- 1Metalloid Transport Group, Department of Physiology and Cell Biology, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany
- 2Experimental Taxonomy, Department of Genebank, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany
- 3Molecular Plant Nutrition, Department of Physiology and Cell Biology, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany
Aquaporins (AQPs) are essential channel proteins that regulate plant water homeostasis and the uptake and distribution of uncharged solutes such as metalloids, urea, ammonia, and carbon dioxide. Despite their importance as crop plants, little is known about AQP gene and protein function in cabbage (Brassica oleracea) and other Brassica species. The recent releases of the genome sequences of B. oleracea and Brassica rapa allow comparative genomic studies in these species to investigate the evolution and features of Brassica genes and proteins. In this study, we identified all AQP genes in B. oleracea by a genome-wide survey. In total, 67 genes of four plant AQP subfamilies were identified. Their full-length gene sequences and locations on chromosomes and scaffolds were manually curated. The identification of six additional full-length AQP sequences in the B. rapa genome added to the recently published AQP protein family of this species. A phylogenetic analysis of AQPs of Arabidopsis thaliana, B. oleracea, B. rapa allowed us to follow AQP evolution in closely related species and to systematically classify and (re-) name these isoforms. Thirty-three groups of AQP-orthologous genes were identified between B. oleracea and Arabidopsis and their expression was analyzed in different organs. The two selectivity filters, gene structure and coding sequences were highly conserved within each AQP subfamily while sequence variations in some introns and untranslated regions were frequent. These data suggest a similar substrate selectivity and function of Brassica AQPs compared to Arabidopsis orthologs. The comparative analyses of all AQP subfamilies in three Brassicaceae species give initial insights into AQP evolution in these taxa. Based on the genome-wide AQP identification in B. oleracea and the sequence analysis and reprocessing of Brassica AQP information, our dataset provides a sequence resource for further investigations of the physiological and molecular functions of Brassica crop AQPs.
Introduction
Arabidopsis thaliana serves frequently as a reference for comparative genomics and gene functions in plants despite its genomic, phylogenetic, and physiological distance to most of the analyzed species. An emerging question from basic research to applied agriculture is to what degree the knowledge on the model plant Arabidopsis matches the biology of crop plants, and thereby to what extent might this knowledge be applicable for breeding strategies.
While a one-to-one transfer of knowledge to distantly related plants such as monocots (e.g., rice, maize, or wheat) is difficult, a transfer of knowledge from Arabidopsis to closely related Brassica crops is imaginable. Brassica crops are used worldwide for animal and human nutrition, as catch and cover crops and for biofuel production. This genus includes important vegetables [Brassica rapa ssp. (e.g., chinese cabbage, pak choi, and turnip), Brassica oleracea ssp. (e.g., broccoli, kohlrabi, kale, cabbage, Brussels sprout, and cauliflower), and Brassica napus ssp. (e.g., rutabaga and Hanover kale)] and oilseed crops (Brassica napus, B. rapa, Brassica juncea, and Brassica carinata) representing the third leading source of vegetable oil in the world, after soybean and palm oil1. The three diploid species B. rapa (A genome), B. nigra (B genome), and B. oleracea (C genome) formed the amphidiploid species B. juncea (A and B genomes), B. napus (A and C genomes), and B. carinata (B and C genomes) probably by independent hybridizations. This interspecific cytogenetic relationship was already described by the ‘U’s triangle theory of Nagaharu (Nagaharu, 1935) stating that the genomes of three ancestral species of Brassica combined to generate three modern vegetables and oilseed crop species. Taxa within the Brassica genus underwent a whole genome triplication around 13–17 million years ago (MYA; Yang et al., 2006) while the Arabidopsis–Brassica lineages split about 20 MYA (Yang et al., 1999). The A. thaliana genome has undergone duplications, deletions, re-arrangements, and a reduction in chromosome number even since the divergence from its close relative Arabidopsis lyrata 5 MYA (Hu et al., 2011).
The recent availability of high quality sequences of the B. rapa and B. oleracea genomes (Wang et al., 2011; Liu et al., 2014) has allowed one to carefully dissect and compare the genomic arrangement between these Brassicaceae species. This comparison confirmed the high level of synteny between their genomes and showed that more than 90% of the genomic sequences are located in 24 large collinear blocks A–X (Wang et al., 2011; Liu et al., 2014) constituting an ancient Brassicaceae karyotype of n = 8 as previously suggested (Parkin et al., 2003; Schranz et al., 2006). These blocks reorganized within the current species-specific numbers of chromosomes found in the Brassica genus.
The B. rapa and B. oleracea genome sequences (Wang et al., 2011; Liu et al., 2014), have also allowed for the identification of homologous genes and comparative analyses of the structural and functional evolution of the major intrinsic protein (MIP) superfamily. MIP channel proteins, also known as aquaporins (AQPs) form a hydrophilic pathway for uncharged molecules across the lipid bilayer of biological membranes (Gomes et al., 2009). They assemble as tetramers, in which each monomer is composed of six transmembrane helices (TMHs) connected by five loops (A–E) and two membrane-embedded half-helices, each containing the conserved MIP signature, the so-called “NPA” motif (asparagine-proline-alanine, or variants thereof). These motifs meet in the center of the membrane, forming a narrow hydrophilic path (Murata et al., 2000). A second selectivity filter, the aromatic/arginine (ar/R) constriction region, is formed by four amino acids (residues R1–R4) located in TMH2 (R1), TMH5 (R2), and loop E (R3 and R4) that contribute to a size exclusion barrier and a hydrogen bond environment necessary for the efficient transport of a substrate (Murata et al., 2000). While up to 13 MIP isoforms have been identified in microbes and mammals (Agre and Kozono, 2003), 35 have been found in A. thaliana (Johanson et al., 2001), 55 in poplar (Populus trichocarpa; Gupta and Sankararamakrishnan, 2009), 66 in soybean (Glycine max; Zhang et al., 2013), 41 in potato (Solanum tuberosum; Venkatesh et al., 2013), 47 in tomato (Solanum lycopersicum; Sade et al., 2009; Reuscher et al., 2013), 71 in cotton (Gossypium hirsutum; Park et al., 2010), 33 in rice (Oryza sativa; Sakurai et al., 2005), and at least 36 in maize (Zea mays; Chaumont et al., 2001). Even the genomes of contemporary members of early evolved land plants such as Physcomitrella patens and Selaginella moellendorffii encode 23 and 19 different MIPs, respectively (Danielson and Johanson, 2008; Anderberg et al., 2012). All MIPs of higher plants belong to the phylogenetic clade of AQPs and are subdivided into five distinct subfamilies, the plasma membrane intrinsic proteins (PIPs), the tonoplast intrinsic proteins (TIPs), the small basic intrinsic proteins (SIPs), the nodulin26-like intrinsic proteins (NIPs), and the uncharacterized X intrinsic proteins (XIPs; Chaumont et al., 2001; Johanson et al., 2001; Danielson and Johanson, 2008). XIPs are present in numerous eudicot plant species, but were neither detected in Brassicaceae species nor in monocots (Danielson and Johanson, 2008). Physiological and molecular studies proved that PIPs and TIPs play key functions in water homeostasis and are essential for transcellular water uptake from the soil, root-to-shoot transport, and osmo-driven growth control (Maurel et al., 2008; Chaumont and Tyerman, 2014). It was further demonstrated that certain AQPs are indispensable for the uptake, translocation, sequestration, or extrusion of uncharged and physiologically important solutes such as carbon dioxide (Uehlein et al., 2003, 2008), nitric oxide (Herrera et al., 2006), hydrogen peroxide (Bienert et al., 2007; Dynowski et al., 2008), urea (Liu et al., 2003), ammonia (Jahn et al., 2004; Loqué et al., 2005), lactic acid (Tsukaguchi et al., 1998; Choi and Roberts, 2007; Bienert et al., 2013), acetic acid (Mollapour and Piper, 2007), arsenite (Bienert et al., 2008b; Ma et al., 2008; Kamiya et al., 2009), boric acid (Takano et al., 2006; Tanaka et al., 2008; Hanaoka et al., 2014), orthosilicic acid (Ma et al., 2006), antimonite (Bienert et al., 2008b; Kamiya et al., 2009), and selenite (Zhao et al., 2010).
The importance to mechanistically understand water relations, signal transduction, and plant nutritional aspects, in particular nutrient transport processes in Brassica crops, is gaining increasing scientific interest, as is evident, e.g., by the increasing number of publications dealing with Brassica AQPs (Supplementary File S1).
In the present study we have classified all AQPs encoded in the sequenced genome of B. oleracea. Moreover, the recent description of AQPs from B. rapa (Tao et al., 2014) has been complemented with the assembly of six additional full-length AQP genes. Phylogenetic analyses of Arabidopsis, B. oleracea, and B. rapa AQPs were performed. Phylogenetic relationships, gene structures, chromosome locations, subgenome distributions, and protein sequences and features of Brassica AQPs were analyzed. The expression of B. oleracea AQP genes was analyzed in flowers, leaves, and roots. The refinement of database information and the analyses of phylogenetic relationships and selectivity filter compositions will be useful for subsequent studies on the recent AQP evolutionary history, and help to identify channel selectivities and therewith protein functions and regulations of Brassica AQPs.
Materials and Methods
Data Resource
Arabidopsis thaliana, B. oleracea, and B. rapa genomic and annotation data were downloaded from the TAIR10 database2, the Bolbase database3, and the BRAD database4, respectively. The B. rapa and B. oleracea sequences were checked manually in comparison with known AQP sequences for correctness of annotation, and intron–exon borders. Genomic, cDNA as well as protein sequences were corrected when necessary.
Alignment and Phylogenetic Analysis of AQP-Encoding Genes
Multiple sequence alignments for all retrieved AQP sequences from the three species together, and for all PIPs, NIPs, SIPs, and TIPs separately, were built using ClustalW as implemented in GENEIOUS PRO v6.15. The alignments were edited manually if necessary. The number of parsimony informative sites was determined using PAUP∗ (Swofford, 2003). For the DNA sequence alignments the best-fit model of nucleotide substitution was selected using jModelTest2 (Darriba et al., 2012). The Bayesian information criterion (BIC) always selected the HKY + I + G (Hasegawa et al., 1985) out of 24 tested models. Bayesian phylogenetic analyses were done in MrBayes version 3.2 (Ronquist and Huelsenbeck, 2003). MrBayes was run by conducting two parallel Metropolis coupled Monte Carlo Markov chain analyses with four chains for two million generations. Trees were sampled every 500 generations. Convergence of the runs was assessed using the standard deviation of split frequencies being <0.01. The continuous parameter values sampled from the chains were checked for mixing using Tracer v1.6 (Rambaut and Drummond, 2007). Convergence of the topologies was checked using the online application tool AWTY (Nylander et al., 2008). A consensus tree was computed in MrBayes version 3.2 after removal (burn-in) of the first 25% of trees. For the amino acid alignment the best-fit model Cprev (Adachi et al., 2000) of amino acid substitution was selected in MrBayes version 3.2. The phylogenetic inference was done using the same settings as for nucleotide data.
Calculation of Sequence Identities in Exons and Introns
The ClustalW alignment in GENEIOUS PRO v6.1 was used to generate an identity matrix for each intron, exon, and protein sequence for each NIP subgroup separately. As a reference the AtNIP variant was chosen and set to 100%. For the NIP1 and NIP4 subgroup the most likely A. thaliana isoform compared with B. oleracea and B. rapa was used as a reference (AtNIP1;2 and AtNIP4;1).
Gene Expression Analysis
RNA-seq data from B. oleracea flowers (GSM1052959), leaves (GSM1052960), and roots (GSM1052961) were obtained from the Gene Expression Omnibus with the accession number GSE42891. B. oleracea AQP genes were subjected to BlastN analysis against the up-to-date version (B. oleracea version 2.1.25) of B. oleracea cDNAs (transcripts/splice variants) at EnsemblPlants6. Gene identifiers and locations of the identified B. oleracea AQP genes are given in Supplementary File S7. CLC Genomics Workbench 7.5 was used for mapping the transcriptome reads to the B. oleracea reference genome version 2.1.25. Assembly was conducted with default mapping parameters allowing for a maximum of two mismatches and the maximum of 10 hits per read. RPKM (Reads per kilobase of exon model per million mapped reads) was the reporting expression value. Flower, leaf, and root data sets were used for a multi-group-comparison of paired samples. The selection for candidate genes (four AQP subfamilies) and the application of logarithmic transformation (Log10) for the expression values led to a compressed dynamic range of the expression values. Genes were visualized in a heatmap in which the four plant AQP subfamilies appear as groups.
Results and Discussion
Identification and Classification of the Complete Set of Brassica oleracea and Brassica rapa AQPs
The “Browse Gene Families” functions “Major Intrinsic Protein MIP Gene Family, PF00230” and “Aquaporin Families” in Bolbase, and Brassica BRAD database, respectively, were used to identify the complete AQP family of B. oleracea and B. rapa. Five entries in the database were miss-annotated as AQPs (homologs of At1g31880). To investigate whether potential AQP genes were lacking or insufficiently annotated in the databases, the diploid genomes of B. oleracea and B. rapa were BLAST-searched using all A. thaliana AQPs as queries. Identified AQP sequences from each species were thereafter iteratively used as queries. This search additionally identified one AQP isoform in B. oleracea which was not identified in the previous “Browse Gene Families” search. As already shown by Tao et al. (2014), 12 additional AQP sequences were identified in the B. rapa genome. For example, all AQPs belonging to the SIP subfamily were not given in the output data of the “Browse Gene Families” function in BRAD4.
Subsequently, each output AQP gene was manually inspected. This assessment demonstrated that for the database-annotated AQP genes the output coding sequences and exon/intron selections were occasionally resulting in non-satisfactory AQP gene models (Supplementary File S2). For the non-satisfactorily annotated AQP sequences (nine in B. oleracea and eight in B. rapa) we were able to manually curate new gene models encoding complete and more typical AQP features using the available genomic data (corrected gene locations are given in Table 1).
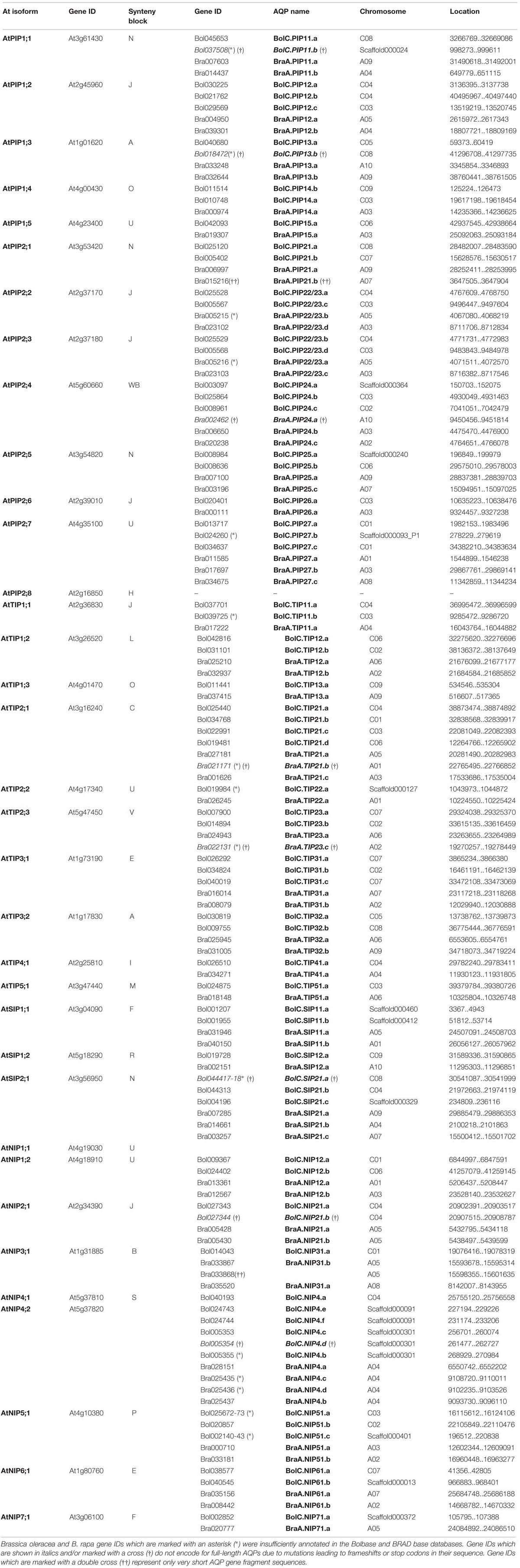
TABLE 1. List of all Arabidopsis thaliana, Brassica oleracea, and Brassica rapa AQP genes and their distribution on chromosomes.
This manual analysis revealed that all nine of the non-satisfactorily annotated B. oleracea AQPs actually do represent full-length AQP gene sequences. Six out of these nine AQPs encode for complete AQP proteins while the remaining three sequences (BolC.SIP21.a, BolC.PIP11.b, and BolC.PIP13.b) carry small deletions or mutations leading to frameshifts or stop codons (see below and Supplementary File S3). Tao et al. (2014) excluded seven short AQP sequences from further analyses in their work. However, a manual inspection showed that six out of these seven sequences represent full-length AQPs (BraA.PIP22/23.b, BraA.PIP22/23.a, BraA.TIP21.b, BraA.NIP4.c, BraA.NIP4.d, and BraA.TIP23.c; corrected gene locations and coding sequences are given in Table 1 and Supplementary File S4). Four out of these six sequences encoded for typical AQP protein sequences (BraA.PIP22/23.b, BraA.PIP22/23.a, BraA.NIP4.c, and BraA.NIP4.d) and should be included in further analyses. The remaining two full-length sequences (BraA.TIP21.b and BraA.TIP23.c) carry point mutations resulting in a stop codon interrupting the reading frame of these water channel genes (see below and Supplementary File S3). Therefore, our manual analysis added four more potentially functional AQP channels to the set of B. rapa AQPs and increases the number of B. rapa AQP isoforms to 57.
In summary, after manual inspection and curating we identified 67 and 59 genomic full-length AQPs nucleotide sequences in the genomes of B. oleracea and B. rapa, respectively. Thereof, we identified 10 sequences which were deemed to be pseudo-genes (Supplementary File S3) of which two were only gene fragments [Bra015216 and Bra033868 – marked with a double cross (††) in Table 1; Supplementary File S3]. The others were “full-length” AQP sequences carrying small mutations leading to frameshifts or stop codons [written in italics and marked with a cross (†) in Table 1] resulting in genes encoding for incomplete AQP-like proteins. Since the sequences carrying mutations covered the full-length genomic nucleotide sequences of potential AQPs they were included in the following phylogenetic analysis. It is important to consider these isoforms as these gene sequences potentially encode for functional channels in other Brassica cultivars or subspecies. We hypothesize that it is unlikely that these mutations are fixed in all the diverse genomes of Brassica morphotypes which evolved in various environments across the world. Finally, 62 and 57 full-length AQP genomic sequences encode for full-length typical water channel proteins in B. oleracea and B. rapa, respectively.
All AQPs from B. oleracea and B. rapa have been (re-)named according to the standardized gene nomenclature for the Brassica genus suggested by Ostergaard and King (2008). In short, the nomenclature obey the following format: genus (one letter) – species (two letters) – genome (one letter) – gene name (three to six letter code; priority should be given to orthologs from Arabidopsis rather than more distantly related species) – locus (one letter) – allele (integer). This nomenclature is unambiguous, directly naming the orthologous gene of A. thaliana and will allow the origins of orthologous isoforms within the polyploid genomes of other Brassica species forming the ‘U’s triangle to be traced. For the B. rapa AQP isoforms we generally kept the “isoform indexing” recommendation by Tao et al. (2014). Using the identified AQP sequences we screened datasets for already described and cloned Brassica AQPs. All identified AQP sequences were phylogenetically compared to the ones derived from the sequenced B. oleracea and B. rapa genomes. To standardize the nomenclature and to prevent double denotation we assigned and re-named previously described isoforms and alleles to the respective genomic versions (Supplementary File S1) according to the nomenclature of Ostergaard and King (2008).
Chromosomal Distribution and Subgenome Distribution of Brassica AQPs
Fifty one B. oleracea AQP-encoding genes were located on chromosomes (76.1%), and 16 genes (23.9%) were located on unanchored scaffolds (Figure 1). In contrast, all B. rapa AQPs were located to chromosomes (Tao et al., 2014). The triplicated Brassica genomes can be divided into three subgenomes. These subgenomes have been grouped based on the level of gene loss relative to A. thaliana since the whole genome triplication 13–17 MYA ago (Wang et al., 2011; Liu et al., 2014). The subgenomes have been designated as the least fractionated genome (named LF or III) with 30% gene loss, the medium fractionated genome (named MF1 or II) with 54% gene loss and the most fractionated genome (named MF2 or I) with 64% gene loss in, e.g., B. rapa (Wang et al., 2011). One could hypothesize that 3 × 35 AQPs (105 isoforms) exist due to the mesohexapolyloidy of Brassica crops relative to Arabidopsis. In B. oleracea there were 82.9, 48.6, and 57.1% of the theoretical Arabidopsis AQP number identified in LF, MF1, and MF2, respectively, while in B. rapa these figures reduced to 80.0, 45.7, and 45.7%, respectively. These percentages match the average gene loss reported for the B. rapa subgenomes (Wang et al., 2011). The genomes of both Brassica species have undergone almost identical patterns of fractionation between orthologous genome segments since their triplication (Liu et al., 2014). Gene retention rates in the syntenic regions were as follows: 53.7 and 53.4% of ancestor genes occur as one copy, about 35.6 and 35.8% occur as two copies and only about 10.5 and 10.9% occur as three copies in B. rapa and B. oleracea, respectively (Liu et al., 2014). Similar retention rates for Brassica AQPs within the three subgenomes (Table 2). were found. The subgenome distribution of AQP copies within B. oleracea and B. rapa is similar (Table 2).
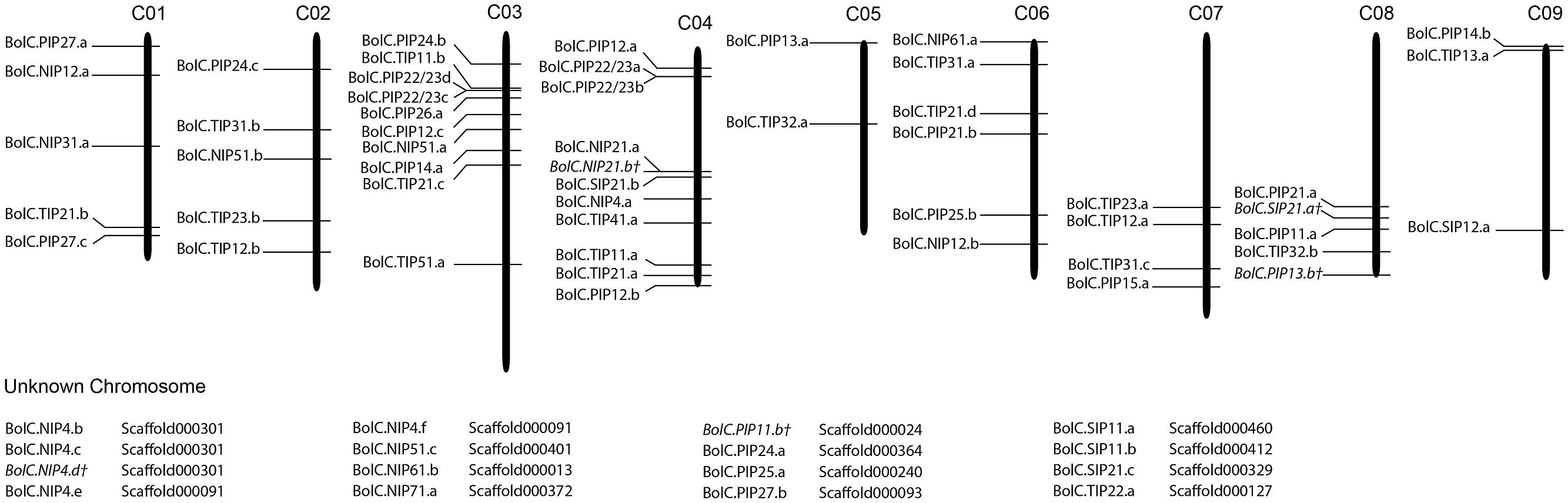
FIGURE 1. Distribution of aquaporin genes on the nine Brassica oleraceae chromosomes. Sixteen genes could not be anchored onto a specific chromosome.
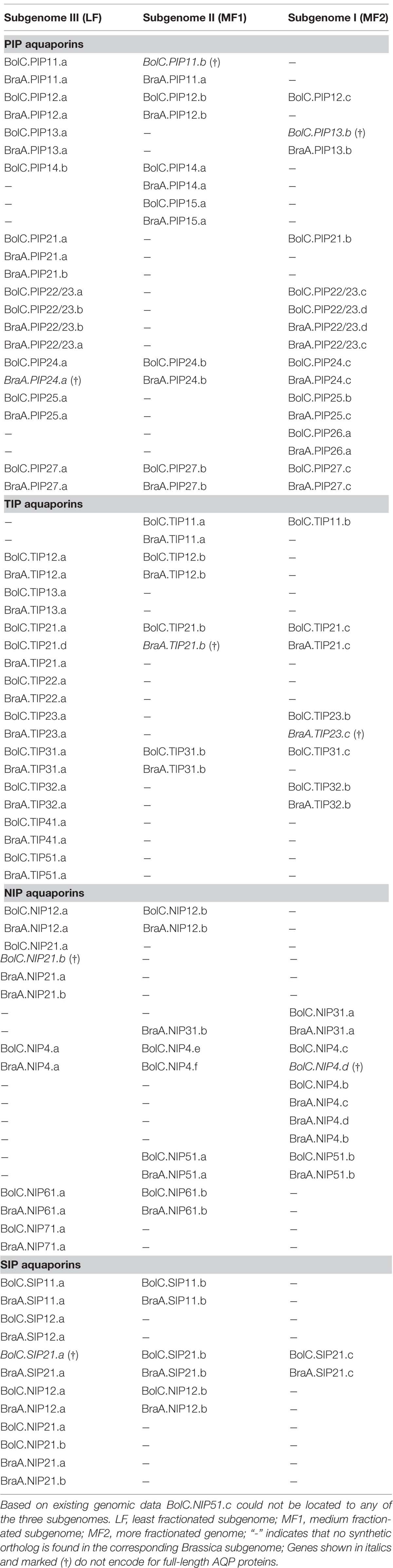
TABLE 2. Distribution of aquaporin genes in the three subgenomes of Brassica oleracea and Brassica rapa.
The association to chromosomes (or unanchored scaffolds), their exact position on the latter (manually corrected for the non-satisfactorily annotated AQPs) and the affiliation to each of the three subgenomes and to the blocks of the ancient Brassica karyotype were determined, listed, and processed (Tables 1 and 2; Figure 1). Only one gene (BolC.NIP51.c) was not assigned to any of the three subgenomes as the surrounding genes on the associated scaffold belonged to different karyotype blocks. A manual assignment was also not successful.
Interestingly, AQP genes which are specifically (AtNIP7;1 and AtTIP5;1) or non-specifically (AtPIP2;6, AtTIP3;1, and AtSIP1;2) expressed in Arabidopsis flowers only possess one single isoform sequence within the triplicated Brassica genomes. Except from BolC.TIP51.a and BolC.NIP71.a, for which no transcripts could be detected at all, all the other orthologous isoforms are also present in B. rapa and B. oleracea flowers (Tao et al., 2014). It will be interesting to analyze whether reproductive organ-specific reduction of redundant genes encoding for transport proteins is also observed for other protein families. One could speculate that the fine regulation of AQP-mediated water and solute transport within a specific tissue of the reproductive organs is hampered by multiple paralogs and that the loss of redundant isoforms therein represents an evolutionary advantage.
Phylogenetic Analysis of Brassica AQPs
The identified full-length Brassica AQPs (67 B. oleracea AQPs and 59 B. rapa AQPs) were subjected to phylogenetic analyses using both the coding nucleotide and corresponding amino acid sequences. Both analyses resulted in similar phylogenetic relationships of all AQP isoforms with only slightly differing supports of specific nodes (Figure 2). Overall, B. oleraceae AQPs followed the phylogenetic pattern of Arabidopsis and clustered into four distinct subfamilies: the PIPs, TIPs, NIPs, and SIPs (Figure 2). Neither in B. oleracea nor in B. rapa were additional phylogenetic AQP subgroups within the four subfamilies formed since the divergence of these species from Arabidopsis. Sequences encoding for XIP proteins were not found. This is in agreement with the present hypothesis that XIPs are absent in Brassicaceae (Danielson and Johanson, 2008).
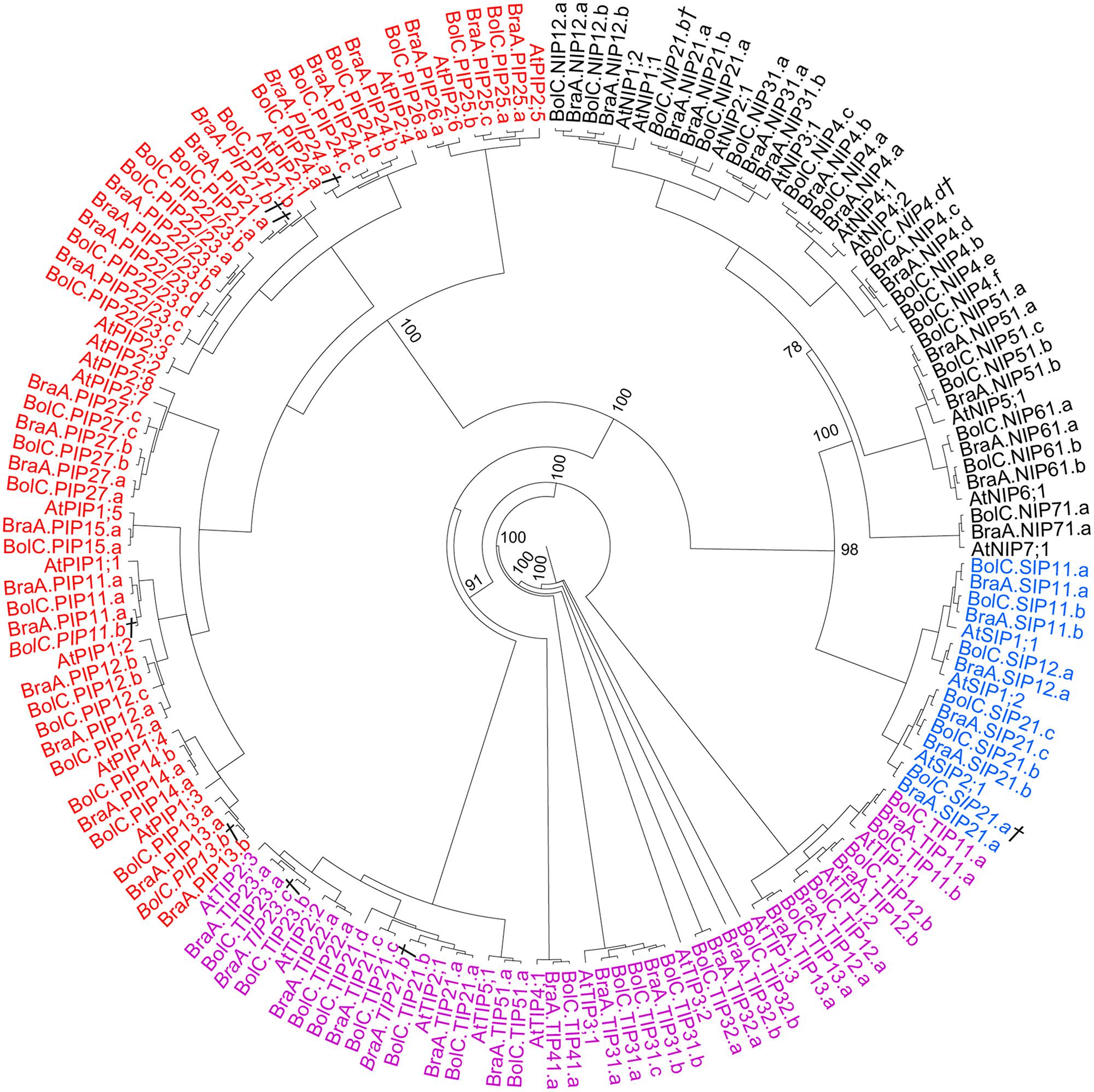
FIGURE 2. Phylogeny of AQPs. Phylogenetic tree derived from AQP cDNA sequences of B. oleracea, B. rapa, and A. thaliana using Bayesian phylogenetic inference. Numbers beside the nodes indicate the posterior probability values >75% for the inner nodes (more details in the separate trees for each AQP subfamily). Genes displayed in black belong to the NIP-, in red to the PIP-, in magenta to the TIP-, and in blue to the SIP-AQP subfamily. Gene IDs which are written in italics and/or marked with a cross (†) do not encode for full-length AQPs due to mutations leading to frameshifts or stop codons in their sequence. Gene IDs which are marked with a double cross (††) represent only very short AQP pseudo-gene fragment sequences.
PIP Subfamily – Phylogeny and Specific Features of PIP Aquaporins
As in other plant species the B. oleracea PIP subfamily is phylogenetically divided into two subgroups, the PIP1s and PIP2s. Brassicaceae PIP2;7/2;8 orthologs seem to form a distinct PIP subgroup. PIP2;7/2;8 isoforms clearly separate basal from the other PIP2 isoforms with a strong node support (Figure 3). To date no unique function or molecular characteristic could be attributed to PIP2;7/2;8 isoforms. In this regard, it is interesting that PIP2;8 orthologs do not exist in either of the Brassica species. This suggests that AtPIP2;8 developed after the Brassica–Arabidopsis lineage split or, less likely, disappeared in all six Brassica subgenomes. The gene duplication event resulting in AtPIP2;2 and AtPIP2;3 occurred before the Brassica–Arabidopsis lineage split as this duplication is also seen in both Brassica genomes. B. oleracea and B. rapa encode for 25 and 23 PIP isoforms respectively. The PIP subfamily is the most homogeneous Brassica AQP subfamily with only 510 informative sites and 354 identical sites (37.8%) in the alignment despite the fact that the subfamily has the highest number of isoforms among all Brassicaceae species (Table 3).

FIGURE 3. Phylogeny of PIPs. Phylogenetic tree derived from PIP cDNA sequences of B. oleracea, B. rapa, and A. thaliana using Bayesian phylogenetic inference. Numbers beside the nodes indicate the posterior probability values >75%. Gene IDs which are written in italics and/or marked with a cross (†) do not encode for full-length AQPs due to mutations leading to frameshifts or stop codons in their sequence. Gene IDs which are marked with a double cross (††) represent only very short AQP pseudo-gene fragment sequences.
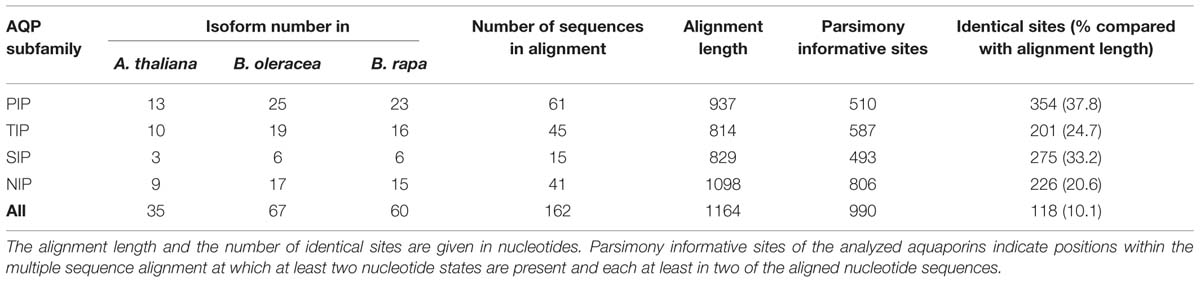
TABLE 3. Alignment statistics of all Arabidopsis thaliana, Brassica oleracea, and Brassica rapa aquaporin nucleotide sequences.
All NPA motifs and ar/R selectivity filters are identical (Table 4). This indicates that a strong selectivity pressure is acting on PIP water channels and that each amino acid divergence will result in a selective disadvantage for the plant, despite the fact that they form the member-richest AQP subfamily within most plants. This might be due to the fact that the maintenance of an efficient temporal and spatial control of the water homeostasis is extremely important for plants and is regulated by the interplay of various isoforms.
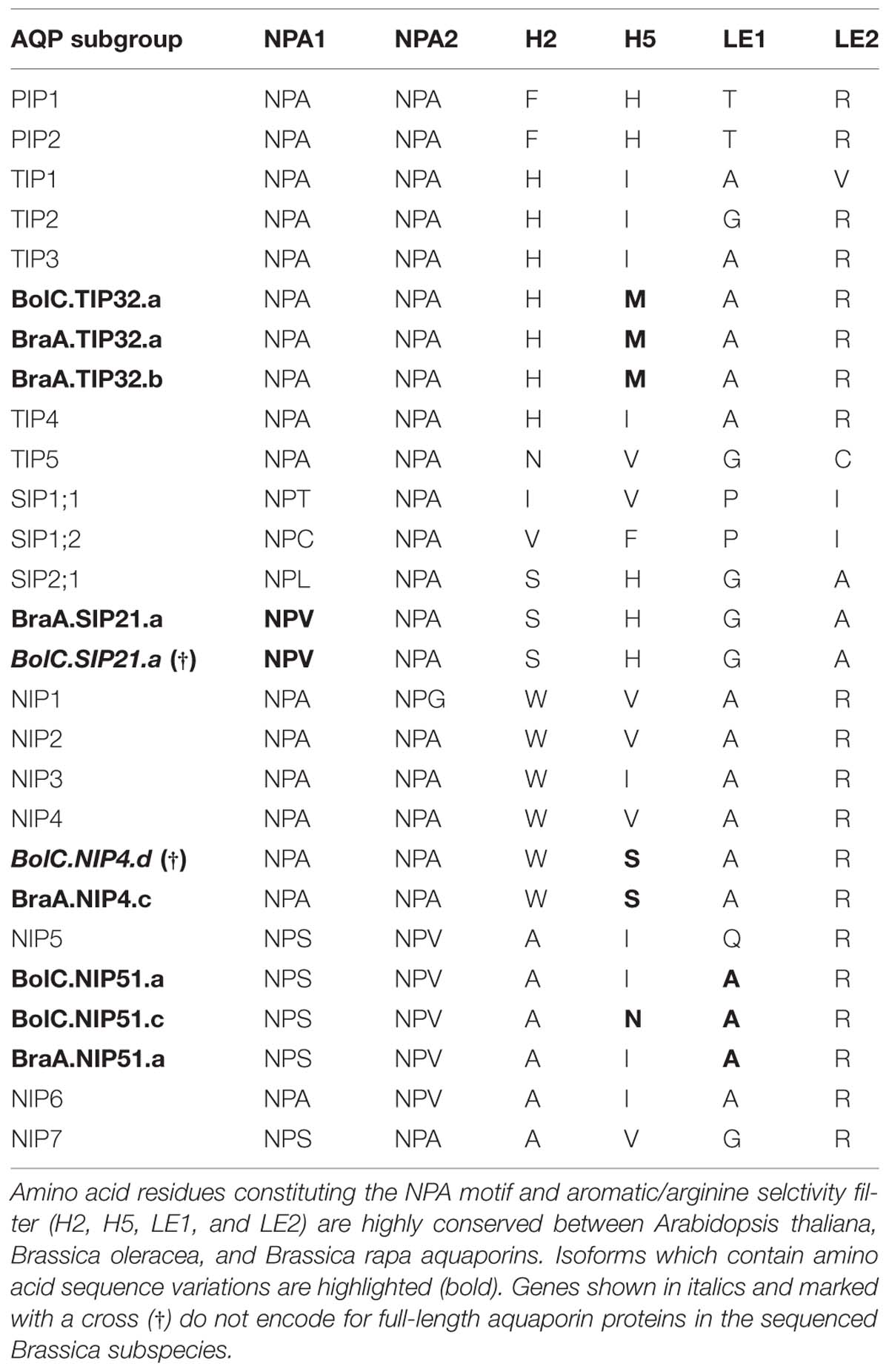
TABLE 4. Amino acid composition of the NPA motif and aromatic/arginine selctivity filters of Arabidopsis thaliana, Brassica oleracea, and Brassica rapa aquaporins.
TIP Subfamily – Phylogeny and Specific Features of TIP Aquaporins
All TIP cDNA sequences included in the study have 24.7% of identical sites. The overall nucleotide sequence identity is lower in comparison to the SIP and PIP subfamilies (Table 3). B. oleracea and B. rapa possess 19 and 16 TIP isoforms, respectively (Table 3). Within each TIP subgroup of both Brassica species the nucleotide sequences of all isoforms are highly conserved (>90%), with the exception of BolC.TIP21.d, which shares only 79% of nucleotide sites with the other TIP21 isoforms. The high degree of TIP sequence conservation is also reflected in the high node support (Figure 4). NPA motifs and the ar/R selectivity filters are identical within the subgroups with the exception of BolC.TIP32.a, BraA.TIP32.a, and BraA.TIP32.b which possess a methionine instead of an isoleucine in the R2 position of the ar/R selectivity filter (Table 4). It is still to be experimentally addressed if this residue exchange results in a differential substrate selectivity.
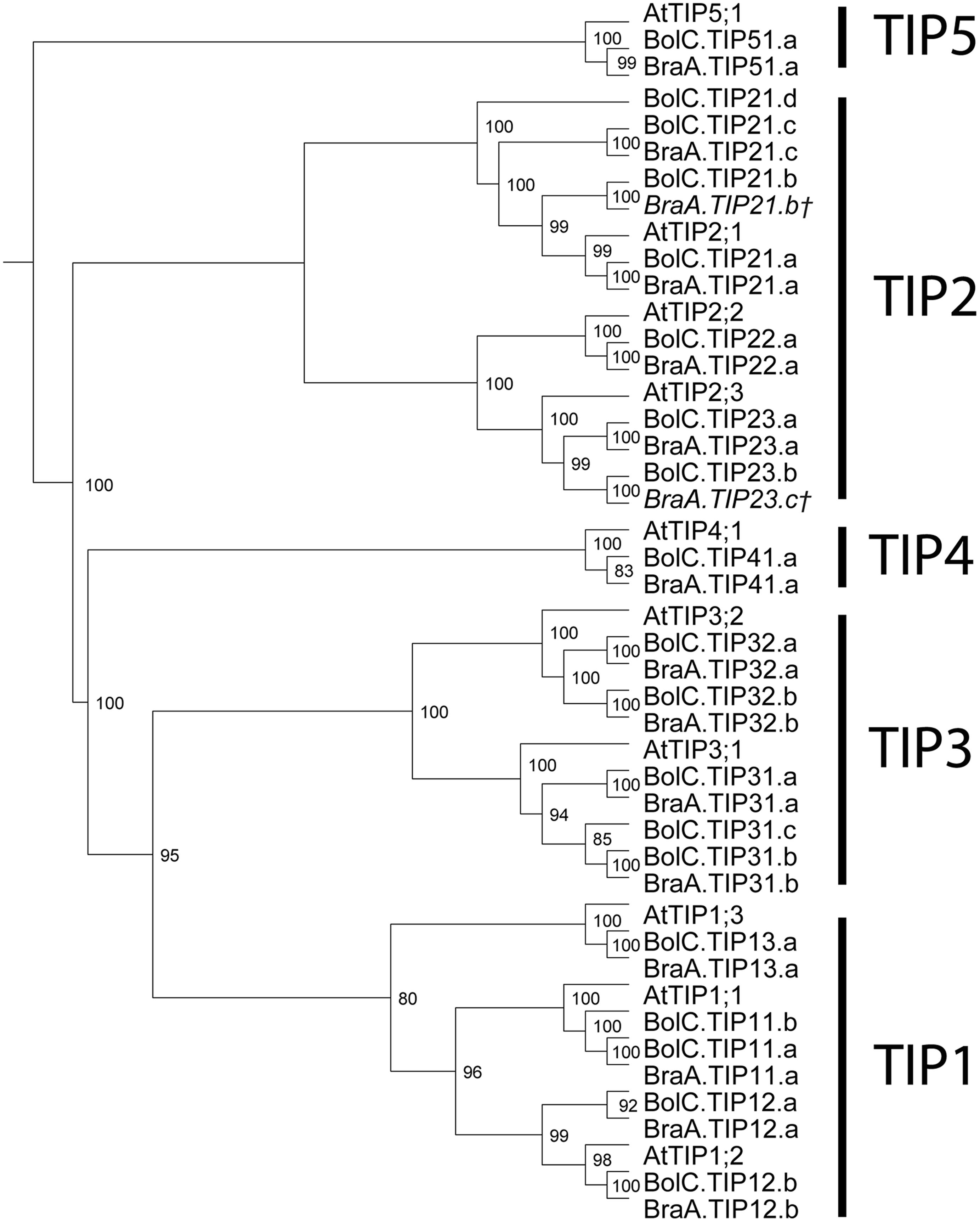
FIGURE 4. Phylogeny of TIPs. Phylogenetic tree derived from TIP cDNA sequences of B. oleracea, B. rapa, and A. thaliana using Bayesian phylogenetic inference. Numbers beside the nodes indicate the posterior probability values >75%. Gene IDs which are written in italics and/or marked with a cross (†) do not encode for full-length AQPs due to mutations leading to frameshifts or stop codons in their sequence.
SIP Subfamily – Phylogeny and Specific Features of SIP Aquaporins
Overall, the SIP cDNA sequences between Arabidopsis and the two Brassica crops are more conserved than the TIPs. They have 33.2% identical nucleotide sites (Table 3). All ar/R selectivity filters are identical (Table 4). Brassica SIP21.a isoforms possess NPV/NPA motifs instead of the NPL/NPA motifs of other SIP21 isoforms (Table 4). Interestingly, the number of SIP subgroup isoforms varies between 1 and 3 (Figure 5). Due to the whole genome triplication within Brassica species three isoforms would have been expected. A deactivation of functional copies of SIP21 paralogs can be observed: BolC.SIP21.a carries mutations leading to several stop codons and therefore encodes for a non-functional protein. The physiological functions of Brassica SIPs, and SIPs in general, have yet be to be uncovered.

FIGURE 5. Phylogeny of SIPs. Phylogenetic tree derived from SIP cDNA sequences of B. oleracea, B. rapa, and A. thaliana using Bayesian phylogenetic inference. Numbers beside the nodes indicate the posterior probability values >75%. Gene IDs which are written in italics and/or marked with a cross (†) do not encode for full-length AQPs due to mutations leading to frameshifts or stop codons in their sequence.
NIP Subfamily – Phylogeny and Specific Features of NIP Aquaporins
Amongst the four AQP subfamilies, which occur in all higher plants, the NIP subfamily encompasses the highest sequence diversity (Table 3; Bansal and Sankararamakrishnan, 2007). We wanted to address whether this diversification can already be observed within the closely related Brassica and Arabidopsis lineages. Indeed, our analysis revealed that NIPs have the highest number of informative sites (73.4%) relative to other AQP subfamilies (Table 3). The number of identical sites in the NIP alignment was rather low compared to the other subfamilies suggesting that NIPs form a highly divergent subfamily at the sequence level. The partly observed weak node support for the division into the different NIP subgroups and polytomies within the different NIP subgroups (Figure 6), confirm previous studies (Danielson and Johanson, 2008). The relation between the different NIP subgroups and the knowledge about which subgroup represents a more ancient group remains to be resolved. In contrast to all other AQP subgroups, NIP4 genes might have amplified within the Brassica genus as three instead of two isoforms (as expected due to the gene duplication in Arabidopsis) have been identified in subgenome I (MF2) of both Brassica species. However, the phylogenetic analysis did not allow us to conclude if one of the B. oleracea or B. rapa NIP4.b, NIP4.c, NIP4.d isoforms (arranged in a triplicate in subgenome I) was originally the paralog of BolC.NIP4.a or BraA.NIP4.a (subgenome III) and subsequently fused to the preexisting gene duplicate, or whether the paralog of Brassica NIP4.a was lost and one of the duplicated isoforms within subgenome I duplicated again. NIP4 sequence information from additional Brassica (sub-) species will be necessary to resolve the exact phylogenetic relationship of these paralogs within and between species. The large number of NIP4 genes within both Brassica genomes is interesting as Brassica NIP4 orthologs are neither present in monocots nor in the genomes of contemporary members of evolutionary old land plants such as P. patens or S. moellendorffii. Furthermore, not all dicots possess NIP4 paralogs or orthologs within their genomes (Abascal et al., 2014). According to Abascal et al. (2014) it is not yet possible to conclude whether the seed plant specific Brassica NIP4 orthologs either got lost in specific species or were newly developed.
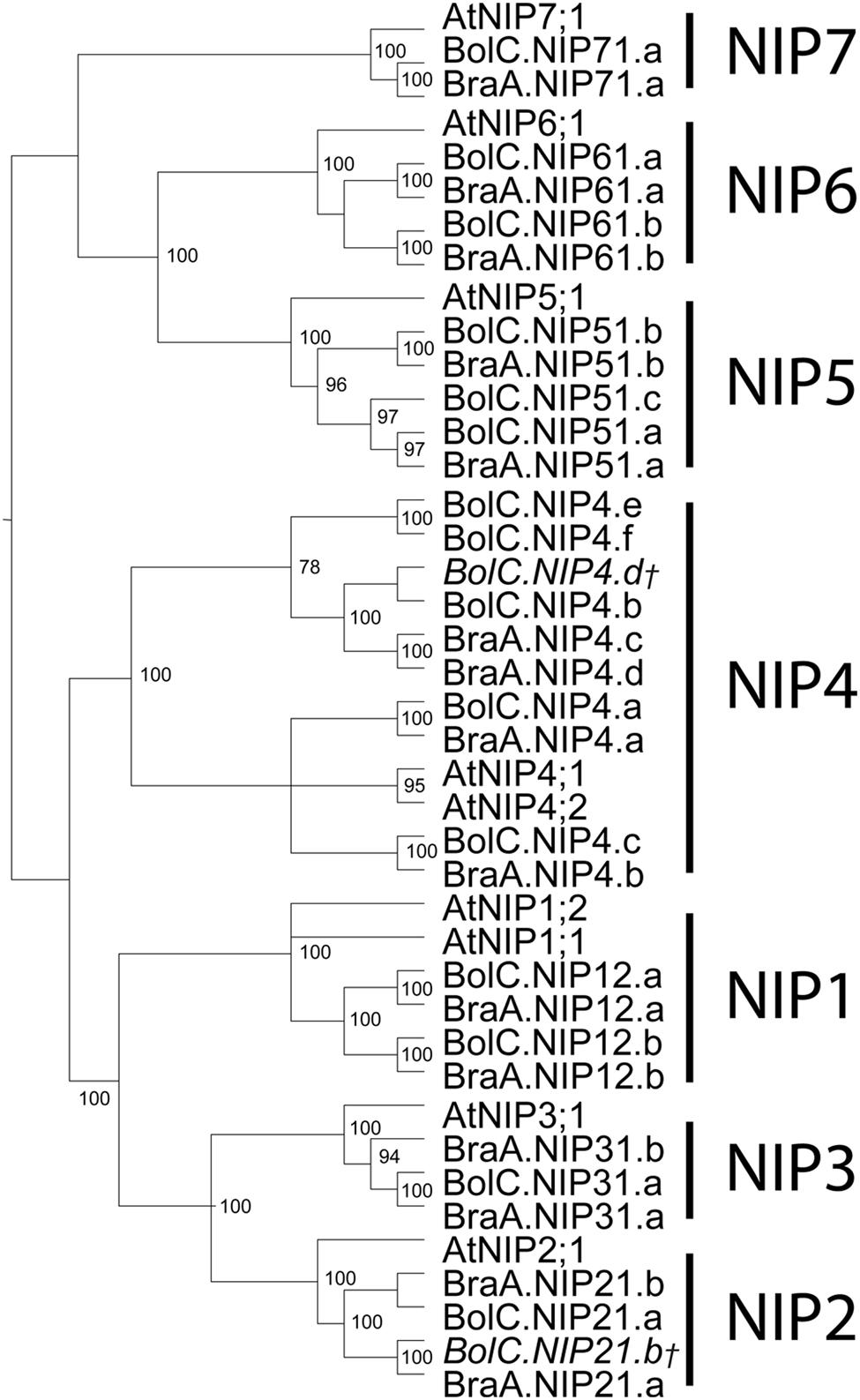
FIGURE 6. Phylogeny of NIPs. Phylogenetic tree derived from NIP cDNA sequences of B. oleracea, B. rapa, and A. thaliana using Bayesian phylogenetic inference. Numbers beside the nodes indicate the posterior probability values >75%. Gene IDs which are written in italics and/or marked with a cross do not encode for full-length AQPs due to mutations leading to frameshifts or stop codons in their sequence.
Orthologous genes of AtNIP1;1 were not found in B. oleracea or in B. rapa (Table 1 and Figure 6). This is interesting as only the Arabidopsis atnip1;1 but not atnip1;2 knockout confers resistance to toxic levels of arsenic and antimony in the growth medium, even though both isoforms are permeable to both metalloids (Kamiya and Fujiwara, 2009; Kamiya et al., 2009). This suggests that only AtNIP1;1 represents an apparently non-controlled uptake pathway for toxic metalloid species. It will be interesting to investigate whether Brassica species which lack NIP1;1 orthologs are more resistant to these compounds.
While most Brassica NIP subgroups have highly conserved residues within the NPA motifs, some isoforms show amino acid residue variation within the ar/R selectivity filter (Table 4). The R2 residue of the ar/R selectivity filter of BraA.NIP4.c is a serine residue. A polar serine residue substitutes a non-polar valine/isoleucine residue in all other NIP subgroups. This substitution probably impacts on substrate selectivity. Two Brassica NIP5 isoforms possess the ar/R selectivity filter composition of NIP6 proteins (Table 4). As AtNIP6;1 was shown to be impermeable to water in contrast to AtNIP5;1 (Wallace and Roberts, 2005; Takano et al., 2006) it will be interesting to test these Brassica NIP5 isoforms for their water permeability.
The sequence identity and genomic structure of NIP6 isoforms are very conserved compared to other NIP subgroups (Figures 6 and 7). In Arabidopsis, AtNIP6;1 is essential for the translocation of the micronutrient boron (Tanaka et al., 2008). AtNIP5;1 and AtNIP6;1 orthologs of other dicot species as well as orthologous monocot NIP3 isoforms transport boric acid in heterologous expression systems and play essential roles in the uptake and distribution of this nutrient in planta (Takano et al., 2006; Tanaka et al., 2008; Durbak et al., 2014; Hanaoka et al., 2014). In dicots NIP5 and NIP6 genes are phylogenetically very closely related (Abascal et al., 2014). Phylogenetic analyses suggest that NIP3 genes of monocots are closer related to Brassicaceae NIP5;1 isoforms and that AtNIP6;1 orthologs are absent in monocots (Abascal et al., 2014; Durbak et al., 2014). The high conservation of NIP6;1 isoforms in Brassica species is striking and suggests a crucial cellular function which is either missing or adopted by other AQPs in monocots.
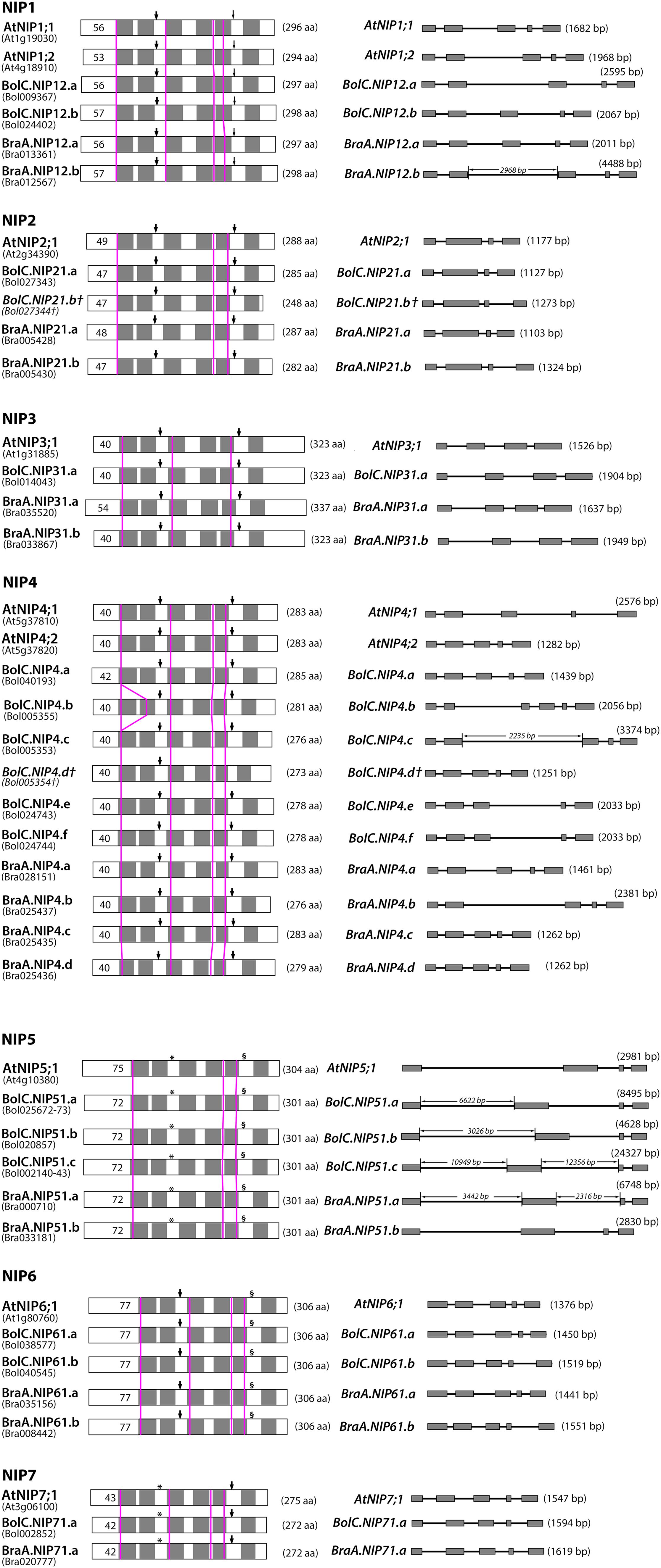
FIGURE 7. Comparison of B. oleracea, B. rapa, and A. thaliana NIP proteins and genes. Left panel: schematic depiction of the hypothetical 2-D structure of NIP proteins. Gray boxes represent predicted transmembrane helixes within the deduced amino acid sequences. Thick arrowheads point toward NPA motifs, thin arrowheads toward NPG motifs (NIP1 only), asterisks (∗) mark NPS motifs (NIP5, NIP7) and section signs (§) mark NPV motifs (NIP5, NIP6). Pink vertical lines indicate the position of the introns in the corresponding nucleotide sequences. Black numbers within boxes indicate the length of the putative N-terminus of each protein and numbers in parentheses behind each protein indicate the putative amino acid chain length. Right panel: intron and exon organization of NIP genes. Gray boxes represent exons, lines between boxes intron sequences. The length of each gene is given in parentheses behind each gene model. The lengths of proteins, exons and introns are size-scaled, so that their absolute lengths can be directly compared to each other (exception: if the intron length is displayed).
Diverse NIP subgroups represent essential metalloid channels within monocots and dicots and play key functions in the regulation of boron, silicon, selenium, arsenic, and antimony transport (Bienert et al., 2008a). On the one hand, all Brassica crops have a very high demand for the essential micronutrient boron, while on the other hand about 25% of the discovered metal and metalloid hyperaccumulating plants (including arsenic, antimony, and selenium hyperaccumulators) belong to the family of Brassicaceae (Rascio and Navari-Izzo, 2011). In this respect, NIP-mediated transport processes should be of special importance for the metalloid metabolism, especially of Brassicaceae plants. A detailed comparative analysis of the gene and protein structure of NIP isoforms within the different species showed that the locations of introns within all NIP sequences were conserved (Figure 7). All NIP1 to NIP7 isoforms possess either three or four introns (Figures 7 and 8). The only exception represents BolC.NIP4.b which possesses an additional intron relative to the other NIP4 members. The first and last intron of all NIP1 to NIP7 isoforms is found in the sequence encoding the beginning of TMH1 and the end of TMH6, respectively (Figure 7).
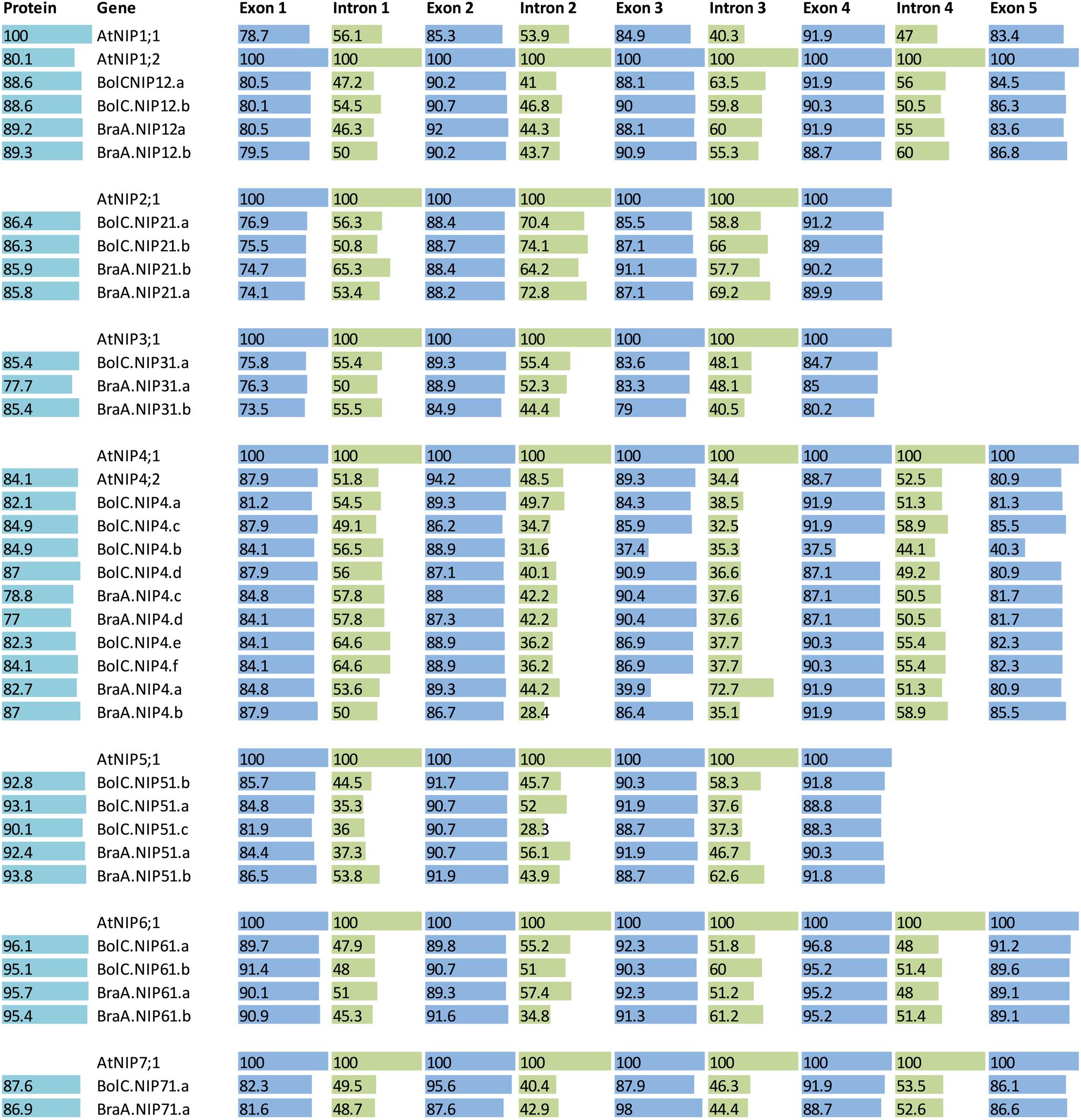
FIGURE 8. Sequence identities of proteins and exons and introns of the different NIP subgroups. A protein and nucleotide identity chart was calculated from an alignment for each exon and intron and each amino acid sequence in each NIP subgroup separately. For each group the AtNIP isoform was set to 100% (bold letters) and the other isoforms were normalized to this. If two At isoforms exist then the most similar isoform (compared with the other genes) was chosen as a standard and set to 100%. Exon and intron identities are displayed in blue and green, respectively and protein identities are displayed in light blue.
The highly conserved intron located in the sequence encoding loop D is missing in all NIP3 isoforms and the otherwise conserved intron separating the exons encoding TMH3 is missing in all NIP2 and NIP5 isoforms. These results show that NIP1, NIP4, NIP6, and NIP7 genes are more similar with respect to their gene structure. The exon and intron lengths within the NIP1, NIP2, NIP3, NIP6, and NIP7 subgroups vary only marginally (Figure 7). Various introns of NIP4 isoforms vary in their length. Interestingly, the size of intron 1 and intron 2 in Brassica NIP5 isoforms varies strongly. While in BraA.NIP51.b the exon and intron lengths are comparable to AtNIP5;1 (about 2900 bp, see Figure 7), all other Brassica NIP5 isoforms possess longer introns. BolC.NIP51.c reached a gene size of more than 24000 bp. Despite these intron elongations, the sequence identities of Brassica NIP5 exons compared to AtNIP5;1 were similar to other NIP subgroups (Figure 8) resulting in highly similar NIP5 protein sequences (Figure 8 and Supplementary File S5). The molecular and evolutionary reasons for this peculiar Brassica NIP5 intron elongation tendency have yet to be resolved.
Another interesting observation was made when comparing the Brassica NIP protein sequences to the orthologous sequences of Arabidopsis (Supplementary File S5). All Brassica NIPs possess certain amino acids within each NIP subgroup which are conserved amongst Brassica NIPs but do not occur in Arabidopsis (Supplementary File S5). These amino acids possess distinct physico-chemical properties compared to the ones in Arabidopsis and are mainly localized to the N and C terminal parts of NIPs. Cytoplasmic termini of NIPs are longer than termini of other plant AQP subfamilies and are hypothesized to be involved in protein regulation, protein trafficking, protein modification, and protein–protein interaction. Distinct termini constitutions suggest that Brassica NIPs possess differential regulative mechanisms additional to those of their Arabidopsis counterparts. Apart from the NIP6 subgroup most other NIP subgroups consist of proteins of varying lengths. The differences in protein length range from 1 to 14 amino acids with the exception of BolC.NIP21.b which is 40 amino acids shorter than AtNIP2;1 due to a stop codon (Figure 7). In most cases these differences are due to a different length of the termini. Loop C of different NIP4s varies by up to two amino acids in length (Figure 7). These variations in sequence length and amino acid composition (Supplementary File S5) are of interest, as loop C was shown to be involved in the substrate selectivity of various AQPs (Beitz et al., 2004; Uzcategui et al., 2008). A single mutation in the extracellular loop C of Leishmania AQP1 resulted in altered substrate selectivity preventing metalloid but not glycerol permeation through the channel (Uzcategui et al., 2008).
The overall similarity between AQPs from Arabidopsis, B. oleracea and B. rapa at the protein level, and especially at the residues constituting the NPA and ar/R selectivity filters, suggests that the knowledge on channel selectivity which was revealed in Arabidopsis might be transferable to Brassica crop isoforms. The next step will be to experimentally verify this hypothesis.
Brassica oleracea Aquaporin Gene Expression in Roots, Leaves, and Flowers
Brassica oleracea illumina RNA-seq data were obtained from the Gene Expression Omnibus database. The transcript reads could be assigned to 58 AQPs of B. oleracea. A heat map displaying the transcript abundance pattern of B. oleracea AQPs in roots, leaves, and flowers was generated (Figure 9A). Forty-nine AQP genes (84.5%) were detected in at least one organ with 41 genes detected in all organs (70.7%) including 23 PIP (95.8% of all detected PIPs), 8 TIP (61.5% of all detected TIPs), 4 SIP (100% of all detected SIPs), and 6 NIP (75% of all detected NIPs) genes.

FIGURE 9. Expression analyses of B. oleracea AQP genes. (A) Visualization of B. oleracea RNA-seq data of flower (GSM1052959), leaf (GSM1052960), and root (GSM1052961) organs. Expression values are given after logarithmic transformation of RPKM (reads per kilobase of exon model per million mapped reads). (B) Assignment of B. oleracea and B. rapa AQP genes to flower, leaf, or root organs based on their expression values. Genes with similar expression patterns than their homologs from A. thaliana are depicted in black. Genes with significant expression in other organs than reported for A. thaliana homologs are depicted in red.
In the PIP subfamily, except for BolC.PIP24.c which was not detected in roots, all other PIPs were detected in all organs with highest expression in the flower. Interestingly, BraA.PIP24 expression was also rarely detected by Tao et al. (2014). In contrast to Brassica PIP24 isoforms, AtPIP2;4 is highly expressed in roots. The high expression levels of most PIPs in all organs of Arabidopsis (Alexandersson et al., 2010), B. rapa (Tao et al., 2014), and B. oleracea (this study) is likely related to their essential function in cellular water transport processes in various physiological conditions.
In the TIP subfamily BolC.TIP11 and BolC.TIP12 isoforms displayed high expression in all organs as it is the case in B. rapa and Arabidopsis. BolC.TIP22.a and BolC.TIP23a/b showed root-specific expression analogous to the orthologs of B. rapa (Tao et al., 2014) and Arabidopsis (Schmid et al., 2005).
In the SIP subfamily all genes were more or less expressed in all organs similar to their orthologs in Arabidopsis. The sequence reads for BolC.SIP21.a and BolC.SIP21.b did not allow discriminating between these two isoforms. As BolC.SIP21.a showed stop -codons in its genomic sequence the data might represent the expression of BolC.SIP21.b.
In the NIP subfamily the transcript reads could not distinguish between BolC.NIP61.a and BolC.NIP61.b. BolC.NIP6 transcript was detected in all three analyzed organs with highest levels in flowers. BolC.NIP6 transcript showed highest expression amongst all BolNIPs. BolC.NIP12 isoforms were lowly expressed in all three organs which is in contrast to the root-specific expression in B. rapa (Tao et al., 2014) but in agreement with expression data from Arabidopsis. NIP3 transcripts were predominantly found in roots of all three Brassicaceae species. Expression of BolC.NIP21 was not detected, which is in line with the exclusive induction of AtNIP2;1 under anoxic conditions (Choi and Roberts, 2007). In B. rapa, BraA.NIP21 transcripts were detected at very low levels (Tao et al., 2014). Expression of NIP4 isoforms, which are pollen-specific in Arabidopsis, was only detected for BolC.NIP4.a at low levels and not detected at all in B. rapa flowers. AtNIP5;1 expression and function is linked to boric acid uptake into the roots of Arabidopsis (Takano et al., 2006). Interestingly, transcripts for all three BolC.NIP5 isoforms were detected in roots but also in leaves and flowers. This suggests that BolC.NIP5 channels play also a role in boron transport in other organs than the root of B. oleracea. In general, members of the NIP subfamily showed lowest expression levels compared to isoforms of the other three plant AQP subfamilies.
Aquaporin orthologs from B. oleracea and B. rapa are mostly expressed in the same organs and at similar levels as far as it can be compared from the RNA-seq data analyzes (Figure 9B). This might reflect the close phylogenetic relationship between these two species. Some AQP isoforms of B. oleracea, exhibit different organ expression patterns compared to their orthologs in Arabidopsis (Figure 9). Either the transcript level in an organ differs substantially or the expression was detected in additional organs. Particularly NIP5, NIP6, and PIP24 isoforms exhibit different organ expression patterns between the three Brassicaceae species. The whole genome triplication in Brassica species resulted in a multiplication of AQP isoform orthologs. While most of the genes for Brassica AQP isoforms are expressed in the same organ-specific manner as their orthologous Arabidopsis isoforms, some of them are expressed in other or additional organs (Figure 9).
Conclusion
The sequence curation and comparative analyses of four plant AQP subfamilies in three Brassicaceae species provide initial insights into AQP evolution in these taxa. Our sequence curation is an important resource for further functional studies on these solute channels in Brassica species. In summary, we identified 67 full-length AQP genes in the cabbage genome belonging to the PIP, TIP, NIP, and SIP subfamilies. We showed that orthologous AQPs generally encode for very similar proteins in the three species and that the overall gene structure is highly conserved. However, AQP isoforms of the different subfamilies and subgroups differ in their copy number. Further knowledge, on genomic sequence variability of non-coding, cis-regulatory, and coding AQP gene elements in a larger number of Brassica species and morphotypes, which have independently developed in diverse environments, and the physiological consequences thereof, will allow to breed cultivars with optimized water and nutrient efficiencies in the future.
Author Contributions
All authors prepared, read, and approved the final manuscript. GB designed the research and wrote the manuscript with the input of all authors. TD and NB performed phylogenetic analyses. TD, BP, AH, and GB collected and analyzed the public datasets.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by an Emmy Noether grant 1668/1-1 from the Deutsche Forschungsgemeinschaft. We thank the consortia for the sequencing of the ‘C’ genome of Brassica oleracea var. capitata cultivar line 02-12 and the ‘A’ genome of B. rapa ssp. pekinensis line Chiifu-401. We are grateful to Jonathan Brassac and Benjamin D. Gruber for helpful comments. We thank the two reviewers for their insightful comments on a previous version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00166/abstract
Footnotes
- ^ http://faostat3.fao.org/
- ^ http://www.arabidopsis.org
- ^ http://www.ocri-genomics.org/bolbase
- ^ http://brassicadb.org
- ^ http://www.geneious.com
- ^ http://plants.ensembl.org
References
Abascal, F., Irisarri, I., and Zardoya, R. (2014). Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 1840, 1468–1481. doi: 10.1016/j.bbagen.2013.12.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Adachi, J., Waddell, P., Martin, W., and Hasegawa, M. (2000). Plastid genome phylogeny and a model of amino acid substitution for proteins encoded by chloroplast DNA. J. Mol. Evol. 50, 348–358.
Agre, P., and Kozono, D. (2003). Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett. 555, 72–78. doi: 10.1016/S0014-5793(03)01083-4
Alexandersson, E., Danielson, J. A., Råde, J., Moparthi, V. K., Fontes, M., Kjellbom, P.,et al. (2010). Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 61, 650–660. doi: 10.1111/j.1365-313X.2009.04087.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Anderberg, H. I., Kjellbom, P., and Johanson, U. (2012). Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sci. 3:33. doi: 10.3389/fpls.2012.00033
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bansal, A., and Sankararamakrishnan, R. (2007). Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Struct. Biol. 7:27. doi: 10.1186/1472-6807-7-27
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Beitz, E., Pavlovic-Djuranovic, S., Yasui, M., Agre, P., and Schultz, J. E. (2004). Molecular dissection of water and glycerol permeability of the aquaglyceroporin from Plasmodium falciparum by mutational analysis. Proc. Natl. Acad. Sci. U.S.A. 101, 1153–1158. doi: 10.1073/pnas.0307295101
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bienert, G. P., Desguin, B., Chaumont, F., and Hols, P. (2013). Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem. J. 454, 559–570. doi: 10.1042/BJ20130388
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bienert, G. P., Møller, A. L., Kristiansen, K. A., Schulz, A., Møller, I. M., Schjoerring, J. K.,et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. doi: 10.1074/jbc.M603761200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bienert, G. P., Schüssler, M. D., and Jahn, T. P. (2008a). Metalloids: essential, beneficial or toxic? Major intrinsic proteins sort it out. Trends Biochem. Sci. 33, 20–26. doi: 10.1016/j.tibs.2007.10.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bienert, G. P., Thorsen, M., Schüssler, M. D., Nilsson, H. R., Wagner, A., Tamás, M. J.,et al. (2008b). A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol. 6:26. doi: 10.1186/1741-7007-6-26
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M. J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. doi: 10.1104/pp.125.3.1206
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chaumont, F., and Tyerman, S. D. (2014). Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 164, 1600–1618. doi: 10.1104/pp.113.233791
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Choi, W. G., and Roberts, D. M. (2007). Arabidopsis NIP2;1, a major intrinsic protein transporter of lactic acid induced by anoxic stress. J. Biol. Chem. 282, 24209–24218. doi: 10.1074/jbc.M700982200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Danielson, J. A., and Johanson, U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8:45. doi: 10.1186/1471-2229-8-45
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Durbak, A. R., Phillips, K. A., Pike, S., O’Neill, M. A., Mares, J., Gallavotti, A.,et al. (2014). Transport of boron by the tassel-less1 aquaporin Is critical for vegetative and reproductive development in maize. Plant Cell 6, 2978–2995. doi: 10.1105/tpc.114.125898
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dynowski, M., Schaaf, G., Loque, D., Moran, O., and Ludewig, U. (2008). Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 414, 53–61. doi: 10.1042/BJ20080287
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gomes, D., Agasse, A., Thiébaud, P., Delrot, S., Gerós, H., and Chaumont, F. (2009). Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochim. Biophys. Acta 1788, 1213–1228. doi: 10.1016/j.bbamem.2009.03.009
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gupta, A. B., and Sankararamakrishnan, R. (2009). Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 9:134. doi: 10.1186/1471-2229-9-134
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hanaoka, H., Uraguchi, S., Takano, J., Tanaka, M., and Fujiwara, T. (2014). OsNIP3;1, a rice boric acid channel, regulates boron distribution and is essential for growth under boron-deficient conditions. Plant J. 78, 890–902. doi: 10.1111/tpj.12511
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hasegawa, M., Kishino, H., and Yano, T. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. doi: 10.1007/BF02101694
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Herrera, M., Hong, N. J., and Garvin, J. L. (2006). Aquaporin-1 transports NO across cell membranes. Hypertension 48, 157–164. doi: 10.1161/01.HYP.0000223652.29338.77
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hu, T. T., Pattyn, P., Bakker, E. G., Cao, J., Cheng, J. F., Clark, R. M.,et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481. doi: 10.1038/ng.807
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jahn, T. P., Møller, A. L., Zeuthen, T., Holm, L. M., Klaerke, D. A., Mohsin, B.,et al. (2004). Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett. 574, 31–36. doi: 10.1016/j.febslet.2004.08.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjövall, S., Fraysse, L.,et al. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369. doi: 10.1104/pp.126.4.1358
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kamiya, T., and Fujiwara, T. (2009). Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant Cell Physiol. 50, 1977–1981. doi: 10.1093/pcp/pcp130
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kamiya, T., Tanaka, M., Mitani, N., Ma, J. F., Maeshima, M., and Fujiwara, T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284, 2114–2120. doi: 10.1074/jbc.M806881200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, L. H., Ludewig, U., Gassert, B., Frommer, W. B., and von Wirén, N. (2003). Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol. 133, 1220–1228. doi: 10.1104/pp.103.027409
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A.,et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Loqué, D., Ludewig, U., Yuan, L., and von Wirén, N. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. doi: 10.1104/pp.104.051268
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ma, J. F., Tamai, K., Yamaji, N., Mitani, N., Konishi, S., Katsuhara, M.,et al. (2006). A silicon transporter in rice. Nature 440, 688–691. doi: 10.1038/nature04590
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ma, J. F., Yamaji, N., Mitani, N., Xu, X. Y., Su, Y. H., and McGrath, S. P.,et al. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U.S.A. 105, 9931–9935. doi: 10.1073/pnas.0802361105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Maurel, C., Verdoucq, L., Luu, D. T., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant. Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mollapour, M., and Piper, P. W. (2007). Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell Biol. 27, 6446–6456. doi: 10.1128/MCB.02205-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Murata, K., Mitsuoka, K., Hirai, T., Walz, T., Agre, P., Heymann, J. B.,et al. (2000). Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605. doi: 10.1128/MCB.02205-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nagaharu, U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot. 7, 389–452. doi: 10.1038/35036519
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ostergaard, L., and King, G. J. (2008). Standardized gene nomenclature for the Brassica genus. Plant Methods 4:10. doi: 10.1186/1746-4811-4-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Park, W., Scheffler, B. E., Bauer, P. J., and Campbell, B. T. (2010). Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 10:142. doi: 10.1186/1471-2229-10-142
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Parkin, I. A., Sharpe, A. G., and Lydiate, D. J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome 46, 291–303. doi: 10.1139/g03-006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rambaut, A., and Drummond, A. (2007). TRACER, Vo1. 5. Available at: http://beast.bio.ed.ac.uk/Tracer [accessed June 11, 2011].
Rascio, N., and Navari-Izzo, F. (2011). Heavy metal hyperaccumulating plants: how and why do they do it? And what makes them so interesting? Plant Sci. 180, 169–181. doi: 10.1016/j.plantsci.2010.08.016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reuscher, S., Akiyama, M., Mori, C., Aoki, K., Shibata, D., and Shiratake, K. (2013). Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 8:e79052. doi: 10.1371/journal.pone.0079052
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sade, N., Vinocur, B. J., Diber, A., Shatil, A., Ronen, G., Nissan, H.,et al. (2009). Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 181, 651–661. doi: 10.1111/j.1469-8137.2008.02689.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sakurai, J., Ishikawa, F., Yamaguchi, T., Uemura, M., and Maeshima, M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. doi: 10.1093/pcp/pci172
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M.,et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schranz, M. E., Lysak, M. A., and Mitchell-Olds, T. (2006). The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. doi: 10.1016/j.tplants.2006.09.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Swofford, D. L. (2003). PAUP∗. Phylogenetic Analysis Using Parsimony (∗ and Other Methods). Version 4. Sunderland, MA: Sinauer Associates.
Takano, J., Wada, M., Ludewig, U., Schaaf, G., von Wirén, N., and Fujiwara, T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18, 1498–1509. doi: 10.1105/tpc.106.041640
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tanaka, M., Wallace, I. S., Takano, J., Roberts, D. M., and Fujiwara, T. (2008). NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20, 2860–2875. doi: 10.1105/tpc.108.058628
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tao, P., Zhong, X., Li, B., Wang, W., Yue, Z., Lei, J.,et al. (2014). Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics 289, 1131–1145. doi: 10.1007/s00438-014-0874-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tsukaguchi, H., Shayakul, C., Berger, U. V., Mackenzie, B., Devidas, S., Guggino, W. B.,et al. (1998). Molecular characterization of a broad selectivity neutral solute channel. J. Biol. Chem. 273, 24737–24743. doi: 10.1074/jbc.273.38.24737
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Uehlein, N., Lovisolo, C., Siefritz, F., and Kaldenhoff, R. (2003). The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737. doi: 10.1038/nature02027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Uehlein, N., Otto, B., Hanson, D. T., Fischer, M., McDowell, N., and Kaldenhoff, R. (2008). Function of Nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell 20, 648–657. doi: 10.1105/tpc.107.054023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Uzcategui, N. L., Zhou, Y., Figarella, K., Ye, J., Mukhopadhyay, R., and Bhattacharjee, H. (2008). Alteration in glycerol and metalloid permeability by a single mutation in the extracellular C-loop of Leishmania major aquaglyceroporin LmAQP1. Mol. Microbiol. 70, 1477–1486. doi: 10.1111/j.1365-2958.2008.06494.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Venkatesh, J., Yu, J. W., and Park, S. W. (2013). Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 73, 392–404. doi: 10.1016/j.plaphy.2013.10.025
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wallace, I. S., and Roberts, D. M. (2005). Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44, 16826–16834. doi: 10.1021/bi0511888
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S.,et al. (2011). Brassica rapa genome sequencing project consortium: the genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yang, T. J., Kim, J. S., Kwon, S. J., Lim, K. B., Choi, B. S., Kim, J. A.,et al. (2006). Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18, 1339–1347. doi: 10.1105/tpc.105.040535
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yang, W. Y., Lai, K. N., Pon-Yean, T., and Wen-Hsiung, L. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48, 597–604. doi: 10.1007/PL00006502
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, D. Y., Ali, Z., Wang, C. B., Xu, L., Yi, J. X., Xu, Z. L.,et al. (2013). Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8:e56312. doi: 10.1371/journal.pone.0056312
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, X. Q., Mitani, N., Yamaji, N., Shen, R. F., and Ma, J. F. (2010). Involvement of silicon influx transporter OsNIP2;1 in selenite uptake in rice. Plant Physiol. 153, 1871–1877. doi: 10.1104/pp.110.157867
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: aquaporin, major intrinsic protein, nodulin26-like intrinsic proteins, Brassica oleracea, Brassica rapa, Arabidopsis thaliana, phylogeny, gene structure
Citation: Diehn TA, Pommerrenig B, Bernhardt N, Hartmann A and Bienert GP (2015) Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front. Plant Sci. 6:166. doi: 10.3389/fpls.2015.00166
Received: 20 December 2014; Accepted: 02 March 2015;
Published online: 07 April 2015
Edited by:
Yi-Fang Tsay, Academia Sinica, TaiwanReviewed by:
Christophe Maurel, Institut National de la Recherche Agronomique, FranceChung-Jui Tsai, University of Georgia, USA
Copyright © 2015 Diehn, Pommerrenig, Bernhardt, Hartmann and Bienert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerd P. Bienert, Metalloid Transport Group, Department of Physiology and Cell Biology, Leibniz Institute of Plant Genetics and Crop Plant Research, Corrensstrasse 3, 06466 Gatersleben, Germany bienert@ipk-gatersleben.de
 Till A. Diehn
Till A. Diehn Benjamin Pommerrenig
Benjamin Pommerrenig Nadine Bernhardt
Nadine Bernhardt Anja Hartmann
Anja Hartmann Gerd P. Bienert
Gerd P. Bienert