1
Computer Science, Yale University, New Haven, CT, USA
2
Unité de Neurosciences Intégratives et Computationelles, CNRS, Gif sur Yvette, France
3
Laboratory for Computational Neuroscience, Ecole Polytechnique Fédérale de Lausanne, Switzerland
The NEURON simulation program now allows Python to be used, alone or in combination with NEURON's traditional Hoc interpreter. Adding Python to NEURON has the immediate benefit of making available a very extensive suite of analysis tools written for engineering and science. It also catalyzes NEURON software development by offering users a modern programming tool that is recognized for its flexibility and power to create and maintain complex programs. At the same time, nothing is lost because all existing models written in Hoc, including graphical user interface tools, continue to work without change and are also available within the Python context. An example of the benefits of Python availability is the use of the
xml module in implementing NEURON's Import3D and CellBuild tools to read MorphML and NeuroML model specifications.
The NEURON simulation environment has become widely used in the field of computational neuroscience, with more than 700 papers reporting work employing it as of April, 2008. In large part this is because of its flexibility and the fact that it is continually being extended to meet the evolving research needs of its user community. Experience shows that most of these needs have a software solution that has already been implemented elsewhere in the domain of scientific computing. The problem is one of interfacing an existing package with NEURON’s interpreter. Some cases demand intimate knowledge of NEURON’s internals and considerable effort; examples include network parallelization with MPI, and adoption of Sundials for adaptive integration. There are many more cases in which existing packages could potentially be employed by NEURON users. Few people, however, have the specialized expertise required to manually interface an existing software package and the creation of such interfaces is tedious. Instead of laborious piecemeal adoption of individual packages that requires intervention by a handful of experts, a better approach is to offer Python as an alternative interpreter so that a huge number of resources becomes available at the cost of only minimal interface code that most users can write for themselves.
Since 1984, the NEURON simulation environment has used the Hoc interpreter (Kernighan and Pike, 1984
) for setup and control of neural simulations. Hoc has a syntax for expressions and control flow vaguely similar to the C language. Hoc is not exactly an interpreted language since, analogous to Pascal, Java, or Python, Hoc statements are first dynamically compiled to an internal stack machine representation using a
yacc parser and then the stack machine statements are executed. A fundamental extension to Hoc syntax was made in the late 80’s in order to represent the notion of continuous cables, called sections. Sections are connected to form a tree shaped structure and their principle purpose is to allow the user to specify the physical properties of a neuron without regard for the purely numerical issue of how many compartments are used to represent each of the cable sections. In the early 90’s, Hoc syntax was again extended to provide some limited support for classes and objects, that is, data encapsulation and polymorphism, but not inheritance.Though Hoc has served well, continuing development and maintenance of a general programming language steals significant time and effort from neurobiology domain-specific improvements. Furthermore, Hoc has turned out to be an orphan language limited to NEURON users. What is desirable is a modern programming language such as Python, which provides expressive syntax, powerful debugging capabilities, and support for modularity, facilitating the construction and maintenance of complex programs. Python has proved its utility by giving rise to a large and diverse community of software developers who are making reusable tools that are easy to plug-in to the user’s code, the so-called “batteries included” (Dubois, 2007
). In the domain of scientific computing, some examples include Numpy (Oliphant, 2007
) and Scipy (Jones et al., 2001
) for core scientific functionality, Matplotlib (Hunter, 2007
) for 2-D plotting, and IPython (Prez and Granger, 2007
) for a convenient interactive environment.
There are three distinct ways to use NEURON with Python. One is to run the NEURON program with Python as the interpreter accepting interactive commands in the terminal window. Another is to run NEURON with Hoc as the interactive interpreter and access Python functionality through Hoc objects and function calls. These first two cases we will refer to as embedded Python. The third way is to dynamically import NEURON in a running Python or IPython instance, which we will refer to as using NEURON as an extension module for Python.
In the sections to follow, we describe the steps required to use NEURON with Python, from a user’s point of view, and the techniques employed to enable NEURON and Python to work together, from a developer’s point of view. We begin in Section “Getting Started Using NEURON with Python” by describing how to install and run NEURON with Python. We then demonstrate how modelling is carried out using Python by comparing it side-by-side with Hoc syntax in Section “Writing NEURON Models in Python”. In Section “Using Python Code from Hoc”, we describe how Python can be accessed from the Hoc interpreter. In Section “Technical Aspects”, we discuss some technical aspects of the implementation of the Python-NEURON interaction. Finally, in Section “Importing MorphML Files — A Practical Example” we give a detailed, practical example, from the current NEURON distribution, of combining Python and Hoc.
The code listings in Figures 1
–3
are available for public download from the ModelDB model repository of the Senselab database,
http://senselab.med.yale.edu
(accession number 116491).Figure 1. Code listing for a simple model neuron: building the neuron. The Python code is on the left and the equivalent Hoc code on the right.
Figure 2. Code listing for a simple model neuron (continued from Figure 1
): instrumenting and running the model. The Python code is on the left and the equivalent Hoc code on the right.
Figure 3. Code listing demonstrating the use of ref and plotting.
Installation
NEURON works with Python on Windows, Mac OS X, Linux, andmany other platforms such as the IBM Blue Gene/L/P and Cray XT3 supercomputers. Detailed installation information can be found at
http://www.neuron. yale.edu
by following the “Download and Install” link.Binary installers are available for Windows, OS X and RPM-based Linux systems. The Windows installer contains a large portion of Cygwin Python 2.5. On OS X and Linux, the latest version of Python 2.3–2.5 previously or subsequently installed is dynamically loaded when NEURON is launched. The binary installers provide Python embedded in NEURON, but do not support using NEURON as an extension module for Python or IPython.
If you would like to use NEURON as an extension module for Python or IPython, if no installer for your platform exists, or if you need to customize the installation (e.g. enable parallel/MPI support, or change the location of binaries), you should instead get the source code for the standard distribution, also available from the above “Download and Install” link, and compile it for your machine. Further instructions for this are given in the Appendix.
Basic Use
NEURON may be started without the graphical user interface (GUI) using
nrniv or with the GUI using nrngui. To use Python as the interpreter, rather than Hoc, use the -python option:
If there are any NEURON NMODL extension mechanisms (Hines and Carnevale, 2000
) in the working directory, and they have been compiled with
nrnivmodl, they will be loaded automatically.Alternatively, you may wish to use NEURON as an extension to the normal Python interpreter, or to IPython (Prez and Granger, 2007
), a more interactive variant. To do so, you must build NEURON from source and install the NEURON shared library for Python, as described in the Appendix. In Python (or IPython) then, NEURON is started (and any NMODL mechanisms loaded) when you
import neuron:
and the NEURON GUI is started by importing the
neuron.gui module: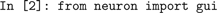
The
h object that we import from the neuron module is the principal interface to NEURON’s functionality. h is a HocObject instance, and has two main functions. First, it gives access to the top-level of the Hoc interpreter, e.g.:
Second, it makes any of the classes defined in Hoc available to Python:
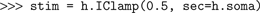
Note that the
soma section created through the Hoc interpreter appears in Python as a Section object. We can also create Sections directly in Python, e.g.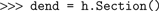
These two section objects are entirely equivalent, the only difference being that the name “dend” is not accessible within the Hoc interpreter. In addition to the
HocObject class (and through it, any class defined in Hoc) and the Section class, the Python neuron module also provides the Segment, Mechanism and RangeVariable classes. More in-depth examples of using NEURON from Python are given in Section “Writing NEURON Models in Python”, while using Python code from Hoc is introduced in Section “Using Python Code from Hoc”.Starting Parallel Neuron
Assuming NEURON was built with parallel support as discussed in the Appendix, suitably parallelized Hoc scripts are started using the MPI job execution command, typically
mpiexec (Hines and Carnevale, 2008
) or the equivalent for your MPI implementation. When Python is used rather than Hoc, the same parallelism features are supported, with only slight changes in the execution model. Both embedded Python (nrniv -python) and NEURON as an extension module to Python are supported. MPI job execution for embedded Python is the same as standard NEURON/Hoc, except that an extra -python command line option must be passed to nrniv: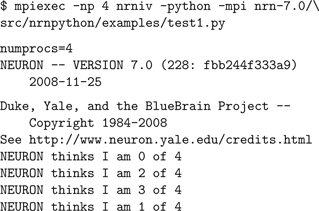
For users who prefer to use NEURON as an extension module to Python or IPython, execution is as follows:
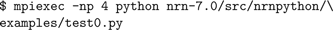

However, there is one important caveat: The NEURON extension module does not initialize MPI itself, but rather delegates this job to Python. To initialize MPI in Python, one must import a Python MPI module, such as “MPI for Python” (
mpi4py) (Dalcín et al., 2008
), prior to importing neuron:
The module
mpi4py is available from the Python Package Index (http://pypi.python.org
).Online Help
For new users of NEURON with Python, a convenient starting place for
help is Python online help, provided through the global function help, which takes one argument, the object on which you would like help:
For commonly used Hoc classes, such as
Vector, APCount, NetCon, etc., helpful reminders of constructor arguments, attributes and units with Python syntax examples are available at the Python prompt:
In IPython, the ? symbol is a quick shorthand roughly equivalent to online help:

To show how a model neuron is implemented using Python, we repeat the example described in Chapter 6 of the NEURON Book (Carnevale and Hines, 2006
), but using Python rather than Hoc. The code listing is given in Figures 1 and 2
, and has Python code on the left and the equivalent Hoc code on the right.
There are only a few syntax and conceptual differences between the Python and Hoc versions, and we expect that Hoc users will have little difficulty transitioning to Python, should they wish to do so (Hoc will continue to be supported, of course). We now comment on the most significant differences.
First are the
import statements, absent from the Hoc listing, although Hoc does have the xopen() function that has similar functionality. Since NEURON is now only one of potentially many modules living within the Python interpreter, it must live in its own namespace, so that the names of NEURON-specific classes and variables do not interfere with those from other modules. Of particular importance is the object h, which is the top-level Hoc interpreter, and gives access to Hoc classes, functions and variables.While sections are created using the
create keyword in Hoc, in Python we instantiate a Section object. Hence the important distinction in Hoc between sections and objects is removed: Everything in Python is an object. Similarly, the connect keyword in Hoc is replaced by a method call of the child section object in Python.In NEURON, each cable section is made up one or more segments, and the diameter is a property of each segment. Hoc’s shorthand, allowing the
diam attribute to be set on all segments by setting it on the section is also available in Python. Inhomogeneous values for range variables such as diam can also be set on the specific Segment object, returned by calling the Section object as a function.The
forall keyword in Hoc, which iterates over all sections, is replaced by the allsec() method of the top-level Hoc interpreter object h. Here again we see, in setting the membrane capacitance cm, the Hoc and Python shorthands to set the value for all segments at once, without having to explicitly iterate over all Segments.In instrumenting the model, we see that Python and Hoc objects have very similar behaviours. In general, all Hoc classes (
Vector, List, NetCon, etc) are accessible within Python via the h object. Hoc object references must be declared using the objref keyword, and objects created using new, but once created, attribute access and method calls have near-identical syntax in Python and Hoc. There are three major exceptions to this rule. First, many functions and methods act in the context of the ‘currently-accessed section’. To support this in Python, these functions take a keyword argument sec. Second, certain method calls take Hoc expressions as arguments, so, for example, in adding the membrane potential of the soma section to the list of variables to plot, in Hoc we use g.addvar(“soma.v(0.5)”), but in the Python version the variable soma does not exist on the Hoc side, and so we have to pass the soma Section object as the sec keyword argument so that the Hoc expression is evaluated in the context of that section. Third, a number of functions/methods take Hoc variable references (indicated by preceding the variable name with the ‘&’ character) as arguments, the most important being Vector.record(&var) and NetCon(&var, target). The equivalent syntax in Python is to precede the variable name with _ref_, e.g.: Vector.record(_ref_var). For example, given ‘pre’ and ‘post’ Section objects and a dictionary of Hoc Vector objects addressed by a mnemonic string, recording the voltage at the centres of those sections is activated by the statements:
Figure 3
shows the complete listing with the above fragment in context and also illustrates the ease with which NEURON code can be mixed with third-party code such as the powerful Pylab/Matplotlib plotting package (
http://matplotlib.sourceforge.net/
): NEURON Vector objects work just as well as Python lists or arrays as arguments to the plot() function.Using User-Defined Mechanisms
One of NEURON’s most powerful features is the ability to write new mechanisms using the NMODL language, and then compile these mechanisms into the executable or into dynamic libraries (DLLs). The standard behaviour of NEURON is to load any mechanisms that have been compiled in the working directory. It is also possible to load DLLs from elsewhere in the filesystem using the Hoc function
nrn_load_dll(). This has the disadvantage that the full path to the shared library file must be provided, which can be hard to determine, since the file is within a hidden folder which itself is within a folder with a platform-specific name. To simplify this, the neuron Python module adds a function load_mechanisms(), which takes as an argument the path to the directory containing the NMODL source files, and searches for shared library files below this directory. Furthermore, in analogy to the PYTHONPATH environment variable which contains a list of paths to search for importable Python modules, if you have defined a NRN_NMODL_PATH environment variable, NEURON will search these paths for shared libraries and load them at import time.Using User-Defined Classes
One of the principal advantages of writing NEURON programs in Python rather than Hoc, especially for large, complex programs, is that Python is a fully object-oriented language, supporting encapsulation, polymorphism and inheritance, whereas Hoc supports only encapsulation and a limited form of polymorphism.
Just as with built-in Hoc classes, access to attributes and methods of user-defined Hoc classes (using the
begintemplate/endtemplate keywords) uses the same syntax in Python as in Hoc. For example, if we have the following user-defined Hoc class in the file string.hoc:
then we can use it as follows:

It is also possible to subclass both built-in and user-defined Hoc classes in Python, although with the restriction that multiple inheritance from Hoc-derived classes is not possible. Subclassing requires the use of the
hclass class factory:
Numerical Data Transfer Between Hoc and Python
The Hoc
Vector object provides NEURON with a convenient and efficient container for storing and manipulating collections of numerical values, such as membrane potential traces or spike-times.In Python, Hoc
Vector objects expose iterator and indexing methods, such that they can be used in most cases where Numpy (Oliphant, 2007
), Scipy (Jones et al., 2001
), and Matplotlib (Hunter, 2007
), the most important scientific modules, accept lists.To benefit from the elegant and expressive notation of Numpy for N-dimensional array manipulation, and from results computed using the large and growing repertoire of scientific packages available for Python, which largely return Numpy arrays, several optimized methods are available for the conversion of Hoc
Vectors to and from Numpy arrays.Transferring one-dimensional Numpy arrays and non-nested lists with float or integer items to Hoc
Vectors is straightforward, as the Hoc Vector constructor accepts an array or list as an argument: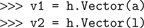
Transferring a Hoc
Vector to an array or list is equally straight forward:
If you would like to transfer between an existing Numpy array and a Hoc Vector, there are the Hoc
Vector “in-place” member functions to_python and from_python:
For interacting with Python, Hoc provides the
nrnpython() function and the PythonObject class. nrnpython() takes as its one argument a string that can be any Python statement, e.g.: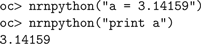
PythonObject has two main uses. Creating an instance using new returns an object that encapsulates the top-level Python interpreter, e.g.
Strings and float/double values move back and forth between Python and Hoc (although Python integers become double values in Hoc and remain doubles if they are passed back to Python). All other Python objects become instances of the
PythonObject class:
For objects (such as lists and tuples) that take integer indices or are callable as functions, there is a special method named ‘_’ (underscore):

The only other trap for the unwary is that both single and double quotes are valid for string definitions in Python, but only double quotes are accepted by Hoc!
A detailed example of using Python from Hoc, and of the value of being able to access its large standard library, is given in Section “Importing MorphML Files — A Practical Example” for the case of importing 3D morphology from a MorphML file.
Tools for building Python extensions, such as BOOST.Python (Abrahams and Grosse-Kunstleve, 2003
) or SWIG (Beazley, 1996
) might have been useful in more expert hands. However, the ability of users to declare variables, objects, and classes in Hoc, the fact that many existing C++ classes and class methods were not generally meant to be directly visible to the user except through the intermediation of Hoc syntax, and the fact that the Hoc connection to the internal NEURON code was already reasonably uniform, of reasonable size, and understood by us in depth, suggested to us that a Python interface written using the Python C-API (
http://docs.python.org/c-api/
) that reused as much as possible the existing Hoc connection to internal data and functions would give us the general control we needed, and allow us to accomplish the project in reasonable time. It should be emphasized that this design decision to reuse a few of the C functions that manipulate the Hoc runtime stack neither hinders nor assists any future work on development of APIs for major NEURON components, such as the numerical solvers, which may be useful to other simulators. However, our interface implementation does provide a compact example of how an application can communicate with NEURON within a shared address space and therefore makes the the process of dynamically linking NEURON into a user application much simpler.Since double precision variables, arrays, constant strings, functions, and objects have very similar syntax and semantics in Hoc and Python, a single
PyTypeObject structure called HocObjectType associated with a PyHocObject structure for a Python object instance containing Hoc Symbol and Object fields was sufficient to allow Python access to all these Hoc data-types. When a name is given to an attribute method of the HocObjectType (the reflexive self PyHocObject is also an argument to the method), the name is looked up in Hoc’s symbol table for the PyHocObject Hoc Object field, and the symbol along with the Hoc object calls the same function that the Hoc interpreter would call to resolve the attribute at runtime. The attribute, which is typically a number, string, or HocObject, is then wrapped in a Python object of the proper type and returned. Function calls from Python into Hoc consist of pushing the function arguments onto the Hoc runtime stack and, again, calling the same function the Hoc interpreter would call at runtime. Thus, Python statements involving PyHocObject objects end up generating and executing the same Hoc stack machine code at runtime that would be accomplished by the corresponding Hoc statement. It should be noted that a great deal of interpreter efficiency can be gained in loop body statements by factoring out as much as possible the precursor objects. For example:
can be optimized by avoiding the
x: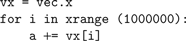
The former takes 1.3 s on a 3 GHz machine, while the latter takes 1.0 s.
A critical requirement was to have as natural a correspondence as possible in Python for the special Hoc syntax for Sections, position along a Section, membrane mechanisms, and Range Variables. This was achieved through the C++ definition of corresponding types in Python to create instances for:
NPySecObj, NPySegObj, NPyMechObj, and NPyRangeVar. For example, the NPySegObj segment (compartment) object points to the NPySecObj of which it is a part, specifies its location, x, and also contains a field to help in iterating over the mechanisms that exist at that location. An NPyRangeVar has, in practice, required only a pointer to the compartment (NPySegObj) where it exists and a pointer to its Hoc Symbol. A Section represents a continuous cable and evaluation of or assignment to a variable associated with a particular location always involves specifying both which Section and the relative arc length location (0 ≤ x ≤1) along the Section. Internally, NEURON employs a Section stack to determine the working Section and Hoc syntax provided three ways to specify the top of the Section stack. The Hoc Section.variable(x) syntax has a direct correspondence to the Python Section(x).variable syntax and the latter perhaps has more clarity. The Hoc Section { Hoc statements } syntax is unique to NEURON and for the Python side we were reduced to explicit management of the Section stack with Section.push() with an explicit h.pop_section() as the final statement. This gets tedious for single function calls and so in Python we allow the keyword argument, sec=Section, to push and pop the Section during the scope of the Hoc function call. The Hoc access Section statement does not require a Python counterpart. However, the Python statement, sec = h.cas(), returns the top of the Section stack.There were several cases of syntax mismatch which could only be overcome by the addition of new idioms. Hoc syntax does not allow an object to be treated as a function, so in Hoc we use
po._( …). Python does not allow call by reference arguments. Therefore, when a Hoc function called from Python requires a reference argument, the variable name must be prefixed by ‘_ref_’. Of course, such variables can only be Hoc variables but that is not a difficulty in practice since either the need is to pass a Hoc RangeVariable or the Python program can construct a Hoc variable for use in these cases. Since all numbers in Hoc are double precision, type errors are raised when Python expects an integer. For the case of array arguments, the Hoc-to-Python interface converts the doubles to integers automatically. Unfortunately, one cannot in general call the __getitem__(int) method explicitly but must use the [expr] Hoc syntax. If this becomes a problem in practice, it will be necessary to supply a set of cast functions that can be explicitly invoked by the user.We have encountered only one problem with freeing object memory that has proved resistant to a solution. In some cases there is an ambiguity in regard to whether the Hoc or the Python side owns a reference to an object. When this situation occurs, a reference to the object is kept in a list for a deferred call to
Py_DECREF when it is guaranteed that it is safe to do so.Assignment of a constant value to a range variable in a Section is far more common than assignment of different values within the segments of a Section and Hoc provides a simple syntax for that case which avoids writing an explicit loop. The latest extension of the NEURON Python interface mimics that behavior in Python by interpreting Section.RangeVariableName in that fashion instead of raising an “AttributeError”. We are also considering extending the implicit iteration idea to SectionLists and Cells to allow not only assignment of constants but also application of inhomogeneous functions.
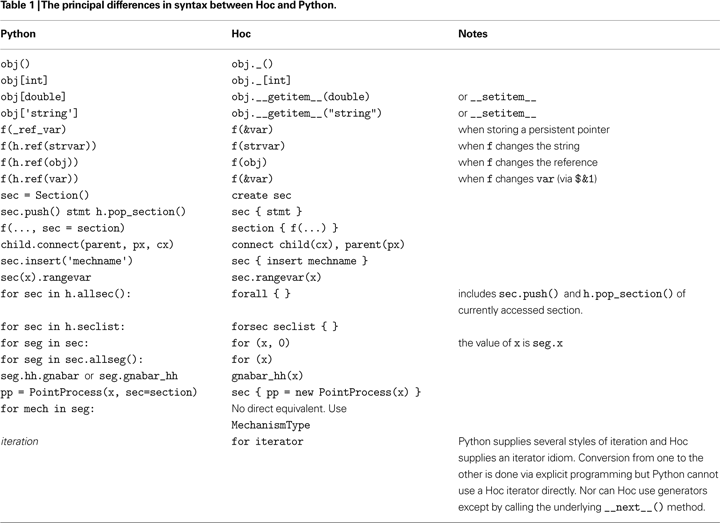
A list of the principal differences in syntax between Hoc and Python is given in Table 1
.
Our first serious use of the NEURON Python interface was to extend the Import3D GUI tool to read MorphML specification files. Import3D is structured around a graphical view of a list of
Import3d_Section objects defined in Hoc. Among many method and field attributes, the principle data field of the Import3d_Section object is the raw x, y, z, diam information along an unbranched cable and a list index indicating the parent Import3d_Section. The list of Import3d_Section objects is constructed by various file reader objects that understand a specific file format such as Eutectic, SWC, or NeuroLucida versions 1 or 3. Since MorphML is an XML format, it was opportune to employ the XML reader module in the standard Python distribution.The problem of parsing and analyzing the MorphML format is similar in difficulty to that for NeuroLucida V3 files. We divided the problem into Hoc and Python code portions. In contrast to a file size of 1180 lines for the NeuroLucida V3 file reader, the
read_morphml.hoc file size is 78 lines and the Python portion of the problem is carried out by rdxml.py with a file size of 370 lines. Since these files are located in the NEURON package default search path – …/nrn/lib/hoc for the read_morphml.hoc file and …/nrn/lib/python for the rdxml.py file – the MorphML reader extension works wherever the NEURON Python interface is installed.The
read_morphml.hoc file defines an Import3d_MorphML Hoc template (class) which interacts with Import3d_GUI in exactly the same manner as the other format readers.When an
Import3d_MorphML instance is created, the Python helper module we wrote to parse the input file is imported with nrnpython(“import rdxml”) and p = new PythonObject() is defined in order to allow access to Python functions.The
proc input() {…} procedure defines a sections list and populates it with Import3dSection objects indirectly via p.rdxml.rdxml($s1, this) which passes the filename selected earlier by the user along with a reference to the Import3dMorphML instance to allow callback from the Python code.The
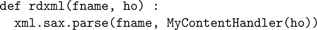
module function calls the xml parser with the filename and a new instance of

The reference to the
Import3d_MorphML instance is stored by the initializer for later use at the end of parsing. During file reading there is no interaction between Hoc and Python, so let it suffice that the xml parsing style is, at the beginning and end of every xml element, to call the MyContentHandler methods
where the
elements literal map associates all possible element names with a MyContentHandler method. E.g.

The methods construct Python lists of
Point, Cable, etc, as well as maps associating identifiers with list indices. At the end of parsing, the MyContentHandler method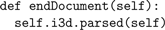
is called by the xml parser.
At this point we find ourselves back in the Hoc world with an argument that references the
MyContentHandler. Through that we can obtain the information saved by the MyContentHandler in various maps and lists and copy it into new Import3d_Section instances.

Note the ‘._’ idiom for accessing a Python list element since, in Hoc,
cables[i] is syntax implying an object reference array created with objref cables[n]. Also, cableid2index is a Python map which associates the cable identifier read from the xml input file, with the cables list.Python makes available within NEURON a very extensive suite of analysis tools written for the general science and engineering communities. All existing models written in Hoc, including GUI tools, continue to work without change. All NEURON objects are accessible to Python via an instance of the HocObject. Within the Hoc interpreter, all Python objects are accessible via the PythonObject. Binary installation remains straightforward for the usage case of launching NEURON with Python embedded: The MS Windows installer contains a large subset of the 2.5 version of Python, and the Linux RPM and Mac OS X dmg installations will use the latest version of Python, if any, that is already present or subsequently installed. The usage case of launching Python, e.g. using IPython, and dynamically importing NEURON also works but presently requires the extra installation steps described in the Appendix. Numpy is not a prerequisite but, if present, copying of vectors between Numpy and NEURON is very efficient. The Python
xml module is used in the present standard distribution to extend NEURON’s Import3D and CellBuild tools to allow reading of MorphML (Crook et al., 2007
) and NeuroML (Goddard et al., 2001
) model specifications. The Hoc portion of the xml readers makes heavy use of Python maps and lists.With the release of NEURON version 7.0, the Python interface has largely stabilized, and is ready for general use. We recommend that new users of NEURON and those already familiar with Python should use Python rather than Hoc to develop new models. Those with considerable expertise in Hoc but without Python knowledge are likely to be more productive by continuing to develop models with Hoc, but accessing Python’s powerful data structures, large standard library and external numerical/plotting packages through
nrnpython() and the PythonObject class. There is no need to rewrite legacy code in Python, as it will continue to work using the Hoc interpreter or mixed in with new Python code and accessed via the h object.Users are encouraged to submit bug reports and feature requests at the NEURON forum (
http://www.neuron.yale.edu/phpBB
) in the “NEURON+Python” sub-section, so that we can continue to improve the Python interface in response to users’ experiences.Here we give detailed instructions for building and installing NEURON as a Python extension. Note that, as mentioned earlier, to use NEURON with Python embedded you can use one of the binary installers.
The following assumes a standard GNU build environment, and a bash shell. You will need both NEURON (
nrn-VERSION.tar.gz) and InterViews (iv-VERSION.tar.gz) sources, available through the “Download and Install” link at http://www.neuron.yale.edu.
First, build and install Interviews:
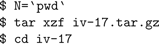
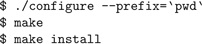
Then build and install NEURON:

Here, the “\” at the end of the fourth line, indicates it is continued on the fifth. If you want to run parallel NEURON (Hines et al., 2008
; Migliore et al., 2006
), add
--with-paranrn to the configure options. This requires a version of MPI to be installed, for example MPICH2 (Gropp, 2002
) or openMPI (Gabriel et al., 2004
).Now add the NEURON
bin directory to your PATH: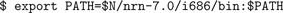
(Here
i686 will be different for different CPU architectures).Now build and install the NEURON shared library for Python:
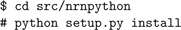
This command installs the neuron package to the Python site-packages directory, which usually requires root access. If you don’t have root access, you can install it locally using
--prefix to specify a location under your home directory: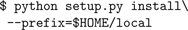
This will install the neuron package to
$HOME/local/lib/python/site-packages under your home directory. You will then have to add this directory to the PYTHONPATH environment variable: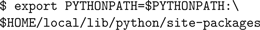
The authors declare that the research presented in this paper was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by NIH grant NS11613, by the European Union under the Bio-inspired Intelligent Information Systems program, project reference IST-2004-15879 (FACETS), and by a grant from the Swiss National Science Foundation.
Abrahams, D., and Grosse-Kunstleve, R. W. (2003). Building hybrid systems with Boost.Python. C/C++ Users J. 21. http://www.ddj.com/cpp/184401666.
Gabriel, E., Fagg, G. E., Bosilca, G., Angskun, T., Dongarra, J. J., Squyres, J M., Sahay, V., Kambadur, P., Barrett, B., Lumsdaine, A., Castain, R. H., Daniel, D. J., Graham, R. L., and Woodall T. S. (2004). Open MPI: goals, concept, and design of a next generation MPI implementation. In Proceedings, 11th European PVM/MPI Users’ Group Meeting, D. Kranzlmüller, P. Kacsuk and J. Dongara, eds (Budapest, Springer), pp. 97–104.
Jones, E., Oliphant, T., Peterson, P., et al. (2001). SciPy: open source scientific tools for Python. URL
http://www.scipy.org/.