Introduction: Medical implants are often used with high success rates. However, some implants fail due to e.g. lack of integration or infection. Resolving the biological events at the implant-tissue interface is essential for proper understanding of the mechanisms behind the success or failure. Laser microdissection (LMD) provides possibilities to harvest specific tissue sites or cells around the implant for subsequent downstream analysis. In the present study, we have analysed the gene expression of cells at increasing distances from titanium (Ti) implants – with or without the presence of Staphylococcus epidermidis.
Materials and Methods: Ti disks ± 106 CFU dose of S. epidermidis (ATCC 35984) were implanted subcutaneously in rats. After 3 days, implants were removed and the surrounding tissue was embedded in paraffin. Sections were stained and subjected to LMD (PALM Microbeam microscope, Carl Zeiss MicroImaging). Three tissue zones were dissected at increasing distance from the implant (Zone 1 < Zone 2 < Zone 3) and processed for subsequent qPCR analysis (Fig. 1). Relative gene expression was analysed for TNF-α, IL-6, IL-8, MCP-1, IL-8R, TLR2 and MMP9, using 18S as reference gene. Five full paraffin sections served as an overall tissue reference. The Mann-Whitney U Test was used for statistical analysis (n=4-8). The study was approved by the local ethical committee for laboratory animals (Dnr 254/11).
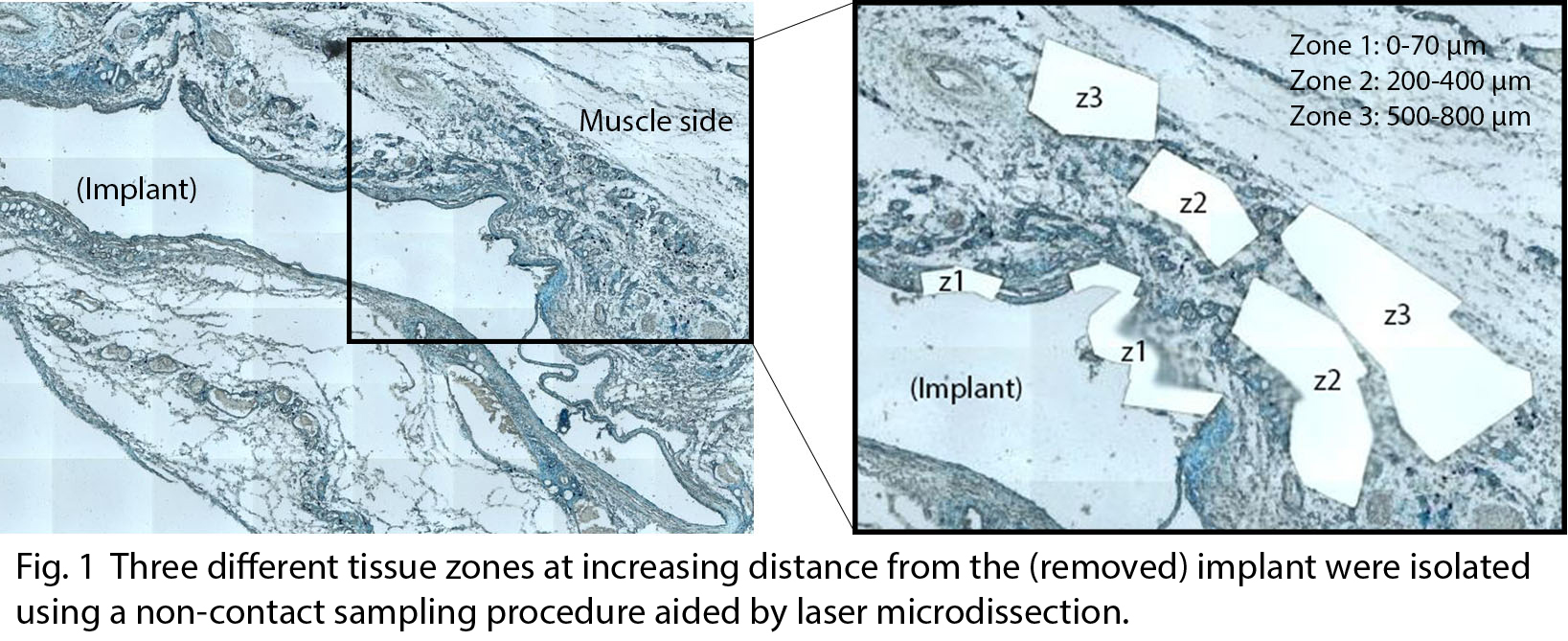
Results and Discussion: Previous results from this soft tissue infection model have shown large differences in the recruited cell number, cell type and gene expression in the implant-surrounding exudate, depending on bacterial presence[1]. The present results on the interface tissue extend the previous findings confirming higher expression levels of inflammatory cytokines and chemokines at infected sites compared with control (Fig. 2). Moreover, a spatial difference in the molecular activity was demonstrated. The gene expression was generally more pronounced at the implant-adjacent interface zone compared with zones at further distance from the implant. Furthermore, high expression of IL-6 and IL-8 was also detected in the distant zones in the infected sites, suggesting a possible chemotaxis gradient.
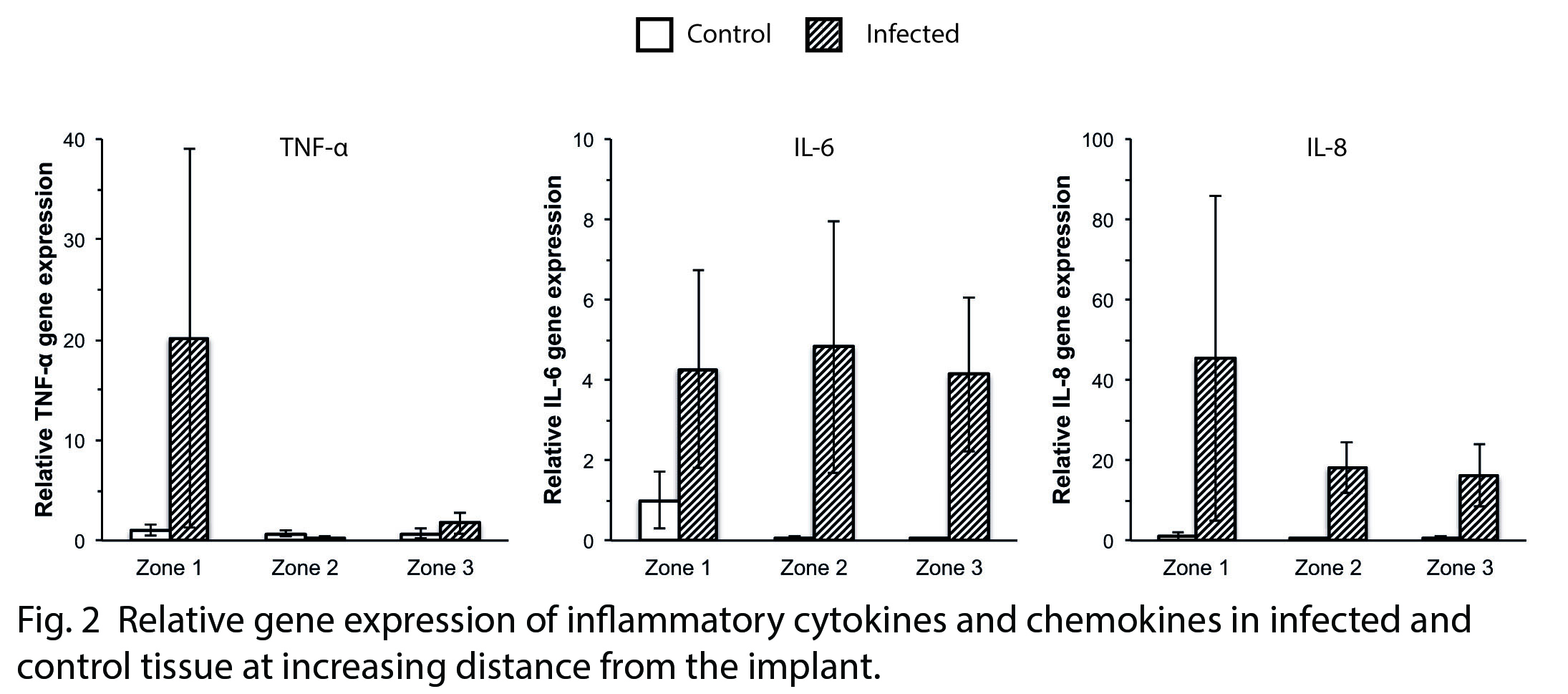
Analysis of the overall “black box” tissue response consistently showed higher gene expression in the infected sites, without providing any details of the spatial distribution of the cellular activity triggered by the presence of bacteria.
Conclusion: The results show spatial differences in host cell gene expression depending on the distance from an implant surface. Furthermore, the study demonstrates the use of LMD as a powerful tool for molecular dissection of the implant-tissue interface during normal healing or in association with infection.
BIOMATCELL Vinn Excellence Center of Biomaterials and Cell Therapy; Swedish Research Council (grant (K2015-52X-09495-28-4); Hjalmar Svensson Research Foundation; Birgitta Norlindh
References:
[1] Svensson S et al. Biomaterials 2015; 41:106-121