Saccharification of Agricultural Lignocellulose Feedstocks and Protein-Level Responses by a Termite Gut-Microbe Bioreactor
- Department of Entomology, Purdue University, West Lafayette, IN, USA
This study investigated saccharification and protein-level responses to the candidate biofuel feedstocks corn stover (CS) and soybean residue (SR) by the gut of a lower termite. The focus termite was Reticulitermes flavipes, which is a highly efficient digester of wood lignocellulose that houses a mixture of prokaryotic and eukaryotic microbes in its gut. Our specific objectives were to (i) measure saccharification potential of the CS and SR feedstocks by termite gut protein extracts, (ii) identify specific proteins in the termite gut responding to feeding on CS and SR diets, and (iii) evaluate gut lignocellulase and accessory enzyme activity responses to CS and SR feeding. Cellulose paper was the control diet. Although CS was saccharified at higher levels, termite gut protein extracts saccharified both CS and SR irrespective of feedstock loading. Consumption of the CS and SR feedstocks by termites resulted in surprisingly few differences in gut protein profiles, with the main exception being elevated myosin abundance with SR feeding. Activity of potential lignocellulases and accessory enzymes was generally similar between CS and SR fed guts as well; however, cellobiohydrolase/exoglucanase activity was higher with CS feeding and glutathione peroxidase activity with SR feeding. These findings have significance from two perspectives. First, SR feeding/digestion appears to cause physiological stress in the termite gut that likely would extend to other types of microbial environments including those within industrial bioreactors. Second, because termites can survive on exclusive CS and SR diets and their guts exhibit clear CS and SR saccharification activity, this validates the R. flavipes system as a potential source for CS and SR degrading enzymes; in particular, cellobiohydrolases/exoglucanases and glutathione peroxidases from this system may play roles in CS and SR breakdown.
Introduction
Lignocellulosic biomass offers promise for biofuel production due to its abundance and availability. Lignocellulose is a complex structure with its three main components of lignin, cellulose, and hemicellulose. It is crucial to break the lignin and hemicellulose polymers encapsulating the cellulose polymer, which is a source for many renewable bioproducts. In nature, organisms with physiological abilities to degrade lignocellulose occur across the tree of life (Cragg et al., 2015). Physiological degradation of lignocellulose is conferred mainly by cellulases and hemicellulases belonging to the glycoside hydrolase (GH) superfamily, and other accessory enzymes (e.g., Horn et al., 2012; Levasseur et al., 2013). Breakdown and/or dissociation of lignin is important to increase access to cellulose and hemicellulose. Cellulose depolymerization occurs with the synergistic action of endoglucanases, cellobiohydrolases (or exoglucanases), and β-glucosidases. Hemicellulose is degraded not only by endo- and exo-xylanases from GH family 5 (Aspeborg et al., 2012) but also in the focus termite of the current study by GH9 endoglucanases that target internal glucose residues (Scharf et al., 2011b). In recent years, wood feeding insects have gained attention for biofuel production due to their inherent ability to enzymatically degrade recalcitrant lignocellulose. Specifically, two groups of wood-feeding insects, wood-boring beetles, and termites (both higher and lower) have been studied for the purpose of deciphering lignocellulose unlocking mechanisms (Geib et al., 2008; Sun and Scharf, 2010; Scharf, 2015a).
Termites along with their gut symbiota efficiently degrade lignocellulose. Higher termites including Nasutitermes takasagoensis and lower termites including Coptotermes formosanus, C. gestroi, and Reticulitermes flavipes have been extensively studied from genes to function to dissect their lignocellulose digestive physiology (Tokuda et al., 1997; Cairo et al., 2011; Zhang et al., 2012; Scharf, 2015a). There is ample evidence of termites having endogenous cellulases that aid in cellulose digestion independent of symbiont action (Watanabe et al., 1998; Zhou et al., 2007), but clearly a synergistic collaboration exists between the termite and its symbiota that achieves efficient lignocellulose digestion (Scharf et al., 2011a,b). Several GH families have been identified in termites with activity toward various cellulosic substrates (Tokuda et al., 1997; Zhou et al., 2007, 2010; Scharf et al., 2010). In contrast, little is known about lignin breakdown mechanisms of wood-feeding insects (Geib et al., 2008), which is a limiting step in biofuel production (Yang and Wyman, 2008).
Reticulitermes flavipes termites provide a model system for studying lignocellulose digestive mechanisms (Scharf, 2015b). In addition to secreting host-derived digestive enzymes, this lower termite harbors a diversity of eukaryotic (protist) and prokaryotic (bacteria) symbionts in its hindgut, which is analogous to a fermentation chamber (Watanabe and Tokuda, 2010; Brune, 2014). Comparison of metatranscriptome profiles for wood and paper (cellulose)-fed termites has shed much light on putative lignocellulose degrading enzymes and detoxification enzymes potentially involved in lignin and metabolite degradation (Raychoudhury et al., 2013). In addition, detoxification and antioxidant enzymes including phenoloxidases, laccases, esterase, cytochrome P450s, catalases, superoxide dismutases, and glutathione peroxidases (GPX) were previously found upregulated in termites fed on lignin phenolics (Tartar et al., 2009; Coy et al., 2010; Sethi et al., 2013). In vitro digestion of a complex lignocellulose substrate (pine sawdust) using recombinant enzymes corresponding to termite lignocellulases conclusively demonstrated their role in lignocellulose saccharification (Sethi et al., 2013). In an extension of the above research to investigate breakdown of second-generation agricultural feedstocks, the gut metatranscriptomes of termites fed on corn stover (CS) and soybean residue (SR) were characterized (Rajarapu et al., 2015). This prior study revealed decreases in microbial abundance with CS and SR feeding relative to cellulose paper, but few differences in functional enzymatic profiles.
The present study investigated protein-level impacts of CS and SR feeding within the R. flavipes gut. Our objectives were as follows: (i) to determine saccharification potential of CS and SR by termite gut homogenates in vitro, (ii) define impacts of CS and SR feeding on gut protein expression, and (iii) identify candidate CS and SR digestive mechanisms in termite guts using enzyme activity assays. Our hypotheses in relation to these objectives were that the termite gut can saccharify CS and SR in vitro, protein profiles of termite guts fed on CS and SR would be similar, and key enzyme activities would be optimized to CS and SR diets according to the “diet-adaptation hypothesis” (Karl and Scharf, 2015). Our findings demonstrate the ability of termite guts to saccharify CS and SR. Additionally, we identified apparently deleterious physiological effects of SR feeding/digestion on total gut protein profiles, along with candidate enzymes that potentially can synergize the saccharification of CS and SR feedstocks.
Materials and Methods
Feedstocks and Chemicals
Corn stover was “specialty 3557 variety” and SR was “Williams 82 variety.” Both CS and SR were grown without chemical insecticides and transgenic toxins. CS and SR were donated by Dr. Nathan Mosier of Purdue University and Dr. Karen Hudson of the USDA-ARS. CS contained 45% glucan, 29% xylan, and 7.1 g lignin/100 g cell wall. SR contained 44% glucan, 33% xylan, and 6.4 g lignin/100 g cell wall (Rajarapu et al., 2015). All substrates used for enzyme activity assays were from Sigma-Aldrich (USA) and Carbosynth Limited (UK). Rat monoclonal myosin antibody was purchased from Abcam and was a gift from Dr. Chris Mattison, USDA-ARS. Soybean Kuntiz trypsin inhibitor was purchased from Cayman Chemicals (USA).
Termites and Feeding Assays
Reticulitermes flavipes workers from lab colonies were used in all studies and were maintained at 22°C in darkness with 70% relative humidity. Feeding assays were performed with the CS and SR feedstocks as described previously (Rajarapu et al., 2015). Feedstocks were ground to fine powder in a coffee grinder (Cuisinart) to prepare “cookies.” 40 termites per treatment were fed these diet cookies for 7 days at 28°C. Each treatment was replicated three times. Feeding bioassays with Kunitz protease inhibitor were performed similarly; fifteen termites were fed six different concentrations (10–0.001 mg) of protease inhibitor on filter paper for 7 days. Controls were fed filter paper without protease inhibitor. Filter paper was moistened every other day with 50 µl sodium phosphate buffer (0.1M, pH 7.0).
Dissections and Protein Extraction
After 7-day feeding assays, whole guts were collected along with salivary glands in 100mM sodium phosphate buffer (pH 7.0). Gut tissues were homogenized in ice-cold phosphate buffer and centrifuged at 10,000 × g for 10 min at 4°C. Supernatant was used for enzyme activity assays and for separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein content was estimated at 562 nm using the Bicinchoninic acid method with bovine serum albumin standard following the manufacturer’s protocol (Thermo Scientific) using a BioTek PowerWave microplate reader (Winooski, VT, USA).
In Vitro Saccharification Assays
Approximately 50 worker termites were dissected per experimental replicate to collect guts. Whole guts from replicate termite lab colonies were isolated and homogenized in 1.5 ml phosphate buffer (0.1M, pH 7.0), centrifuged at 10,000 × g for 10 min at 4°C, and the clarified supernatant was saved for use in in vitro assays (Scharf et al., 2011a). Feedstocks were milled to 0.5 mm particle size in a UDY cyclone sample mill for use in assays. Saccharification assays were done in 750 µl volumes containing 150 µl gut supernatant (i.e., five gut equivalents) and 600 µl HEPES buffer (0.1M, pH 7.0). The assays were contained within vented 1.5 ml Eppendorf tubes in a shaking incubator set to 37°C and 200 rpm. Tubes were vented by melting a pinhole in the assay tube lids. All assay conditions are based on extensive preliminary work showing circum-neutral gut pH in R. flavipes, digestive enzyme pH optima around 7, and enzyme stability at 37°C (Zhou et al., 2007, 2010; Tartar et al., 2009; Coy et al., 2010; Scharf et al., 2010, 2011a; Sethi et al., 2013). A range of feedstock amounts was compared (0.2, 0.4, 0.8, 1, and 2% w/v). Assays were incubated for 4 h and reactions stopped with a final concentration of 4mM ethylenediaminetetraacetic acid (EDTA). EDTA was added to inhibit metalloenzymes, like laccases (Coy et al., 2010), and it also stabilizes color formation for the glucose detection reagent, which includes hydrogen peroxide that could be acted upon by gut metalloenzymes (Scharf et al., 2011a). Liberated glucose was quantified using a colorimetric glucose detection kit that utilizes glucose mutarotase biochemistry (Wako Chemicals USA; Catalog No. 298-65701) against a glucose standard curve (5–0.3125mM). Two experimental replicates were performed for each feedstock using termites from the same colony. Blanks that included feedstock without gut homogenate for each w/v% were run in parallel and used to correct for non-enzymatic saccharification (negligible). Gut preparations were also tested for the presence glucose and it was negligible (Scharf et al., 2011a and results not shown).
SDS-PAGE and Protein Identification by LC-MS/MS
Gut homogenates from termites fed on paper, CS, and SR were combined 1:1 with sample loading buffer, loaded at 40 μg/lane and separated on 8% or 4–20% SDS-PAGE gels at a constant voltage of 120 V for 1.0 or 1.5 h. Gels were stained in 0.3% Coomassie blue for 1–2 h and destained in 40% methanol with 10% acetic acid for 1 h and 10% Methanol + 5% acetic acid overnight. Stained gels were photographed using ChemiDoc™ XRS + system (Bio-Rad, Hercules, SA). Coomassie stained bands were quantified using ImageJ (Rasband, 2014). Bands of interest were cut from gels and sequenced at the Indiana University–Purdue University proteomics core, Indianapolis, IN, USA. Briefly, gel bands were destained, reduced with 10mM dithiothreitol in 10mM ammonium bicarbonate, and then alkylated with 55mM iodoacetamide (prepared in 10mM ammonium bicarbonate). Alkylated samples were digested with trypsin (Promega) overnight at 37°C. Digested peptides were extracted from gel slices first with 50% ACN (acetonitrile) + 49.9%water + 0.1% TFA (trifluoro acetic acid), and then 99.9% ACN + 0.1% TFA. Trypsin-cleaved peptides were injected onto a C18 column (Thermo-Fisher Scientific LTQ and Surveyor HPLC system). Peptides were eluted with a linear gradient of 5–35% ACN (in water with 0.1% FA) developed over 60 min at room temperature, at a flow rate of 50 µl min−1, and effluent was electro-sprayed into the LTQ mass spectrometer. Blanks were run prior to samples to ensure there was no significant signal from solvents or the column. Band identification was done using the Sequest™ algorithm against the translated host and symbiont metatranscriptomes of R. flavipes (Genbank accession Nos. PRJNA275308, FL634956-FL640828, and FL641015-FL645753) (Tartar et al., 2009; Rajarapu et al., 2015) and the predicted proteome of Zootermopsis nevadensis (UP000027135).
Immunoblotting
For western blotting, 8% SDS-gels were run at the same conditions mentioned above and proteins transferred to nitrocellulose membrane at 100 V for 1 h. For dot blots, five serial dilutions of gut protein (20–0.032 µg) per sample were spotted directly onto nitrocellulose membranes and allowed to dry. Membranes were blocked with 3% non-fat dry milk overnight, incubated with primary rat myosin antibody in a ratio of 1:4,000 for 1 h, washed 3× for 10 min with Tris buffered saline (TBS), incubated with horse radish peroxidase conjugated anti-rat G goat antibody secondary antibody (Bio-Rad) in a ratio of 1:5,000 for 30 min, washed 3× for 10 min with TBS; and stained with SuperSignal™ West Pico chemiluminescent substrate (Life Technologies). Blots were visualized using a Chemi-Doc XRS system (Bio-Rad).
Enzyme Assays
Carbohydrate Degrading Enzymes
Cellulose and Hemicellulose Substrates
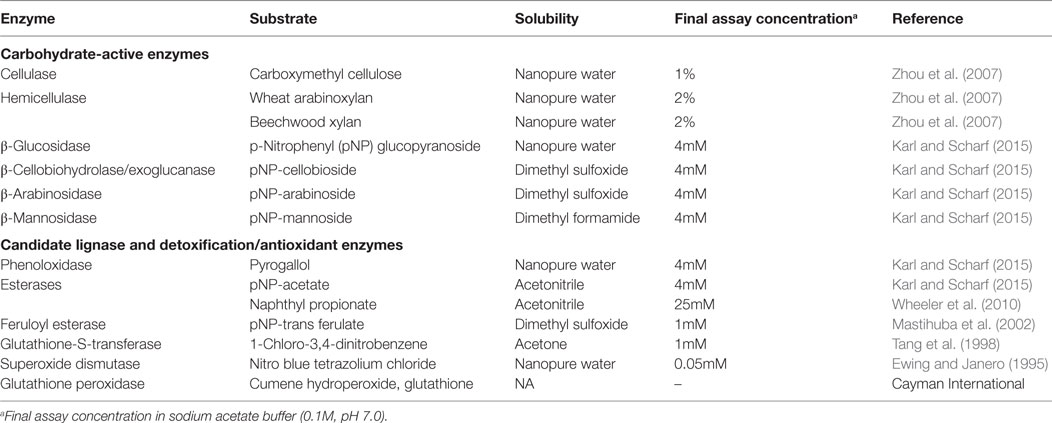
Assays to estimate the activity of potential cellulases and hemicellulases were performed using seven model substrates (Table 1). Endoglucanase and hemicellulase activities were assayed using carboxymethyl cellulose, beech xylan, and wheat arabinan using the dinitrosalicylate (DNSA) detection method as described (Zhou et al., 2007; Karl and Scharf, 2015). Reaction mixtures containing 10 µl gut supernatant and 90 µl of substrate solutions were incubated for 1 h at 30°C. Reactions were stopped by adding 100 µl DNSA solution and incubated at 100°C for 20 min. Absorbance was measured at 540 nm after cooling plates on ice for 15 min. Glucose released was determined against a glucose standard curve (5–0.625mM).
p-Nitrophenyl Substrate Assays
All p-nitrophenyl carbohydrate substrates were assayed as described earlier with slight modifications (Karl and Scharf, 2015). Briefly, stock solutions of p-nitrophenyl substrates were prepared in their respective solvents (Table 1), and 4mM working solutions were prepared fresh in sodium acetate buffer (0.1M, pH 7.0). Reaction mixtures had 10 µl of gut supernatant + 90 µl of substrate solution in a reaction volume that was made to 200 µl with sodium acetate buffer (0.1M, pH 7.0). Reactions were read kinetically for 20 min at 410 nm. Mean velocities were calculated using Gen5 data analysis software (BioTek) and were used for calculating specific activity as micromole p-nitrophenol released per minute per milligram of protein. The molar extinction coefficient of p-nitrophenol is 0.6605mM OD−1 (Zhou et al., 2007).
Accessory Enzymes
Esterases
Esterase activity was measured using two different substrates, naphthyl propionate and p-nitrophenyl acetate. Naphthyl propionate assays were performed as described earlier (Wheeler et al., 2010). Reaction mixtures consisted of 10 µl gut supernatant, 2 µl of 25mM naphthyl propionate in ACN and 188 µl sodium phosphate buffer (100mM, pH 7.5). Reactions were incubated for 10 min at room temperature and stopped with 50 µl of 0.3% Fast Blue BB dissolved in nanopure water containing 3.5% SDS. Reactions proceeded at 30°C for 15 min and were read as endpoints at 600 nm. Specific activity was measured as the amount of α-naphthol released against a naphthol standard curve (5–0.625mM). p-Nitrophenyl acetate assays were performed as detailed in the preceding section. Feruloyl esterase activity was measured using p-nitrophenyl trans-ferulate as described by Mastihuba et al. (2002) with slight modifications. The assay was adapted to a 96-well microplate wherein the reaction mixture consisted of 10 µl gut supernatant and 190 µl of 1mM substrate. Working solution of 1mM substrate was prepared in an emulsion of phosphate buffer with Tween-20 (3 µl ml−1) and DMSO (12 µl ml−1). The phosphate buffer emulsion was added to the appropriate amount of substrate stock and mixed immediately to prevent precipitation. The substrate solution was prepared fresh and used within an hour. Absorbance was read kinetically at 410 nm for every 10 s for 5 min. Substrate reactions and enzyme blanks were run in parallel.
Phenoloxidase, Superoxide Dismutase, Glutathione Peroxidase, and Glutathione-S-Transferase
Phenoloxidase activity was measured using the substrate pyrogallol as described previously (Coy et al., 2010). Superoxide dismutase was assayed as detailed previously (Ewing and Janero, 1995). Briefly, the reaction mixture consisted of 25 µl of gut homogenate, 50µM nitro-blue tetrazolium (NBT) chloride, and 98µM NADH in 50mM sodium phosphate buffer (pH 7.4) containing 0.1mM EDTA. Reactions were initiated by the addition of 25 µl of 33µM phenazine methylsulfate in 50mM phosphate buffer. Absorbance of reduced NBT was measured kinetically at 560 nm for 5 min, with the extinction coefficient for reduced NBT of 15 × 106 mM−1 cm−1. Glutathione-S-transferase activity was measured using 1-chloro-2,4-dinitrobenzene (CDNB) as the substrate (Tang et al., 1998). Assay mixtures of 1mM CDNB, 5mM reduced glutathione in sodium phosphate buffer (100mM, pH 7.4) were prepared fresh. The reaction was started by adding 225 µl assay mixture to 10 µl gut supernatant. Absorbance of the resultant conjugate, dinitrophenyl glutathione, was read kinetically at 324 nm for 10 min. The dinitrophenol extinction coefficient used was 9.5 mM−1 cm−1. Glutathione peroxidase activity was measured using a glutathione peroxidase assay kit (Cayman Chemical, Ann Arbor, MI, USA) following manufacturer protocols.
Statistics
Significant differences in glucose liberation from saccharification assays were determined by non-parametric Kruskal–Wallis tests. Statistical differences in protein band densities were determined by comparing means of three replicates by Kruskal–Wallis tests. Activities of different enzymes were tested for significant differences using one way ANOVA with Tukey mean separation tests to identify diet effects. Statistics were performed at α of 0.05 in JMP statistical software.
Results
In Vivo Feeding and In Vitro Saccharification Assays
Termites consumed similar amounts of cellulose paper, CS, and SR diets but their body weights did not change significantly on any of the diets (p > 0.05; Figure S1 in Supplementary Material). Termite gut homogenates were able to saccharify CS and SR at significant detectable levels; however, CS was saccharified at 150–220× greater levels than SR across the loading rates tested (Figures 1A,B). Background glucose levels in termite gut homogenates were negligible and therefore did not interfere with glucose detection. Glucose release was not significantly affected by the % w/v of feedstock present in assays for both CS and SR (p > 0.05; Figure S2 in Supplementary Material). There was also an unexpected reduction in detectable glucose levels in incubations lasting longer than 4 h for both feedstocks (Figures S2 and S3A in Supplementary Material), and co-incubation of assays with the antimicrobial compound sodium azide did not significantly impact glucose release levels over time (Figure S3B in Supplementary Material).
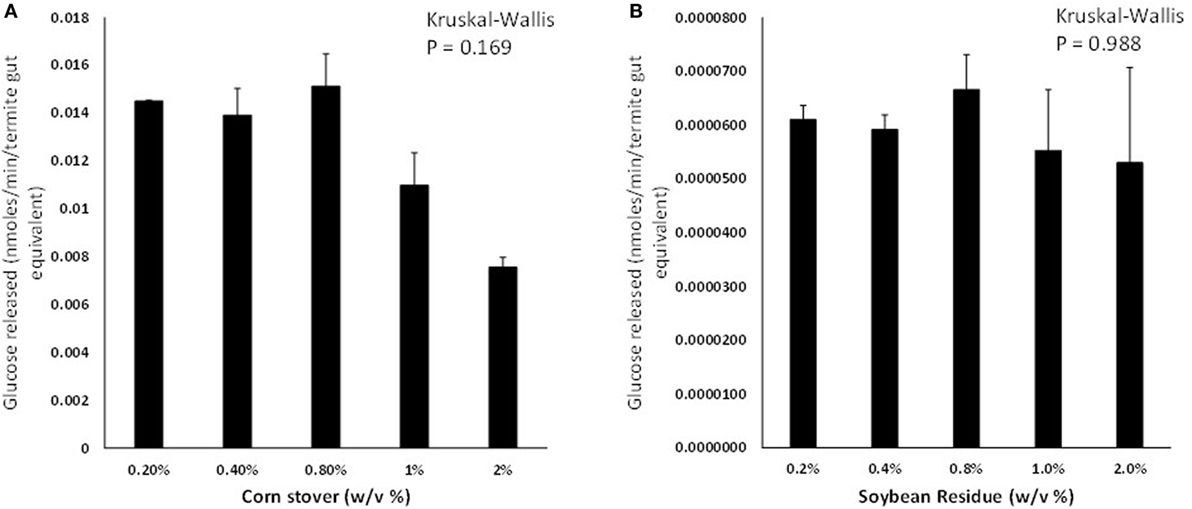
Figure 1. Glucose liberation (nanomoles of glucose liberated per minute per termite gut equivalent) from (A) corn stover and (B) soybean residue by termite gut protein extracts in vitro. Data points represent the average ± SE of two experimental replicates using termite gut preparations from the same colony. Results of global Kruskal–Wallis analyses are shown indicating no effects of feedstock w/v% loading rates.
Protein Profiles of Termite Guts Fed on CS and SR
sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein profiles of termite guts fed on paper, CS, and SR were similar, except for 250, 15, and 10 kDa protein bands that were uniquely associated with SR feeding (Figure 2A) These PAGE results were verified by densitometry analyses of band intensities from independent biological replicate gels (Figure 2B), which showed only the 250 kDa band (myosin heavy chain) was significantly different across treatments. Tandem mass spectrometry was performed to identify protein bands of interest. Proteins were identified by querying against termite transcript and protein databases previously generated from our lab and elsewhere. Identity was confirmed based on coverage scores of sequenced peptides relative to reference proteins, with higher coverage scores indicating higher confidence of identity (Table 2). The significant SR-associated proteins were identified as myosin heavy chain (250 kDa), Cu–Zn superoxide dismutase (15 kDa), and lipocalin/fatty acid binding protein (10 kDa). Bands near the predicted molecular weight of putative lignocellulose degrading enzymes that were relatively abundant after CS and SR feeding were also sequenced and corroborated with previous proteomic studies in R. flavipes (Sethi et al., 2013) (Table 2). However, densitometry analyses of SDS-PAGE separated proteins showed only the 250 kDa (myosin heavy chain) band was significantly different across treatments.
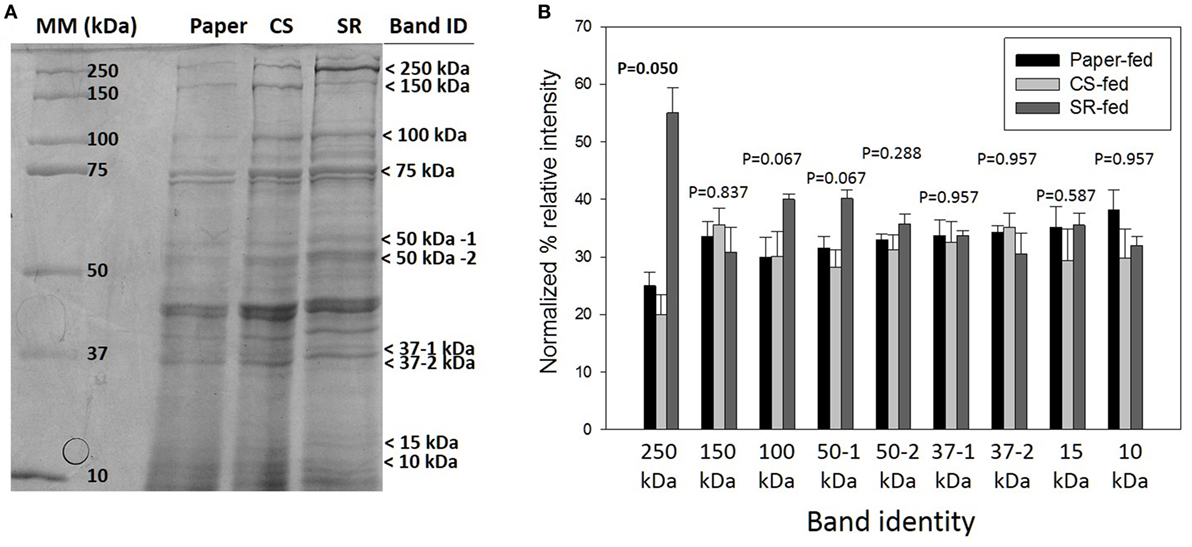
Figure 2. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole gut protein extracts from termites fed on paper, corn stover (CS) or soybean residue (SR). A representative 4–20% gradient gel stained with 0.1% Coomassie blue R-250 is shown. Arrows indicate bands sequenced for identification (Table 1). (B) Densitometry quantification represented as mean density of three biological replicates (±SEM) of SDS-PAGE separated proteins indicated by arrows in panel (A).
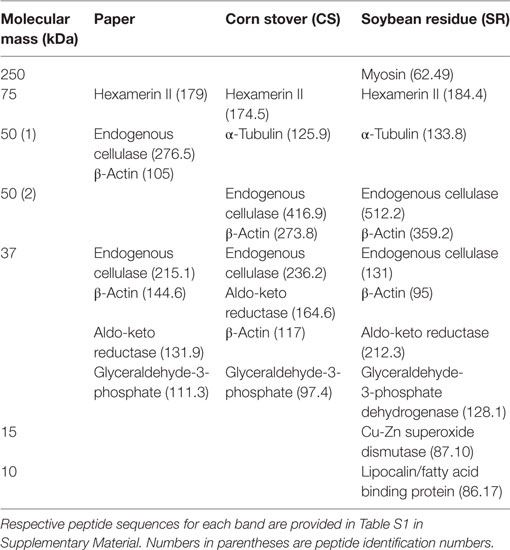
Table 2. Identification of sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein bands by tandem MS analysis.
Myosin Immunoblotting
The identity of the 250 kDa SR-abundant protein identified above as myosin by tandem MS was further verified by immunoblotting (Figure 3). Western and dot blots with a myosin-specific antibody together showed more intense signals in guts from termites that had fed on SR. Myosin patterns in guts of termites fed with soybean Kunitz protease inhibitor were different than those of SR-fed guts (Figure S4 in Supplementary Material), indicating the observed myosin result is not linked to soybean-associated protease inhibition.
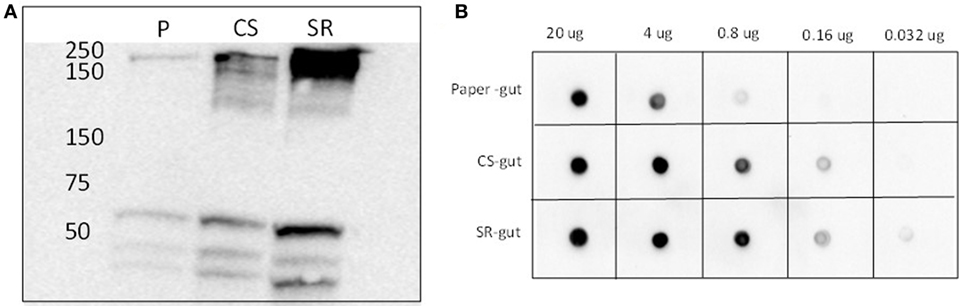
Figure 3. (A) Western blots and (B) dot blots of whole gut protein extracts from termites fed on paper (P), corn stover (CS) and soybean residue (SR). Blots were incubated with primary rat monoclonal myosin antibody (1:4,000) and secondary goat anti-rat horse radish peroxidase-conjugated antibody (1:5,000). Hybridizations were visualized via chemiluminescence. Numbers across the top in panel (B) indicate protein loadings per dot.
Impacts of CS and SR Feeding on Activity of Candidate Lignocellulase and Accessory Enzymes
In vitro gut lignocellulase activities were mostly similar across the CS, SR, and paper-feeding treatments (Table 3). However, among carbohydrate-active enzymes, cellobiohydrolase/exoglucanase (CBH) activity was higher in CS relative to SR and paper-fed guts (p = 0.0006, one way ANOVA). β-Mannosidase activity was lower in SR relative to paper-fed guts (p = 0.047, one way ANOVA), and there were no differences in β-xylosidase and β-arabinosidase activities across feeding treatments. Among the accessory enzymes studied, esterases had lower activity in SR relative to paper-fed guts (p = 0.0003, one way ANOVA); whereas, glutathione peroxidase had higher activity relative to both paper- and CS-fed guts (p = 0.0125, one way ANOVA). No differences were observed for other accessory enzyme activities investigated.
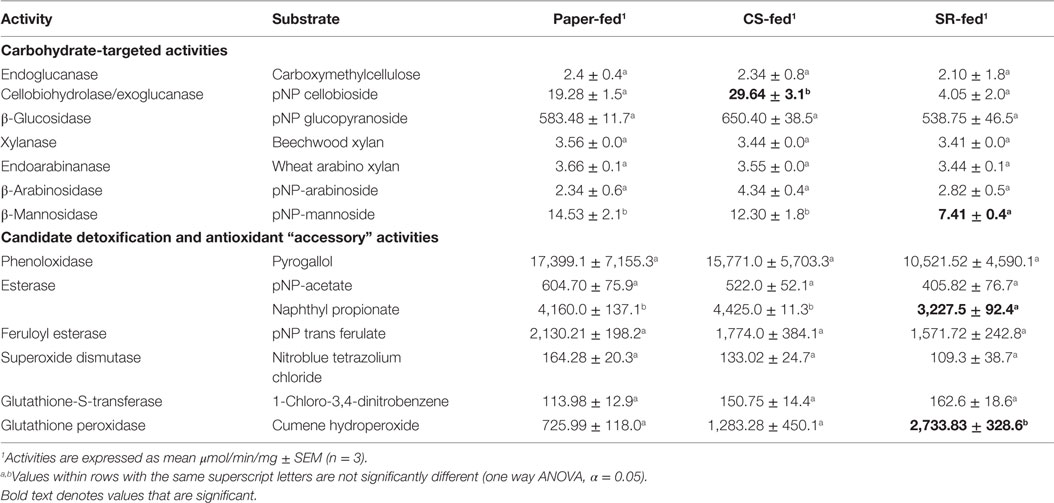
Table 3. Activities of carbohydrate-active and accessory enzymes (μmol/min/mg) in whole-gut homogenates prepared from termites fed for 7 days on paper, corn stover (CS), or soybean residue (SR).
Discussion
Saccharification is the major limiting step in biofuel production from lignocellulosic biomass. Existing pretreatment strategies to degrade the lignocellulose matrix include biological, physical, chemical, and physio-chemical methods that are expensive and in many cases produce hazardous wastes. Due to these disadvantages, there has been a need to develop efficient and environmentally favorable pretreatment methods (Yang and Wyman, 2008). Enzymatic strategies modeled after enzymatic mechanisms of organisms specializing on wood diets have utility for pretreatment of lignocellulosic biomasses (Ke et al., 2011). The termite studied here, R. flavipes, has been an excellent model system for identifying potential enzymes involved in the breakdown of both forest and agricultural feedstocks via the process of diet adaptation (Scharf, 2015a). Through preceding work, we studied the metatranscriptome profiles of R. flavipes guts fed on CS and SR relative to paper in an attempt to identify novel transcripts responding to the respective diets; however, while results showed shifts in gut microbe profiles, few differences in the functional profiles of termite guts fed on CS and SR relative to paper were identifiable (Rajarapu et al., 2015). In the current study, we evaluated protein-level saccharification and changes in response to feeding on the same CS and SR feedstocks. Similar to our prior metratranscriptome study, termites remained active and survived well on the diets for a 7-day period, displaying their flexibility to accept novel lignocellulosic diets. Saccharification assays showing gut-dependent release of glucose from the CS and SR feedstocks agree with the high degrees of survivability seen in bioassays. Overall gut protein profiles and activity of potential lignocellulose degrading enzymes were mostly similar across the CS, SR, and paper-feeding treatments, but some key differences as detailed below were still observed.
Termite Guts Saccharify Feedstocks In Vitro
Findings showed a clear ability of the R. flavipes gut to saccharify CS and SR in vitro. These findings are supported by in vivo bioassays in which termites had essentially 100% survival for 1 week when held with CS and SR as their only food sources [present study and Rajarapu et al. (2015)]. However, in in vitro studies, CS released considerably more glucose than SR, which correlates with the apparent proteolytic impacts of SR on protein composition (discussed in the next section), as well as disappearance of gut microbiota in response to SR feeding (Rajarapu et al., 2015). There also was an unexpected reduction in detectable glucose levels in incubations lasting longer than 4 h for both feedstocks, but co-incubation of assays with the antimicrobial compound sodium azide did not significantly impact glucose release levels over time. These findings point toward conversion of the released glucose and/or inhibition of the saccharification process with longer incubation times, rather than microbial consumption of released glucose in assay reactions. Released glucose can be further converted to byproducts not detectable by the colorimetric method we used to detect glucose (Popoff and Theander, 1972, 1976) or glucose might be conjugated with secondary plant chemicals present in the feedstocks. Production of inhibitory products during pretreatment is another major concern in biofuel production. Several inhibitory compounds have been identified in physical and chemical pretreatments (Jönsson et al., 2013). Additionally, lignin monomers have the potential to inhibit cellulases (Qin et al., 2016). Further research studying metabolite profiles of the saccharification assay mixture can shed more light on potential inhibitory compounds formed during of CS and SR processing.
Gut Protein Profiles
The rationale behind protein profiling studies was that differentially expressed proteins identified would potentially be host (termite) proteins responsive to dietary components, or symbiont proteins showing differential expression in correlation with fluctuating symbiont populations. Overall gut protein profiles of termites fed on CS and SR assessed by SDS-PAGE were very similar except for minor differences in 10 and 15 kDa bands (putatively identified as a lipocalin and a superoxide dismutase), and a 250-kDa band identified as myosin that was unique to SR-fed guts. The lack of changes in carbohydrate-active proteins here is in strong agreement with prior metatranscriptome sequencing performed after CS and SR feeding that also did not identify changes in carbohydrate-active enzyme encoding transcripts (Rajarapu et al., 2015). Myosin belongs to a large superfamily of proteins involved in muscle contraction and cytoskeletal structure (Weiss and Leinwand, 1996). While there is little information on the functional roles of myosin in insects, significant upregulation of myosin in the soldier castes of R. flavipes and C. formosanus termites was previously observed, suggesting a role for myosin in soldier musculature or cell differentiation (Tarver et al., 2012). Also, in Drosophila melanogaster, the myosin heavy chain protein is involved in cell shape and motility during development (Young et al., 1993). Given the structural role of myosin, it is possible that abundance of myosin in SR-fed termite guts might be due to breakdown of termite tissue or symbiota in the hindgut, as suggested by previous research (Rajarapu et al., 2015). Alternatively, myosin liberation might result from direct or indirect effects of soybean anti-herbivory defense mechanisms, including protease inhibitors. We tested this hypothesis by feeding commercially available Kunitz trypsinase inhibitor to termites in a 7-day bioassay. Total protein profiles of termite guts fed on protease inhibitor were different than those of guts fed a SR diet (Figure S4 in Supplementary Material) suggesting that complex proteolysis-related mechanisms, or their inhibition (Chu et al., 2015), might be involved in increasing the myosin detectability in SR fed termite guts. Nonetheless, this investigation of protein profiles demonstrates an effect of SR digestion on termite gut tissue and/or microbial physiology, suggesting possible implications for SR processing by microbes in bioreactor environments. This “degradation” hypothesis may explain the discrepancy between saccharification potential of CS and SR by termite gut protein preparations, but not in most model substrate activities as detailed in the following section. Another possible explanation is the relative recalcitrance of SR and CS feedstocks relative to each other and the model substrates tested.
Impacts on Potential Lignocellulase Activities
Carbohydrate-Active Enzymes
All carbohydrate degrading activities investigated except cellobiohydrolase/exoglucanase were similar across the CS, SR, and paper-feeding treatments. These results are similar to earlier findings wherein R. flavipes was fed five different diets including CS and SR (Karl and Scharf, 2015). Cellulose is synergistically degraded in R. flavipes by endogenous (host) endoglucanase and β-glucosidase activities, with additive effects by protist (symbiont) CBH/exoglucanase activity (Scharf et al., 2011a; Sethi et al., 2013). Cellulose exists in crystalline and amorphous forms due to differences in hydrogen bonds within the polymer. Endoglucanases hydrolyze internal glycosidic bonds of the cellulose polymer whereas CBHs (also known as exoglucanases) act on amorphous cellulose. Nevertheless, CBH from yeast, fungi, and marine invertebrates can also hydrolyze crystalline cellulose (Teeri, 1997). β-glucosidase acts on the hydrolyzates of CBH, releasing monomeric glucose units. Cellulose substrates from different sources vary in amorphous content and crystallinity, as well as size and shape of crystallites (Montanari et al., 2005). The higher CBH activity measured in CS-fed guts might be due to qualitative differences in cellulose composition between CS and SR. Additionally, CBH (GHF7) enzymes in R. flavipes are primarily produced by gut symbionts (Tartar et al., 2009), and higher activity of CBH in CS-fed guts therefore suggests significant participation by symbionts in CS saccharification. In contrast, endoglucanase and β-glucosidase, which are primarily host-derived (Tartar et al., 2009; Scharf et al., 2010; Zhou et al., 2010), had similar activity across guts from termites fed on different diets. An alternative explanation for the observed differences in CBH activity between feedstocks is that, as noted above, protein degradation/inhibition was more substantial with SR feeding and/or SR impacts on symbiont mortality were more significant.
Access to cellulose in lignocellulose is increased with degradation of hemicellulose surrounding the cellulose polymer. Hemicellulose is a heterogeneous polysaccharide frequently containing mannose, xylose, glucose, galactose, and arabinose. Interestingly, gut hemicellulase activity was similar in termites fed on CS and SR, except for β-mannosidase that was reduced with SR feeding. In lower termites like R. flavipes, β-mannosidases are likely contributed by hindgut protists (Brune, 2014). A decrease in β-mannosidase activity could be thus due to a decrease in protist numbers in termite guts fed on SR (Rajarapu et al., 2015). It was hypothesized that xylanase, endoarabinase, and β-arabinosidase activities would be higher in CS-fed termite guts, as hemicellulose in the Poaceae is generally composed of a xylanopyranosyl backbone with arabinofuranosyl side branches. However, there were no significant differences observed in these activities across feeding treatments.
Detoxification and Antioxidant (Accessory) Enzymes
Unlike cellulose and hemicellulose, lignin is a polymer of phenyl propane units linked by C–C or C–O bonds between the three monomers coumaryl, coniferyl, and sinapyl alcohol (Chen, 2014). There is significant independent evidence suggesting that termites can degrade lignin polymers, but the mechanisms of degradation remain elusive (Cookson, 1987, 1988; Geib et al., 2008). From previous studies, enzyme-coding transcripts responding to lignin diets were identified, which included esterase, feruloyl esterase, laccase/phenoloxidase, glutathione-S-transferase, superoxide dismutase, and glutathione peroxidase (Sethi et al., 2013), which were activities tested in this study. Among these activities, glutathione peroxidase activity was higher in SR-fed relative to paper- and CS-fed guts; whereas, naphthyl propionate-specific esterases had lowest activity in SR-fed guts. Lignin is esterified onto the sugar side chains of hemicellulose within the lignocellulose structure. Esterases can thus play a role in breaking the bonds between the lignin polymer and hemicellulose. However, the CS and SR tested in this study have similar lignin quantities around 6–7 mg/100 mg cell wall (Rajarapu et al., 2015), and thus, the difference in esterase activity measured between CS and SR feeding treatments might be due to qualitative differences in lignin composition between CS and SR, or possibly due to aromatics distinct to soybeans (e.g., isoflavones, anthocyanins, phenolics). Alternatively, esterases in R. flavipes are produced by both host and symbiota (Karl and Scharf, 2015). Consequently, the decrease in esterase activity with SR feeding might be due to a loss of gut symbiota. Along with esterases, peroxidases might be involved in lignin degradation either directly by radicalizing bonds in the lignin polymer or indirectly by quenching the potential oxidative stress generated by lignin disassociation/depolymerization (Sethi et al., 2013). Lastly, GPXs are cytosolic enzymes involved in detoxification of hydrogen peroxide with assistance from reduced glutathione. GPX could therefore neutralize oxidative stress created during saccharification. Alternatively, it is possible that GPX could potentially saccharify SR in vitro, but this is not consistent with the known mechanism of action for GPX enzymes.
Conclusion
This study assessed the simultaneous enzymatic pretreatment and saccharification of agricultural feedstocks in vivo and in vitro by the termite gut. Our overall objective was to identify termite gut proteins that might specifically degrade lignocellulose in CS and SR to fermentable glucose. Our findings demonstrate the ability of the lower termite gut to saccharify CS and SR, verifying the utility of this system as a source of enzymes for enzymatic pretreatment and/or saccharification of agricultural feedstocks. Additionally, we identified lignocellulase activities that might be responsible for degrading both CS and SR feedstocks, also supporting the diet-adaptation hypothesis as proposed earlier (Karl and Scharf, 2015). Among the potential lignocellulase and accessory activities assayed, CBH and glutathione peroxidase activity were higher, respectively, in termite guts fed on CS and SR. The cytoskeletal/structural protein myosin was also more abundant in termite guts fed on SR. Myosin-dependent degradation of SR does not seem likely, and its increased presence is more likely indicative of a physiological stress response to deleterious soybean secondary chemicals known to interfere with a number of physiological processes (Weiss and Leinwand, 1996). This outcome highlights the potential effects of SR digestion on biological systems that likely would extend to fermentation chamber microenvironments.
Author Contributions
SR performed the research, analyzed the data, and wrote the manuscript. MS designed the research, interpreted the data, and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Indiana University Purdue University proteomics core for sequencing and identification of protein bands and Jesse Hoteling for maintaining termite colonies.
Funding
This work was supported by National Science Foundation grant no. CBET 1233484 awarded to MS, and the O.W. Rollins/Orkin Endowment in the Department of Entomology at Purdue University.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fenrg.2017.00005/full#supplementary-material.
References
Aspeborg, H., Coutinho, P. M., Wang, Y., Brumer, H., and Henrissat, B. (2012). Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. doi: 10.1186/1471-2148-12-186
Brune, A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180. doi:10.1038/nrmicro3182
Cairo, J., Leonardo, F. C., Alvarez, T. M., Ribeiro, D. A., Büchli, F., Costa-Leonardo, A. M., et al. (2011). Functional characterization and target discovery of glycoside hydrolases from the digestome of the lower termite Coptotermes gestroi. Biotechnol. Biofuels 4, 50. doi:10.1186/1754-6834-4-50
Chen, H. (ed.) (2014). “Chemical composition and structure of natural lignocellulose,” in Biotechnology of Lignocellulose (Beijing: Springer), 25–71.
Chu, C. C., Zavala, J. A., Spencer, J. L., Curzi, M. J., Fields, C. J., Drnevich, J., et al. (2015). Patterns of differential gene expression in adult rotation-resistant and wild-type western corn rootworm digestive tracts. Evol. Appl. 8, 692–704. doi:10.1111/eva.12278
Cookson, L. (1987). 14C-lignin degradation by three Australian termite species. Wood Sci. Technol. 21, 11–25.
Cookson, L. (1988). The site and mechanism of 14 C-lignin degradation by Nasutitermes exitiosus. J. Insect Physiol. 34, 409–414. doi:10.1016/0022-1910(88)90111-4
Coy, M., Salem, T., Denton, J., Kovaleva, E., Liu, Z., Barber, D., et al. (2010). Phenol-oxidizing laccases from the termite gut. Insect Biochem. Mol. Biol. 40, 723–732. doi:10.1016/j.ibmb.2010.07.004
Cragg, S. M., Beckham, G. T., Bruce, N. C., Bugg, T. D., Distel, D. L., Dupree, P., et al. (2015). Lignocellulose degradation mechanisms across the Tree of Life. Curr. Opin. Chem. Biol. 29, 108–119. doi:10.1016/j.cbpa.2015.10.018
Ewing, J. F., and Janero, D. R. (1995). Microplate superoxide dismutase assay employing a nonenzymatic superoxide generator. Anal. Biochem. 232, 243–248. doi:10.1006/abio.1995.0014
Geib, S. M., Filley, T. R., Hatcher, P. G., Hoover, K., Carlson, J. E., del Mar Jimenez-Gasco, M., et al. (2008). Lignin degradation in wood-feeding insects. Proc. Natl. Acad. Sci. U.S.A. 105, 12932–12937. doi:10.1073/pnas.0805257105
Horn, S. J., Vaaje-Kolstad, G., Westereng, B., and Eijsink, V. G. (2012). Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5, 45. doi:10.1186/1754-6834-5-45
Jönsson, L. J., Alriksson, B., and Nilvebrant, N.-O. (2013). Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol. Biofuels 6, 16. doi:10.1186/1754-6834-6-16
Karl, Z. J., and Scharf, M. E. (2015). Effects of five diverse lignocellulosic diets on digestive enzyme biochemistry in the termite Reticulitermes flavipes. Arch. Insect Biochem. Physiol. 90, 89–103. doi:10.1002/arch.21246
Ke, J., Laskar, D. D., Singh, D., and Chen, S. (2011). In situ lignocellulosic unlocking mechanism for carbohydrate hydrolysis in termites: crucial lignin modification. Biotechnol. Biofuels 4, 17. doi:10.1186/1754-6834-4-17
Levasseur, A., Drula, E., Lombard, V., Coutinho, P. M., and Henrissat, B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6, 41. doi:10.1186/1754-6834-6-41
Mastihuba, V. R., Kremnický, L. R., Mastihubová, M., Willett, J., and Côté, G. L. (2002). A spectrophotometric assay for feruloyl esterases. Anal. Biochem. 309, 96–101. doi:10.1016/S0003-2697(02)00241-5
Montanari, S., Roumani, M., Heux, L., and Vignon, M. R. (2005). Topochemistry of carboxylated cellulose nanocrystals resulting from TEMPO-mediated oxidation. Macromolecules 38, 1665–1671. doi:10.1021/ma048396c
Popoff, T., and Theander, O. (1972). Formation of aromatic compounds from carbohydrates: part 1. Reaction of d-glucuronic Acid, d-glacturonic Acid, d-xylose, and l-arabinose in slightly acidic, aqueous solution. Carbohydr. Res. 22, 135–149. doi:10.1016/S0008-6215(00)85733-X
Popoff, T., and Theander, O. (1976). Formation of aromatic compounds from carbohydrates. Acta Chem. Scand 30, 397–402. doi:10.3891/acta.chem.scand.30b-0397
Qin, L., Li, W.-C., Liu, L., Zhu, J.-Q., Li, X., Li, B.-Z., et al. (2016). Inhibition of lignin-derived phenolic compounds to cellulase. Biotechnol. Biofuels 9, 1–10. doi:10.1186/s13068-016-0485-2
Rajarapu, S. P., Shreve, J. T., Bhide, K. P., Thimmapuram, J., and Scharf, M. E. (2015). Metatranscriptomic profiles of Eastern subterranean termites, Reticulitermes flavipes (Kollar) fed on second generation feedstocks. BMC Genomics 16:332. doi:10.1186/s12864-015-1502-8
Rasband, W. S. (2014). ImageJ. Bethesda, MD: U. S. National Institutes of Health. Available at: http://imagej.nih.gov/ij/
Raychoudhury, R., Sen, R., Cai, Y., Sun, Y., Lietze, V. U., Boucias, D., et al. (2013). Comparative metatranscriptomic signatures of wood and paper feeding in the gut of the termite Reticulitermes flavipes (Isoptera: Rhinotermitidae). Insect Mol. Biol. 22, 155–171. doi:10.1111/imb.12011
Scharf, M. E. (2015a). Omic research in termites: an overview and a roadmap. Front. Genet. 6:76. doi:10.3389/fgene.2015.00076
Scharf, M. E. (2015b). Termites as targets and models for biotechnology. Annu. Rev. Entomol. 60, 77–102. doi:10.1146/annurev-ento-010814-020902
Scharf, M. E., Karl, Z. J., Sethi, A., and Boucias, D. G. (2011a). Multiple levels of synergistic collaboration in termite lignocellulose digestion. PLoS ONE 6:e21709. doi:10.1371/journal.pone.0021709
Scharf, M. E., Karl, Z. J., Sethi, A., Sen, R., Raychoudhury, R., and Boucias, D. G. (2011b). Defining host-symbiont collaboration in termite lignocellulose digestion: “The view from the tip of the iceberg”. Commun. Integr. Biol. 4, 761–763. doi:10.4161/cib.17750
Scharf, M. E., Kovaleva, E. S., Jadhao, S., Campbell, J. H., Buchman, G. W., and Boucias, D. G. (2010). Functional and translational analyses of a beta-glucosidase gene (glycosyl hydrolase family 1) isolated from the gut of the lower termite Reticulitermes flavipes. Insect Biochem. Mol. Biol. 40, 611–620. doi:10.1016/j.ibmb.2010.06.002
Sethi, A., Slack, J. M., Kovaleva, E. S., Buchman, G. W., and Scharf, M. E. (2013). Lignin-associated metagene expression in a lignocellulose-digesting termite. Insect Biochem. Mol. Biol. 43, 91–101. doi:10.1016/j.ibmb.2012.10.001
Sun, J.-Z., and Scharf, M. (2010). Exploring and integrating cellulolytic systems of insects to advance biofuel technology. Insect Sci. 17, 163. doi:10.1111/j.1744-7917.2010.01348.x
Tang, J., Siegfried, B. D., and Hoagland, K. D. (1998). Glutathione-S-transferase and in vitro metabolism of atrazine in freshwater algae. Pestic. Biochem. Physiol. 59, 155–161. doi:10.1006/pest.1998.2319
Tartar, A., Wheeler, M. M., Zhou, X., Coy, M. R., Boucias, D. G., and Scharf, M. E. (2009). Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol. Biofuels 2, 25. doi:10.1186/1754-6834-2-25
Tarver, M. R., Florane, C. B., Mattison, C. P., Holloway, B. A., and Lax, A. (2012). Myosin gene expression and protein abundance in different castes of the Formosan subterranean termite (Coptotermes formosanus). Insects 3, 1190–1199. doi:10.3390/insects3041190
Teeri, T. T. (1997). Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 15, 160–167. doi:10.1016/S0167-7799(97)01032-9
Tokuda, G., Watanabe, H., Matsumoto, T., and Noda, H. (1997). Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): distribution of cellulases and properties of endo-β-1, 4-gIucanase. Zoolog. Sci. 14, 83–93. doi:10.2108/zsj.14.83
Watanabe, H., Noda, H., Tokuda, G., and Lo, N. (1998). A cellulase gene of termite origin. Nature 394, 330–331. doi:10.1038/28527
Watanabe, H., and Tokuda, G. (2010). Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632. doi:10.1146/annurev-ento-112408-085319
Weiss, A., and Leinwand, L. A. (1996). The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 12, 417–439. doi:10.1146/annurev.cellbio.12.1.417
Wheeler, M. M., Tarver, M. R., Coy, M. R., and Scharf, M. E. (2010). Characterization of four esterase genes and esterase activity from the gut of the termite Reticulitermes flavipes. Arch. Insect Biochem. Physiol. 73, 30–48. doi:10.1002/arch.20333
Yang, B., and Wyman, C. E. (2008). Pretreatment: the key to unlocking low-cost cellulosic ethanol. Biofuels Bioprod. Biorefin. 2, 26–40. doi:10.1002/bbb.49
Young, P., Richman, A., Ketchum, A., and Kiehart, D. (1993). Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 7, 29–41. doi:10.1101/gad.7.1.29
Zhang, D., Lax, A. R., Henrissat, B., Coutinho, P., Katiya, N., Nierman, W. C., et al. (2012). Carbohydrate-active enzymes revealed in Coptotermes formosanus (Isoptera: Rhinotermitidae) transcriptome. Insect Mol. Biol. 21, 235–245. doi:10.1111/j.1365-2583.2011.01130.x
Zhou, X., Kovaleva, E. S., Wu-Scharf, D., Campbell, J. H., Buchman, G. W., Boucias, D. G., et al. (2010). Production and characterization of a recombinant beta-1,4-endoglucanase (glycohydrolase family 9) from the termite Reticulitermes flavipes. Arch. Insect Biochem. Physiol. 74, 147–162. doi:10.1002/arch.20368
Keywords: Reticulitermes flavipes, second-generation feedstock, corn stover, soybean residue, cellulase, ligninase
Citation: Rajarapu SP and Scharf ME (2017) Saccharification of Agricultural Lignocellulose Feedstocks and Protein-Level Responses by a Termite Gut-Microbe Bioreactor. Front. Energy Res. 5:5. doi: 10.3389/fenrg.2017.00005
Received: 16 December 2016; Accepted: 10 March 2017;
Published: 07 April 2017
Edited by:
Jalel Labidi, University of the Basque Country, SpainReviewed by:
Bing-Zhi Li, Tianjin University, ChinaShishir P. S. Chundawat, Rutgers University, USA
Zheng-Jun Li, Beijing University of Chemical Technology, China
Copyright: © 2017 Rajarapu and Scharf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael E. Scharf, mscharf@purdue.edu
†Present address: Swapna Priya Rajarapu, Department of Entomology, Ohio State University-OARDC, Wooster, OH, USA
 Swapna Priya Rajarapu
Swapna Priya Rajarapu Michael E. Scharf
Michael E. Scharf