Tracking Bioluminescent ETEC during In vivo BALB/c Mouse Colonization
- 1Laboratorio de Investigación en Bacteriología Intestinal, Hospital Infantil de México Federico Gómez, Ciudad de México, Mexico
- 2Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Ciudad de México, Mexico
- 3Departamento de Ecología de Agentes Patógenos, Hospital General “Dr. Manuel Gea González”, Ciudad de México, Mexico
Enterotoxigenic Escherichia coli (ETEC) is a leading cause of diarrhea worldwide. Adhesion to the human intestinal tract is crucial for colonization. ETEC adhesive structures have been extensively studied; however, colonization dynamics remain uncharacterized. The aim of this study was to track bioluminescent ETEC during in vivo infection. The promoter region of dnaK was fused with the luc gene, resulting in the pRMkluc vector. E. coli K-12 and ETEC FMU073332 strains were electroporated with pRMkluc. E. coli K-12 pRMkluc was bioluminescent; in contrast, the E. coli K-12 control strain did not emit bioluminescence. The highest light emission was measured at 1.9 OD600 (9 h) and quantified over time. The signal was detected starting at time 0 and up to 12 h. Streptomycin-treated BALB/c mice were orogastrically inoculated with either ETEC FMU073332 pRMkluc or E. coli K-12 pRMkluc (control), and bacterial colonization was determined by measuring bacterial shedding in the feces. ETEC FMU073332 pRMkluc shedding started and stopped after inoculation of the control strain, indicating that mouse intestinal colonization by ETEC FMU073332 pRMkluc lasted longer than colonization by the control. The bioluminescence signal of ETEC FMU073332 pRMkluc was captured starting at the time of inoculation until 12 h after inoculation. The bioluminescent signal emitted by ETEC FMU073332 pRMkluc in the proximal mouse ileum was located, and the control signal was identified in the cecum. The detection of maximal light emission and bioluminescence duration allowed us to follow ETEC during in vivo infection. ETEC showed an enhanced colonization and tropism in the mouse intestine compared with those in the control strain. Here, we report the first study of ETEC colonization in the mouse intestine accompanied by in vivo imaging.
Introduction
Enterotoxigenic Escherichia coli (ETEC) is a leading etiologic agent of diarrhea worldwide, causing millions of diarrheic episodes and approximately 120,000 deaths every year (Qadri et al., 2005; Lozano et al., 2012). Adhesion to the human intestinal tract is one of the most important features of ETEC and represents a crucial step toward colonization. In recent decades, ETEC adhesive structures have been studied (Gaastra and Svennerholm, 1996; Wolf, 1998); however, the colonization dynamics and colonic receptor interactions of this human pathogen remain largely uncharacterized (Guevara et al., 2013). Moreover, the lack of suitable animal models has hampered the thorough evaluation of ETEC virulence factors (Allen et al., 2006). Conventional mice display natural resistance to colonization by pathogenic E. coli, but the oral administration of streptomycin, which alters the intestinal microbiota, permits colonization of the mouse intestine (Bhinder et al., 2014).
Bioluminescence imaging (BLI) has rapidly progressed in the field of bacterial pathogenesis to facilitate the visualization and quantitation of host-pathogen interactions in live animals (Hutchens and Lurken, 2007; Rhee et al., 2011). Bioluminescent images permit the extent of pathogenic infection to be determined in real time in living animals, providing temporal and spatial information regarding labeled bacteria and their metabolic activities (Jawhara and Mordon, 2004; Coombes and Robey, 2010). Bioluminescence is an enzymatic process by which the enzyme luciferase produces visible light in the presence of a specific substrate, oxygen and an energy source (Wiles et al., 2009). Luciferase has been used extensively to monitor bacterial infections in living mice, including characterization of the tissue distribution exhibited by Salmonella enterica serovar Typhimurium, evaluation the effects of antibiotic compounds on Staphylococcus aureus in a deep wound model, bacterial dissemination tracking of Yersinia pestis, and assessment of the role of virulence factors during E. coli O104:H4 colonization (Contag et al., 1995; Kuklin et al., 2003; Gonzalez et al., 2012; Torres et al., 2012). The application bioluminescence technology to study ETEC under in vivo conditions may elucidate the behavior of this bacterium in the gastrointestinal tract in further detail.
The aim of this study was to evaluate ETEC colonization by performing bioluminescent tracking during in vivo mouse infection. We generated a vector harboring the luc gene under the regulation of the dnaK gene promoter. Light emission by and duration of light-emitting bacteria were determined in vitro, and bioluminescent ETEC colonization was studied during in vivo mouse infection. Ex vivo tissue imaging indicated ETEC exhibited a tropism for the mouse ileum.
Materials and Methods
Bacterial Strains and Culture Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. ETEC strain FMU073332 is a clinical isolate; it belongs to sequence type 4 and the ST215 clonal group according to PubMLST and the Pasteur system, respectively (Saldaña-Ahuactzi et al., 2017). ETEC strain FMU073332 carries the classic virulence genes eltA, eltB, sta2, and cstH; and the nonclassic etpA and etpB genes (Cruz-Córdova et al., 2014). Bacterial strains were stored at −70°C in Luria-Bertani (LB) broth (Dibico; CDMX, México) and 20% glycerol (v/v). Bacteria were grown on LB agar plates or in LB broth at 37°C. Antibiotics were added as required at the following concentrations: ampicillin at 100 μg/mL and tetracycline at 100 μg/mL (Sigma; MI, USA).
Plasmid DNA Extraction, Transformation, and Purification
Bacteria were cultured in LB broth overnight at 37°C, and 5–10 mL of culture were pelleted at 250 rpm. Plasmid DNA was purified with a QIAprep® Spin Miniprep Kit (Qiagen; H, Germany). Mobilization of DNA into E. coli K-12 was performed via electroporation (BMC Harvard Apparatus; MA, USA) at 1800 V. DNA amplification was carried out using PCR Master Mix (Promega; WI, USA) or Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, California, USA) in a Verity 96-well thermal cycler (ThermoFisher Scientific; MA, USA). Purification of digested fragments was performed using a DNA Clean & Concentrator™ Kit (Zymo Research; CA, USA). DNA electrophoresis was carried out in 1% agarose gel with TAE 1x buffer, following by ethidium bromide staining and analysis using a BIORAD Chemi Doc® (Bio-Rad Laboratories; CA, USA).
Construction of the Luciferase Reporter Vector
A 400-bp DNA region upstream of the start codon of the dnaK gene was amplified with primers containing BamHI and HindIII restriction sites (Table 1). The reporter gene luc, which codes for the luciferase enzyme, was obtained from the pGEM®-luc plasmid (Promega; WI, USA). The pGEM®-luc plasmid (4931 bp) and dnaK promoter were digested with the BamHI and HindIII restriction enzymes (Promega; WI, USA), ligated with T4 DNA ligase (ThermoFisher; MA, USA) and transformed into E. coli K-12. Clones were selected on 100 μg/mL ampicillin LB agar plates and screened for the dnaK promoter region by PCR.
The dnaK promoter ligated to the luc gene fragment was extracted from the pGEM®-luc vector with the HindIII and SalI restriction enzymes (Promega; WI, USA) and ligated into the pBR322 vector, which was digested with these same restriction enzymes. Resulting clones were cultured on LB agar plates containing 100 μg/mL tetracycline or 100 μg/mL ampicillin for negative and positive selection, respectively, followed by PCR assays. Luciferin (VivoGlo™ Luciferin, In Vivo Grade) (Promega; WI, USA) was used as the luciferase substrate. E. coli strains were transformed with the pRMkluc vector prior to in vitro and in vivo assays.
Bioluminescence Emission Measurements
A 250-mL glass flask containing LB was inoculated with E. coli pRMkluc to a final absorbance of 0.05. The flask was incubated at 250 rpm and 37°C, and the bacterial culture was measured each hour to determine absorbance by placing an aliquot in a well of a 96-well polystyrene plate. Fifty microliters (15 μg/μL) (Foucault et al., 2010) of luciferin was added to each well. The bioluminescence of each well was captured with a Fusion FX imaging system (Vilber Lourmat; SU, Germany) and analyzed with FusionCapt Advance FX7 software (Vilber Lourmat; SU, Germany).
In vitro, In vivo, and Ex vivo Bioluminescence Assays
For in vitro agar media assays, 200 μL of luciferin (15 ng/μL) were spread on LB agar plates before the bacteria were cultured. Strains harboring the luciferase reporter vector were cultured overnight (12 h) at 37°C on LB agar media with luciferin. In vitro broth media assays were carried out in 96-well polystyrene plates, strains were grown overnight (12 h) in LB broth at 37°C, and 100-μL aliquots of each strain were placed individually in wells with 50 μL (15 ng/μL) of luciferin. LB agar plates and 96-well polystyrene plates were analyzed using a petri dish bioluminescence application. The light emission of the samples was captured for 2–10 min using a Fusion FX imaging system (Vilber Lourmat; SU, Germany). Animal procedures were performed according to the guidelines of the Hospital Infantil de México “Federico Gómez” bioethics committee. Three sets of 2- to 4-week-old BALB/c mice were intraperitoneally administered 200 μL (15 ng/μL) of luciferin prior to orogastric or intraperitoneal inoculation at different times. Animals were anesthetized with ketamine/acepromazine at a dosage of 0.3 IU (100-2.5 mg/kg ratio) per g of weight. Mouse gastrointestinal tracts were dissected following euthanization. Complete gastrointestinal tract tissue was placed in 1x PBS and washed, and tissues were immediately placed into the Fusion FX for imaging. Images of mice, petri dishes, and polystyrene plates are presented as pseudocolor images indicating light intensity (red being the most intense and blue the least intense). The colors are superimposed over grayscale reference images. The signal is expressed as the total number of photons emitted per second (photons/s). Images were captured and analyzed using a Fusion FX imaging system (Vilber Lourmat; SU, Germany).
In vivo Colonization Assays
For colonization assays, 3 sets of 2 4-week-old BALB/c mice were orally gavaged with 0.1 mL of streptomycin (20 mg/mL) diluted in 1x PBS to eliminate the intestinal microbiota (Bhinder et al., 2014). Bacteria in the feces of antibiotic-treated mice were screened to ensure “cleaning” of the mouse gut. Treated mice were orally inoculated (gavaged) with 1 × 108 colony forming units (CFU) of wild-type strains diluted in 0.1 M carbonate buffer, pH 9. Mouse feces were collected, homogenized, diluted in 1x cold PBS, and spotted on MacConkey agar plates. The CFU were counted and plotted, and the values obtained for each replicate were statistically analyzed with a paired t-test using GraphPad Prism software.
Results
Construction of the pRMkluc Vector
The plasmid pRMkluc was obtained by cloning the dnaK promoter region (data not shown) into pGEM®-luc. Digestion of pGEM-luc-dnaK with the HindIII, BamHI, and SalI restriction enzymes was performed, and the products were subjected to agarose gel electrophoresis. The following fragments were observed: the 400-bp dnaK promoter region (McCarty and Walker, 1994), the 1649-bp luc region, and the 3282-bp fragment corresponding to the rest of the pGEM®-luc plasmid (Figure S1). The dnaK promoter region ligated to luc was digested with the HindIII and SalI restriction enzymes and cloned into the pBR322 vector, producing pRMkluc. The pRMkluc was digested with the HindIII, BamHI, and SalI restriction enzymes, resulting in the following fragments: the 400-bp dnaK promoter region, the 1649-bp luc region and 3,745 bp corresponding to the pBR322 vector (Figure S1).
E. coli K-12 Harboring the pRMkluc Construct is Bioluminescent
Bioluminescence is a naturally occurring phenomenon in organisms from different genera, such as bacteria (Vibrio cholerae) (Doyle et al., 2004), and it has been used as a tool to understand bacterial behavior. In vitro bioluminescence assays were carried out to measure reporter gene activity prior to animal infection. The strains were cultured overnight in petri dishes containing LB agar with luciferin substrate. E. coli K-12 pBR322 did not emit bioluminescence (Figures 1D–F), indicating that neither E. coli K-12 or the pBR322 vector carried elements that confer bioluminescence (Figures 1D–F). In contrast, E. coli K-12 harboring the pRMkluc vector emitted a bioluminescent signal (Figures 1A–C).
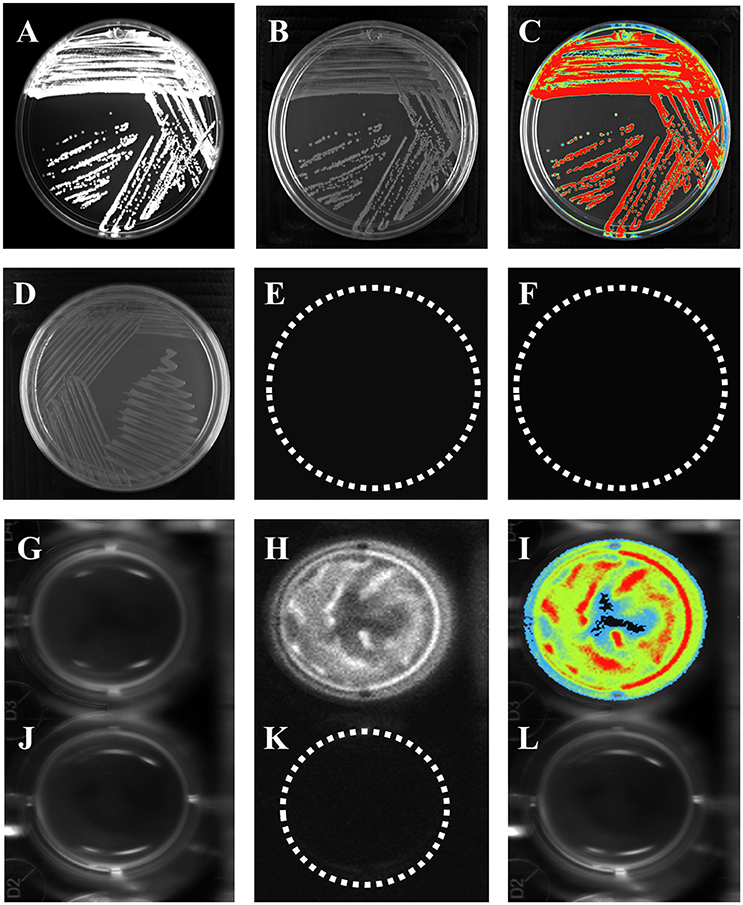
Figure 1. E. coli K-12 harboring pRMkluc generates bioluminescence. (A–C,G–I) E. coli K-12 harboring pRMkluc. (D–F,J–L) E. coli K-12 harboring pBR322 (negative control). (A,D,G,J) Petri dish/96-well polystyrene plate under white light. (B,E,H,K) Light captured from Petri dish/96-well polystyrene plates. (C,F,I,L) Pseudocolor representation from Petri dish/96-well polystyrene plates. (Red = intense, blue = less intense).
To measure light emission intensity and duration, assays were carried out in polystyrene 96-well plates. Bacterial cultures (100-μL aliquots) were incubated for 1 h in stationary phase with luciferin (15 ng/100 μL) and were protected from light. After incubation, bioluminescence signals were captured (Figures 1G–I). In contrast to E. coli K-12 harboring pBR322 (Figures 1J–L). Light emission was captured either from solid or liquid media.
Bioluminescence Generated by E. coli Harboring pRMkluc is Visible for a Long Duration
E. coli K-12 harboring pRMkluc generated bioluminescence after overnight culture and 1 h of incubation with luciferin; however, we sought to explore the bacterial growth density at which light emission was optimal (Figure 2). A kinetic growth curve was constructed, and light emission from each sample was measured every hour. One hundred-microliter bacterial culture aliquots and 50 μL of luciferin were together placed in individual wells of a 96-well polypropylene plate, and light emission was measured. Light was emitted starting at 0.129 optical density (OD600) (1.146 relative light units, RLU); however, at 1.9 OD600, light emission was highest (3.9 RLU) among all time points (Figure 2A). An OD600 of 1.9 was selected to determine the duration of light emission over time. E. coli K-12 harboring pRMkluc was cultured to 1.9 OD600 and incubated with 50 μL (15 ng/μL) of luciferin. Light emission from the samples was quantified. Light was emitted starting at time 0 (3.901 RLU) and up to 12 h (2.06 RLU). However, the maximum intensity of light emission occurred from 0 to 6 h (3.901-2.496 RLU), and after that period of time, bioluminescence emission faded significantly (Figure 2B).
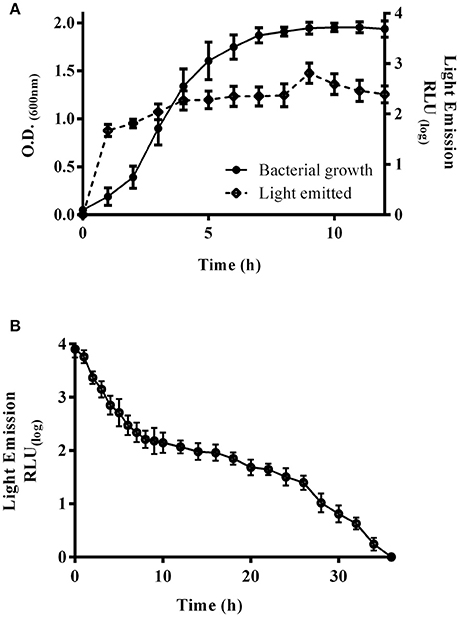
Figure 2. Bioluminescence emission peaks and duration. (A) Bacterial growth curve of E. coli K-12 harboring pRMkluc. The OD600 was measured each hour, and 100-μL aliquots were incubated with luciferin to capture and measure light emission. (B) A 100-μL aliquot of 1.9-OD600 cultured E. coli K-12 harboring pRMkluc was incubated with luciferin (50 μL) and placed in a 96-well polystyrene plate. Light emission was captured and quantified each hour.
Mouse In vivo Assay Utilizing E. coli K-12 Harboring pRMkluc
In vivo tracking of bacteria allows us to understand the role of ETEC during bacterial infection. Strains were cultured to 1.9 OD600. A 100-μL aliquot was obtained and incubated for 5 min with 50 μL of luciferin (15 μg/μL). BALB/c mice were intraperitoneally inoculated with E. coli K-12 harboring pRMkluc or E. coli K-12 harboring pBR322 (incubated with luciferin). After 1 h of infection, bioluminescent signal emission from the animals was captured. Mice infected with E. coli K-12 harboring pBR322 did not exhibit bioluminescent emission (Figure 3B). However, mice infected with E. coli harboring pRMkluc emitted bioluminescent signals in the mouse inoculation zone (Figure 3A).
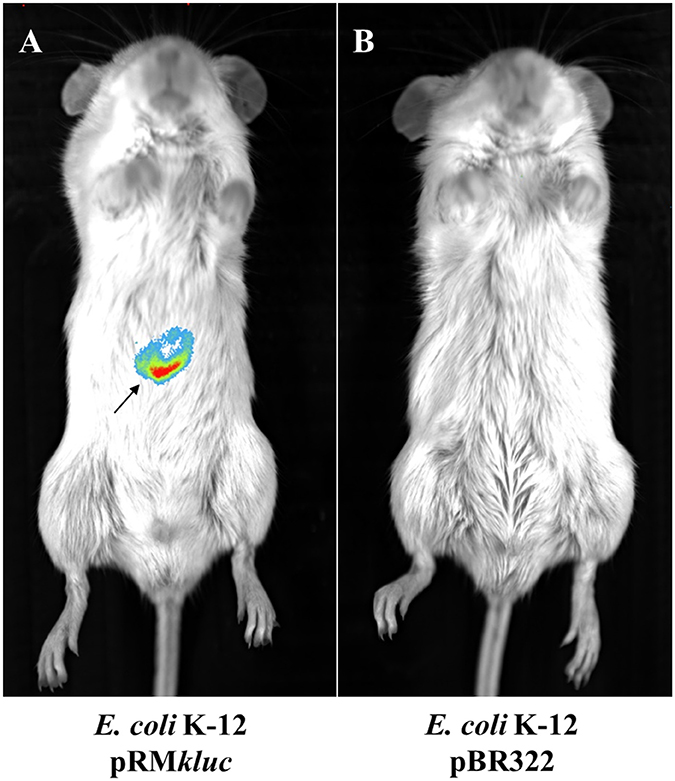
Figure 3. In vivo bioluminescence emission of E. coli K-12 harboring pRMkluc. (A) E. coli K-12 harboring pRMkluc. (B) E. coli K-12 harboring pBR322 (negative control). E. coli strains (1 × 108 bacteria) were incubated with luciferin for 1 h and intraperitoneally inoculated into 4-week-old BALB/c mice. The bioluminescent signal is depicted as pseudocolor (red = intense, blue = less intense). The arrow indicates the bioluminescent signal.
ETEC Shedding through the Mouse Gastrointestinal Tract Occurs at Lower Levels than E. coli K-12
ETEC colonizes the small intestine; however, there have been no in vivo studies investigating ETEC passage through the animal intestine. We studied ETEC colonization of the streptomycin-treated mouse intestine by comparing bacterial shedding of ETEC FMU073332 vs. E. coli K-12 wild-type over time. Inoculation of 1 × 108 bacteria of each strain was achieved via gastric gavage, and bacterial shedding of E. coli K-12 wild-type began 10 h after gastric gavage and continued for 120 h. In contrast, ETEC FMU073332 wild-type shedding started 22 h after gastric gavage and continued for 120 h. A total of 34.6 × 106 CFU/mL of wild-type ETEC FMU073332 were present at 22 h and 8.3 × 106 CFU/mL after 120 h (Figure 4). There were 4.6 × 106 CFU/mL of wild-type E. coli K-12 after 10 h and 1.8 × 106 CFU/mL on 120 h.
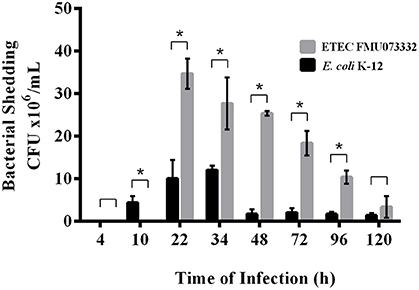
Figure 4. Bacterial shedding after bacterial infection. Streptomycin-treated BALB/c mice were orogastrically gavaged with E. coli strains. Feces were collected, and CFU were quantified. Black bar: E. coli K-12 harboring pRMkluc (negative control). Gray bar: ETEC FMU073332 harboring pRMkluc. The asterisk indicates a significant difference (p ≥ 0.005) when comparing ETEC FMU073332 vs. E. coli K-12 shedding. n = 3.
In vivo and Ex vivo Mouse Infections to Investigate ETEC Bioluminescence Emission
Bacterial passage through the mouse intestine determines the colonization dynamics of intestinal pathogens such as ETEC. Streptomycin-treated BALB/c mice were intraperitoneally administered with 200 μL (15 ng/μL) of luciferin and gastrically gavaged with 1 × 108 CFU ETEC FMU073332 harboring pRMkluc and E. coli K-12 harboring pRMkluc. Following gastric inoculation, mice were anesthetized and immobilized to capture bioluminescent emissions. Light capture began following gastric inoculation (Figure 5A), and the light signals in mice inoculated with ETEC FMU073332 harboring pRMkluc at this time point confirmed bacterial inoculation (0 h) (Figures 5A, 6A). After inoculation, the bioluminescent signals displaced toward the small intestine. Forty-eight hours after gastric inoculation, the bioluminescent signals indicated bacterial passage through the mouse intestine. The bacterial bioluminescent signals remained in the mouse intestine after the 120 h post-inoculation (Figures 5A, 120 h and 6C), which corresponded to the bacterial shedding data. However, no signals were recovered from the distal portion of the mouse intestine. E. coli K-12 bioluminescent signals were localized in the cecum (Figures 5, 6).
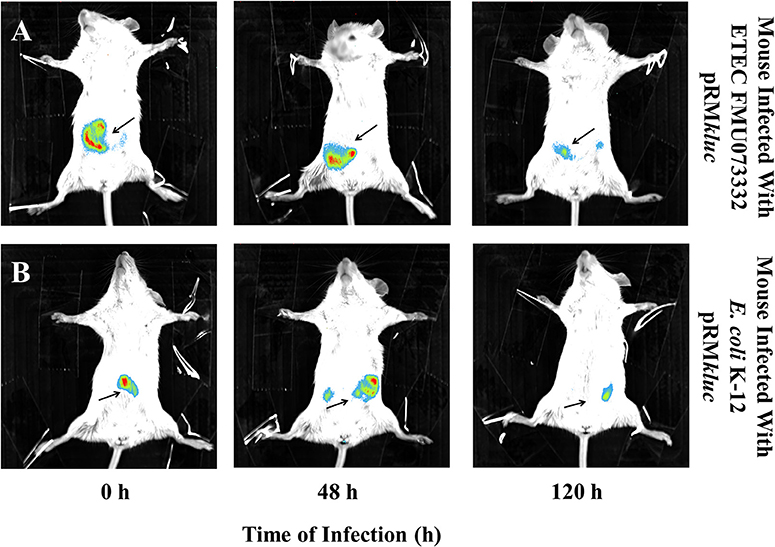
Figure 5. Bioluminescence tracking of in vivo bacterial colonization. Streptomycin-pretreated BALB/c mice were orogastrically inoculated with (A) ETEC FMU073332 harboring pRMkluc (B) E. coli K-12 harboring pRMkluc. Bioluminescent signals were captured upon initial inoculation (0 h), 48 h and 120 h post-inoculation. The bioluminescent signal is depicted as pseudocolor (red = intense, blue = less intense). Arrows indicate the bioluminescent signal. n = 3.
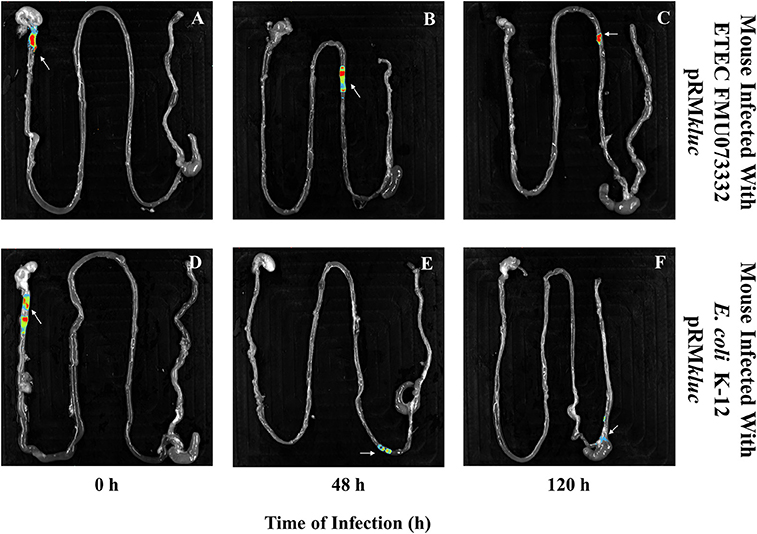
Figure 6. Bioluminescence imaging of bacterial passage through the mouse gastrointestinal tract. Bioluminescence capture of (A) ETEC FMU073332 harboring pRMkluc at 0 h, (B) ETEC FMU073332 harboring pRMkluc at 48 h, (C) ETEC FMU073332 harboring pRMkluc at 120 h, (D) E. coli K-12 harboring pRMkluc at 0 h, (E) E. coli K-12 harboring pRMkluc at 48 h, and (F) E. coli K-12 harboring pRMkluc at 120 h. (Red = intense, blue = less intense). The arrow indicates the bioluminescent signal. n = 3. Each image is representative of 3 mice.
After 120 h of E. coli infection, mouse gastrointestinal tracts were extracted to perform ex vivo imaging. Intestinal tract dissection comprised the esophagus to the rectum. The bioluminescent signals emitted by ETEC FMU073332 harboring pRMkluc were located in the proximal mouse ileum approximately 6 cm from the cecum, whereas the control signals were identified in the cecum and in the proximal colon (Figure 6).
Discussion
Luciferase reporters have been extensively used for imaging in small animals (Doyle et al., 2004), and a luciferase reporter gene under the control of a constitutive promoter allows us to track bacteria during different stages of infection. The burden left by ETEC infection during previous decades is immeasurable, and this pathogen still represents a threat to human health (Petri et al., 2008; Lozano et al., 2012). Children are particularly vulnerable to diarrhea caused by ETEC, and they struggle with the dehydration caused by enterotoxin activity (Qadri et al., 2005). ETEC virulence factors, such as enterotoxins and colonization factors have been extensively studied. However, ETEC colonization dynamics remain uncharacterized and understudied (Luo et al., 2014). Colonization studies may help us understand ETEC pathogenicity and propose novel strategies to prevent ETEC colonization.
Here, we developed a vector permitting the use of bioluminescence to follow ETEC during in vivo infection. The dnaK promoter region was chosen to promote constitutive expression of the luc gene; DnaK is involved in bacterial chromosome division and in the bacterial stress response (McCarty and Walker, 1994). The dnaK promoter region fused to the luc gene enables bacteria to emit bioluminescence. Other similar systems exist, such as the luxCDABE system, in which an entire operon is cloned into a vector or inserted into the bacterial chromosome. However, bacterial metabolic stress has reported (Jawhara and Mordon, 2004), and given our aim to use a bioluminescence system to track and study the highly adherent clinical isolate ETEC FMU073332 and its colonization factors, we wanted to avoid unnecessary bacterial metabolic stress.
In vitro assays were performed to determine whether our transcriptional fusion permitted bacteria harboring pRMkluc to emit light; however, images recovered from solid LB agar plates permitted us to determine only whether our strains were bioluminescent. Light emitted in the petri dish polystyrene assays allowed us perform signal quantification (Foucault et al., 2010), and we observed it was possible to reduce the incubation time with luciferin in the polystyrene assay from 1 h to 5 min. We used the dnak gene promoter region as a constitutive promoter and observed bioluminescence emission by E. coli K-12 harboring pRMkluc beginning during the initial stages of kinetic growth (0.129 OD600 and 1 h); light emission peaked at 1.9 OD600 (9 h) during the late exponential phase of kinetic growth. These data indirectly indicate the dnak promoter region successfully induced luc gene expression, resulting in light production due to luciferase activity.
The mouse intestinal tract is covered with connective tissue, muscle, skin and fur, and superimposition of this tissue makes light exposition and imaging difficult (Doyle et al., 2004). In vitro light emission assays allowed us to measure light duration over time, and we determined that the first minutes are essential for light emission, which decreases by the hour. Light-emitting bacterial signals enabled detection following intraperitoneal inoculation. Luciferase was administered intraperitoneally to avoid decreased light emission during in vivo infection assays; this luciferase administration permitted us to track bacteria during passage through the mouse intestine (Foucault et al., 2010).
Streptomycin treatment eliminates the resident facultative anaerobic microbiota to permit colonization by E. coli (Myhal et al., 1982; Bhinder et al., 2014). The biochemical characteristics and virulence factors implicated in host infectivity determine whether a bacterial strain colonizes the intestinal tract (Bernier-Fébreau et al., 2004; Kumar et al., 2016). Colonization assays permit the effects of virulence factors on colonization ability to be assessed; strains lacking essential virulence and colonization factors are shed for a significantly shorter duration and at a lower magnitude than wild-type strains (Mundy et al., 2006). ETEC FMU073332 shedding appeared 22 h after gastric inoculation, but declined over time. Nevertheless, ETEC FMU073332 shedding values remained higher than E. coli K-12 values. Based on these data, we suggest that ETEC FMU073332 colonizes the mouse intestinal tract, and its virulence characteristics may enhance adhesion to intestinal tissue. Our future aim is to determine which colonization factors mediate ETEC colonization.
BLI imaging suggests that passage of ETEC FMU073332 through the intestinal tract is slower than the movement of E. coli K-12 and other E. coli pathotypes, such as enteropathogenic and enterohemorrhagic E. coli, which are found in the cecum after 180 min (Rhee et al., 2011). The appearance of BLI signals may explain the results observed in the colonization assay. ETEC FMU073332 is likely retained in the intestine due to the presence of its adhesive structures and biochemical characteristics that facilitate intestinal colonization. Ex vivo imaging allowed us to locate the region of the intestine colonized by ETEC. Similar to a report describing colonization of the mouse ileum by ETEC H10407 (Allen et al., 2006), we found a BLI signal in the proximal ileum 120 h after infection, indicating ETEC FMU073332 colonizes the mouse ileum as previously reported for other ETEC strains. This is the first study investigating ETEC pathogenesis to employ BLI technology. Our vector is suited for BLI imaging during in vitro, in vivo, and ex vivo assays. In summary, ETEC colonizes the mouse intestine and exhibits tropism for the mouse ileum.
Ethics Statement
This study was reviewed and approved by the Research Committee (Dr. Onofre Muñoz Hernández), Ethics Committee (Dr. Amparo Faure Fontenla), and Biosecurity Committee (Dr. Herlinda Vera Hermosillo) of HIMFG (permit numbers HIM/2015/034 and HIM/2016/028).
Author Contributions
Designed and conceived the experiments: GR, FM, and JX. Performed the experiments: GR and FM. Analyzed the data: GR, FM, ZS, AC, KE, RH, and JX. Contributed reagents/materials/analytical tools: AC, SO, and JX. Wrote and reviewed the manuscript: GR, FM, ZS, AC, and JX.
Funding
This work was supported by grant number CONACYT 133451 and Public Federal Funds (HIM/2015/034 and HIM/2016/028) from the Hospital Infantil de México Federico Gómez (HIMFG) supported this work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GR is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM), and received fellowship 272718 from CONACYT. We thank Dr. Genaro Patiño and MVZ Raul Castro Luna for their technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fcimb.2017.00187/full#supplementary-material
Figure S1. Construction of the pMRkluc vector. (1) Molecular weight, (2) pGem-luc-dnaK plasmid, (3) Linearization of pGem-luc-dnaK with BamHI, (4) Triple digestion with SalI, HindIII, and BamHI, (5) pRMkluc plasmid, (6) Linearization of pRMkluc with BamHI, (7) Triple digestion with SalI, HindIII, and BamHI.
References
Allen, K. P., Randolph, M. M., and Fleckestein, J. M. (2006). Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74, 869–875. doi: 10.1128/IAI.74.2.869-875.2006
Bernier-Fébreau, C., Du Merle, L., Turlin, E., Labas, V., Ordonez, J., Gilles, A. M., et al. (2004). Use of deosyribose by intestinal and extraintestinal pathogenic Escherichia coli strains: a metabolic adaptation involved in competitiveness. Infect. Immun. 72, 6151–6156. doi: 10.1128/IAI.72.10.6151-6156.2004
Bhinder, G., Stahl, M., Sham, H. P., Crowley, S. M., Morampudi, V., Dalwadi, U., et al. (2014). Intestinal epithelium-specific MyD88 signaling impacts host susceptibility to infectous colitis by promoting protective goblet cell and antimicrobial responses. Infect. Immun. 82, 3753–3763. doi: 10.1128/IAI.02045-14
Bolivar, F., Rodriguez, R. L., Greene, P. J., Betlach, M. C., Heyneker, H. L., Boyer, H. W., et al. (1997). Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2, 95–113. doi: 10.1016/0378-1119(77)90000-2
Contag, C. H., Contag, P. R., Mulling, J. I., Spilman, S. D., Stevenson, D. K., and Benaron, D. A. (1995). Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18, 593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x
Coombes, J. L., and Robey, E. A. (2010). Dynamic imaging of host-pathogen interactions in vivo. Nat. Rev. Immunol. 10, 353–364. doi: 10.1038/nri2746
Cravioto, A., Reyes, R. E., Trujillo, F., Uribe, F., Navarro, A., De la Roca, J. M., et al. (1990). Risk of diarrhea during the first year of life associated with initial and subsequent colonization by specific enteropathogens. Am. J. Epidemiol. 131, 886–904. doi: 10.1093/oxfordjournals.aje.a115579
Cruz-Córdova, A., Espinoza-Mazariego, K., Ochoa, S. A., Saldaña-Ahuactzi, Z., Rodea, G. E., Cázares-Domínguez, V., et al. (2014). CS21 positive multidrug-resistant ETEC clinical isolate from children with diarrhea are associated with self-aggregation, and adherence. Front. Microbiol. 5:709. doi: 10.3389/fmicb.2014.00709
Doyle, T. C., Burns, S. M., and Contag, C. H. (2004). In vivo bioluminescence imaging for integrated studies of infection. Cell. Microbiol. 6, 303–317. doi: 10.1111/j.1462-5822.2004.00378.x
Foucault, M.-L., Thomas, L., Goussard, S., Branchini, B. R., and Grillot-Courvalin, C. (2010). In vivo bioluminescence imaging for the study of intestinal colonization by Escherichia coli in mice. Appl. Environ. Microbiol. 76, 264–274. doi: 10.1128./AEM.01686-09
Gaastra, W., and Svennerholm, A.-M. (1996). Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4, 444–452. doi: 10.1016/0966-842X(96)10068-8
Gonzalez, R. J., Weening, E. H., Frothingham, R., Sempowski, G. D., and Miller, V. L. (2012). Bioluminescence imaging to track bacterial dissemination of Yersinia pestis using different routes of infection in mice. BMC Microbiol. 12:147. doi: 10.1186/1471-2180-12-147
Guevara, C. P., Luiz, W. B., Sierra, A., Cruz, C., Qadri, F., Kaushik, R. S., et al. (2013). Enterotoxigenic Escherichia coli ETEC CS21 pilus contributes to adhesión to intestinal cells and to pathogenesis under in vivo conditions. Microbiology 159(Pt 8), 1725–1735. doi: 10.1099/mic.0.065532-0
Hutchens, M., and Lurken, G. D. (2007). Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 9, 2315–2322. doi: 10.1111/j.1462-5822.2007.00995.x
Jawhara, S., and Mordon, S. (2004). In vivo imaging of bioluminescent Escherichia coli in a cutaenous wound infection model for evaluation of an antibiotic therapy. Antimicrob. Agents Chemother. 48, 3436–3441. doi: 10.1128/AAC.48.9.3436-3441.2004
Kuklin, N. A., Pancari, G. D., Tobery, T. W., Cope, L., Jackson, J., Gill, C., et al. (2003). Real-time monitoring of bacterial infection in vivo: development of bioluminescent staphylococcal foreign-body and deep-thigh-wound mouse infection models. Antimicrob. Agents Chemother. 47, 2740–2748. doi: 10.1128/AAC.47.9.2740-2748.2003
Kumar, P., Kuhlmann, F. M., Bhullar, K., Yang, H., Vallance, B. A., Xia, L., et al. (2016). Dynamic interactions of a conserved enterotoxigenic Escherichia coli adhesion with intestinal mucin govern epithelial engagement and toxin delivery. Infect. Immun. 84, 3608–3617. doi: 10.1128/IAI.00692-16
Lozano, R., Naghavi, M., Foreman, K., Lim, S., Shibuya, K., Aboyans, V., et al. (2012). Global and regional mortality from 235 causes of death for age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128. doi: 10.1016/s0140-6736(12)61728-0
Luo, Q., Kumar, P., Vickers, T. J., Sheikh, A., Lewis, W. G., Rasko, D. A., et al. (2014). Enterotoxigenic Escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect. Immun. 82, 509–521. doi: 10.1128/IAI.01106-13
McCarty, J. S., and Walker, G. C. (1994). DnaK mutants defective in ATPase activity are defective in negative regulation of the heat shock response: expression of mutant DnaK proteins results in filamentation. J. Bacteriol. 176, 764–780. doi: 10.1128/jb.176.3.764-780.1994
Mundy, R., Girard, F., Fitzgerald, A. J., and Frankel, G. (2006). Comparison of colonization dynamics and pathology of mice infected with enteropathogenic Escherichia coli, enterohaemorragic E. coli and Citrobacter rodentium. FEMS Microbiol. Lett. 265, 126–132. doi: 10.1111/j.1574-6968.2006.00481.x
Myhal, M. L., Laux, D. C., and Cohen, P. S. (1982). Relative colonizing abilities of human fecal and K12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur. J. Clin. Microbiol. 1, 186–192. doi: 10.1007/BF02019621
Petri, W. A. Jr., Miller, M., Binder, H. J., Levine, M. M., Dillinham, R., and Guerrant, R. L. (2008). Enteric infections, diarrea, and their impact on function and development. J. Clin. Invest. 118, 1277–1290. doi: 10.1172/JCI34005
Qadri, F., Svennerholm, A.-M., Faruque, A. S. G., and Sack, R. B. (2005). Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment and prevention. Clin. Microbiol. Rev. 18, 465–483. doi: 10.1128/CMR.18.3.465-483.2005
Rhee, K.-J., Cheng, H., Harris, A., Morin, C., Kaper, J. B., and Hecht, G. A. (2011). Determination of spatial and temporal colonization of enteropathogenic E. coli and enterohemorrhagic E. coli in mice using bioluminescent in vivo imaging. Gut Microbes 2, 34–41. doi: 10.4161/gmic.2.1.14882
Saldaña-Ahuactzi, Z., Cruz-Córdova, A., Rodea, G. E., Porta, H., Navarro-Ocaña, A., Eslava-Campos, C., et al. (2017). Genome sequence of enterotoxigenic Escherichia coli strain FMU073332. Genome Announc. 5:e01600-16. doi: 10.1128/genomeA.01600-16
Torres, A. G., Cieza, R. J., Rojas-Lopez, M., Bluentritt, C. A., Souza, C. S., Johnston, R. K., et al. (2012). In vivo bioluminescence imaging of Escherichia coli O104:H4 and role of aerobactin during colonization of a mouse model of infection. BMC Microbiol. 12:112. doi: 10.1186/1471-2180-12-112
Keywords: ETEC, bioluminescence, colonization, in vivo, infection
Citation: Rodea GE, Montiel-Infante FX, Cruz-Córdova A, Saldaña-Ahuactzi Z, Ochoa SA, Espinosa-Mazariego K, Hernández-Castro R and Xicohtencatl-Cortes J (2017) Tracking Bioluminescent ETEC during In vivo BALB/c Mouse Colonization. Front. Cell. Infect. Microbiol. 7:187. doi: 10.3389/fcimb.2017.00187
Received: 03 March 2017; Accepted: 28 April 2017;
Published: 16 May 2017.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Philip R. Hardwidge, Kansas State University, USAJ. Daniel Dubreuil, Université de Montréal, Canada
Copyright © 2017 Rodea, Montiel-Infante, Cruz-Córdova, Saldaña-Ahuactzi, Ochoa, Espinosa-Mazariego, Hernández-Castro and Xicohtencatl-Cortes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Xicohtencatl-Cortes, juanxico@yahoo.com
 Gerardo E. Rodea
Gerardo E. Rodea Francisco X. Montiel-Infante
Francisco X. Montiel-Infante Ariadnna Cruz-Córdova
Ariadnna Cruz-Córdova Zeus Saldaña-Ahuactzi
Zeus Saldaña-Ahuactzi Sara A. Ochoa
Sara A. Ochoa Karina Espinosa-Mazariego
Karina Espinosa-Mazariego Rigoberto Hernández-Castro
Rigoberto Hernández-Castro Juan Xicohtencatl-Cortes
Juan Xicohtencatl-Cortes