genes and their known Avr Determinants
While for fungi and bacteria many resistance genes have been cloned and characterized, resistance genes against plant viruses have received growing interest during the last two decades, still only few of the latter have been cloned so far. Table S1.1 gives an up-to-date summary of all R genes against plant viruses, known or currently under investigation. For some of these genes the viral Avr determinant has been identified. From this large, extensive list of R genes (>200), only 22 have been cloned and characterized. Some R genes have functional alleles in other plant species, often showing a similar Avr recognition. The majority of the known R gene products are of the CC-NB-LRR type, whereas only a small group belongs to the TIR-NB-LRR group (Table 1).
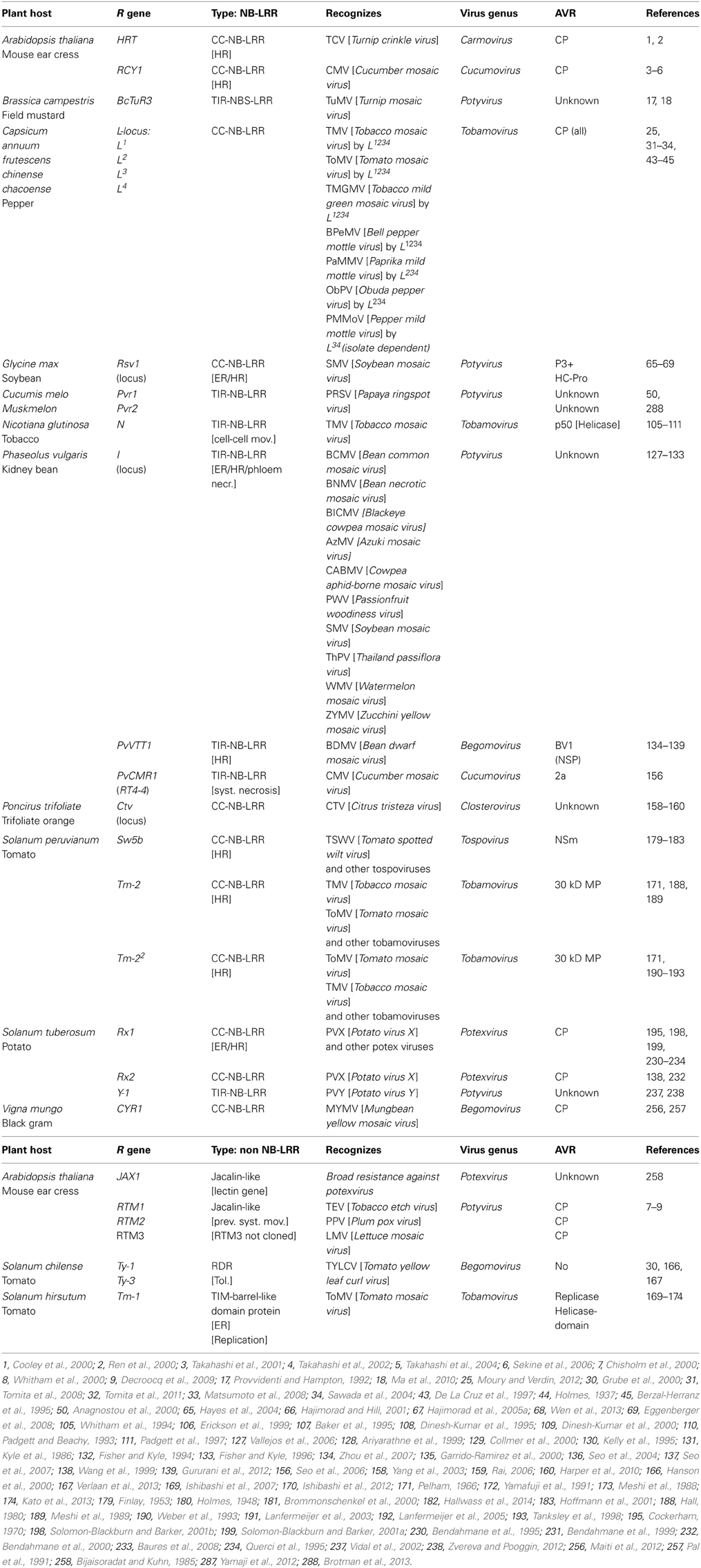
Table 1. Cloned dominant resistance genes against plant viruses, organized into the NB-LRRs and the non-NB-LRRs, and their Avr determinants (when identified).
A few dominant R genes against viruses have been described that do not belong to the NB-LRR type of genes, e.g., RTM1, RTM2, and RTM3. Latter resistance genes have been identified from A. thaliana and prevent the systemic spread of several potyviruses. In those cases the virus is not able to upload into the phloem to systemically disseminate into the host. In addition, there is also no induction of HR or production of salicylic acid (SA), as commonly observed with NB-LRR mediated resistance responses (Cosson et al., 2012). No direct interaction occurs between the RTM proteins with the potyvirus CP (Avr) protein. A resistance gene recently identified is JAX, a lectin gene that resembles the RTM gene based resistance and works broadly against potexviruses in A. thaliana, indicating an important role for lectins in plant immunity (Yamaji et al., 2012). Another type of a distinct R gene is Tm-1, found in the wild tomato species S. hirsutum, encoding a protein that contains a TIM-barrel. This barrel binds the replication proteins of Tomato mosaic virus (ToMV) and thereby inhibits RNA replication (Ishibashi et al., 2007). Also here, no typical NB-LRR type-associated response, like HR, is induced. Many homologs of Tm-1 are found in other organisms from different kingdoms, like fungi, archae, and bacteria, suggesting that this gene (originally) presents a housekeeping gene (Ishibashi et al., 2012). Both RTM and Tm-1 seem to play a role in the inhibition of a specific step required for successful infection by the virus. Whether these present a new class of dominant resistance genes remains to be determined.
From only a 1/3 of the total number of R genes directed against plant viruses, the virus Avr determinant is identified (Tables 1 and S1.1). Interestingly, functionally quite different viral proteins act as Avr determinants. Several R genes belong to the same locus (for instance the L-proteins in Capsicum spec.) or clearly act as homologs (Rx1 and Rx2 from S. tuberosum) and recognize the same Avr protein from overlapping virus species, indicating that these conserved R proteins are able to recognize similar structures but with an adapted spectrum (Bendahmane et al., 1995, 2000; Moury and Verdin, 2012). For several viruses, their corresponding R genes have not been identified yet, but their single dominant nature is deduced from the observation that an HR is being triggered. In some of these cases, the viral gene responsible for the induction of resistance, as indirectly monitored by HR, has been identified.
As described before and clear from Table 1, many different viral proteins can act as Avr determinants; whether it is the coat protein (e.g., L-locus from Capsicum against Tobamoviruses), the movement protein (e.g., Tm-2/Tm-22 from tomato against Tobamoviruses), the replicase protein (e.g., Tm-1 from tomato against Tobacco mosaic virus) or the RNAi suppressor protein (e.g., HRT from A. thaliana against Turnip crinkle virus), all potentially can act as elicitor of resistance (Meshi et al., 1989; Ishibashi et al., 2012; Moury and Verdin, 2012). Interestingly, for a majority of cases the ability to induce the resistance, as monitored by visual HR, could be uncoupled from the endogenous function of the viral protein but exceptions exist.
While the function of a viral protein is not a selective criterium to act as Avr-determinant, the “Zig-zag-model” by Jones and Dangl (2006) (Figure 1) implies that ETI (R gene mediated resistance) is a response to ETS and governed by effectors, i.e., molecules that act as virulence factors and contribute to (enhance) pathogen fitness. It is obvious that in case of RNAi as a PTI response against viruses, viral Avr proteins containing RSS activity contribute to virus fitness as a result of PTI suppression and thereby initiating ETS. On the other hand, some viral Avr proteins lack RSS activity which would indicate that effectors not necessarily would have to suppress PTI (RNAi) to contribute to virulence, as observed with several bacterial effectors. Unless, instead of RNAi, another innate immune response is being counteracted that is triggered via the activation of different intracellular PAMP receptors (e.g., in analogy to animal TLRs, see below).
Model of R Gene Recognition
Although the mode of action of resistance genes still remains a matter of debate, models have been proposed for the triggering of the largest and most studied group of the NB-LRR type of dominant R genes. One of the most commonly accepted models is the “guard hypothesis” (Van Der Biezen and Jones, 1998; Jones and Dangl, 2006). In this model the resistance gene product guards a certain host protein, the “guardee,” and perceives alterations of this protein upon interaction with the Avr determinant to subsequently initiate a resistance response. It is possible that multiple R genes guard the same guardee, possibly vice versa as well, which thereby broadens the resistance spectrum of (a limited number of) R genes to a wide range of various pathogens; e.g., Rx1 and GPa2 both interact with the same guardee RanGAP2 (Tameling and Baulcombe, 2007; Moffett, 2009). Unfortunately, this model does not explain how resistance breaking virus isolates maintain their virulence. For this reason, alternative models have been postulated. According to the “decoy model” (Van Der Hoorn and Kamoun, 2008), a (proteinaceous) decoy evolved to act as a molecular sensor to only detect a pathogen without having any other role in the household machinery of the host. The “bait and switch model” and the similar “mousetrap model” have been more recently postulated and proposes that the R gene product in an “OFF” state forms a complex together with the guardee/decoy protein, that upon interaction of the Avr protein with the complex leads to a conformational switch (“ON”) and activates a downstream signaling pathway leading to resistance (Collier and Moffett, 2009; Lukasik and Takken, 2009). Recent studies on the resistance gene Rx from potato against PVX have shown that indeed intramolecular interactions keep the R gene product in an inactive state, while interaction of the effector protein releases these interactions and thereby activating the resistance downstream (Bendahmane et al., 2002; Moffett et al., 2002; Lukasik and Takken, 2009; Slootweg et al., 2013).
The downstream mechanism after resistance induction still remains unclear. However, one described case of the R gene from tobacco, the N gene, has revealed some of the downstream ways of controlling virus replication and obtaining resistance. The N gene encodes a TIR-NB-LRR protein and confers resistance against TMV and, upon transient co-expression with the p50 elicitor (helicase), an HR is induced in N. tabacum, a response that does not occur in N. benthamiana. Bhattacharjee et al. (2009) employed this observation in a series of experiments to dissect and assign downstream signaling of defense responses, related to the R gene. The studies indicated that the N gene based antiviral response leads to a translational arrest of viral transcripts by a process that involves Argonaute 4 (AGO4). As a result, synthesis of viral proteins is inhibited, ultimately preventing virus accumulation and spread. Whether this mechanism is generic to all R genes against plant-viruses remains to be investigated.
More recently, two independent studies were published that showed that the translation of R genes is tightly controlled through the activity of miRNAs. One study showed the miR482/2118 superfamily negatively controlled the translation of NB-LRR proteins by targeting its P-loop motif (Shivaprasad et al., 2012), while Li et al. (2012) showed that other miRNA families controlled the translation of NB-LRR proteins as well, with the TIR-NB-LRR protein N as example. During the on-going “arms race” between virus and host, viruses counter-defend against PTI/antiviral RNAi by their RSS proteins, some of which exhibit strong affinity to bind small (si- and mi-)RNAs. As a consequence such RSS proteins may suppress the miRNA induced silencing of R genes, and lead to enhanced expression of the R genes and induction of ETI. Considering that high expression levels of R genes lead to auto-immunity (Xia et al., 2013), viral RSS proteins with affinity to sRNAs thus may play a major role in the induction of HR. However, viral RSS are clearly not the only criterium as observed by the virus-specific activation of R genes and final HR, which indicates that a more complex interplay between viral effectors and R gene products is (additionally) required.
Downstream Defense Responses
Dominant R genes trigger a hypersensitive response (HR) or an extreme response (ER) in case the reaction occurs in a single cell. Both involve a programmed cell death (PCD) response that rapidly kills infected cells and prevents systemic spread of the (virus) pathogen. An induced HR is quite characteristic and involves the activation and expression of SA, jasmonic acid (JA), nitride oxide (NO), ethylene, reactive oxygen species (ROS), and Ca2+, and expression of Pathogenesis Related (PR)-genes. While each component has a specificity toward certain pathogens, only SA, ROS, and Ca2+ seem to be effective against viruses (Loebenstein, 2009; Carr et al., 2010).
In the past, an HR was considered to be part of the resistance response, however, recent insights into R protein downstream signaling indicate that programmed cell death (HR) and resistance are distinct physiological pathways (Bendahmane et al., 1999; Bai et al., 2012). One of the best examples in support of this comes from studies on Rx-based resistance against PVX. The Rx gene product is a CC-NB-LRR protein from potato that is triggered by the PVX structural CP protein. The Rx protein localizes in the cytoplasm while shuttling to and from the nucleus thereby triggering resistance (Slootweg et al., 2010). Although an HR is monitored, this response can be knocked out without affecting Rx-mediated resistance against PVX (Bendahmane et al., 1999). Another example is the N-gene mediated resistance against TMV as described above in section Model of R Gene Recognition (Bhattacharjee et al., 2009). Similar observations have been made by others (Cole et al., 2001; Cawly et al., 2005; Genger et al., 2008; Bulgarelli et al., 2010; Bai et al., 2012) and indicate that the actual resistance response is different from an HR, although both mostly are triggered and may act in concert to clear viral invasions. Whether both are triggered by a pathogens' Avr determinant or whether HR is sequentially triggered following the R gene response is not clear.
While several interacting proteins have been identified that control R protein activity in the absence of pathogens (e.g., RAR1, SGT1, WRKY1, TPR1, Hsp90), more recently it has been found that there are also proteins that modulate the strength of defense responses (RanGAP, EDS1-PAD4) (Lu et al., 2003; Wiermer et al., 2005; Sacco et al., 2009). The benefit for the plant in a modulated fine-tuning of the ETI response to specific pathogens lies in improved effector sensing and minimizing the fitness costs involved with certain defense responses (free radical production, defense protein synthesis, cell death) (Padmanabhan and Dinesh-Kumar, 2010). While R gene mediated defense is taking place locally at the site of entry, it is also able to induce defense signaling responses in distally located tissues, known as systemic acquired resistance (SAR) (Vlot et al., 2008). For both the N gene in tobacco and Rx1 in potato, SAR has been demonstrated (Delaney et al., 1994; Liu et al., 2010) and in both cases this response is mediated by the SA-dependent pathway as a mobile signal. SAR also prevents infection by other pathogens in the host by activating PR genes in the systemic tissue, which are used as a hallmark of SAR and were shown to have antimicrobial activity, although a direct inhibition on virus replication has not been shown (Durrant and Dong, 2004; Loebenstein, 2009; Carr et al., 2010).
Functional and Structural Homology of Plant- and Animal-Sensors of Innate Immunity
Viruses are pathogens to many different organisms and, irrespective of the host species they infect, often share similarities in genome organization and functions of encoded proteins. A good example of this is exemplified by viruses from the Bunyaviridae family where all members infect animals with the exception of those from the Tospovirus genus that, besides infecting their thrips vector, are plant pathogenic and are postulated to have evolved from a common ancestor. Likewise, as a result of co-evolution driven by host-pathogen interactions, plants and animals show some similarities in their innate immune sensory systems. While in plants the aforementioned R genes are important in mounting an ETI response, in animals two major classes are distinguished that (partially) share similarity to these R genes, however both function as PRRs in the PTI response. The first major class present the “nucleotide-binding domain and leucine-rich repeat”-proteins (NLRs) and the second class is that of Toll like receptors (TLRs), which are all found to function as PRRs in the PTI response. Both are immune receptors aimed at detecting “foreign” structures and activating downstream defense responses. The family of NLRs share the most homology, as evidenced when looking at R genes from plants and NACHT-LRR encoding genes from the animal kingdom (NAIP–CIITA–HET-E–TP1 domain) (Leipe et al., 2004; Takken et al., 2006; Maekawa et al., 2011b). They both contain a nucleotide binding domain and a LRR (Maekawa et al., 2011b) (Figure 2). Additionally, plant R proteins also share homology at their N-termini with animal TLRs, membrane-bound immune receptors that function as sensors in pathogen recognition across membranes.
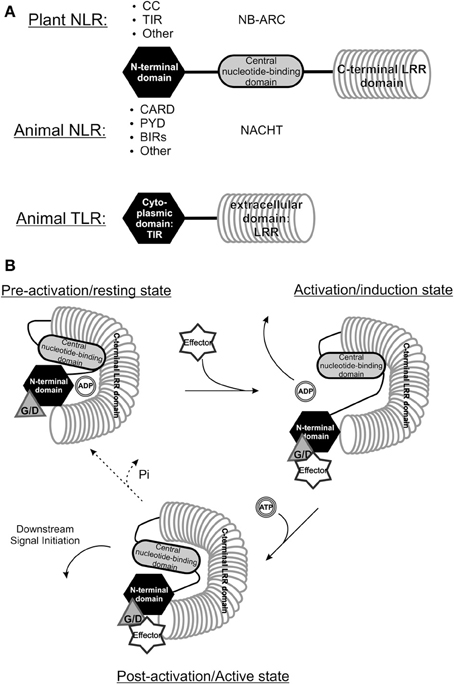
Figure 2. Comparison between the structure of plant and animal NLRs. (A) The structure of “Nucleotide binding and leucine rich repeat proteins” (NLRs) from the animal and plant kingdom share highest homology, as all proteins belonging to this class have a C-terminal leucine rich repeat (LRR), a central nucleotide binding domain and a varying N-terminal domain (modified from Maekawa et al., 2011b). Animal TLRs also contain an (extracellular) LRR domain and possess a TIR-domain, they do however, lack a nucleotide binding domain. CC, Coiled-coil; TIR, Toll-interleukin receptor; CARD, Caspase-activation and recruitment domain; PYR, Pyrin domain; BIR, Baculovirus inhibitor-of-apoptosis repeats; NB-ARC, Nucleotide binding and Apaf1-R protein-CED4 domain; NACHT, NAIP – CIITA - HET-E - TP1 domain. (B) A model of NB-LRR R protein recognizing a specific Avr protein through a guardee or decoy host protein. Upon interaction with the Avr protein the R protein conformationally changes and the ADP can be exchanged for ATP, leading to a second conformational change triggering downstream resistance (Modified from Lukasik and Takken, 2009). Whether the R protein returns to its resting state is not known yet. G/D, Guardee/Decoy.
The NLRs of both plant and animal kingdom share homology through the presence of the Leucine-rich repeats (LRR) in these proteins. The most prevalent type of R proteins in plants belong to the NB-LRR protein structural class, from which the central nuclear binding domain (NBS) exhibits similarity to the nucleotide binding domain in several metazoan apoptosis regulating proteins like Apaf-1 from mammals and CED-4 from C. elegans. Due to the latter the NBS domain is also often referred to as NB-ARC domain (from Apaf1–R-protein–CED4) (Van Der Biezen and Jones, 1998; Takken et al., 2006). The N-terminal domain furthermore separates different classes of R genes; TIR-NB-LRRs harbor a Toll/Interleukin-1 Receptor domain with similarity to metazoan TLRs (Burch-Smith et al., 2007; Bernoux et al., 2011; Maekawa et al., 2011b; Hao et al., 2013). CC-NB-LRRs contain coiled coil domain forming the more irregular shaped intertwined alpha-helices (Lupas, 1997). Parallel to the discovery of many NB-LRR encoding R genes in plants in the recent years, the search for homology to Apaf-1 and CED-4 resulted in the recognition of the NACHT-LRR protein family in vertebrates (Koonin and Aravind, 2000; Leipe et al., 2004). Animal NLRs activate caspase-1 leading to activation and release of the cytokine interleukin-1 beta (Case, 2011), which subsequently induces local and systemic immune reactions. Similar to plant NB-LRR proteins, NLRs were found to act as higher-order active complexes, e.g., NLRP1-3 and NLRC4 form a complex often termed the inflammasome (Maekawa et al., 2011b).
TLRs represent the best studied family of PRRs in mammals so far. They are transmembrane glycoprotein receptors with an extracellular PAMP-binding domain consisting of multiple LRR that fold into a “horseshoe” structure. Additionally, it possesses intracellular signaling regions that have similarity to the intracellular domain of the Interleukin-I receptor ((IL-1R), which is referred to as Toll/IL-1R (TIR) domain that mediates downstream signaling upon activation of the receptor. TLRs initiate signal cascades involving the activation of nuclear factor kappa b (NF-κ B), mitogen-activated protein kinase (MAPK), and interferon regulatory factors (IRFs). This subsequently leads to a concerted expression of interferons, cytokines, and chemokines. Finally, inflammatory processes, cell cycle arrest, and cell death are induced (Honda et al., 2005; Kaisho and Akira, 2006). In humans, 10 TLRs have been identified of which TLR2, -3, -4, -7, and -8 are involved in sensing structural components of RNA viruses like double-stranded RNA, single-stranded RNA and viral glycoproteins (Bowie and Unterholzner, 2008). While most TLRs are involved in extracellular recognition of PAMPs, TLR3, -7, and -8 are primarily restricted to intracellular compartments (endoplasmic-reticulum (ER), endosomes etc.) where they sense structural components of viral RNA. Besides TLRs cytosolic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs; RIG-I, MDA5, and LSP2) have been identified as sensors of RNA viruses and are involved in the very early response of some RNA viruses (Bowie and Unterholzner, 2008; Gerlier and Lyles, 2011; Jensen and Thomsen, 2012).
Pathogen recognition in both animal and plant kingdoms involves the LRR domain, which binds the ligand in its horseshoe shape, often followed by activation of a signaling cascade through kinase phosphorylation. Structural similarities between animal TLR/NLR and plant NB-LRR proteins point to a convergent evolution of these defense-related pathways (Yue et al., 2012). However innate immunity in animals and plants differs substantially in their downstream defense response, with interleukin/interferon-activated inflammatory responses combined with activating the adaptive immune system in mammalian systems and a resistance response (as explained before) often seen as a programmed cell death response in form of HR in plants.
Outlook
Dominant resistance against plant viruses are of increasing interest to breeders and scientists in the past years. More and more is known about the molecular mechanisms behind R gene mediated defense and the induction of the HR. However, the number of well-studied examples is still very limited, and therefore makes it difficult to extrapolate to other less studied R genes. In depth studies on other R genes from different crops providing resistance against different phytopathogens will have to show whether there is common mechanism of defense shared between all R genes, or whether specialization between different classes of R genes occurs. Knowledge on mammalian innate immunity sensors and their mode of action may provide interesting and cross pollinating views in this. The rapid generation of resistance breaking virus isolates against dominant resistance genes already indicates the importance for alternative resistance genes, to provide more durable, and effective resistances.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00307/abstract
References
Acosta-Leal, R., Bryan, B. K., Smith, J. T., and Rush, C. M. (2010). Breakdown of host resistance by independent evolutionary lineages of Beet necrotic yellow vein virus involves a parallel C/U mutation in its p25 gene. Phytopathology 100, 127–133. doi: 10.1094/PHYTO-100-2-0127
Amiri, R., Moghaddam, M., Mesbah, M., Sadeghian, S. Y., Ghannadha, M. R., and Izadpanah, K. (2003). The inheritance of resistance to Beet necrotic yellow vein virus (BNYVV) in B. vulgaris subsp maritima, accession WB42: Statistical comparisons with Holly-1-4. Euphytica 132, 363–373. doi: 10.1023/A:1025063625350
Anagnostou, K., Jahn, M., and Perl-Treves, R. (2000). Inheritance and linkage analysis of resistance to Zucchini yellow mosaic virus, Watermelon mosaic virus, Papaya ringspot virus and powdery mildew in melon. Euphytica 116, 265–270. doi: 10.1023/A:1004005716806
Angel, C. A., Hsieh, Y.-C., and Schoelz, J. E. (2011). Comparative analysis of the capacity of Tombusvirus P22 and P19 proteins to function as avirulence determinants in Nicotiana species. Mol. Plant Microbe Interact. 24, 91–99. doi: 10.1094/MPMI-04-10-0089
Angel, C. A., and Schoelz, J. E. (2013). A survey of resistance to Tomato bushy stunt virus in the genus Nicotiana reveals that the hypersensitive response is triggered by one of three different viral proteins. Mol. Plant Microbe Interact. 26, 240–248. doi: 10.1094/MPMI-06-12-0157-R
Ariyarathne, H. M., Coyne, D. P., Jung, G., Skroch, P. W., Vidaver, A. K., Steadman, J. R., et al. (1999). Molecular mapping of disease resistance genes for halo blight, common bacterial blight, and bean common mosaic virus in a segregating population of common bean. J. Am. Soc. Hortic. Sci. 124, 654–662.
Baerecke, M. L. (1967). Hypersensitivity to the S-virus of potato in a Bolivian andigena clone. Der Züchter 37, 281–286.
Bagnall, R. H., and Young, D. A. (1972). Resistance to virus S in potato. Am. Potato J. 49, 196. doi: 10.1007/Bf02864739.
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Baker, B., Dineshkumar, S. P., Corr, C., and Whitham, S. (1995). Isolation and characterization of the Tobacco mosaic virus resistance gene N. Plant Physiol. 108, 2.
Barker, H. (1996). Inheritance of resistance to potato viruses Y and A in progeny obtained from potato cultivars containing gene Ry: evidence for a new gene for extreme resistance to PVA. Theor. Appl. Genet. 93, 710–716. doi: 10.1007/BF00224066
Barker, H. (1997). Extreme resistance to Potato virus V in clones of Solanum tuberosum that are also resistant to Potato viruses Y and A: evidence for a locus conferring broad-spectrum potyvirus resistance. Theor. Appl. Genet. 95, 1258–1262. doi: 10.1007/s001220050690
Bashir, M., Iqbal, M. S., Ghafoor, A., Ahmad, Z., and Qureshi, A. S. (2002). Variability in cowpea germplasm for reaction to virus infection under field conditions. Pak. J. Bot. 34, 47–48.
Baures, I., Candresse, T., Leveau, A., Bendahmane, A., and Sturbois, B. (2008). The Rx gene confers resistance to a range of potexviruses in transgenic Nicotiana plants. Mol. Plant Microbe Interact. 21, 1154–1164. doi: 10.1094/MPMI-21-9-1154
Ben Chaim, A., Grube, R. C., Lapidot, M., Jahn, M., and Paran, I. (2001). Identification of quantitative trait loci associated with resistance to Cucumber mosaic virus in Capsicum annuum. Theor. Appl. Genet. 102, 1213–1220. doi: 10.1007/s001220100581
Bendahmane, A., Farnham, G., Moffett, P., and Baulcombe, D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. doi: 10.1046/j.1365-313X.2002.01413.x
Bendahmane, A., Kanyuka, K., and Baulcombe, D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. doi: 10.1105/tpc.11.5.781
Bendahmane, A., Kohn, B. A., Dedi, C., and Baulcombe, D. C. (1995). The coat protein of Potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 8, 933–941. doi: 10.1046/j.1365-313X.1995.8060933.x
Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D. C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. doi: 10.1046/j.1365-313x.2000.00654.x
Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., et al. (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211. doi: 10.1016/j.chom.2011.02.009
Berzal-Herranz, A., De La Cruz, A., Tenllado, F., Diaz-Ruiz, J. R., Löpez, L., Sanz, A. I., et al. (1995). The Capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 209, 498–505. doi: 10.1006/viro.1995.1282
Bhattacharjee, S., Zamora, A., Azhar, M. T., Sacco, M. A., Lambert, L. H., and Moffett, P. (2009). Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 58, 940–951. doi: 10.1111/j.1365-313X.2009.03832.x
Bijaisoradat, M., and Kuhn, C. W. (1985). Nature of resistance in soybean to Cowpea chlorotic mottle virus. Phytopathology 75, 351–355. doi: 10.1094/Phyto-75-351
Boerma, H. R., and Kuhn, C. W. (1976). Inheritance of resistance to Peanut mottle virus in soybeans. Crop Sci. 16, 533–534. doi: 10.2135/cropsci1976.0011183X001600040024x
Bowie, A. G., and Unterholzner, L. (2008). Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8, 911–922. doi: 10.1038/nri2436
Brigneti, G., Garciamas, J., and Baulcombe, D. C. (1997). Molecular mapping of the Potato virus Y resistance gene Ry(sto) in potato. Theor. Appl. Genet. 94, 198–203. doi: 10.1007/s001220050400
Brommonschenkel, S. H., Frary, A., Frary, A., and Tanksley, S. D. (2000). The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 13, 1130–1138. doi: 10.1094/MPMI.2000.13.10.1130
Brotman, Y., Normantovich, M., Goldenberg, Z., Zvirin, Z., Kovalski, I., Stovbun, N., et al. (2013). Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to Papaya ring spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 6, 235–238. doi: 10.1093/mp/sss121
Brown, R. N., Bolanos-Herrera, A., Myers, J. R., and Jahn, M. M. (2003). Inheritance of resistance to four cucurbit viruses in Cucurbita moschata. Euphytica 129, 253–258. doi: 10.1023/A:1022224327064
Bruening, G. (2011). Not as they seem. Annu. Rev. Phytopathol. 49, 1–16. doi: 10.1146/annurev-phyto-072910-095329
Bruening, G., Ponz, F., Glascock, C., Russell, M. L., Rowhani, A., and Chay, C. (1987). Resistance of cowpeas to Cowpea mosaic virus and to Tobacco ringspot virus. Ciba Found. Symp. 133, 23–37.
Bulgarelli, D., Biselli, C., Collins, N. C., Consonni, G., Stanca, A. M., Schulze-Lefert, P., et al. (2010). The CC-NB-LRR-type Rdg2a resistance gene confers immunity to the seed-borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS ONE 5:e12599. doi: 10.1371/journal.pone.0012599
Burch-Smith, T. M., Schiff, M., Caplan, J. L., Tsao, J., Czymmek, K., and Dinesh-Kumar, S. P. (2007). A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5:e68. doi: 10.1371/journal.pbio.0050068
Burgyan, J., and Havelda, Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. doi: 10.1016/j.tplants.2011.02.010
Buss, G. R., Roane, C. W., Tolin, S. A., and Vinardi, T. A. (1985). A second dominant gene for resistance to Peanut mottle virus in soybeans. Crop Sci. 25, 314–316. doi: 10.2135/cropsci1985.0011183X002500020025x
Cadman, C. H. (1942). Autotetraploid-inheritance in the potato: some new evidence. J. Genet. 44, 33–52. doi: 10.1007/BF02982766
Caranta, C., Pflieger, S., Lefebvre, V., Daubeze, A. M., Thabuis, A., and Palloix, A. (2002). QTLs involved in the restriction of Cucumber mosaic virus (CMV) long-distance movement in pepper. Theor. Appl. Genet. 104, 586–591. doi: 10.1007/s001220100753
Caranta, C., Thabuis, A., and Palloix, A. (1999). Development of a CAPS marker for the Pvr4 locus: a tool for pyramiding potyvirus resistance genes in pepper. Genome 42, 1111–1116. doi: 10.1139/gen-42-6-1111
Carr, J. P., Lewsey, M. G., and Palukaitis, P. (2010). Signaling in induced resistance. Adv. Virus Res. 76, 57–121. doi: 10.1016/S0065-3527(10)76003-6
Case, C. L. (2011). Regulating caspase-1 during infection: roles of NLRs, AIM2, and ASC. Yale J. Biol. Med. 84, 333–343. doi: 10.1128/mBio.00117-11
Cawly, J., Cole, A. B., Kiraly, L., Qiu, W. P., and Schoelz, J. E. (2005). The plant gene CCD1 selectively blocks cell death during the hypersensitive response to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 18, 212–219. doi: 10.1094/MPMI-18-0212
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Chalhoub, B. A., Sarrafi, A., and Lapierre, H. D. (1995). Partial resistance in the Barley (Hordeum vulgare L.) cultivar ‘Chikurin Ibaraki 1’ to two PAV-like isolates of Barley yellow dwarf virus: allelic variability at the Yd2 gene Locus. Plant Breed. 114, 303–307. doi: 10.1111/j.1439-0523.1995.tb01238.x
Chelkowski, J., Tyrka, M., and Sobkiewicz, A. (2003). Resistance genes in barley (Hordeum vulgare L.) and their identification with molecular markers. J. Appl. Genet. 44, 291–309.
Chen, H. Y., Yang, J., Lin, C., and Yuan, Y. A. (2008). Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760. doi: 10.1038/embor.2008.118
Chen, Q., Conner, R. L., Ahmad, F., Laroche, A., Fedak, G., and Thomas, J. B. (1998b). Molecular characterization of the genome composition of partial amphiploids derived from Triticum aestivum x Thinopyrum ponticum and T. aestivum x Th. intermedium as sources of resistance to Wheat streak mosaic virus and its vector, Aceria tosichella. Theor. Appl. Genet. 97, 1–8. doi: 10.1007/s001220050860
Chen, Q., Friebe, B., Conner, R. L., Laroche, A., Thomas, J. B., and Gill, B. S. (1998a). Molecular cytogenetic characterization of Thinopyrum intermedium derived wheat germplasm specifying resistance to Wheat streak mosaic virus. Theor. Appl. Genet. 96, 1–7. doi: 10.1007/s001220050701
Chen, Y., Liu, Z., and Halterman, D. A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 8:e1002595. doi: 10.1371/journal.ppat.1002595
Chisholm, S. T., Mahajan, S. K., Whitham, S. A., Yamamoto, M. L., and Carrington, J. C. (2000). Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of Tobacco etch virus. Proc. Natl. Acad. Sci. U.S.A. 97, 489–494. doi: 10.1073/pnas.97.1.489
Chung, H., Jeong, Y.-M., Mun, J.-H., Lee, S.-S., Chung, W.-H., and Yu, H.-J. (2014). Construction of a genetic map based on high-throughput SNP genotyping and genetic mapping of a TuMV resistance locus in Brassica rapa. Mol. Genet. Genomics 289, 149–160. doi: 10.1007/s00438-013-0798-9
Cockerham, G. (1943). VIII. Potato breeding for virus resistance. Ann. Appl. Biol. 30, 105–107. doi: 10.1111/j.1744-7348.1943.tb06166.x
Cockerham, G. (1955). “Strains of Potato virus X,” in Proceedings of the Second Conference on Potato Virus Diseases (Lisse-Wageningen).
Cockerham, G. (1958). “Experimental breeding in relation to virus resistance,” in Proceedings of the Third Conference on Potato Virus Diseases (Lisse-Wageningen).
Cockerham, G. (1970). Genetical studies on resistance to Potato viruses X and Y. Heredity 25, 309–348. doi: 10.1038/hdy.1970.35
Cole, A. B., Kiraly, L., Ross, K., and Scheolz, J. E. (2001). Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 14, 31–41. doi: 10.1094/MPMI.2001.14.1.31
Collier, S. M., Hamel, L. P., and Moffett, P. (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 24, 918–931. doi: 10.1094/MPMI-03-11-0050
Collier, S. M., and Moffett, P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. doi: 10.1016/j.tplants.2009.08.001
Collins, N. C., Paltridge, N. G., Ford, C. M., and Symons, R. H. (1996). The Yd2 gene for Barley yellow dwarf virus resistance maps close to the centromere on the long arm of barley chromosome 3. Theor. Appl. Genet. 92, 858–864. doi: 10.1007/BF00221898
Collmer, C. W., Marston, M. F., Taylor, J. C., and Jahn, M. (2000). The I gene of bean: a dosage-dependent allele conferring extreme resistance, hypersensitive resistance, or spreading vascular necrosis in response to the potyvirus Bean common mosaic virus. Mol. Plant Microbe Interact. 13, 1266–1270. doi: 10.1094/MPMI.2000.13.11.1266
Cooley, M. B., Pathirana, S., Wu, H. J., Kachroo, P., and Klessig, D. F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. doi: 10.1105/tpc.12.5.663
Cosson, P., Schurdi-Levraud, V., Le, Q. H., Sicard, O., Caballero, M., Roux, F., et al. (2012). The RTM resistance to potyviruses in Arabidopsis thaliana: natural variation of the RTM genes and evidence for the implication of additional genes. PLoS ONE 7:e39169. doi: 10.1371/journal.pone.0039169
Cruz, S. S., and Baulcombe, D. C. (1993). Molecular analysis of Potato virus X isolates in relation to the potato hypersensitivity gene Nx. Mol. Plant Microbe Interact. 6, 707–714. doi: 10.1094/MPMI-6-707
Csorba, T., Pantaleo, V., and Burgyan, J. (2009). RNA silencing: an antiviral mechanism. Adv. Virus Res. 75, 35–71. doi: 10.1016/S0065-3527(09)07502-2
Cui, Y., Lee, M. Y., Huo, N. X., Bragg, J., Yan, L. J., Yuan, C., et al. (2012). Fine mapping of the Bsr1 Barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS ONE 7:e38333. doi: 10.1371/journal.pone.0038333
Danin-Poleg, Y., Tadmor, Y., Tzuri, G., Reis, N., Hirschberg, J., and Katzir, N. (2002). Construction of a genetic map of melon with molecular markers and horticultural traits, and localization of genes associated with ZYMV resistance. Euphytica 125, 373–384. doi: 10.1023/A:1016021926815
Dardick, C. D., Taraporewala, Z., Lu, B., and Culver, J. N. (1999). Comparison of tobamovirus coat protein structural features that affect elicitor activity in pepper, eggplant, and tobacco. Mol. Plant Microbe Interact. 12, 247–251. doi: 10.1094/MPMI.1999.12.3.247
Decroocq, V., Salvador, B., Sicard, O., Glasa, M., Cosson, P., Svanella-Dumas, L., et al. (2009). The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N-terminal region of the coat protein. Mol. Plant Microbe Interact. 22, 1302–1311. doi: 10.1094/MPMI-22-10-1302
De La Cruz, A., López, L., Tenllado, F., Díaz-Ruíz, J. R., Sanz, A. I., Vaquero, C., et al. (1997). The coat protein is required for the elicitation of the Capsicum L2 gene-mediated resistance against the tobamoviruses. Mol. Plant Microbe Interact. 10, 107–113. doi: 10.1094/MPMI.1997.10.1.107
Delaney, T. P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). A central role of salicylic-acid in plant-disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
Delogu, G., Cattivelli, L., Snidaro, M., and Stanca, A. M. (1995). The Yd2 gene and enhanced resistance to Barley yellow dwarf virus (BYDV) in winter barley. Plant Breed. 114, 417–420. doi: 10.1111/j.1439-0523.1995.tb00823.x
De Ronde, D., Butterbach, P., Lohuis, D., Hedil, M., Van Lent, J. W. M., and Kormelink, R. (2013). Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 14, 405–415. doi: 10.1111/mpp.12016
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100, 8024–8029. doi: 10.1073/pnas.1230660100
De Zeeuw, D. J., and Ballard, J. C. (1959). Inheritance in cowpea of resistance to Tobacco ringspot virus. Phytopathology 49, 332–334.
Dinesh-Kumar, S. P., Tham, W. H., and Baker, B. J. (2000). Structure-function analysis of the Tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. U.S.A. 97, 14789–14794. doi: 10.1073/pnas.97.26.14789
Dinesh-Kumar, S. P., Whitham, S., Choi, D., Hehl, R., Corr, C., and Baker, B. (1995). Transposon tagging of Tobacco mosaic virus resistance gene N: its possible role in the TMV-N-mediated signal-transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 92, 4175–4180. doi: 10.1073/pnas.92.10.4175
Ding, J. Q., Li, H. M., Wang, Y. X., Zhao, R. B., Zhang, X. C., Chen, J. F., et al. (2012). Fine mapping of Rscmv2, a major gene for resistance to Sugarcane mosaic virus in maize. Mol. Breed. 30, 1593–1600. doi: 10.1007/s11032-012-9741-8
Ding, S. W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
Ding, S.-W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Diveki, Z., Salanki, K., and Balazs, E. (2004). The necrotic pathotype of the Cucumber mosaic virus (CMV) Ns strain is solely determined by amino acid 461 of the 1a protein. Mol. Plant Microbe Interact. 17, 837–845. doi: 10.1094/MPMI.2004.17.8.837
Dodds, P. N., Lawrence, G. J., Catanzariti, A.-M., Teh, T., Wang, C.-I. A., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Dogimont, C., Palloix, A., Daubze, A. M., Marchoux, G., Selassie, K. G., and Pochard, E. (1996). Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L). Euphytica 88, 231–239. doi: 10.1007/BF00023895
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Dussle, C. M., Quint, M., Xu, M. L., Melchinger, A. E., and Lubberstedt, T. (2002). Conversion of AFLP fragments tightly linked to SCMV resistance genes Scmv1 and Scmv2 into simple PCR-based markers. Theor. Appl. Genet. 105, 1190–1195. doi: 10.1007/s00122-002-0964-7
Dziewonska, M. A., and Ostrowska, K. (1978). Resistance to Potato virus M in certain wild potato species. Potato Res. 21, 129–131. doi: 10.1007/BF02361611
Edwards, M. C., Gonsalves, D., and Provvidenti, R. (1983). Genetic analysis of Cucumber mosaic virus in relation to host resistance: location of determinants for pathogenicity to certain legumes and Lactuca saligna. Phytopathology 73, 269–273. doi: 10.1094/Phyto-73-269
Edwards, M. C., and Steffenson, B. J. (1996). Genetics and mapping of Barley stripe mosaic virus resistance in barley. Phytopathology 86, 184–187. doi: 10.1094/Phyto-86-184
Eggenberger, A. L., Hajimorad, M. R., and Hill, J. H. (2008). Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 21, 931–936. doi: 10.1094/MPMI-21-7-0931
Erickson, F. L., Holzberg, S., Calderon-Urrea, A., Handley, V., Axtell, M., Corr, C., et al. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18, 67–75. doi: 10.1046/j.1365-313X.1999.00426.x
Fahim, M., Mechanicos, A., Ayala-Navarrete, L., Haber, S., and Larkin, P. J. (2012). Resistance to Wheat streak mosaic virus - a survey of resources and development of molecular markers. Plant Pathol. 61, 425–440. doi: 10.1111/j.1365-3059.2011.02542.x
Fan, Q. L., Niroula, M., Feldstein, P. A., and Bruening, G. (2011). Participation of the Cowpea mosaic virus protease in eliciting extreme resistance. Virology 417, 71–78. doi: 10.1016/j.virol.2011.04.022
Finlay, K. W. (1953). Inheritance of spotted wilt resistance in the tomato. II. Five genes controlling spotted wilt resistance in four tomato types. Aust. J. Biol. Sci. 6, 153–163.
Fisher, M. L., and Kyle, M. M. (1994). Inheritance of resistance to potyviruses in Phaseolus vulgaris L. III. Cosegregation of phenotypically similar dominant responses to nine potyviruses. Theor. Appl. Genet. 89, 818–823.
Fisher, M. L., and Kyle, M. M. (1996). Inheritance of resistance to potyviruses in Phaseolus vulgaris L. IV. Inheritance, linkage relations, and environmental effects on systemic resistance to four potyviruses. Theor. Appl. Genet. 92, 204–212. doi: 10.1007/BF00223377
Flis, B., Hermig, J., Strzelczyk-Zyta, D., Gebhardt, C., and Marezewski, W. (2005). The Ry-f(sto) gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122(718) in PVY resistant potato cultivars. Mol. Breed. 15, 95–101. doi: 10.1007/s11032-004-2736-3
Ford, C. M., Paltridge, N. G., Rathjen, J. P., Moritz, R. L., Simpson, R. J., and Symons, R. H. (1998). Rapid and informative assays for Yd2, the Barley yellow dwarf virus resistance gene, based on the nucleotide sequence of a closely linked gene. Mol. Breed. 4, 23–31. doi: 10.1023/A:1009686932457
Francki, M. G., Ohm, H. W., and Anderson, J. M. (2001). Novel germplasm providing resistance to Barley yellow dwarf virus in wheat. Aust. J. Agric. Res. 52, 1375–1382. doi: 10.1071/AR01020
Fribourg, C. E., and Nakashima, J. (1984). Characterization of a new potyvirus from potato. Phytopathology 74, 1363–1369. doi: 10.1094/Phyto-74-1363
Garrido-Ramirez, E. R., Sudarshana, M. R., Lucas, W. J., and Gilbertson, R. L. (2000). Bean dwarf mosaic virus BV1 protein is a determinant of the hypersensitive response and avirulence in Phaseolus vulgaris. Mol. Plant Microbe Interact. 13, 1184–1194. doi: 10.1094/MPMI.2000.13.11.1184
Gawehns, F., Cornelissen, B. J. C., and Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microb. Biotechnol. 6, 223–229. doi: 10.1111/1751-7915.12023
Genger, R. K., Jurkowski, G. I., Mcdowell, J. M., Lu, H., Jung, H. W., Greenberg, J. T., et al. (2008). Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol. Plant Microbe Interact. 21, 1285–1296. doi: 10.1094/MPMI-21-10-1285
Gerlier, D., and Lyles, D. S. (2011). Interplay between innate immunity and negative-strand RNA viruses: towards a rational model. Microbiol. Mol. Biol. Rev. 75, 468–490, second page of table of contents. doi: 10.1128/MMBR.00007-11
Ghazala, W., and Varrelmann, M. (2007). Tobacco rattle virus 29K movement protein is the elicitor of extreme and hypersensitive-like resistance in two cultivars of Solanum tuberosum. Mol. Plant Microbe Interact. 20, 1396–1405. doi: 10.1094/MPMI-20-11-1396
Gilbert, R. Z., Kyle, M. M., Munger, H. M., and Gray, S. M. (1994). Inheritance of resistance to Watermelon mosaic virus in Cucumis melo L. HortScience 29, 107–110.
Gilbert-Albertini, F., Lecoq, H., Pitrat, M., and Nicolet, J. L. (1993). Resistance of Cucurbita moschata to Watermelon mosaic virus type 2 and its genetic relation to resistance to Zucchini yellow mosaic virus. Euphytica 69, 231–237. doi: 10.1007/BF00022369
Giner, A., Lakatos, L., García-Chapa, M., López-Moya, J. J., and Burgyán, J. (2010). Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6:e1000996. doi: 10.1371/journal.ppat.1000996
Goodrick, B. J., Kuhn, C. W., and Boerma, H. R. (1991). Inheritance of nonnecrotic resistance to Cowpea chlorotic mottle virus in soybean. J. Hered. 82, 512–514.
Grube, R. C., Wintermantel, W. M., Hand, P., Aburomia, R., Pink, D. A. C., and Ryder, E. J. (2005). Genetic analysis and mapping of resistance to lettuce dieback: a soilborne disease caused by tombusviruses. Theor. Appl. Genet. 110, 259–268. doi: 10.1007/s00122-004-1825-3
Grube, R. C., Zhang, Y. P., Murphy, J. F., Loaiza-Figueroa, F., Lackney, V. K., Provvidenti, R., et al. (2000). New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis. 84, 885–891. doi: 10.1094/PDIS.2000.84.8.885
Grumet, R., Kabelka, E., Mcqueen, S., Wai, T., and Humphrey, R. (2000). Characterization of sources of resistance to the watermelon strain of Papaya ringspot virus in cucumber: allelism and co-segregation with other potyvirus resistances. Theor. Appl. Genet. 101, 463–472. doi: 10.1007/s001220051504
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., and Park, S. W. (2012). Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78, 51–65. doi: 10.1016/j.pmpp.2012.01.002
Hajimorad, M., Eggenberger, A., and Hill, J. (2005a). Absence of Soybean mosaic virus elicitor functions provoking Rsv1-mediated resistance response is insufficient for virulence on Rsv1-genotype soybean. Phytopathology 95:S39.
Hajimorad, M. R., Eggenberger, A. L., and Hill, J. H. (2005b). Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1-mediated lethal systemic hypersensitive response maps to P3. J. Virol. 79, 1215–1222. doi: 10.1128/JVI.79.2.1215-1222.2005
Hajimorad, M. R., and Hill, J. H. (2001). Rsv1-mediated resistance against Soybean mosaic virus-N is hypersensitive response independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 14, 587–598. doi: 10.1094/MPMI.2001.14.5.587
Hall, T. J. (1980). Resistance at the Tm-2 locus in the tomato to Tomato mosaic virus. Euphytica 29, 189–197. doi: 10.1007/BF00037266
Hallwass, M., Silva Da Oliveiro, A., Dianese, E., Lohuis, D., Boiteux, L. S., Inoue-Nagata, A. K., et al. (2014). The Tomato spotted wilt virus cell-to-cell movement protein (NSm) triggers a hypersensitive response in Sw-5 containing resistant tomato lines and Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. doi: 10.1111/mpp.12144. [Epub ahead of print].
Hamalainen, J. H., Sorri, V. A., Watanabe, K. N., Gebhardt, C., and Valkonen, J. P. T. (1998). Molecular examination of a chromosome region that controls resistance to potato Y and A potyviruses in potato. Theor. Appl. Genet. 96, 1036–1043. doi: 10.1007/s001220050836
Hanson, P. M., Bernacchi, D., Green, S., Tanksley, S. D., Muniyappa, V., Padmaja, S., et al. (2000). Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. J. Am. Soc. Hortic. Sci. 125, 15–20.
Hao, W., Collier, S. M., Moffett, P., and Chai, J. (2013). Structural basis for the interaction between the Potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating Protein 2 (RanGAP2). J. Biol. Chem. 288, 35868–35876. doi: 10.1074/jbc.M113.517417
Hao, Y. F., Wang, Y. Y., Chen, Z. B., Bland, D., Li, S. S., Brown-Guedira, G., et al. (2012). A conserved locus conditioning Soil-borne wheat mosaic virus resistance on the long arm of chromosome 5D in common wheat. Mol. Breed. 30, 1453–1464. doi: 10.1007/s11032-012-9731-x
Harper, S. J., Dawson, T. E., and Pearson, M. N. (2010). Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch. Virol. 155, 471–480. doi: 10.1007/s00705-010-0604-5
Hassan, S., and Thomas, P. E. (1983). Two types of immunity to Tomato yellow top virus identified in Lycopersicon peruvianum and its tomato hybrids. Phytopathology 73, 959–959.
Hassan, S., and Thomas, P. E. (1984a). Discovery of resistance to infection and translocation of Tomato yellow top virus (TYTV) in Lycopersicon peruvianum and some of its tomato hybrids. Phytopathology 74, 1138.
Hassan, S., and Thomas, P. E. (1984b). Etiological distinctions between Tomato yellow top virus and Potato leafroll and Beet western yellows viruses. Plant Dis. 68, 684–685. doi: 10.1094/PD-69-684
Hassan, S., and Thomas, P. E. (1988). Extreme resistance to Tomato yellow top virus and Potato leaf roll virus in Lycopersicon peruvianum and some of its tomato hybrids. Phytopathology 78, 1164–1167. doi: 10.1094/Phyto-78-1164
Hayes, A. J., Jeong, S. C., Gore, M. A., Yu, Y. G., Buss, G. R., Tolin, S. A., et al. (2004). Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to Soybean mosaic virus in soybeans. Genetics 166, 493–503. doi: 10.1534/genetics.166.1.493
Hayes, A. J., Ma, G. R., Buss, G. R., and Maroof, M. A. S. (2000). Molecular marker mapping of RSV4, a gene conferring resistance to all known strains of Soybean mosaic virus. Crop Sci. 40, 1434–1437. doi: 10.2135/cropsci2000.4051434x
Hikida, H. R., and Raymer, W. B. (1972). Sources and inheritance of peru tomato virus tolerance in tomato. Phytopathology 62, 764.
Hobbs, H. A., Jossey, S., Wang, Y., Hartman, G. L., and Domier, L. L. (2012). Diverse soybean accessions identified with temperature-sensitive resistance to Tobacco streak virus. Crop Sci. 52, 738–744. doi: 10.2135/cropsci2011.05.0265
Hobbs, H. A., Kuhn, C. W., Papa, K. E., and Brantley, B. B. (1987). Inheritance of non-necrotic resistance to Southern bean mosaic virus in cowpea. Phytopathology 77, 1624–1629. doi: 10.1094/Phyto-77-1624
Hoffmann, K., Qiu, W. P., and Moyer, J. W. (2001). Overcoming host- and pathogen-mediated resistance in tomato and tobacco maps to the M RNA of Tomato spotted wilt virus. Mol. Plant Microbe Interact. 14, 242–249. doi: 10.1094/MPMI.2001.14.2.242
Holmes, F. O. (1937). Inheritance of resistance to tobacco mosaic disease in the pepper. Phytopathology 24, 984–1002.
Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., et al. (2005). IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777. doi: 10.1038/nature03464
Hu, Z., Zhang, T., Yao, M., Feng, Z., Miriam, K., Wu, J., et al. (2012). The 2a protein of Cucumber mosaic virus induces a hypersensitive response in cowpea independently of its replicase activity. Virus Res. 170, 169–173. doi: 10.1016/j.virusres.2012.10.007
Hughes, S. L., Green, S. K., Lydiate, D. J., and Walsh, J. A. (2002). Resistance to Turnip mosaic virus in Brassica rapa and B. napus and the analysis of genetic inheritance in selected lines. Plant Pathol. 51, 567–573. doi: 10.1046/j.1365-3059.2002.00755.x
Hughes, S. L., Hunter, P. J., Sharpe, A. G., Kearsey, M. J., Lydiate, D. J., and Walsh, J. A. (2003). Genetic mapping of the novel Turnip mosaic virus resistance gene TuRB03 in Brassica napus. Theor. Appl. Genet. 107, 1169–1173. doi: 10.1007/s00122-003-1363-4
Hussain, M., Mansoor, S., Iram, S., Fatima, A. N., and Zafar, Y. (2005). The nuclear shuttle protein of Tomato leaf curl New Delhi virus is a pathogenicity determinant. J. Virol. 79, 4434–4439. doi: 10.1128/JVI.79.7.4434-4439.2005
Incarbone, M., and Dunoyer, P. (2013). RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392. doi: 10.1016/j.tplants.2013.04.001
Ishibashi, K., Masuda, K., Naito, S., Meshi, T., and Ishikawa, M. (2007). An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. U.S.A. 104, 13833–13838. doi: 10.1073/pnas.0703203104
Ishibashi, K., Mawatari, N., Miyashita, S., Kishino, H., Meshi, T., and Ishikawa, M. (2012). Coevolution and hierarchical interactions of Tomato mosaic virus and the resistance gene Tm-1. PLoS Pathog. 8:e1002975. doi: 10.1371/journal.ppat.1002975
Jahn, M., Paran, I., Hoffmann, K., Radwanski, E. R., Livingstone, K. D., Grube, R. C., et al. (2000). Genetic mapping of the Tsw locus for resistance to the Tospovirus Tomato spotted wilt virus in Capsicum spp. and its relationship to the Sw-5 gene for resistance to the same pathogen in tomato. Mol. Plant Microbe Interact. 13, 673–682. doi: 10.1094/MPMI.2000.13.6.673
Janzac, B., Fabre, F., Palloix, A., and Moury, B. (2009). Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol. Plant Pathol. 10, 599–610. doi: 10.1111/j.1364-3703.2009.00554.x
Janzac, B., Montarry, J., Palloix, A., Navaud, O., and Moury, B. (2010). A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant Microbe Interact. 23, 823–830. doi: 10.1094/MPMI-23-6-0823
Jefferies, S. P., King, B. J., Barr, A. R., Warner, P., Logue, S. J., and Langridge, P. (2003). Marker-assisted backcross introgression of the Yd2 gene conferring resistance to Barley yellow dwarf virus in barley. Plant Breed. 122, 52–56. doi: 10.1046/j.1439-0523.2003.00752.x
Jenner, C. E., Sanchez, F., Nettleship, S. B., Foster, G. D., Ponz, F., and Walsh, J. A. (2000). The cylindrical inclusion gene of Turnip mosaic virus encodes a pathogenic determinant to the Brassica resistance gene TuRB01. Mol. Plant Microbe Interact. 13, 1102–1108. doi: 10.1094/MPMI.2000.13.10.1102
Jennings, D. L. (1964). Studies on inheritance in red raspberry of immunities from three nematode-borne viruses. Genetica 35, 152–164. doi: 10.1007/BF01804883
Jensen, S., and Thomsen, A. R. (2012). Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86, 2900–2910. doi: 10.1128/JVI.05738-11
Jeong, S. C., Kristipati, S., Hayes, A. J., Maughan, P. J., Noffsinger, S. L., Gunduz, I., et al. (2002). Genetic and sequence analysis of markers tightly linked to the Soybean mosaic virus resistance gene, Rsv3. Crop Sci. 42, 265–270. doi: 10.2135/cropsci2002.0265
Jia, Y., Mcadams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jin, M., Lee, S.-S., Ke, L., Kim, J., Seo, M.-S., Sohn, S.-H., et al. (2014). Identification and mapping of a novel dominant resistance gene, TuRB07 to Turnip mosaic virus in Brassica rapa. Theor. Appl. Genet. 127, 509–519. doi: 10.1007/s00122-013-2237-z
Jones, A. T., Murant, A. F., Jennings, D. L., and Wood, G. A. (1982). Association of Raspberry bushy dwarf virus with raspberry yellows disease; reaction of Rubus species and cultivars, and the inheritance of resistance. Ann. Appl. Biol. 100, 135. doi: 10.1111/j.1744-7348.1982.tb07200.x
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, M. W., Redinbaugh, M. G., and Louie, R. (2007). The Mdm1 locus and maize resistance to Maize dwarf mosaic virus. Plant Dis. 91, 185–190. doi: 10.1094/PDIS-91-2-0185
Jones, R. A. C. (1990). Strain group specific and virus specific hypersensitive reactions to infection with potyviruses in potato cultivars. Ann. Appl. Biol. 117, 93–105. doi: 10.1111/j.1744-7348.1990.tb04198.x
Jones, R. A. C. (2012). Virus diseases of annual pasture legumes: incidences, losses, epidemiology, and management. Crop Pasture Sci. 63, 399–418. doi: 10.1071/CP12117
Kai, H., Takata, K., Tsukazaki, M., Furusho, M., and Baba, T. (2012). Molecular mapping of Rym17, a dominant and rym18 a recessive Barley yellow mosaic virus (BaYMV) resistance genes derived from Hordeum vulgare L. Theor. Appl. Genet. 124, 577–583. doi: 10.1007/s00122-011-1730-5
Kaisho, T., and Akira, S. (2006). Toll-like receptor function and signaling. J. Allergy Clin. Immunol. 117, 979–987. doi: 10.1016/j.jaci.2006.02.023
Kaneko, Y. H., Inukai, T., Suehiro, N., Natsuaki, T., and Masuta, C. (2004). Fine genetic mapping of the TuNI locus causing systemic veinal necrosis by Turnip mosaic virus infection in Arabidopsis thaliana. Theor. Appl. Genet. 110, 33–40. doi: 10.1007/s00122-004-1824-4
Kang, B. C., Yeam, I., and Jahn, M. M. (2005). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. doi: 10.1146/annurev.phyto.43.011205.141140
Kang, W. H., Hoang, N. H., Yang, H. B., Kwon, J. K., Jo, S. H., Seo, J. K., et al. (2010). Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor. Appl. Genet. 120, 1587–1596. doi: 10.1007/s00122-010-1278-9
Kang, W. H., Seo, J. K., Chung, B. N., Kim, K. H., and Kang, B. C. (2012). Helicase domain encoded by Cucumber mosaic virus RNA1 determines systemic infection of Cmr1 in Pepper. PLoS ONE 7:e43136. doi: 10.1371/journal.pone.0043136
Karasawa, A., Okada, I., Akashi, K., Chida, Y., Hase, S., Nakazawa-Nasu, Y., et al. (1999). One amino acid change in Cucumber mosaic virus RNA polymerase determines virulent/avirulent phenotypes on cowpea. Phytopathology 89, 1186–1192. doi: 10.1094/PHYTO.1999.89.12.1186
Kato, M., Ishibashi, K., Kobayashi, C., Ishikawa, M., and Katoh, E. (2013). Expression, purification, and functional characterization of an N-terminal fragment of the Tomato mosaic virus resistance protein Tm-1. Protein Expr. Purif. 89, 1–6. doi: 10.1016/j.pep.2013.02.001
Kelley, K. B., Whitworth, J. L., and Novy, R. G. (2009). Mapping of the Potato leafroll virus resistance gene, Rlr (etb), from Solanum etuberosum identifies interchromosomal translocations among its E-genome chromosomes 4 and 9 relative to the A-genome of Solanum L. sect. Petota. Mol. Breed. 23, 489–500. doi: 10.1007/s11032-008-9251-x
Kelly, J. D., Afanador, L., and Haley, S. D. (1995). Pyramiding genes for resistance to Bean common mosaic virus. Euphytica 82, 207–212. doi: 10.1007/BF00029562
Khatabi, B., Fajolu, O. L., Wen, R. H., and Hajimorad, M. R. (2012). Evaluation of north American isolates of Soybean mosaic virus for gain of virulence on Rsv-genotype soybeans with special emphasis on resistance-breaking determinants on Rsv4. Mol. Plant Pathol. 13, 1077–1088. doi: 10.1111/j.1364-3703.2012.00817.x
Kim, B. M., Suehiro, N., Natsuaki, T., Inukai, T., and Masuta, C. (2010). The P3 protein of Turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Mol. Plant Microbe Interact. 23, 144–152. doi: 10.1094/MPMI-23-2-0144
Kim, C. H., and Palukaitis, P. (1997). The plant defense response to Cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects virus accumulation in single cells. EMBO J. 16, 4060–4068. doi: 10.1093/emboj/16.13.4060
Kiraly, L., Cole, A. B., Bourque, J. E., and Schoelz, J. E. (1999). Systemic cell death is elicited by the interaction of a single gene in Nicotiana clevelandii and gene VI of Cauliflower mosaic virus. Mol. Plant Microbe Interact. 12, 919–925. doi: 10.1094/MPMI.1999.12.10.919
Knight, V. H., and Barbara, D. J. (1981). Susceptibility of red raspberry varieties to Raspberry bushy dwarf virus and its genetic control. Euphytica 30, 803–811. doi: 10.1007/BF00038809
Knorr, D. A., and Dawson, W. O. (1988). A point mutation in the Tobacco mosaic virus capsid protein gene induces hypersensitivity in Nicotiana sylvestris. Proc. Natl. Acad. Sci. U.S.A. 85, 170–174. doi: 10.1073/pnas.85.1.170
Koonin, E. V., and Aravind, L. (2000). The NACHT family – a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem. Sci. 25, 223–224. doi: 10.1016/S0968-0004(00)01577-2
Kopisch-Obuch, F. J., Koval, N. C., Mueller, E. M., Paine, C., Grau, C. P., and Diers, B. W. (2008). Inheritance of resistance to Alfalfa mosaic virus in soybean PI 153282. Crop Sci. 48, 933–940. doi: 10.2135/cropsci2007.08.0454
Kørner, C. J., Klauser, D., Niehl, A., Domínguez-Ferreras, A., Chinchilla, D., Boller, T., et al. (2013). The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 26, 1271–1280. doi: 10.1094/MPMI-06-13-0179-R
Krasileva, K. V., Dahlbeck, D., and Staskawicz, B. J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22, 2444–2458. doi: 10.1105/tpc.110.075358
Kyle, M. M., and Dickson, M. H. (1988). Linkage of hypersensitivity to five viruses with the B-locus in Phaseolus vulgaris L. J. Hered. 79, 308–311.
Kyle, M. M., and Provvidenti, R. (1987). Inheritance of resistance to potato Y viruses in Phaseolus vulgaris L.I. Two independent genes for resistance to Watermelon mosaic virus 2. Theor. Appl. Genet. 74, 595–600. doi: 10.1007/BF00288858
Kyle, M. M., and Provvidenti, R. (1993). Inheritance of resistance to potyviruses in Phaseolus vulgaris L.II. Linkage relations and utility of a dominant gene for lethal systemic necrosis to Soybean mosaic virus. Theor. Appl. Genet. 86, 189–196.
Kyle, M. M., Provvidenti, R., and Munger, H. M. (1986). A major gene for broad-spectrum virus resistance in Phaseolus vulgaris L. HortScience 21, 874.
Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E. J., Carrington, J. C., Liu, Y. P., et al. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. doi: 10.1038/sj.emboj.7601164
Lanfermeijer, F. C., Dijkhuis, J., Sturre, M. J. G., De Haan, P., and Hille, J. (2003). Cloning and characterization of the durable Tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol. Biol. 52, 1037–1049. doi: 10.1023/A:1025434519282
Lanfermeijer, F. C., Warmink, J., and Hille, J. (2005). The products of the broken Tm-2 and the durable Tm-2(2) resistance genes from tomato differ in four amino acids. J. Exp. Bot. 56, 2925–2933. doi: 10.1093/jxb/eri288
Larkin, P. J., Young, M. J., Gerlach, W. L., and Waterhouse, P. M. (1991). The Yd2 resistance to Barley yellow dwarf virus is effective in barley plants but not in their leaf protoplasts. Ann. Appl. Biol. 118, 115–125. doi: 10.1111/j.1744-7348.1991.tb06090.x
Lecoq, H., Pochard, E., Pitrat, M., Laterrot, H., and Marchoux, G. (1982). Research and utilization of virus-resistance in vegetables. Cryptogam. Mycol. 3, 333–345.
Lee, M. Y., Yan, L. J., Gorter, F. A., Kim, B. Y. T., Cui, Y., Hu, Y., et al. (2012). Brachypodium distachyon line Bd3-1 resistance is elicited by the Barley stripe mosaic virus triple gene block 1 movement protein. J. Gen. Virol. 93, 2729–2739. doi: 10.1099/vir.0.045880-0
Lehmann, P., Petrzik, K., Jenner, C., Greenland, A., Spak, J., Kozubek, E., et al. (1997). Nucleotide and amino acid variation in the coat protein coding region of Turnip mosaic virus isolates and possible involvement in the interaction with the Brassica resistance gene TuRBO1. Physiol. Mol. Plant Pathol. 51, 195–208. doi: 10.1006/pmpp.1997.0116
Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343, 1–28. doi: 10.1016/j.jmb.2004.08.023
Lewellen, R. T. (1973). Inheritance of Beet mosaic virus resistance in sugarbeet. Phytopathology 63, 877–881. doi: 10.1094/Phyto-63-877
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, H. W., Lucy, A. P., Guo, H. S., Li, W. X., Ji, L. H., Wong, S. M., et al. (1999). Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18, 2683–2691. doi: 10.1093/emboj/18.10.2683
Lim, W. L., Wang, S. H., and Ng, O. C. (1978). Resistance in chinese cabbage to Turnip mosaic virus. Plant Dis. Rep. 62, 660–662.
Liu, P. P., Bhattacharjee, S., Klessig, D. F., and Moffett, P. (2010). Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Mol. Plant Pathol. 11, 155–160. doi: 10.1111/j.1364-3703.2009.00564.x
Loebenstein, G. (2009). Local Lesions and Induced Resistance. Adv. Virus Res. 75, 73–117. doi: 10.1016/S0065-3527(09)07503-4
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546
Lukasik, E., and Takken, F. L. W. (2009). STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12, 427–436. doi: 10.1016/j.pbi.2009.03.001
Lupas, A. (1997). Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7, 388–393. doi: 10.1016/S0959-440X(97)80056-5
Ma, J. F., Hou, X. L., Xiao, D., Qi, L., Wang, F., Sun, F. F., et al. (2010). Cloning and characterization of the BcTuR3 gene related to resistance to Turnip mosaic virus (TuMV) from Non-heading chinese cabbage. Plant Mol. Biol. Rep. 28, 588–596. doi: 10.1007/s11105-010-0183-3
Maekawa, T., Cheng, W., Spiridon, L. N., Töller, A., Lukasik, E., Saijo, Y., et al. (2011a). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. doi: 10.1016/j.chom.2011.02.008
Maekawa, T., Kufer, T. A., and Schulze-Lefert, P. (2011b). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12, 817–826. doi: 10.1038/ni.2083
Maiti, S., Paul, S., and Pal, A. (2012). Isolation, characterization, and structure analysis of a non-TIR-NBS-LRR encoding candidate gene from MYMIV-resistant Vigna mungo. Mol. Biotechnol. 52, 217–233. doi: 10.1007/s12033-011-9488-1
Makkouk, K. M., Comeau, A., and Stpierre, C. A. (1994). Screening for Barley yellow dwarf Luteovirus resistance in barley on the basis of virus movement. J. Phytopathol. 141, 165–172. doi: 10.1111/j.1439-0434.1994.tb01458.x
Malcuit, I., Marano, M. R., Kavanagh, T. A., De Jong, W., Forsyth, A., and Baulcombe, D. C. (1999). The 25-kDa movement protein of PVX elicits Nb-mediated hypersensitive cell death in potato. Mol. Plant Microbe Interact. 12, 536–543. doi: 10.1094/MPMI.1999.12.6.536
Mallor, C., Alvarez, J. M., and Luis-Arteaga, M. (2003). A resistance to systemic symptom expression of Melon necrotic spot virus in melon. J. Am. Soc. Hortic. Sci. 128, 541–547.
Marczewski, W., Flis, B., Syller, J., Schafer-Pregl, R., and Gebhardt, C. (2001). A major quantitative trait locus for resistance to Potato leafroll virus is located in a resistance hotspot on potato chromosome XI and is tightly linked to N-gene-like markers. Mol. Plant Microbe Interact. 14, 1420–1425. doi: 10.1094/MPMI.2001.14.12.1420
Marczewski, W., Flis, B., Syller, J., Strzelczyk-Zyta, D., Hennig, J., and Gebhardt, C. (2004). Two allelic or tightly linked genetic factors at the PLRV.4 locus on potato chromosome XI control resistance to Potato leafroll virus accumulation. Theor. Appl. Genet. 109, 1604–1609. doi: 10.1007/s00122-004-1780-z
Marczewski, W., Hennig, J., and Gebhardt, C. (2002). The Potato virus S resistance gene Ns maps to potato chromosome VIII. Theor. Appl. Genet. 105, 564–567. doi: 10.1007/s00122-002-0976-3
Marczewski, W., Ostrowska, K., and Zimnoch-Guzowska, E. (1998). Identification of RAPD markers linked to the Ns locus in potato. Plant Breed. 117, 88–90. doi: 10.1111/j.1439-0523.1998.tb01455.x
Marczewski, W., Strzelczyk-Zyta, D., Hennig, J., Witek, K., and Gebhardt, C. (2006). Potato chromosomes IX and XI carry genes for resistance to Potato virus M. Theor. Appl. Genet. 112, 1232–1238. doi: 10.1007/s00122-006-0224-3
Maroof, M. A. S., Jeong, S. C., Gunduz, I., Tucker, D. M., Buss, G. R., and Tolin, S. A. (2008). Pyramiding of Soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci. 48, 517–526. doi: 10.2135/cropsci2007.08.0479
Matsumoto, K., Johnishi, K., Hamada, H., Sawada, H., Takeuchi, S., Kobayashi, K., et al. (2009). Single amino acid substitution in the methyltransferase domain of Paprika mild mottle virus replicase proteins confers the ability to overcome the high temperature-dependent Hk gene-mediated resistance in Capsicum plants. Virus Res. 140, 98–102. doi: 10.1016/j.virusres.2008.11.011
Matsumoto, K., Sawada, H., Matsumoto, K., Hamada, H., Yoshimoto, E., Ito, T., et al. (2008). The coat protein gene of tobamovirus P-0 pathotype is a determinant for activation of temperature-insensitive L-1a-gene-mediated resistance in Capsicum plants. Arch. Virol. 153, 645–650. doi: 10.1007/s00705-008-0032-y
Melchinger, A. E., Kuntze, L., Gumber, R. K., Lubberstedt, T., and Fuchs, E. (1998). Genetic basis of resistance to sugarcane mosaic virus in European maize germplasm. Theor. Appl. Genet. 96, 1151–1161. doi: 10.1007/s001220050851
Meshi, T., Motoyoshi, F., Adachi, A., Watanabe, Y., Takamatsu, N., and Okada, Y. (1988). Two concomitant base substitutions in the putative replicase genes of Tobacco mosaic virus confer the ability to overcome the effects of a tomato resistance gene, Tm-1. EMBO J. 7, 1575–1581.
Meshi, T., Motoyoshi, F., Maeda, T., Yoshiwoka, S., Watanabe, H., and Okada, Y., (1989). Mutations in the Tobacco mosaic virus 30 kD protein gene overcome Tm-2 resistance in tomato. Plant Cell 1, 515–522. doi: 10.1105/tpc.1.5.515
Mestre, P., Brigneti, G., and Baulcombe, D. C. (2000). An Ry-mediated resistance response in potato requires the intact active site of the NIa proteinase from Potato virus Y. Plant J. 23, 653–661. doi: 10.1046/j.1365-313x.2000.00834.x
Mestre, P., Brigneti, G., Durrant, M. C., and Baulcombe, D. C. (2003). Potato virus Y NIa protease activity is not sufficient for elicitation of Ry-mediated disease resistance in potato. Plant J. 36, 755–761. doi: 10.1046/j.1365-313X.2003.01917.x
Mignouna, H. D., Abang, M. M., Onasanya, A., Agindotan, B., and Asiedu, R. (2002). Identification and potential use of RAPD markers linked to Yam mosaic virus resistance in white yam (Dioscorea rotundata). Ann. Appl. Biol. 140, 163–169. doi: 10.1111/j.1744-7348.2002.tb00169.x
Miklas, P. N., Seo, Y. S., and Gilbertson, R. L. (2009). Quantitative resistance to Bean dwarf mosaic virus in common bean is associated with the Bct gene for resistance to Beet curly top virus. Plant Dis. 93, 645–648. doi: 10.1094/PDIS-93-6-0645
Modawi, R. S., Heyne, E. G., Brunetta, D., and Willis, W. G. (1982). Genetic studies of field reaction to Wheat soilborne mosaic virus. Plant Dis. 66, 1183–1184. doi: 10.1094/PD-66-1183
Moffett, P. (2009). Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 75, 1–33. doi: 10.1016/S0065-3527(09)07501-0
Moffett, P., Farnham, G., Peart, J., and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. doi: 10.1093/emboj/cdf453
Montesclaros, L., Nicol, N., Ubalijoro, E., Leclercpotvin, C., Ganivet, L., Laliberte, J. F., et al. (1997). Response to potyvirus infection and genetic mapping of resistance loci to potyvirus infection in Lactuca. Theor. Appl. Genet. 94, 941–946. doi: 10.1007/s001220050499
Montes-Garcia, C. E., Garza-Ortega, S., and Brown, J. K. (1998). “Inheritance of the resistance to Squash leaf curl virus in Cucurbita pepo,” in Cucurbitaceae '98. Evaluation and Enhancement of Cucurbit Germplasm, ed J. D. McCreight (Alexandria, VA: ASHS Press), 328–330.
Moury, B., Caromel, B., Johansen, E., Simon, V., Chauvin, L., Jacquot, E., et al. (2011). The helper component proteinase cistron of Potato virus Y induces hypersensitivity and resistance in Potato genotypes carrying dominant resistance genes on chromosome IV. Mol. Plant Microbe Interact. 24, 787–797. doi: 10.1094/MPMI-10-10-0246
Moury, B., and Verdin, E. (2012). Viruses of pepper crops in the mediterranean basin: a remarkable stasis. Adv. Virus Res. 84, 127–162. doi: 10.1016/B978-0-12-394314-9.00004-X
Mubin, M., Amin, I., Amrao, L., Briddon, R. W., and Mansoor, S. (2010). The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 11, 245–254. doi: 10.1111/j.1364-3703.2009.00601.x
Munoz, F. J., Plaisted, R. L., and Thurston, H. D. (1975). Resistance to Potato virus Y in Solanum tuberosum Spp Andigena. Am. Potato J. 52, 107–115. doi: 10.1007/BF02852043
Mysore, K. S., and Ryu, C. M. (2004). Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. doi: 10.1016/j.tplants.2003.12.005
Neuhaus, G., Werner, K., Weyen, J., Friedt, W., and Ordon, F. (2003). First results on SNP-scanning in fragments linked to resistance genes against the Barley yellow mosaic virus complex. J. Plant Dis. Prot. 110, 296–303.
Niks, R. E., Habekuss, A., Bekele, B., and Ordon, F. (2004). A novel major gene on chromosome 6H for resistance of barley against the Barley yellow dwarf virus. Theor. Appl. Genet. 109, 1536–1543. doi: 10.1007/s00122-004-1777-7
Nono-Womdim, R., Marchoux, G., Pochard, E., Palloix, A., and Gebre-Selassie, K. (1991). Resistance of pepper lines to the movement of Cucumber mosaic virus. J. Phytopathol. 132, 21–32. doi: 10.1111/j.1439-0434.1991.tb00090.x
Novy, R. G., Gillen, A. M., and Whitworth, J. L. (2007). Characterization of the expression and inheritance of Potato leafroll virus (PLRV) and Potato virus Y (PVY) resistance in three generations of germplasm derived from Solanum etuberosum. Theor. Appl. Genet. 114, 1161–1172. doi: 10.1007/s00122-007-0508-2
Osorno, J. M., Munoz, C. G., Beaver, J. S., Ferwerda, F. H., Bassett, M. J., Miklas, P. N., et al. (2007). Two genes from Phaseolus coccineus confer resistance to Bean golden yellow mosaic virus in common bean. J. Am. Soc. Hortic. Sci. 132, 530–533.
Ovesna, J., Vacke, J., Kucera, L., Chrpova, J., Novakova, I., Jahoor, A., et al. (2000). Genetic analysis of resistance in barley to Barley yellow dwarf virus. Plant Breed. 119, 481–486. doi: 10.1046/j.1439-0523.2000.00522.x
Padgett, H. S., and Beachy, R. N. (1993). Analysis of a Tobacco mosaic virus strain capable of overcoming N gene-mediated resistance. Plant Cell 5, 577–586. doi: 10.1105/tpc.5.5.577
Padgett, H. S., Watanabe, Y., and Beachy, R. N. (1997). Identification of the TMV replicase sequence that activates the N gene-mediated hypersensitive response. Mol. Plant Microbe Interact. 10, 709–715. doi: 10.1094/MPMI.1997.10.6.709
Padmanabhan, M. S., and Dinesh-Kumar, S. P. (2010). All hands on deck-the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant Microbe Interact. 23, 1368–1380. doi: 10.1094/MPMI-05-10-0113
Pal, S. S., Dhaliwal, H. S., and Bains, S. S. (1991). Inheritance of resistance to Yellow mosaic virus in some Vigna species. Plant Breed. 106, 168–171. doi: 10.1111/j.1439-0523.1991.tb00496.x
Paltridge, N. G., Collins, N. C., Bendahmane, A., and Symons, R. H. (1998). Development of YLM, a codominant PCR marker closely linked to the Yd2 gene for resistance to barley yellow dwarf disease. Theor. Appl. Genet. 96, 1170–1177. doi: 10.1007/s001220050853
Palukaitis, P. (2012). Resistance to viruses of potato and their vectors. Plant Pathol. J. 28, 248–258. doi: 10.5423/PPJ.RW.06.2012.0075
Palukaitis, P., and Garcia-Arenal, F. (2003). Cucumoviruses. Adv. Virus Res. 62, 241–323. doi: 10.1016/S0065-3527(03)62005-1
Park, S. J., and Tu, J. C. (1991). Inheritance and allelism of resistance to a severe strain of Bean yellow mosaic virus in common bean. Can. J. Plant Pathol. 13, 7–10. doi: 10.1080/07060669109500967
Parrella, G., Moretti, A., Gognalons, P., Lesage, M. L., Marchoux, G., Gebre-Selassie, K., et al. (2004). The Am gene controlling resistance to Alfalfa mosaic virus in tomato is located in the cluster of dominant resistance genes on chromosome 6. Phytopathology 94, 345–350. doi: 10.1094/PHYTO.2004.94.4.345
Patel, P., Mligo, J., Leyna, H., Kuwite, C., and Mmbaga, E. (1982). Sources of resistance, inheritance, and breeding of cowpeas for resistance to a strain of Cowpea aphid-borne mosaic virus from Tanzania. Indian J. Genet. Plant Breed. 42, 221–229.
Peiro, A., Carmen Canizares, M., Rubio, L., Lopez, C., Moriones, E., Aramburu, J., et al. (2014). The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. doi: 10.1111/mpp.12142. [Epub ahead of print].
Pelham, J. (1966). Resistance in tomato to Tobacco mosaic virus. Euphytica 15, 258. doi: 10.1007/Bf00022331
Pitrat, M., and Lecoq, H. (1980). Inheritance of resistance to Cucumber mosaic virus transmission by Aphis gossypii in Cucumis melo. Phytopathology 70, 958–961. doi: 10.1094/Phyto-70-958
Pitrat, M., and Lecoq, H. (1983). Two alleles for Watermelon mosaic virus 1 resistance in muskmelon. Cucurbit Genet. Coop. Rep. 6, 52–53.
Pitrat, M., and Lecoq, H. (1984). Inheritance of Zucchini yellow mosaic virus resistance in Cucumis melo L. Euphytica 33, 57–61. doi: 10.1007/BF00022750
Ponz, F., Russell, M. L., Rowhani, A., and Bruening, G. (1988). A cowpea line has distinct genes for resistance to Tobacco ringspot virus and Cowpea mosaic virus. Phytopathology 78, 1124–1128. doi: 10.1094/Phyto-78-1124
Provvide, R. (1974). Inheritance of resistance to Watermelon mosaic virus 2 in Phaseolus vulgaris. Phytopathology 64, 1448–1450. doi: 10.1094/Phyto-64-1448
Provvidenti, R. (1988). Inheritance of resistance to Broad bean wilt virus in bean. HortScience 23, 895–896.
Provvidenti, R. (2000). Inheritance of resistance to Passionfruit woodiness virus in common bean (Phaseolus vulgaris L.). HortScience 35, 880–881.
Provvidenti, R., Gonsalves, D., and Ranalli, P. (1982). Inheritance of resistance to Soybean mosaic virus in Phaseolus vulgaris. J. Hered. 73, 302–303.
Provvidenti, R., Gonsalves, D., and Taiwo, M. A. (1983). Inheritance of resistance to Blackeye cowpea mosaic and Cowpea aphid-borne mosaic viruses in Phaseolus vulgaris. J. Hered. 74, 60–61.
Provvidenti, R., and Hampton, R. O. (1992). Sources of resistance to viruses in the Potyviridae. Arch. Virol. Suppl. 5, 189–211. doi: 10.1007/978-3-7091-6920-9_17
Provvidenti, R., Robinson, R. W., and Shail, J. W. (1980). A source of resistance to a strain of Cucumber mosaic virus in Lactuca saligna L. HortScience 15, 528–529.
Querci, M., Baulcombe, D. C., Goldbach, R. W., and Salazar, L. F. (1995). Analysis of the resistance-breaking determinants of Potato virus X (PVX) strain Hb on different potato genotypes expressing extreme resistance to Pvx. Phytopathology 85, 1003–1010. doi: 10.1094/Phyto-85-1003
Quint, M., Mihaljevic, R., Dussle, C. M., Xu, M. L., Melchinger, A. E., and Lubberstedt, T. (2002). Development of RGA-CAPS markers and genetic mapping of candidate genes for Sugarcane mosaic virus resistance in maize. Theor. Appl. Genet. 105, 355–363. doi: 10.1007/s00122-002-0953-x
Rai, M. (2006). Refinement of the Citrus tristeza virus resistance gene (Ctv) positional map in Poncirus trifoliata and generation of transgenic grapefruit (Citrus paradisi) plant lines with candidate resistance genes in this region. Plant Mol. Biol. 61, 399–414. doi: 10.1007/s11103-006-0018-7
Ren, T., Qu, F., and Morris, T. J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to Turnip crinkle virus. Plant Cell 12, 1917–1926. doi: 10.1105/tpc.12.10.1917
Robbins, M. A., Witsenboer, H., Michelmore, R. W., Laliberte, J. F., and Fortin, M. G. (1994). Genetic mapping of Turnip mosaic virus resistance in Lactuca sativa. Theor. Appl. Genet. 89, 583–589. doi: 10.1007/BF00222452
Roman, M. A., Castaneda, A. M., Sanchez, J. C. A., Munoz, C. G., and Beaver, J. S. (2004). Inheritance of normal pod development in bean golden yellow mosaic resistant common bean. J. Am. Soc. Hortic. Sci. 129, 549–552.
Ross, A. F. (1961). Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology 14, 329. doi: 10.1016/0042-6822(61)90318-X
Ruge, B., Linz, A., Pickering, R., Proeseler, G., Greif, P., and Wehling, P. (2003). Mapping of Rym14(Hb), a gene introgressed from Hordeum bulbosum and conferring resistance to BaMMV and BaYMV in barley. Theor. Appl. Genet. 107, 965–971. doi: 10.1007/s00122-003-1339-4
Ruge-Wehling, B., Linz, A., Habekuss, A., and Wehling, P. (2006). Mapping of Rym16(Hb), the second soil-borne virus-resistance gene introgressed from Hordeum bulbosum. Theor. Appl. Genet. 113, 867–873. doi: 10.1007/s00122-006-0345-8
Sacco, M. A., Koropacka, K., Grenier, E., Jaubert, M. J., Blanchard, A., Goverse, A., et al. (2009). The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2 and RanGAP2 dependent plant cell death. PLoS Pathog. 5:e1000564. doi: 10.1371/journal.ppat.1000564
Saito, T., Meshi, T., Takamatsu, N., and Okada, Y. (1987). Coat protein gene sequence of Tobacco mosaic virus encodes a host response determinant. Proc. Natl. Acad. Sci. U.S.A. 84, 6074–6077. doi: 10.1073/pnas.84.17.6074
Salanki, K., Gellert, A., Naray-Szabo, G., and Balazs, E. (2007). Modeling-based characterization of the elicitor function of amino acid 461 of Cucumber mosaic virus 1a protein in the hypersensitive response. Virology 358, 109–118. doi: 10.1016/j.virol.2006.08.014
Sawada, H., Takeuchi, S., Hamada, H., Kiba, A., Matsumoto, M., and Hikichi, Y. (2004). A new tobamovirus-resistance gene, L-1a, of sweet pepper (Capsicum annuum L.). J. Jpn. Soc. Hortic. Sci. 73, 552–557. doi: 10.2503/jjshs.73.552
Sawada, H., Takeuchi, S., Matsumoto, K., Hamada, H., Kiba, A., Matsumoto, M., et al. (2005). A new tobamovirus-resistance gene, Hk, in Capsicum annuum. J. Jpn. Soc. Hortic. Sci. 74, 289–294. doi: 10.2503/jjshs.74.289
Schoelz, J., Shepherd, R. J., and Daubert, S. (1986). Region VI of Cauliflower mosaic virus encodes a host range determinant. Mol. Cell. Biol. 6, 2632–2637. doi: 10.1128/mcb.6.7.2632
Schroeder, W. T., and Provvidenti, R. (1968). Resistance of bean (Phaseolus vulgaris) to PV2 strain of Bean yellow mosaic virus conditioned by single dominant gene By. Phytopathology 58, 1710.
Seifers, D. L., Perumal, R., and Little, C. R. (2012). New sources of resistance in sorghum (Sorghum bicolor) germplasm are effective against a diverse array of potyvirus spp. Plant Dis. 96, 1775–1779. doi: 10.1094/PDIS-03-12-0224-RE
Sekine, K. T., Ishihara, T., Hase, S., Kusano, T., Shah, J., and Takahashi, H. (2006). Single amino acid alterations in Arabidopsis thaliana RCY1 compromise resistance to Cucumber mosaic virus, but differentially suppress hypersensitive response-like cell death. Plant Mol. Biol. 62, 669–682. doi: 10.1007/s11103-006-9048-4
Seo, Y. S., Gepts, P., and Gilbertson, R. L. (2004). Genetics of resistance to the geminivirus, Bean dwarf mosaic virus, and the role of the hypersensitive response in common bean. Theor. Appl. Genet. 108, 786–793. doi: 10.1007/s00122-003-1504-9
Seo, Y. S., Jeon, J. S., Rojas, M. R., and Gilbertson, R. L. (2007). Characterization of a novel Toll/interleukin-1 receptor (TIR)-TIR gene differentially expressed in common bean (Phaseolus vulgaris cv. Othello) undergoing a defence response to the geminivirus Bean dwarf mosaic virus. Mol. Plant Pathol. 8, 151–162. doi: 10.1111/j.1364-3703.2007.00379.x
Seo, Y. S., Rojas, M. R., Lee, J. Y., Lee, S. W., Jeon, J. S., Ronald, P., et al. (2006). A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. U.S.A. 103, 11856–11861. doi: 10.1073/pnas.0604815103
Sharma, N., Sahu, P. P., Puranik, S., and Prasad, M. (2013). Recent advances in plant-virus interaction with emphasis on small interfering RNAs (siRNAs). Mol. Biotechnol. 55, 63–77. doi: 10.1007/s12033-012-9615-7
Sharma, P., and Ikegami, M. (2010). Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 396, 85–93. doi: 10.1016/j.virol.2009.10.012
Shepherd, D. N., Martin, D. P., Van Der Walt, E., Dent, K., Varsani, A., and Rybicki, E. P. (2010). Maize streak virus: an old and complex ‘emerging’ pathogen. Mol. Plant Pathol. 11, 1–12. doi: 10.1111/j.1364-3703.2009.00568.x
Shivaprasad, P. V., Chen, H.-M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., et al. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. doi: 10.1016/S0092-8674(01)00576-1
Simko, I., Pechenick, D. A., Mchale, L. K., Truco, M. J., Ochoa, O. E., Michelmore, R. W., et al. (2009). Association mapping and marker-assisted selection of the lettuce dieback resistance gene Tvr1. BMC Plant Biol. 9:135. doi: 10.1186/1471-2229-9-135
Sinclair, J. B., and Walker, J. C. (1955). Inheritance of resistance to Cucumber mosaic virus in cowpea. Phytopathology 45, 563–564.
Singh, A. K., and Singh, A. K. (1987). Chemical composition of cowpea seeds as influenced by Southern bean mosaic virus and Cowpea mosaic virus. Phyton 26, 165–170.
Singh, R. P. (1993). Genetic association of gene Bdv1 for tolerance to Barley yellow dwarf virus with genes Lr34 and Yr18 for adult-plant resistance to rusts in bread wheat. Plant Dis. 77, 1103–1106. doi: 10.1094/PD-77-1103
Singh, S. P., and Schwartz, H. F. (2010). Breeding common bean for resistance to diseases: a review. Crop Sci. 50, 2199–2223. doi: 10.2135/cropsci2009.03.0163
Slootweg, E. J., Spiridon, L. N., Roosien, J., Butterbach, P., Pomp, R., Westerhof, L., et al. (2013). Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 162, 1510–1528. doi: 10.1104/pp.113.218842
Slootweg, E., Roosien, J., Spiridon, L. N., Petrescu, A. J., Tameling, W., Joosten, M., et al. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22, 4195–4215. doi: 10.1105/tpc.110.077537
Solomon-Blackburn, R. M., and Barker, H. (2001a). Breeding virus resistant potatoes (Solanum tuberosum): a review of traditional and molecular approaches. Heredity 86, 17–35. doi: 10.1046/j.1365-2540.2001.00799.x
Solomon-Blackburn, R. M., and Barker, H. (2001b). A review of host major-gene resistance to potato viruses X, Y, A and V in potato: genes, genetics and mapped locations. Heredity 86, 8–16. doi: 10.1046/j.1365-2540.2001.00798.x
Stamova, B. S., and Chetelat, R. T. (2000). Inheritance and genetic mapping of Cucumber mosaic virus resistance introgressed from Lycopersicon chilense into tomato. Theor. Appl. Genet. 101, 527–537. doi: 10.1007/s001220051512
Stewart, L. R., Haque, M. A., Jones, M. W., and Redinbaugh, M. G. (2013). Response of maize (Zea mays L.) lines carrying Wsm1, Wsm2, and Wsm3 to the potyviruses Johnsongrass mosaic virus and Sorghum mosaic virus. Mol. Breed. 31, 289–297. doi: 10.1007/s11032-012-9789-5
Stoutjesdijk, P., Kammholz, S. J., Kleven, S., Matsay, S., Banks, P. M., and Larkin, P. J. (2001). PCR-based molecular marker for the Bdv2 Thinopyrum intermedium source of Barley yellow dwarf virus resistance in wheat. Aust. J. Agric. Res. 52, 1383–1388. doi: 10.1071/AR01083
Swiezynski, K. M., Krusiec, J., Osiecka, M., Sieczka, M. T., and Zarzycka, H. (1993). Potato-tuber resistance to Phytophthora infestans and its relation to maturity. Plant Breed. 110, 161–164. doi: 10.1111/j.1439-0523.1993.tb01229.x
Szajko, K., Chrzanowska, M., Witek, K., Strzelczyk-Zyta, D., Zagorska, H., Gebhardt, C., et al. (2008). The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor. Appl. Genet. 116, 297–303. doi: 10.1007/s00122-007-0667-1
Taiwo, M. A., Provvidenti, R., and Gonsalves, D. (1981). Inheritance of resistance to Blackeye cowpea mosaic virus in Vigna unguiculata. J. Hered. 72, 433–434.
Takacs, A., Kazinczi, G., Horvath, J., and Hadzsi, M. (2006). Natural virus infection of tomato plants in Hungary. Cereal Res. Commun. 34, 689–691. doi: 10.1556/CRC.34.2006.1.172
Takacs, A. P., Kazinczi, G., Horvath, J., and Gaborjanyi, R. (2003). Reaction of Lycopersicon species and varieties to Potato virus Y (PVY(NTN)) and Tomato mosaic virus (ToMV). Commun. Agric. Appl. Biol. Sci. 68, 561–565.
Takahashi, H., Kanayama, Y., Zheng, M. S., Kusano, T., Hase, S., Ikegami, M., et al. (2004). Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to Cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. doi: 10.1093/pcp/pch085
Takahashi, H., Miller, J., Nozaki, Y., Takeda, M., Shah, J., Hase, S., et al. (2002). RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to Cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 32, 655–667. doi: 10.1046/j.1365-313X.2002.01453.x
Takahashi, H., Suzuki, M., Natsuaki, K., Shigyo, T., Hino, K., Teraoka, T., et al. (2001). Mapping the virus and host genes involved in the resistance response in Cucumber mosaic virus infected Arabidopsis thaliana. Plant Cell Physiol. 42, 340–347. doi: 10.1093/pcp/pce039
Takken, F. L. W., Albrecht, M., and Tameling, W. I. L. (2006). Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390. doi: 10.1016/j.pbi.2006.05.009
Tameling, W. I., and Baulcombe, D. C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19, 1682–1694. doi: 10.1105/tpc.107.050880
Tanksley, S. D., Bernachi, D., Beck-Bunn, T., Emmatty, D., Eshed, Y., Inai, S., et al. (1998). Yield and quality evaluations on a pair of processing tomato lines nearly isogenic for the Tm2(a) gene for resistance to the Tobacco mosaic virus. Euphytica 99, 77–83. doi: 10.1023/A:1018320232663
Tao, X. R., Zhou, X. P., Li, G. X., and Yu, J. L. (2002). The pathogenicity on legumes of Cucumber mosaic virus was determined by 243 nucleotides on 2a polymerase gene of viral RNA2. Chin. Sci. Bull. 47, 748. doi: 10.1360/02tb9169
Taylor, S., and Martin, R. R. (1999). Sequence comparison between common and resistance breaking strains of Raspberry bushy dwarf virus. Phytopathology 89:576.
Tian, Y. P., and Valkonen, J. P. T. (2013). Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene Ny in potato. Mol. Plant Microbe Interact. 26, 297–305. doi: 10.1094/MPMI-09-12-0219-R
Tomita, R., Murai, J., Miura, Y., Ishihara, H., Liu, S., Kubotera, Y., et al. (2008). Fine mapping and DNA fiber FISH analysis locates the tobamovirus resistance gene L(3) of Capsicum chinense in a 400-kb region of R-like genes cluster embedded in highly repetitive sequences. Theor. Appl. Genet. 117, 1107–1118. doi: 10.1007/s00122-008-0848-6
Tomita, R., Sekine, K. T., Mizumoto, H., Sakamoto, M., Murai, J., Kiba, A., et al. (2011). Genetic basis for the hierarchical interaction between tobamovirus spp. and L resistance gene alleles from different pepper species. Mol. Plant Microbe Interact. 24, 108–117. doi: 10.1094/MPMI-06-10-0127
Troutman, J. L., and Fulton, R. W. (1958). Resistance in tobacco to Cucumber mosaic virus. Virology 6, 303–316. doi: 10.1016/0042-6822(58)90084-9
Truniger, V., and Aranda, M. A. (2009). Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–159. doi: 10.1016/S0065-3527(09)07504-6
Ueda, H., Yamaguchi, Y., and Sano, H. (2006). Direct interaction between the Tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45. doi: 10.1007/s11103-005-5817-8
Uma, B., Rani, T. S., and Podile, A. R. (2011). Warriors at the gate that never sleep: non-host resistance in plants. J. Plant Physiol. 168, 2141–2152. doi: 10.1016/j.jplph.2011.09.005
Valkonen, J. P. T. (1994). Natural genes and mechanisms for resistance to viruses in cultivated and wild potato species (Solanum Spp). Plant Breed. 112, 1–16. doi: 10.1111/j.1439-0523.1994.tb01270.x
Valkonen, J. P. T., Koivu, K., Slack, S. A., and Pehu, E. (1995). Modified resistance of Solanum brevidens to Potato Y potyvirus and Tobacco mosaic tobamovirus following genetic-transformation and explant regeneration. Plant Sci. 106, 71–79. doi: 10.1016/0168-9452(95)04065-3
Vallejos, C. E., Astua-Monge, G., Jones, V., Plyler, T. R., Sakiyama, N. S., and Mackenzie, S. A. (2006). Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172, 1229–1242. doi: 10.1534/genetics.105.050S15
Van Der Biezen, E. A., and Jones, J. D. G. (1998). The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R227. doi: 10.1016/S0960-9822(98)70145-9
Van Der Hoorn, R. A. L., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Van Koevering, M., Haufler, K. Z., Fulbright, D. W., Isleib, T. G., and Everson, E. H. (1987). Heritability of resistance in winter wheat to Wheat spindle streak mosaic virus. Phytopathology 77, 742–744. doi: 10.1094/Phyto-77-742
Vaucheret, H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13, 350–358. doi: 10.1016/j.tplants.2008.04.007
Verlaan, M. G., Hutton, S. F., Ibrahem, R. M., Kormelink, R., Visser, R. G., Scott, J. W., et al. (2013). The Tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-Class RNA-dependent RNA polymerases. PLoS Genet. 9:e1003399. doi: 10.1371/journal.pgen.1003399
Vidal, S., Cabrera, H., Andersson, R. A., Fredriksson, A., and Valkonen, J. P. T. (2002). Potato gene Y-1 is an N gene homolog that confers cell death upon infection with Potato virus Y. Mol. Plant Microbe Interact. 15, 717–727. doi: 10.1094/MPMI.2002.15.7.717
Vlot, A. C., Klessig, D. F., and Park, S. W. (2008). Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442. doi: 10.1016/j.pbi.2008.05.003
Wade, B. L., and Zaumeyer, W. J. (1940). Genetic studies of resistance to Alfalfa mosaic virus and of stringiness in Phaseolus Vulgaris. Agron. J. 32, 127–134. doi: 10.2134/agronj1940.00021962003200020005x
Wai, T., and Grumet, R. (1995). Inheritance of resistance to the watermelon strain of Papaya ringspot virus in the cucumber line Tmg-1. HortScience 30, 338–340.
Walsh, J. A., Rusholme, R. L., Hughes, S. L., Jenner, C. E., Bambridge, J. M., Lydiate, D. J., et al. (2002). Different classes of resistance to Turnip mosaic virus in Brassica rapa. Eur. J. Plant Pathol. 108, 15–20. doi: 10.1023/A:1013962911906
Wang, H. L., Sudarshana, M. R., Gilbertson, R. L., and Lucas, W. J. (1999). Analysis of cell-to-cell and long-distance movement of a bean dwarf mosaic geminivirus-green fluorescent protein reporter in host and nonhost species: identification of sites of resistance. Mol. Plant Microbe Interact. 12, 345–355. doi: 10.1094/MPMI.1999.12.4.345
Wang, Y., Hobbs, H. A., Hill, C. B., Domier, L. L., Hartman, G. L., and Nelson, R. L. (2005). Evaluation of ancestral lines of US soybean cultivars for resistance to four soybean viruses. Crop Sci. 45, 639–644. doi: 10.2135/cropsci2005.0639
Ward, J. A., Boone, W. E., Moore, P. P., and Weber, C. A. (2012). Developing molecular markers for marker assisted selection for resistance to Raspberry bushy dwarf virus (RBDV) in Red Raspberry. Acta Hortic. 946, 61–66.
Was, M., and Dziewonska, M. A. (1984). “Reaction of PVM and PVS in potato clones with the genes Gm and Ns,” in 9th Triennial Conference of the European Association of Potato Researchers (Interlaken).
Weber, H., Schultze, S., and Pfitzner, A. J. (1993). Two amino acid substitutions in the Tomato mosaic virus 30-kilodalton movement protein confer the ability to overcome the Tm-2(2) resistance gene in the tomato. J. Virol. 67, 6432–6438.
Weeden, N. F., and Provvidenti, R. (1988). A marker locus, Adh-1, for resistance to Pea enation mosaic virus in Pisum sativum. J. Hered. 79, 128–131.
Welz, H. G., Schechert, A., Pernet, A., Pixley, K. V., and Geiger, H. H. (1998). A gene for resistance to the Maize streak virus in the African CIMMYT maize inbred line CML202. Mol. Breed. 4, 147–154. doi: 10.1023/A:1009602620244
Wen, R. H., Khatabi, B., Ashfield, T., Saghai Maroof, M. A., and Hajimorad, M. R. (2013). The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 26, 203–215. doi: 10.1094/MPMI-06-12-0156-R
Whitham, S. A., Anderberg, R. J., Chisholm, S. T., and Carrington, J. C. (2000). Arabidopsis RTM2 gene is necessary for specific restriction of Tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12, 569–582. doi: 10.1105/tpc.12.4.569
Whitham, S., Dineshkumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the Tobacco mosaic virus resistance gene N: similarity to Toll and the Interleukin-1 receptor. Cell 78, 1101–1115. doi: 10.1016/0092-8674(94)90283-6
Wiermer, M., Feys, B. J., and Parker, J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. doi: 10.1016/j.pbi.2005.05.010
Xia, S., Cheng, Y. T., Huang, S., Win, J., Soards, A., Jinn, T.-L., et al. (2013). Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162, 1694–1705. doi: 10.1104/pp.113.214551
Xia, X. C., Melchinger, A. E., Kuntze, L., and Lubberstedt, T. (1999). Quantitative trait loci mapping of resistance to Sugarcane mosaic virus in maize. Phytopathology 89, 660–667. doi: 10.1094/PHYTO.1999.89.8.660
Xu, M. L., Melchinger, A. E., and Lubberstedt, T. (2000). Origin of Scm1 and Scm2 - two loci conferring resistance to Sugarcane mosaic virus (SCMV) in maize. Theor. Appl. Genet. 100, 934–941. doi: 10.1007/s001220051373
Xu, Q. F., Ma, Y. Z., Xin, Z. Y., Chen, X., and Lin, Z. S. (1999). In situ hybridization and RAPD analyses of disomic Thinopyrum intermedium addition lines in wheat with Barley yellow dwarf virus resistance. Acta Genet. Sin. 26, 52–53.
Yamafuji, R., Watanabe, Y., Meshi, T., and Okada, Y. (1991). Replication of TMV-L and Lta1 RNAs and their recombinants in TMV-resistant Tm-1 tomato protoplasts. Virology 183, 99–105. doi: 10.1016/0042-6822(91)90122-R
Yamaji, Y., Maejima, K., Komatsu, K., Shiraishi, T., Okano, Y., Himeno, M., et al. (2012). Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 24, 778–793. doi: 10.1105/tpc.111.093658
Yang, Z. N., Ye, X. R., Molina, J., Roose, M. L., and Mirkov, T. E. (2003). Sequence analysis of a 282-kilobase region surrounding the Citrus tristeza virus resistance gene (Ctv) locus in Poncirus trifoliata L. Raf. Plant Physiol. 131, 482–492. doi: 10.1104/pp.011262
Yu, J., Gu, W. K., Provvidenti, R., and Weeden, N. F. (1995). Identifying and mapping 2 DNA markers linked to the gene conferring resistance to pea enation mosaic-virus. J. Am. Soc. Hortic. Sci. 120, 730–733.
Yu, Y. G., Maroof, M. A. S., Buss, G. R., Maughan, P. J., and Tolin, S. A. (1994). RFLP and microsatellite mapping of a gene for Soybean mosaic virus resistance. Phytopathology 84, 60–64. doi: 10.1094/Phyto-84-60
Yue, J.-X., Meyers, B. C., Chen, J.-Q., Tian, D., and Yang, S. (2012). Tracing the origin and evolutionary history of plant nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes. New Phytol. 193, 1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x
Zaumeyer, W. J., and Meiners, J. P. (1975). Disease resistance in beans. Annu. Rev. Phytopathol. 13, 313–334. doi: 10.1146/annurev.py.13.090175.001525
Zhang, Q. P., Li, Q., Wang, X., Wang, H. Y., Lang, S. P., Wang, Y. N., et al. (2005). Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS center dot 4DL conferring resistance to Wheat spindle streak mosaic virus. Euphytica 145, 317–320. doi: 10.1007/s10681-005-1743-8
Zhang, X., Yuan, Y. R., Pei, Y., Lin, S. S., Tuschl, T., Patel, D. J., et al. (2006). Cucumber mosaic virus encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. doi: 10.1101/gad.1495506
Zhang, Z. Y., Lin, Z. S., and Xin, Z. Y. (2009). Research progress in BYDV resistance genes derived from wheat and its wild relatives. J. Genet. Genomics 36, 567–573. doi: 10.1016/S1673-8527(08)60148-4
Zhang, Z. Y., Xin, Z. Y., and Larkin, P. J. (2001). Molecular characterization of a Thinopyrum intermedium Group 2 chromosome (2Ai-2) conferring resistance to Barley yellow dwarf virus. Genome 44, 1129–1135. doi: 10.1139/gen-44-6-1129
Zhang, Z. Y., Xin, Z. Y., Lin, Z. S., Chen, X., and Wang, X. P. (2000). Identification of molecular markers for the Thinopyrum intermedium chromosome 2Ai-2 with resistance to Barley yellow dwarf virus. Acta Bot. Sin. 42, 1051–1056.
Zhang, Z. Y., Xu, J. S., Xu, Q. J., Larkin, P., and Xin, Z. Y. (2004). Development of novel PCR markers linked to the BYDV resistance gene Bdv2 useful in wheat for marker-assisted selection. Theor. Appl. Genet. 109, 433–439. doi: 10.1007/s00122-004-1649-1
Zheng, Y., and Edwards, M. C. (1990). Expression of resistance to Barley stripe mosaic virus in barley and oat protoplasts. J. Gen. Virol. 71, 1865–1868. doi: 10.1099/0022-1317-71-8-1865
Zhou, Y. C., Garrido-Ramirez, E. R., Sudarshana, M. R., Yendluri, S., and Gilbertson, R. L. (2007). The N-terminus of the Begomovirus nuclear shuttle protein (BV1) determines virulence or avirulence in Phaseolus vulgaris. Mol. Plant Microbe Interact. 20, 1523–1534. doi: 10.1094/MPMI-20-12-1523
Keywords: R genes, PTI, PAMP-triggered immunity, ETI, RNAi, avirulence, hypersensitive response
Citation: de Ronde D, Butterbach P and Kormelink R (2014) Dominant resistance against plant viruses. Front. Plant Sci. 5:307. doi: 10.3389/fpls.2014.00307
Received: 13 April 2014; Accepted: 10 June 2014;
Published online: 27 June 2014.
Edited by:
Corné M. J. Pieterse, Utrecht University, NetherlandsReviewed by:
Peter Moffett, Université de Sherbrooke, CanadaFrank L. W. Takken, University of Amsterdam, Netherlands
Copyright © 2014 de Ronde, Butterbach and Kormelink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard Kormelink, Laboratory of Virology, Department of Plant Sciences, Wageningen University, Droevendaalsesteeg 1, 6708 PB Wageningen, Netherlands e-mail: richard.kormelink@wur.nl
 Dryas de Ronde
Dryas de Ronde Patrick Butterbach
Patrick Butterbach Richard Kormelink
Richard Kormelink