- 1Food and Feed Safety Research Unit, Agricultural Research Service, United States Department of Agriculture, College Station, TX, United States
- 2Diamond V Mills, Inc., Cedar Rapids, IA, United States
Salmonella is a significant food safety concern in commercial beef production, and some contamination is thought to occur by inclusion of Salmonella-infected peripheral lymph nodes (LN) in ground beef and through fecal contamination. Surveillance in processing plants assists packers in risk management of Salmonella by understanding seasonal trends and risks associated with different cattle types. Approximately 25 fecal samples and 20 LN were collected from animals representing each of five cattle types (cull beef cattle, cull dairy cows, conventional feedlot cattle, all-natural feedlot cattle raised without pharmaceuticals, and grass-finished cattle) and each of five climate regions (mixed-temperatures and dry, mixed-temperatures and humid, hot and humid, hot and dry, cold) during each of three seasons (summer, fall, winter) to better characterize Salmonella inputs into a commercial cattle processing facility. In total, 1,840 fecal samples and 1,550 LN samples were collected. Fecal samples and LN were cultured for Salmonella, and select isolates were serogrouped and screened for antimicrobial resistance. Conventional feedlot cattle had the highest LN Salmonella concentrations (1.17 log10 CFU/g LN) in this data set, while cull dairy cows had the highest fecal Salmonella concentrations (1.96 log10 CFU/g feces). Conventional feedlot cattle and cull dairy cows had the greatest Salmonella prevalence in both LN (32 and 18%, respectively) and feces (37 and 49%, respectively), while all-natural feedlot cattle had the lowest prevalence in the LN (3%) and feces (7%). As expected, Salmonella prevalence and concentration was lowest for all cattle types during winter compared to warmer seasons. When examined by climate region, a greater Salmonella prevalence in both feces and LN was observed in climate region 4 (hot-dry), than the other regions. Only 21 of 50 Salmonella isolates examined for antimicrobial susceptibility were identified as multidrug resistant (MDR); cull dairy cows were responsible for 48% of MDR isolates, cull beef cattle were responsible for 38%, and conventional feedlot, grass-fed, and all-natural feedlot cattle were each responsible for 4.8%. These results indicate that different production schemes, season, and climate region may influence which cattle are most likely to introduce Salmonella to the abattoir, allowing for greater risk awareness during the slaughter process.
Introduction
Despite decades of advancements in food safety, Salmonella is still occasionally found in retail beef. Peripheral lymph nodes (LN) harboring Salmonella are thought to be a primary source of ground beef contamination, as LN are sheltered within adipose or muscle tissue from post-harvest carcass sanitation procedures. Research has shown differences in prevalence of Salmonella-positive LN depending on season, region, and cattle source (Webb et al., 2017; Nickelson et al., 2019), with prevalence often increased in feedlot cattle compared to cull breeding animals, animals raised in southern latitudes compared to northern regions, and in warmer months compared to colder months. More recently, anecdotal observations from within the meat packing industry suggested that these trends may not always hold true. Regardless, research is scarce on the effects of other cattle types and climate regions on prevalence patterns.
Commercial beef processors that slaughter multiple cattle types from different regions within the U.S. are aware that differences in Salmonella burden coming into their plants may exist, and these patterns should be elucidated to the greatest extent possible. The results of a longitudinal study requested by a commercial beef processer are presented herein. Fecal samples and subiliac LN were collected for the culture of Salmonella at slaughter from a commercial cattle processing facility, representing cattle of different type, sourced from various climate regions of the southern United States, across seasons. Serogroup and antimicrobial resistance were also evaluated on a portion of the Salmonella isolates.
Materials and Methods
Sample Collection
Samples were collected by plant personnel from a single commercial beef processing plant in the southwestern U.S. Collections were made with the goal of obtaining 25 fecal samples and 20 LN from animals representing each cattle type and climate region (detailed below) during each season of the year. Overall, 1,840 fecal samples and 1,550 subiliac LN were collected. Paired fecal samples and LN were not intentionally collected, however some samples may have been collected from the same animal.
Fecal samples (~100 g) were collected from the rectum of individual cattle using a clean palpation sleeve, then placed into a sterile specimen cup for shipment. The subiliac LN, within the surrounding adipose tissue, were placed in individual sterile sample collection bags. Following collection, samples were shipped on ice overnight to the USDA-ARS laboratory in College Station, TX for bacterial culture as described below.
Cattle Type and Climate Regions
Five cattle types were examined: conventional feedlot cattle (raised with pharmaceuticals and technologies which increase feed efficiency, digestive health, and carcass quality); all-natural feedlot cattle (raised without antibiotics, exogenous growth-promoting hormones, or in-feed technologies such as ionophores and beta-agonists); grass-fed cattle (raised to market weight with varying amounts of pasture access); cull dairy cows (primiparous and multiparous); and cull beef cattle (cows and bulls). The host packing plant provided information on cattle source for this study. Five climate regions where cattle originated were represented in the sample collections (Figure 1; adapted from Building America climate map from the Office of Energy Efficiency and Renewable Energy, 2012): Region 1, mixed temperatures and dry; 2, mixed temperatures and humid; 3, hot and humid; 4, hot and dry; 5, cold. Cattle originated from feedlots, farms, and ranches within ~970 km of the Texas panhandle. Samples were collected from these cattle types and regions in the summer, fall, and winter of 2019 but due to the development of the COVID-19 pandemic, were not collected in the spring of 2020.
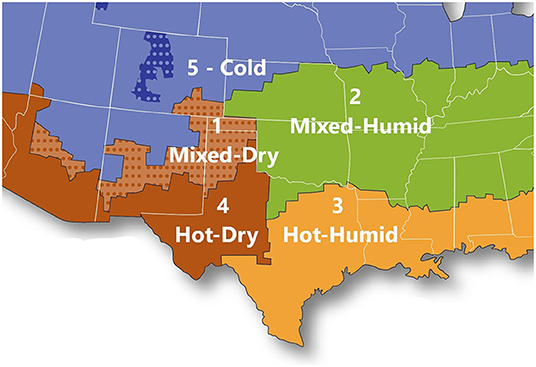
Figure 1. Climate regions from which cattle originated. Adapted from (https://www.energy.gov/eere/buildings/building-america-climate-specific-guidance).
Salmonella Culture
Unless otherwise stated, all media and agar were sourced from Difco Laboratories (Detroit, MI), and reagents and antibiotics were obtained from Sigma Chemical Co. (St. Louis, MO). Fecal samples were analyzed as described previously (Brichta-Harhay et al., 2007). Briefly, 10 g of each fecal sample was enriched in 90 mL tetrathionate broth containing 1.8 mL of iodine solution for 24 h at 37°C. For enumeration of Salmonella, a 50 μL aliquot of the pre-incubation mixture was plated onto XLD using a spiral plater, incubated for 24 h at 37°C, and then an additional 24 h at room temperature. Following incubation of the feces-tetrathionate mixture, a 100 μL aliquot was transferred to 5 mL Rapport-Vassiliadis R10 (RV) broth and incubated an additional 24 h at 42°C. After this selective enrichment, samples were dual-plated onto brilliant green agar (BGA) with novobiocin (25 μg/mL) and BGA with tetracycline (30 μg/mL) to assist in the identification of isolates for multidrug resistance (MDR) testing. All plates were incubated for 24 h at 37°C, then up to three suspect colonies (pink with distinct round border) per plate were biochemically confirmed using lysine iron and triple sugar iron agars.
Lymph nodes were processed and cultured quantitatively and qualitatively for Salmonella as described by Brichta-Harhay et al. (2012). Lymph nodes were trimmed of excess fat and fascia, then surface sterilized by immersion in boiling water for 3–5 s. The surface-sterilized LN was placed in a filtered stomacher bag, weighed, and pulverized using a rubber mallet. Next, 80 mL of tryptic soy broth (TSB) was added and the mixture homogenized for 30 s using a laboratory stomacher. For enumeration, 1 mL of the homogenized mixture was applied to Petrifilm EB (3M Health Care, St. Paul, MN) in duplicate and incubated at 37°C overnight. Films with bacterial growth were transferred to XLD plates containing 10 μg/mL cefsoludin and 15 μg/mL novobiocin and incubated (24 h, 37°C). Black colonies were counted and converted to log10 CFU/g LN. For prevalence analysis, the original LN-TSB mixture was incubated at room temperature for 2 h, then for 12 h at 42°C. Next, 1 mL from each enrichment culture was subjected to anti-Salmonella immunomagnetic separation with 20 μL of anti-Salmonella beads (Invitrogen, Waltham, MA) and incubated with shaking at 800 rpm at room temperature for 15 min. The beads were extracted from the enrichment samples and washed twice in PBS-Tween 20, then were transferred to 3 mL of RV broth and incubated (24 h, 42°C). Finally, 100 μL of RV broth was plated each to BGA with sulfadiazine (80 μg/mL), and BGA with tetracycline (30 μg/mL), then incubated at 37°C overnight. Lastly, suspect colonies were biochemically confirmed using lysine iron and triple sugar iron agars.
Serogrouping and Antimicrobial Susceptibility Testing
Isolates which grew on BGA with tetracycline were re-streaked on tryptic soy agar with 5% sheep blood (Becton, Dickenson and Co., Sparks, MD) for serogrouping using slide agglutination with Salmonella antiserum and antimicrobial susceptibility testing. Serogroups were confirmed by traditional slide agglutination (O typing) methods, using commercial antisera (Becton, Dickinson and Co.) following the manufacturer's guidelines.
Susceptibility to 14 antimicrobial agents was determined by use of an automated broth microdilution method (Sensititre Gram Negative NARMS Plates, TREK Diagnostics Inc., Oakwood Village, OH) according to the manufacturer's recommendations. Minimum inhibitory concentrations (MICs) were determined using the breakpoints established by the Clinical and Laboratory Standards Institute (CLSI, 2017) or by National Antimicrobial Resistance Monitoring System (NARMS, 2019) when CLSI criteria were not established, in order to classify isolates as susceptible or resistant. Isolates that were resistant to three or more classes of antimicrobials were considered MDR.
Statistical Analysis
Data were compiled and organized using Excel (Microsoft Corp., Redmond, WA), and then were analyzed using JMP 15 (SAS Institute Inc., Cary, NC). Fecal samples that were below the limit of detection (~10 CFU/g) during direct plating for enumeration but were positive for Salmonella after enrichment, were assigned a concentration of 1 log10 CFU/g feces (Brichta-Harhay et al., 2007). Similarly, lymph nodes that were below the limit of detection (~1 CFU/g) but were positive following enrichment, were assigned a concentration of 0.1 log10 CFU/g lymph node.
To examine effects of season and cattle production type on Salmonella recovery in feces and peripheral LN, season and cattle type were included in the model as fixed effects, and the sample ID within cattle type was included as a random effect. To examine effect of climate region on Salmonella recovery in feces and peripheral LN, climate region was included as a fixed effect while cattle type was included as a random effect, due to imbalance of cattle type across regions. Quantitative concentration data were log-transformed prior to analysis to stabilize variances. Concentration data were analyzed using ANOVA, with Student's t-test for pairwise comparisons of treatment means when warranted. Prevalence data were analyzed using nominal logistic regression, with chi-square tests for significance. Significance was declared at P ≤ 0.05.
Results
A total of 1,550 subiliac LN and 1,840 fecal samples were collected across the five cattle types, in five climate regions, across three seasons. Overall, concentration ranged from 0.1 to 3.8 log10 CFU/g in LN, with 79% of positive samples having concentrations < 1 log10 CFU/g. In fecal samples, concentrations ranged from 1.0 to 6.2 log10 CFU/g, with 72% of positive samples' concentrations equal to 1.0 log10 CFU/g. There were 8 and 4.8% of samples quantitatively positive (detectable prior to enrichment) for Salmonella in feces and LN, respectively, and prevalence was 29.3 and 13.9% feces and LN, respectively.
Season and Cattle Type
Results of the Salmonella culture from the subiliac LN of cattle are presented in Table 1 by cattle type, across all seasons and for each season. Across all seasons, conventional feedlot cattle had the greatest (P < 0.01) Salmonella concentration (1.17 log10 CFU/g LN), while grass-fed cattle had the least concentration (0.12 log10 CFU/g LN). Conventional feedlot cattle and cull dairy cows had a greater percentage of quantitatively positive LN and a greater prevalence rate compared to other cattle types (P < 0.01). Summer represented the season with the highest prevalence for each cattle type except cull dairy cows and conventional feedlot cattle, for which autumn was the season of highest prevalence. In all-natural feedlot cattle, autumn represented the time of lowest prevalence, but in all other cattle types, the lowest prevalence occurred through winter. During winter, there were no differences (P > 0.70) in concentration or prevalence between cattle types.
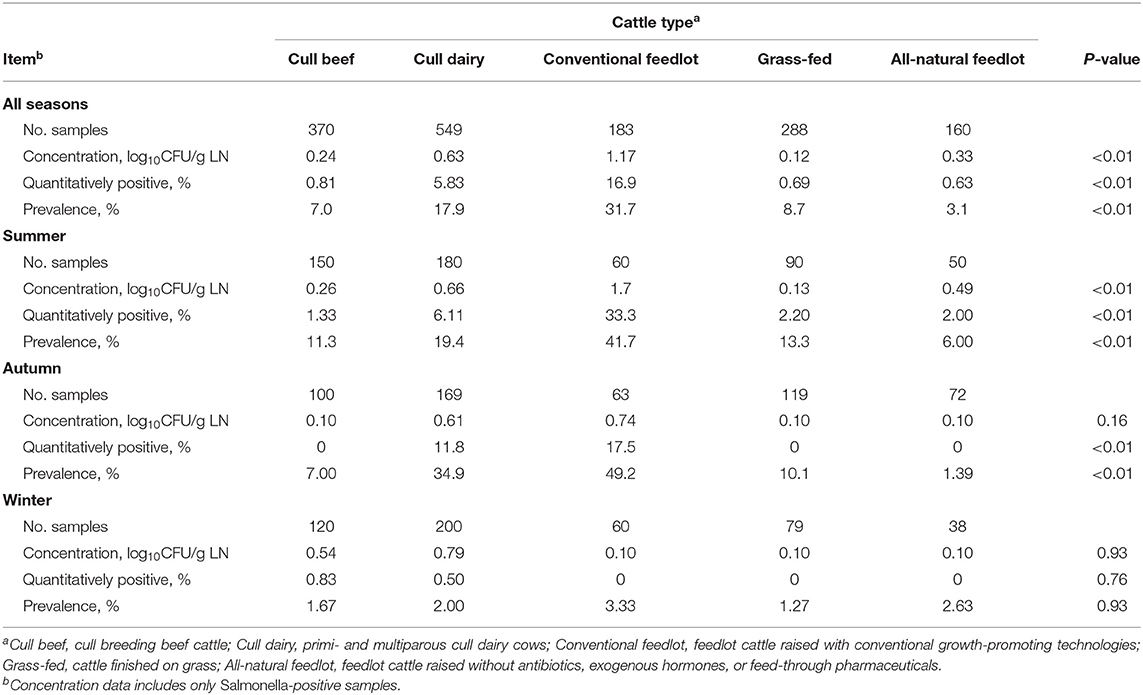
Table 1. Characterization of Salmonella-positive subiliac lymph nodes (LN) collected from different cattle types by and across three seasonal collections.
Results of the Salmonella recovery from fecal samples of different cattle types across season are presented in Table 2 by cattle type, across all seasons and for each season. Across all seasons, cull dairy cows had the greatest (P < 0.01) Salmonella concentration (1.9 log10 CFU/g feces), compared to all other cattle types (≤1.6), though that difference may not be biologically significant. Across all seasons, the prevalence rate in fecal samples was greater than that of the LN for each cattle type. The greatest (P < 0.01) percent quantitatively positive fecal samples and prevalence rate again occurred in cull dairy and conventional feedlot cattle, while all-natural feedlot cattle had the lowest. In autumn, cull dairy cows had the greatest (P < 0.01) Salmonella concentration (2.24 log10 CFU/g feces), compared to all other cattle types (≤1.2 log10 CFU/g). Winter persisted as the season with the least qualitative prevalence in feces within each cattle type, but cull dairy cows had substantially greater (P < 0.01) prevalence rate (35.8%) during that season compared to other types (≤10%).
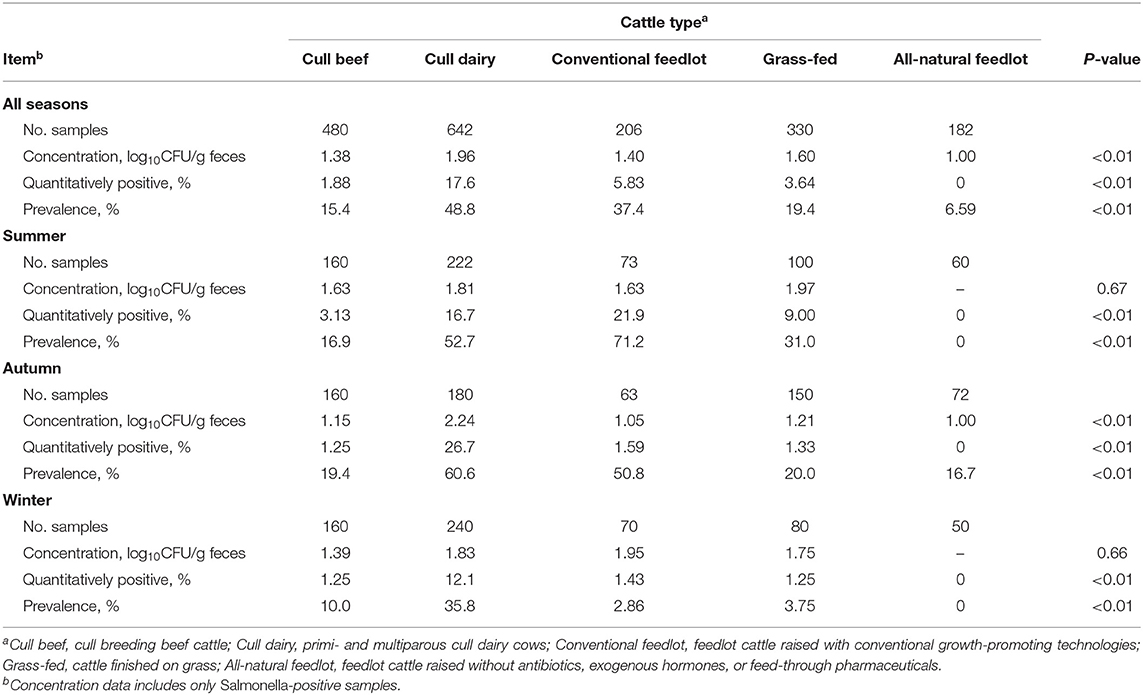
Table 2. Characterization of Salmonella-positive fecal samples collected from different cattle types by and across three seasonal collections.
Climate Region
Differences in Salmonella culture by climate region of origin for subiliac LN and fecal samples are presented in Table 3. Differences in Salmonella concentration among the LN and fecal samples across region are not likely of a biologically-significant magnitude, despite statistical significance, as all means were within 1 log10 CFU/g. Region 4 (hot-dry) presented the greatest prevalence of Salmonella in the LN, and region 2 (mixed temperature-humid) presented the lowest (P < 0.05). Region 4 also had the greatest Salmonella prevalence in fecal samples, while region 5 (cold) presented the lowest (P < 0.01).
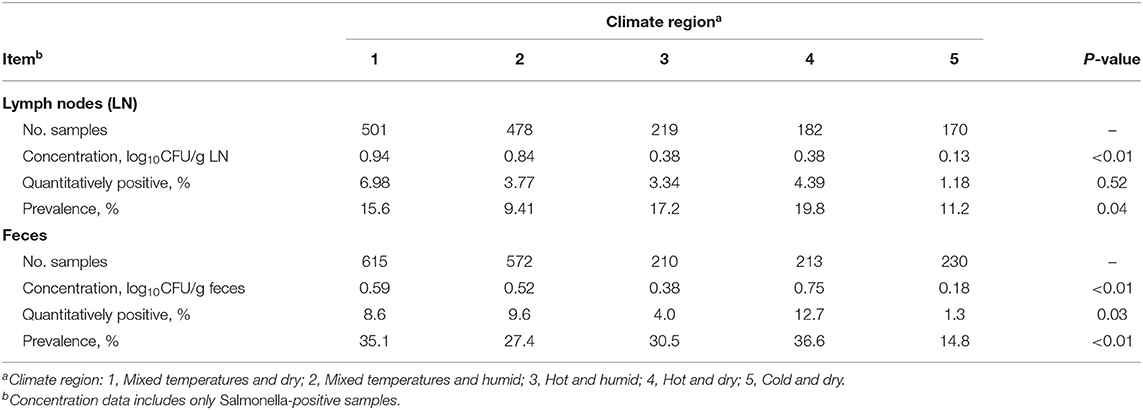
Table 3. Prevalence of Salmonella in subiliac lymph nodes and feces in cattle at harvest by climate region.
Multi-Drug Resistance
Isolates from positive BGAtet plates (n = 50) were serogrouped and subjected to antimicrobial susceptibility testing, and classified as MDR if resistance to three or more classes of antimicrobials was exhibited. Resistance patterns and source cattle types of MDR Salmonella isolates, by serogroup, are presented in Table 4. Twenty-one MDR isolates were identified, and only 4 isolates originated from LN (all of which were from cull dairy cows) while the rest were of fecal origin. Further, cull dairy cows were responsible for 48% (10/21) of all the MDR isolates, while cull beef cattle were responsible for 38% (8/21), and conventional feedlot, grass-fed, and all-natural feedlot cattle were each responsible for 4.8% (each 1/21). Serogroups C1 and B represented the most frequent serogroups among MDR isolates (43 and 33%, respectively), though this may just reflect greater prevalence of these serogroups in general, as pansusceptible isolates were not serogrouped for comparison.
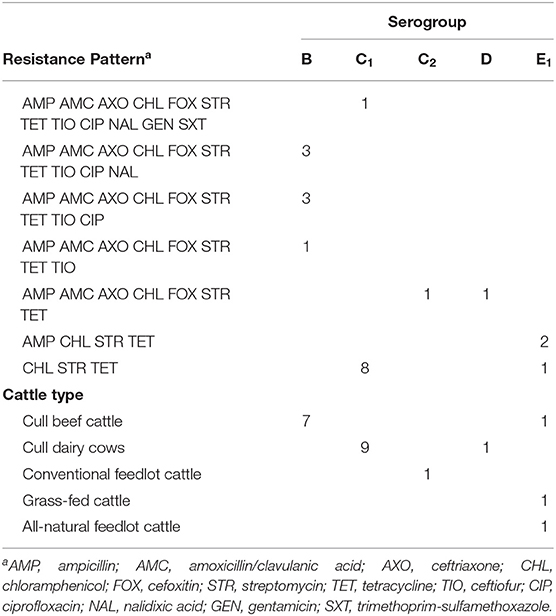
Table 4. Multi-drug resistant Salmonella isolates by serogroup isolated from subiliac lymph nodes (n = 4) and feces (n = 17) of cattle, by antimicrobial resistance pattern and source.
Discussion
It is well-established that cattle may harbor Salmonella in the intestinal tract, thereby presenting the opportunity for fecal contamination of carcasses at slaughter, a significant food-safety risk (Koohmaraie et al., 2012; Muñoz-Vargas et al., 2018). Further, as Salmonella sequestered in the peripheral LN of cattle is protected from in-plant interventions, it may provide a source for potential ground beef contamination (Brichta-Harhay et al., 2012; Webb et al., 2017; Nickelson et al., 2019; Gutiérrez et al., 2020).
Cattle Type
Cattle types are not always clearly defined in study reports. For this study, conventional feedlot cattle were those raised with pharmaceuticals and technologies which increase feed efficiency, digestive health, and carcass quality (e.g., growth-promoting implants, antibiotics, ionophores), while all-natural feedlot cattle were raised in a similar environment but without exogenous pharmaceuticals. Grass-fed cattle were raised to market weight with varying amounts of access to pasture. Cull dairy cows included primiparous and multiparous cows removed from production for unrecorded reasons. Cull beef cattle included cows and bulls, removed from production for unrecorded reasons. In the present study, conventional feedlot cattle presented the greatest prevalence of Salmonella in LN (32%) followed by cull dairy cattle (18%), while the other cattle types had substantially lower prevalence. Koohmaraie et al. (2012) also observed 18% prevalence rate in the cervical LN of 100 cull dairy cattle at harvest. Webb et al. (2017) reported a lower Salmonella prevalence of 7% in LN of conventional feedlot cattle and of 1.8% in LN from cull (mixed beef and dairy) cattle, and Brichta-Harhay et al. (2012) reported a mere 0.8% Salmonella prevalence in the LN of cull beef and dairy cattle. Alternatively, Gragg et al. (2013a) and Levent et al. (2019) each reported ~75% prevalence rate in LN from fed cattle at slaughter. More recently, a study of 400 Mexican feedlot cattle reported a Salmonella prevalence of 9.7% on the peripheral LN (Gutiérrez et al., 2020). As it is posited that Salmonella primarily reaches the peripheral LN by escaping the gut, it is reasonable to assume that diet may have an effect on Salmonella prevalence within the LN, as it does in liver abscesses (Amachawadi and Nagaraja, 2015; Sanz-Fernandez et al., 2020). Further, cattle in feedlots and dairy systems are typically confined such that animal concentration per land area is greater, compared to pasture-based systems, which likely results in increased exposure to other animals, feces, and mud. Regardless, more research is needed to further investigate correlations between live animal behavior, diet and management, and Salmonella prevalence in the LN.
Research is limited regarding Salmonella culture from LN of all-natural feedlot cattle; however, previous research by the authors found relatively high Salmonella concentrations and prevalence in this cattle type (Edrington, unpublished data). Based on this previously-collected data and the fact that all-natural feedlot cattle typically receive a ration very similar to that of conventional feedlot cattle simply without antibiotics and other growth-promoting supplements, it was expected that similar Salmonella prevalence would be observed among the two types of feedlot cattle. Generally, the only difference between all-natural and conventional feedlot cattle is the use of antibiotics and growth-promoting compounds outlined above. Often, both types of cattle may be found within the same feedlot, managed and housed similarly with the above exceptions. The very low Salmonella concentration and prevalence observed in feces and LN from all-natural feedlot cattle and young grass-fed cattle in the present study is intriguing, and warrants further investigation. Continued research comparing these cattle types will help elucidate what factors, such as pen density and use of growth-promoting technologies, influence Salmonella burden in market cattle.
Research regarding Salmonella prevalence in feces varies widely across and within cattle types. Muñoz-Vargas et al. (2018) reported that periparturient dairy cows had increased fecal Salmonella shedding 1 wk prior and 1 wk following parturition (~54%) compared to 3 wk prior to and 3 wk post-parturition (~37%), evidencing that increased physiological stress may lead to increased Salmonella prevalence. Similarly, Edrington et al. (2004) reported an overall average of 39% Salmonella fecal prevalence rate in healthy lactating dairy cows, but prevalence varied widely across farm and season. Manishimwe et al. (2021) recently reported a fecal Salmonella prevalence rate of 18.8% among healthy dairy cows in Texas. The fecal samples in the present study were collected at time of harvest and a much greater prevalence rate (49%) in cull dairy cows was observed. Kunze et al. (2008) collected feces from fresh fecal pats at a feedlot and reported prevalence of 32%, which is similar to what was observed in the present study in conventional feedlot cattle. Dargatz et al. (2016) reported a fecal Salmonella prevalence rate of 9% in cattle in feedlots across 12 states which is substantially lower than what was observed in the present study (37%), but the following year Gragg et al. (2013a) reported a 94% fecal Salmonella prevalence rate in fed cattle at slaughter. In a similar trend to what was observed in the present study, Fegan et al. (2004) reported 4.5% fecal Salmonella prevalence in grass-fed cattle at slaughter, compared to 9% in grain-fed cattle. However, Barlow et al. (2015) reported 13% fecal Salmonella prevalence in grass-fed cattle at slaughter, compared to 10% in grain-fed cattle. Looper et al. (2009) reported just 2% Salmonella prevalence in grazing beef cows. It is likely that part of the discrepancy between studies is the time of sample collection, as Salmonella prevalence seems to increase following transportation and lairage. Indeed, Schmidt et al. (2015) observed a 5.4% fecal Salmonella prevalence rate in cattle at a feedlot, but prevalence increased to 44.6% at time slaughter. Further, differences exist between farms of the same type, within the same region (Edrington et al., 2004, 2008a; Haneklaus et al., 2012; Loneragan et al., 2012). These differences could be due in part to bacterial culture preferences utilized by different laboratories, however they are more likely a function of the wide variability in Salmonella shedding frequently observed among farms due to factors yet to be elucidated. Clearly, prevalence is a multifaceted phenomenon with substantial variation between year, farm, and group of cattle. Further research investigating the effect of diet, stress, and management is warranted to understand differences in Salmonella prevalence within and across cattle types.
Season
It has been commonly reported that warmer months of the year are associated with increased Salmonella prevalence in cattle compared to cooler periods (Kunze et al., 2008; Webb et al., 2017; Levent et al., 2019; Nickelson et al., 2019), such as was observed in the present research. The causal mechanism has yet to be fully explained and is generally thought to be a function of elevated temperatures producing more hospitable environmental conditions for Salmonella growth and persistence. However, other factors such as seasonal insect vectors and the influence of day length on hormone production are likely involved (Edrington et al., 2006; Pangloli et al., 2008). In the present study, winter presented the time of lowest Salmonella prevalence in LN and feces. The seasonal differences in LN and fecal Salmonella prevalence is stark, particularly for conventional feedlot and cull dairy cows, suggesting that whatever the effect of warmer weather, it is more impactful on those cattle types. Indeed, Edrington et al. (2004) reported fecal Salmonella prevalence of 8% in four dairy farms during in the winter of 2001, but that increased to 41% that summer. Similarly, Edrington et al. (2008b) reported fecal Salmonella prevalence of 96% in the summer of 2004 which decreased to 19% that autumn. There may also be a differential effect of season on different breed types, as Nickelson et al. (2019) reported that though they were fattened in the same feedlot, U.S.-raised cattle had a 41% cool season vs. 59% warm season LN Salmonella prevalence, while cattle of Mexican origin were unaffected by season (52 and 56%, respectively). It has been shown that heat stress affects tight junction integrity in the intestine (Koch et al., 2019), which may explain a facet of the increased prevalence in LN during warmer months.
Climate Region
Likely by the same mechanism that results in decreased Salmonella prevalence during winter, Region 5 (cold) and Region 2 (mixed temperature—humid) had lower Salmonella prevalence in the LN while Region 4 (hot—dry) had the greatest in the present study. Comparing LN Salmonella carriage between geographical monitoring regions, Gragg et al. (2013b) reported that the Midwest U.S. (cold) was highest for cull cows (4.2%) and Southwest U.S. (hot–dry) was highest for feedlot cattle (19%). In a similar study comparing LN Salmonella carriage across three regions [East U.S. (mixed temperatures and humid), Southwest U.S., and West U.S. (mixed temperatures and dry)] in feedlot and cull cattle, Webb et al. (2017) reported that feedlot cattle in the Southwest U.S. had greater prevalence (19%) than in other areas, while cull cattle from the East U.S. had greater prevalence (3.2%) than other areas. Those authors cautioned against inferring too much from regional trends, as weather within a region is unpredictable year-to-year and probably contributes more to Salmonella carriage than region does, however it is compelling that such similar results were observed by Gragg et al. (2013b) and Webb et al. (2017), and the present study, in that hot and dry climate regions seem to present conditions for elevated prevalence.
Multi-Drug Resistance
Though overall isolation of MDR Salmonella was rare in this study, the greatest incidence of MDR Salmonella came from cull beef and dairy cattle. As they are culled animals, it is possible that antimicrobial therapy was administered to the animals prior to culling. Additionally, the increased age of these cattle types compared to feedlot and grass-fed cattle provided more opportunity for them to receive antimicrobial therapy throughout life and therefore more prospect to develop resistance. Supporting the theory of recent or increased antibiotic use in animal classes presenting greater MDR Salmonella, Edrington et al. (2008a) reported 45% of fecal samples from sick dairy cows and 58% of fecal samples from hutch calves had tetracycline-resistant Salmonella, compared to 10% in healthy heifers and cows. Other studies have reported 5–10% occurrence of MDR Salmonella in cull dairy cow fecal samples (Loneragan et al., 2012; Rodriguez-Rivera et al., 2016). Manishimwe et al. (2021) reported only 1% of dairy cattle fecal samples were positive for MDR Salmonella, which is more similar to what was found in the present study (1.6%; 10/642). Prevalence of MDR isolates may increase from the time cattle are transported from the feedlot or ranch to the abattoir; Schmidt et al. (2015) reported a 0.5% prevalence rate of 3rd generation cephalosporin-resistant Salmonella in fecal samples collected at a feedlot, but that number increased to 1.6% at time of slaughter. More research is needed to understand physiologic changes that occur in times of stress, such as transportation and lairage, that may lead to a rapid increase in prevalence of MDR Salmonella in cattle immediately prior to harvest.
Conclusion
Similar to other research, results reported herein found that conventional feedlot cattle and cull dairy cows had the highest prevalence of Salmonella in both the LN and feces, while surprisingly, all-natural feedlot cattle had the lowest. A primary limitation of this study was that samples were unable to be paired from the same subjects, which could perhaps strengthen future research. In general, results such as these should aid cattle processors that harvest multiple cattle types in a single establishment in risk assessment and abatement. While variation in Salmonella is known to exist among similar cattle operations within a region, it is reasonable to assume that concentration of animals on dairies and feedlots, as well as differing feedstuffs and ration formulation, may partly explain why feedlot and dairy animals have the highest Salmonella incidence. Future research is underway to better understand the dynamics of Salmonella within these cattle types and assess potential pre-harvest intervention strategies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LW organized the data, performed statistical analysis, and wrote the manuscript. TE conceptualized and designed the study, gathered data, and revised the manuscript. RA assisted with conceptualization and data interpretation, and revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
TE was employed by Diamond V Mills, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Kate Andrews for her outstanding technical service to complete this research. We also extend our gratitude to the processing plant and its personnel for their hard work in collecting and documenting samples.
Abbreviations
LN, Lymph node; MDR, Multidrug-resistance.
References
Amachawadi, R. G., and Nagaraja, T. G. (2015). First report of anaerobic isolation of Salmonella enterica from liver abscesses of feedlot cattle. J. Clin. Microbiol. 53, 3100–3101. doi: 10.1128/JCM.01111-15
Barlow, R. S., McMillan, K. E., Duffy, L. L., Fegan, N., Jordan, D., and Mellor, G. E. (2015). Prevalence and antimicrobial resistance of Salmonella and Escherichia coli from Australian cattle populations at slaughter. J. Food Prot. 78, 912–920. doi: 10.4315/0362-028X.JFP-14-476
Brichta-Harhay, D. M., Arthur, T. M., Bosilevac, J. M., Guerini, M. N., Kalchayanand, N., and Koohmaraie, M. (2007). Enumeration of Salmonella and Escherichia coli O157:H7 in ground beef, cattle carcass, hide and fecal samples using direct plating methods. J. Appl. Microbiol. 103, 1657–1668. doi: 10.1111/j.1365-2672.2007.03405.x
Brichta-Harhay, D. M., Arthur, T. M., Bosilevac, J. M., Kalchayanand, N., Schmidt, J. W., Wang, R., et al. (2012). Microbiological analysis of bovine lymph nodes for the detection of Salmonella enterica. J. Food Prot. 75, 854–858. doi: 10.4315/0362-028X.JFP-11-434
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. CLSIsupplement M100. Wayne, PA: Clinical and Laboratory Standards Institute.
Dargatz, D. A., Kopral, C. A., Erdman, M. M., and Fedorka-Cray, P. J. (2016). Prevalence and antimicrobial resistance of Salmonella isolated from cattle feces in United States feedlots in 2011. Foodborne Pathog. Dis. 13, 483–489. doi: 10.1089/fpd.2016.2128
Edrington, T. S., Callaway, T. R., Anderson, R. C., and Nisbet, D. J. (2008a). Prevalence of multidrug-resistant Salmonella on commercial dairies utilizing a single heifer raising facility. J. Food Prot. 71, 27–34. doi: 10.4315/0362-028X-71.1.27
Edrington, T. S., Callaway, T. R., Ives, S. E., Engler, M. J., Looper, M. L., Anderson, R. C., et al. (2006). Seasonal shedding of Escherichia coli O157:H7 in ruminants: a new hypothesis. Foodborne Pathog. Dis. 3, 413–421. doi: 10.1089/fpd.2006.3.413
Edrington, T. S., Hume, M. E., Looper, M. L., Schultz, C. L., Fitzgerald, A. C., Callaway, T. R., et al. (2004). Variation in the fecal shedding of Salmonella and E. coli O157:H7 in lactating dairy cattle and examination of Salmonella genotypes using pulsed-field gel electrophoresis. Lett. Appl. Microbiol. 38, 366–372. doi: 10.1111/j.1472-765X.2004.01495.x
Edrington, T. S., Ross, T. T., Callaway, T. R., Martinez, C. H., Hume, M. E., Genovese, K. J., et al. (2008b). Investigation into the seasonal salmonellosis in lactating dairy cattle. Epidemiol. Infect. 136, 381–390. doi: 10.1017/S0950268807008680
Fegan, N., Vanderlinde, P., Higgs, G., and Desmarchelier, P. (2004). Quantification and prevalence of Salmonella in beef cattle presenting at slaughter. J. Appl. Microbiol. 97, 892–898. doi: 10.1111/j.1365-2672.2004.02380.x
Gragg, S. E., Loneragan, G. H., Brashears, M. M., Arthur, T. M., Bosilevac, J. M., Kalchayanand, N., et al. (2013b). Cross-sectional study examining Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at harvest. Foodborne Pathog Dis. 10, 368–374. doi: 10.1089/fpd.2012.1275
Gragg, S. E., Loneragan, G. H., Nightingale, K. K., Brichta-Harhay, D. M., Ruiz, H., Elder, J. R., et al. (2013a). Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl. Environ. Microbiol. 79, 4744–4750. doi: 10.1128/AEM.01020-13
Gutiérrez, T. P., Lozano, M. S. R., Suárez, E. J. D., Guzmán, N. R., Ramos, O. S., Pérez, C. F. H., et al. (2020). Lymph nodes and ground beef as public health importance reservoirs of Salmonella spp. Rev. Mex. Cienc. Pecuarias. 11, 795–810. doi: 10.22319/rmcp.v11i3.5516
Haneklaus, A. N., Harris, K. B., Griffin, D. B., Edrington, T. S., Lucia, L. M., and Savell, J. W. (2012). Salmonella prevalence in bovine lymph nodes differs among feedyards. J. Food Prot. 75, 1131–1133. doi: 10.4315/0362-028X.JFP-11-530
Koch, F., Thom, U., Albrecht, E., Weikard, R., Nolte, W., Kuhla, B., et al. (2019). Heat stress directly impairs gut integrity and recruits distinct immune cell populations into the bovine intestine. Proc. Natl. Acad. Sci. U. S. A. 116, 10333–10338. doi: 10.1073/pnas.1820130116
Koohmaraie, M., Scanga, J. A., De La Zerda, M. J., Koohmaraie, B., Tapay, L., Beskhlebnaya, V., et al. (2012). Tracking the sources of Salmonella in ground beef produced from nonfed cattle. J. Food Prot. 75, 1464–1468. doi: 10.4315/0362-028X.JFP-11-540
Kunze, D. J., Loneragan, G. H., Platt, T. M., Miller, M. F., Besser, T. E., Koohmaraie, M., et al. (2008). Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl. Environ. Microbiol. 74, 345–351. doi: 10.1128/AEM.02076-07
Levent, G., Schlochtermeier, A., Ives, S. E., Norman, K. N., Lawhon, S. D., Loneragan, G. H., et al. (2019). Population dynamics of Salmonella enterica within beef cattle cohorts followed from single-dose metaphylactic antibiotic treatment until slaughter. Appl. Environ. Microbiol. 85, e01386-19. doi: 10.1128/AEM.01386-19
Loneragan, G. H., Thomson, D. U., McCarthy, R. M., Webb, H. E., Daniels, A. E., Edrington, T. S., et al. (2012). Salmonella diversity and burden in cows on and culled from dairy farms in the Texas high plains. Foodborne Pathog. Dis. 9, 549–555. doi: 10.1089/fpd.2011.1069
Looper, M. L., Edrington, T. S., and Rosenkrans, C. F Jr. (2009). Influence of body condition and forage type on prevalence of Escherichia coli O157:H7 and Salmonella in grazing beef cows. Lett. Appl. Microbiol. 49, 361–365. doi: 10.1111/j.1472-765X.2009.02669.x
Manishimwe, R., Moncada, P. M., Bugarel, M., Scott, H. M., and Loneragan, G. H. (2021). Antibiotic resistance among Escherichia coli and Salmonella isolated from dairy cattle feces in Texas. PLoS ONE 16, e0242390. doi: 10.1371/journal.pone.0242390
Muñoz-Vargas, L., Opiyo, S. O., Digianantonio, R., Williams, M. L., Wijeratne, A., and Habing, G. (2018). Fecal microbiome of periparturient dairy cattle and associations with the onset of Salmonella shedding. PLoS ONE 13, e0196171. doi: 10.1371/journal.pone.0196171
NARMS (2019). Antibiotics Tested by NARMS. National Antimicrobial Resistance Monitoring System. Available online at: https://www.cdc.gov/narms/antibiotics-tested.html (accessed July 9, 2021).
Nickelson, K. J., Taylor, T. M., Griffin, D. B., Savell, J. W., Gehring, K. B., and Arnold, A. N. (2019). Assessment of Salmonella prevalence in lymph nodes of U.S. and Mexican cattle presented for slaughter in Texas. J. Food Prot. 82, 310–315. doi: 10.4315/0362-028X.JFP-18-288
Office of Energy Efficiency Renewable Energy (2012). Building America Climate-Specific Guidance. Available online at: https://www.energy.gov/eere/buildings/building-america-climate-specific-guidance (accessed June 1, 2021).
Pangloli, P., Dje, Y., Ahmed, O., Doane, C. A., Oliver, S. P., and Draughon, F. A. (2008). Seasonal incidence and molecular characterization of Salmonella from dairy cows, calves, and farm environment. Foodborne Pathog. Dis. 5, 87–96. doi: 10.1089/fpd.2007.0048
Rodriguez-Rivera, L. D., Cummings, K. J., Loneragan, G. H., Rankin, S. C., Hanson, D. L., Leone, W. M., et al. (2016). Salmonella prevalence and antimicrobial susceptibility among dairy farm environmental samples collected in Texas. Foodborne Pathog. Dis. 13, 205–211. doi: 10.1089/fpd.2015.2037
Sanz-Fernandez, M. V., Daniel, J. B., Seymour, D. J., Kvidera, S. K., Bester, Z., Doelman, J., et al. (2020). Targeting the hindgut to improve health and performance in cattle. Animals 10, 1817. doi: 10.3390/ani10101817
Schmidt, J. W., Agga, G. E., Bosilevac, J. M., Brichta-Harhay, D. M., Shackelford, S. D., Wang, R., et al. (2015). Occurrence of antimicrobial-resistant Escherichia coli and Salmonella enterica in the beef cattle production and processing continuum. Appl. Environ. Microbiol. 81, 713–725. doi: 10.1128/AEM.03079-14
Keywords: cattle, climate region, feces, lymph nodes, Salmonella, season
Citation: Wottlin LR, Edrington TS and Anderson RC (2022) Salmonella Carriage in Peripheral Lymph Nodes and Feces of Cattle at Slaughter Is Affected by Cattle Type, Region, and Season. Front. Anim. Sci. 3:859800. doi: 10.3389/fanim.2022.859800
Received: 21 January 2022; Accepted: 23 February 2022;
Published: 21 March 2022.
Edited by:
Todd Riley Callaway, University of Georgia, United StatesReviewed by:
Elisa Cabrera-Díaz, University of Guadalajara, MexicoKim Stanford, University of Lethbridge, Canada
Copyright © 2022 Wottlin, Edrington and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tom S. Edrington, dGVkcmluZ3RvbkBkaWFtb25kdi5jb20=
 Lauren R. Wottlin
Lauren R. Wottlin Tom S. Edrington
Tom S. Edrington Robin C. Anderson
Robin C. Anderson