Biochemical and structural properties of a thermostable mercuric ion reductase from Metallosphaera sedula
- 1Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT, USA
- 2Department of Chemistry and Geochemistry, Colorado School of Mines, Golden, CO, USA
- 3Department of Microbiology and Immunology, Montana State University, Bozeman, MT, USA
- 4Thermal Biology Institute, Montana State University, Bozeman, MT, USA
Mercuric ion reductase (MerA), a mercury detoxification enzyme, has been tuned by evolution to have high specificity for mercuric ions (Hg2+) and to catalyze their reduction to a more volatile, less toxic elemental form. Here, we present a biochemical and structural characterization of MerA from the thermophilic crenarchaeon Metallosphaera sedula. MerA from M. sedula is a thermostable enzyme, and remains active after extended incubation at 97°C. At 37°C, the NADPH oxidation-linked Hg2+ reduction specific activity was found to be 1.9 μmol/min⋅mg, increasing to 3.1 μmol/min⋅mg at 70°C. M. sedula MerA crystals were obtained and the structure was solved to 1.6 Å, representing the first solved crystal structure of a thermophilic MerA. Comparison of both the crystal structure and amino acid sequence of MerA from M. sedula to mesophillic counterparts provides new insights into the structural determinants that underpin the thermal stability of the enzyme.
Introduction
The ionic form of mercury, which is one of the most toxic metals known to biology (Gertrud et al., 1989; Nies, 2003; Vetriani et al., 2005), is naturally present at elevated concentrations in many hydrothermal vents, hot springs, and acid mine drainage fluids (Batten and Scow, 2003; Simbahan et al., 2005; Vetriani et al., 2005; King et al., 2006; Boyd et al., 2009; Wang et al., 2011). In these environments, biology utilizes a finely tuned protein catalyst termed the mercuric reductase (MerA) (encoded by the merA gene) in order to reduce toxic ionic mercury (Hg2+) to the far less toxic, volatile, and elemental form (Hg0). The reaction catalyzed by MerA follows the reaction scheme of NADPH + Hg2+ → NADP+ + Hg0 (Barkay et al., 2003). MerAs, which are part of the disulfide oxidoreductase (DSOR) family (Fox and Walsh, 1982), are ancient enzymes, having arisen in high temperature environments after the great oxidation event ~2.4 billion years ago (Barkay et al., 2010). Since that time, evolution has finely tuned MerA through recruitment of regulatory and transport proteins (Boyd and Barkay, 2012) to serve a diversity of organisms, including both Archaea and Bacteria, which encounter Hg2+ ions in less extreme mesophilic settings, while retaining extremely high stability and substrate specificity. These characteristics of mercuric reductases lend them to possible sensor applications, wherein the redox properties of the enzyme could be coupled to an amplifiable electrical signal (Adami et al., 1995; Han et al., 2001; Zhang et al., 2011). A stable mercuric reductase may also be used to potentially mitigate mercury contamination (Nascimento and Chartone-Souza, 2003).
Metallosphaera sedula (Mse), isolated previously from Pisciarelli Solfatara in Naples, Italy (Gertrud et al., 1989), has a minimum and maximum temperature for growth range of 50–80°C (Auernik et al., 2008a). Pisciarelli Solfatara itself contains a variety of thermal features that range in temperature from ~30°C to nearly 100°C, and a pH range of 1.5 to around 6.0 with elevated concentrations of heavy metals, including Hg2+ at concentrations up to 0.005 g/kg (Huber et al., 2000). The genome sequence of Mse was completed in 2008, (Auernik et al., 2008b), making it possible to identify mechanisms of Hg2+ tolerance at the genomic level. The mer operon in Mse includes both MerA and MerH, where MerH may aid metal trafficking to the MerR transcription factor (Schelert et al., 2013).
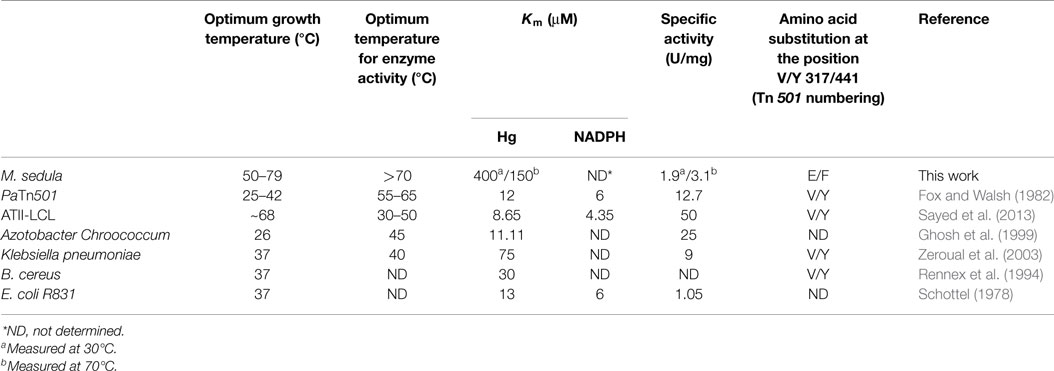
A variety of MerAs have been characterized previously, most notably a protein encoded on a transposon isolated from Pseudomonas aeruginosa, which is termed Tn501 (Fox and Walsh, 1982), as well as MerA from Bacillus cereus (BcMerA) (Schiering et al., 1991) and a MerA from a deep brine environment, termed ATII-LCL (Sayed et al., 2013). Collectively, these biochemical studies have revealed MerAs that exhibit Km values for Hg2+ that range from 9–70 μM and specific activities that range from 1.05–50 μmol/min⋅mg. Structural characterization was first carried out on BcMerA (Schiering et al., 1991) and later on Tn501 (Ledwidge et al., 2005). Most recently, the Tn501 structure has been solved in complex with Hg2+(Lian et al., 2014). Structural characterization confirmed that MerA is a member of the DSOR protein family, which adopts a βαββαβ fold, and which is known to catalyze pyridine-dependent substrate reduction with a characteristic active site CXXXXC motif (Argyrou and Blanchard, 2004). Some MerAs also harbor an additional N-terminal GMTCXXC motif (Boyd and Barkay, 2012) that assists in metal recruitment (Ledwidge et al., 2005). A third pair of conserved cysteines are located in a flexible region on the C-terminal domain, and are responsible for delivering mercuric ions to the active site of the opposing monomer (Lian et al., 2014).
Despite these advances, the structural characterization of a MerA from a thermophile has yet to be conducted, even though this is critical for understanding the properties of enzymes involved in mercury detoxification of high-temperature environments where mercury concentrations are very high. Structural characterization is important for both understanding the thermophilic origins of the protein (Barkay et al., 2010; Boyd and Barkay, 2012) as well as for possible incorporation into stable biotechnologies. Here, we report biochemical and structural characterization of a thermostable MerA from the aerobic thermoacidophilic Crenarchaeon Mse (MseMerA).
Materials and Methods
Bioinformatics
MerA homologs were compiled from the Department of Energy-Integrated Microbial Genomes database using BLASTp and the Tn501 MerA as a query. Representative homologs were screened for conserved residues that define MerA (as described above), and those protein sequences with these residues were aligned using CLUSTALX (version 2.0.8) specifying the Gonnet 250 protein substitution matrix and default gap extension and opening penalties (Larkin et al., 2007), with dihydrolipoamide dehydrogenase from Magnetospirillum magneticum AMB-1 (YP_423326), Thermus thermophilus HB27 (YP_005669), and Pseudomonas fluorescens Pf0-1 (YP_351398) serving as outgroups. N-terminal “NmerA” sequence was trimmed from the alignment block as previously described (Barkay et al., 2010) and the phylogeny of MerA was evaluated with PhyML (ver. 3.0.1) (Guindon et al., 2010) using the LG amino acid substitution matrix with a discrete four category gamma substitution model and a defined proportion of invariant sites. A consensus phylogenetic tree was projected from 100 bootstrap replications using FigTree (ver. 1.2.2) (http://tree.bio.ed.ac.uk/software/figtree/).
Structural superimpositions were generated by the program UCSF Chimera (Pettersen et al., 2004). The protein sequence of MseMerA was blasted with NCBI BLASTp. The top eight hits were compared with mesophilic mercuric reductases from Staphylococcus aureus, B. cereus, P. aeruginosa, and a sequence from a hydrothermal deep-sea brine environment, ATII-LCL (Sayed et al., 2013). It should be noted that while the ATII-LCL sequence was isolated from a hydrothermal vent system with a temperature of 68°C, the optimum temperature for activity was shown to be 30–50°C (Sayed et al., 2013), indicating that it is not adapted to the thermal regime from where it was isolated or that the environment from where it was isolated is variable with respect to temperature. VADAR was used to evaluate the surface area and charged residue percentage of MerA homologs (Willard et al., 2003), while the ProtParam tool available from ExPASy was used to calculate the aliphatic index of MerA homologs (Gasteiger et al., 2005).
Expression and Purification
MseMerA DSM 5348 sequence was codon-optimized and synthesized by GenScript USA Inc. with an N-terminal 6× His-tag (Data Sheet 1 in Supplementary Material). The gene was cloned into MCS1 of pETDuet-1 and transformed into Escherichia coli BL21DE3 cells (Novagen, EMD Millipore, USA). Sequence-based confirmation of MseMerA transformation was performed by Davis Sequencing, Inc. (1450 Drew Ave, Suite 100, Davis, CA, USA).
Fifty milliliters of Luria-Bertani (LB) broth, supplemented with 0.5 mM riboflavin and 0.1 g/L ampicillin, were inoculated with recombinant E. coli cells containing MseMerA and shaken at 250 rpm at room temperature overnight. One liter of LB medium, as described above, was inoculated with 2 mL from the overnight culture, and shaken at 250 rpm until an OD600 of 0.5–0.7 was reached. About 2 mM IPTG was added and expression was carried out for 4 h, after which the cultures were centrifuged at 6000 × g for 10 min (4°C), with the resultant cell pellet immediately being flash frozen in liquid nitrogen and stored at −80°C. Each liter of cell culture yielded 3.0–3.5 g of cell paste.
Cell paste was subjected to three freeze/thaw cycles to facilitate lysis, after which cells were re-suspended in 5 mL Buffer A (100 mM NaCl, 50 mM MOPS with a pH of 6.7, 25 mM imidazole) per gram of cells. Lysozyme and deoxyribonuclease (DNase) were added to final concentrations of 0.1 mg/mL along with phenylmethylsulfonyl fluoride (PMSF)-saturated isopropanol to a final concentration of 0.1% v/v, and this mixture was incubated for 30 min at room temperature. Triton X-100 was then added to a final concentration of 1% v/v, and this was mixed for 30 min. The crude lysate was then clarified by centrifugation at 100,000 × g for 1 h (4°C). The resulting clarified lysate was observed to have a yellow color.
Purification of MseMerA was carried out using a 75 mL gradient from 100% Buffer A to 100% Buffer B (100 mL NaCl, 50 mM MOPS with a pH 6.7, 250 mM imidazole) on a 2 mL Ni-NTA column (Qiagen) at 3 mL/min. Seven milliliter fractions were collected and further analyzed with an SDS-PAGE gel. Fractions containing pure protein were combined and concentrated to 10 mg/mL, buffer exchanged to Buffer C (10 mM MOPS pH of 6.7), and the protein was then concentrated to 30 mg/mL and flash-frozen in liquid nitrogen. Purity of the protein was confirmed by SDS- and Native PAGE (Figure S1 in Supplementary Material). A yield of 1.5 mg of pure protein per liter of growth culture was achieved.
Activity Assay
Activity assays were carried out in 100 mM NaCl, 50 mM MOPS with a pH of 6.7, 0.2 mg/mL MseMerA, and 1 mM HgCl2, and these were initiated by the addition of 0.2 mM NADPH, similar to previously established procedures (Fox and Walsh, 1982). For kinetic studies, the concentration of Hg2+ ranged from 28.6 μM to 2.77 mM. NADPH oxidation was monitored at 338 nm using a Cary 6000 UV/Vis spectrometer equipped with a 1 × 1 Peltier. Assays were conducted from 37 to 70°C, above which temperature the rate of non-enzymatic NADPH oxidation was too high to accurately measure enzymatic activity. In order to determine the thermostability of MseMerA, an aliquot of the enzyme was assayed at 37°C and the remaining protein was boiled at 97°C for 100 min, after which the enzymatic activity was once again measured at 37°C.
Crystallization and Structure Determination
MseMerA crystals were obtained using the hanging drop method. Crystallization drops contained 0.085M TRIS (pH 8.5), 15% v/v glycerol, 14% w/v PEG400, 0.19M LiSO4, and 20 mg/mL protein. Crystals were obtained after 2 weeks, mounted on cryo loops, and shipped to the Stanford Synchrotron Radiation Lightsource for X-ray data collection. Diffraction data were collected at 100 K using the 12-2 beamline. Diffraction images were indexed, integrated, and scaled using HKL2000 (Otwinowski and Minor, 1997).
The structure of MseMerA was solved to 1.6 Å using CCP4 molecular replacement (Cowtan et al., 2011) of Tn501MerA (PDB ID: 1ZK7), which shares 37% amino acid identity with MseMerA. Model building was performed in Coot (Emsley et al., 2010) and coordinates were refined to reasonable stereochemistry at a resolution 1.6 Å (Figure S3 in Supplementary Material) using REFMAC5 (Murshudov et al., 1997). The structure was validated using MolProbity (Chen et al., 2010) and all molecular images were calculated in PyMol (Delano, 2002). Structural superimpositions were generated both with 1ZK7 (Ledwidge et al., 2005) and 4K7Z (Lian et al., 2014), in which the active site cysteines were substituted by alanines and could be solved in complex with the Hg2+ ion.
Results
Thermal Adaptation of MseMerA
Phylogenetic reconstruction of representative core (NmerA trimmed) MerA sequences revealed a number of deeply branching lineages from thermophilic taxa, consistent with previous analyses that indicate MerA likely originated in a high temperature environment (Schelert et al., 2004; Barkay et al., 2010; Boyd and Barkay, 2012). MseMerA clustered among MerA from thermophilic crenarchaeota (Figure 1). Sequence alignments reveal both the active site CXXXXC motif and C-terminal cysteines that are conserved among all MerA sequences. However, several key differences were observed that may be involved in conferring thermotolerance (Figure 2). Specifically, the thermophilic enzymes are missing regions corresponding to amino acids 66–71 and 130–134 Tn501 (Tn501MerA numbering), suggesting a reduction in loop regions in comparison to the mesophilic enzymes (Figure 2). Two sets of residues, V317 and Y441, are within putative coordination distance of the active-site mercury. These residues are substituted for an E and F, respectively, in MseMerA and other thermophiles with the exception of Hydrogenobacter thermophilus TK-6 (YNP_003432979) and Hydrogenobaculum sp. Y04AAS1 (YNP_002121876).
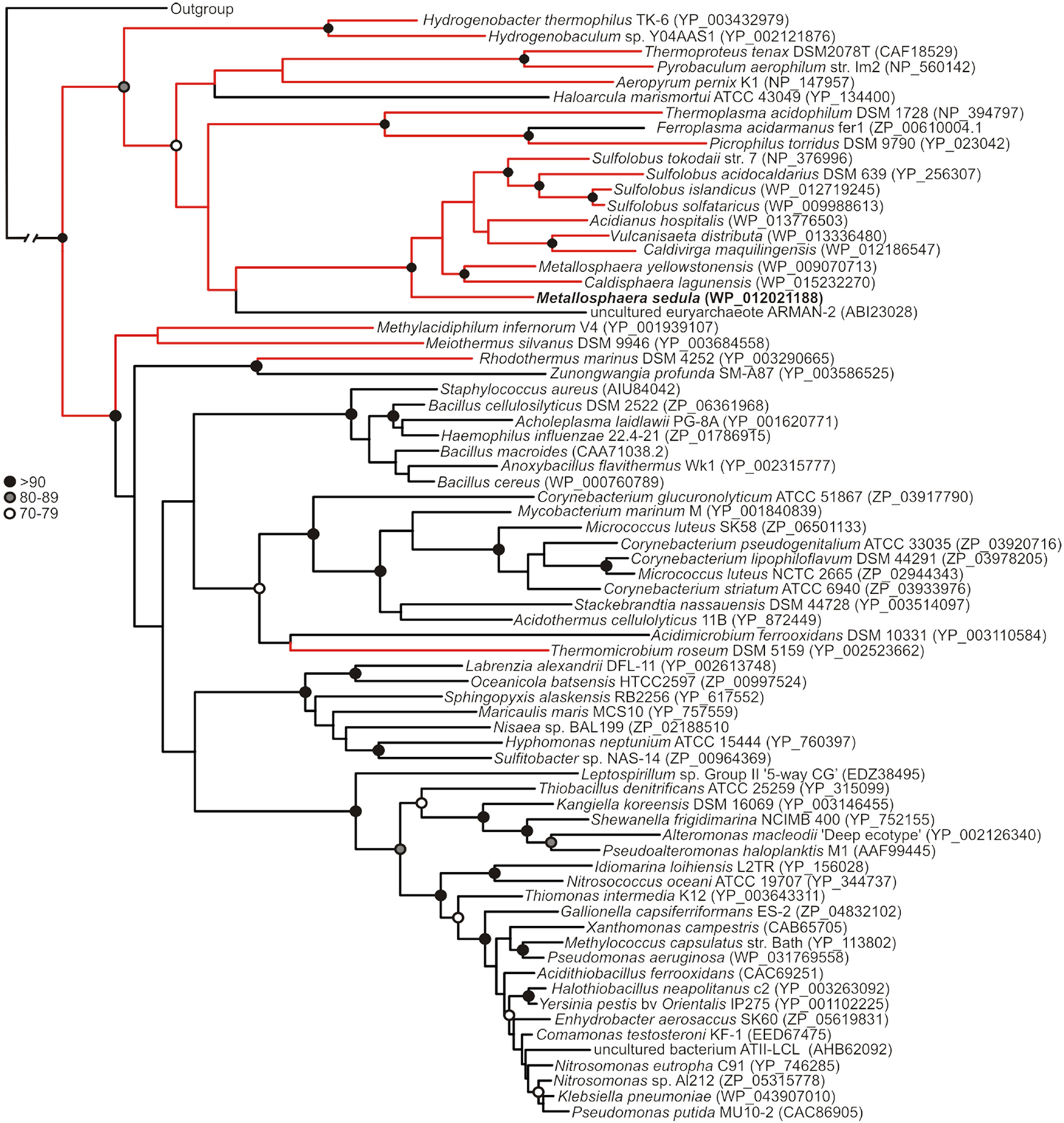
Figure 1. Maximum-likelihood phylogenetic reconstruction of MerAs, with homologs from thermophilic taxa highlighted in red. MseMerA is boldfaced. Bootstrap support is indicated by black (>90), gray (80–89), and open (70–79) circles. Nodes with no symbol exhibited bootstrap values of <70.
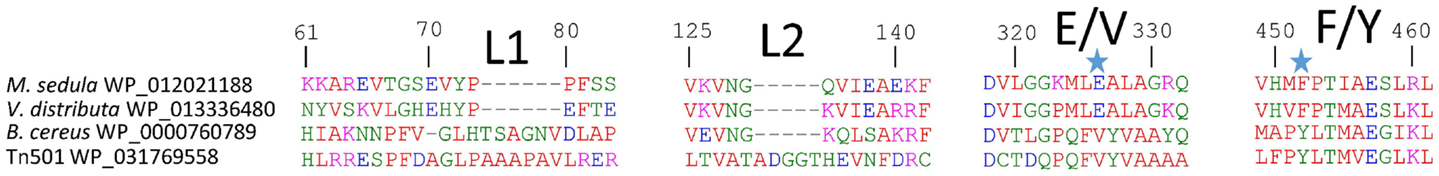
Figure 2. MseMerA aligned with other MerAs reveals two loop regions, L1 and L2, which may be involved in conferring thermostability, and two positions at 326 and 452 (highlighted with stars), where the active site region is different between thermophiles and mesophiles.
A comparison of the MseMerA crystal structure to the previously determined Tn501MerA structure (PDB: 1ZK7) (Ledwidge et al., 2005) reveals that the two structures are highly similar, with an overall C-alpha deviation of 1.5 Å rmsd as calculated by Dali Lite (McWilliam et al., 2013). Two particular loop regions are shorter in MseMerA (Figure 3A). This was further supported by VADAR (Willard et al., 2003), which calculated a 4% decrease in coil regions in MseMerA. The calculated surface area of MseMerA, 19,966.5 Å2, is slightly reduced in comparison to Tn501MerA, with a surface area of 21,217.4 Å2.
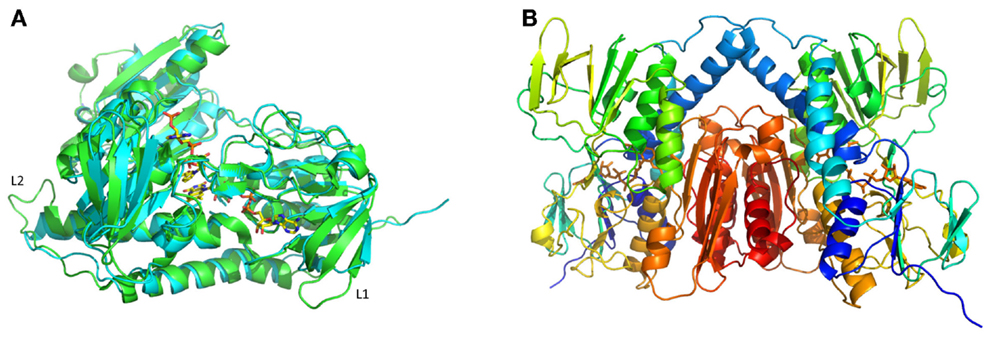
Figure 3. (A) Structural superimposition of MseMerA monomer (cyan) with Tn501MerA (green) reveals a decrease in loop regions (labeled L1 and L2) in MseMerA. (B) Cartoon representation of a dimer of MseMerA with bound FAD.
MEGA (Tamura et al., 2013) was used to compile an amino acid composition chart for the sequences examined. The thermophiles were observed to have a larger number of positively charged amino acids. VADAR calculated the total charged residues in MseMerA to be 25% of residues compared to 21% of residues in Tn501MerA, and 24% in BcMerA. An increase in ionic interactions may therefore represent a factor contributing to MerAs thermal stability (Szilágyi and Závodszky, 2000). The aliphatic index of Mse, Tn501, and Bc MerAs were calculated by ExPASy’s ProtParam tool (Gasteiger et al., 2005), and found to be 101.63, 98.65, and 97.86, respectively, again in agreement with MseMerA having higher thermostability (Ikai, 1980).
Biochemical Characterization
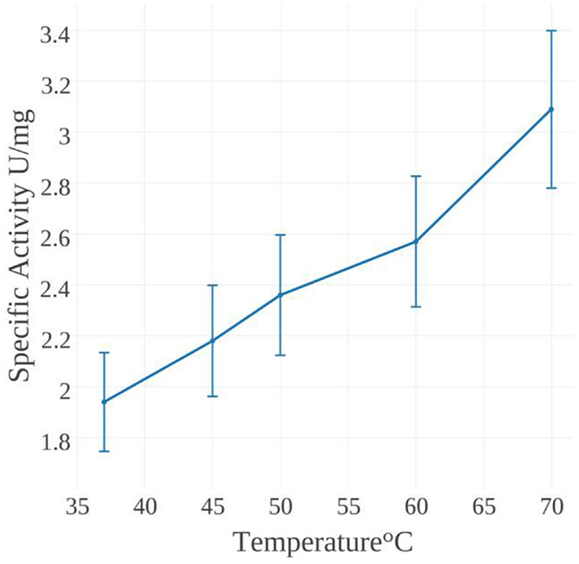
The specific activity of MseMerA was examined from 37 to 70°C (Figure 4). One unit of activity was defined as 1 μmol NADPH oxidized per minute. At 37°C, the specific activity was found to be 1.9 U/mg, increasing up to 3.1 U/mg at 70°C. Mercury dependence of MseMerA was determined, with Km values of 400 and 150 μM at 37 and 70°C, respectively. Specific activity was not determined above 70°C due to the difficulty of discriminating between enzymatic and non-enzymatic NADPH oxidation at high temperatures. The thermal stability of MseMerA was tested by incubating the enzyme at 97°C for up to 100 min, followed by assessment of enzymatic activity at 37°C. Even after 100 min of incubation at 97°C, no decrease in overall activity was observed when compared to the untreated enzyme (Figure S2 in Supplementary Material). The Kcat at 70°C was found to be 23 s−1, with a Kcat/Km of 0.15 μM−1 s−1.
Structural Characterization of MseMerA
MseMerA crystals were obtained using vapor diffusion in a precipitating solution of 14% polyethylene glycol 4000 and 0.19M lithium sulfate. These crystals belonged to space group P22121 and contained two monomers per asymmetric unit, assembled into one homodimer (Figure 3B). The crystal structure of MseMerA was solved to 1.6 Å, with R and Rfree values of 16.9 and 19.6%, respectively. Bound FAD was observed, suggesting that these molecules act to stabilize the structure. No mercury was observed in the active site. As expected based on the sequence alignment, a clear reduction in loop regions was observed in comparison to Tn501MerA (Figure 3A). No electron density for the carboxy terminus of MseMerA was identified from 440 to 448, including the conserved pair of cysteines at residues 446 and 447. This is in agreement with the carboxy terminus being able to undergo conformational changes during the catalytic cycle (Lian et al., 2014). The solved structure has been deposited in the Protein Data Bank with the accession code 4YWO.
Discussion
Bioinformatic and phylogenetic data overwhelmingly support MseMerA being a thermostable protein, as illustrated by features consistent with other enzymes from thermophiles, including a reduction in loop regions, a greater percent of charged amino acids, and an overall reduced surface area in comparison to its mesophilic counterpart. Collectively, these strategies are likely to interact synergistically to convey the high degree of thermostability observed. Retention of 100% activity after incubation at 97°C for 100 min further confirms the highly thermostable nature of MseMerA.
Though practical constraints made measuring specific activity above 70°C impossible, catalytic activity was found to increase over the range of 37–70°C, with a Vmax of 3.1 U/mg at 70°C. This places MseMerA in the range of average activity when compared to other MerAs (Table 1). The Km for Hg2+of MseMerA was found to decrease from 400 μM at 37°C to 150 μM at 70°C, indicating a higher affinity for Hg2+ ions at elevated temperatures. The Km of MseMerA is around an order of magnitude higher than that found for other MerAs (Table 1), and may be an adaptive strategy to cope with elevated Hg2+ concentrations commonly encountered in the acidic, high temperature environments where Mse resides (King et al., 2006; Boyd et al., 2009; Wang et al., 2011).
The Kcat of MseMerA is 23 s−1, which is very similar to the Kcat of ATII-LCL at 22.5 s−1 (Sayed et al., 2013) and also similar to BcMerA at 12 s−1 (Rennex et al., 1994). The higher Km value observed in MseMerA translates to the lowest overall catalytic efficiency, with a Kcat/Km of 0.15 μM−1 s−1.
Though P. aeruginosa (Pa) from which the Tn501 transposon was isolated is a mesophilic organism, the MerA enzyme was found to have optimal activity at 55–65°C, and retained full activity at 37°C even following a 10-min incubation at 100°C (Nakahara et al., 1985; Vetriani et al., 2005). Intriguingly, phylogenetic analysis indicates that Tn501MerA groups closely with the mesophiles (Figure 1). Conversely, phylogenetic analysis of MerA from a high temperature brine pool, ATII-LCL (Sayed et al., 2013), was found to group with MerA sequences from mesophilic organisms (Figure 1). While the environment from which ATII-LCL was isolated is at 68°C, the enzyme has maximum activity over a range of 30–50°C and, when measured at 37°C, was found to be half inactivated after a 10-min incubation at 75°C (Sayed et al., 2013). The ATII-LCLMerA is therefore not nearly as thermostable as MseMerA, and is not adapted to its local environment, with respect to the thermal regime, but is adapted with respect to salinity regime.
The structure of MseMerA reveals a dimeric biological assembly, as has been shown with previous structures (Schiering et al., 1991; Ledwidge et al., 2005; Lian et al., 2014). With this architecture, the active site cleft on one monomer interacts with the C-terminal domain of the opposing monomer (Figure 5; Table 1). This style of interaction is generally conserved among enzymes of the DSOR family. For example in glutathione reductase, His467, located near the C-terminus of one monomer, is necessary for catalytic function of the opposing monomer (Misra et al., 1985). In MerA, this has been substituted to a catalytically important tyrosine (Rennex et al., 1994).
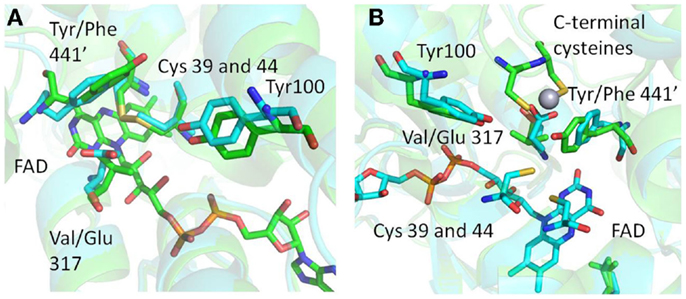
Figure 5. (A) Structural superimposition of MseMerA with Tn501MerA 1ZK7 shows the Y441′/V317 amino acids conserved in mesophiles and the F441/E317 amino acids conserved in thermophiles, suggesting an alternative Hg2+ coordination strategy in MseMerA. (B) An alternative angle of the active site environment of MseMerA superimposed onto Tn501MerA 4K7Z, which depicts the Hg2+ ion bound to the c-terminal cysteines. The monomer with the C-terminal cysteines is noted by a “prime” designation.
Structural superimposition of MseMerA (described here) with the recently solved Tn501MerA structure with bound mercury (4K7Z) reveals two specific amino acid substitutions, V317 to E, and Y441′ to F′, in the active site of MseMerA compared to Tn501MerA (numbering is by Tn501MerA 4K7Z) (Figure 5). Another residue thought to be involved in metal coordination, Y100 (in Bc structure is Y264) (Schiering et al., 1991), is strictly conserved. For Tn501MerA and BcMerA, the hydroxyl groups of Y441′ and Y100 likely act in concert to facilitate metal transfer from the C-terminal cysteines to the active site cysteines. In contrast, in MseMerA, the F441′ in the position of tyrosine in Tn501MerA lacks a hydroxyl group to coordinate the Hg2+ ion, but a glutamic acid in place of the Tn501MerA V317 provides a different residue with which the Hg2+ ion could potentially be coordinated.
The conservation of either the V/Y′ in mesophiles or the E/F′ amino acid pair in thermophiles, along with the observed positions of the amino acids, is suggestive of an alternative metal binding strategy for Hg2+ ion transfer from the C′ cysteine pair to the active site cysteines C42 and C47. In Tn501MerA and BcMerA, upon Hg2+ ion binding to the C′ cysteines, the C′ terminal region folds into the catalytic cleft, delivering the mercuric ion (Lian et al., 2014) to the conserved Y100 and Y441′, which facilitate transfer to the active site cysteines. Given that MseMerA lacks the Y441 with which to coordinate the Hg2+ion during active site delivery, the E317 is the most rational alternative.
Rennex et al. (1994) have previously substituted individual amino acids Y441F and Y100F in BcMerA. The Km for Hg2+ increased from 30 to 39 μM in the case of the Y441F variant, and decreased to 6 μM in the case of the Y100F variant. However, in both cases, the Kcat/Km was decreased around 15-fold. It is therefore likely that the observed low catalytic efficiency of the variant enzymes is due in part to a lack of a residue to coordinate the Hg2+ ion, such as the glutamic acid found in MseMerA and other thermophiles. Moreover, Sayed et al. (2013) previously demonstrated that glutamic acid residues may play a role in Hg2+ ion coordination and transfer. However, the active site glutamic acid found in MseMerA is a different site from what Sayed et al. (2013) have previously characterized. Furthermore, sequence alignment shows that the ATII-LCL enzyme has the V/Y amino acid pair (Table 1).
Both the Tn501MerA Y441′ and the MseMerA E300 are about 5 Å from the active site cysteines, although they coordinate from different positions, with the Y441′ coordinating the Hg2+ ion almost perpendicular to E317. The different placement and nature of these side chains may help explain the higher Km observed in MseMerA relative to homologs from mesophilic organisms. Since the high Hg2+ concentrations are common features of high temperature environments, these differences may reflect adaptations to function at elevated Hg2+ concentrations and as such represent the structural determinants of specificity for mercuric reductases. Highly specific stable enzymes, especially those that catalyze oxidation-reduction reactions coupled to the specific molecular recognition, could potentially be used as chemical sensors in which the redox chemistry could be coupled to produce an amplifiable electrical signal.
In conclusion, here we present a characterization of the thermostable mercuric reductase from M. sedula. We show that the enzyme is highly resistant to heat treatment while retaining similar catalytic rates to other characterized MerAs. The enzyme appears to have a potentially different way of coordinating Hg2+ and has a lower affinity for Hg2+ ions than previously characterized enzymes. Considering that Mse is a thermophile and its MerA is likely to harbor properties more similar to those of primitive MerA that evolved in a high temperature environments (Barkay et al., 2010), these results may indicate that the activity of MerA has been refined through evolutionary time to successfully detoxify environmental Hg2+ at lower concentrations than those that are naturally present in thermal environments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by a grant from the Air Force Office of Scientific Research (FA9550-14-110147) to JP, MP, and EB. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology program is supported by the US Department of Energy, Office of Biological and Environmental Research, the US National Institutes of Health, National Center for Research Resources, Biomedical Technology program, and the US National Institute of General Medical Sciences. EB acknowledges support from a grant from the National Science Foundation (EAR-1123689).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fbioe.2015.00097
References
Adami, M., Martini, M., and Piras, L. (1995). Characterization and enzymatic application of a redox potential biosensor based on a silicon transducer. Biosens. Bioelectron. 10, 633–638. doi: 10.1016/0956-5663(95)96939-V
Argyrou, A., and Blanchard, J. S. (2004). Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 78, 89–142. doi:10.1016/s0079-6603(04)78003-4
Auernik, K. S., Cooper, C. R., and Kelly, R. M. (2008a). Life in hot acid: pathway analyses in extremely thermoacidophilic archaea. Curr. Opin. Biotechnol. 19, 445–453. doi:10.1016/j.copbio.2008.08.001
Auernik, K. S., Maezato, Y., Blum, P. H., and Kelly, R. M. (2008b). The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74, 682–692. doi:10.1128/aem.02019-07
Barkay, T., Kritee, K., Boyd, E., and Geesey, G. (2010). A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 12, 2904–2917. doi:10.1111/j.1462-2920.2010.02260.x
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi:10.1016/S0168-6445(03)00046-9
Batten, K. M., and Scow, K. M. (2003). Sediment microbial community composition and methylmercury pollution at four mercury mine-impacted sites. Microb. Ecol. 46, 429–441. doi:10.1007/s00248-003-1005-z
Boyd, E. S., and Barkay, T. (2012). The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 3:349. doi:10.3389/fmicb.2012.00349
Boyd, E. S., King, S., Tomberlin, J. K., Nordstrom, D. K., Krabbenhoft, D. P., Barkay, T., et al. (2009). Methylmercury enters an aquatic food web through acidophilic microbial mats in Yellowstone National Park, Wyoming. Environ. Microbiol. 11, 950–959. doi:10.1111/j.1462-2920.2008.01820.x
Chen, V. B., Arendall, W. B. III, Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. doi:10.1107/S0907444909042073
Cowtan, K., Emsley, P., and Wilson, K. S. (2011). From crystal to structure with CCP4. Acta Crystallogr. D Biol. Crystallogr. 67, 233–234. doi:10.1107/S0907444911007578
Delano, W. L. (2002). The PyMOL Molecular Graphics System. San Carlos, CA: PyMol. Available at: http://www.pymol.org
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi:10.1107/S0907444910007493
Fox, B., and Walsh, C. T. (1982). Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J. Biol. Chem. 257, 2498–2503.
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. E., Wilkins, M., Appel, R., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. Walker (Totowa, NJ: Humana Press), 571–607.
Gertrud, H., Carola, S., Agata, G., and Karl, O. S. (1989). Metallosphaera sedula gen, and sp. nov. Represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst. Appl. Microbiol. 12, 38–47. doi:10.1016/S0723-2020
Ghosh, S., Sadhukhan, P. C., Chaudhuri, J., Ghosh, D. K., and Mandal, A. (1999). Purification and properties of mercuric reductase from Azotobacter chroococcum. J. Appl. Microbiol. 86, 7–12. doi:10.1046/j.1365-2672.1999.00605.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Han, S. B., Zhu, M., Yuan, Z. B., and Li, X. (2001). A methylene blue-mediated enzyme electrode for the determination of trace mercury(II), mercury(I), methylmercury, and mercury-glutathione complex. Biosens. Bioelectron. 16, 9–16. doi:10.1016/s0956-5663(00)00114-7
Huber, R., Huber, H., and Stetter, K. O. (2000). Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev. 24, 615–623. doi:10.1111/j.1574-6976.2000.tb00562.x
Ikai, A. (1980). Thermostability and aliphatic index of globular proteins. J. Biochem. 88, 1895–1898.
King, S. A., Behnke, S., Slack, K., Krabbenhoft, D. P., Nordstrom, D. K., Burr, M. D., et al. (2006). Mercury in water and biomass of microbial communities in hot springs of Yellowstone National Park, USA. Appl. Geochem. 21, 1868–1879. doi:10.1016/j.apgeochem.2006.08.004
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Ledwidge, R., Patel, B., Dong, A., Fiedler, D., Falkowski, M., Zelikova, J., et al. (2005). NmerA, the metal binding domain of mercuric ion reductase, removes Hg2+ from proteins, delivers it to the catalytic core, and protects cells under glutathione-depleted conditions†,‡. Biochemistry 44, 11402–11416. doi:10.1021/bi050519d
Lian, P., Guo, H.-B., Riccardi, D., Dong, A., Parks, J. M., Xu, Q., et al. (2014). X-ray structure of a Hg2+ complex of mercuric reductase (MerA) and quantum mechanical/molecular mechanical study of Hg2+ Transfer between the c-terminal and buried catalytic site cysteine pairs. Biochemistry 53, 7211–7222. doi:10.1021/bi500608u
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600. doi:10.1093/nar/gkt376
Misra, T. K., Brown, N. L., Haberstroh, L., Schmidt, A., Goddette, D., and Silver, S. (1985). Mercuric reductase structural genes from plasmid R100 and transposon Tn501: functional domains of the enzyme. Gene 34, 253–262. doi:10.1016/0378-1119(85)90134-9
Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. doi:10.1107/S0907444996012255
Nakahara, H., Schottel, J. L., Yamada, T., Miyakawa, Y., Asakawa, M., Harville, J., et al. (1985). Mercuric reductase enzymes from Streptomyces species and group B Streptococcus. J. Gen. Microbiol. 131, 1053–1059.
Nascimento, A. M., and Chartone-Souza, E. (2003). Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet. Mol. Res. 2, 92–101.
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi:10.1016/S0168-6445(03)00048-2
Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326. doi:10.1016/s0076-6879(97)76066-x
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Rennex, D., Pickett, M., and Bradley, M. (1994). In vivo and in vitro effects of mutagenesis of active site tyrosine residues of mercuric reductase. FEBS Lett. 355, 220–222. doi:10.1016/0014-5793(94)01180-X
Sayed, A., Ghazy, M. A., Ferreira, A. J. S., Setubal, J. C., Chambergo, F. S., Ouf, A., et al. (2013). A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red Sea. J. Biol. Chem. 289, 1675–1687. doi:10.1074/jbc.M113.493429
Schelert, J., Dixit, V., Hoang, V., Simbahan, J., Drozda, M., and Blum, P. (2004). Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186, 427–437. doi:10.1128/JB.186.2.427-437.2004
Schelert, J., Rudrappa, D., Johnson, T., and Blum, P. (2013). Role of MerH in mercury resistance in the archaeon Sulfolobus solfataricus. Microbiology 159, 1198–1208. doi:10.1099/mic.0.065854-0
Schiering, N., Kabsch, W., Moore, M. J., Distefano, M. D., Walsh, C. T., and Pai, E. F. (1991). Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature 352, 168–172. doi:10.1038/352168a0
Schottel, J. L. (1978). The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J. Biol. Chem. 253, 4341–4349.
Simbahan, J., Kurth, E., Schelert, J., Dillman, A., Moriyama, E., Jovanovich, S., et al. (2005). Community analysis of a mercury hot spring supports occurrence of domain-specific forms of mercuric reductase. Appl. Environ. Microbiol. 71, 8836–8845. doi:10.1128/aem.71.12.8836-8845.2005
Szilágyi, A., and Závodszky, P. (2000). Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8, 493–504. doi:10.1016/S0969-2126(00)00133-7
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi:10.1093/molbev/mst197
Vetriani, C., Chew, Y. S., Miller, S. M., Yagi, J., Coombs, J., Lutz, R. A., et al. (2005). Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71, 220–226. doi:10.1128/aem.71.1.220-226.2005
Wang, Y. P., Boyd, E., Crane, S., Lu-Irving, P., Krabbenhoft, D., King, S., et al. (2011). Environmental conditions constrain the distribution and diversity of archaeal merA in Yellowstone National Park, Wyoming, U.S.A. Microb. Ecol. 62, 739–752. doi:10.1007/s00248-011-9890-z
Willard, L., Ranjan, A., Zhang, H., Monzavi, H., Boyko, R. F., Sykes, B. D., et al. (2003). VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31, 3316–3319. doi:10.1093/nar/gkg565
Zeroual, Y., Moutaouakkil, A., Dzairi, F. Z., Talbi, M., Chung, P. U., Lee, K., et al. (2003). Purification and characterization of cytosolic mercuric reductase from Klebsiella pneumoniae. Ann. Microbiol. 53, 149–160.
Keywords: mercuric reductase, mercury detoxification, thermophile, thermostability, structure, biosensor, MerA
Citation: Artz JH, White SN, Zadvornyy OA, Fugate CJ, Hicks D, Gauss GH, Posewitz MC, Boyd ES and Peters JW (2015) Biochemical and structural properties of a thermostable mercuric ion reductase from Metallosphaera sedula. Front. Bioeng. Biotechnol. 3:97. doi: 10.3389/fbioe.2015.00097
Received: 10 April 2015; Accepted: 19 June 2015;
Published: 13 July 2015
Edited by:
Noha M. Mesbah, Suez Canal University, EgyptCopyright: © 2015 Artz, White, Zadvornyy, Fugate, Hicks, Gauss, Posewitz, Boyd and Peters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John W. Peters, Department of Chemistry and Biochemistry, Montana State University, 224 Chemistry and Biochemistry Building, Bozeman, MT 59717, USA, john.peters@chemistry.montana.edu
 Jacob H. Artz
Jacob H. Artz Spencer N. White
Spencer N. White Oleg A. Zadvornyy
Oleg A. Zadvornyy Corey J. Fugate1
Corey J. Fugate1
 Danny Hicks
Danny Hicks Matthew C. Posewitz
Matthew C. Posewitz Eric S. Boyd
Eric S. Boyd John W. Peters
John W. Peters