Production of H5N1 Influenza Virus Matrix Protein 2 Ectodomain Protein Bodies in Tobacco Plants and in Insect Cells as a Candidate Universal Influenza Vaccine
- 1Biopharming Research Unit, Department of Molecular and Cell Biology, University of Cape Town, Rondebosch, South Africa
- 2Institute of Infectious Disease and Molecular Medicine, Faculty of Heath Science, University of Cape Town, Cape Town, South Africa
The spread of influenza A viruses is partially controlled and prevented by vaccination. The matrix protein 2 ectodomain (M2e) is the most conserved sequence in influenza A viruses, and is therefore a good potential target for a vaccine to protect against multiple virus subtypes. We explored the feasibility of an M2e-based universal influenza A vaccine candidate based on the highly pathogenic avian influenza A virus, H5N1. A synthetic M2e gene was human- and plant-codon optimized and fused in-frame with a sequence encoding the N-terminal proline-rich domain (Zera®) of the γ-zein protein of maize. Zera®M2e was expressed transiently in Nicotiana benthamiana and Sf21 baculovirus/insect cell expression systems, and Zera®M2e protein bodies (PBs) were successfully produced in both expression systems. The plant-produced Zera®M2e PBs were purified and injected into Balb/c mice. Western blot analysis using insect cell-produced Zera®M2e PBs and multiple tandem M2e sequences (5xM2e) fused with the avian influenza H5N1 transmembrane and cytosolic tail (5xM2e_tHA) confirmed the presence of M2e-specific antibodies in immunized mice sera. The immunogenicity of the Zera®M2e indicates that our plant-produced protein has potential as an inexpensive universal influenza A vaccine.
Introduction
Influenza A viruses can be highly contagious, causing acute viral respiratory diseases seasonally in the human population (Cox and Subbarao, 1999; Thompson et al., 2003). Currently, vaccination with selected inactivated influenza virus strains is the most effective way of reducing the morbidity and mortality caused by these viruses (Cox and Subbarao, 1999). Influenza A viruses are divided into subtypes by their two surface glycoproteins, the hemagglutinin (HA) and neuraminidase (NA). HA and NA are the primary targets for vaccine development as they elicit neutralizing immune responses (Johansson et al., 1989; Fiore et al., 2009). Unfortunately, due to the high mutation rate of these glycoproteins, influenza vaccines need to be manufactured seasonally in order to be effective against the current circulating strains (Webster et al., 1992). It would therefore be ideal to develop a universal vaccine that is cross-protective against multiple influenza A virus strains as well as against subtypes (Price et al., 2010; Andersson et al., 2012).
There is a type III integral membrane protein (M2) present on the surface of the influenza A virus particle (Lamb et al., 1985). It functions as a pH-activated ion channel (viroporin) and is required for viral infection (Black et al., 1993). It also prevents low pH-induced structural changes in HA during maturation (Sugrue and Hay, 1991), and thus plays a role in viral assembly (Chen et al., 2008; Rossman et al., 2010). M2 (97 amino acids) consists of an N-terminal ectodomain (M2e) (23 amino acids), a lipid bilayer spanning single transmembrane domain (19 amino acids) and a C-terminal cytosolic tail (54 amino acids), and polymerizes into homotetramers in the virion envelope (Pinto and Lamb, 2006). The M2e sequence is highly conserved in all influenza A viruses (Black et al., 1993; Betakova, 2007). It also plays an important role in the incorporation into virions (Park et al., 1998), and it can elicit antibodies that can neutralize virion infectivity (Fiers et al., 2004; Feng et al., 2006). Thus, this domain has the potential to be used as an influenza A virus universal vaccine. However, M2e is covered by the HA and NA proteins in intact virions, and it is therefore unable to react effectively with immune effector cells, making it poorly immunogenic (Lamb et al., 1985; Jegerlehner et al., 2004; Feng et al., 2006). In attempts to enhance its immunogenicity, M2e has been linked to different carrier molecules, such as the TLR5 ligand flagellin (Huleatt et al., 2007; Mardanova et al., 2015), the surface of virus-like particles (VLPs) (Matic et al., 2011), and as a fusion peptide on β-glucuronidase (Firsov et al., 2015) and HA (Stanekova et al., 2011). De Filette et al. (2005) used hepatitis B virus core protein (HBc) as a carrier, and fused M2e to either the C-terminal of HBc or inserted in the immune dominant loop of HBc. Immunization of mice with this HBc-M2e candidate vaccine resulted in 100% protection against lethal challenge (Fiers et al., 2004; De Filette et al., 2005, 2008).
Influenza antigens have been successfully produced using insect cell expression systems. Baculovirus-expressed influenza vaccines can be produced rapidly, which is necessary when taking into account that currently circulating strains need to be assessed annually. The resultant insect cell-expressed product is considered to be both safe and of a high standard (Safdar and Cox, 2007; Cox, 2008). Producing M2e tetramers in baculovirus insect cell expression systems and accumulating them into nanoclusters results in increased humoral and cellular immunogenicity (Wang et al., 2014).
As an alternative approach, numerous researchers have successfully produced influenza antigens in tobacco plants – and in particular HA (D’Aoust et al., 2008; Shoji et al., 2008; Mortimer et al., 2012). Plant expression systems are advantageous due to their ability to carry out post-translational modifications similar to other eukaryotes, and in rapidly producing large quantities of antigen. This system is more economical since plants do not need expensive material for growth and maintenance, and it reduces concerns over human pathogens contaminating vaccine preparations (Nemchinov and Natilla, 2007; Gomez et al., 2009; Rybicki, 2009). Nemchinov and Natilla (2007) developed a candidate plant-based universal influenza vaccine by displaying the M2e epitope on the capsid protein (CP) of cucumber mosaic virus Ixora strain (CMV-Ix) in a potato virus X (PVX)-based vector. The resulting plant-based chimeric CMV capsids reacted specifically to antibodies raised against the synthetic M2e, indicating the potential of this system.
This study forms part of an ongoing initiative to investigate and establish a rapid-response vaccine production platform to deal with future influenza pandemics in South Africa. The highly pathogenic H5N1 influenza A virus, with a mortality rate of up to 60% in humans (http://www.who.int/), was chosen for this purpose. To date, human-to-human transmissions are limited but the likelihood of H5N1 mutating into a strain that facilitates transfer necessitates efficient pandemic vaccination preparedness strategies and awareness (Webster and Govorkova, 2006; Imai et al., 2013; Kaplan and Webby, 2013). To date, potential plant-produced subunit HA vaccines (Mortimer et al., 2012) as well as HA DNA vaccine candidates (Mortimer et al., 2013) have been created as part of this South African initiative.
Fusion of small or soluble proteins to a signal sequence that drives the assembly and sequestration of the protein bodies (PBs) (Torrent et al., 2009) can significantly increase the immunogenicity of the protein. Accordingly, for this study, we investigated the fusion of a consensus M2e sequence to a signal tag (Zera®, ERA Biotech) that targets the recombinant protein to form PBs. This tag has previously been shown to dramatically improve yields of non-structural papillomavirus protein (E7SH) in plants as well as to have adjuvant properties (Whitehead et al., 2014). Zera® had adjuvant activity, whether fused to E7SH or simply added to it, which could be highly advantageous in a vaccine candidate. The N-terminal proline-rich domain of maize γ-zein (Zera® tag) is characterized by 4 domains: these are a 19 amino acid signal peptide, a repeat domain containing 8 repeats of the sequence PPPVHL, a Pro-X domain including numerous proline residues as well as a hydrophobic cysteine rich C-terminal domain, and lastly, a sequence that retains it in the endoplasmic reticulum (ER). This allows for the formation of membrane-bound PBs, thereby protecting the recombinant protein from proteolytic degradation inside the host cells, and concentrating and sequestering the recombinant protein. PBs are easily concentrated and partially purified by simple centrifugation, and the polypeptide is generally water soluble in the presence of reducing agents, which greatly facilitates and simplifies recombinant protein purification (Torrent et al., 2009).
For this study, we determined the feasibility of creating an immunogenic M2e candidate vaccine by transiently expressing Zera®M2e PBs in tobacco plants and in insect cells. The protein expressed by recombinant baculovirus cells Sf21 was used as an experimental control reagent. M2e was fused to Zera® to enable protein purification and to increase the immunogenicity of the protein (Torrent et al., 2009; Whitehead et al., 2014). Codon optimization has been widely used to enhance protein expression in heterologous systems (Gouy and Gautier, 1982). The Zera®M2e gene was codon optimized for this study such that it either displayed characteristics of abundantly expressed plant genes (Nicotiana benthamiana codon optimized) or human genes (human-codon optimized), as we have found it necessary to empirically determine codon preferences in other studies (Maclean et al., 2007). Immunogenicity of the PBs isolated from plants was established by immunization of mice, and analysis of the immune sera for the presence of antibodies against M2e.
Materials and Methods
Identification and Synthesis of Zera®M2e Peptide
Multiple avian and human influenza A H5N1 virus M2e sequences were retrieved from GenBank and aligned using Clustal X (Larkin et al., 2007). From these, four sequences were selected (EU590690, EU590684, EU146698, and EU263984) to create a consensus sequence, SLLTEVETPTRNEWECRCSDSSD, which corresponded exactly to the EU263984 sequence [A/human/China/GD02/2006(H5N1)] (Figure 1). To create the Zera®M2e sequence, the Zera® sequence (ERA Biotech), including an enterokinase cleavage site (DDDDK) (Whitehead et al., 2014), was synthesized and inserted upstream of the M2e consensus sequence. The Zera®M2e nucleotide sequence was both plant- and human-codon optimized, and synthesized by GeneArt (Germany).

Figure 1. Avian and human influenza A H5N1 virus M2e sequences retrieved from GenBank and aligned using Clustal X. EU590690 turkey, EU590684 houbara bustard, EU263984 human, and EU146698 human. The 23 amino acid ectodomain is indicated by the red square. Differences in the amino acid sequence are indicated in different colors.
Construction Plant Recombinant Vector
For plant expression, both plant- and human-codon optimized Zera®M2e were cloned into the plant expression vector pTRAc (GenBank ID: AY027531) using AflIII and XhoI restriction enzyme sites (pTRAc-Zera®M2e). The pTRAc vector allows protein expression in the cytoplasm (Maclean et al., 2007), with subsequent targeting to the ER by the Zera® sequence. The plasmids were transformed into E. coli DH5α and recombinant bacterial colonies were confirmed by PCR using Zera®M2e primers (Fw: 5′-ATGCGGGTGCTGCTGGTC-3′ and Rev: 5′-TGGGTGTCTCCACCTCGGTC-3′). The integrity of the plasmids was confirmed by restriction digest mapping with XhoI and AflIII restriction enzymes as well as sequencing. The pTRAc-Zera®M2e plasmids were subsequently transformed into Agrobacterium tumefaciens GV3101 via electroporation (Maclean et al., 2007).
Expression and Purification of Zera®M2e in Nicotiana benthamiana
Agrobacterium tumefaciens-mediated transient expression in N. benthamiana plants was performed according to Mortimer et al. (2012). In short, recombinant plant- and human-codon optimized pTRAc-Zera®M2e plasmids were vacuum infiltrated into 6-week-old plants, with co-infiltration of A. tumefaciens LBA4404 (pBIN-NSs) containing the NSs gene silencing suppressor of tomato-spotted wilt virus (TSWV) (Marcel Prins, Laboratory of Virology, Wageningen, The Netherlands); this enhances gene expression by suppressing post-translational gene silencing (Takeda et al., 2002).
Infiltrated plant tissue was harvested 8 days post infiltration (dpi), followed by grinding in liquid nitrogen with a mortar and pestle, after which the extract was homogenized in the Zera® extraction buffer [100 mM Tris (pH 8), 0.5M NaCl, 50 mM MgCl2, and 10 mM EDTA]. The homogenate was filtered through two layers of Miracloth (Merck) and purified by ultracentrifugation (Beckman SW32Ti rotor) at 21,600 × g for 2 h through a 60% sucrose cushion.
Protein expression was assessed by western blot analysis, with proteins resolved on 15% SDS-PAGE gels. The primary antibody, rabbit anti-Zera® polyclonal antibody (provided by ERA Biotech, Spain), was used at a dilution of 1:7000 together with a secondary goat anti-rabbit antibody (Sigma, Steinheim, Germany) at 1:7000 dilution. Nitro blue tetrazolium chloride/5-bromo-4 chloro-3-indolyl phosphate (NBT/BCIP) phosphate substrate (KPL, Gaithersburg, MD, USA) was used for detection. Plant-produced Zera®M2e was quantified by comparing band intensities of the Zera®M2e to known bovine serum albumin (BSA) concentrations by gel densitometry (Gene Genius Bioediting system, Syngene).
Construction and Expression of Zera®M2e in Insect Cells
For insect cell expression, plant- and human-codon optimized Zera®M2e was cloned into the pFastBac Dual vector (InVitrogen, Carlsbad, CA, USA) between the polyhedrin (PPH) promoter and Tn7L terminator using EcoRI and PstI restriction sites, resulting in pFastBac-Zera®M2e. Recombinant plasmids screened by PCR with pFastBac primers (Fw: 5′-GATGGTGGGACGGTATGAATAATCC-3′ and Rev: 5′-GGTATTGTCTCCTTCCGTGTTTGA-3′). The integrity of the plasmids was confirmed by plasmid mapping with EcoRI and PstI restriction enzymes and sequencing. Recombinant bacmid DNA was obtained by transposition of pFastBac-Zera®M2e into E. coli DH10Bac according to the manufactures instructions (InVitrogen, Carlsbad, CA, USA).
Recombinant baculoviruses (rBV) Sf 21 cells containing plant- and human-codon optimized Zera®M2e were generated, and plaque assays to determine rBV titers were performed according to the Bac-to-Bac© baculovirus expression system manufacturer’s protocols (InVitrogen, Carlsbad, CA, USA). TC Plates were stained with 1 g/ml neutral red solution (Sigma, Steinheim, Germany) to visualize individual plaques. Protein expression and purification analysis are as described for the plant-produced proteins.
Animal Trials and Serum Analysis
Only the plant-produced Zera®M2e PB yields were judged to be sufficient for animal trials. Accordingly, 20 female Balb/c mice (7 weeks old) were divided into two groups: (a) plant-produced Zera®M2e PB and (b) PBS negative control group. A dose of 4.5 μg Zera®M2e PB was administered intramuscularly (I.M.) to mice, into each anterior tibialis muscle. Four doses were administered at 2-week intervals on days 0, 14, 28, and 31. Pre-vaccination serum was collected 3 days prior to vaccination. Following vaccination, sera were collected before each boost (on days 14, 28, and 31), and stored at −20°C until for further analysis. Eleven days after the final dose, animals were euthanized (day 42). The animal experiments were approved by the University of Cape Town’s (UCT) Animal Ethics Committee (HSFAEC 009/001).
Western blots were performed to determine the presence of Zera®M2e-specific antibodies in the mouse sera. Our insect cell- and plant-produced Zera®M2e PB samples were resolved separately on 15% SDS-PAGE gels, followed by transfer onto a nylon membrane (Armersham, Bioscience, UK) by semi-dry blotting (BioRad Hercules, CA, USA). The membranes were cut into individual strips and were incubated in 1:5000 dilutions of serum from mice injected with pTRAc-Zera®M2e PB and PBS, respectively. The secondary goat anti-mouse antibody (Sigma, Steinheim, Germany) was used at 1:7000 dilutions. As a positive control, the commercial rabbit polyclonal anti-M2 antibody (ab65086) (Abcam, Cambridge, UK) was used at a 1:5000 dilution followed by the secondary goat anti-rabbit antibody (Sigma), at a 1: 7000 dilution.
To assess if the sera did not only bind the Zera® but also the M2e, western blots were performed with a construct encoding multiple M2e (5xM2e) fused with the avian influenza H5N1 (A/Vietnam/1194/2004 H5N1) transmembrane and cytosolic tail (5xM2e_tHA). Crude plant extract containing the 5xM2e_tHA protein was resolved on 12% SDS-PAGE gels, and the protein was probed on blots with a 1:100 sera dilution. As a positive control, 5xM2e_tHA was probed with 1:5000 anti-M2 monoclonal antibody (14C2). Alkaline phosphate-conjugated goat anti-mouse IgG was used as a secondary antibody at a 1:10,000 dilution.
Results
Expression of Recombinant Protein in N. benthamiana
Gene-codon optimization has been shown to significantly enhance gene expression in plants, and especially if the genes have a high GC content. The Zera®M2e gene was plant- and human-codon optimized and successfully cloned into plant expression vector pTRAc. Protein expression analysis revealed that both plant- and human-codon optimized Zera®M2e PB were successfully expressed in N. benthamiana 8 dpi. In western blots, an expected band of 17 kDa corresponding to the M2e epitope fused with Zera® tag was observed. There were no differences in the expression levels between the plant- and human-codon optimized Zera®M2e, indicating that codon optimization did not influence gene translation and expression in the plant expression system (Figure 2A).
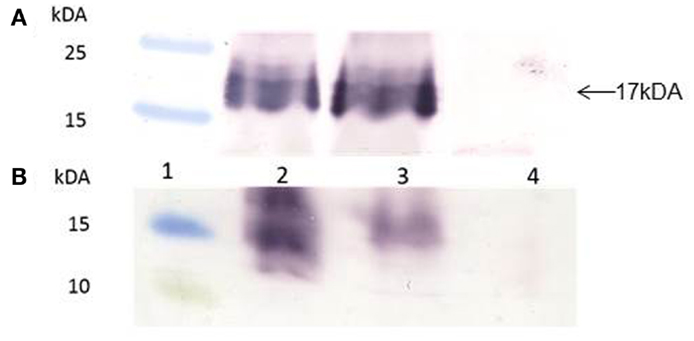
Figure 2. Western blots of human- and plant-codon optimized recombinant Zera®M2e protein bodies (PBs). Protein expression was detected with anti-Zera® antibody and goat anti-rabbit antibody. (A) Transient expression in N. benthamiana. Plants were harvested 8 days post infiltration. Lane 1 contained PageRuler™ Prestained protein ladder (Fermentas), lane 2 contained the Zera®M2e plant-codon optimized protein, lane 3 contained the Zera®M2e human-codon optimized, and lane 4 contained non-infiltrated control plant. (B) Expression of recombinant baculovirus in Sf21 insect cells, 72 h post infection (hpi), using the Bac-to-Bac© baculovirus expression system. Lane 1 contained the PageRuler™ Prestained protein ladder (Fermentas), lane 2 contained the recombinant human-codon-optimized Zera®M2e PB, lane 3 is the recombinant plant-codon-optimized Zera®M2e, and lane 4 contained the negative cell lysate transfection control.
Expression of Recombinant Protein in Insect Cells
For insect cell expression, plant- and human-codon optimized Zera®M2e were successfully cloned into pFastBac Dual vector under the control of the pH10 promoter and expressed in Sf21 insect cells 72 h post infection (hpi) (Figure 2B). The plant-codon optimized gene expression was weak as assessed by western blot compared to the human-codon optimized gene. In this case, human-codon usage was more favorable for the insect cell expression system.
Purification of Recombinant Protein
The PBs can be easily purified by centrifugation through a sucrose cushion (Torrent et al., 2009). Ultracentrifugal concentration through a sucrose cushion showed that the Zera®M2e PBs formed insoluble pellets. The pelleted insect cell-produced Zera®M2e was detectable on western blots but was undetectable on Coomassie-stained SDS-PAGE gels, indicating low protein concentration. The concentrated and partially purified plant-produced Zera®M2e PB was visible on Coomassie-stained SDS-PAGE gels and was quantified by comparing it to known BSA concentrations run on stained gels (Figure 3). Bands corresponding to the monomeric (17 kDa), dimeric (34 kDa), and tetrameric (51 kDa) forms of the Zera®M2e PB were observed (Figure 3). Only the band corresponding to the putative monomeric form was used for quantitation, to give estimated expression levels ranging from 125 to 205 mg Zera®M2e PB/kg fresh weight (FW) as measured by densitometry. The higher yielding plant-produced Zera®M2e PB was used for animal trials.
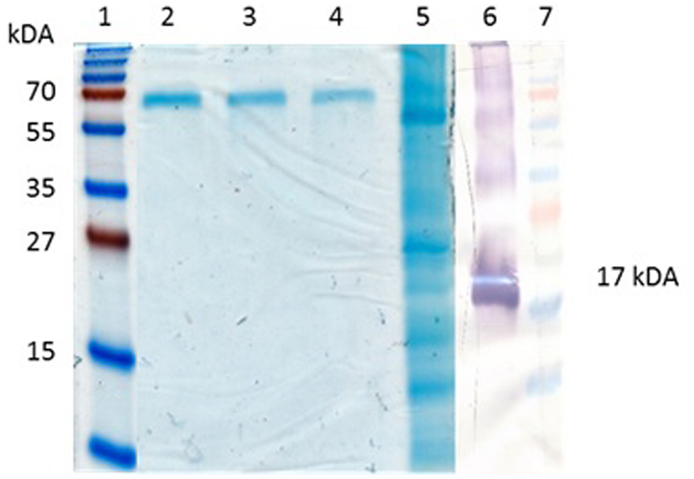
Figure 3. Quantification of Zera®M2e produced in plants. Bovine serum albumin (BSA) was used as a standard. Lane 1 and 7 contained PageRuler™ Prestained protein ladder (Fermentas), lane 2 contained 6.26 mg/ml, lane 3 contained 3.13 mg/ml, lane 4 contained 1.56 mg/ml, and lane 5 contained partially purified Zera®M2e plant-codon optimized protein. While lane 6 is a western blot containing the partially purified Zera®M2e plant-codon optimized protein detected with anti-Zera®antibody.
Animal Serum Analysis
Ten mice were immunized with the plant-produced Zera®M2e PB. No clinical manifestation was observed after the injection of the PBs in any of the mice. After the fourth immunization, the sera were analyzed for the presence of Zera®M2e-specific antibodies. Western blots indicated that the immune sera successfully reacted with the plant-produced Zera®M2e PB at a dilution of 1:5000, indicating a high Zera®M2e-specific antibody titer. Sera from PBS-inoculated control mice did not bind Zera®M2e PB (Figure S1 in Supplementary Material).
When the mouse sera were tested with our plant-produced Zera®M2e PB, a high level of background was observed on the western blots even when the sera were diluted 1:40,000, which made it difficult to identify the expected band sizes (Figure S1 in Supplementary Material). This is because our candidate vaccine was produced in plants, and therefore the sera also reacted with high specificity to plant proteins contaminating the preparations. In an attempt to lower the background for more accurate results, the sera were then analyzed using our insect cell-produced Zera®M2e PB (Figure 4). The immune sera reacted far more specifically, with a distinct band corresponding to the monomeric Zera®M2e PB.
Figure 4. Detection of Zera®M2e-specific antibodies in plant-produced Zera®M2e immunized mice sera. Insect cell-produced Zera®M2e protein bodies (PBs) were loaded in each lane, and then the membrane was cut into strips and probed with individual mouse serum. Lane 1 contains PageRuler™ Prestained protein ladder (Fermentas), lane 2 contains the positive control, i.e., Zera®M2e PB detected with a commercial M2 primary antibody (1:5000) (ab65086, Abcam, Cambridge, UK). Lane 3–6 were detected with mice sera from mice immunized with plant-produced Zera®M2e PB (1:5000) and Lane 7 contains negative control sera: mice immunized with PBS.
To be certain whether the mouse sera produced reacted specifically with the M2e peptide, the sera were tested by western blot against the plant-produced 5xM2e_tHA, which contains no Zera® sequence (Figure 5). The serum strongly reacted with the 35-kDa multiple protein dimer and a 70-kDa trimeric form, affirming that indeed the mouse sera did not only have antibodies against Zera® peptide but also against the M2e sequence.
Figure 5. Confirmation of specificity of the Zera®M2e mice sera against the fused M2e plant-produced protein. Equal volume of plant-produced 5xM2e_tHA proteins were loaded in each lane. The membranes were cut in the middle and each half was probed either with the commercial M2 antibody or mice serum from mice immunized with plant-produced Zera®M2e PB. Lanes 1 and 5 contain Agrobacterium-infiltrated crude plant extract (negative control), Lanes 2 and 4 contain the 5xM2e_tHA plant crude extract, and Lane 3 contains PageRuler™ Prestained protein ladder (Fermentas). Lanes 1 and 2 were detected with commercial M2 (14C2) primary antibody (1:5000). Lanes 4 and 5 were detected with mice sera from mice immunized with plant-produced Zera®M2e PB (1:100).
Discussion
In this study, we attempted to overcome the limitations of the current influenza vaccines with regards to antigenic shift and drift associated with HA and NA (Webster et al., 1992), by focusing on the influenza A virus M2e peptide as a universal vaccine candidate, due to its high degree of conservation since the emergence of the highly virulent Spanish flu pandemic strain of 1918 (Fiers et al., 2004). Generally, M2e vaccine candidates are produced either as chemically conjugated molecules or by genetic fusion to a variety of carrier proteins, such as virion- or VLP-forming proteins (Mozdzanowska et al., 2003; Ionescu et al., 2006; Tompkins et al., 2007; Denis et al., 2008; Matic et al., 2011; Stanekova et al., 2011; Ravin et al., 2012). In our study, we fused the M2e with a self-aggregating signal tag, Zera®. The Zera® fusion tag has the ability to segregate the protein from the plant secretory pathway into membrane-delimited organelles in the ER: the retention and the self-assembly of γ-zein lead to PB formation, stabilizing the protein inside vesicles formed by invagination of the ER lumen (Mainieri et al., 2004; Torrent et al., 2009). This is known to facilitate protein purification, as shown by the result of ultracentrifugation of the plant-produced Zera®M2e PBs through a 60% sucrose cushion: PBs were concentrated in the pellet fraction, with very little soluble protein present.
As the 2009 H1N1 “swine flu” pandemic illustrated, South Africa will have to rely on developed countries for vaccine supplies during an outbreak (Mortimer et al., 2012, 2013). We are therefore systematically investigating the feasibility of establishing rapid-response platforms to produce influenza virus vaccine candidates in South Africa, by implementing novel strategies that are both cost-effective and can be more readily up-scaled than traditional egg-based vaccines (Mortimer et al., 2012).
In the present work, we were able to express Zera®M2e in both plants and insect cells. The protein was expressed as monomers, dimers, and tetramers. This is in line with previous work, where M2e was expressed in baculovirus expression system (Holsinger and Lamb, 1991; Sugrue and Hay, 1991). In plants, we achieved a yield of 125–205 mg/kg FW for the Zera®M2e. These are generally higher yields than those that have been achieved by other researchers. Nemchinov and Natilla (2007) previously reported yields of avian influenza A CMV-M2e fusion protein expression in N. benthamiana of 6–8 mg/kg leaf tissue. Matic et al. (2011) expressed influenza M2e epitopes on chimeric HPV VLPs in plants and obtained 78–120 mg/kg plant material. Most recently, Firsov et al. (2015) expressed M2e fused with β-glucuronidase in transgenic duckweed, and obtained yields from 90 to 970 mg/kg plant FW.
When the serum from mice vaccinated with plant-produced Zera®M2e PB was used to detect the same protein produced in insect cells on western blots, the serum specifically detected only the Zera®M2e protein (Figure 4), indicating that the plant-produced antigen was immunogenic. Use of the plant-produced protein, however, showed that the sera also reacted with other plant proteins that co-purified with the Zera®M2e PBs. Zera® fusions are known to form large polymers that are resistant to degradation (Torrent et al., 2009). The contaminating proteins could include chloroplastic, ribosomal, cytoplasmic, cytoskeleton, and mitochondrial plant proteins (Joseph et al., 2012). It is clear that using a different expression system for antibody detection from that which was used for the antigen production, allowed for more efficient and clear detection of the protein. Future work will include ultracentrifugation on a sucrose density step gradient (Whitehead et al., 2014) to remove unwanted plant protein: this should then lead to more specific immune responses only to the Zera®M2e PB, with reduced reaction against other plant proteins. Testing the Zera®M2e antigen for protection against multiple strains of influenza would be advantageous but was not possible in this investigation.
While we successfully produced Zera®M2e PB in insect cells in this work, and this was valuable as a reagent, our plant-produced Zera®M2e PB had by far the highest yield, with the different soluble forms (monomeric, dimeric, trimeric, and tetrameric) probably contributing significantly to the immunogenicity of the candidate vaccine. Bands corresponding to the monomeric, dimeric, and tetrameric forms have also been detected in previous M2e studies (Holsinger and Lamb, 1991; Sugrue and Hay, 1991).
The production of M2e as a fusion product in plants is not new; Nemchinov and Natilla (2007) expressed M2e in N. benthamiana via a plant viral vector as an internal fusion in the CP of Cucumber mosaic virus; Ravin et al. (2012) used a similar vector to express M2e fused to the HBc and showed protection against lethal challenge in mice. Matic et al. (2011) used the pEAQ-HT vector to transiently express M2e and a shortened version (M2e2–9) as fusions to HPV-16 L1 protein; the longer peptide was presented on capsomers and VLPs, and reacted with anti-M2e antibodies. Petukhova et al. (2013) produced recombinant tobacco mosaic virus particles presenting M2e on their surfaces and showed these were highly immunogenic and protective.
However, our use of the Zera® peptide is novel – moreover, the high yield and the immunogenicity of the product, coupled with what is almost certainly a far easier purification protocol than for any of the fusions detailed above, make it a valuable addition to our rapid-response armory against pandemic influenza. Our work is therefore further proof that plants are a viable vehicle for high-level expression of a peptide vaccine known to elicit broad-spectrum protection against influenza A viruses.
To conclude, we successfully expressed Zera®M2e PB in both insect cell and plant expression systems. Our plant-produced Zera®M2e elicited M2e-specific antibodies in mice, which indicates that it has potential as a candidate universal influenza vaccine. Future work will look at determining the efficacy of these antibodies and their potential of broad-spectrum protection against influenza strains.
Author Contributions
SM created the expression constructs, carried out transient expression experiments, and drafted the manuscript; LM carried out insect cell expression, supervised the work, and participated in drafting of the manuscript, FP created and expressed 5xM2e-tHA in plants; IH designed, coordinated, and supervised the study and participated in drafting of the manuscript; ER initiated study and participated in drafting of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Rodney Lucas and Noel Markgraaff at UCT for their assistance in the animal trial and Pau Marzabal for critical reading of the manuscript. Rainer Fischer is thanked for kindly providing the pTRA vectors (Fraunhofer Institute, Germany), ERA Biotech, Spain for Zera® sequence and anti-Zera® antibody. Owen Karimanzira is thanked for producing the recombinant insect cell expression vectors.
Funding
This research was funded by the Poliomyelitis Research Foundation (PRF 07/02) and Medical Research Foundation (MRC), South Africa. EM was funded by the National Research Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fbioe.2015.00197
References
Andersson, A. M., Hakansson, K. O., Jensen, B. A., Christensen, D., Andersen, P., Thomsen, A. R., et al. (2012). Increased immunogenicity and protective efficacy of influenza M2e fused to a tetramerizing protein. PLoS One 7:e46395. doi:10.1371/journal.pone.0046395
Betakova, T. (2007). M2 protein-a proton channel of influenza A virus. Curr. Pharm. Des. 13, 3231–3235. doi:10.2174/138161207782341295
Black, R. A., Rota, P. A., Gorodkova, N., Cramer, A., Klenk, H. D., and Kendal, A. P. (1993). Production of the M2 protein of influenza A virus in insect cells is enhanced in the presence of amantadine. J. Gen. Virol. 74(Pt 8), 1673–1677. doi:10.1099/0022-1317-74-1-143
Chen, B. J., Leser, G. P., Jackson, D., and Lamb, R. A. (2008). The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82, 10059–10070. doi:10.1128/JVI.01184-08
Cox, M. M. (2008). Progress on baculovirus-derived influenza vaccines. Curr. Opin. Mol. Ther. 10, 56–61.
Cox, N. J., and Subbarao, K. (1999). Influenza. Lancet 354, 1277–1282. doi:10.1016/S0140-6736(99)01241-6
D’Aoust, M. A., Lavoie, P. O., Couture, M. M., Trepanier, S., Guay, J. M., Dargis, M., et al. (2008). Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6, 930–940. doi:10.1111/j.1467-7652.2008.00384.x
De Filette, M., Martens, W., Smet, A., Schotsaert, M., Birkett, A., Londono-Arcila, P., et al. (2008). Universal influenza A M2e-HBc vaccine protects against disease even in the presence of pre-existing anti-HBc antibodies. Vaccine 26, 6503–6507. doi:10.1016/j.vaccine.2008.09.038
De Filette, M., Min Jou, W., Birkett, A., Lyons, K., Schultz, B., Tonkyro, A., et al. (2005). Universal influenza A vaccine: optimization of M2-based constructs. Virology 337, 149–161. doi:10.1016/j.virol.2005.04.004
Denis, J., Acosta-Ramirez, E., Zhao, Y., Hamelin, M. E., Koukavica, I., Baz, M., et al. (2008). Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine 26, 3395–3403. doi:10.1016/j.vaccine.2008.04.052
Feng, J., Zhang, M., Mozdzanowska, K., Zharikova, D., Hoff, H., Wunner, W., et al. (2006). Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 3, 102. doi:10.1186/1743-422X-3-102
Fiers, W., De Filette, M., Birkett, A., Neirynck, S., and Min Jou, W. (2004). A “universal” human influenza A vaccine. Virus Res. 103, 173–176. doi:10.1016/j.virusres.2004.02.030
Fiore, A. E., Bridges, C. B., and Cox, N. J. (2009). Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 333, 43–82. doi:10.1007/978-3-540-92165-3_3
Firsov, A., Tarasenko, I., Mitiouchkina, T., Ismailova, N., Shaloiko, L., Vainstein, A., et al. (2015). High-yield expression of M2e peptide of avian influenza virus H5N1 in transgenic duckweed plants. Mol. Biotechnol. 57, 653–661. doi:10.1007/s12033-015-9855-4
Gomez, E., Zoth, S. C., and Berinstein, A. (2009). Plant-based vaccines for potential human application: a review. Hum. Vaccin. 5, 738–744. doi:10.4161/hv.5.11.9879
Gouy, M., and Gautier, C. (1982). Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10, 7055–7074. doi:10.1093/nar/10.22.7055
Holsinger, L. J., and Lamb, R. A. (1991). Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 183, 32–43. doi:10.1016/0042-6822(91)90115-R
Huleatt, J. W., Jacobs, A. R., Tang, J., Desai, P., Kopp, E. B., Huang, Y., et al. (2007). Vaccination with recombinant fusion proteins incorporating toll-like receptor ligands induces rapid cellular and humoral immunity. Vaccine 25, 763–775. doi:10.1016/j.vaccine.2006.08.013
Imai, M., Herfst, S., Sorrell, E. M., Schrauwen, E. J. A., Linster, M., De Graaf, M., et al. (2013). Transmission of influenza A/H5N1 viruses in mammals. Virus Res. 178, 15–20. doi:10.1016/j.virusres.2013.07.017
Ionescu, R. M., Przysiecki, C. T., Liang, X., Garsky, V. M., Fan, J., Wang, B., et al. (2006). Pharmaceutical and immunological evaluation of human papillomavirus virus-like particle as an antigen carrier. J. Pharm. Sci. 95, 70–79. doi:10.1002/jps.20493
Jegerlehner, A., Schmitz, N., Storni, T., and Bachmann, M. F. (2004). Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 172, 5598–5605. doi:10.4049/jimmunol.172.9.5598
Johansson, B. E., Bucher, D. J., Pokorny, B. A., Mikhail, A., and Kilbourne, E. D. (1989). Identification of PR8 M1 protein in influenza virus high-yield reassortants by M1-specific monoclonal antibodies. Virology 171, 634–636. doi:10.1016/0042-6822(89)90638-7
Joseph, M., Ludevid, M. D., Torrent, M., Rofidal, V., Tauzin, M., Rossignol, M., et al. (2012). Proteomic characterisation of endoplasmic reticulum-derived protein bodies in tobacco leaves. BMC Plant Biol. 12:36. doi:10.1186/1471-2229-12-36
Kaplan, B. S., and Webby, R. J. (2013). The avian and mammalian host range of highly pathogenic avian H5N1 influenza. Virus Res. 178, 3–11. doi:10.1016/j.virusres.2013.09.004
Lamb, R. A., Zebedee, S. L., and Richardson, C. D. (1985). Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40, 627–633. doi:10.1016/0092-8674(85)90211-9
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Maclean, J., Koekemoer, M., Olivier, A. J., Stewart, D., Hitzeroth, I. I., Rademacher, T., et al. (2007). Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. J. Gen. Virol. 88, 1460–1469. doi:10.1099/vir.0.82718-0
Mainieri, D., Rossi, M., Archinti, M., Bellucci, M., De Marchis, F., Vavassori, S., et al. (2004). Zeolin. A new recombinant storage protein constructed using maize gamma-zein and bean phaseolin. Plant Physiol 136, 3447–3456. doi:10.1104/pp.104.046409
Mardanova, E. S., Kotlyarov, R. Y., Kuprianov, V. V., Stepanova, L. A., Tsybalova, L. M., Lomonosoff, G. P., et al. (2015). Rapid high-yield expression of a candidate influenza vaccine based on the ectodomain of M2 protein linked to flagellin in plants using viral vectors. BMC Biotechnol. 15:42. doi:10.1186/s12896-015-0164-6
Matic, S., Rinaldi, R., Masenga, V., and Noris, E. (2011). Efficient production of chimeric human papillomavirus 16 L1 protein bearing the M2e influenza epitope in Nicotiana benthamiana plants. BMC Biotechnol. 11:106. doi:10.1186/1472-6750-11-106
Mortimer, E., Hitzertoth, I., Buys, A., Mbewana, S., and Rybicki, E. (2013). An H5N1 influenza DNA vaccine for South Africa. S. Afr. J. Sci. 109, 4. doi:10.1590/sajs.2013/20120053
Mortimer, E., Maclean, J. M., Mbewana, S., Buys, A., Williamson, A. L., Hitzeroth, I. I., et al. (2012). Setting up a platform for plant-based influenza virus vaccine production in South Africa. BMC Biotechnol. 12:14. doi:10.1186/1472-6750-12-14
Mozdzanowska, K., Feng, J., Eid, M., Kragol, G., Cudic, M., Otvos, L. Jr., et al. (2003). Induction of influenza type A virus-specific resistance by immunization of mice with a synthetic multiple antigenic peptide vaccine that contains ectodomains of matrix protein 2. Vaccine 21, 2616–2626. doi:10.1016/S0264-410X(03)00040-9
Nemchinov, L. G., and Natilla, A. (2007). Transient expression of the ectodomain of matrix protein 2 (M2e) of avian influenza A virus in plants. Protein Expr. Purif. 56, 153–159. doi:10.1016/j.pep.2007.05.015
Park, E. K., Castrucci, M. R., Portner, A., and Kawaoka, Y. (1998). The M2 ectodomain is important for its incorporation into influenza A virions. J. Virol. 72, 2449–2455.
Petukhova, N. V., Gasanova, T. V., Stepanova, L. A., Rusova, O. A., Potapchuk, M. V., Korotkov, A. V., et al. (2013). Immunogenicity and protective efficacy of candidate universal influenza A nanovaccines produced in plants by tobacco mosaic virus-based vectors. Curr. Pharm. Des. 19, 5587–5600. doi:10.2174/13816128113199990337
Pinto, L. H., and Lamb, R. A. (2006). Influenza virus proton channels. Photochem. Photobiol. Sci. 5, 629–632. doi:10.1039/b517734k
Price, G. E., Soboleski, M. R., Lo, C.-Y., Misplon, J. A., Quirion, M. R., Houser, K. V., et al. (2010). Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS One 5:e13162. doi:10.1371/journal.pone.0013162
Ravin, N. V., Kotlyarov, R. Y., Mardanova, E. S., Kuprianov, V. V., Migunov, A. I., Stepanova, L. A., et al. (2012). Plant-produced recombinant influenza vaccine based on virus-like HBc particles carrying an extracellular domain of M2 protein. Biochemistry (Mosc) 77, 33–40. doi:10.1134/S000629791201004X
Rossman, J. S., Jing, X., Leser, G. P., Balannik, V., Pinto, L. H., and Lamb, R. A. (2010). Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84, 5078–5088. doi:10.1128/JVI.00119-10
Rybicki, E. P. (2009). Plant-produced vaccines: promise and reality. Drug Discov. Today 14, 16–24. doi:10.1016/j.drudis.2008.10.002
Safdar, A., and Cox, M. M. (2007). Baculovirus-expressed influenza vaccine. A novel technology for safe and expeditious vaccine production for human use. Expert Opin. Investig. Drugs 16, 927–934. doi:10.1517/13543784.16.7.927
Shoji, Y., Chichester, J. A., Bi, H., Musiychuk, K., de la Rosa, P., Goldschmidt, L., et al. (2008). Plant-expressed HA as a seasonal influenza vaccine candidate. Vaccine 26, 2930–2934. doi:10.1016/j.vaccine.2008.03.045
Stanekova, Z., Kiraly, J., Stropkovska, A., Mikuskova, T., Mucha, V., Kostolansky, F., et al. (2011). Heterosubtypic protective immunity against influenza A virus induced by fusion peptide of the hemagglutinin in comparison to ectodomain of M2 protein. Acta Virol. 55, 61–67. doi:10.4149/av_2011_01_61
Sugrue, R. J., and Hay, A. J. (1991). Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 180, 617–624. doi:10.1016/0042-6822(91)90075-M
Takeda, A., Sugiyama, K., Nagano, H., Mori, M., Kaido, M., Mise, K., et al. (2002). Identification of a novel RNA silencing suppressor, NSs protein of tomato spotted wilt virus. FEBS Lett 532, 75–79. doi:10.1016/S0014-5793(02)03632-3
Thompson, W. W., Shay, D. K., Weintraub, E., Brammer, L., Cox, N., Anderson, L. J., et al. (2003). Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289, 179–186. doi:10.1001/jama.289.2.179
Tompkins, S. M., Zhao, Z. S., Lo, C. Y., Misplon, J. A., Liu, T., Ye, Z., et al. (2007). Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerging Infect. Dis. 13, 426–435. doi:10.3201/eid1303.061125
Torrent, M., Llompart, B., Lasserre-Ramassamy, S., Llop-Tous, I., Bastida, M., Marzabal, P., et al. (2009). Eukaryotic protein production in designed storage organelles. BMC Biol. 7:5. doi:10.1186/1741-7007-7-5
Wang, L., Hess, A., Chang, T. Z., Wang, Y. C., Champion, J. A., Compans, R. W., et al. (2014). Nanoclusters self-assembled from conformation-stabilized influenza M2e as broadly cross-protective influenza vaccines. Nanomedicine 10, 473–482. doi:10.1016/j.nano.2013.08.005
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M., and Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179.
Webster, R. G., and Govorkova, E. A. (2006). H5N1 influenza – continuing evolution and spread. N. Engl. J. Med. 355, 2174–2177. doi:10.1056/NEJMp068205
Keywords: influenza A virus, M2e, plant expression, insect cell expression, vaccine
Citation: Mbewana S, Mortimer E, Pêra FFPG, Hitzeroth II and Rybicki EP (2015) Production of H5N1 Influenza Virus Matrix Protein 2 Ectodomain Protein Bodies in Tobacco Plants and in Insect Cells as a Candidate Universal Influenza Vaccine. Front. Bioeng. Biotechnol. 3:197. doi: 10.3389/fbioe.2015.00197
Received: 28 September 2015; Accepted: 23 November 2015;
Published: 08 December 2015
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Inge Broer, University of Rostock, GermanyBasavaprabhu L. Patil, ICAR-National Research Centre on Plant Biotechnology, India
Copyright: © 2015 Mbewana, Mortimer, Pêra, Hitzeroth and Rybicki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inga Isabel Hitzeroth, inga.hitzeroth@uct.ac.za
 Sandiswa Mbewana
Sandiswa Mbewana Elizabeth Mortimer1
Elizabeth Mortimer1
 Francisco F. P. G. Pêra
Francisco F. P. G. Pêra Inga Isabel Hitzeroth
Inga Isabel Hitzeroth Edward P. Rybicki
Edward P. Rybicki