Bioprocess Optimization for the Production of Aromatic Compounds With Metabolically Engineered Hosts: Recent Developments and Future Challenges
- Centre of Biological Engineering, University of Minho, Braga, Portugal
The most common route to produce aromatic chemicals – organic compounds containing at least one benzene ring in their structure – is chemical synthesis. These processes, usually starting from an extracted fossil oil molecule such as benzene, toluene, or xylene, are highly environmentally unfriendly due to the use of non-renewable raw materials, high energy consumption and the usual production of toxic by-products. An alternative way to produce aromatic compounds is extraction from plants. These extractions typically have a low yield and a high purification cost. This motivates the search for alternative platforms to produce aromatic compounds through low-cost and environmentally friendly processes. Microorganisms are able to synthesize aromatic amino acids through the shikimate pathway. The construction of microbial cell factories able to produce the desired molecule from renewable feedstock becomes a promising alternative. This review article focuses on the recent advances in microbial production of aromatic products, with a special emphasis on metabolic engineering strategies, as well as bioprocess optimization. The recent combination of these two techniques has resulted in the development of several alternative processes to produce phenylpropanoids, aromatic alcohols, phenolic aldehydes, and others. Chemical species that were unavailable for human consumption due to the high cost and/or high environmental impact of their production, have now become accessible.
Introduction
The increasing demand for “natural” labeled products, the adoption of a healthy life style associated with growing concerns about global warming and limited supplies of fossil fuels, promote the development of alternative ways for producing fuels and commodity chemicals using renewable feedstocks in eco-friendly processes. In this scenario, the use of biotechnological platforms for their production is becoming a promising alternative (Sun et al., 2015; Braga et al., 2018a; Milke et al., 2018; Park et al., 2018).
An important class of petrochemical compounds that have been considered as promising targets for biotechnological production are aromatic compounds (Knaggs, 2003; Lee and Wendisch, 2017; Noda and Kondo, 2017). They are typically produced employing fossil feedstocks as raw materials and have a wide range of industrial and commercial applications as building blocks for the synthesis of polymer materials like functional plastics and fibers, food and feed additives, nutraceuticals and pharmaceuticals (Krömer et al., 2013; Averesch and Krömer, 2018). The economic importance of these compounds is quite significant; in 2017 their global market size was USD185.9 billion and it is expected that, in 2025, their global production volume will reach 168,733.35 thousand tons (Averesch and Kayser, 2014), with the demand for aromatic compounds for gasoline, pharmaceuticals and detergents as main driving force.
In the last decades, microorganisms have emerged as attractive platforms for producing former petroleum-derived compounds from renewable starting materials (Borodina and Nielsen, 2014; Krivoruchko and Nielsen, 2015; Noda and Kondo, 2017). Until now, several derivatives of BTX (benzene, toluene, and the three isomers of xylene), such as styrene, hydroxystyrene, phenol and vanillin, have been produced using microbial hosts by direct bioconversion of precursors or via de novo synthesis (Wierckx et al., 2005; Vannelli et al., 2007; Mckenna and Nielsen, 2011; Ni et al., 2015). However, only a few compounds, such as vanillin and resveratrol, have reached bio-based production at commercial scale (Nakamura and Whited, 2003; Yim et al., 2011; Paddon et al., 2013; Van Dien, 2013). Nevertheless, despite the efforts that have been made until now, the production of benzene, toluene or xylene in a renewable way has not been reported.
Microorganisms can grow with high growth rates and achieve high biomass yields, in scalable cultivation and production processes. They are also able to grow in diverse media, from abundant and inexpensive feedstocks. However, they do not naturally (over-)produce these compounds or, if they do, the yields are very low. In order to enable production, it is necessary to functionally integrate heterologous pathways or genetically modify the microbial hosts (Rodrigues et al., 2015; Chouhan et al., 2017; Gottardi et al., 2017; Milke et al., 2018; Wang J. et al., 2018). Aromatic compounds are produced by microbial hosts via the shikimate pathway, which leads to the production of aromatic amino acids as well as other aromatic precursors (Herrmann, 1995; Maeda and Dudareva, 2012; Averesch and Krömer, 2018). This can be achieved by the functional reconstruction of naturally occurring pathways or by de novo pathway engineering (Dhamankar and Prather, 2011; Wu et al., 2018). Escherichia coli and Saccharomyces cerevisiae are the most commonly employed microorganisms for aromatic compound production. However, more recently, other hosts have also been explored due to their peculiarities, such as Corynebacterium glutamicum, Lactococcus lactis, Pseudomonas putida, and Streptomyces lividans (Sachan et al., 2006; Gosset, 2009; Verhoef et al., 2009; Gaspar et al., 2016; Kallscheuer et al., 2016, 2019; Dudnik et al., 2017, 2018; Braga et al., 2018a; Tilburg et al., 2019).
This review presents an overview of recent advances in microbial production of the most relevant aromatic compounds, including vanillin, salicylic acid, p-hydroxybenzoic acid and others strategies for strain design are compared with an emphasis on the development of biosynthetic pathways, the application of protein engineering, carbon flux redirection, use of alternative substrates, engineering substrate uptake and optimization of culture conditions. We present and explain some of the current challenges and gaps that in our knowledge, must be overcome in order to render the biotechnological production of aromatic compounds, in an attractive and feasible way for the commercial scale. Table 1 presents a summary of the recent reports (last 4 years) regarding the production of aromatic compounds in engineered microbial hosts, comparing the used carbon source, organism and strain, (over-)expressed and/or knocked out genes.
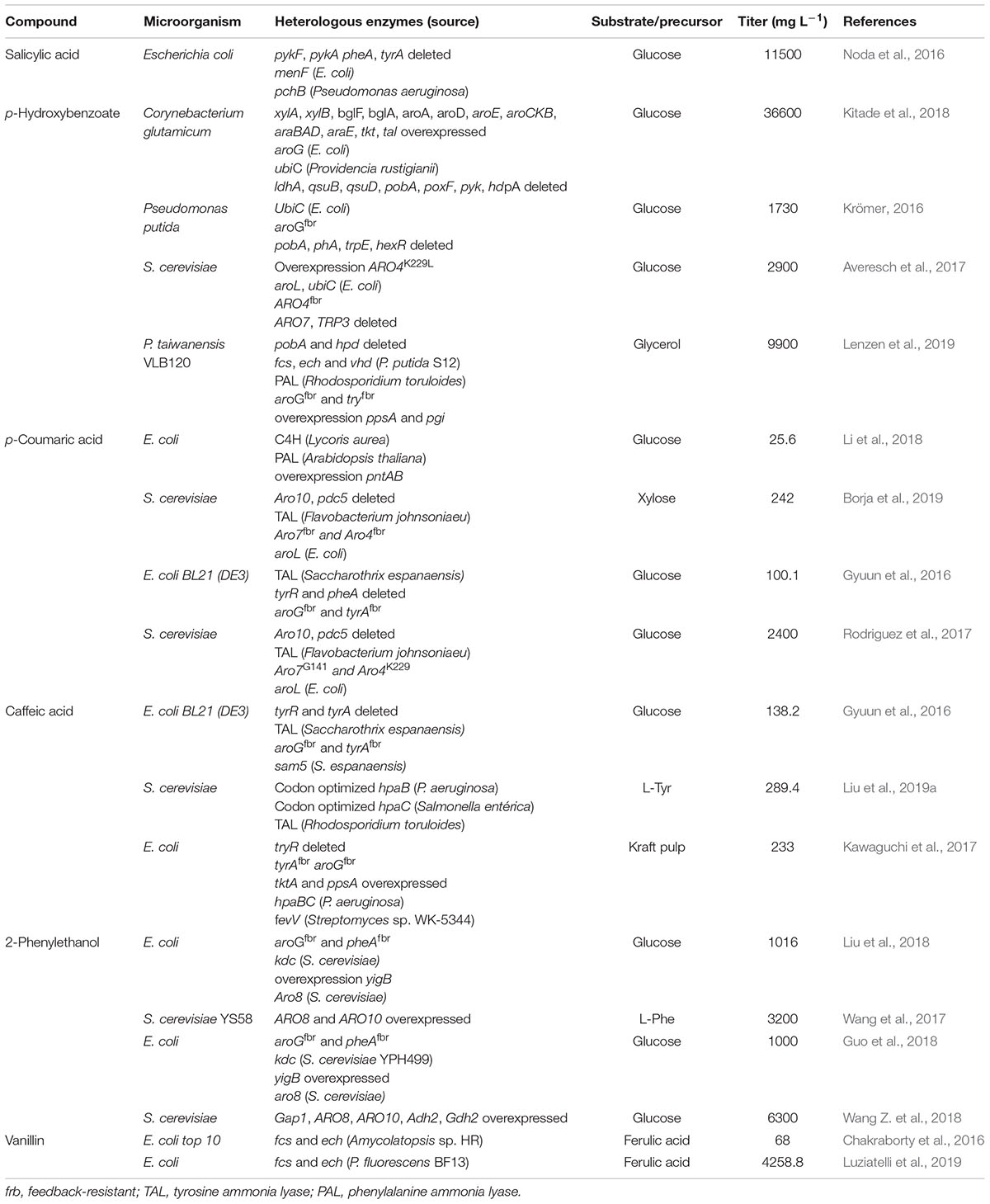
Table 1. Summary of the literature on the production titer of some aromatic compounds obtained through metabolic engineering in microorganisms, in the last 4 years.
The Shikimate Pathway: a Path for Aromatic Compounds Production
In microorganisms, the production of aromatic compounds is almost always obtained via the shikimate (SKM) pathway. This route leads to the biosynthesis of aromatic amino acids, L-tyrosine (L-Tyr), L-tryptophan (L-Trp) and L-phenylalanine (L-Phe), and a wide range of aromatic precursors (Knaggs, 2003; Noda et al., 2016; Lai et al., 2017). The first reaction in the shikimate pathway is the condensation of the central carbon metabolism intermediates, phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P), to yield 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP). After that, six successive enzymatic reactions lead to the production of chorismate (CHO), the end product of the SKM pathway (Figure 1) and the starter unit for the production of aromatic amino acids as well as different aromatic compounds (phenylpropanoids, salicylic acid, p-hydroxybenzoic acid, aromatic alcohols, vanillin, among others) (Noda et al., 2016).
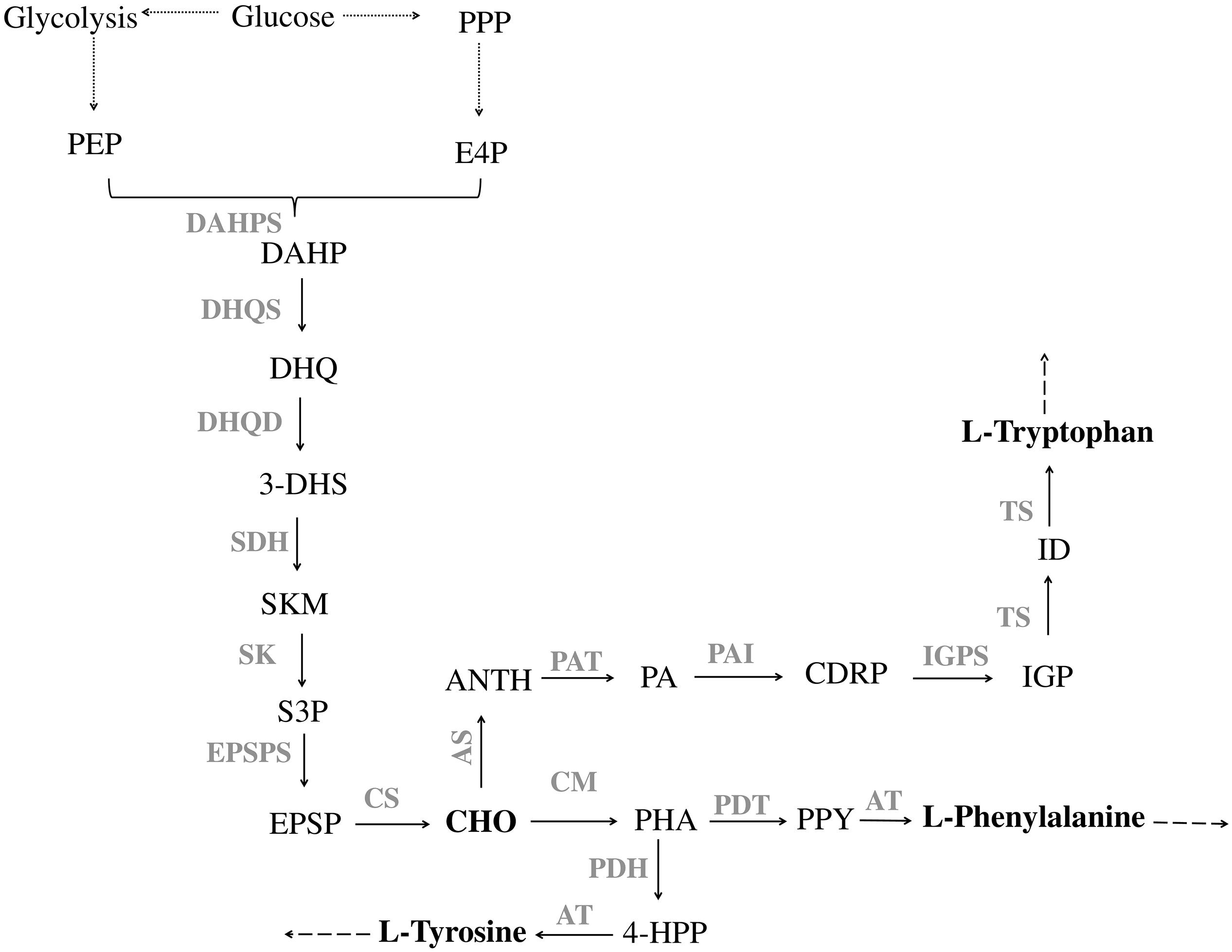
Figure 1. Pathway of aromatic amino acid biosynthesis. PPP, pentose phosphate pathway; E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate; DAHPS, DAHP synthase; DAHP, 3-Deoxy-D-arabinoheptulosonate 7-phosphate; DHQS, 3-dehydroquinate synthase; DHQ, 3-dehydroquinate; DHQD, 3-dehydroquinate dehydratase; 3-DHS, 3-dehydroshikimate; SDH, shikimate 5-dehydrogenase; SKM, shikimate; SK, shikimate kinase; S3P, shikimate 3-phosphate; EPSPS, 5-enolpyruvylshikimate 3-phosphate synthase; EPSP, 5-enolpyruvylshikimate-3-phosphate; CS, chorismate synthase; CHO, chorismate; CM, chorismate mutase; PHA, prephenate; PDH, prephenate dehydrogenase; 4-HPP, 4-hydroxyphenylpyruvate; AT, aminotransferase; PDT, prephenate dehydratase; PPY, phenylpyruvate; AS, anthranilate synthase; ANTH, anthranilate; PAT, phosphoribosylanthranilate transferase; PA, phosphoribosylanthranilate; PAI, phosphoribosylanthranilate isomerase; CDRP, l-(O-carboxyphenylamino)-l-deoxyribulose-5-phosphate; IGPS, indole-3-glycerol phosphate synthase; IGP, indole-3-glycerol phosphate; TS, tryptophan synthase; ID, indole. Solid lines indicate a single step; dotted lines indicate multiple steps.
The first step for L-Phe and L-Tyr production is catalyzed by chorismate mutase (CM), which converts CHO to prephenate (PHA). After that, PHA undergoes decarboxylation and dehydration yielding phenylpyruvate (PPY) or is oxidatively decarboxylated to 4-hydroxyphenylpyruvate (4-HPP). The reactions are catalyzed by prephenate dehydratase (PDT) and prephenate dehydrogenase (PDH), respectively. The last step comprises the transamination of PPY to L-Phe and of 4-HPP to L-Tyr that is catalyzed by an aminotransferase (AT) (Tzin et al., 2001; Figure 1). The pathway for L-Trp production from CHO requires six steps. The first one is catalyzed by anthranilate synthase (AS) that converts CHO to anthranilate (ANTH), which is further converted to phosphoribosylanthranilate (PA) by anthranilate phosphoribosyl transferase (PAT). The third step in this pathway leads to the production of l-(O-carboxyphenylamino)-l-deoxyribulose-5–phosphate (CDRP) by phosphoribosylanthranilate isomerase (PAI). The fourth enzyme of L-Trp biosynthesis is indole-3-glycerol phosphate synthase (IGPS), which catalyzes the conversion of CDRP to indole-3-glycerol phosphate (IGP). In the last two steps, IGP is cleaved by tryptophan synthase (TS) into indole (ID) that is ligated to L-serine to yield L-Trp (Figure 1; Priya et al., 2014).
One of the main bottlenecks in the microbial production of aromatic compounds is the availability of the precursors PEP (produced during glycolysis) and E4P (derived from the pentose phosphate pathway – PPP) (Suástegui et al., 2016; Noda and Kondo, 2017; Averesch and Krömer, 2018; Wu et al., 2018). Different strategies have been described in order to engineer the central carbon metabolism into this direction (Leonard et al., 2005; Papagianni, 2012; Nielsen and Keasling, 2016). In fact, the available fluxes of both precursors differ considerably. Suástegui et al. (2016) studied the E4P and PEP flux in S. cerevisiae using metabolic flux analysis and observed that E4P was clearly the limiting precursor. Therefore, establishing a balance between the ratio of both precursors and increasing their availability appeared to be the two main strategies to follow in order to increase aromatic compounds production.
E4P can be produced from PPP or from sedoheptulose-1,7-bisphosphate in a reaction that is probably favored when the intracellular levels of sedoheptulose-7-phosphate (S7P)are high (Nagy and Haschemi, 2013). S7P is an intermediate in non-oxidative part of PPP, that is produced from xylulose 5-phosphate and ribose 5-phosphate by transketolase. The most common approaches to increase E4P production are the overexpression of transaldolase and transketolase genes, to promote the conversion of S7P and glyceraldehyde-3-phosphate (G3P) to E4P and fructose 6-phosphate (F6P) (Bongaerts et al., 2001; Noda and Kondo, 2017; Averesch and Krömer, 2018) and to enhance the supply of E4P (Lütke-Eversloh and Stephanopoulos, 2007; Bulter et al., 2003). Following this strategy, Knop et al. (2001) observed an increase in the shikimic acid titer, an intermediate of the SMK pathway, from 38 to 52 g L–1, after overexpression of transketolase gene (tktA) in E. coli. The role of transaldolase for the production of the PPP was analyzed by Lu and Liao (1997) and Sprenger et al. (1998). They observed that the overexpression of talB increases the production of DAHP from glucose. Moreover, Lu and Liao (1997) concluded that transketolase is more effective in directing the carbon flux to the aromatic pathway than transaldolase. In fact, the overexpression of the transaldolase gene in strains which already overexpress the transketolase gene did not show a further increase in production of aromatic compounds. This result may be related with the saturation of E4P supply when tktA was overexpressed (Lu and Liao, 1997). Nevertheless, the overexpression of the transketolase gene proved to have a limited impact in the E4P poll which can be correlated with the preference of this enzyme for catalyzing the E4P consuming reaction (Curran et al., 2013). Other efforts to increase the carbon flux in the PPP include the overexpression of the gene coding for glucose-6-phosphate dehydrogenase, that has been shown to increase the availability of ribulose-5-phosphate (R5P) and E4P (Yakandawala et al., 2008; Rodriguez et al., 2013) or the deletion of genes that encode the phosphoglucose isomerase, that forces the cell to metabolize the substrate completely via PPP (Mascarenhas et al., 1991). However, the later approach blocks the oxidative shunt of the PPP, which is the main source of the redox cofactor NADPH, required by the shikimate dehydrogenase as well as by many enzymes in downstream pathways (Zhang J. et al., 2015). The use of other carbon sources that have different transporters, such as hexoses (as sucrose and gluconate), pentoses (xylose and arabinose) and glycerol (Kai Li and Frost, 1999; Ahn et al., 2008; Martínez et al., 2008; Chen et al., 2012), is also an alternative way to increase the E4P poll. In the last year, Liu et al. (2019b) proposed a different strategy to increase the scarcity of E4P in S. cerevisiae. They investigated a heterologous phosphoketolase (PHK) pathway, including a phosphoketolase from Bifidobacterium breve (Bbxfpk) and a phosphotransacetylase from Clostridium kluyveri (Ckpta). Phosphoketolase is able to split fructose-6-phosphate into E4P and acetyl-phosphate, and the introduction of this pathway could, theoretically, divert part of the carbon flux from glycolysis directly toward E4P. The authors observed a 5.4-fold enhance in E4P concentration in the BbXfpk-expressing strain. When compared with the overexpression of the transketolase-encoding gene, this approach resulted in a lower E4P availability (Liu et al., 2019b). However, none of these strategies were able to efficiently divert carbon flux from glycolysis toward E4P, to provide sufficient levels for the biosynthesis of aromatic compounds.
The availability of the other precursor, PEP, is also an important factor that needs to be considered when designing a strategy to construct a strain able to produce aromatic compounds (Rodriguez et al., 2013; Noda and Kondo, 2017). PEP is required for the simultaneous uptake and phosphorylation of glucose (PEP:glucose phosphotransferase system – PTS) and it is also involved in reactions catalyzed by the enzymes phosphoenolpyruvate carboxylase and pyruvate kinase that catalyzes the ATP-producing conversion of PEP to pyruvate. The glucose transport by PTS is the main PEP consuming activity and for this reason the construction of PTS-deficient strains is one of the most common approaches to increase PEP availability and therefore, aromatic compounds yield from glucose (Gu et al., 2012, 2013). However, the main problem of this strategy is the low cellular growth rate. The use of a non-PTS system which does not consume PEP is an alternative way to allow high PEP availability (Sprenger et al., 1998; Chandran et al., 2003). For example, glucose can be transported by galactose permease (encoded by galP) and further phosphorylated by glucokinase (encoded by glk) (Yi et al., 2003; Balderas-hernández et al., 2009). Additionally, Yi et al. (2003) described the utilization of a glucose facilitator from Zymomonas mobilis (encoded by glf), that transports glucose by facilitated diffusion, in combination with plasmid-localized Z. mobilis glk (encoded glucokinase), attaining a 3-dehydroshikimic acid (a key intermediate for aromatic compounds production) production of 60 g L–1, in E. coli. In 2011, Ikeda et al. (2011) identified a new non-PTS system, a myo-inositol-induced transporter (encoded by iolT1) in C. glutamicum. Furthermore, an increase in the PEP availability has been achieved by modulation of the carbon flux from PEP to the tricarboxylic acid cycle (TCA) by inactivation of pyruvate kinase genes (Gosset et al., 1996; Chandran et al., 2003; Escalante et al., 2010) and PEP carboxylase (Tan et al., 2013). On the other hand, the overexpression of the genes that encode PEP synthetase which catalyzes the conversion of pyruvate into PEP, enhanced its level (Patnaik and Liao, 1994; Tatarko and Romeo, 2001). PEP carboxykinase catalyzes the formation of PEP from oxaloacetate. The overexpression of the gene that encodes it – pckA – has also been proposed as a strategy to increase the yield of aromatic amino acids (Gulevich et al., 2006). An interesting approach to enhance the PEP availability is the attenuation of CsrA, a protein that regulates transcription of genes involved in carbon metabolism and energy metabolism (Wang et al., 2013). It was found that the absence of CsrA could enhance the metabolic flow of gluconeogenesis, contributing to the accumulation of PEP. Tatarko and Romeo (2001) knocked out the csraA gene and observed an increase in the PEP concentration. The overexpression of csrB, a small untranslated RNA from the carbon storage regulator, also improves the availability of PEP in E. coli (Yakandawala et al., 2008).
Notwithstanding the progress achieved, the industrial application of these strategies still poses some problems. The redirection of the carbon flux into a desired pathway usually results in a reduced cell growth rate and/or production of unwanted by-products (Patnaik et al., 1992) which usually ruins the economic viability of an eventual industrial process as will be further discussed (section “Discussion”).
After the establishment of an adequate supply of precursors it is essential to redirect this carbon toward the SKM pathway and remove limiting steps to increase the production of target compounds. The SKM pathway is highly complex. It is mainly regulated at the transcription and enzymatic activity level. As previously described, the first step of the SKM pathway is the DAHP production, catalyzed by DAHP synthases. This is one of the most strictly regulated steps in this route (Figure 1). In fact, DAHP synthase activity is regulated by the concentration of the downstream reaction products of the SKM pathway, the aromatic amino acids. This mechanism is a clever way for cells to make just the right amount of product. When the concentration of aromatic amino acids is high, they will block the DAHP synthase activity, preventing its production until the existing supply has been used up (Bongaerts et al., 2001; Gosset, 2009; Gottardi et al., 2017). In S. cerevisiae, two DAHP synthase isozymes (encoded by ARO3 and ARO4 genes) are feedback inhibited by L-Phe and L-Tyr, respectively (Paravicini et al., 1989). E. coli has three different DAHP synthase isozymes (encoded by aroF, aroG, aroH), and each one is vulnerable to inhibition by an aromatic amino acids: L-Phe, L-Tyr and L-Trp, respectively (Hu et al., 2003; Gu et al., 2012). In addition to the allosteric inhibition, it is also necessary to take into account the transcriptional repression mediated by the protein TyrR (tyrosine repressor). This can repress aroF and aroG, whereas the transcription of aroH is controlled by the protein TrpP (tryptophan repressor) (Pittard et al., 2005; Keseler et al., 2013). In C. glutamicum, the DAHP synthase isozymes are encoded by aroG and aroF. AroG is feedback inhibited by L-Phe, chorismate and prephenate, whereas aroF is feedback inhibited by L-Tyr and L-Trp (Lee et al., 2009).
To overcome this natural limitation, different strategies have been described, such as the use of DAHP synthase which is not sensitive to feedback-inhibition (feedback-resistant – fbr) (Frost and Draths, 1995). Shumilin et al. (1999) determined a 3D structure of DAHP synthase co-crystallized with PEP, demonstrating the possible nine binding sites of L-Phe for feedback-inhibition. Random or directed mutagenesis at these specific amino acids residues, such as Asp146Asn, and Pro150Leu (Kikuchi et al., 1997), is the most common approach used to generate feedback-resistant variants of DAHP synthase. Hartmann et al. (2003) determined the crystal structure of Aro4p and demonstrated that with a single lysine-to-leucine substitution at position 229, the protein is L-Phe and L-Tyr insensitive. In combination with the deletion of ARO3, this strategy led to a 4-fold increase in the flux through the aromatic amino acid-forming pathway (Luttik et al., 2008). Similarly, the introduction of a tyrosine-insensitive ARO4 allele (ARO4 G226S) was also reported by Schnappauf et al. (1998). In E. coli a similar approach was also described with the introduction of feedback-resistant derivatives of aroFfbr and aroGfbr, using either plasmids or chromosomal integration for expression of the modified encoding genes (Ger et al., 1994; Jossek et al., 2001). In this context, the reactions catalyzed by 3-dehydroquinate synthase, shikimate kinase and shikimate 5-dehydrogenase are also considered rate-limiting (Dell and Frost, 1993; Kramer et al., 2003; Oldiges et al., 2004; Juminaga et al., 2012). Different strategies have been applied to overcome these limitations, such as: the overexpression of the genes that encode these enzymes by plasmid-cloned genes, their chromosomal integration, promoter engineering by chromosomal evolution, or co-expression of the genes in a modular operon under control of diverse promoters (Chandran et al., 2003; Lütke-Eversloh and Stephanopoulos, 2005; Escalante et al., 2010; Rodriguez et al., 2013; Cui et al., 2014).
Another regulatory point is present at the chorismate branch, at which the chorismate mutase and prephenate dehydratase are feedback regulated by the end products, L-Phe and L-Try (Lütke-Eversloh and Stephanopoulos, 2005; Reifenrath et al., 2018). The most common strategies to overcome this bottleneck are the application of mutations that confer feedback resistance to chorismate mutase-prephenate dehydratase or the utilization of evolved genes (pheAev) (Báez-Viveros et al., 2004; Lütke-Eversloh and Stephanopoulos, 2005; Ikeda, 2006; Sprenger, 2007; Luttik et al., 2008). Backman et al. (1990) used a recombinant E. coli strain carrying pheAfbr and aroFfbr for L-Phe production and achieved a titer of 50 g L–1 with a yield of 0.25 (mol L-Phe mol glucose–1) after 36 h. This is the highest titer reported so far. Furthermore, Báez-Viveros et al. (2004) observed that the overexpression of evolved genes (pheAev) had a positive and significant impact on L-Phe production in E. coli, showing a 3–4-fold improvement, when compared with equivalent strains expressing pheAfbr. The use of L-Tyr- or L-Phe-overproducing strains for the production of some aromatic compounds, derived from aromatic amino acids, has been reported by many authors. For their construction, the most common approaches include overexpression of aroGfbr and tyrAfbr and in some cases ppsA, tktA and the deletion of tyrR (Kang et al., 2012; Lin and Yan, 2012; Santos et al., 2012; Huang et al., 2013), achieving L-Phe and L-Try titers of 50 and 55 g L–1, respectively (Patnaik and Liao, 1994; Ikeda, 2006; Sprenger, 2007). However, most of the studies that have been performed, reported the expression and/or regulation of key genes, under the control of constitutively expressed or inducible promoters in plasmid-cloned operons. Nevertheless, this approach has several drawbacks, ranging from structural and segregational instability to metabolic burden of plasmid replication (Noack et al., 1981; Bentley et al., 1990). To overcome these drawbacks, Cui et al. (2014) developed a plasmid free methodology for shikimic acid production, an important intermediate of the SKM pathway, in E. coli. AroGfbr, aroB, aroE, and tktA genes were chromosomally integrated by tuning the copy number and expression using chemically induced chromosomal evolution with triclosan. They also overexpressed the ppsA and csrB genes to enhance the PEP/pyruvate pool. Finally, pntAB or nadK genes were also chromosomally overexpressed in order to increase the NADPH pull. The final strain was able to produce 3.12 g L–1 of shikimic acid with a glucose yield of 0.33 mol mol–1. They also demonstrated that the overexpression of pntAB or nadK genes increase the NADPH availability. This is the first report of an engineered shikimic acid producing strain of E. coli that lacks both a plasmid and an antibiotic marker.
Despite the efforts that have been made, it is also important to study different strategies to minimize carbon loss to competing pathways. Gu et al. (2012) reported an increase in L-Trp concentration after a knock out in the gene tnaA, which codes for a tryptophanase to avoid product degradation. On the other hand, the modification of aromatic compounds transport system, as the inactivation of permease genes aroP, mtr and tnaB to avoid product re-internalization, or the overexpression of genes that encodes exporter proteins (e.g., yddG), can also be used as interesting approaches to increase its production (Liu et al., 2012; Wang et al., 2013). Rodriguez et al. (2013) demonstrated that the inactivation of ydiB (coding for shikimate dehydrogenase/quinate dehydrogenase) leads to a decrease in byproduct formation, improving the carbon flux toward the desired aromatic compound production.
Strategies for Production of Aromatic Compounds
In the last decade, several attempts to implement the production of aromatic compounds in cells have been reported (Bongaerts et al., 2001; Dias et al., 2017; Lee and Wendisch, 2017; Beata et al., 2019). The first studies focused on the identification of the microorganisms that are able to produce, natively, aromatic compounds, such as 2-phenylethanol, and/or metabolites that are biosynthetic precursors or derivatives of aromatic compounds, such as SKM, chorismate (CHO), and aromatic amino acids (L-Phe, L-Tyr and Trp), with high efficiency. Then, engineered strains were developed and the production processes optimized in order to raise the product titer to g L–1-scale. Nowadays, microbial hosts are able to produce a large spectrum of target products, including chemicals they do not naturally produce (Pandey et al., 2016; Wang et al., 2016).
In this section, we will focus on illustrating the current strategies described for producing aromatic compounds, beginning with the products that are considered industrial building blocks, as salicylic acid, p-hydroxybenzoic acid, p-coumaric acid, cinnamic acid, ferulic acid and 2-phenylethanol. A brief overview of some relevant aromatic compounds that are widely used as fine chemicals, such as vanillin, will be further presented.
Salicylic acid (SLA) (2-hydroxybenzoic acid) is a valuable aromatic compound that can be obtained from CHO (Lin et al., 2014; Jiang and Zhang, 2016). SLA is an important drug precursor mainly used to produce acetylsalicylic acid, widely applied as a non-steroidal anti-inflammatory drug, in the treatment of fever, pain, aches and inflammations (Vane and Botting, 2003). Isochorismate synthase (ICS) converts CHO to isochorismate and then isochorismate pyruvate lyase (IPL) converts isochorismate into SLA (Serino et al., 1997; Figure 2). Lin et al. (2013) attained an SLA titer of 158 mg L–1 in E. coli after the expression of entC from E. coli (ICS step) and pfpchB from P. fluorescens (IPL step), as an operon. Lin et al. (2014) further improved the metabolic flux toward SLA using a medium copy number plasmid, pCA-APTA, to express aroL, ppsA, tktA and aroGfbr, under the control of an IPTG-inducible promoter (PLlacO1), attaining an SLA titer of 1.2 g L–1, using glycerol as carbon source. Noda et al. (2016) reported the highest SLA titer to the date, 11.5 g L–1 (Table 1), with a yield of 41.1 % from glucose, after enhancing the availability of PEP in E. coli. They removed the endogenous PEP consuming PTS, that was replaced by GalP/Glk system, as well as the genes responsible for the conversion of PEP to pyruvate (pykF and pykA). Finally, the strain was further modified by the introduction of menF from E. coli (ICS step) and pchB from P. aeruginosa (IPL step). In that report, an 8-fold increase in SLA concentration (from 1.4 to 11. 5 g L–1) was attained after the process scale up to 1-L jar fermenter. These findings demonstrate the importance of balancing the plasmid copy number and the impact of deleting the genes from SKM pathway in cell growth and production titers. However, the SA toxicity toward the producing cell remains a challenge, and it is necessary to develop more resistant strains or explore alternative chassis that naturally exhibit high tolerance toward toxic compounds, as Pseudomonas aeruginosa (Jiménez et al., 2002; Nikel and de Lorenzo, 2018). The microbial production of p-hydroxybenzoic acid (PHBA) can also be achieved from CHO by chorismate pyruvate lyase (Figure 2). This aromatic compound is used as a building block for liquid crystal polymers and for antibacterial parabens – a key group of compounds used as food preservatives (Barker and Frost, 2001), with an estimated market value of $150 million per year (Krömer et al., 2013). Nowadays, PHBA is chemically synthesized from benzene via cumene and phenol (Heinz-Gerhard and Jurgen, 2012). The biotechnological production of PHBA has already been described in plants, like tobacco (Nicotiana tabacum L.) and potato (Solanum tuberosum L.) (Köhle et al., 2003), in E. coli (Barker and Frost, 2001), Klebsiella pneumoniae (Müller et al., 1995), C. glutamicum (Kallscheuer and Marienhagen, 2018), and P. putida (Verhoef et al., 2007), using glucose as carbon source or with complex mixtures such as sugar cane (Mcqualter et al., 2005). Barker and Frost (2001) reported PHBA production in E. coli after overexpression of aroFfbr (feedback-inhibition resistant DAHP synthase), as well as the genes involved in the SKM pathway (tktA, aroA, aroL, aroC, and aroB) and encoding chorismate pyruvate lyase (ubiC, that was expressed in a plasmid under the control of a tac promoter). A PHBA yield of 12 g L–1 was obtained in a fed-batch fermentation. The application of an E. coli–E. coli co-culture system was recently reported by Zhang H. et al. (2015) for PHBA production from glucose and xylose, a sugar mixture that can be derived from lignocellulose. The authors reported a 8.6-fold improvement in PHBA production when its biosynthesis was switched from the monoculture strategy to the co-culture strategy. Finally, a fed-batch bioreactor was used to scale up the PHBA production and under this new condition its titer was improved to 2.3 g L–1. The production of this aromatic compound in yeast was described for the first time by Krömer et al. (2013). The authors reported a PHBA titer of 90 mg L–1 in S. cerevisiae after overexpression of an ubiC from E. coli and deletion of ARO7 and TRPp3, avoiding the biosynthesis of aromatic amino acids. To increase the flux to chorismate, they expressed ARO4K229L and aroL. This strain was then used and allowed a PHBA formation from CHO, with a titer of 2.9 g L–1 and yield of 3.1 mg gglucose–1, in a fed-batch process (Averesch et al., 2017). To date, the highest PHBA titer was reported by Kitade et al. (2018) in C. glutamicum (Table 1). This was achieved by chromosomal integration of aroG from E. coli and wild-type aroCKB from C. glutamicum, encoding chorismate synthase, shikimate kinase, and 3-dehydroquinate synthase. In order to convert CHO to HPBA a highly HPBA-resistant chorismate pyruvate lyase (encoded by ubiC) from the intestinal bacterium Providencia rustigianii was used. In order to increase product formation, the synthesis of by-products was also reduced by deleting hdpA and pyk. The final strain produced 36.6 g L–1 of PHBA from glucose after 24 h, with a glucose yield of 40 % (mol mol–1). Despite the efforts that have been made to increase the production yields and concentration of PHBA, the obtained results still fall behind those benchmarks from an industrial perspective. In fact, PHBA itself could also be toxic to the cells at high concentrations and the application of in situ product removal (ISPR) strategies will lead to a continuous PHBA removal from the fermentation broth. An interesting approach was presented by Johnson et al. (2000) that applied an in situ product removal technique for PHBA production with E. coli using Amberlite IRA-400 as adsorbent, and observed an increase in the PHBA titer from 6 to 22.9 g L–1. In the future, further improvements in the biotechnological process will be necessary, such as the utilization of low-cost substrates, as residues and wastes, as well as the development of a “green” downstream process, to pique the industry interest in these processes.
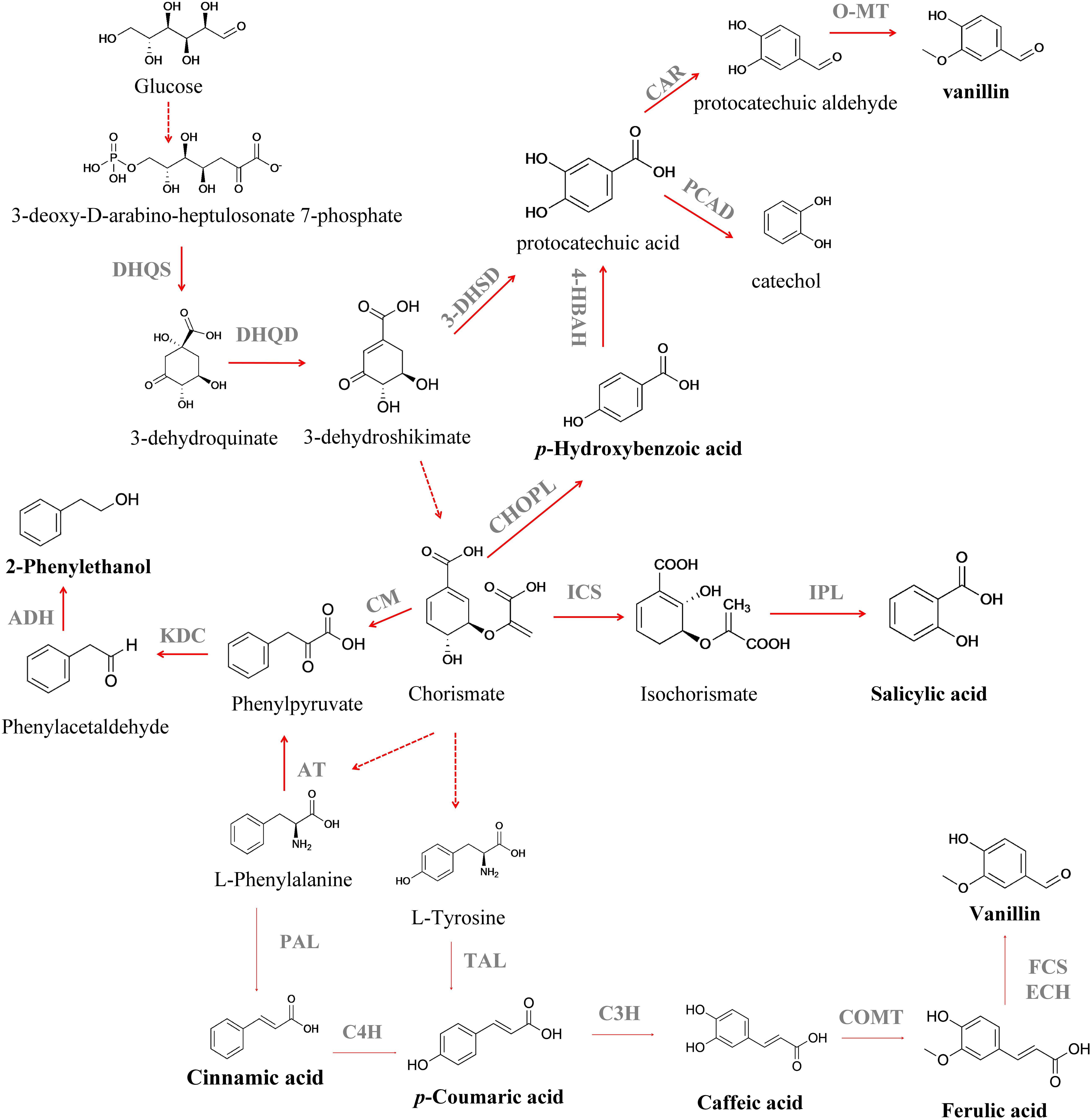
Figure 2. Biosynthesis of different aromatic compounds derived from the extended shikimate pathway. DHQS, 3-dehydroquinate synthase; DHQD, 3-dehydroquinate dehydratase; 3-DHSD, 3-dehydroshikimate dehydratase; CAR, carboxylic acid reductase; PCAD, protocatechuic acid decarboxylase; O-MT, O-methyltransferase; ICS, isochorismate synthase; CHOPL, chorismate pyruvate lyase; IPL, isochorismate pyruvate lyase; 4-HBAH, 4-hydroxybenzoic acid hydroxylase; CM, chorismate mutase; KDC, phenylpyruvate decarboxylase; ADH, alcohol dehydrogenase; PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; TAL, tyrosine ammonia lyase; FCS, feruloyl-CoA synthetase; ECH, feruloyl-CoA hydratase/lyase; C3H, p-coumarate 3-hydroxylase; AT, aminotransaminase. Solid lines indicate a single step; dotted lines indicate multiple steps.
Hydroxycinnamic acids are an important class of hydroxylated aromatic acids that contain a phenol ring and at least one organic carboxylic acid group. This group of compounds includes p-coumaric acid, caffeic acid and ferulic acid, among others. p-Coumaric acid is an important platform chemical used as monomer of liquid crystal polymers for electronics (Kaneko et al., 2006), as well as precursor for the synthesis of polyphenols (Rodriguez et al., 2015). The route for their production starts with L-Phe and L-Tyr deamination to further produce the phenylpropanoid cinnamic acid and p-coumaric acid, respectively, by the activity of phenylalanine ammonia lyase (PAL) (MacDonald and D’Cunha, 2007) and tyrosine ammonia lyase (TAL) (Nishiyama et al., 2010; Figure 2). The enzyme P450 monooxygenase cinnamate 4-hydroxylase (C4H) can further oxidize the cinnamic acid yielding the p-coumaric acid (Rasmussen et al., 1999; Achnine et al., 2004). The heterologous expression of TAL and PAL/TAL encoding genes allowed the p-coumaric acid production in E. coli, S. cerevisiae, Streptomyces lividans and P. putida (Table 1; Nijkamp et al., 2007; Trotman et al., 2007; Vannelli et al., 2007; Kawai et al., 2013; Rodriguez et al., 2015; Vargas-Tah and Gosset, 2015). However, due to their low activity, first studies reported its production from culture medium supplemented with L-Phe or L-Tyr (Ro and Douglas, 2004; Hwang et al., 2003; Watts et al., 2004; Jendresen et al., 2015; Mao et al., 2017). Efforts have also been made to find enzymes from different sources with higher PAL or TAL activity (Nijkamp et al., 2005, 2007; Vannelli et al., 2007; Kang et al., 2012; Jendresen et al., 2015). p-Coumaric acid production from a simple carbon source, such as glucose, is desirable. In order to achieve this, S. cerevisiae was genetically modified (Vannelli et al., 2007). The encoding PAL/TAL gene from Rhodotorula glutinis (expressed under the control of the galactose promoter) was used due to its higher affinity toward L-Tyr compared to L-Phe. The heterologous expression of a C4H gene from Helianthus tuberosus allowed its production via the PAL route. More recently, Li et al. (2018) proposed the p-coumaric acid production from glucose via phenylalanine in E. coli. They expressed the C4H-encoding gene from Lycoris aurea and PAL1 of Arabidopsis thaliana, under a trc promoter induced by IPTG, attaining a titer of 25.6 mg L–1 in shake flasks, after the regulation of the intracellular level of NADPH. The authors observed that the level of intracellular NADPH has a strong impact on the conversion of trans-cinnamic acid into p-coumaric acid and different strategies were tested in order to increase the level of intracellular NADPH. When pntAB, that encodes a membrane-bound transhydrogenase that catalyzes the NADH to NADPH conversion, was overexpressed under the control of a T7 constitutive promoter and the synthetic small regulatory RNA (srRNA) anti(SthA) was used to specifically repress the translation of the soluble transhydrogenase SthA, a synergetic positive effect was observed on the de novo production of p-coumaric acid. To date, the highest p-coumaric acid titer, 2.4 g L–1, has been achieved in S. cerevisiae after overexpression of the encoding TAL gene from Flavobacterium johnsoniae; overexpression of aroL from E. coli, under control of the P-TEF promoter; overexpression of Aro7G141S and Aro4K229L from S. cerevisiae under control of the promoters P-TEF and P-PGK1, respectively and deletion of Aro10 and Pdc5 genes (Rodriguez et al., 2017; Table 1). However, it is also important to explore the application of other raw materials derived from biomass as carbon sources – see section “Discussion.” Vargas-Tah and Gosset (2015) managed to produce cinnamic acid and p-coumaric acid in E. coli using lignocellulosic hydrolysates as complex carbon source. However, other strains that grow naturally on complex carbon sources were also used in the production of these hydroxycinnamic acids, such as Streptomyces lividans (Noda et al., 2011, 2012; Kawai et al., 2013) with product concentrations ranging from 130 to 736 mg L–1. In the last year, Borja et al. (2019) constructed a S. cerevisiae strain that uses xylose as sole carbon source for p-coumaric acid production, attaining a titer of 242 mg L–1, that represents a 45-fold increase over their condition with glucose (5.25 mg L–1) (Table 1). To construct this strain, they knocked out Aro10 and Pdc5 genes, in order to reduce the byproduct formation, and overexpressed the encoding TAL gene from F. johnsoniae and aroL from E. coli. To increase the carbon flux through the aromatic amino acid pathway they overexpress Aro7fbr (feedback-inhibition resistant DAHP synthase) and Aro4fbr (feedback-inhibition resistant chorismite mutase). Another important issue detected in these studies is the toxic effect of p-coumaric acid to the producing cells. An alternative strategy was proposed by Huang et al. (2013), that involves pulse feeding to avoid the accumulation of p-coumaric acid to a toxic level. The resistance to toxic compounds can also be increased using membrane transport engineering as a method to decrease the intracellular concentration of a toxic compound. In E. coli, the overexpression of aaeXAB gene, that encodes an efflux pump for several aromatic compounds, resulted in a twofold increase in tolerance to p-coumaric acid (Dyk et al., 2004; Sariaslani, 2007). Caffeic acid is another important intermediate of the phenylpropanoid metabolism. It serves as a precursor for the synthesis of caffeoyl alcohol and 3,4-dihydroxystyrene (monomer for plastic synthesis) (Zhang and Stephanopoulos, 2013). Caffeic acid is biosynthesized by hydroxylation of p-coumaric acid through p-coumarate 3-hydroxylase (C3H) (Berner et al., 2006; Figure 2). First studies have reported microbial caffeic acid production with medium supplementation of the precursors such as L-Tyr and p-coumaric acid (Sachan et al., 2006; Choi et al., 2011). In order to produce it directly from p-coumaric acid, it is necessary to express the genes that encode enzymes with suitable ring 3-hydroxylation activity (Berner et al., 2006; Furuya et al., 2012). In fact, one of the major difficulties in the heterologous expression of genes of the plant phenylpropanoid pathway in E. coli is the lack of cytochrome P450 reductase activity making the search for alternative strategies essential. Choi et al. (2011) identified that the sam5 gene from Saccharothrix espanaensis encodes C3H, that is a FAD-dependent enzyme. This enzyme was then used to produce caffeic acid in E. coli, demonstrating the feasibility of this two-step pathway to produce caffeic acid using alternative enzymes. It was also demonstrated that a bacterial cytochrome P450 CYP199A2, from Rhodopseudomonas palustris, was able to efficiently convert p-coumaric acid to caffeic acid (Hernández-Chávez et al., 2019). Some microorganisms, harboring genes encoding the two sub-units of the enzyme 4-hydroxyphenylacetate 3-hydroxylase (4HPA3H), have proved to be able to act on aromatic compounds (Galán et al., 2000). It was also observed that 4HPA3H is able to convert p-coumaric acid into caffeic acid. Based on this Furuya and Kino (2014) expressed the 4HPA3H gene from Pseudomonas aeruginosa in E. coli and a caffeic acid production of 10.2 g L–1 was obtained after repeated additions of p-coumaric acid (20 mM each pulse), in a medium with glycerol as carbon source. For caffeic acid biosynthesis from L-Tyr an additional step of non-oxidative deamination, catalyzed by TAL (Rodrigues et al., 2015) is needed, and the first approach is the search for alternative enzymes from different sources. TAL from R. glutinis proved to be the most active TAL identified (Vannelli et al., 2007; Santos et al., 2011). However, the production of this compound from simple carbon sources is much more desirable and the production of caffeic acid from glucose and xylose was described (Lin and Yan, 2012; Zhang and Stephanopoulos, 2013). The use of renewable feedstocks, such as lignocellulosic biomass, was also evaluated using E. coli as producing host (Kawaguchi et al., 2017). A maximum caffeic acid concentration of 233 mg L–1 (Table 1) was produced from kraft pulp using a tyrosine-overproducing E. coli strain harboring the hpaBC gene from P. aeruginosa and fevV gene from Streptomyces sp. WK-5344. Ferulic acid is a component of lignocellulose (De Oliveira et al., 2015). This O-methylated hydroxycinnamic acid is biosynthesized from caffeic acid by caffeic acid O-methyltransferase (COMT) (Figure 2). However, there are few reports about the heterologous production of ferulic acid in microbial hosts. Choi et al. (2011) reported the production of ferulic acid in E. coli by expression of sam5, a TAL gene from S. espanaensis and a COMT gene from A. thaliana, under the control of a T7 promoter, attaining a titer of 0.1 mg L–1 from L-Tyr. Later, Kang et al. (2012) engineered an E. coli strain capable of producing 196 mg L–1 of ferulic acid from glucose, after introduction of COMT from A. thaliana in a caffeic acid-over-producing strain, containing a codon optimized tal gene, under the control of a T7 promoter. This aromatic acid can also be used as substrate for vanillin and coniferyl alcohol biosynthesis (Hua et al., 2007; Lee et al., 2009; Chen et al., 2017). Research efforts have been made in order to make the microbial production of hydroxycinnamic acids a competitive process.
Another important class of aromatic compounds is the aromatic flavors class. The two most popular benzenoid flavors are 2-phenylethanol (2-PE) and vanillin. 2-PE is an aromatic alcohol with a delicate fragrance of rose petals widely used in flavor and fragrances industries (Burdock, 2010; Carlquist et al., 2015). It was recently identified as a potent next generation biofuel (Keasling and Chou, 2008). Furthermore, 2-PE can also be used as raw material to produce other flavor compounds (2-phenylethyl acetate and phenylacetaldehyde) and styrene (Etschmann et al., 2002). Microorganisms can naturally produce 2-PE as part of their amino acid metabolism (Etschmann et al., 2002). This benzoid flavor can be produced through the SKM pathway or via Ehrlich pathway from L-Phe (Albertazzi et al., 1994; Carlquist et al., 2015; Figure 2). Through the Ehrlich pathway, L-Phe is firstly converted to phenylpyruvate by transamination, which is then transformed to phenylacetaldehyde by decarboxylation. Then, the derivative aldehyde is reduced to 2-PE by an alcohol dehydrogenase (Etschmann et al., 2002; Figure 2). This is the fastest pathway to produce 2-PE, however cheaper precursors than L-Phe should be used to achieve a more competitive process (ÄYräpää, 1965; Kim T.Y. et al., 2014; Zhang et al., 2014). Thus, de novo production of 2-PE has been described in different microorganisms: Kluyveromyces marxianus (Kim T.Y. et al., 2014), E. coli (Guo et al., 2018; Liu et al., 2018), and Enterobacter sp. (Zhang et al., 2014; Table 1). The 2-PE production from the SKM pathway is achieved from its end product phenylpyruvate, that is decarboxylated to phenylacetaldehyde, followed by a dehydrogenation that leads to 2-PE production (Etschmann et al., 2002; Carlquist et al., 2015; Figure 2).
Nonetheless, the de novo synthesis is inefficient since glycolysis and PPP are mainly used for cell growth, producing very low 2-PE concentrations. The most common strategies employed for strain constructions focus on increasing phenylpyruvate decarboxylase and alcohol dehydrogenase activities, which are the rate-limiting enzymes in de novo synthesis pathway, in combination with feedback-resistant DAHP synthase and chorismate mutase. Kim B. et al. (2014) overexpressed the Aro10, that encodes a transaminated amino acid decarboxylase and Adh2, encoding an alcohol dehydrogenase, from S. cerevisiae in K. marxianus BY25569, under the control of the constitutive promoter ScPGK1/ScTEF1. Then, serial subcultures with an L-Phe analog, p-fluorophenylalanine, were conducted in order to obtain an evolved strain resistant to the L-Phe analog. Finally, the expression of aroGfbr from Klebsiella pneumoniae, that encodes a feedback-resistant mutant of DAHP synthase, was also performed. This genetically modified strain was able to produce 1.3 g L–1 of 2-PE from glucose without addition of L-Phe. More recently, Liu et al. (2018) constructed a heterologous pathway from Proteus mirabilis in E. coli and the recombinant strain was able to produce 1.2 g L–1 of 2-PE without L-Phe supplementation (Table 1).
Another challenge in the biosynthesis of 2-PE is its toxicity. Different strategies were investigated in order to improve the process yield and productivity, being the most common the optimization of medium composition, operational conditions and application of ISPR techniques (Chung et al., 2000; Hua et al., 2010; Cui et al., 2011; Celińska et al., 2013; Mihal’ et al., 2014). 2-PE production is highly dependent on media composition and culture conditions (Garavaglia et al., 2007). The utilization of an interesting alternative carbon source was reported by Celińska et al. (2013). They use glycerol as carbon source in bioconversion of L-Phe to 2-PE by Yarrowia lipolytica NCYC3825, reaching a 2-PE production of 0.77 g L–1 after 54 h. Another recent approach for this flavor production was reported by Martínez-Avila et al. (2018). In the proposed system, K. marxianus ATCC10022 used the available nutrients from a residue-substrate (sugarcane bagasse) supplemented with L-Phe, achieving a 2-PE production of 10.21 mg g–1 (mass of product per mass of solid) in a fed-batch system. The application of alternative modes of operation, such as fed-batch and continuous, that allow the possible removal or dilution of 2-PE in the medium, are also interesting approaches recently reported. Last year, de novo production of 2-PE by Metschnikowia pulcherrima NCYC373 reached higher titers in continuous mode operation, than in batch and fed-batch cultures (Chantasuban et al., 2018). In continuous fermentation, 2-PE concentration levels reached 1.5 g L–1, before it became too toxic and caused the flush out (Table 1). Even with the efforts to optimize the culture medium and cultivation conditions, and choose the most producing microorganism, product inhibition is still the major problem of 2-PE biosynthesis (Carlquist et al., 2015). Some strategies, such as ISPR techniques, have been developed to reduce the 2-PE toxicity in the fermentation medium, increasing its production (Carlquist et al., 2015). Recently, Chantasuban et al. (2018) reported the application of oleyl alcohol as an extraction phase in the 2-PE production by M. pulcherrima NCYC373. The production levels were enhanced with the application of this ISPR technique, achieving a 2-PE concentration of 1.96 g L–1 in the aqueous phase and an overall production of 3.13 g L–1. Gao and Daugulis (2009) reported a highly significant enhancement in the 2-PE production, using a solid-liquid two-phase partition bioreactor with polymer beads as the sequestering immiscible phase. The batch mode system reached a final 2-PE concentration of 13.7 g L–1 (88.74 g L–1 in the polymer phase and 1.2 g L–1 in the aqueous phase), whereas the fed-batch achieved an overall titer of 20.4 g L–1 (97.0 g L–1 in the polymer phase and 1.4 g L–1 in the aqueous phase). During the last years, great improvements have been achieved in the bioproduction of 2-PE leaving its industrial application closer. In fact, 2-PE concentrations of 21 g L–1 were reached (Mihal’ et al., 2014), in an hybrid system that consists of a fed-batch stirred tank bioreactor and a hollow fiber membrane module immersed at the bottom of the bioreactor, where 2-PE is continuously extracted from the fermentation broth using pentane as the organic phase.
Vanillin (4-hydroxy-3-methoxybenzaldehyde), a widely used flavor compound in different industries, is the primary component of the extract of the vanilla bean. The economic importance of this plant natural product is quite significant; it was reported that synthetic vanillin has a price of around US$ 11 kg–1, while biotech vanillin is sold for a price of around US$ 1000 kg–1 (Schrader et al., 2004). Over the last years, vanillin production through biotransformation of ferulic acid, isoeugenol, lignin, was reported, with vanillin titers that range from 0.13 to 32.5 g L–1 (Huang et al., 1993; Priefert et al., 2001; Zhao et al., 2005; Kaur and Chakraborty, 2013). Furuya et al. (2015) reported a vanillin concentration of 7.8 g L–1 from ferulic acid in a two-stage process with an E. coli carrying two expression plasmids harboring fdc from Bacillus pumilus and cso2 from Caulobacter segnis. However, the vanillin production through these pathways has several bottlenecks that include the price of precursors, the formation of undesired side-products and the cytotoxicity of the precursors (Gallage and Møller, 2015). Based on this, its production by de novo biosynthesis from cheap and more available carbon sources is much more attractive.
In the pathway from 3-dehydroshikimate (3-DHS) to vanillin, the first step is the dehydration of 3-DHS to protocatechuic acid, catalyzed by 3-dehydroshikimate dehydratase, that is further converted to protocatechuic aldehyde, by carboxylic acid reductase; and, the final step is catalyzed by an O-methyltransferase leading to vanillin (Hansen et al., 2009; Figure 2). Li and Frost (1998) were the first to report an engineered pathway for vanillin production in E. coli. Here, protocatechuic acid was converted to vanillic acid by catechol-O-methyltransferase, and further reduced to vanillin by an aryl aldehyde dehydrogenase. Hansen et al. (2009) explored for the first time the vanillin production in the yeasts Schizosaccharomyces pombe and S. cerevisiae. The authors introduced a DHS dehydratase (3DSD) from Podospora pausiceta, an aromatic carboxylic acid reductase (ACAR) from Nocardia sp., a phosphopantetheinyl transferase (PPTase) from C. glutamicum to activate ACAR, and an O-methyl transferase (OMT) from Homo sapiens, allowing a vanillin production of 45 mg L–1. More recently, Kunjapur et al. (2014) used an E. coli strain with decreased aromatic aldehyde reduction activity as a host for the biosynthesis of vanillin. A vanillin titer of 119 mg L–1 was achieved using an E. coli strain expressing a Bacillus thuringiensis 3-dehydroshikimate dehydrogenase gene (asbF), a H. sapiens O-methyltransferase gene (Hs-S-COMT) and Nocardia iowensis carboxylic acid reductase gene (car), that are codon optimized and expressed in a plasmid. They also introduced a feedback-resistant DAHP synthase (encoded by aroG) and a phosphopantetheinyl transferase (encoded by sfp) from B. subtilis, which have been shown to activate CAR.
However, de novo vanillin production in recombinant bacteria and yeasts still has challenges that are not only related with product formation itself but also the product toxicity. The major hurdle in the biotechnological production of vanillin is the strong inhibitory effect that this flavor has on microorganism growth (Gallage et al., 2014; Ma and Daugulis, 2014). An interesting approach was proposed by Brochado et al. (2010), in which the natural pathway for vanillin production in plants was mimicked and assembled in S. cerevisiae. To overcome the toxicity of vanillin, a gene encoding an uridine diphosphate–glucose glycosyltransferase (UGT) from Arabidopsis thaliana was expressed in S. cerevisiae. This UGT catalyzes the glycosylation of vanillin and produces a less toxic final product, vanillin-β-d-glucoside (VG). The same strategy was implemented by Ni et al. (2015) allowing a VG production of 500 mg L–1, with a yield of 32 mg gglucose–1, that is 5-fold higher than the 45 mg L–1 reported by Hansen et al. (2009). A different strategy was presented by Yoon et al. (2007) for vanillin production from ferulic acid using an E. coli strain harboring a plasmid with fcs (feruloyl-CoA synthase) and ech (enoyl-CoA hydratase/aldolase) genes from Amycolatopsis sp. strain. To reduce the vanillin toxicity, they improved the vanillin-resistance of this strain using NTG mutagenesis as well as a XAD-2 resin to remove the vanillin from the medium. When 50 % (w/v) of XAD-2 resin was used with 10 g L–1 of ferulic acid, the vanillin production with the NTG-VR1 mutant strain was 2.9 g L–1, which was 2-fold higher than that obtained without resin. Recently, Luziatelli et al. (2019) reported, for the first time, the vanillin production from ferulic acid using a plasmid free E. coli strain, after chromosomal integration of fcs and ech genes from Pseudomonas. In addition, they also performed an optimization of the bioconversion conditions (namely stirring speed and initial substrate concentration) using a response surface methodology. At the same time, the authors used a two-phase (solid-liquid) system where the substrate was incorporated in a gel matrix (agarose-gel) in order to perform a fixed volume fed-batch approach, for controlled release of ferulic acid. Using this two-phase system, a vanillin titer of 4.3 g L–1 was attained in the liquid phase – one of the highest found in the literature for recombinant E. coli strains (Table 1).
Discussion
The production of aromatic compounds through plant extraction or chemical synthesis is a profitable business that’s been in place for quite a few years, now. In order to replace these industrial processes by fermentation based biotechnological processes, these must have clear economic, environmental and/or product wise advantages. From an economic point of view, the advantages have to be significant enough to justify the investment on new industrial equipment.
In general, these processes have a lower environment impact and are able to produce high quality final products when compared to their plant extraction and/or chemical synthesis counterparts (Thompson et al., 2015; Dudnik et al., 2017; Kallscheuer et al., 2019).
One of the advantages of fermentation based processes is the low-cost and abundance of the raw materials – low added value sugars. However, in order to achieve this advantage, the processes must not consider the supply of expensive precursors, antibiotics, inducers, etc… The optimization of the microorganisms in order to produce the desired aromatic compound directly form the substrate is usually a requirement to achieve economic feasibility. This is supported by Li et al. (2018) and Liu et al. (2018) studies – among other examples previously presented (Table 1) – showing that the strain engineering toward deregulation of the aromatic amino acids metabolism and an optimal connection of the heterologous pathways to the host metabolism enable aromatic compounds production starting from glucose without any need for supplementation of precursor metabolites.
In recent years, some research effort has been put into replacing the hydrocarbon source for these fermentation processes by residues and waste materials (Vargas-Tah and Gosset, 2015; Martínez-Avila et al., 2018; Borja et al., 2019). Sugars like glucose are abundant low-cost raw materials. On the other the use of waste materials in large scale industrial processes implies assuring a reliable constant supply and usually a pre-treatment step that may add a significant cost to the process. Nevertheless, whenever there is a need to process these wastes in order avoid their environmental impact, a fermentation process that converts them to higher added value products is an alternative worth considering.
Another possible bottleneck for the industrialization of these processes is the cost of the purification step. Microorganisms tend to produce a mixture of by-products where the compound of interest may be in higher or lower concentration. Although the amount of research put into this field is not always as significant as one would expect, the search for higher titers and the use of ISPR techniques has direct impact on the economic sustainability of these processes (Van Hecke et al., 2014). Moreover, it will also be interesting to study the synergetic application of engineered strains, able to use wastes and residues as substrates, in systems using ISPR techniques, as well as the product recovery and purification (Braga et al., 2018b; Kallscheuer et al., 2019).
Different microorganisms have been used to produce aromatic compounds. The most commonly used hosts are E. coli, S. cerevisiae and C. glutamicum. However, tolerant and thermotolerant microorganisms, as well as a broad spectrum of bacteria were also employed as aromatic compound producers (Table 1). The choice of the microorganism employed is always a determinant factor for the outcome of any research project in this field. The exploitation of well established platform organisms, for which metabolic engineering tools are available, is the most common approach. However, it is also important to explore non-model organisms that can naturally produce the desired compounds, even though the available genetic tools are still scarce. For example, the metabolic versatility of Pseudomonas as well as it inherent tolerance to toxic compounds, offers an excellent starting point for suppressing the hurdles of using and producing toxic compounds of natural or heterogeneous origin (Krömer, 2016; Lenzen et al., 2019; Table 1).
Some aromatic compounds are toxic for the producing host. In order to tackle this obstacle, several alternative strategies have been proposed such as: starting the fermentation with a lower substrate concentration and further additions following its consumption rate (step-wise fed-batch); the use of in situ product removal strategies and the application of adaptive evolution to obtain strains with enhanced resistance to toxic products. Aiming at industrial scale operation, the reduction of the production costs is always crucial. In order to reduce the medium cost, chromosomal integration of heterologous genes avoid the use of expensive antibiotics and inducers for plasmid maintenance and inducible expression (Cui et al., 2014).
Nowadays, the microbial production of aromatic compounds is already implemented at industrial scale in economically viable process. Evolva, for instance, has launched a process for vanillin production from glucose with a genetically modified Schizosaccharomyces pombe (Vanilla, 2014). Similarly, Solvay has a process for vanillin production with Streptomyces setonii from ferulic acid (Muheim et al., 2001). In these cases, the product titers attained are in the order of a hundred g per L, and the methodologies for product separation and purification are well established. However, despite the efforts that have been made to increase the production titers with microbial hosts, the concentrations obtained for more complex compounds are still too low (mg and μg per L) for an industrial process (Table 1), and these molecules are still produced by extraction from natural sources or by chemical synthesis. In fact, it has been predicted that the market value of a bulk chemical is less than $10 kg–1. On the other hand, fine chemicals are produced in limited volumes (<1000 tons per year) but at relatively high prices (>$10kg–1) (Joshi and Ranade, 2016). Based on this, at least for now, the commercialization of fine aromatic compounds will emerge with more successes (Cao et al., 2019). The current challenges that still need to be addressed are the metabolic imbalances in the producer strain, the availability of the metabolites required for biomass formation and the product extraction and purification. In a near future it can be expected that with the application of novel synthetic biology approaches, such as CRISPR/Cas9, rational strain engineering, adaptive laboratory evolution and high-throughput screening approaches, it will be possible to render the microbial production of additional aromatic compounds and its derivatives economically viable.
Final Remarks
Production cost is, by far, the main obstacle to overcome in order to industrialize the production of aromatic compounds through fermentation. This explains the two main research goals mentioned throughout this review: the maximization of product titers and the removal of expensive fermentation media ingredients.
The main challenges to address are still the low availability of precursor molecules by the microbial metabolism, the elimination of complex pathway regulations, disruption of competing pathways and the low activity of heterologous enzymes in the microbial hosts. It is expected that with recent technological innovations the engineering of microbial host strains will be faster providing more precursor molecules to increase aromatic compound synthesis. Identification of the most suitable enzymes and their further improvement will also be an important step toward the production of different aromatic chemicals, avoiding the accumulation of undesired intermediates that can be toxic to the microbial host and leading to an increase in the final product titer. Metabolic engineering, system and synthetic biology tools for strain design, together with process engineering strategies have been and will continue to be the main resources applied. In addition, the identification of novel enzymes that catalyze non-natural reactions, or novel synthetic pathways not found in nature, allow the production of the desired molecule with a high yield or a non-natural compound with possibly superior or new therapeutic properties.
Overall, the paradigm moves toward the development of better microbial chassis and new metabolic pathways that allow the shift from producing aromatic compounds from fossil resources to a bio-based production. The authors consider that now, the biotechnological production of aromatics is not a question of whether or not it is theoretically possible, but of when will it become technically and economically feasible.
Author Contributions
AB and NF contributed to the conception and design of the study. AB reviewed the literature, extracted the data, and drafted the manuscript. NF edited the manuscript. Both authors read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Achnine, L., Blancaflor, E. B., Rasmussen, S., Dixon, R. A., Division, P. B., Roberts, S., et al. (2004). Colocalization of L -Phenylalanine ammonia-lyase and cinnamate 4-Hydroxylase for metabolic channeling in Phenylpropanoid biosynthesis. Plant Cell 16, 3098–3109. doi: 10.1105/tpc.104.024406.pathways
Ahn, J. O., Lee, H. W., Saha, R., Park, M. S., Jung, J. K., and Lee, D. Y. (2008). Exploring the effects of carbon sources on the metabolic capacity for shikimic acid production in Escherichia coli using in silico metabolic predictions. J. Microbiol. Biotechnol. 18, 1773–1784. doi: 10.4014/jmb.0700.705
Albertazzi, E., Cardillo, R., Servi, S., and Zucchi, G. (1994). Biogeneration of 2-phenylethanol and 2-phenylethylacetate important aroma components. Biotechnol. Lett. 16, 491–496. doi: 10.1007/BF01023331
Averesch, N. J. H., and Kayser, O. (2014). Assessing heterologous E xpression of Hyoscyamine 6-β-Hydroxylase - a feasibility study. Proc. Chem. 13, 69–78. doi: 10.1016/j.proche.2014.12.008
Averesch, N. J. H., and Krömer, J. O. (2018). Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds—present and future strain construction strategies. Front. Bioeng. Biotechnol. 6:32. doi: 10.3389/fbioe.2018.00032
Averesch, N. J. H., Prima, A., and Kro, J. O. (2017). Enhanced production of para -hydroxybenzoic acid by genetically engineered Saccharomyces cerevisiae. Bioprocess Biosyst. Eng. 40, 1283–1289. doi: 10.1007/s00449-017-1785-z
ÄYräpää, T. (1965). Tthe formation of phenethyl alcohol from 14C-labelled Phenylalanine. J. Inst. Brew. 71, 341–347. doi: 10.1002/j.2050-0416.1965.tb02068.x
Backman, K., Oconnor, M. J., Maruya, A., Rudd, E., McKay, D., Balakrishnan, R., et al. (1990). Genetic engineering of metabolic pathways applied to the production of phenylalanine. Ann. N. Y. Acad. Sci. 589, 16–24. doi: 10.1111/j.1749-6632.1990.tb24231.x
Báez-Viveros, J., Osuna, J., Hernández-Chávez, G., Soberón, X., Bolívar, F., and Gosset, G. (2004). Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli. Biotechnol. Bioeng. 87, 516–524. doi: 10.1002/bit.20159
Balderas-hernández, V. E., Sabido-ramos, A., Silva, P., Cabrera-valladares, N., Hernández-chávez, G., Báez-, J. L., et al. (2009). Metabolic engineering for improving anthranilate synthesis from glucose in Escherichia coli. Microb. Cell Fact. 12, 1–12. doi: 10.1186/1475-2859-8-19
Barker, J. L., and Frost, J. W. (2001). Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol. Bioeng. 76, 376–339.
Beata, Ż, Niemczyk, E., and Lipok, J. (2019). Metabolic relation of cyanobacteria to aromatic compounds. Appl. Microbiol. Biotechnol. 103, 1167–1178. doi: 10.1007/s00253-018-9568-2
Bentley, W. E., Mirjalili, N., Andersen, D. C., Davis, R. H., and Kompala, D. S. (1990). Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol. Bioeng. 35, 668–681. doi: 10.1002/bit.260350704
Berner, M., Krug, D., Bihlmaier, C., Vente, A., Mu, R., and Bechthold, A. (2006). Genes and enzymes involved in Caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J. Bacteriol. 188, 2666–2673. doi: 10.1128/JB.188.7.2666
Bongaerts, J., Krämer, M., Müller, U., Raeven, L., and Wubbolts, M. (2001). Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3, 289–300. doi: 10.1006/mben.2001.0196
Borja, G. M., Rodriguez, A., Campbell, K., Borodina, I., Chen, Y., and Nielsen, J. (2019). Metabolic engineering and transcriptomic analysis of Saccharomyces cerevisiae producing p-coumaric acid from xylose. Microb. Cell Fact. 18, 1–14. doi: 10.1186/s12934-019-1244-1244
Borodina, I., and Nielsen, J. (2014). Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 9, 609–620. doi: 10.1002/biot.201300445
Braga, A., Ferreira, P., Oliveira, J., Rocha, I., and Faria, N. (2018a). Heterologous production of resveratrol in bacterial hosts: current status and perspectives. World J. Microbiol. Biotechnol. 34:122. doi: 10.1007/s11274-018-2506-2508
Braga, A., Silva, M., Oliveira, J., Silva, A. R., Ferreira, P., Ottens, M., et al. (2018b). An adsorptive bioprocess for production and recovery of resveratrol with Corynebacterium glutamicum. J. Chem. Technol. Biotechnol. 93, 1661–1668. doi: 10.1002/jctb.5538
Brochado, A., Matos, C., Møller, B. L., Hansen, J., Mortensen, U. H., and Patil, K. (2010). Improved vanillin production in baker’s yeast through in silico design. Microb. Cell Fact. 9:84. doi: 10.1186/1475-2859-9-84
Bulter, T., Bernstein, J. R., and Liao, J. C. (2003). A perspective of metabolic engineering strategies: moving up the systems hierarchy. Biotechnol. Bioeng. 84, 815–821. doi: 10.1002/bit.10845
Cao, M., Gao, M., Suastegui, M., Mei, Y., and Shao, Z. (2019). Building microbial factories for the production of aromatic amino acid pathway derivatives: from commodity chemicals to plant-sourced natural products. Metab. Eng. doi: 10.1016/j.ymben.2019.08.008
CrossRef Full Text [Epub ahead of print],
Carlquist, M., Gibson, B., Yuceer, Y. K., Paraskevopoulou, A., Sandell, M., Angelov, A. I., et al. (2015). Process engineering for bioflavour production with metabolically active yeasts – a mini-review. Yeast 32, 123–143. doi: 10.1002/yea.3058
Celińska, E., Kubiak, P., Białas, W., Dziadas, M., and Grajek, W. (2013). Yarrowia lipolytica: the novel and promising 2-phenylethanol producer. J. Ind. Microbiol. Biotechnol. 40, 389–392. doi: 10.1007/s10295-013-1240-1243
Chakraborty, D., Gupta, G., and Kaur, B. (2016). Metabolic engineering of E. coli top 10 for production of vanillin through FA catabolic pathway and bioprocess optimization using RSM. Protein Expr. Purif. 128, 123–133. doi: 10.1016/j.pep.2016.08.015
Chandran, S., Yi, J., Draths, K., Von Daeniken, R., Weber, W., Frost, J. W., et al. (2003). Phosphoenolpyruvate availability and the biosynthesis of Shikimic acid. Biotechnol. Prog. 19, 808–814. doi: 10.1021/bp025769p
Chantasuban, T., Santomauro, F., Gore-lloyd, D., Parsons, S., Henk, D., Scott, J., et al. (2018). Elevated production of the aromatic fragrance molecule, 2-phenylethanol, using Metschnikowia pulcherrima through both de novo and ex novo conversion in batch and continuous modes. J. Chem. Technol. Biotechnol. 93, 2118–2130. doi: 10.1002/jctb.5597
Chen, K., Dou, J., Tang, S., Yang, Y., Wang, H., Fang, H., et al. (2012). Deletion of the aroK gene is essential for high shikimic acid accumulation through the shikimate pathway in E. coli. Bioresour. Technol. 119, 141–147. doi: 10.1016/j.biortech.2012.05.100
Chen, Z., Sun, X., Li, Y., Yan, Y., and Yuan, Q. (2017). Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab. Eng. 39, 102–109. doi: 10.1016/j.ymben.2016.10.021
Choi, O., Wu, C.-Z., Kang, S. Y., Ahn, J. S., Uhm, T.-B., and Hong, Y.-S. (2011). Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J. Ind. Microbiol. Biotechnol. 38, 1657–1665. doi: 10.1007/s10295-011-0954-953
Chouhan, S., Sharma, K., Zha, J., Guleria, S., and Koffas, M. A. G. (2017). Recent advances in the recombinant biosynthesis of polyphenols. Front. Microbiol. 8:2259. doi: 10.3389/fmicb.2017.02259
Chung, H., Lee, S. L., and Chou, C. C. (2000). Production and molar yield of 2-phenylethanol by Pichia fermentans L-5 as affected by some medium components. J. Biosci. Bioeng. 90, 142–147. doi: 10.1016/S1389-1723(00)80101-80102
Cui, Y., Ling, C., Zhang, Y., Huang, J., and Liu, J. (2014). Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering. Microb. Cell Fact. 13, 1–11. doi: 10.1186/1475-2859-13-21
Cui, Z., Yang, X., Shen, Q., Wang, K., and Zhu, T. (2011). Optimisation of biotransformation conditions for production of 2-phenylethanol by a Saccharomyces cerevisiae CWY132 mutant. Nat. Prod. Res. 25, 754–759. doi: 10.1080/14786419.2010.529441
Curran, K. A., Leavitt, J. M., Karim, A. S., and Alper, H. S. (2013). Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 15, 55–66. doi: 10.1016/j.ymben.2012.10.003
De Oliveira, D. M., Finger-teixeira, A., Mota, T. R., Salvador, V. H., Moreira-vilar, C., Bruno, H., et al. (2015). Ferulic acid: a key component in grass lignocellulose recalcitrance to hydrolysis. Plant Biotechnol. J. 13, 1224–1232. doi: 10.1111/pbi.12292
Dell, K. A., and Frost, J. W. (1993). Identification and removal of impediments to biocatalytic synthesis of aromatics from D-Glucose: rate-limiting enzymes in the common pathway of aromatic amino acid biosynthesis. J. Am. Chem. Soc. 115, 11581–11589. doi: 10.1021/ja00077a065
Dhamankar, H., and Prather, K. L. J. (2011). Microbial chemical factories: recent advances in pathway engineering for synthesis of value added chemicals. Curr. Opin. Struct. Biol. 21, 488–494. doi: 10.1016/j.sbi.2011.05.001
Dias, F. M. S., Gomez, J. G. C., and Silva, L. F. (2017). Exploring the microbial production of aromatic fine chemicals to overcome the barriers of traditional methods. Adv. Appl. Sci. Res. 8, 94–109. doi: 10.1038/nmeth.1297
Dudnik, A., Almeida, A. F., Andrade, R., Avila, B., Bañados, P., Barbay, D., et al. (2017). BacHBerry: BACterial Hosts for production of Bioactive phenolics from bERRY fruits. Phytochem. Rev. 17, 291–326. doi: 10.1007/s11101-017-9532-9532
Dudnik, A., Gaspar, P., Neves, A. R., and Forster, J. (2018). Engineering of microbial cell factories for the production of plant polyphenols with health-beneficial properties. Curr. Pharm. Des. 24, 2208–2225. doi: 10.2174/1381612824666180515152049
Dyk, T. K., Van Templeton, L. J., Cantera, K. A., Sharpe, P. L., and Sariaslan, F. S. (2004). Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186, 7196–7204. doi: 10.1128/JB.186.21.7196
Escalante, A., Calderón, R., Valdivia, A., de Anda, R., Hernández, G., Ramírez, O. T., et al. (2010). Metabolic engineering for the production of shikimic acid in an evolved Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system. Microb. Cell Fact. 9, 1–12. doi: 10.1186/1475-2859-9-21
Etschmann, M., Bluemke, W., Sell, D., and Schrader, J. (2002). Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 59, 1–8. doi: 10.1007/s00253-002-0992-x
Frost, J. W., and Draths, K. M. (1995). Biocatalytic syntheses of aromatics from D-glucose: renewable microbial sources of aromatic compounds. Annu. Rev. Microbiol. 49, 557–579. doi: 10.1146/annurev.mi.49.100195.003013
Furuya, T., Arai, Y., and Kino, K. (2012). Biotechnological production of caffeic acid by bacterial cytochrome. Appl. Environ. Microbiol. 78, 6087–6094. doi: 10.1128/AEM.01103-1112
Furuya, T., and Kino, K. (2014). Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 98, 1145–1154. doi: 10.1007/s00253-013-4958-y
Furuya, T., Miura, M., Kuroiwa, M., and Kino, K. (2015). High-yield production of vanillin from ferulic acid by a coenzyme-independent decarboxylase/oxygenase two-stage process. N. Biotechnol. 32, 335–339. doi: 10.1016/j.nbt.2015.03.002
Galán, B., Díaz, E., Prieto, M. A., and García, J. L. (2000). Functional analysis of the small component of the 4-Hydroxyphenylacetate 3-Monooxygenase of Escherichia coli W: a prototype of a new flavin: NAD (P)H reductase subfamily. J. Bacteriol. 182, 627–636. doi: 10.1128/jb.182.3.627-636.2000
Gallage, N. J., Hansen, E. H., Kannangara, R., Olsen, C. E., Motawia, M. S., Jørgensen, K., et al. (2014). Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme. Nat. Commun. 5:5037. doi: 10.1038/ncomms5037
Gallage, N. J., and Møller, B. L. (2015). Vanillin-bioconversion and bioengineering of the most popular plant flavor and its de novo biosynthesis in the vanilla orchid. Mol. Plant 8, 40–57. doi: 10.1016/j.molp.2014.11.008
Gao, F., and Daugulis, A. J. (2009). Bioproduction of the aroma compound 2-phenylethanol in a solid-liquid two-phase partitioning bioreactor system by Kluyveromyces marxianus. Biotechnol. Bioeng. 104, 332–339. doi: 10.1002/bit.22387
Garavaglia, J., Flôres, S. H., Pizzolato, T. M., Peralba, M. D. C., and Ayub, M. A. Z. (2007). Bioconversion of L-phenylalanine into 2-phenylethanol by Kluyveromyces marxianus in grape must cultures. World J. Microbiol. Biotechnol. 23, 1273–1279. doi: 10.1007/s11274-007-9361-9363
Gaspar, P., Dudnik, A., Neves, A. R., and Föster, J. (2016). “Engineering Lactococcus lactis for stilbene production,” in Proceedings of the Abstract from 28th International Conference on Polyphenols 2016, Vienna.
Ger, Y. M., Chen, S. L., Chiang, H. J., and Shiuan, D. (1994). A single ser-180 mutation desensitizes feedback inhibition of the phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthetase in Escherichia coli. J. Biochem. 116, 986–990. doi: 10.1093/oxfordjournals.jbchem.a124657
Gosset, G. (2009). Production of aromatic compounds in bacteria. Curr. Opin. Biotechnol. 20, 651–658. doi: 10.1016/j.copbio.2009.09.012
Gosset, G., Yong-Xiao, J., and Berry, A. (1996). A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli. J. Ind. Microbiol. 17, 47–52. doi: 10.1007/BF01570148
Gottardi, M., Reifenrath, M., Boles, E., and Tripp, J. (2017). Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: bioconversion from glucose. FEMS Yeast Res. 17:fox035. doi: 10.1093/femsyr/fox035
Gu, P., Kang, J., Yang, F., and Wang, Q. (2013). The improved L -tryptophan production in recombinant Escherichia coli by expressing the polyhydroxybutyrate synthesis pathway. Appl. Microbiol. Biotechnol. 97, 4121–4127. doi: 10.1007/s00253-012-4665-4660
Gu, P., Yang, F., Kang, J., Wang, Q., and Qi, Q. (2012). One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 11, 1–9. doi: 10.1186/1475-2859-11-30
Gulevich, A., Biryukova, I., Zimenkov, D., Skorokhodova, A., Kivero, A., Belareva, A., et al. (2006). Method for Producing An L-amino Acid Using A Bacterium Having Enhanced Expression of the pckA Gene. United States Patent Application 20060035348.
Guo, D., Zhang, L., Kong, S., Liu, Z., Li, X., and Pan, H. (2018). Metabolic engineering of Escherichia coli for production of 2-Phenylethanol and 2-Phenylethyl acetate from glucose. J. Agric. Food Chem. 66, 5886–5891. doi: 10.1021/acs.jafc.8b01594
Gyuun, D., Mi, A., Cha, N., and Prabhu, S. (2016). Bacterial synthesis of four hydroxycinnamic acids. Appl. Biol. Chem. 59, 173–179. doi: 10.1007/s13765-015-0137-134
Hansen, E. H., Møller, B. L., Kock, G. R., Bünner, C. M., Kristensen, C., Jensen, O. R., et al. (2009). De novo biosynthesis of Vanillin in fission yeast (schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 75, 2765–2774. doi: 10.1128/AEM.02681-2688
Hartmann, M., Schneider, T. R., Pfeil, A., Heinrich, G., Lipscomb, W. N., and Braus, G. H. (2003). Evolution of feedback-inhibited/barrel isoenzymes by gene duplication and a single mutation. Proc. Natl. Acad. Sci. U.S.A. 100, 862–867. doi: 10.1073/pnas.0337566100
Heinz-Gerhard, F., and Jurgen, S. (2012). Industrial Aromatic Chemistry: Raw Materials⋅ Processes⋅ Products. Berlin: Springer Science & Business Media.
Hernández-Chávez, G., Martinez, A., and Gosset, G. (2019). Metabolic engineering strategies for caffeic acid production in Escherichia coli. Electr. J. Biotechnol. 38, 19–26. doi: 10.1016/j.ejbt.2018.12.004
Herrmann, K. M. (1995). The Shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell 7:907. doi: 10.2307/3870046
Hu, C., Jiang, P., Xu, J., Wu, Y., and Huang, W. (2003). Mutation analysis of the feedback inhibition site of phenylalanine- sensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase of Escherichia coli. J. Basic Microbiol. 43, 399–406. doi: 10.1002/jobm.200310244
Hua, D., Lin, S., Li, Y., Chen, H., Zhang, Z., Du, Y., et al. (2010). Enhanced 2-phenylethanol production from L-phenylalanine via in situ product adsorption. Biocatal. Biotrans. 28, 259–266. doi: 10.3109/10242422.2010.500724
Hua, D., Ma, C., Song, L., Lin, S., Zhang, Z., Deng, Z., et al. (2007). Enhanced vanillin production from ferulic acid using adsorbent resin. Appl. Microbiol. Biotechnol. 74, 783–790. doi: 10.1007/s00253-006-0735-735
Huang, Q., Lin, Y., and Yan, Y. (2013). Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 110, 3188–3196. doi: 10.1002/bit.24988
Huang, Z., Dostal, L., and Rosazza, J. P. N. (1993). Microbial transformations of ferulic acid by Saccharomyces cerevisiae and Pseudomonas fluorescens. Appl. Environ. Microbiol. 59, 2244–2250. doi: 10.1128/aem.59.7.2244-2250.1993
Hwang, E. Il, Hwang, E. Il, Ohnishi, Y., Ohnishi, Y., Horinouchi, S., and Horinouchi, S. (2003). Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69, 2699–2706. doi: 10.1128/AEM.69.5.2699
Ikeda, M. (2006). Towards bacterial strains overproducing L -tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 69, 615–626. doi: 10.1007/s00253-005-0252-y
Ikeda, M., Mizuno, Y., and Awane, S. (2011). Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 90, 1443–1451. doi: 10.1007/s00253-011-3210-x
Jendresen, C. B., Stahlhut, S. G., Li, M., Gaspar, P., Siedler, S., Förster, J., et al. (2015). Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 4458–4476. doi: 10.1128/AEM.00405-415
Jiang, M., and Zhang, H. (2016). Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli. Curr. Opin. Biotechnol. 42, 1–6. doi: 10.1016/j.copbio.2016.01.016
Jiménez, J. I., Miñambres, B., García, J. L., and Díaz, E. (2002). Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4, 824–841. doi: 10.1046/j.1462-2920.2002.00370.x
Johnson, B., Amaratunga, M., and Lobos, J. H. (2000). Method for Increasing Total Production of 4-Hydroxybenzoic Acid by Biofermentation. US patent application US 0 129.
Joshi, S. S., and Ranade, V. V. (2016). Industrial Catalytic Processes for Fine and Specialty Chemicals. Amsterdem: Elsevier.
Jossek, R., Bongaerts, J., and Sprenger, G. A. (2001). Characterization of a new feedback-resistant 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli. FEMS Microbiol. Lett. 202, 145–148. doi: 10.1016/S0378-1097(01)00311-311
Juminaga, D., Baidoo, E. E. K., Redding-Johanson, A. M., Batth, T. S., Burd, H., Mukhopadhyay, A., et al. (2012). Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78, 89–98. doi: 10.1128/AEM.06017-6011
Kallscheuer, N., and Marienhagen, J. (2018). Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb. Cell Fact. 17:70. doi: 10.1186/s12934-018-0923-x
Kallscheuer, N., Menezes, R., Foito, A., Henriques da Silva, M. D., Braga, A., Dekker, W., et al. (2019). Identification and microbial production of the raspberry phenol salidroside that is active against Huntingtons disease. Plant Physiol. 179, 969–985. doi: 10.1104/pp.18.01074
Kallscheuer, N., Vogt, M., Stenzel, A., Gätgens, J., Bott, M., and Marienhagen, J. (2016). Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab. Eng. 38, 47–55. doi: 10.1016/j.ymben.2016.06.003
Kaneko, T., Thi, T. H., Shi, D. J., and Akashi, M. (2006). Environmentally degradable, high-performance thermoplastics from phenolic phytomonomers. Nat. Mater. 5, 966–970. doi: 10.1038/nmat1778
Kang, S., Choi, O., Lee, J. K., Hwang, B. Y., and Uhm, T. (2012). Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Fact. 11:153. doi: 10.1186/1475-2859-11-153
Kaur, B., and Chakraborty, D. (2013). Biotechnological and molecular approaches for vanillin production: a review. Appl. Biochem. Biotechnol. 169, 1353–1372. doi: 10.1007/s12010-012-0066-61
Kawaguchi, H., Katsuyama, Y., Danyao, D., Kahar, P., Yoshihara, K., Minami, H., et al. (2017). Caffeic acid production by simultaneous saccharification and fermentation of kraft pulp using recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 101, 5279–5290. doi: 10.1007/s00253-017-8270-8270
Kawai, Y., Noda, S., Ogino, C., Takeshima, Y., Okai, N., and Tanaka, T. (2013). p -Hydroxycinnamic acid production directly from cellulose using endoglucanase- and tyrosine ammonia lyase-expressing Streptomyces lividans. Microb. Cell Fact. 12:45. doi: 10.1186/1475-2859-12-45
Keasling, J. D., and Chou, H. (2008). Metabolic engineering delivers next-generation biofuels Tiny tiles, tiny targets. Nat. Biotechnol. 26, 298–299. doi: 10.1038/ng.85
Keseler, I. M., Mackie, A., Peralta-gil, M., Santos-zavaleta, A., Kothari, A., Krummenacker, M., et al. (2013). EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41, 605–612. doi: 10.1093/nar/gks1027
Kikuchi, Y., Tsujimoto, K., and Kurahashi, O. (1997). Mutational analysis of the feedback sites of phenylalanine-sensitive synthase of Escherichia coli. Appl. Environ. Microbiol. 63, 761–762. doi: 10.1128/aem.63.2.761-762.1997
Kim, B., Cho, B. R., and Hahn, J. S. (2014). Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol. Bioeng. 111, 115–124. doi: 10.1002/bit.24993
Kim, T. Y., Lee, S. W., and Oh, M. K. (2014). Biosynthesis of 2-phenylethanol from glucose with genetically engineered Kluyveromyces marxianus. Enzyme Microb. Technol. 6, 44–47. doi: 10.1016/j.enzmictec.2014.04.011
Kitade, Y., Hashimoto, R., Suda, M., Hiraga, K., and Inuia, M. (2018). Production of 4-Hydroxybenzoic acid by an aerobic growth- arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl. Environ. Microbiol. 84, 1–12. doi: 10.1128/AEM.02587-17
Knaggs, A. R. (2003). The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 20, 119–136. doi: 10.1039/b100399m
Knop, D. R., Draths, K. M., Chandran, S. S., Barker, J. L., Daeniken, R., Von Weber, W., et al. (2001). Hydroaromatic equilibration during biosynthesis of shikimic acid. J Am. Chem. Soc. 123, 10173–10182. doi: 10.1021/ja0109444
Köhle, A., Sommer, S., Li, S., Schilde-rentschler, L., Ninnemann, H., and Heide, L. (2003). Secondary metabolites in transgenic tobacco and potato: high accumulation of 4-hydroxybenzoic acid glucosides results from high expression of the bacterial gene ubi C. Mol. Breed. 11, 15–24.
Kramer, M., Bongaerts, ÃJ., Bovenberg, R., Kremer, S., Mu, U., Orf, S., et al. (2003). Metabolic engineering for microbial production of shikimic acid. Metab. Eng. 5, 277–283. doi: 10.1016/j.ymben.2003.09.001
Krivoruchko, A., and Nielsen, J. (2015). Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 35, 7–15. doi: 10.1016/j.copbio.2014.12.004
Krömer, J. O. (2016). Metabolic engineering of Pseudomonas putida KT2440 for the production of para -hydroxy benzoic acid. Front. Bioeng. Biotechnol. 4:90. doi: 10.3389/fbioe.2016.00090
Krömer, J. O., Nunez-Bernal, D., Averesch, N. J. H., Hampe, J., Varela, J., and Varela, C. (2013). Production of aromatics in Saccharomyces cerevisiae-A feasibility study. J. Biotechnol. 163, 184–193. doi: 10.1016/j.jbiotec.2012.04.014
Kunjapur, A. M., Tarasova, Y., and Prather, K. L. J. (2014). Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J. Am. Chem. Soc. 136, 11644–11654. doi: 10.1021/ja506664a
Lai, B., Plan, M. R., Averesch, N. J. H., Yu, S., Kracke, F., Lekieffre, N., et al. (2017). Quantitative analysis of aromatics for synthetic biology using liquid chromatography. Biotechnol. J. 12:1600269. doi: 10.1002/biot.201600269
Lee, E., Yoon, S., Das, A., Lee, S., Li, C., Kim, J., et al. (2009). Directing vanillin production from ferulic acid by increased acetyl-CoA consumption in recombinant Escherichia coli. Biotechnol. Bioeng. 102, 200–208. doi: 10.1002/bit.22040
Lee, J., and Wendisch, V. (2017). Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257, 211–221. doi: 10.1016/j.jbiotec.2016.11.016
Lenzen, C., Wynands, B., Otto, M., Bolzenius, J., Mennicken, P., Blank, L. M., et al. (2019). High-Yield production of 4-Hydroxybenzoate from glucose or glycerol by an engineered Pseudomonas taiwanensis VLB120. Front. Bioeng. Biotechnol. 7:130. doi: 10.3389/fbioe.2019.00130
Leonard, E., Yan, Y., Lim, K. H., Koffas, M. A. G., Al, L. E. T., and Icrobiol, A. P. P. L. E. N. M. (2005). Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol 71, 8241–8248. doi: 10.1128/AEM.71.12.8241
Li, K., and Frost, J. W. (1998). Synthesis of vanillin from glucose [2]. J. Am. Chem. Soc. 120, 10545–10546. doi: 10.1021/ja9817747
Li, K., and Frost, J. W. (1999). Microbial synthesis of 3-dehydroshikimic acid: a comparative analysis of D-xylose, L-arabinose, and D-glucose carbon sources. Biotechnol. Prog. 15, 876–883. doi: 10.1021/bp990095c
Li, Y., Li, J., Qian, B., Cheng, L., Xu, S., and Wang, R. (2018). De Novo Biosynthesis of p-Coumaric Acid in E. coli with a trans-Cinnamic Acid 4-Hydroxylase from the Amaryllidaceae Plant Lycoris aurea. Molecules 23:3185. doi: 10.1007/s00253-005-0197-191
Lin, Y., Shen, X., Yuan, Q., and Yan, Y. (2013). Microbial biosynthesis of the anticoagulant precursor 4-hydroxycoumarin. Nat. Commun. 4:3603. doi: 10.1038/ncomms3603
Lin, Y., Sun, X., Yuan, Q., and Yan, Y. (2014). Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 23, 62–69. doi: 10.1016/j.ymben.2014.02.009
Lin, Y., and Yan, Y. (2012). Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Fact. 11, 3–11. doi: 10.1186/1475-2859-11-42
Liu, C., Zhang, K., Cao, W., Zhang, G., Chen, G., Yang, H., et al. (2018). Genome mining of 2 - phenylethanol biosynthetic genes from Enterobacter sp. CGMCC 5087 and heterologous overproduction in Escherichia coli. Biotechnol. Biofuels 11:305. doi: 10.1186/s13068-018-1297-1293
Liu, L., Liu, H., Zhang, W., Yao, M., Li, B., Liu, D., et al. (2019a). Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering 5, 287–295. doi: 10.1016/j.eng.2018.11.029
Liu, Q., Yu, T., Li, X., Chen, Y., Campbell, K., Nielsen, J., et al. (2019b). Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10, 1–13. doi: 10.1038/s41467-019-12961-12965
Liu, Q., Cheng, Y., Xie, X., Xu, Q., and Chen, N. (2012). Modification of tryptophan transport system and its impact on production of L -tryptophan in Escherichia coli. Bioresour. Technol. 114, 549–554. doi: 10.1016/j.biortech.2012.02.088
Lu, J. L., and Liao, J. C. (1997). Metabolic engineering and control analysis for production of aromatics: role of transaldolase. Biotechnol. Bioeng. 53, 132–138.
Lütke-Eversloh, T., and Stephanopoulos, G. (2005). Feedback inhibition of Chorismate Mutase / Prephenate Dehydrogenase (TyrA) of Escherichia coli: generation and characterization of Tyrosine-Insensitive mutants. Appl Env. Microbiol. 71, 7224–7228. doi: 10.1128/AEM.71.11.7224
Lütke-Eversloh, T., and Stephanopoulos, G. (2007). L-Tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110. doi: 10.1007/s00253-006-0792-799
Luttik, M. A. H., Vuralhan, Z., Suir, E., Braus, G. H., Pronk, J. T., and Daran, J. M. (2008). Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 10, 141–153. doi: 10.1016/j.ymben.2008.02.002
Luziatelli, F., Brunetti, L., Ficca, A. G., and Ruzzi, M. (2019). Maximizing the efficiency of vanillin production by biocatalyst enhancement and process optimization. Front. Bioeng. Biotechnol. 7:279. doi: 10.3389/fbioe.2019.00279
Ma, X. K., and Daugulis, A. J. (2014). Transformation of ferulic acid to vanillin using a fed-batch solid-liquid two-phase partitioning bioreactor. Biotechnol. Prog. 30, 207–214. doi: 10.1002/btpr.1830
MacDonald, M. J., and D’Cunha, G. B. (2007). A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 85, 273–282. doi: 10.1139/O07-018
Maeda, H., and Dudareva, N. (2012). The Shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Mao, J., Liu, Q., Song, X., Wang, H., Feng, H., Xu, H., et al. (2017). Combinatorial analysis of enzymatic bottlenecks of l-tyrosine pathway by p-coumaric acid production in Saccharomyces cerevisiae. Biotechnol. Lett. 39, 977–982. doi: 10.1007/s10529-017-2322-2325
Martínez, K., de Anda, R., Hernández, G., Escalante, A., Gosset, G., Ramírez, O. T., et al. (2008). Co-utilization of glucose and glycerol enhances the production of aromatic compounds in an Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system. Microb. Cell Fact. 7:1. doi: 10.1186/1475-2859-7-1
Martínez-Avila, O., Sánchez, A., and Font, X. (2018). Bioproduction of 2-phenylethanol and 2-phenethyl acetate by Kluyveromyces marxianus through the solid-state fermentation of sugarcane bagasse. Appl. Microbiol. Biotechnol. 102, 4703–4716. doi: 10.1007/s00253-018-8964-y
Mascarenhas, D., Ashworth, D. J., and Chen, C. S. (1991). Deletion of pgi alters tryptophan biosynthesis in a genetically engineered strain of Escherichia coli. Appl. Environ. Microbiol. 57, 2995–2999. doi: 10.1177/1077546315573916
Mckenna, R., and Nielsen, D. R. (2011). Styrene biosynthesis from glucose by engineered E. coli. Metab. Eng. 13, 544–554. doi: 10.1016/j.ymben.2011.06.005
Mcqualter, R. B., Chong, B. F., Meyer, K., Dyk, D. E., Van Shea, M. G. O., Walton, N. J., et al. (2005). Initial evaluation of sugarcane as a production platform for p -hydroxybenzoic acid. Plant Biotechnol. J. 31, 29–41. doi: 10.1111/j.1467-7652.2004.00095.x
Mihal’, M., Goncalves, R. F., and Markoš, J. (2014). Intensive 2-phenylethanol production in a hybrid system combined of a stirred tank reactor and an immersed extraction membrane module. Chem. Pap. 68, 1656–1666. doi: 10.2478/s11696-014-0575-571
Milke, L., Aschenbrenner, J., Marienhagen, J., and Kallscheuer, N. (2018). Production of plant-derived polyphenols in microorganisms: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 1575–1585. doi: 10.1007/s00253-018-8747-8745
Muheim, A., Müller, B., Münch, T., and Wetli, M. (2001). Microbiological Process for Producing Vanillin. Karnataka: GIVAUDAN-ROURE.
Müller, R., Wagener, A., Schmidt, K., and Leistner, E. (1995). Microbial production of specifically ring-13C-labelled 4-hydroxybenzoic acid. Appl. Microbiol. Biotechnol. 43, 985–988. doi: 10.1007/bf00166913
Nagy, C., and Haschemi, A. (2013). Sedoheptulose kinase regulates cellular carbohydrate metabolism by sedoheptulose 7-phosphate supply. Biochem. Soc. Trans. 41, 674–680. doi: 10.1042/BST20120354
Nakamura, C. E., and Whited, G. M. (2003). Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459. doi: 10.1016/j.copbio.2003.08.005
Ni, J., Tao, F., Du, H., and Xu, P. (2015). Mimicking a natural pathway for de novo biosynthesis: natural vanillin production from accessible carbon sources. Sci. Rep. 5:13670. doi: 10.1038/srep13670
Nielsen, J., and Keasling, J. D. (2016). Engineering cellular metabolism. Cell 164, 1185–1197. doi: 10.1016/j.cell.2016.02.004
Nijkamp, K., Van Luijk, N., De Bont, J. A. M., and Jan, W. (2005). The solvent-tolerant Pseudomonas putida S12 as host for the production of cinnamic acid from glucose. Appl. Microbiol. Biotechnol. 69, 170–177. doi: 10.1007/s00253-005-1973-1977
Nijkamp, K., Westerhof, R. G. M., Ballerstedt, H., Bont, J. A. M., and De Wery, J. (2007). Optimization of the solvent-tolerant Pseudomonas putida S12 as host for the production of p -coumarate from glucose. Appl. Microbiol. Biotechnol. 74, 617–624. doi: 10.1007/s00253-006-0703-700
Nikel, P. I., and de Lorenzo, V. (2018). Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 50, 142–155. doi: 10.1016/j.ymben.2018.05.005
Nishiyama, Y., Yun, C. S., Matsuda, F., Sasaki, T., Saito, K., and Tozawa, Y. (2010). Expression of bacterial tyrosine ammonia-lyase creates a novel p-coumaric acid pathway in the biosynthesis of phenylpropanoids in Arabidopsis. Planta 232, 209–218. doi: 10.1007/s00425-010-1166-1161
Noack, D., Roth, M., Geuther, R., Müller, G., Undisz, K., Hoffmeier, C., et al. (1981). Maintenance and genetic stability of vector plasmids pBR322 and pBR325 in Escherichia coli K12 strains grown in a chemostat. MGG Mol. Gen. Genet. 184, 121–124. doi: 10.1007/BF00271207
Noda, S., and Kondo, A. (2017). Recent advances in microbial production of aromatic chemicals and derivatives. Trends Biotechnol. 35, 785–796. doi: 10.1016/j.tibtech.2017.05.006
Noda, S., Miyazaki, T., and Miyoshi, T. (2011). Cinnamic acid production using Streptomyces lividans expressing phenylalanine ammonia lyase. J. Ind. Microbiol. Biotechnol. 38, 643–648. doi: 10.1007/s10295-011-0955-952
Noda, S., Miyazaki, T., Tanaka, T., Ogino, C., and Kondo, A. (2012). Production of Streptoverticillium cinnamoneum transglutaminase and cinnamic acid by recombinant Streptomyces lividans cultured on biomass-derived carbon sources. Bioresour. Technol. 104, 648–651. doi: 10.1016/j.biortech.2011.10.045
Noda, S., Shirai, T., Oyama, S., and Kondo, A. (2016). Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 33, 119–129. doi: 10.1016/j.ymben.2015.11.007
Oldiges, M., Kunze, M., Degenring, D., Sprenger, G. A., and Takors, R. (2004). Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Prog. 20, 1623–1622.
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528. doi: 10.1038/nature12051
Pandey, R. P., Parajuli, P., Koffas, M. A. G., and Sohng, J. K. (2016). Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 34, 634–662. doi: 10.1016/j.biotechadv.2016.02.012
Papagianni, M. (2012). Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb. Cell Fact. 11, 1–13. doi: 10.1186/1475-2859-11-50
Paravicini, G., Mosch, H., Schmidheini, T., and Braus, G. (1989). The general control activator protein GCN4 is essential for a basal level of AR03 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 144–151. doi: 10.1128/mcb.9.1.144
Park, S. Y., Yang, D., Ha, S. H., and Lee, S. Y. (2018). Metabolic engineering of microorganisms for the production of natural compounds. Adv. Biosyst. 2:1700190. doi: 10.1002/adbi.201700190
Patnaik, R., and Liao, J. C. (1994). Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl. Environ. Microbiol. 60, 3903–3908. doi: 10.7550/rmb.31286
Patnaik, R., Roof, W. D., Young, R. F., and Liao, J. C. (1992). Stimulation of glucose catabolism in Escherichia coli by a potential futile cycle. J. Bacteriol. 174, 7527–7532. doi: 10.1128/jb.174.23.7527-7532.1992
Pittard, J., Camakaris, H., and Yang, J. (2005). The TyrR regulon. Mol. Microbiol. 55, 16–26. doi: 10.1111/j.1365-2958.2004.04385.x
Priefert, H., Rabenhorst, J., and Steinbüchel, A. (2001). Biotechnological production of vanillin. Appl. Microbiol. Biotechnol. 56, 296–314. doi: 10.1007/s002530100687
Priya, V. K., Sarkar, S., and Sinha, S. (2014). Evolution of tryptophan biosynthetic pathway in microbial genomes: a comparative genetic study. Syst. Synth. Biol. 8, 59–72. doi: 10.1007/s11693-013-9127-9121
Rasmussen, S., Dixon, R. A., Division, P. B., Roberts, S., Foundation, N., and Parkway, S. N. (1999). Transgene-mediated and Elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11, 1537–1551. doi: 10.1105/tpc.11.8.1537
Reifenrath, M., Bauer, M., Oreb, M., and Boles, E. (2018). Bacterial bifunctional chorismate mutase-prephenate dehydratase PheA increases flux into the yeast phenylalanine pathway and improves mandelic acid production. Metab. Eng. Commun. 7, 1–9. doi: 10.1016/j.mec.2018.e00079
Ro, D., and Douglas, C. J. (2004). Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae). J. Biol. Chem. 279, 2600–2607. doi: 10.1074/jbc.M309951200
Rodrigues, J. L., Prather, K. L. J., Kluskens, L. D., and Rodrigues, L. R. (2015). Heterologous production of curcuminoids. Microbiol. Mol. Biol. Rev. 79, 39–60. doi: 10.1128/MMBR.00031-14
Rodriguez, A., Chen, Y., Khoomrung, S., Özdemir, E., and Borodina, I. (2017). Comparison of the metabolic response to over-production of p -coumaric acid in two yeast strains. Metab. Eng. 44, 265–272. doi: 10.1016/j.ymben.2017.10.013
Rodriguez, A., Kildegaard, K. R., Li, M., Borodina, I., and Nielsen, J. (2015). Establishment of a yeast platform strain for production of p- coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 31, 181–188. doi: 10.1016/j.ymben.2015.08.003
Rodriguez, A., Martínez, J. A., Báez-Viveros, J. L., Flores, N., Hernández-Chávez, G., Ramírez, O. T., et al. (2013). Constitutive expression of selected genes from the pentose phosphate and aromatic pathways increases the shikimic acid yield in high-glucose batch cultures of an Escherichia coli strain lacking PTS and pykF. Microb. Cell Fact. 12:86. doi: 10.1186/1475-2859-12-86
Sachan, A., Ghosh, S., Sen, S. K., and Mitra, A. (2006). Co-production of caffeic acid and p-hydroxybenzoic acid from p-coumaric acid by Streptomyces caeruleus MTCC 6638. Appl. Microbiol. Biotechnol. 71, 720–727. doi: 10.3390/molecules23123185
Santos, C. N. S., Koffas, M., and Stephanopoulos, G. (2011). Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab. Eng. 13, 392–400. doi: 10.1016/j.ymben.2011.02.002
Santos, C. N. S., Xiao, W., and Stephanopoulos, G. (2012). Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 13538–13543. doi: 10.1073/pnas.1206346109
Sariaslani, F. S. (2007). Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annu. Rev. Microbiol. 61, 51–69. doi: 10.1146/annurev.micro.61.080706.093248
Schnappauf, G., Hartmann, M., Künzler, M., and Braus, G. H. (1998). The two 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase isoenzymes from Saccharomyces cerevisiae show different kinetic modes of inhibition. Arch. Microbiol. 169, 517–524. doi: 10.1007/s002030050605
Schrader, J., Etschmann, M. M. W., Sell, D., Hilmer, J., and Rabenhorst, J. (2004). Applied biocatalysis for the synthesis of natural flavour compounds. Biotechnol. Lett. 26, 463–472. doi: 10.1023/b:bile.0000019576.80594.0e
Serino, L., Reimmann, C., Visca, P., Beyeler, M., Chiesa, V. D., Haas, D., et al. (1997). Biosynthesis of pyochelin and Dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 179, 248–257. doi: 10.1128/jb.179.1.248-257.1997
Shumilin, I. A., Kretsinger, R. H., and Bauerle, R. H. (1999). Crystal structure of phenylalanine-regulated 3-deoxy- D - arabino - heptulosonate-7-phosphate synthase from Escherichia coli. Structure 7, 865–875. doi: 10.1016/S0969-2126(99)80109-80109
Sprenger, G., Siewe, R., Martin, K., and Sonke, T. (1998). Microbial Preparation of Substances from Aromatic Metabolism. Patent No.US6316232B1 United States.
Sprenger, G. A. (2007). From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 75, 739–749. doi: 10.1007/s00253-007-0931-y
Suástegui, M., Guo, W., Feng, X., and Shao, Z. (2016). Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae. Biotechnol. Bioeng. 113, 2676–2685. doi: 10.1002/bit.26037
Sun, X., Shen, X., Jain, R., Lin, Y., Wang, J., Sun, J., et al. (2015). Synthesis of chemicals by metabolic engineering of microbes. Chem. Soc. Rev. 44, 3760–3785. doi: 10.1039/c5cs00159e
Tan, Z., Zhu, X., Chen, J., and Li, Q. (2013). Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl. Environ. Microbiol. 79, 4838–4844. doi: 10.1128/AEM.00826-813
Tatarko, M., and Romeo, T. (2001). Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli. Curr. Microbiol. 43, 26–31. doi: 10.1007/s002840010255
Thompson, B., Machas, M., and Nielsen, D. R. (2015). Creating pathways towards aromatic building blocks and fine chemicals. Curr. Opin. Biotechnol. 36, 1–7. doi: 10.1016/j.copbio.2015.07.004
Tilburg, A. Y., Van Cao, H., Meulen, S. B., and Van Der Kuipers, O. P. (2019). Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories. Curr. Opin. Biotechnol. 59, 1–7. doi: 10.1016/j.copbio.2019.01.007
Trotman, R. J., Camp, C. E., Ben-bassat, A., Dicosimo, R., Huang, L., Crum, G. A., et al. (2007). Calcium alginate bead immobilization of cells containing tyrosine ammonia lyase activity for use in the production of p -hydroxycinnamic acid. Biotechnol. Prog. 23, 638–644. doi: 10.1021/bp060379e
Tzin, V., Galili, G., and Ahroni, A. (2001). Shikimate pathway and aromatic amino acid biosynthesis. eLS 1–10. doi: 10.1002/9780470015902.a0001315.pub2
Van Dien, S. (2013). From the first drop to the first truckload: commercialization of microbial processes for renewable chemicals. Curr. Opin. Biotechnol. 24, 1061–1068. doi: 10.1016/j.copbio.2013.03.002
Van Hecke, W., Kaur, G., and De Wever, H. (2014). Advances in in-situ product recovery (ISPR) in whole cell biotechnology during the last decade. Biotechnol. Adv. 32, 1245–1255. doi: 10.1016/j.biotechadv.2014.07.003
Vane, J. R., and Botting, R. M. (2003). The mechanism of action of aspirin. Thromb. Res. 110, 255–258. doi: 10.1016/S0049-3848(03)00379-377
Vanilla (2014). A Sustainable Production Route. Evolva A/S. Available at: https://www.evolva.com/vanillin/ (accessed April 26, 2019).
Vannelli, T., Qi, W. W., Sweigard, J., Gatenby, A. A., and Sariaslani, F. S. (2007). Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metab. Eng. 9, 142–151. doi: 10.1016/j.ymben.2006.11.001
Vargas-Tah, A., and Gosset, G. (2015). Production of cinnamic and p-hydroxycinnamic acids in engineered microbes. Front. Bioeng. Biotechnol. 3:116. doi: 10.3389/fbioe.2015.00116
Verhoef, S., Ruijssenaars, H. J., Bont, J. A. M., and De Wery, J. (2007). Bioproduction of p -hydroxybenzoate from renewable feedstock by solvent-tolerant Pseudomonas putida S12. J. Biotechnol. 132, 49–56. doi: 10.1016/j.jbiotec.2007.08.031
Verhoef, S., Wierckx, N., Westerhof, R. G. M., Winde, J. H., and De Ruijssenaars, H. J. (2009). Bioproduction of p -hydroxystyrene from glucose by the solvent-tolerant bacterium Pseudomonas putida S12 in a two-phase water-decanol fermentation. Appl. Microbiol. Biotechnol. 75, 931–936. doi: 10.1128/AEM.02186-2188
Wang, J., Cheng, L., Wang, J., and Liu, Q. (2013). Genetic engineering of Escherichia coli to enhance production of L -tryptophan. Appl. Microbiol. Biotechnol. 97, 7587–7596. doi: 10.1007/s00253-013-5026-5023
Wang, J., Guleria, S., Koffas, M. A. G., and Yan, Y. (2016). Microbial production of value-added nutraceuticals. Curr. Opin. Biotechnol. 37, 97–104. doi: 10.1016/j.copbio.2015.11.003
Wang, J., Yang, Y., and Yan, Y. (2018). “Bioproduction of Resveratrol,” in Biotechnology of Natural Products, eds W. Schwab, B. M. Lange, and M. Wüst, (Cham: Springer International Publishing), 61–79. doi: 10.1007/978-3-319-67903-7_3
Wang, Z., Jiang, M., Guo, X., Liu, Z., and He, X. (2018). Reconstruction of metabolic module with improved promoter strength increases the productivity of 2 - phenylethanol in Saccharomyces cerevisiae. Microb. Cell Fact. 17:60. doi: 10.1186/s12934-018-0907-x
Wang, Z., Bai, X., Guo, X., and He, X. (2017). Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 44, 129–139. doi: 10.1007/s10295-016-1852-1855
Watts, K. T., Lee, P. C., and Schmidt-dannert, C. (2004). Exploring recombinant flavonoid biosynthesis in metabolically engineered Escherichia coli. ChemBioChem 5, 500–507. doi: 10.1002/cbic.200300783
Wierckx, N. J. P., Ballerstedt, H., Bont, J. A. M., and De Wery, J. (2005). Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Env. Microbiol. 71, 8221–8227. doi: 10.1128/AEM.71.12.8221
Wu, F., Cao, P., Song, G., Chen, W., and Wang, Q. (2018). Expanding the repertoire of aromatic chemicals by microbial production. J. Chem. Technol. Biotechnol. 93, 2804–2816. doi: 10.1002/jctb.5690
Yakandawala, N., Romeo, T., Friesen, A. D., and Madhyastha, S. (2008). Metabolic engineering of Escherichia coli to enhance phenylalanine production. Appl. Microbiol. Biotechnol. 78, 283–291. doi: 10.1007/s00253-007-1307-z
Yi, J., Draths, K. M., Li, K., and Frost, J. W. (2003). Altered Glucose Transport and Shikimate Pathway Product Yields in E. coli. Biotechnol. Prog. 19, 1450–1459. doi: 10.1021/bp0340584
Yim, H., Haselbeck, R., Niu, W., Pujol-baxley, C., Burgard, A., Boldt, J., et al. (2011). Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 7, 444–445. doi: 10.1038/nchembio.580
Yoon, S. H., Lee, E. G., Das, A., Lee, S. H., Li, C., Ryu, H. K., et al. (2007). Enhanced vanillin production from recombinant E. coli using NTG mutagenesis and adsorbent resin. Biotechnol. Prog. 23, 1143–1148. doi: 10.1021/bp070153r
Zhang, H., Cao, M., Jiang, X., Zou, H., Wang, C., Xu, X., et al. (2014). De-novo synthesis of 2-phenylethanol by Enterobacter sp. CGMCC 5087. BMC Biotechnol. 14:30. doi: 10.1186/1472-6750-14-30
Zhang, H., Pereira, B., Li, Z., and Stephanopoulos, G. (2015). Engineering Escherichia coli coculture systems for the production of biochemical products. Proc. Natl. Acad. Sci. U.S.A. 112, 8266–8271. doi: 10.1073/pnas.1506781112
Zhang, J., Pierick, A. Ten, Van Rossum, H. M., Maleki Seifar, R., Ras, C., Daran, J. M., et al. (2015). Determination of the cytosolic NADPH/NADP ratio in Saccharomyces cerevisiae using shikimate dehydrogenase as sensor reaction. Sci. Rep. 5, 12846. doi: 10.1038/srep12846
Zhang, H., and Stephanopoulos, G. (2013). Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl. Microbiol. Biotechnol. 97, 3333–3341. doi: 10.1007/s00253-012-4544-4548
Keywords: aromatic compounds, metabolic engineering, microorganisms, process optimization, synthetic biology, shikimate pathway
Citation: Braga A and Faria N (2020) Bioprocess Optimization for the Production of Aromatic Compounds With Metabolically Engineered Hosts: Recent Developments and Future Challenges. Front. Bioeng. Biotechnol. 8:96. doi: 10.3389/fbioe.2020.00096
Received: 25 June 2019; Accepted: 03 February 2020;
Published: 20 February 2020.
Edited by:
Nils Jonathan Helmuth Averesch, Stanford University, United StatesReviewed by:
Nicolai Kallscheuer, Radboud University, NetherlandsXiangzhao Mao, Ocean University of China, China
Mattijs K. Julsing, Wageningen University and Research, Netherlands
Copyright © 2020 Braga and Faria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adelaide Braga, abraga@deb.uminho.pt
 Adelaide Braga
Adelaide Braga Nuno Faria
Nuno Faria