- 1Center for Alternatives to Animal Testing (CAAT), Department of Environmental Health and Engineering, Bloomberg School of Public Health and Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States
- 2Department of Biostatistics, Johns Hopkins Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, United States
- 3Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD, United States
- 4Department of Materials Science and Engineering, Johns Hopkins University, Baltimore, MD, United States
- 5Department of Chemistry, Johns Hopkins University, Baltimore, MD, United States
- 6Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 7Laboratory for Computational Sensing and Robotics (LCSR), Johns Hopkins University, Baltimore, MD, United States
- 8Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 9Department of Ophthalmology, Johns Hopkins Hospital, Baltimore, MD, United States
- 10Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 11Berman Institute of Bioethics, Johns Hopkins University, Baltimore, MD, United States
- 12Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, United States
- 13Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, United States
- 14Research and Exploratory Development Department, Johns Hopkins University Applied Physics Laboratory, Laurel, MD, United States
- 15Cortical Labs, Melbourne, VIC, Australia
- 16Department of Health Policy and Management, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, United States
- 17Department of Pediatrics, Stem Cell Program, University of California, San Diego, San Diego, CA, United States
- 18Department of Cellular & Molecular Medicine, Stem Cell Program, University of California, San Diego, San Diego, CA, United States
- 19Applied Physics Laboratory, Johns Hopkins University, Baltimore, MD, United States
- 20Luxembourg Centre for Systems Biomedicine (LCSB), University of Luxembourg, Luxembourg, Luxembourg
- 21Department of Computer Science, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States
- 22Department of Physics and Astronomy, Krieger School of Arts & Sciences, Johns Hopkins University, Baltimore, MD, United States
- 23Mark Foundation Center for Advanced Genomics and Imaging, Johns Hopkins University, Baltimore, MD, United States
- 24Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 25CAAT-Europe, University of Konstanz, Konstanz, Germany
Abstract
Recent advances in human stem cell-derived brain organoids promise to replicate critical molecular and cellular aspects of learning and memory and possibly aspects of cognition in vitro. Coining the term “organoid intelligence” (OI) to encompass these developments, we present a collaborative program to implement the vision of a multidisciplinary field of OI. This aims to establish OI as a form of genuine biological computing that harnesses brain organoids using scientific and bioengineering advances in an ethically responsible manner. Standardized, 3D, myelinated brain organoids can now be produced with high cell density and enriched levels of glial cells and gene expression critical for learning. Integrated microfluidic perfusion systems can support scalable and durable culturing, and spatiotemporal chemical signaling. Novel 3D microelectrode arrays permit high-resolution spatiotemporal electrophysiological signaling and recording to explore the capacity of brain organoids to recapitulate the molecular mechanisms of learning and memory formation and, ultimately, their computational potential. Technologies that could enable novel biocomputing models via stimulus-response training and organoid-computer interfaces are in development. We envisage complex, networked interfaces whereby brain organoids are connected with real-world sensors and output devices, and ultimately with each other and with sensory organ organoids (e.g. retinal organoids), and are trained using biofeedback, big-data warehousing, and machine learning methods. In parallel, we emphasize an embedded ethics approach to analyze the ethical aspects raised by OI research in an iterative, collaborative manner involving all relevant stakeholders. The many possible applications of this research urge the strategic development of OI as a scientific discipline. We anticipate OI-based biocomputing systems to allow faster decision-making, continuous learning during tasks, and greater energy and data efficiency. Furthermore, the development of “intelligence-in-a-dish” could help elucidate the pathophysiology of devastating developmental and degenerative diseases (such as dementia), potentially aiding the identification of novel therapeutic approaches to address major global unmet needs.
Key points
- Biological computing (or biocomputing) could be faster, more efficient, and more powerful than silicon-based computing and AI, and only require a fraction of the energy.
- ‘Organoid intelligence’ (OI) describes an emerging multidisciplinary field working to develop biological computing using 3D cultures of human brain cells (brain organoids) and brain-machine interface technologies.
- OI requires scaling up current brain organoids into complex, durable 3D structures enriched with cells and genes associated with learning, and connecting these to next-generation input and output devices and AI/machine learning systems.
- OI requires new models, algorithms, and interface technologies to communicate with brain organoids, understand how they learn and compute, and process and store the massive amounts of data they will generate.
- OI research could also improve our understanding of brain development, learning, and memory, potentially helping to find treatments for neurological disorders such as dementia.
- Ensuring OI develops in an ethically and socially responsive manner requires an ‘embedded ethics’ approach where interdisciplinary and representative teams of ethicists, researchers, and members of the public identify, discuss, and analyze ethical issues and feed these back to inform future research and work.
Introduction
Human brains are slower than machines at processing simple information, such as arithmetic, but they far surpass machines in processing complex information as brains deal better with few and/or uncertain data. Brains can perform both sequential and parallel processing (whereas computers can do only the former), and they outperform computers in decision-making on large, highly heterogeneous, and incomplete datasets and other challenging forms of processing. The processing power of the brain is illustrated by the observation that in 2013, the world’s fourth-largest computer took 40 minutes to model 1 second of 1% of a human’s brain activity (1). Moreover, each brain has a storage capacity estimated at 2,500 TB, based on its 86–100 billion neurons having more than 1015 connections (2, 3). In this article, we describe the emerging field that we term “organoid intelligence” (OI), which aims to leverage the extraordinary biological processing power of the brain.
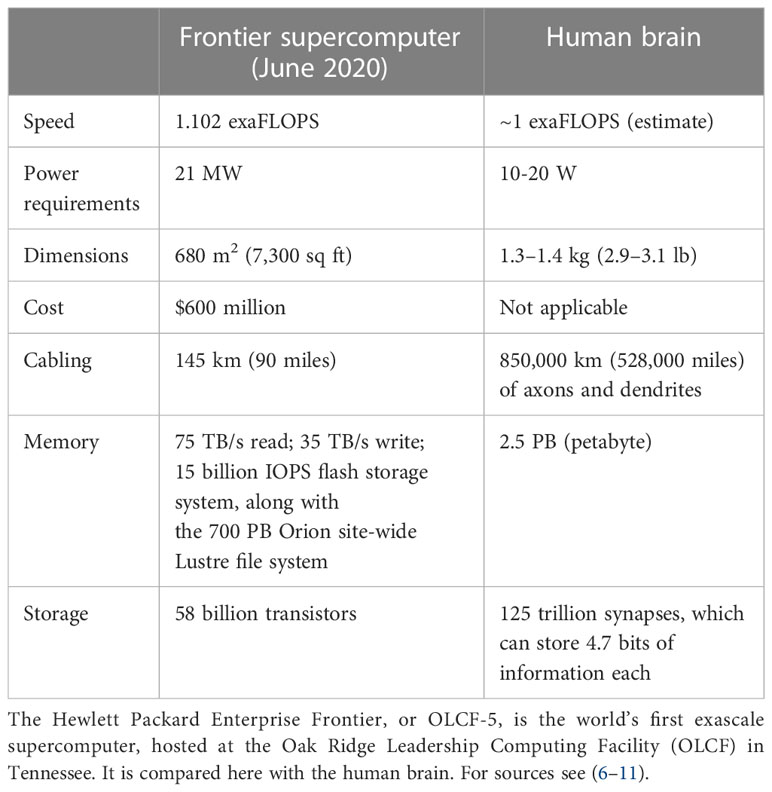
Superficially, both biological learning and machine learning/AI by an intelligent agent build internal representations of the world to improve their performance in conducting tasks. However, fundamental differences between biological and machine learning in the mechanisms of implementation and their goals result in two drastically different efficiencies. First, biological learning uses far less power to solve computational problems. For example, a larval zebrafish navigates the world to successfully hunt prey and avoid predators (4) using only 0.1 microwatts (5), while a human adult consumes 100 watts, of which brain consumption constitutes 20% (6, 7). In contrast, clusters used to master state-of-the-art machine learning models typically operate at around 106 watts. Since June 2022, the USA’s Frontier has been the world’s most powerful supercomputer, reaching 1102 petaFlops (1.102 exaFlops) on the LINPACK benchmarks. The power consumption of the new supercomputer is 21 megawatts, while the human brain operates at the estimated same 1 exaFlop and consumes only 20 watts (Table 1) (8–11). Thus, humans operate at a 106-fold better power efficiency relative to modern machines albeit while performing quite different tasks.
Second, biological learning uses fewer observations to learn how to solve problems. For example, humans learn a simple “same-versus-different” task using around 10 training samples (12); simpler organisms, such as honeybees, also need remarkably few samples (~102) (13). In contrast, in 2011, machines could not learn these distinctions even with 106 samples (14) and in 2018, 107 samples remained insufficient (15). Thus, in this sense, at least, humans operate at a >106 times better data efficiency than modern machines. The AlphaGo system, which beat the world champion at the complex game Go, offers a concrete illustration (16, 17). AlphaGo was trained on data from 160,000 games (17); a human playing for five hours/day would have to play continuously for more than 175 years to experience the same number of training games – indicating the far higher efficiency of the brain in this complex learning activity. The implication is that AI and machine-learning approaches have limited usefulness for tasks requiring real-time learning and dynamic actions in a changing environment. The power and efficiency advantages of biological computing over machine learning are multiplicative. If it takes the same amount of time per sample in a human or machine, then the total energy spent to learn a new task requires 1010 times more energy for the machine. AlphaGo was trained for 4 weeks using 50 graphics processing units (GPUs) (17), requiring approximately 4 ×1010 J of energy – about the same amount of energy required to sustain the metabolism of an active adult human for a decade. This high energy consumption prevents AI from achieving many aspirational goals, for example matching or exceeding human capabilities for complex tasks such as driving (18). Even large multinational corporations are beginning to reach the limits of machine learning owing to its inefficiencies (19), and the associated exponential increase in energy consumption is unsustainable (20), especially if technology companies are to adhere to their commitments to become carbon negative by 2030 (21, 22). At a national level, already in 2016 it took the equivalent of 34 coal-powered plants, each generating 500 megawatts, to meet the power demands of US-based data centers (23). Being much more energy efficient than current computers, human brains could theoretically meet the same US data storage capacity using only 1,600 kilowatts of energy. Notably, the power demands of any current or future implementation of OI is very different from the energy consumption of the human body, especially considering the energy footprint of modern cell culture relative to small organoids today. These comparisons of brains and computers serve only as illustrations of the high efficiency of the human brain.
Together, these observations have created high expectations for biological, brain-directed computing (24–26) as an alternative to silicon-based computing, with the potential for unprecedented advances in computing speed, processing power, data efficiency, and storage capabilities – all with lower energy needs. However, realizing the potential of biocomputing has proved challenging, and most research remains in its infancy. To date, the term “biological computing” has been used mainly to describe the use of DNA to store digital data (27, 28). An exception to this is the recent work by Kagan et al. (29), which uses the term “synthetic biological intelligence” (SBI) to describe the use of synthetic biology to generate intelligent systems through brain-directed computing, albeit using only simple 2D monolayer cell cultures (which poorly replicate the complexity of the in vivo brain) as a proof-of-concept.
We have coined the term “organoid intelligence” (OI) to describe an emerging field aiming to expand the definition of biocomputing toward brain-directed OI computing, i.e. to leverage the self-assembled machinery of 3D human brain cell cultures (brain organoids) to memorize and compute inputs. Brain organoids recapitulate organ histoarchitecture and functionality far more closely than traditional 2D cultures. They can contain myelinated axons (30–32) and not only show spontaneous electrophysiological activity (33) but also demonstrate complex oscillatory behavior (34), and exhibit high cell density and layering patterns, all of which make brain organoids superior to traditional monolayer cultures (34–36). The question is: can we learn from and harness the computing capacity of these organoids? Achieving this will require major advances in the interfacing of brain cell cultures and computers. We envision using biofeedback to systematically train organoids with increasingly complex sensory inputs and output opportunities – interfacing the brain organoids with computers, sensors, and machine interfaces to facilitate supervised and unsupervised learning. We use the term “OI” for this approach to stress its complementarity to AI – where computers aim to perform tasks done by brains, often by modeling our understanding of learning. However, while AI aims to make computers more brain-like, OI research will explore how a 3D brain cell culture can be made more computer-like.
The many possible applications of this work include a new generation of biological and hybrid (biological-electronic) computing technologies, together with advances in our understanding of the physiology of cognition, learning, and memory, and the pathophysiological effects of developmental and degenerative diseases, intoxication, and infection – which in turn could stimulate drug development and other interventions. OI also has the potential to unlock new neuromimetic AI algorithms (with the potential to overcome current AI limitations) and aid the development of new brain-computer-interface technology.
The concept of brain-machine interfacing emerged around five decades ago. Before the advent of more complex human neuronal cultures and brain organoids, pioneering work on learning and memory was carried out using primitive animals such as the lamprey, showing long-term potentiation (37). This led to brain-machine interaction studies establishing bidirectional communications between the nervous system and external devices (38). Others used brain slices of different species to study the basic phenomena of learning on a cellular level (39). Neuron cultures were later shown to perform simple robotic tasks or demonstrate increased plasticity within a delayed closed-loop environment (40, 41). The combination of brain cell cultures and computers has also been attempted: 2D cultured rat neurons displayed evidence of self-organized activity in a computational task (blind source separation) when supplied with electrical information (42). A different study showed that these cultures learned to respond in the form of distinct electrophysiological patterns to low-frequency focal stimuli (43, 44).
To the best of our knowledge, however, no relevant approach using brain organoids as learning systems has been reported. Previous research has demonstrated spontaneous electrophysiological signals and synchronous neural network activity of dissociated organoids (45), sliced organoids (46), or developing full organoids (34, 47), and advanced manipulation of neural circuits within assembloids (organoids merging two distinct brain regions) was recently published (48). The only study resembling our vision is the recent work by Kagan et al. (29), which embedded monolayers of cortical neurons in a real-time closed-loop environment via electrophysiological stimulation and recording. These cultures self-organized to rapidly alter their activity to display goal-directed behavior in a simulated game environment. Similarly, the European Union-funded NEU-CHiP project aims to demonstrate the growth of layered networks of brain stem cells on microchips (49). In addition, the Human Brain Project models similar (but virtual) input-brain output machine models (50).
Obviously, terms such as “cognition,” “intelligence,” “sentience,” and “consciousness,” describing human capabilities, cannot be directly translated to simple cell culture models; they are used here to describe the realization of basic functions underlying these higher-order functionalities. A workshop to define adequate terminology for the field is in preparation. Please see the glossary included, which attempts to provide definitions in the context of this manuscript.
In this article, we present an architecture (Figure 1) and blueprint for an OI development and implementation program designed to:
● Determine the biofeedback characteristics of existing human brain organoids caged in microelectrode shells, potentially using AI to analyze recorded response patterns to electrical and chemical (neurotransmitters and their corresponding receptor agonists and antagonists) stimuli.
● Empirically test, refine, and, where needed, develop neurocomputational theories that elucidate the basis of in vivo biological intelligence and allow us to interact with and harness an OI system.
● Further scale up the brain organoid model to increase the quantity of biological matter, the complexity of brain organoids, the number of electrodes, algorithms for real-time interactions with brain organoids, and the connected input sources and output devices; and to develop big-data warehousing and machine learning methods to accommodate the resulting brain-directed computing capacity.
● Explore how this program could improve our understanding of the pathophysiology of neurodevelopmental and neurodegenerative disorders toward innovative approaches to treatment or prevention.
● Establish a community and a large-scale project to realize OI computing, taking full account of its ethical implications and developing a common ontology.
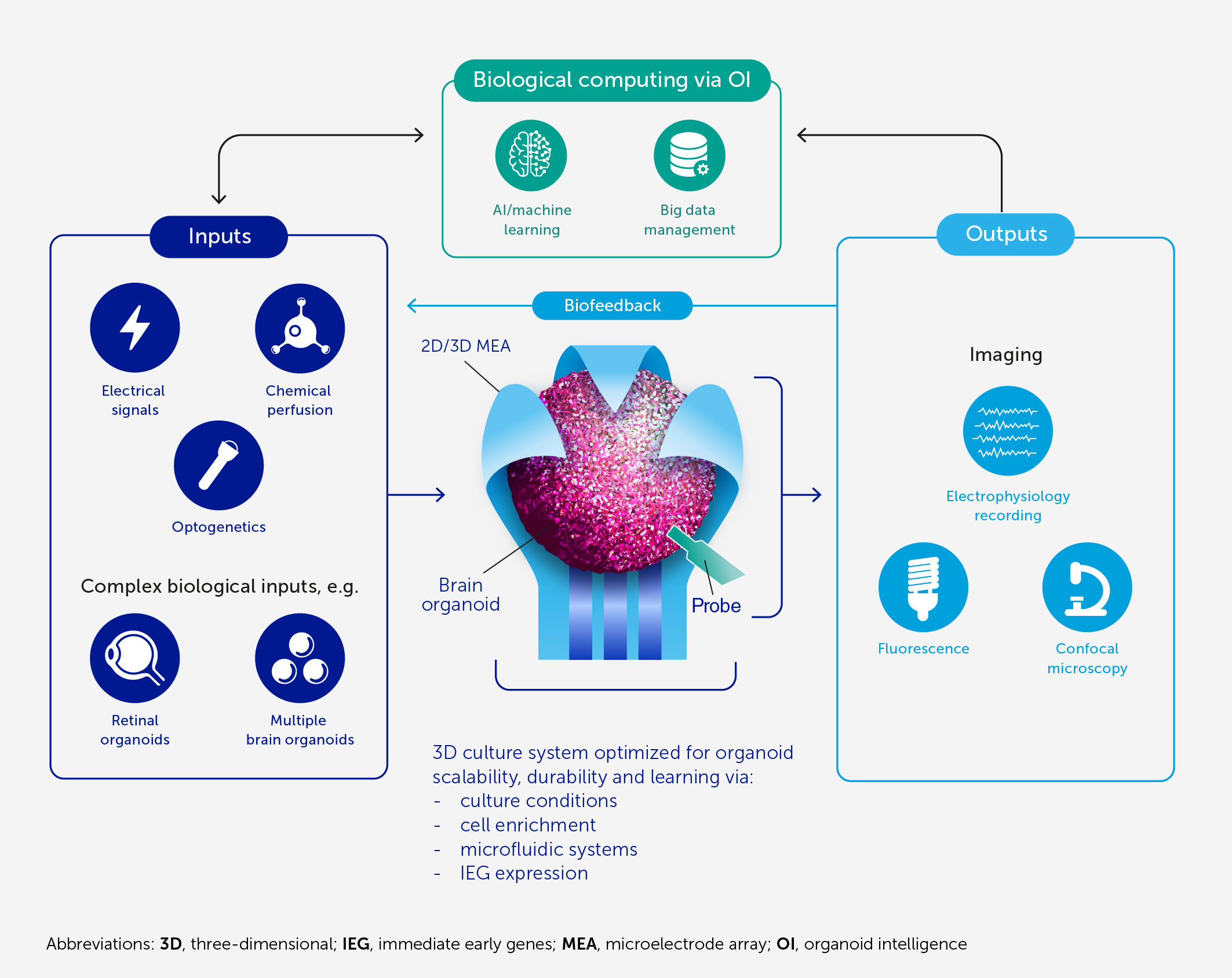
Figure 1 Architecture of an OI system for biological computing. At the core of OI is the 3D brain cell culture (organoid) that performs the computation. The learning potential of the organoid is optimized by culture conditions and enrichment by cells and genes critical for learning (including IEGs). The scalability, viability, and durability of the organoid are supported by integrated microfluidic systems. Various types of input can be provided to the organoid, including electrical and chemical signals, synthetic signals from machine sensors, and natural signals from connected sensory organoids (e.g. retinal). We anticipate high-resolution output measurement both by electrophysiological recordings obtained via specially designed 2D or 3D (shell) MEA, and potentially from implantable probes, and imaging of organoid structural and functional properties. These outputs can be used directly for computation purposes and as biofeedback to promote organoid learning. AI and machine learning are used throughout to encode and decode signals and to develop hybrid biocomputing solutions, in conjunction with a suitable big-data management system.
To the latter point, a community-forming workshop was held in February 2022 (51), which gave rise to the Baltimore Declaration Toward OI (52). It provides a statement of vision for an OI community that has led to the development of the program outlined here.
Prerequisites for biocomputing models
Culture and bioengineering technologies to advance 3D brain organoids
The advent of two biological technologies makes our OI approach possible: the groundbreaking work to reprogram human somatic cells back to stem cells [i.e. induced pluripotent stem cells (iPSC)] (53); and the more recent development of 3D brain organoids from iPSC.
Advances in 3D organoid culture
The past decade has seen a revolution in brain cell cultures, moving from traditional monolayer cultures to more organ-like, organized 3D cultures – i.e. brain organoids (Figure 2A). These can be generated either from embryonic stem cells or from the less ethically problematic iPSC typically derived from skin samples (54). The Johns Hopkins Center for Alternatives to Animal Testing, among others, has produced such brain organoids with high levels of standardization and scalability (32) (Figure 2B). Having a diameter below 500 μm, and comprising fewer than 100,000 cells, each organoid is roughly one 3-millionth the size of the human brain (theoretically equating to 800 MB of memory storage). Other groups have reported brain organoids with average diameters of 3–5 mm and prolonged culture times exceeding 1 year (34–36, 55–59).
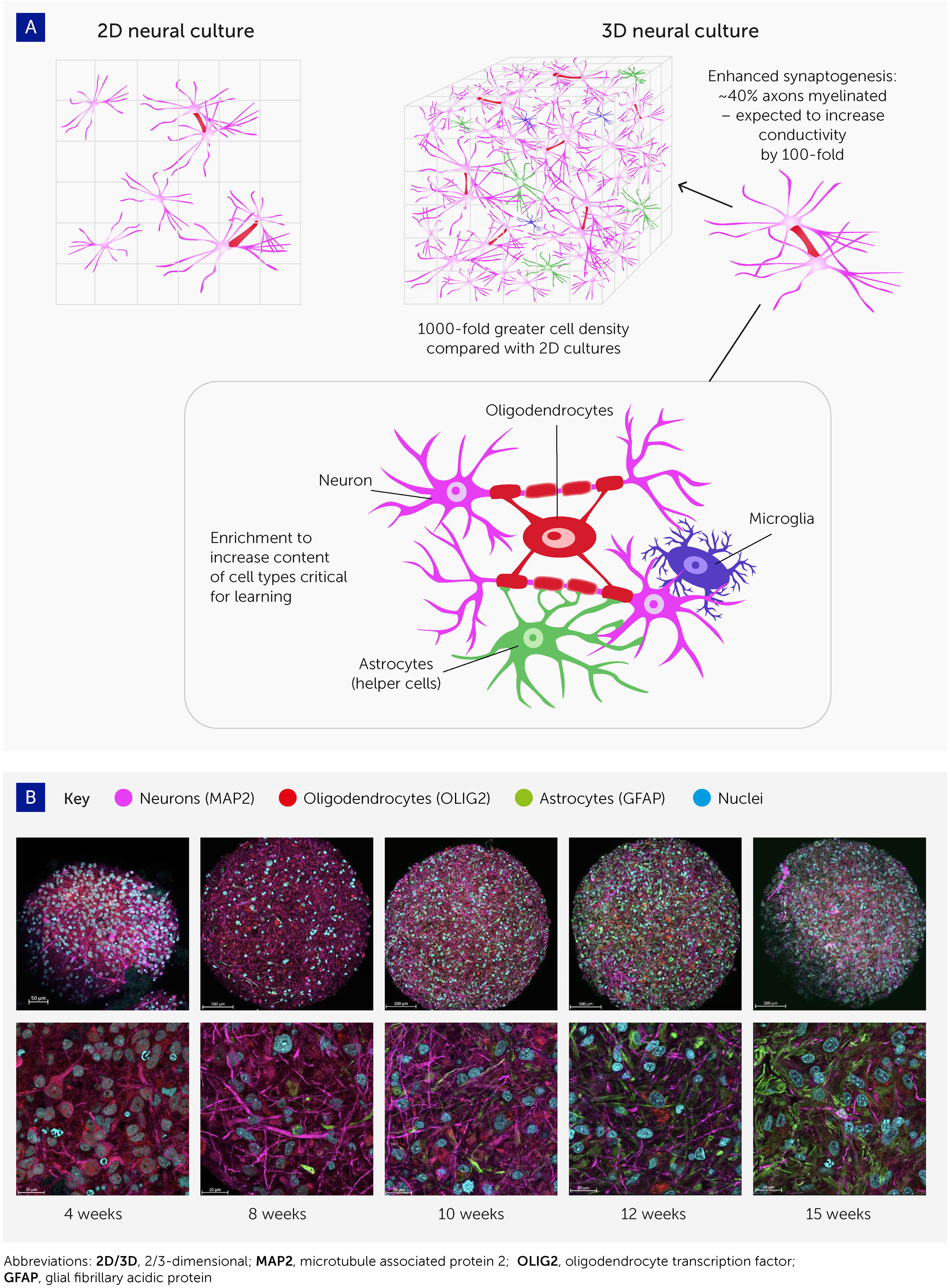
Figure 2 Advances in 3D cell culturing provide the foundation for systems to explore organoid intelligence. (A) 3D neural cell cultures have important advantages for biological learning, compared with conventional 2D monolayers – namely a far greater density of cells, enhanced synaptogenesis, high levels of myelination, and enrichment by cell types essential to learning. (B) Brain organoid differentiation over time from 4 to 15 weeks, showing neurons (microtubule associated protein 2 [MAP2]; pink), oligodendrocytes (oligodendrocyte transcription factor [OLIG2]; red), and astrocytes (glial fibrillary acidic protein [GFAP]; green). Nuclei are stained with Hoechst 33342 (blue). Images were taken with an LCM 880 confocal microscope with 20x and 63x magnification. Scale bars are 100 μm and 20 μm, respectively. The images show the presence of MAP2-positive neurons as early as 4 weeks, while glial cells emerge at 8 weeks and there is a continuous increase in the number of astrocytes over time.
These organoids show various attributes that should improve their potential for biocomputing (Figure 2). First, cell density in these 3D models is similar to the in vivo cell density, and much higher than in monolayer cultures; the ratio of cells to media volume is also much higher compared to monolayers. Second, most of these brain organoids show spontaneous electrophysiological activity and reactivity to electrical stimulation (via evoked field potentials) (32), confirming the presence of active synapses. Trujilio et al. have shown patterning of cortex layers and oscillation waves comparable to electroencephalograms (EEGs) from human preterm babies’ brains (34).
Third, axons in these organoids show extensive myelination. Pamies et al. were the first to develop a 3D human brain model showing significant myelination of axons (32). About 40% of axons in the brain organoids were myelinated (30, 31), which approaches the 50% found in the human brain (60, 61). Myelination has since been reproduced in other brain organoids (47, 62). Myelin reduces the capacitance of the axonal membrane and enables saltatory conduction from one node of Ranvier to the next. As myelination increases electrical conductivity approximately 100-fold, this promises to boost biological computing performance, though its functional impact in this model remains to be demonstrated.
Finally, these organoid cultures can be enriched with various cell types involved in biological learning, namely oligodendrocytes, microglia, and astrocytes. Glia cells are integrally important for the pruning of synapses in biological learning (63–65) but have not yet been reported at physiologically relevant levels in brain organoid models. Preliminary work in our organoid model has shown the potential for astroglia cell expansion to physiologically relevant levels (47). Furthermore, recent evidence that oligodendrocytes and astrocytes significantly contribute to learning plasticity and memory suggests that these processes should be studied from a neuron-to-glia perspective, rather than the neuron-to-neuron paradigm generally used (63–65). In addition, optimizing the cell culture conditions to allow the expression of immediate early genes (IEGs) is expected to further boost the learning and memory capacities of brain organoids since these are key to learning processes and are expressed only in neurons involved in memory formation – as detailed below.
Scaling up these 3D organoids is a key early aim. We set out to produce brain organoids with about 10 million neural cells (66). Existing differentiation protocols for scaling up the cultures, and starting 3D differentiation directly from iPSC (bypassing the intermediate generation of neuroprogenitor cells), are very promising (67, 68). These developments benefit from general progress in microphysiological systems (MPS), which include organoids, and aim to establish organ architecture and functionality such as OI as proposed here. Notably, some coauthors are involved in spearheading quality assurance guidance for Good Cell Culture Practice, expanding earlier guidance to stem cell-based models, MPS, organ-on-chip models (69), and establishing an annual MPS World Summit series (70) and international society.
Microfluidic perfusion systems
While brain organoids may recapitulate spatiotemporal molecular signatures, gene expression networks (71), certain histoarchitectures (e.g. cortex patterning), and neuron phenotypes within the human brain, they do not reflect its regional organization and the complexity of its neuronal circuitry to levels allowing for higher-order brain function (35, 72, 73). Part of the human brain’s complexity stems from its size and the vasculature that supports its growth (74, 75). Although brain vasculature models are under development (76, 77), most brain organoid models so far are still avascular and rely on passive diffusion to deliver nutrients; the average scope of diffusion is approximately 300 μm before starving-derived necrosis occurs at the core (78) (Figure 3A). Thus, the lack of a perfusable vasculature is a major limitation for improving biological complexity and in vivo-like functionality (78).
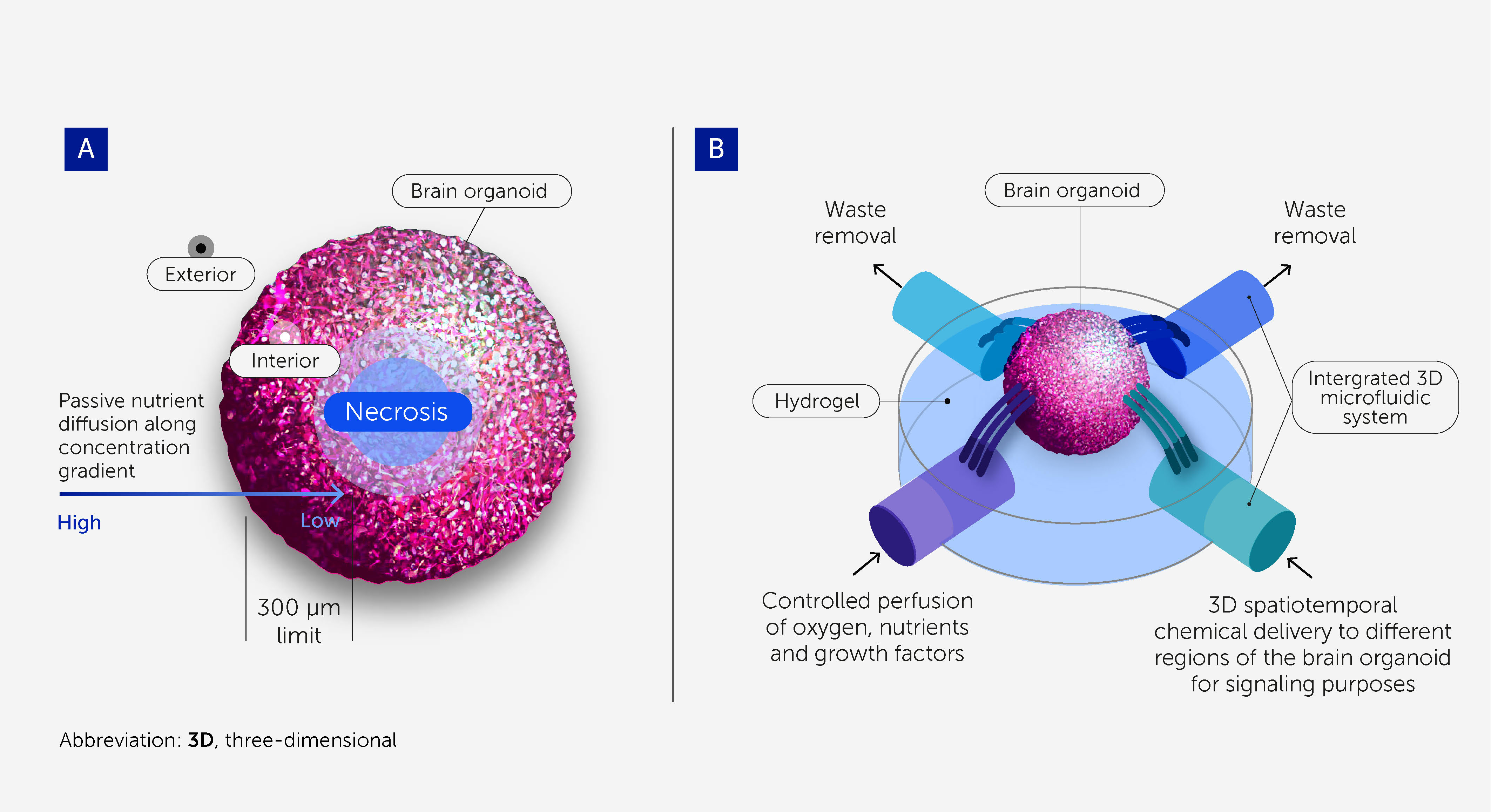
Figure 3 3D microfluidic devices to support scalability and long-term homeostasis of brain organoids. (A) Cells within brain organoids require perfusion with oxygen, nutrients, and growth factors, as well as the removal of waste products, to provide conditions approximating physiologic homeostasis. Passive diffusion penetrates to a depth of only around 300 μm, and so necrosis occurs at the core of larger organoids owing to starvation. This prevents brain organoids from being scaled up to the size and complexity required for OI research and limits their durability. (B) 3D microfluidic systems enable greater scalability and durability by providing controlled perfusion throughout larger organoids. They also enable 3D spatiotemporal dosing of chemicals for signaling purposes.
Microfluidic systems that substitute for vasculature – allowing controlled perfusion of oxygen, nutrients, and growth factors and the removal of waste products – will be critical to the scalable and durable culturing of brain organoids (Figure 3B) (79, 80). These will support the homeostasis and viability of the organoids, allowing a more physiologic-like differentiation toward a more complex, sophisticated, and “in vivo-like” model. Flexible, self-folding microfluidics can already deliver chemicals with 3D spatiotemporal control (81), and recent advances in 3D printing with sacrificial materials offer the potential to create perfusable scaffolds for organoids (82, 83).
These microfluidic systems will also support chemical signaling to organoids. The importance of spatiotemporal chemical patterns to encode information is well established in neuroscience and behavioral science (84–86). Significant advances in micropatterning and microfluidics over the past two decades have already allowed 2D chips to offer significant tunability of the chemical microenvironment around neurons, and tree-like microfluidic gradient generators have been widely used to create chemical patterns (87). Importantly, 3D spatiotemporal microfluidic interfaces (88) now enable localized dosing and replication of chemical environments with neurotransmitters, neuropeptides, and other neurochemicals. A comprehensive list of the agonists and antagonists is available (89).
High-resolution recording of complex neuronal networks
3D microelectrode arrays for brain organoids
Robust and reproducible systems to record electrophysiological outputs from brain organoids are critical to developing OI systems and will need to address various challenges in reading and writing to complex neural assemblies. Brain-machine interface technologies have been in progress for at least two decades (90) but remain primitive. Microelectrode arrays (MEAs) form the heart of many such interfaces since they can be used to both stimulate and record, and offer unprecedented parallelism and individual addressability. However, most are predominantly in a 2D chip-based format, being designed for use with monolayer cell cultures (91). This represents a likely problem as brain organoids are spherical 3D structures that make limited contact with a 2D MEA chip. Furthermore, most 2D electrode chip interfaces are rigid, and a mismatch in the stiffness of the recording interface and cell system compromises performance (92, 93).
Therefore, we and others are developing novel 3D MEA interfaces specifically designed for organoids (93–96) and inspired by the EEG caps used to record brain electrical patterns from the scalp. Organoids are grown inside flexible, ultra-soft-coated, self-folding, and buckled shells, covered with patterned nanostructures and probes (92, 93, 97–99) (Figure 4). This model allows multichannel stimulation and recording spatiotemporally across the entire surface of the organoid with unprecedented resolution and high signal-to-noise ratio, resulting from the greatly enhanced recording surface areas (92, 93). After the spontaneous signal is stabilized and synchronized, the response to repeated chemical stimuli from neurotransmitter gradients [glutamate, γ-Aminobutyric acid (GABA), dopamine, serotonin, acetylcholine] and main receptors agonists/antagonists [e.g. kainic acid, kynurenic acid, γ-Amino-β-hydroxybutyric acid (GABOB), bicuculline, haloperidol, nicotine, methylbromide (89)] can be recorded to address and modulate the synaptic plasticity.
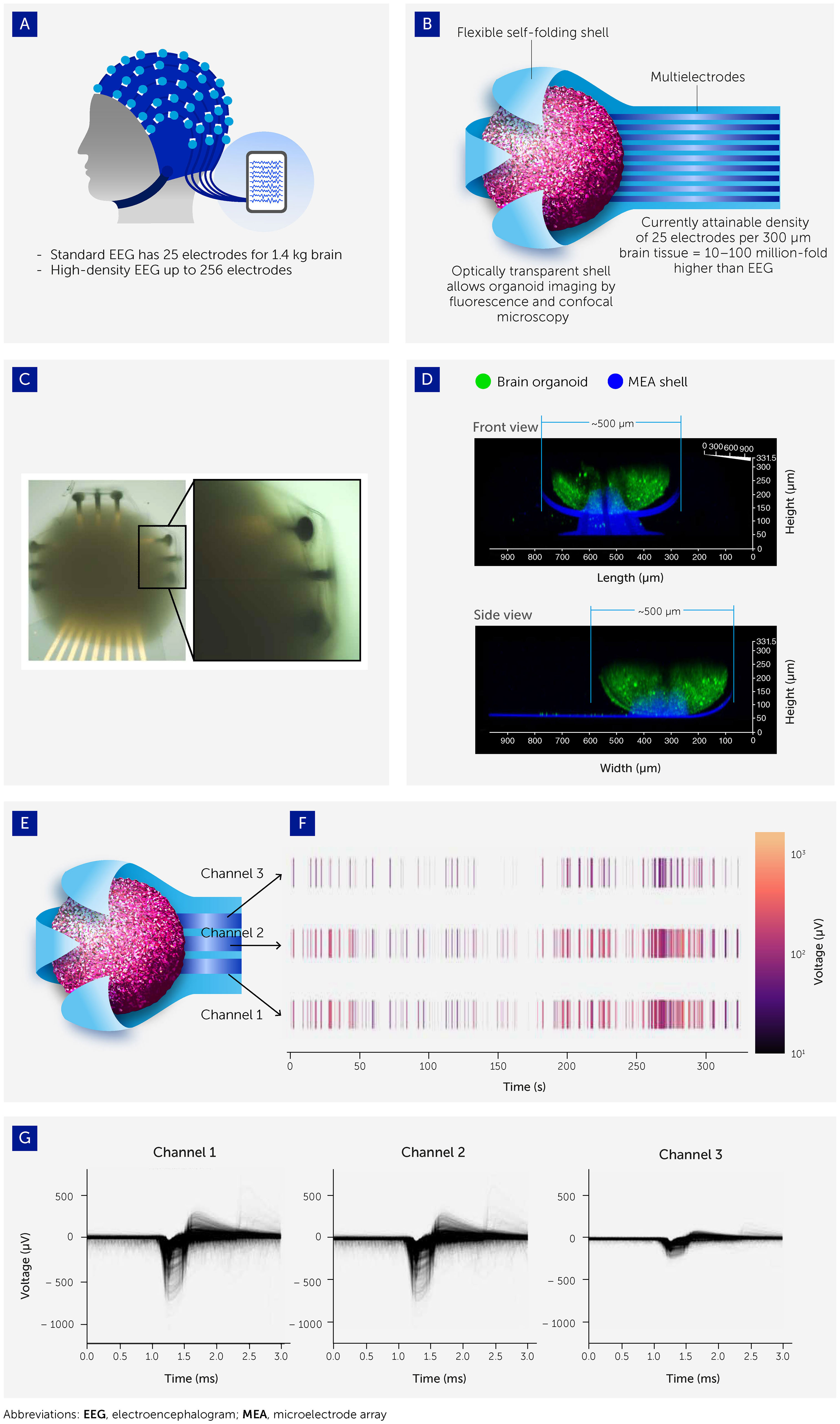
Figure 4 Interfacing organoids with 3D microelectrode arrays (MEAs) to allow electrophysiological output recording. (A) Organoid-MEA interfaces were inspired by the electroencephalograph (EEG) used to take electrophysiological recordings from the human brain. (B) Organoids are grown inside flexible, ultrasoft-coated, self-folding, and buckled shells covered with patterned multielectrode nanostructures and probes. These interfaces allow ultra-high-resolution 3D spatiotemporal stimulation and recording of electrophysiological patterns across the entire organoid surface (see also 93). (C) Brightfield image of brain organoid captured inside the shell. (D) Confocal image showing the side view (projected confocal stack) of a brain organoid (green; Fluo-4 calcium dye) with a diameter around 500 μm encapsulated in the electrodes of the 3D shell (blue). (E, F) Three channels of electrodes are distributed across the shell with representative raster plot showing the spontaneous electrical activity of the brain organoid. (G) Overlaid spike waveform from channels 1, 2, and 3.
These shell MEA interfaces can be integrated with the aforementioned 3D microfluidic systems, supporting the scalability and durability of the system and chemical signaling via spatial patterning and gradients. Together, they create a robust platform to gain an iterative, in-depth understanding of organoid behavior and responses to a range of highly modifiable environmental and input stimuli, which in turn will allow us to explore their capacity to recapitulate the molecular mechanisms of learning and memory formation and ultimately their computational potential.
High-resolution implantable electrophysiology devices
We consider that the shell MEAs described above strike an appropriate balance by providing comprehensive, high-resolution electrophysiological recordings with minimal disruption to the organoid. However, future systems might permit organoids to be grown around implantable electrodes to further enhance signal resolution and to access the inside of the organoid. The efficiency of such systems must be balanced by their invasiveness since any damage to neuronal networks could alter the behavior of the organoid.
Neuropixels are silicon probes developed for extracellular recording in animals (mostly mice and rats) (100). They lend themselves to direct integration with brain organoids, though in principle, Neuropixels can also be integrated into shells. These large (10 mm), dense (100 sites/mm) implantable neural devices allow the recording of hundreds of well-resolved single neuron signal traces. They can be combined with light sources, electrical stimulation, and photometry, dramatically increasing the input and output opportunities and the mapping of activity in the learning organoid. These devices also appear uniquely capable of long-lived exposure to neural tissue. Chronic implants in rats and mice frequently last for 150 days or more with little degradation in recorded neural activity, indicating suitable compatibility and stability (100, 101). Recently, the Kosik laboratory (University of California Santa Barbara, CA, USA) used such CMOS shank probes in parallel with 2D high-density MEAs to record intrinsic network activity in brain organoids (46).
Mounting and inserting such probes into the organoid is a complex challenge, and work is ongoing to develop a new generation of suitable (more flexible) probes. If the organoid is grown in well plates, mounting of the probe would be like skull mounting in a rodent. If the growth medium needs to be exchanged periodically for organoid health without disturbing the probe-organoid interface, then an unconventional means for removing and adding media to the wells would need to be developed. The minimum size of the probe base (4.2 mm × 1 mm) allows sufficient “headroom” for this fluidic machinery. Once a microfluidic growth chamber is adopted, a membrane interface to the probe shank would be needed. Neuropixels are routinely used with a Kwik-Sil (WPI) silicon layer over the brain while recording and sealing chronic implants. Further investigation may be required to perfect this geometry, but interfacing with a low-pressure fluidic chamber does not present any fundamental problem. Finally, a Neuropixels 2.0 probe (101) has four shanks, each 10 mm long with a cross-section of 70 μm × 24 μm. For a 1 mm diameter organoid, approximately 500 electrodes would contact the cells, giving the capacity to record from 384 switch-selectable sites at a time. The probe electrodes would displace ~1.5% of the current organoid volume, likely an acceptable perturbation. Yields of 200–600 “units” are typical in recordings from rodents, limited by activity. The probe would be within detection distance of ~14% of the cells in a 1 mm diameter organoid.
Among the challenges facing brain-machine interface technologies is the scale of connectivity. Cortical neurons each have in the order of 104 input synapses and connect to in the order of 103 cells, some across many millimeters – even in small brains such as those of mice. It is not yet clear if organoids have similar or reduced synaptic counts. Current brain-machine interfaces have many unresolved cells per input (reading) for each electrode (all within 20–80 μm) and a largely unknown number of cells for output (writing or stimulation). Except for special cases in the visual cortex, the cellular understanding of writing remains difficult, while reading has enabled the control of prosthetic robots.
How might traction be achieved to begin harnessing organoid computing power? We propose that two paths be explored.
All-optical
An all-optical path for organoids would allow cell-by-cell excitation and whole organoid reading (again cell-resolved). While optical imaging is not likely a terminal technology for harnessing OI, it allows exploration of the system behavior, and the kinds of computation that could be initially (102) performed. There are rapidly developing technologies for large-volume imaging that make this attractive. Techniques such as Bessel holography can image volumes with hundreds of μm diameter at kHz rates, with an accurate cellular resolution if the activity is relatively sparse (102). Directly writing with opsins is also well established, and holographic methods are again exploited (103), but the cell write rate is significantly lower than the cell read rate. These methods are immediately available and would help inform the design of electrophysiological systems necessary to progress OI.
High-throughput electrophysiology
Electrophysiology recording capacity is impeded by the “dark matter” observation resulting from the fact that most neurons are not firing most of the time. As such, an electrophysiology channel sensitive to reading and writing individual local neurons can be inefficient: even the best-performing probes “see” <1% of the neurons within their detection range (104). It seems unnecessary to achieve a synapse level of interpretability of these systems if the base principles are elucidated, empirically tested, and used to control the system as a whole. Notably, bionic implants for humans have managed to convey significant information with relatively few input electrodes; e.g. the bionic eye (105) used 24 electrodes, and current cochlear implants use between 10 and 22 (106, 107). Therefore, different resolutions may be required for input vs. output.
Although organoid EEG and implantable neural devices are feasible to investigate the scale of required input/output contacts, both technologies need to be explored further to assess the recording patterns and learning potential of brain organoids.
Memory and learning in organoids: training using biofeedback, big data, and AI/machine learning
Understanding the capacity of brain organoids to learn is fundamental to determining whether they can be used computationally by harnessing the advantages of biological learning. At this stage, learning is identified as an increased frequency to show and memorize a response pattern to a stimulatory pattern. We aim to use the iterations of technologies described above to interface organoids and computers to initiate supervised learning simulations (i.e. trained stimulus response patterns). To achieve this goal, the brain organoids should be exposed to spatiotemporal patterns of electrical and chemical stimulation, with the associated recordings delineating relationships between inputs and outputs. A feedback loop is required to train a learning system. Changes in the brain organoid architecture and functionality (synaptic connections and electrophysiology) due to such training cycles can then be analyzed. These two factors affect synaptic plasticity – the main mechanism of memory formation and learning. Hence, the recorded responses to electrical or chemical stimuli should demonstrate whether and how learning may occur in the organoids.
Ultimately, robust, high-resolution encoding and decoding of signals going into and out of human cortical tissue is required. Recent achievements using intracortical microstimulation (ICMS) and decoding of motor and sensory cortices using MEA in human subjects offer promise (108, 109), though further advances in scale and resolution will be necessary.
AI analysis of responses
Organoid-MEAs will generate massive recording datasets that will themselves need to be analyzed by statistical and machine learning techniques. Given the recording density and volume, this will necessitate a novel big-data infrastructure and supercomputing capacity tailored to the sophisticated needs of this form of modern biological data.
Fundamentally, the two major challenges for AI analysis in this context are : (a) how to decode the input provided to an organoid (e.g. the game Pong) (29) to relate to changes with its architecture and/or functionality; and (b) how to relate these organoid changes to certain outputs (e.g. the improvement in playing Pong). In other words, biological computing includes OI as a mediating mechanistic process between the inputs and outputs. To answer these two challenges, we foresee the use of interdisciplinary tools integrating machine learning, statistics, signal processing, information theory, and optimization. We also believe that the questions raised will motivate new methodological developments in these fields.
Specifically, we believe the following three paths must be explored to relate OI inputs to outputs:
1. Machine learning and statistical algorithms are needed to quantify organoid function changes. This involves: (a) sensor integration to accelerate processing based on unsupervised learning and dimension reduction, such as principal component analysis (PCA), independent component analysis (ICA), and autoencoders including hierarchical versions (110–112); (b) signal detection using sequencing and time series (e.g. state-space) models (113–115), which are often used in brain imaging analysis; (c) pattern recognition to identify the real signal pattern and deconvolute it from the background noises (116).
2. Algorithms are also needed to quantify organoid architecture changes. Challenges include pinpointing the exact parts of the organoid that respond to the input [e.g. using mixture models (117)] and then quantifying these changes by monitoring their physical appearances. The application of multiscale unsupervised structure learning methods to the recorded responses can identify discrete, statistically distinguishable, observer-unbiased response phenotypes.
3. Models must then be trained to relate the quantified organoid changes to the output variables via multivariate causal models (118–120). OI will require novel developments integrating AI/machine learning and both space- and time-dependent causal modeling (121–123).
Clearly, inferring or estimating the connectivity of organoids will be a core endeavor. Connectivity, in neuronal circuits, is usually divided into structural, functional, and effective connectivity (124). The distinction between functional and effective connectivity is particularly prescient here: functional connectivity refers to the statistical correlations between neuronal fluctuations in different populations, while effective connectivity refers to the causal and directed connectivity between populations. The corresponding data analysis techniques can be parsed into frontal connectivity methods such as coherence analyses and Granger causality. These can be contrasted with inferences about directed (effective) connectivity, usually using some form of dynamic causal modeling (125).
Applying these statistical and computational methods and modern big and complex data tools, such as those from brain imaging and computational biology, will allow us to map input and outputs from organoids’ neurological connections. As an example of a relevant technique, a fundamentally different approach to neuron behavior mapping was developed in Drosophila: optogenetic activation of neurons and the application of multiscale unsupervised structure learning methods to the recorded responses to identify discrete, statistically distinguishable, observer-unbiased response phenotypes (126). This could be a starting point for connectivity- and activity-mapping studies to further investigate the mechanisms through which neurons mediate diverse behaviors. However, with respect to its application to brain organoid recordings, we are entering terra incognita.
Machine learning and other mathematical models are increasingly applied to certain parts of organoid research (127–129). However, machine learning, in the sense of deep learning and supervised learning, deserves further comment. This is because it presupposes that supervised learning is an appropriate theoretical formulation of self-organization in organoids, in the sense that the opportunities afforded by organoid research transcend questions about how to engineer a particular behavior through supervised learning (130). The opportunities probably require a more generic theoretical framework within which to formalize self-organization and active exchange between an organoid and its external milieu (131, 132). Practically, if one wanted to train an organoid to do this or that, it would be impossible to implement the procedures for supervised learning in machine learning (i.e. either backpropagation of errors or local energy-based schemes). Current developments favoring reinforcement learning, where self-organization is met by feedback on functionality, lend themselves to such problems. Strategically, if correct, this means that the direction of travel of organoid research may be either toward reinforcement machine learning or more aligned with some of the foundational questions posed in neurobiology (133) or, indeed, the physics of nonequilibrium self-organization (134, 135).
Big-data infrastructures
Providing suitable infrastructure for the storage, curation, and processing of OI big data is a scientific challenge of its own. Analytic datasets need to be stored in efficient shared memory structures; appropriate technology will be needed where manipulations, such as standard matrix or tensor calculations, can be obtained without partial or streaming access to the full dataset. Furthermore, a fast, robust, and scalable computational analytic and curation infrastructure for the resulting data will be needed. A likely requirement will be efficient dimension-reducing transforms of the 3D sensor arrays applicable for streaming data; e.g. by “scattering transform” (136), which has been particularly successful in analyzing audio streams (137).
Each deep biological network computes some function, transforming inputs to outputs depending on many variables, chief among them: (1) the weights and biases and (2) the activation functions. In both AI and OI, it is reasonably safe to assume that the activation functions are essentially static; i.e. any given node at any given time is activated only upon reaching a certain threshold. However, Sinapayen et al. (138) suggest that neural networks can autonomously change activity to avoid external stimulation. The key difference between AI and OI networks is that in OI networks, the weights and biases may dynamically change over time – for example, owing to growth and/or maintaining homeostatic equilibrium – however, with unclear changes in the network function. Indeed, vastly different neural networks can implement approximately the same function (139). A similar result is well established in biological networks (140). We, therefore, hypothesize that although the precise weights and architecture of the OI may be dynamic, the memories may be stable over time.
The amount of data and respective curation, compression, and processability will dramatically increase with boosted electrophysiological recordings from OI systems. Additional challenges come from spatiotemporal recording and the combination of electrophysiology and high-content imaging. In addition, the training and experimental data created will require efficient frameworks for storage and analyses. This could include a combination of in vivo proto-neural networks developed in brain organoids and in silico analog/digital hybrid, and/or neuromorphic computing (41, 90, 141, 142). Transmedia progressive learning will therefore exhibit the advantages of both biological and machine computing and learning, while mitigating the limitations of each. The main aspect of our strategy for storage is to develop a scheme resembling the Large Hadron Collider experiment at CERN, where sophisticated triggers are used to detect events in real time and only events with a potential discovery value are kept – greatly decimating the data rate. We envisage a similar event-driven way of analyzing the data: we will send discrete stimuli to the brain organoids, look for coordinated responses in many channels, and store only the discrete events related to these intervals. We will use insights gained through multiple versions of experiment reduction to develop a sensor correlation model and optimal event triggers that optimize the trade-offs between data reduction and discovery value.
Many large ongoing efforts aim to create data-processing tools, storage solutions, and standards to handle the scale of data generated by modern neuroscience experiments, e.g. the US BRAIN Initiative (143). Open-source community solutions, which the OI community could leverage, are being developed for terabyte- and even petabyte-scale data across multiple modalities. As high-throughput MEAs represent the most promising initial technology for interfacing with organoids, solutions from the invasive and in vitro electrophysiology communities can likely serve as the basis for the OI community. Standardized community-processing pipelines exist for this application, including machine learning tools such as DataJoint Elements (144). Many individual tools and techniques for electrode arrays have been developed for spike sorting (145) and analysis of local field potentials (146). Community cloud-based archives are available for publishing such electrophysiological data in an open and accessible manner, such as the OpenNeuro Archive (147) and DANDI (148). These archives are hosting an ever-increasing range of data from varied experimental paradigms. Several standards initiatives could be leveraged to facilitate reuse, reanalysis, and meta-analysis, such as the Brain Imaging Data Standard (BIDS) (149) or Neurodata Without Borders (NWB) (150). By leveraging the standards, processing pipelines, storage, and dissemination techniques developed by the larger electrophysiology community, the OI community can rapidly establish a robust and reproducible big-data infrastructure.
Looking to the future, additional imaging modalities may become critical to providing further insights into organoid function and learning. Other processing tools and archives exist for relevant modalities such as fluorescence microscopy and also other forms of microscopy; for example, the CaImAn pipeline for calcium imaging data analysis (151) and the Brain Imaging Library (152) for archiving and storage. In the invasive in vivo neuroimaging community, functional and structural connectivity (or connectomics) is also being studied with improved resolution and larger volumes (153, 154). Cloud-based processing pipelines for deriving connectivity are under development (155), and the Brain Observatory Storage Service and Database (BossDB) ecosystem has been developed to host and archive large connectivity datasets at the neuron-synapse level (156). Many emerging graph analysis tools enable improved insight, such as statistical characterization of connectomics (157) and identification of repeated motifs (158). In time, multimodal datasets and infrastructure may play an important role in the development of the OI community.
Ultimately, the OI community should seek to build on these tools to establish standardized analysis and storage infrastructures. Open data sharing can be a powerful approach to grow the community and maximize the reuse of experimental data. Establishing a large-scale, standardized set of experimental data may rapidly improve processing tools, provide theoretical insight, and generate hypotheses for future experiments. An interesting model approach is the Human Connectome Project (159), which used standardized approaches to high-resolution magnetic resonance imaging to produce a gold-standard dataset for the emerging field of human connectomics. A similar data and infrastructure effort for the OI community could provide invaluable insights.
To summarize, a big-data ecosystem necessary to study OI will require:
● Standardization of experimental data and metadata, building on existing standards such as BIDS or NWB
● Robust, repeatable, and standardized processing pipelines that scale to large electrophysiological datasets
● Efficient, accessible, and open data storage, possibly leveraging existing cloud archives such as OpenNeuro or DANDI
● The potential development of multimodal OI datasets
● The establishment of standard, reference datasets for the community
In the foregoing discussion, we have anticipated a need for the collation, storage, and dissemination of big data, under the assumption that organoid research will recapitulate developments in neuroscience (and in particular imaging neuroscience). However, there is an alternative path, which reflects a move from “big data” to “smart data.” In other words, we need only informative data that enables people to answer well-posed questions. In effect, this would represent a pushback against big data and a return to carefully designed experiments that elicit the right kind of data to make inferences about the dynamics, plasticity, and functional architectures in brain organoids. In short, the experiences of the neuroscience community may usefully inform the kind of resources needed to realize the full potential of organoid research over the ensuing years.
Advancing biocomputing complexity
Optimized algorithms for organoid-in silico interactions
Realizing the potential of OI requires more than interfacing a computer with an organoid. In OI, the organoid can take on the role of an embodied agent that interacts with an environment through the organoid-in silico interface. This will require optimized algorithms for organoid-in silico interactions as well as research into theoretical frameworks for learning and adaptations in organoids drawing from the theoretical neuroscience literature. The viability of OI is dependent on optimized algorithms for organoid-in silico interactions, in addition to the fast data storage and retrieval described above.
There are two broad environments in which OI may operate: open-loop or closed-loop:
● Open-loop involves feeding information into cells and measuring the response. Recent work leveraging this approach has found that neural systems can alter their activity to perform various tasks, including context-dependent encoding (160) and blind-source separation (42), and to display features such as adaption toward scale-free dynamics (161) or long-term activity-dependent plasticity (41).
● Closed-loop extends the open-loop environment to include feedback to the neural systems about the result of the system activity. Examples of this are limited owing to technical difficulties, but high-latency/low-temporal resolution analysis suggests that under these conditions, cultures can demonstrate changes in neural dynamics (41) and arbitrary learning (44). Moreover, real-time implementations have recently resulted in goal-directed learning displayed through a change in neural culture activity when applied in line with neurocomputational theories (29).
Exploring, applying, and refining empirically supported theories of what fundamentally drives learning intelligence is a critical factor. So far, numerous theories have been proposed to explain how, at the fundamental level, neural systems process and respond to information.
The first branch of theories focuses on how neural systems are organized, both structurally and functionally. Key notions include neural criticality (162), neural Darwinism (163), cell assembly and Hebbian plasticity (164), rule-based learning (165), and core ideas behind population coding approaches (166, 167). Generally, these theories aim to explain how the incredibly complex organization within the brain results in its ultimate functioning. Thus, they offer generally compatible frameworks for analyzing and interacting with brain organoids, providing the opportunity for optimized input and decoding of output.
A second “optimization” category of theories generally focuses on how a system or agent works to maintain homeostasis in a dynamic environment. Broadly, this can be achieved either through maximizing a utility or reward or by minimizing surprise or uncertainty (165, 168). Key theories include the Bayesian brain hypothesis (169), efficient coding hypothesis (170), value-dependent learning (171), optimal control theory (172), and learning by stimulation avoidance (138). Attempts have been made to unify these theories, recently by exploring the underlying compatibilities, most notably through the proposal of the free energy principle (173), where the system or agent may engage in active inference to construct a generative model of the external environment and act in a manner to minimize the difference between the internal model and the perceived external world. At present, it is difficult to empirically test many of these theories in a controlled manner because in vivo organisms possess compensatory mechanisms that hamper the interpretation of results. OI offers a pathway for highly controlled experiments to empirically test these theories.
In addition to frameworks for learning in neural systems, the OI community will require methods to assess embodied intelligence; i.e. computational approaches to understand intelligent behavior in organoids for both open-loop and closed-loop environments. Previously, in vitro experiments have demonstrated the ability of cell cultures to control both physical robotic systems and simulated video games (29, 174). Experience within the AI community suggests that the OI community will likely benefit from standardized testing environments and conditions, accounting for variability and constraints in input/output interfacing in terms of channel count and bandwidth. The AI reinforcement learning community has produced a huge range of games, simulation environments, and physical systems that could be adapted to OI evaluation. Of particular interest to the OI community may be the field of continual or lifelong learning (175, 176), where embodied agents are assessed in learning that occurs over a sequence of experiences (often referred to as a “lifetime”). Such testing environments may serve the OI community by providing important benchmarks for understanding functional activity and learning in organoids.
Incorporating complex biological inputs in OI
Previous sections describe how we intend to interface organoids and computers. Combining organoids with various types of more complex inputs and output stimulation and recording interfaces (177, 178) will allow us to understand the potential for real-time controls. The consequences of such interconnections can be explored, starting with two brain organoids, one with complex input and one with complex output connections. A sensory organ, such as a retinal organoid, could then be connected with a brain organoid. Eventually, networks of organoids will be interconnected to implement more complex OI. The organoid will be interfaced with electrical and fluidic-sensing and simple outputs controlling machines through biofeedback on the cellular level; i.e. giving the brain organoid control by feeding back the results of its induced actions.
By connecting retinal and brain organoids we can determine whether signals can transfer between these different neuronal organoids and how this exogenous biological signaling will be interpreted by the brain organoid – establishing the initial baseline of organoid–organoid communication. Retinal organoids containing laminated retinas with outgrowth of outer segment-like structures and synaptic ribbons have been developed (179, 180). Synapse formation between two organoids is complex and was demonstrated recently in assembloids (48). Retinal and brain organoids can be connected either through electrodes or directly. Since methods to generate mature, endogenous, light-sensing retina are preliminary and very inefficient, engineered photosensitive ion channels, which are expressed under the control of a photoreceptor-specific promoter (181–183), can substitute as a proxy for light-reactive photoreceptors. Optimization of retinal organoid culture conditions to promote more robust signaling has recently been published (184).
Understanding and perhaps even influencing the connectivity of retinal and brain neurons would be extremely exciting. Although we and others are actively investigating ways to establish and modulate these connections, we are still quite far away from a system that can demonstrate robustly modifying neuronal connections in a directed manner. Ultimately, we aim to build on this simplified approximation or representation of visual input toward a system that more fully approximates vision.
Leveraging advances in the molecular basis of biological learning
Advances in the molecular biology of synaptic plasticity will be critical for optimizing the capacity of organoid systems for learning and OI. Growth conditions can now be optimized to allow organoid neurons to optimally express genes essential for learning in the human brain.
First, they need to express genes coding for the neurotransmitter receptors that mediate synaptic transmission. We have already characterized the expression dynamics of different subunits of the N-methyl-D-aspartate (NMDA) glutamate receptor during differentiation (unpublished observation). The long-term organoid maturation and age based on gene expression and switch of the main receptor subunits were recently extensively characterized (185). The machinery of the organoid’s biochemistry, especially of neurotransmitters, will be an important part of understanding the signaling cascades and synaptic plasticity necessary for learning; to this end, a thorough characterization of protein expression is warranted.
Secondly, it will be important for organoids to express IEGs. IEGs mediate synaptic processes essential to memory consolidation and are rapidly transcribed in adult neurons as they process information (186). Moreover, evidence indicates that IEGs are required for circuits capable of gamma frequency rhythmicity (187), which is associated with attention and information processing. Using RNA-sequencing to compare organoids (at different stages of maturation and grown under different culture conditions) with fetal, adolescent, and adult brains will allow the identification of growth conditions that optimize expression of these essential genes. It is critical to ensure IEG expression that is robust and dynamically responsive to activity evoked by a pharmacological block of GABAA inhibition (e.g. using bicuculline). Dynamic, activity-dependent IEG expression would assure the presence of molecular substrates necessary for organoids to establish circuits with balanced excitatory and inhibitory neurons and synapses. The cell culture conditions should be optimized so that 10–30% of neurons express IEGs in response to informational inputs. This level of sparsity is typical of brain regions involved in learning and memory (including the hippocampus and neocortex) and may ensure that multiple ensembles can be created to uniquely encode different streams of data. This contrasts with the global activation of IEGs that may occur with non-informational activity, such as a seizure. IEG expression and/or associated reporters can also provide a means to assess the formation of ensembles of neurons that are stably linked to specific inputs. In the brain, such ensembles are thought to represent memory engrams. EEG data can then be correlated with dynamic activity reporters (Ca2+ or voltage) and IEG expression data. Ultimately, neuronal activity can be stimulated and recorded optogenetically, as recently described (48). To our knowledge, IEG expression has not previously been utilized as an endpoint for optimizing organoid growth conditions. Successful use of this parameter would establish a biological basis for OI and address concerns regarding the uncertainty of the developmental state of organoids and their potential utility as information-processing memory units.
OI-led advances in medical research and innovation
In addition to pioneering the use of human brain organoids for computing and learning, OI research will also allow the exploration of inter-individual neurodevelopmental and neurodegenerative differences between stem cell donors. Alzheimer’s disease and other dementias could represent one particularly important priority for research. Globally, more than 55 million people are living with dementia, and this number is projected to exceed 150 million by 2050 (188, 189). Dementia is among the top 10 leading causes of death (190) and globally costs at least $1 trillion annually (191, 192). Clinical trials of novel Alzheimer’s disease therapies have shown very poor success rates, in part owing to premature translation of successful results in animal models that mirror only limited aspects of the pathology in humans (193, 194). The adaptation of OI research models to neurodegenerative diseases would offer the first human-based preclinical model to help us understand and develop effective treatments for these devastating diseases.
In addition to neurodegenerative diseases, neurodevelopmental disorders also lend themselves to OI, given that the different stages of brain development are reflected in the bioengineering of these models from stem cells. Conditions such as autism are major concerns owing to the enormous increase in prevalence. In the US, autism spectrum disorder (ASD) was diagnosed in the 1970s in 1 in 10,000 children, but according to the Centers for Disease Control (CDC), in 2021 it was 1 in 44 (195). While the disorder has heritable aspects, environmental influences are an increasing focus. The brain organoid model has shown promise for both developmental neurotoxicity (196, 197) and gene × environment (198) studies of autism. A varying combination of cognitive impairments characterizes this complex spectrum of diseases (199). Similarly, leukodystrophies are a diverse group of rare genetic disorders affecting white matter and linked to cognitive impairment (200). Using OI to explore the genetic basis of autism or leukodystrophies appears to represent an important path to understanding these disorders and to allowing screening of potential drugs that might boost underdeveloped cognitive functions.
Schizophrenia affects around 1% of the population worldwide and is one of the top 10 illnesses contributing to the global burden of disease (201). Schizophrenia has a prominent genetic basis, and it has been suggested that it may be neurodevelopmental in origin (202). Prenatal complications are an important contributor to the condition (203), while cognitive dysfunction is a hallmark (204), with a strong similarity to autism (205). Human iPSC lines, e.g. with genetic backgrounds associated with disorders, are available and continuously growing [e.g. SFARI base for autism (206)]. Organoids developed from iPSCs from individuals with various conditions could be compared against control samples to help identify differences that may elucidate the pathogenesis, risk factors, and treatments. The application of an OI approach using these cell lines would thus be very promising to aid further understanding and characterization of the etiology of the neurodegeneration, neurodevelopmental, and psychiatric disorders. This enables a multitude of applications, from de-risking (pediatric) drugs for adverse effects on cognitive development (207), the identification of toxicants and illicit drugs with long-term effects on cognitive capabilities, and the optimization of lead drug candidates acting on respective pharmacological targets.
The study of memory, learning, and cognition – and the impact of neurodegeneration on these functions – will require physiologically relevant neuron-to-glia ratios and high levels of biological complexity and interregional communication. Besides assembloids (48, 208, 209), aspects of ventral and dorsal regions in the absence of external morphogens or growth factors have been recapitulated (210–212), highlighting an innate ability of self-organization in both cases. Recently, Cederquist et al. (213) demonstrated that a chimeric assembloid of an early organoid and a cluster of sonic hedgehog (SHH)-secreting cells resulted in dorsal-ventral and anterior-posterior positional axes. However, organoids do not have predictable anatomy nor defined topography, and generally do not reflect the characteristic developmental asymmetry (208, 213). Nonetheless, brain organoids do show a remarkable self-organizational capacity. This might be further enhanced by functional demands, which may be harnessed when scaling up the size and inducing regional polarization, thereby increasing the biological complexity of neural networks and interregional connections to better reflect brain architecture and function (48, 208–213). Bioprinting approaches might also help here.
Ethics of biological computing with organoids
Creating a human brain model with input and output as well as learning capabilities raises complex ethical questions. At 12 weeks of fetal development, a human brain has a weight of approximately 3 g, a volume of approximately 3.5 ml, and 3 × 109 cells in the neocortical part of the fetal telencephalon (214–216). An adult mouse brain weighs approximately 0.4 g. In comparison, current brain organoids have a diameter below 500 μm in culture, with less than 100,000 cells. The maturation of the brain organoid is accelerated by growth factors so that at 10 weeks of culture, organoids show some features (such as myelination) that start in the fetus after 20 weeks of gestation (217). Furthermore, the stimulation with information input might lead to very different organoid development, and much longer culture periods would be envisaged for training organoids, together possibly augmenting cognitive capabilities.
The ethical concerns raised by brain organoid research have mainly focused on questions about creating entities that could potentially exhibit consciousness (45). Could organoids experience pain and, if so, would they suffer – even in rudimentary ways? These concerns will mount during the development of OI, as the organoids become structurally more complex, receive inputs, generate outputs, and – at least theoretically – process information about their environment and build a primitive memory. This will require deeper analysis and research regarding the morally salient neurobiological features that contribute to human capacities, including consciousness, and the implications for OI research and implementation when some or all of these are met. Articulating the physiological conditions that are necessary and sufficient for consciousness is one of the most difficult puzzles of neuroscience (143, 218). To assess whether organoids exhibit the criteria for consciousness will require some consensus to be reached about what those criteria are (219). Work underway to uncover the neural basis of consciousness will inform the evaluation of the ethical issues raised by OI. However, it will also be important to distinguish “consciousness” from “sentience,” formally considered as “awareness to stimuli,” i.e. response to sensory input (220, 221). Such use of terminology can be debated and represents a critical challenge for the forming OI community. We use the term “sentience” in its most basic way, similar to the way in which many aspects of cognition have to be understood as very basic cellular mechanisms, not human-level brain functions. Even recent proposals (222) for the use of the perturbational complexity index (PCI) assume the more complex idea of phenomenological consciousness when the behavior could be explained by simpler sentience (174). Notably, the suggested OI program does not aim to recreate human consciousness, but rather functional aspects related to learning, cognition, and computing.
Nevertheless, as advances in the structural and functional complexity of OI systems begin to recapitulate aspects of human neurobiological (sub)-processes, such as learning and cognition, researchers will inevitably encounter the Greely Dilemma: a situation whereby incremental successes in modelling aspects of the human brain will raise the same kind of ethical concerns that originally motivated their development (223). Sufficient advances in OI will raise questions about the moral status of these entities and concerns for their welfare. Frameworks have been proposed to address these ethical concerns in research practices (224, 225) but it remains unknown whether these proposals adequately attend to moral concerns held by the public. For example, harm reduction policies are often unsuccessful in gaining public support when the underlying attitude is based on a moral conviction (226) with implications for public discourse (227). Comprehensive ethical analysis of OI will require input from diverse publics and relevant stakeholder groups (228), in order to (i) prevent misunderstandings from creating unintended moral appraisals, and (ii) and foster trust, confidence, and inclusion through responsible public engagement. Notably, moral attitudes toward OI may depend less on epistemological concerns mentioned above, such as the role of specific cognitive capacities in assessments of moral status, and more on ontological arguments of what constitutes a human being. Perceptions of (re)creating ‘human-like’ entities in the lab are likely to evoke concerns about infringing on human dignity that could reflect secular or theological beliefs about the ‘essential’ nature of the human being (229, 230). Our approach to embedded ethics in OI will seek to identify and attend to these ethical concerns by informing future public engagement and deliberation on OI.
Other issues anticipated to require attention include privacy concerns on the part of iPSC donors and aspects of intellectual property. What does the organoid exhibit about the cell donor? Is there a moral obligation to inform the donor if, for example, something relevant to their health is identified during research? Do donors have rights that extend beyond the donation?
In common with other scientific and bioengineering aspects of OI, this is truly uncharted territory. The ethical considerations and viewpoints can be expected to evolve with an increased understanding of organoid systems. It is therefore critical to frame the ethical considerations at the onset of this research in a manner that encompasses all anticipated issues, and which continually reflects on progress and new lessons. We propose to use an “embedded ethics” approach whereby an ethics team will identify, discuss, and analyze ethical issues as they arise in the course of this work. Embedded ethics is a standard approach in interdisciplinary ethics research, whereby expert ethicists join and collaborate integrally with research and development teams to consider and address ethical issues via an iterative and continuous process as the research evolves (231). Box 1 offers a preliminary framework of ethical considerations informed by discussions at the “First organoid intelligence (OI) workshop to form an OI community” workshop (22–24 February 2022) (51).
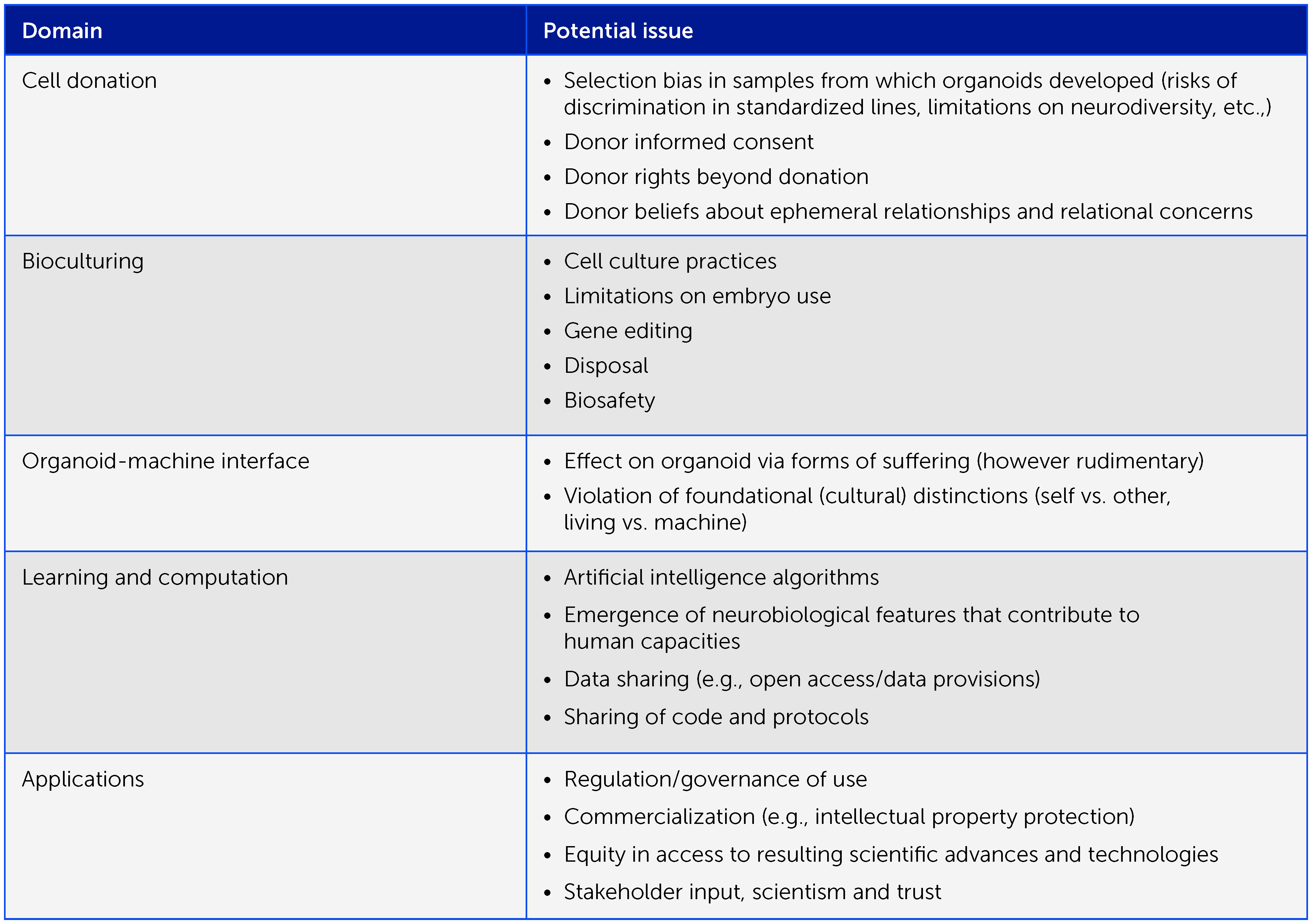
Box 1 Preliminary framework of ethical considerations in organoid intelligence research. Organoid intelligence research entails many important ethical aspects that warrant an iterative, collaborative ethical process as the field develops, involving all relevant stakeholders. Here we offer a preliminary framework of issues for consideration.
While the embedded ethics approach provides a mechanism for investigating the philosophical and scientific conditions relevant to the moral status of brain organoids, it has no inherent mechanisms for seeking, identifying, or incorporating public values in the development of OI. It is important to understand public perceptions of OI, and this cannot be delegated to ethicists alone. It needs to be embedded within the field as a three-way feedback loop involving researchers, ethicists, and members of the public, including stakeholders who could be especially impacted by advances in OI (e.g. neurodiversity advocates). This feedback loop will enable specific applications of OI to be articulated by researchers, analyzed by ethicists based on theoretical principles, and evaluated by members of the public with diverse moral perspectives. The views expressed by the public then inform the work of both scientists and ethicists seeking to advance OI in a socially responsive manner. This call for public dialogue has been echoed by a National Academy of Sciences report on human neural organoids (232), the recommendations on innovation in neurotechnology by the Organisation for Economic Co-operation and Development (233), and various neuroethics committees. Moreover, researchers in science communication and deliberative democracy have demonstrated that deliberative techniques are one of the most effective mechanisms for informing the public and mitigating the risk of polarization on contentious issues. Finally, public engagement on OI will not only be necessary to prevent adverse public reactions but will also maximize the future impact of the field and serve as an exemplar of how to embed society within science.
Of relevance in this context, coauthor JK co-chaired the neuroethics subcommittee for the new strategic plan for NIH BRAIN 2.0 (234). The only project we know of with relevant parallel aspects is the Brainstorm Project led by Insoo Hyun at Case Western University (235). We will be further actively promoting discussions between biologists, ethicists, and philosophers who are interested in brain organoids and navigating brain organoid ethics in research, as we did before (219). A recommended way forward would include: (1) an agreement on commonly used language, (2) the need for research on the neural basis for consciousness (as above); and (3) the development of best practice guidelines that consider the views of all relevant stakeholders.
Conclusion: an action plan for OI research
We present a collaborative, iterative multidisciplinary program aiming to establish OI as a form of genuine biological computing that harnesses brain organoids using the scientific and bioengineering approaches described here in an ethically responsible manner (Figure 5). Ultimately, we aim toward a revolution in biological computing that could overcome many of the limitations of silicon-based computing and AI and have significant implications worldwide. Specifically, we anticipate OI-based biocomputing systems to allow faster decision-making (including on massive, incomplete, and heterogenous datasets), continuous learning during tasks, and greater energy and data efficiency. Furthermore, the development of “intelligence-in-a-dish” offers unparalleled opportunities to elucidate the biological basis of human cognition, learning, and memory, together with various disorders associated with cognitive deficits – potentially aiding the identification of novel therapeutic approaches to address major global unmet needs.
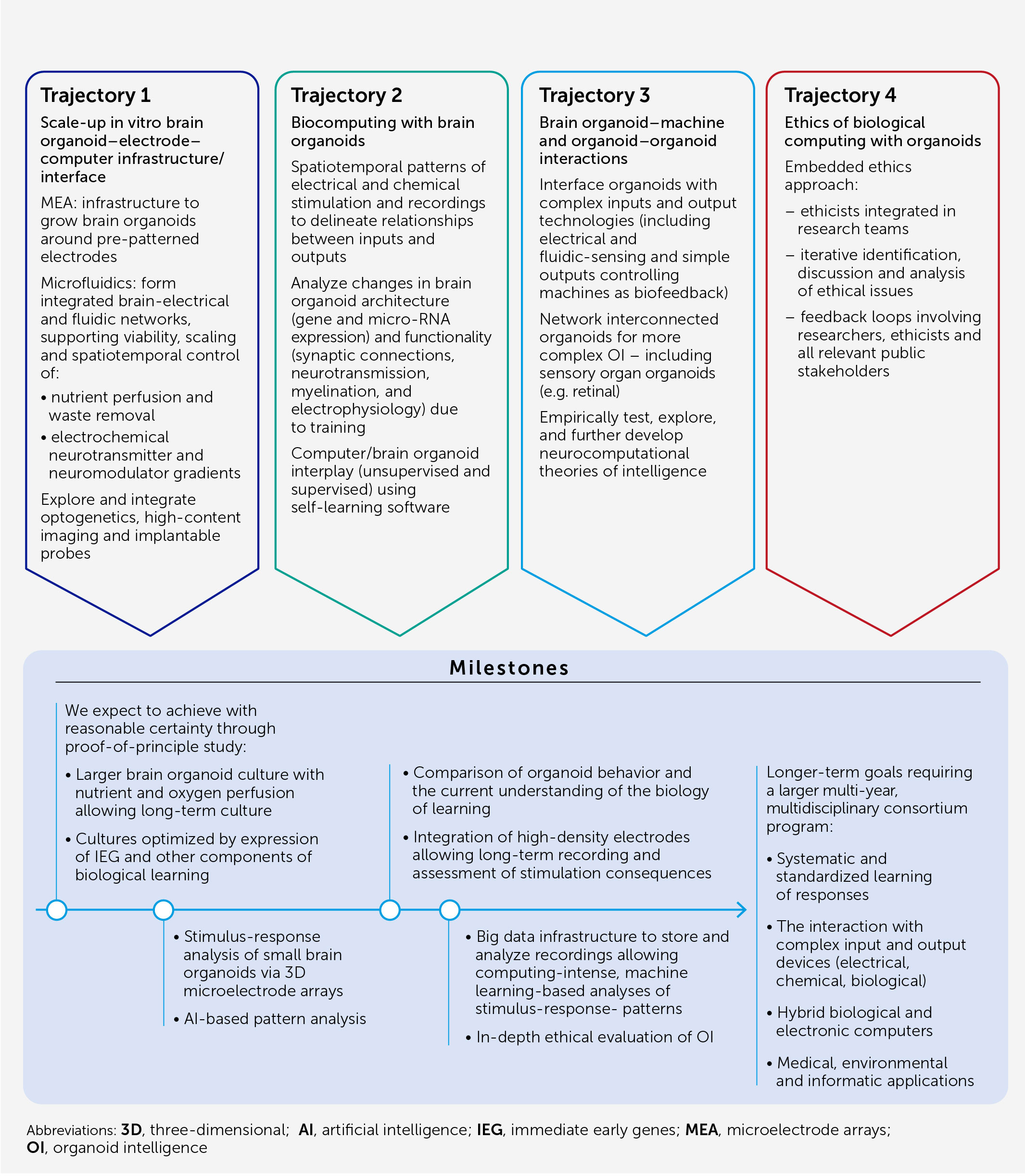
Figure 5 Organoid intelligence (OI) action plan and research trajectories. Our program foresees assembling multidisciplinary teams working in four research and development directions (or “trajectories”) employing the scientific and bioengineering advances described to establish OI as a genuine biological computing field harnessing brain organoids in an iterative and ethically responsible manner.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors are most grateful to Lee Baker and colleagues at Frontiers for the professional editing of the manuscript. An early version of this manuscript was shared with the participants of the “First organoid intelligence (OI) workshop to form an OI community” (22–24 February 2022) organized by the Transatlantic Think Tank for Toxicology (t4) at the Johns Hopkins Center for Alternatives to Animal Testing, and financed by the Doerenkamp-Zbinden Foundation, Switzerland, the Johns Hopkins Whiting School of Engineering, and Frontiers. While the presentations and discussions led to a parallel report (51), they have also shaped this article and the authors are most grateful for the stimulating discussions. The work was supported by a Johns Hopkins Discovery award (to TH) and a Johns Hopkins SURPASS award (to TH and EJ).
Conflict of interest
TH is named inventor on a patent by Johns Hopkins University on the production of brain organoids, which is licensed to AxoSim, New Orleans, LA, United States, and receives royalty shares. TH and LS consult AxoSim. JS is named as inventor on a patent by the University of Luxembourg on the production of midbrain organoids, which is licensed to OrganoTherapeutics SARL, Esch-sur-Alzette, Luxembourg. JS is also co-founder and shareholder of OrganoTherapeutics SARL. AM is a co-founder and has equity interest in TISMOO, a company dedicated to genetic analysis and human brain organogenesis, focusing on therapeutic applications customized for autism spectrum disorders and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. BK is an inventor on patents for technology related to this paper along with being employed at and holding shares in Cortical Labs Pty Ltd, Melbourne, Australia. No specific funding or other incentives were provided for involvement in this publication.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Biological computation: calculation (not necessarily as mathematical operations) carried out by a biological system.
Biological computing: tasks typically done by computers carried out by biological systems.
Brain organoid: induced pluripotent stem cell (iPSC)-derived 3-dimensional neural cultures, recapitulating aspects of brain cellular composition, architecture, and functionality.
Cognition: the human mental action or process of acquiring knowledge and understanding through thought, experience, and the senses (236).
Cognition-in-a-dish: a basic ability to process an input and provide a measurably output; a learned adequate response to the stimuli which is enabled by the presence of the necessary molecular machinery and physiological features such as learning circuits of long-term memory.
Consciousness: the human state of being aware of and responsive to one’s surroundings (236); a hypothetical organoid’s state of being responsive to and “aware of” the environment.
Embodied intelligence: the computational approach to the design and understanding of intelligent behavior in embodied and situated agents through the consideration of the strict coupling between the agent and its environment (situatedness), mediated by the constraints of the agent’s own body, perceptual and motor system, and brain (embodiment) (237).
Intelligence: the human ability to acquire and apply knowledge and skills (238).
Intelligence-in-a-dish: vision of OI-implementing cell models to perform computer functions (238) and to test substances (e.g., for toxicological or pharmacological purposes).
Learning and memory: in the context of OI, learning is identified as an increased frequency to show and memorize a response pattern to a stimulatory pattern.
Organoid intelligence (OI): describes an emerging field aiming to expand the definition of biocomputing toward brain-directed OI computing, i.e., to leverage the self-assembled machinery of three-dimensional (3D) human brain cell cultures (brain organoids) to memorize and compute inputs.
Sentience: in humans, the simplest or most primitive form of cognition, consisting of a conscious awareness of stimuli without association or interpretation (220); for OI, basic responsiveness to sensory input, e.g. light, heat, etc.
References
1. Hornyak T. Fujitsu Supercomputer simulates 1 second of brain activity. CNET (2013). Available at: https://www.cnet.com/culture/fujitsu-supercomputer-simulates-1-second-of-brain-activity/.
2. Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc Natl Acad Sci USA (2012) 109:10661–8. doi: 10.1073/pnas.1201895109
3. Reber P. What is the memory capacity of the human brain? Sci Am (2010). https://www.scientificamerican.com/article/what-is-the-memory-capacity/.
4. Orger MB, de Polavieja GG. Zebrafish behavior: opportunities and challenges. Annu Rev Neurosci (2017) 40:125–47. doi: 10.1146/annurev-neuro-071714-033857
5. Webb PW. The swimming energetics of trout. II. oxygen consumption and swimming efficiency. J Exp Biol (1971) 55:521–40. doi: 10.1242/jeb.55.2.521
6. Raichle ME, Gusnard DA. Appraising the brain’s energy budget. Proc Natl Acad Sci USA (2002) 99(16):10237–9. doi: 10.1073/pnas.172399499
8. Hewlett Packard Enterprise. Hewlett Packard Enterprise ushers in new era with world’s first and fastest exascale supercomputer “Frontier” for the U.S. department of energy’s oak ridge national laboratory (2022). Available at: https://www.hpe.com/us/en/newsroom/press-release/2022/05/hewlett-packard-enterprise-ushers-in-new-era-with-worlds-first-and-fastest-exascale-supercomputer-frontier-for-the-us-department-of-energys-oak-ridge-national-laboratory.html.
9. Vora H, Kathiria P, Agrawal S, Patel U. Neuromorphic computing: review of architecture, issues, applications and research opportunities. In: Singh. Springer, Singapore (2022).
10. Lapedus M. Chiplets enter the supercomputer race. In: Semiconductor engineering (2022). Available at: https://semiengineering.com/chiplets-enter-the-supercomputer-race/.
11. Grace K. A summary of AI surveys. AI Impacts (2015). Available at: https://aiimpacts.org/transmitting-fibers-in-the-brain-total-length-and-distribution-of-lengths/a.
12. Wu Z, Liu Z, Lin J, Lin Y, Han S. Lite transformer with long-short range attention. arXiv:2004.11886 (2020). doi: 10.48550/arXiv.2004.11886
13. Giurfa M, Zhang S, Jenett A, Menzel R, Srinivasan MV. The concepts of “sameness” and “difference” in an insect. Nature (2001) 410:930–3. doi: 10.1038/35073582
14. Fleuret F, Li T, Dubout C, Wampler EK, Yantis S, Geman D. Comparing machines and humans on a visual categorization test. Proc Natl Acad Sci USA (2011) 108:17621–5. doi: 10.1073/pnas.1109168108
15. Junkyung K, Matthew R, Thomas S. Not-So-CLEVR: learning same–different relations strains feedforward neural networks. Interface Focus (2018) 8(20180011). doi: 10.1098/rsfs.2018.0011
16. Borowiec S. AlphaGo seals 4-1 victory over go grandmaster Lee sedol. In: The guardian (2016). Available at: https://www.theguardian.com/technology/2016/mar/15/googles-alphago-seals-4-1-victory-over-grandmaster-lee-sedol.
17. Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature (2016) 529:484–9. doi: 10.1038/nature16961
18. Strubell E, Ganesh A, Mccallum A. Energy and policy considerations for deep learning in NLP. arXiv: [cs CL] (2015) 10:4. doi: 10.48550/arXiv.1906.02243
19. Knight W. Facebook’s head of AI says the field will soon “hit the wall”. In: Wired (2019). Available at: https://www.wired.com/story/facebooks-ai-says-field-hit-wall/.
20. Thompson NC, Greenewald K, Lee K, Manso GF. The computational limits of deep learning. arXiv:2007.05558 (2020). doi: 10.48550/arXiv.2007.05558
21. Smith B. Microsoft Will be carbon neutral by 2030. Official Microsoft Blog (2020). Available at: https://news.microsoft.com/2020/01/16/microsoftannounces-it-will-be-carbon-negative-by-2030/.
22. Apple. Apple commits to be 100 percent carbon neutral for its supply chain and products by 2030. Available at: https://www.apple.com/uk/newsroom/2020/07/apple-commits-to-be-100-percent-carbon-neutral-for-its-supply-chain-and-products-by-2030/.
23. Masanet E, Shehabi A, Lei N, Smith S, Koomey J. Recalibrating global data center energy-use estimates. Science (2020) 367(6481):984–6. doi: 10.1126/science.aba3758
24. Grozinger L, Amos M, Gorochowski TE, Carbonell P, Oyarzún DA, Stoof R. Pathways to cellular supremacy in biocomputing. Nat Commun (2019) 10(1):5211–50. doi: 10.1038/s41467-019-13232-z
25. Schmidt E. Former Google CEO Eric Schmidt believes biology is the next frontier in computing. CNBC (2019). Available at: https://www.cnbc.com/2019/10/02/ericschmidt-says-hes-eyeing-biology-for-the-next-computing-frontier.html.
26. van Hooidjonk R. How close are we to organic computers? (2019). Available at: https://blog.richardvanhooijdonk.com/en/how-close-are-we-to-organic-computers/.
27. Lee HH, Kalhor R, Goela N. Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat Commun (2019) 10:2383. doi: 10.1038/s41467-019-10258-1
28. Fu Y, Tsai TH, Mao C, Mun SK, Ressom HW, Wang M, et al. “Biological computing”. Feng DD, editor. Biomedical Information Technology (2020) 81–104.
29. Kagan BJ, Kitchen AC, Tran NT, Habibollahi F, Khajehnejad M, Parker BJ, et al. In vitro neurons learn and exhibit sentience when embodied in a simulated game-world. Neuron (2022) 110(23):3952–69. doi: 10.1016/j.neuron.2022.09.001
30. Chesnut M, Hartung T, Hogberg HT, Pamies D. Human oligodendrocytes and myelin in vitro to evaluate developmental neurotoxicity. Int J Mol Sci (2021) 22(15):7929. doi: 10.3390/ijms22157929
31. Chesnut M, Paschoud H, Repond C, Smirnova L, Hartung T, Zurich MG, et al. Human 3D iPSC-derived brain model to study chemical-induced myelin disruption. Int J Mol Sci (2021) 22(17):9473. doi: 10.3390/ijms22179473
32. Pamies D, Barreras P, Block K, Makri G, Kumar A, Wiersma D, et al. Human brain microphysiological system derived from iPSC to study central nervous system toxicity and disease. ALTEX (2017) 34:362–76. doi: 10.14573/altex.1609122
33. Anderson WA, Bosak A, Hogberg HT, Hartung T, Moore MJ. Advances in 3D neuronal microphysiological systems: towards a functional nervous system on a chip. In Vitro Cell Dev Biol Anim (2021) 57(2):191–206. doi: 10.1007/s11626-020-00532-8
34. Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell (2019) 25(4):558–69. doi: 10.1016/j.stem.2019.08.002
35. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature (2013) 501(7467):373–9. doi: 10.1038/nature12517
36. Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods (2015) 3(7):671–8. doi: 10.1038/nmeth.3415
37. Alford S, Zompa I, Dubuc R. Long-term potentiation of glutamatergic pathways in the lamprey brainstem. J Neurosci (1995) 15(11):7528–38. doi: 10.1523/JNEUROSCI15-11-07528.1995
38. Kositsky M, Chiappalone M, Alford ST, Mussa-Ivaldi FA. Brain-machine interactions for assessing the dynamics of neural systems. Front Neurorobot (2009) 3:1.2009. doi: 10.3389/neuro.12.001.2009
39. Teyler TJ. Use of brain slices to study long-term potentiation and depression as examples of synaptic plasticity. Methods (1999) 18(2):109–16. doi: 10.1006/meth.1999.0764
40. Tessadori J, Chiappalone M. Closed-loop neuro-robotic experiments to test computational properties of neuronal networks. J Vis Exp (2015) 97(52341). doi: 10.3791/52341
41. Bakkum DJ, Chao ZC, Potter SM. Spatio-temporal electrical stimuli shape behavior of an embodied cortical network in a goal-directed learning task. J Neural Eng (2008) 5:310–23. doi: 10.1088/1741-2560/5/3/004
42. Isomura T, Kotani K, Jimbo Y. Cultured cortical neurons can perform blind source separation according to the free-energy principle. PloS Comput Biol (2015) 11(12):e1004643. doi: 10.1371/journal.pcbi.1004643
43. Marom S, Shahaf G. Development, learning and memory in large random networks of cortical neurons: lessons beyond anatomy. Q Rev Biophys (2002) 35(1):63–87. doi: 10.1017/S0033583501003742
44. Shahaf G, Marom S. Learning in networks of cortical neurons. J Neurosci (2001) 21(22):8782–8. doi: 10.1523/JNEUROSCI21-22-08782.2001
45. Sawai T, Sakaguchi H, Thomas E, Takahashi J, Fujita M. The ethics of cerebral organoid research: being conscious of consciousness. Stem Cell Rep (2019) 13(3):440–7. doi: 10.1016/j.stemcr.2019.08.003
46. Sharf T, van der Molen T, Guzman E, Glasauer SMK, Luna G, Cheng Z, et al. Intrinsic network activity in human brain organoids. SSRN Electronic J (2021) 73. doi: 10.2139/ssrn.3797268
47. Smits LM, Magni S, Kinugawa K, Grzyb K, Luginbuehl J, Sabate-Soler S, et al. Single-cell transcriptomics reveals multiple neuronal cell types in human midbrain-specific organoids. Cell Tissue Res (2020) 382:463–76. doi: 10.1007/s00441-020-03249-y
48. Miura Y, Li M-Y, Revah O, Yoon S-J, Narazaki G, Paşca SP. Engineering brain assembloids to interrogate human neural circuits. Nat Protoc (2022) 17:15–35. doi: 10.1038/s41596-021-00632-z
49. NEUCHIP.EU. (2022). NEUCHIP.EU: Biological AI. Available at: https://neuchip.eu.
50. Falotico E, Vannucci L, Ambrosano A, Albanese U, Ulbrich S, Tieck V, et al. Connecting artificial brains to robots in a comprehensive simulation framework: the neurorobotics platform. Front Neurorobot (2017) 11(2). doi: 10.3389/fnbot.2017.00002
51. Morales Pantoja IE, Smirnova L, Muotri AR, Wahlin KJ, Kahn J, Boyd JL, et al. First Organoid Intelligence (OI) workshop to form an OI community. Front Artif Intell (2023) 6:1116870. doi: 10.3389/frai.2023.1116870
52. Hartung T, Smirnova L, Morales Pantoja IE, Akwaboah A, Alam El Din D-M, Berlinicke CA, et al. The Baltimore declaration toward the exploration of organoid intelligence. Front Sci (2023) 1:1068159. doi: 10.3389/fsci2022.1068159
53. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. CellNov (2007) 30(131):5. doi: 10.1016/j.cell.2007.11.019
54. Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell (2008) 3(5):519–32. doi: 10.1016/j.stem.2008.09.002
55. Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. CellJul (2015) 16(162):2. doi: 10.1016/j.cell.2015.06.034
56. Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell (2016) 19:248–57. doi: 10.1016/j.stem.2016.07.005
57. Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature (2017) 545(7652):48–53. doi: 10.1038/nature22047
58. Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron (2017) 95(4):779–90. doi: 10.1016/j.neuron.2017.07.035
59. Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci (2019) 22(3):484–91. doi: 10.1038/s41593-018-0316-9
60. Williamson JM, Lyons DA. Myelin dynamics throughout life: an ever-changing landscape? Front Cell (2018) 12(424):107–27. doi: 10.3389/fncel.2018.00424
61. Buyanova IS, Arsalidou M. Cerebral white matter myelination and relations to age, gender, and cognition: a selective review. Front Hum Neurosci (2021) 15:66203. doi: 10.3389/fnhum.2021.662031
62. Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. A novel approach to derive human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep (2017) 8(5):1144–54. doi: 10.1016/j.stemcr.2017.03.010
63. Wilton DK, Dissing-Olesen L, Stevens B. Neuron-glia signaling in synapse elimination. Annu. Rev Neurosci (2019) 42:107–27. doi: 10.1146/annurev-neuro-070918-050306
64. Huang AY, Woo J, Sardar D, Lozzi B, Bosquez Huerta NA, Lin C-CJ, et al. Region-specific transcriptional control of astrocyte function oversees local circuit activities. Neuron (2020) 106(6):992–1008. doi: 10.1016/j.neuron.2020.03.025
65. Xin W, Chan JR. Myelin plasticity: sculpting circuits in learning and memory. Nat Rev Neurosci (2020) 21(12):682–94. doi: 10.1038/s41583-020-00379-8
66. Keller D, Eroe C, Markram H. Cell densities in the mouse brain: a systematic review. Front Neuroanat (2018) 12. doi: 10.3389/fnana.2018.00083
67. Oh Y, Jang J. Directed differentiation of pluripotent stem cells by transcription factors. Mol Cells (2019) 42(3):200–9. doi: 10.14348/molcells.2019.2439
68. Boussaad I, Cruciani G, Bolognin S, Antony P, Dording CM, Kwon YJ, et al. Integrated, automated maintenance, expansion and differentiation of 2D and 3D patient-derived cellular models for high throughput drug screening. Sci Rep (2021) 11(1):1439. doi: 10.1038/s41598-021-81129-3
69. Pamies D, Leist M, Coecke S, Bowe G, Allen DG, Gstraunthaler G, et al. Guidance document on good cell and tissue culture practice 2.0 (GCCP 2.0). ALTEX (2022) 39:30–70. doi: 10.14573/altex.2111011
70. MPS World Summit. 2nd microphysiological systems world summit (2023). Available at: https://mpsworldsummit.com.
71. Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell (2019) 76(4):743–56. doi: 10.1016/j.cell.2019.01.017
72. Qian X, Song H, Ming GL. Brain organoids: advances, applications and challenges. Dev (2019) 146(8):dev166074. doi: 10.1242/dev.166074
73. Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature (2020) 578(7793):142–8. doi: 10.1038/s41586-020-1962-0
74. Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb Perspect Biol (2009) 1(2):1–17. doi: 10.1101/cshperspect.a002519
75. Hofman MA. Evolution of the human brain: when bigger is better. Front Neuroanat (2014) 8(15):1–12. doi: 10.3389/fnana.2014.00015
76. Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang Y-J, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods (2019) 16:1169–75. doi: 10.1038/s41592-019-0586-5
77. Matsui TK, Tsuru Y, Hasegawa K, Kuwako KI. Vascularization of human brain organoids. Stem Cells (2021) 39(8):1017–24. doi: 10.1002/stem.3368
78. Zhang S, Wan Z, Kamm RD. Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip (2021) 1(3):473–88. doi: 10.1039/D0LC01186J
79. Roth A, MPS-WS Berlin 2019. Human microphysiological systems for drug development. Science (2021) 375(6561):1304–6. doi: 10.1126/science.abc3734
80. Marx U, Akabane T, Andersson TB, Baker E, Beilmann M, Beken S, et al. Biology-inspired microphysiological systems to advance medicines for patient benefit and animal welfare. ALTEX (2020) 37(3):364–94. doi: 10.14573/altex.2001241
81. Jamal M, Zarafshar AM, Gracias DH. Differentially photo-crosslinked polymers enable self-assembling microfluidics. Nat Commun (2011) 2(1):527. doi: 10.1038/ncomms1531
82. Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater (2012) 11(9):768–74. doi: 10.1038/nmat3357
83. Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, et al. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science (2019) 364(6439):458–64. doi: 10.1126/science.aav9750
84. Sabatini BL, Tian L. Imaging neurotransmitter and neuromodulator dynamics in vivo with genetically encoded indicators. Neuron (2020) 108(1):17–32. doi: 10.1016/j.neuron.2020.09.036
85. Liu C, Goel P, Kaeser PS. Spatial and temporal scales of dopamine transmission. Nat Rev Neurosci (2021) 22(6):345–58. doi: 10.1038/s41583-021-00455-7
86. Mei J, Muller E, Ramaswamy S. Informing deep neural networks by multiscale principles of neuromodulatory systems. Trends Neurosci (2022) 45(3):237–50. doi: 10.1016/j.tins.2021.12.008
87. Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem (2001) 73(6):1240–6. doi: 10.1021/ac001132d
88. Berger E, Magliaro C, Paczia N, Monzel A, Antony P, Linster C, et al. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab Chip (2018) 18:3172–83. doi: 10.1039/C8LC00206A
89. Romano G. Neuronal receptor agonists and antagonists. Labome Mater Methods (2019) 9. doi: 10.13070/mm.en.9.2851
90. DeMarse TB, Wagenaar DA, Blau AW, Potter SM. The neurally controlled animat: biological brains acting with simulated bodies. Auton Robots (2001) 11:305–10. doi: 10.1023/A:1012407611130
91. Chen R, Canales A, Anikeeva P. Neural recording and modulation technologies. Nat Rev Mater (2017) 2:2. doi: 10.1038/natrevmats.2016.93
92. Cools J, Jin Q, Yoon E, Alba Burbano D, Luo Z, Cuypers D, et al. A micropatterned multielectrode shell for 3D spatiotemporal recording from live cells. Adv Sci (Weinh) (2018) 5:17007. doi: 10.1002/advs.201700731
93. Huang Q, Tang B, Romero JC, Yang Y, Elsayed SK, Pahapale G, et al. Shell microelectrode arrays (MEAs) for brain organoids. Sci Adv (2022) 8(33). doi: 10.1126/sciadv.abq5031
94. Park Y, Chung TS, Rogers JA. Three dimensional bioelectronic interfaces to small-scale biological systems. Curr Opin Biotechnol (2021) 72:1–7. doi: 10.1016/j.copbio.2021.07.023
95. Park Y, Franz CK, Ryu H, Luan H, Cotton KY, Kim JU, et al. Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Sci Adv (2021) 17:7. doi: 10.1126/sciadv.abf9153
96. Kalmykov A, Huang C, Bliley J, Shiwarski D, Tashman J, Abdullah A, et al. Organon-e-chip: three-dimensional self-rolled biosensor array for electrical interrogations of human electrogenic spheroids. Sci Adv (2019) 23:5.
97. Song E, Li J, Won SM, Bai W, Rogers JA. Materials for flexible bioelectronic systems as chronic neural interfaces. Nat Mater (2020) 19(6):590–603. doi: 10.1038/s41563-020-0679-7
98. Shi Q, Liu H, Tang D, Li Y, Li X, Xu F. Bioactuators based on stimulus-responsive hydrogels and their emerging biomedical applications. NPG Asia Mater (2019) 11(1):64. doi: 10.1038/s41427-019-0165-3
99. Efros AL, Delehanty JB, Huston AL, Medintz IL, Barbic M, Harris TD. Evaluating the potential of using quantum dots for monitoring electrical signals in neurons. Nat Nanotechnol (2018) 13(4):278–88. doi: 10.1038/s41565-018-0107-1
100. Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, et al. Fully integrated silicon probes for high-density recording of neural activity. Nature (2017) 551(7679):232–6. doi: 10.1038/nature24636
101. Steinmetz NA, Aydin C, Lebedeva A, Okun M, Pachitariu M, Bauza M, et al. Neuropixels 2.0: a miniaturized high-density probe for stable, long-term brain recordings. Science (2021) 372(6539). doi: 10.1126/science.abf4588
102. Puppo F, Sadegh S, Trujillo CA, Thunemann M, Campbell EP, Vandenberghe M, et al. All-optical electrophysiology in hiPSC-derived neurons with synthetic voltage sensors. Front Cell Neurosci (2021) 15:671549. doi: 10.3389/fncel.2021.671549
103. Sun S, Zhang G, Cheng Z, Gan W, Cui M. Large-Scale femtosecond holography for near simultaneous optogenetic neural modulation. Opt Express (2019) 27(22):32228–34. doi: 10.1364/OE.27.032228
104. Ovsepian SV. The dark matter of the brain. Brain Struct Funct (2019) 224(3):973–83. doi: 10.1007/s00429-019-01835-7
105. Royal Victorian Eye and Ear Hospital. First implantation of prototype bionic eye with 24 electrodes: “All of a sudden I could see a little flash of light”. ScienceDaily (2012). Available at: www.sciencedaily.com/releases/2012/08/120831065003.htm.
106. Patrick JF, Clark JM. The nucleus 22-channel cochlear implant system. Ear Hear (1991) 12:3S–9S. doi: 10.1097/00003446-199108001-00002
107. Advanced Bionics. Advanced bionics HiRes™ bionic ear system. Available at: https://www.advancedbionics.com/us/en/portals/professional-portal/products/ci-internal-components.html.
108. Osborn LE, Christie BP, McMullen DP, Nickl RW, Thompson MC, Pawar AS, et al. Intracortical microstimulation of somatosensory cortex enables object identification through perceived sensations. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE (2021) 6259–62. Available at: (https://jhu.pure.elsevier.com/en/publications/intracortical-microstimulation-of-somatosensory-cortex-enables-ob).
109. McMullen DP, Thomas TM, Fifer MS, Candrea DN, Tenore FV, Nickl RW, et al. Novel intraoperative online functional mapping of somatosensory finger representations for targeted stimulating electrode placement: technical note. J Neurosurg (2021), 1–8. doi: 10.3171/2020.9.JNS202675
110. Kingma DP, Welling M. Auto-encoding variational bayes. arXiv:1312.6114 (2014). doi: 10.48550/arXiv.1312.6114
111. Kramer MA. Nonlinear principal component analysis using autoassociative neural networks. AIChE J (1991) 37(2):233–43. doi: 10.1002/aic.690370209
112. Di CZ, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. Ann Appl Stat (2009) 3:3. doi: 10.1214/08-AOAS206SUPP
113. Jewell S, Witten D. Exact spike train inference via ℓ0 optimization. Ann Appl Stat (2018) 12(4):2457–82. doi: 10.1214/18-AOAS1162
114. Gao C, Han F, Zhang CH. On estimation of isotonic piecewise constant signals. Ann Stat (2020) 48(2):629–54. doi: 10.1214/18-AOS1792
115. Shen Y, Han Q, Han F. On a phase transition in general order spline regression. IEEE Trans Inf Theory (2022) 68(6):4043–69. doi: 10.1109/TIT.2022.3150253
116. Fan J. On the optimal rates of convergence for nonparametric deconvolution problems. Ann Stat (1991) 19(3):1257–72. doi: 10.1214/aos/1176348248
117. Miao Z, Kong W, Vinayak RK, Sun W, Han F. Fisher-Pitman permutation tests based on nonparametric poisson mixtures with application to single cell genomics. J American Stat Association (2022). doi: 10.1080/01621459.2022.2120401
118. Chén OY, Crainiceanu C, Ogburn EL, Caffo BS, Wager TD, Lindquist MA. High-dimensional multivariate mediation with application to neuroimaging data. Biostatistics (2018) 19(2):121–36. doi: 10.1093/biostatistics/kxx027
119. Zhao Y, Lindquist MA, Caffo BS. Sparse principal component based high-dimensional mediation analysis. Comput Stat Data Anal (2020) 142(10683):5. doi: 10.1016/j.csda.2019.106835
120. Caffo B, Chen S, Stewart W, Bolla K, Yousem D, Davatzikos C, et al. Are brain volumes based on magnetic resonance imaging mediators of the associations of cumulative lead dose with cognitive function? Am J Epidemiol (2008) 167(4):429–37. doi: 10.1093/aje/kwm326
121. Pearl J. The limitations of opaque learning machines. In: Brockman J, editor. Possible minds: Twenty-five ways of looking at AI, Penguin press (2019) 13–9.
122. Pearl J, Mackenzie D. AI Can’t reason why. Wall Street J (2018). Available at: https://www.wsj.com/articles/ai-cant-reason-why-1526657442.
123. Bressler SL, Seth AK. Wiener-Granger causality: a well established methodology. NeuroImage (2011) 58(2):323–9. doi: 10.1016/j.neuroimage.2010.02.059
124. Available at: http://www.scholarpedia.org/article/Brain_connectivity.
125. Friston KJ, Harrison L, Penny W. Dynamic causal modelling. NeuroImage (2003) 19(4):1273–302. doi: 10.1016/S1053-8119(03)00202-7
126. Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, Priebe CE, et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science (2014) 344(6182):386–92. doi: 10.1126/science.1250298
127. Pir P, le Novère N. Mathematical models of pluripotent stem cells: at the dawn of predictive regenerative medicine. Methods Mol Biol (2016) 1386:331–50. doi: 10.1007/978-1-4939-3283-2_15
128. Sharpe J. Computer modeling in developmental biology: growing today, essential tomorrow. Development (2017) 144(23):4214–25. doi: 10.1242/dev.151274
129. Zheng H, Feng Y, Tang J, Ma S. Interfacing brain organoids with precision medicine and machine learning. Cell Rep Phys Sci (2022) 3(7):100974. doi: 10.1016/j.xcrp.2022.100974
130. Libby ARG, Briers D, Haghighi I, Joy DA, Conklin BR, Belta C, et al. Automated design of pluripotent stem cell self-organization. Cell Syst (2019) 9(5):483–495.e10. doi: 10.1016/j.cels.2019.10.008
131. Xavier da Silveira dos Santos A, Liberali P. From single cells to tissue self-organization. FEBS J (2019) 286(8):1495–513. doi: 10.1111/febs.14694
132. Silva GA, Muotri AR, White C. Understanding the human brain using brain organoids and a structure-function theory. BioRxiv (2020) 19. doi: 10.1101/2020.07.28.225631
133. Dresp-Langley B. Seven properties of self-organization in the human brain. Big Data Cognit Comput (2020) 4(2):10. doi: 10.3390/bdcc4020010
134. Game CJA. Non-equilibrium thermodynamics and the brain. In: Pribram KH, editor. Origins: Brain and self-organization. Publisher TBC: Routledge, New York (1994). p. 196. Available at: https://www.taylorfrancis.com/books/mono/10.4324/9781315789347/origins-karl-pribram.
135. Grande-García I. The evolution of brain and mind: a non-equilibrium thermodynamics approach. Ludus Vitalis (2007) XV(27):103–25.
136. Andén J, Mallat S. Deep scattering spectrum. IEEE Trans Signal Process (2014) 62:16: 4114–28. doi: 10.1109/TSP.2014.2326991
137. Baugé C, Lagrange M, Andén J, Mallat S. Representing environmental sounds using the separable scattering transform. In: IEEE International conference on acoustics. speech and signal processing (2013) 8667–71.
138. Sinapayen L, Masumori A, Ikegami T. Learning by stimulation avoidance: a principle to control spiking neural networks dynamics. PloS One (2017) 12:2. doi: 10.1371/journal.pone.0170388
139. Zhu G, Liu Y, Wang Y, Bi X, Baudry M. Different patterns of electrical activity lead to long-term potentiation by activating different intracellular pathways. J Neurosci (2015) 35(2):621–33. doi: 10.1523/JNEUROSCI2193-14.2015
140. Daur N, Nadim F, Bucher D. The complexity of small circuits: the stomatogastric nervous system. Curr Opin NeuroBiol (2016) 41:1–7. doi: 10.1016/j.conb.2016.07.005
141. Schottdorf M. The reconstitution of visual cortical feature selectivity in vitro. Georg-August-Universitaet Goettingen (2017). Available at: https://ediss.uni-goettingen.de/handle/11858/00-1735-0000-002E-E348-B?locale-attribute=en.
142. Seth AK, Bayne T. Theories of consciousness. Nat Rev Neurosci (2022) 23(7):439–52. doi: 10.1038/s41583-022-00587-4
143. Insel TR, Landis SC, Collins FS. The NIH BRAIN initiative. Science (2013) 340(6133):687–8. doi: 10.1126/science.1239276
144. Yatsenko D, Nguyen T, Shen S, Gunalan K, Turner CA, Guzman R. DataJoint elements: data workflows for neurophysiology. bioRxiv (2021) 11. doi: 10.1101/2021.03.30.437358
145. Carlson D, Carin L. Continuing progress of spike sorting in the era of big data. Curr Opin NeuroBiol (2019) 55:90–6. doi: 10.1016/j.conb.2019.02.007
146. Unakafova VA, Gail A. Comparing open-source toolboxes for processing and analysis of spike and local field potentials data. Front Neuroinform (2019) 13:57. doi: 10.3389/fninf.2019.00057
147. Markiewicz CJ, Gorgolewski KJ, Feingold F, Blair R, Halchenko YO, Miller E, et al. The OpenNeuro resource for sharing of neuroscience data. Elife (2021) 10:e71774. doi: 10.7554/eLife.71774
148. DANDI. The DANDI archive. Available at: https://dandiarchive.org/.
149. Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data (2016) 3(160044):1–9. doi: 10.1038/sdata.2016.44
150. Rübel O, Tritt A, Ly R, Dichter BK, Ghosh S, Niu L, et al. The neurodata without borders ecosystem for neurophysiological data science. Elife (2022) 11:e78362. doi: 10.7554/eLife.78362
151. Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, et al. CaImAn an open source tool for scalable calcium imaging data analysis. Elife (2019) 8. doi: 10.7554/eLife.38173
152. Brain Image Library. About the brain image library. Available at: https://www.brainimagelibrary.org/about.html.
153. Lichtman JW, Pfister H, Shavit N. The big data challenges of connectomics. Nat Neurosci (2014) 17(11):1448–54. doi: 10.1038/nn.3837
154. Wang Z, Sair HI, Crainiceanu C, Lindquist M, Bennett A, Landman BA, et al. On statistical tests of functional connectome fingerprinting. Can J Stat (2021) 49:63–88. doi: 10.1002/cjs.11591
155. Johnson EC, Wilt M, Rodriguez LM, Norman-Tenazas R, Rivera C, Drenkow N, et al. Toward a scalable framework for reproducible processing of volumetric, nanoscale neuroimaging datasets. Gigascience (2020) 9:12. doi: 10.1093/gigascience/giaa147
156. Hider R Jr, Kleissas D, Gion T, Xenes D, Matelsky J, Pryor D, et al. The brain observatory storage service and database (BossDB): a cloud-native approach for petascale neuroscience discovery. Front Neuroinform (2022) 16(828787). doi: 10.3389/fninf.2022.828787
157. Chung J, Bridgeford E, Arroyo J, Pedigo BD, Saad-Eldin A, Gopalakrishnan V, et al. Statistical connectomics. Annu Rev Stat Appl (2021) 8:463–92. doi: 10.1146/annurev-statistics-042720-023234
158. Matelsky JK, Reilly EP, Johnson EC, Stiso J, Bassett DS, Wester BA, et al. DotMotif: an open-source tool for connectome subgraph isomorphism search and graph queries. Sci Rep (2021) 11(1):1–14. doi: 10.1038/s41598-021-91025-5
159. van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-minn human connectome project: an overview. NeuroImage (2013) 80:62–79. doi: 10.1016/j.neuroimage.2013.05.041
160. George JB, Abraham GM, Rashid Z, Amrutur B, Sikdar SK. Random neuronal ensembles can inherently do context dependent coarse conjunctive encoding of input stimulus without any specific training. Sci Rep (2018) 8:1403. doi: 10.1038/s41598-018-19462-3
161. Clawson WP, Wright NC, Wessel R, Shew WL. Adaptation towards scale-free dynamics improves cortical stimulus discrimination at the cost of reduced detection. PloS Comput Biol (2017) 13(5):e1005574. doi: 10.1371/journal.pcbi.1005574
162. Beggs JM, Plenz D. Neuronal avalanches in neocortical circuits. J Neurosci (2003) 3(23):11167–77. doi: 10.1523/JNEUROSCI23-35-11167.2003
163. Edelman GM. Neural Darwinism: selection and reentrant signaling in higher brain function. Neuron (1993) 10:115–25. doi: 10.1016/0896-6273(93)90304-A
165. Kangassalo L, Spapé M, Ravaja N, Ruotsalo T. Information gain modulates brain activity evoked by reading. Sci Rep (2020) 10:7671. doi: 10.1038/s41598-020-63828-5
166. Ebitz RB, Hayden BY. The population doctrine in cognitive neuroscience. Neuron (2021) 109:3055–68. doi: 10.1016/j.neuron.2021.07.011
167. Ebitz RB, Tu JC, Hayden BY. Rules warp feature encoding in decision-making circuits. PloS Biol (2020) 18:11. doi: 10.1371/journal.pbio.3000951
168. Schwartenbeck P, FitzGerald T, Mathys C, Dolan R, Kronbichler M, Friston K. Evidence for surprise minimization over value maximization in choice behavior. Sci Rep (2015) 5:16575. doi: 10.1038/srep16575
169. Knill DC, Pouget A. The Bayesian brain: the role of uncertainty in neural coding and computation. Trends Neurosci (2004) 27(12):712–9. doi: 10.1016/j.tins.2004.10.007
170. Barlow HB. Possible principles underlying the transformations of sensory messages. Sensory Communication (1961) 1. doi: 10.7551/mitpress/9780262518420.003.0013
171. Friston KJ, Tononi G, Reeke GN, Sporns O, Edelman GM. Value-dependent selection in the brain: simulation in a synthetic neural model. Neuroscience (1994) 59:229–43. doi: 10.1016/0306-4522(94)90592-4
172. Madhav MS, Cowan NJ. The synergy between neuroscience and control theory: the nervous system as inspiration for hard control challenges. Annu Rev Control Robot Auton Syst (2020) 3(1):243–67. doi: 10.1146/annurev-control-060117-104856
173. Friston KT. The free-energy principle: a unified brain theory? Nat Rev Neurosci (2010) 11:127–38. doi: 10.1038/nrn2787
174. Kagan BJ, Duc D, Stevens I, Gilbert F. Neurons embodied in a virtual world: evidence for organoid ethics? AJOB Neurosci (2022) 13(2):114–7. doi: 10.1080/21507740.2022.2048731
175. Khetarpal K, Riemer M, Rish I, Precup D. Towards continual reinforcement learning: a review and perspectives. arXiv:2012.1349)v2 (2020) 78. doi: 10.48550/arXiv.2012.13490
176. Parisi GI, Kemker R, Part JL, Kanan C, Wermter S. Continual lifelong learning with neural networks: a review. Neural Netw (2019) 13:54–71. doi: 10.1016/j.neunet.2019.01.012
177. Lebedev MA, Nicolelis MAL. Brain machine interface: from basic science to neuroprotheses and neurorehabilitation. Physiol Rev (2017) 97:767–837. doi: 10.1152/physrev.00027.2016
178. Bozhkov L. Deep learning models for brain machine interfaces. Ann Math Artif Intell (2019) 90:1175–90. doi: 10.1007/s10472-019-09668-0
179. Wahlin KJ, Maruotti JA, Sripathi SR, Ball J, Angueyra JM, Kim C, et al. Photoreceptor outer segment-like structures in long-term 3D retinas from human pluripotent stem cells. Sci Rep (2017) 7:1. doi: 10.1038/s41598-017-00774-9
180. Kallman A, Capowski EE, Wang J, Kaushik AM, Jansen AD, Edwards KL, et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun Biol (2020) 3(1):82. doi: 10.1038/s42003-020-0808-5
181. Rost BR, Schneider-Warme F, Schmitz D, Hegemann P. Optogenetic tools for subcellular applications in neuroscience. Neuron (2017) 96(3):572–603. doi: 10.1016/j.neuron.2017.09.047
182. Garita-Hernandez M, Guibbal L, Toualbi L, Routet F, Chaffiol A, Winckler C, et al. Optogenetic light sensors in human retinal organoids. Front Neurosci (2018) 12:789. doi: 10.3389/fnins.2018.00789
183. McGregor JE, Godat T, Dhakal KR, Parkins K, Strazzeri JM, Bateman BA, et al. Optogenetic restoration of retinal ganglion cell activity in the living primate. Nat Commun (2020) 11(1):1703. doi: 10.1038/s41467-020-15317-6
184. Cowan CS, Renner M, de Gennaro M, Gross-Scherf B, Goldblum D, Hou Y, et al. Cell types of the human retina and its organoids at single-cell resolution. Cell (2020) 182(6):1623–40. doi: 10.1016/j.cell.2020.08.013
185. Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci (2021) 24(3):331–42. doi: 10.1038/s41593-021-00802-y
186. Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene arc in hippocampal neuronal ensembles. Nat Neurosci (1999) 2:1120–4. doi: 10.1038/16046
187. Xiao MF, Roh SE, Zhou J, Chien CC, Lucey BP, Craig MT, et al. A biomarker-authenticated model of schizophrenia implicating NPTX2 loss of function. Sci Adv (2021) 7(48). doi: 10.1126/sciadv.abf6935
188. World Health Organization. Dementia (2021). Available at: https://www.who.int/news-room/fact-sheets/detail/dementia.
189. Nichols E, Steinmetz JD, Vollset SE, Fukutaki K, Chalek J, Abd-Allah F, et al. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the global burden of disease study 2019. Lancet Public Health (2022) 7(2):e105–25. doi: 10.1016/S2468-2667(21)00249-8
190. Centers for Disease Control and Prevention. Leading causes of death (2021). Available at: https://www.cdc.gov/nchs/fastats/leadingcauses-of-death.htm.
191. Xu J, Zhang Y, Qiu C, Cheng F. Global and regional economic costs of dementia: a systematic review. Lancet (2017) 390. doi: 10.1016/S0140-6736(17)33185-9
192. World Health Organization. Global status report on the public health response to dementia. WHO Geneva: World Health Organization (2021).
193. Pistollato F, Ohayon EL, Lam A, Langley GR, Novak TJ, Pamies D, et al. Alzheimer Disease research in the 21st century: past and current failures, new perspectives and funding priorities. Oncotarget (2016) 7(26):38999–9016. doi: 10.18632/oncotarget.9175
194. Drummond E, Wisniewski T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol (2017) 133(2):155–75. doi: 10.1007/s00401-016-1662-x
195. Maenner MJ, Shaw KA, Bakian AV, Bilder DA, Durkin MS, Esler A, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years – autism and developmental disabilities monitoring network, 11 sites, united states, 2018. MMWR Surveill Summ (2021) 70(11):1–16. doi: 10.15585/mmwr.ss7011a1
196. Zhong X, Harris G, Smirnova L, Zufferey V, de Cassia da Silveira E Sá R, Baldino Russo F, et al. Antidepressant paroxetine exerts developmental neurotoxicity in an iPSC-derived 3D human brain model. Front Cell Neurosci (2020) 14(25). doi: 10.3389/fncel.2020.00025
197. Pamies D, Block K, Lau P, Gribaldo L, Pardo CA, Barreras P, et al. Rotenone exerts developmental neurotoxicity in a human brain spheroid model. Toxicol Appl Pharmacol (2018) 354:101–14. doi: 10.1016/j.taap.2018.02.003
198. Modafferi S, Zhong X, Kleensang A, Murata Y, Fagiani F, Pamies D, et al. Gene– environment interactions in developmental neurotoxicity: a case study of synergy between chlorpyrifos and CHD8 knockout in human brainspheres. Environ. Health Perspect (2021) 129(7):077001. doi: 10.1289/EHP8580
199. Fóthi Á, Soorya L, Lőrincz A. The autism palette: combinations of impairments explain the heterogeneity in ASD. Front Psychiatry (2020) 11:503462. doi: 10.3389/fpsyt.2020.503462
200. Kehrer C, Groeschel S, Kustermann-Kuhn B, Bürger F, Köhler W, Kohlschütter A, et al. Language and cognition in children with metachromatic leukodystrophy: onset and natural course in a nationwide cohort. Orphanet J Rare Dis (2014) 9(1):18. doi: 10.1186/1750-1172-9-18
201. Charlson FJ, Ferrari AJ, Santomauro DF, Diminic S, Stockings E, Scott JG, et al. Global epidemiology and burden of schizophrenia: findings from the global burden of disease study 2016. Schizophr Bull (2018) 44(6):1195–203. doi: 10.1093/schbul/sby058
202. Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry (1987) 44(7):660. doi: 10.1001/archpsyc.1987.01800190080012
203. Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A, et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat Med (2018) 24(6):792–801. doi: 10.1038/s41591-018-0021-y
204. Bowie CR, Harvey PD. Cognitive deficits and functional outcome in schizophrenia. Neuropsychiatr Dis Treat (2006) 2(4):531–6. doi: 10.2147/nedt.2006.2.4.531
205. Eack SM, Bahorik AL, McKnight SAF, Hogarty SS, Greenwald DP, Newhill CE, et al. Commonalities in social and non-social cognitive impairments in adults with autism spectrum disorder and schizophrenia. Schizophr Res (2013) 148(1–3):24–8. doi: 10.1016/j.schres.2013.05.013
206. Simons Foundation Autism Research Initiative. SFARI base. Available at: https://www.sfari.org/resource/sfari-base/.
207. Beilmann M, Boonen H, Czich A, Dear G, Hewitt P, Mow T, et al. Optimizing drug discovery by investigative toxicology: current and future trends. ALTEX (2019) 36(2):289–313. doi: 10.14573/altex.1808181
208. Miura Y, Paşca SP. Polarizing brain organoids. Nat Biotechnol (2019) 37(4):377–8. doi: 10.1038/s41587-019-0084-4
209. Sloan SA, Andersen J, Paşca AM, Birey F, Paşca SP. Generation and assembly of human brain region–specific three-dimensional cultures. Nat Protoc (2018) 13(9):2062–85. doi: 10.1038/s41596-018-0032-7
210. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc (2014) 9(10):2329–40. doi: 10.1038/nprot.2014.158
211. Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J (2017) 36(10):1316–29. doi: 10.15252/embj.201694700
212. Kim H, Xu R, Padmashri R, Dunaevsky A, Liu Y, Dreyfus CF, et al. Pluripotent stem cell-derived cerebral organoids reveal human oligodendrogenesis with dorsal and ventral origins. Stem Cell Rep (2019) 14(12):5. doi: 10.1016/j.stemcr.2019.04.011
213. Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, et al. Specification of positional identity in forebrain organoids. Nat Biotechnol (2019) 37(4):436–44. doi: 10.1038/s41587-019-0085-3
214. Archie JG, Collins JS, Lebel RR. Quantitative standards for fetal and neonatal autopsy. Am J Clin Pathol (2006) 126(2):256–65. doi: 10.1309/FK9D5WBA1UEPT5BB
215. Holland D, Chang L, Ernst TM, Curran M, Buchthal S, Alicata D, et al. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol (2014) 71(10):1266–74. doi: 10.1001/jamaneurol.2014.1638
216. Samuelsen GB, Larsen KB, Bogdanovic N, Laursen H, Graem N, Larsen JF, et al. The changing number of cells in the human fetal forebrain and its subdivisions: a stereological analysis. Cereb Cortex (2003) 13:115–22. doi: 10.1093/cercor/13.2.115
217. Jakovcevski I, Filipovic R, Mo Z, Rakic S, Zecevic N. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front Neuroanat (2009) 3:5. doi: 10.3389/neuro.05.005.2009
218. Melloni L, Mudrik L, Pitts M, Koch C. Making the hard problem of consciousness easier. Science (2021) 28(372):911–2. doi: 10.1126/science.abj3259
219. Jeziorski J, Brandt R, Evans JH, Campana W, Kalichman M, Thompson E, et al. Brain organoids, consciousness, ethics and moral status. Semin Cell Dev Biol (2022) S1084–9521(22). doi: 10.1016/j.semcdb.2022.03.020
220. American Psychological Association. APA dictionary of psychology website (2023). Available at: https://dictionary.apa.org/.
221. Friston K. Functional integration and inference in the brain. Prog Neurobiol (2002) 68(2):113–43. doi: 10.1016/S0301-0082(02)00076-X
222. Sawai T, Hayashi Y, Niikawa T, Shepherd J, Thomas E, Lee TL, et al. Mapping the ethical issues of brain organoid research and application. AJOB Neurosci (2022) 13(2):81–94. doi: 10.1080/21507740.2021.1896603
223. Greely HT. Human brain surrogates research: the onrushing ethical dilemma. Am J Bioeth (2021) 21(1):34–45. doi: 10.1080/15265161.2020.1845853
224. Koplin JJ, Savulescu J. Moral limits of brain organoid research. J Law Med Ethics (2019) 47(4):760–7. doi: 10.1177/1073110519897789
225. Beauchamp TL, DeGrazia D. Principles of animal research ethics. Oxford: Oxford University Press (2020).
226. MacCoun RJ. Moral outrage and opposition to harm reduction. Criminal Law Philosophy (2013) 7(1):83–98. doi: 10.1007/s11572-012-9154-0
227. Skitka LS, Morgan GS. The social and political implications of moral conviction. Advanced Political Psychol (2014) 35(S1):95–110. doi: 10.1111/pops.12166
228. Boyd JL, Sugarman J. Toward responsible public engagement in neuroethics. AJOB Neurosci (2022) 13(2):103–6. doi: 10.1080/21507740.2022.2048736
229. Haselager DR, Boers SN, Jongsma KR, Vinkers CH, Broekman ML, Bredenoord AL. Breeding brains? patients' and laymen's perspectives on cerebral organoids. Regener Med (2020) 15(12):2351–60. doi: 10.2217/rme-2020-0108
230. Niemelä J. What puts the 'yuck' in the yuck factor? Bioethics (2011) 25(5):267–79. doi: 10.1111/j.1467-8519.2010.01802.x
231. McLennan S, Fiske A, Celi LA, M, ller R, Harder J, et al. An embedded ethics approach for AI development. Nat Mach Intell (2020) 2:488–90. doi: 10.1038/s42256-020-0214-1
232. National Academy of Sciences. The emerging field of human neural organoids, transplants, and chimeras. Washington, D.C: National Academies Press (2021).
233. Organisation for Economic Co-operation and Development. OECD recommendation on responsbile innovation in neurotechnology (2019). Available at: https://www.oecd.org/science/recommendation-on-responsible-innovation-in-neurotechnology.htm.
234. Ngai J. BRAIN 2.0: Transforming neuroscience. Cell (2022) 185(1):4–8. doi: 10.1016/j.cell.2021.11.037
235. National Institutes of Health BRAIN Initiative. The brainstorm project: A collaborative approach to facilitating the neuroethics of bioengineered brain modeling research (2018). Available at: https://braininitiative.nih.gov/funded-awards/brainstorm-project-collaborative-approach-facilitating-neuroethics-bioengineered-brain.
236. Oxford English Dictionary. Oxford University Press (2020). Available at: https://www.oed.com.
237. Cangelosi A, Bongard J, Fischer MH, Nolfi S. Embodied intelligence. In: Kacprzyk J, Pedrycz W, editors. Springer handbook of computational intelligence. Berlin, Heidelberg: Springer (2015). 697–714.
238. Dictionary by Merriam-Webster. Available at: https://www.merriam-webster.com.
Keywords: organoid intelligence, biocomputing, microphysiological systems, electrophysiology, cognition, artificial intelligence
Citation: Smirnova L, Caffo BS, Gracias DH, Huang Q, Morales Pantoja IE, Tang B, Zack DJ, Berlinicke CA, Boyd JL, Harris TD, Johnson EC, Kagan BJ, Kahn J, Muotri AR, Paulhamus BL, Schwamborn JC, Plotkin J, Szalay AS, Vogelstein JT, Worley PF and Hartung T. Organoid intelligence (OI): the new frontier in biocomputing and intelligence-in-a-dish. Front Sci (2023) 1:1017235. doi: 10.3389/fsci.2023.1017235
Received: 11 August 2022; Accepted: 07 February 2023;
Published: 28 February 2023.
Edited by:
Arti Ahluwalia, University of Pisa, ItalyReviewed by:
Karl Friston, University College London, United KingdomGary Miller, Columbia University, United States
Copyright © 2023 Smirnova, Caffo, Gracias, Huang, Morales Pantoja, Tang, Zack, Berlinicke, Boyd, Harris, Johnson, Kagan, Kahn, Muotri, Paulhamus, Schwamborn, Plotkin, Szalay, Vogelstein, Worley and Hartung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Hartung, thartun1@jhu.edu
 Lena Smirnova
Lena Smirnova Brian S. Caffo
Brian S. Caffo David H. Gracias
David H. Gracias Qi Huang
Qi Huang Itzy E. Morales Pantoja
Itzy E. Morales Pantoja Bohao Tang2
Bohao Tang2 Donald J. Zack
Donald J. Zack Cynthia A. Berlinicke
Cynthia A. Berlinicke J. Lomax Boyd
J. Lomax Boyd Timothy D. Harris
Timothy D. Harris Brett J. Kagan
Brett J. Kagan Alysson R. Muotri
Alysson R. Muotri Barton L. Paulhamus
Barton L. Paulhamus Jens C. Schwamborn
Jens C. Schwamborn Jesse Plotkin
Jesse Plotkin Alexander S. Szalay
Alexander S. Szalay Joshua T. Vogelstein
Joshua T. Vogelstein Paul F. Worley
Paul F. Worley Thomas Hartung
Thomas Hartung