Tell Us a Story Granddad: Age and Origin of an Iconic Australian Lungfish
- 1Environomics Future Science Platform, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Indian Ocean Marine Research Centre, Crawley, WA, Australia
- 2Department of Regional Development, Manufacturing and Water, Bundaberg, QLD, Australia
- 3Seqwater, Ipswich, QLD, Australia
The modern discovery of the Australian lungfish (Neoceratodus forsteri) by European settlers in 1870 was considered one of the most important events in natural history by leading international scientists and naturalists of that time. Its distinct evolutionary lineage and unique extant morphological characteristics fostered the romantic zoological moniker “living fossil”. Although N. forsteri were suspected of being long-lived, a reliable estimate of maximum age has remained elusive. Maximum age is critical to inform wildlife management and conservation efforts, including the use of population viability models. To estimate the maximum age for N. forsteri, we sourced DNA from “Granddad”, the presumed longest-living lungfish known in a zoological park and utilised an epigenetic ageing clock developed for N. forsteri. This lungfish specimen was gifted to the Chicago John G. Shedd Aquarium from Australia in 1933 and lived there for 84 years until death in 2017. We estimated the age of Granddad at death to be 109 years (±6 years), confirming N. forsteri as a true centenarian species. Genotyping also revealed the natal origin of Granddad to be the Burnett River, Queensland, Australia, the location of the species’ original discovery in 1870. We demonstrate the application of novel molecular techniques to a unique long-lived and captive-raised specimen, to improve estimates of maximum age for the species, and to identify natal origin. This information will support future conservation efforts for this iconic yet endangered species.
Introduction
The Australian lungfish (Neoceratodus forsteri), known to Australia’s First Nations people by various names including Dhal’la, Djelleh or Theebine (Watson, 1933), was brought to the attention of the scientific community in 1870 as the “Burnett Salmon” (Sclater, 1870), after the river where it was first captured and its characteristic pink flesh. Leading international ichthyologists and naturalists venerated the discovery of this mysterious lungfish with its unique phylogeny that bridged the evolutionary gaps between fish, amphibians and tetrapods (Anon, 1870; Krefft, 1870). “Living fossil” was afforded to N. forsteri based on the discovery of an extant species that connected zoological orders with divergent evolutionary taxonomy (Darwin, 1859); for which fossil remains were discovered before living forms (Cavin and Guinot, 2014); and for which a general morphological stasis was observed compared to the fossil record (Lee et al., 2006). Classified in the genus Neoceratodus (meaning “new” and “horned tooth”) due to unique dentition that was well represented in fossil records dating back 140 million years (Kemp, 1997), N. forsteri sits within the air-breathing Dipnoi order, which flourished throughout the planet during the Devonian period (416–359 Mya) (Pusey et al., 2004). Today, only 6 lungfish species remain (4 African, 1 South American, and 1 Australian) (Tokita et al., 2005; Nelson et al., 2016).
International interest in N. forsteri grew rapidly after their discovery, driving demand for living specimens to be shipped internationally (Anon, 1873, Anon, 1898, Anon, 1912, Anon, 1913, Anon, 1936; O’Connor, 1934). One specimen, which became known as “Granddad” (Figure 1), made the 20-day sea voyage to the United States of America in April 1933 for the opening of Chicago’s World Fair, and remained at the John G. Shedd Aquarium for 84 years until death in 2017. Neoceratodus forsteri have long been suspected of long life span due to their slow growth rates and large maximum adult sizes attained in the wild, with individuals of 100 cm in length shown to range in age from 10 to 75 years (Kind, 2011; Fallon et al., 2019). These highly variable juvenile and adult growth rates combined with a lack of accurate and cost-effective ageing methods, have hindered reliable maximum age estimates for the species. At time of shipment, Granddad was already of adult size, being approximately 109 cm and at death measured 120 cm (Mary-Anne Rodgers Pers Comm) thus providing a rare opportunity to investigate the potential maximum age of N. forsteri.

FIGURE 1. The iconic Australian lungfish specimen resided at the John G. Shedd Aquarium for 84 years before passing, became affectionately known as “Granddad” due to his presumed old age, reported to have been seen by over 100 million visitors, is still contributing to conservation outcomes in death through the information held within his DNA (Image: © Shedd Aquarium).
The maximum age, longevity, lifespan or life expectancy (Mayne et al., 2020) of wildlife is often unknown or difficult to estimate (Baylis et al., 2014), however, it is critical to modern conservation planning and management approaches such as population viability modelling (Todd et al., 2017). Estimates of maximum age can be affected by sampling error and can vary within populations of a species due to many genomic and environmental factors (Todd, 1978; de Magalhães and Costa, 2009). Novel molecular methods have the potential to complement quantitative population modelling tools for conservation planning, including epigenetic clocks that measure levels of DNA methylation to predict age (Field et al., 2018). Epigenetic clocks work by measuring DNA methylation at cytosine-phosphate-guanine (CpG) sites (Bell et al., 2019). As a cell undergoes mitosis, DNA methylation at CpG sites is supposed to be maintained. However, cells can make mistakes and one of the hallmarks of ageing and cell senescence is a decrease in the maintenance of epigenetic modifications including DNA methylation. Several studies use this biological process to make epigenetic clocks to predict the age of a wide variety of vertebrate species (De Paoli-Iseppi et al., 2017). This is particularly important for species such as N. forsteri that cannot be aged using traditional methods (Fallon et al., 2019). The most common form of DNA methylation in vertebrates is the addition of a methyl group to CpG sites (Bird, 1993). DNA methylation has recently been used to develop a “Lungfish clock” to predict the age of N. forsteri (Mayne et al., 2021b).
In Australia, N. forsteri are found in the Burnett, Mary and Brisbane River catchments of south-eastern Queensland (Kind, 2016). Genomic studies indicate extant N. forsteri populations originated from the Burnett and Mary rivers with evidence of genetic inputs from the Burnett to the Mary River from the late Pleistocene to ∼12,000 years ago when sea levels rose (Hughes et al., 2015; Bishop et al., 2017). In 1896, N. forsteri specimens were translocated by The Royal Society of Queensland into various catchments south of their presumed native range, including the Brisbane River, in response to an apparent rarity of juveniles and a perceived high extinction risk (Illidge, 1893; O’Connor, 1896). N. forsteri currently face multiple anthropogenic threats throughout its range (Arthington, 2009), which have elevated risk of population decline and possible extinction, resulting in recent relisting as “endangered” under the IUCN Red List of Threatened Species (Brooks et al., 2019). Concomitantly, numerous studies have focused on N. forsteri, aiming to address key knowledge gaps to inform conservation efforts (Espinoza et al., 2013; Marshall et al., 2014; Roberts et al., 2014; Schmidt et al., 2018; Fallon et al., 2019; Olden et al., 2019; Mayne et al., 2021b; Meyer et al., 2021).
In this study, we aimed to build on conservation efforts by applying novel genomic methods to a unique long-lived specimen of N. forsteri to 1. Produce an updated maximum age estimate for N. forsteri, 2. Assess the maximum age of N. forsteri within the animal kingdom, and 3. Demonstrate methods that reveal the river of origin for this iconic flagship specimen. Results substantially revise the maximum age estimate for N. forsteri, contributing towards conservation and management outcomes for this iconic species in the wild.
Methods
Sample Collection and DNA Extraction
A sample of fin tissue from Granddad was preserved in 70% ethanol after euthanasia. DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN) and RNAse A treated (QIAGEN) using the manufacturer’s protocol.
Age Prediction by DNA Methylation
To estimate the age of Granddad we used a previously developed Lungfish clock (Mayne et al., 2021b). The Lungfish clock uses the DNA methylation levels of 31 cytosine-phosphate-guanine (CpG) sites to determine age. DNA (150 ng) is bisulphite treated as previously described (Clark et al., 2006). Bisulphite treated DNA is then used for multiplex PCR targeting 31 CpG sites (Supplementary Tables S1, S2). The multiplex PCR product is barcoded with Fluidigm barcodes (Supplementary Tables S3, S4) and is sequenced on an Illumina MiSeq. The fastq file was trimmed 15 bp at the 5′ and 3′ ends to remove adaptor sequences (Shen et al., 2016). Trimmed reads were aligned to the zebrafish reference genome (danRer10, Illumina iGenomes) with bismark version 0.20.0 and bowtie2 v2.3.4 with the following options: -bowtie2-N1-L15–bam-p2–score L, −0.6, −0.6 –non_directional (Krueger and Andrews, 2011; Langmead and Salzberg, 2012). DNA methylation levels were called using the bismark_methylation_extractor function with default parameters. The Lungfish clock was developed and validated using a data set of 141 known age Australian lungfish samples ranging from 1 to 77 years old. The method to determine the age of the Australian lungfish is provided in Supplementary Table S5. The revised maximum lifespan estimate for N. forsteri were compared to longevity records maintained in the Animal Ageing and Longevity Database (de Magalhães and Costa, 2009).
River Origin Genotyping
Restriction site-associated DNA sequencing (RAD-seq) data from 92 wild-caught Australian lungfish from a previous research project was used to identify the minimum number of single nucleotide polymorphisms (SNPs) for determining the origin of an individual within the species’ distribution (BioProject Accession# PRJNA477902) (Schmidt et al., 2018). This data set included samples from three populations; the Brisbane River (n = 30); Burnett River (n = 32); and the Mary River (n = 30). Paired-end reads were quality trimmed using trimmomatic v 0.38 with the following options: - PE phred33 ILLUMINACLIP:TruSeq3-PE:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36 (Bolger et al., 2014). Trimmed reads were aligned to the Australian lungfish reference genome (neoFor_v3) with bowtie2 v2.3.4 default settings (Langmead and Salzberg, 2012; Meyer et al., 2021). SNP calling was carried out using the Genome Analysis Toolkit (GATK) v4.1.9.0 best practices pipeline as it has been shown to perform better at differentiating populations compared to other software (McKenna et al., 2010; Van der Auwera et al., 2013; Wright et al., 2019).
SNP Markers for Population Differentiation
Although the three populations can be differentiated by 15,201 loci as previously described, it is not cost-effective to run RAD-seq on one individual (Schmidt et al., 2018). Here, the challenge is to identify the minimum number of SNPs to identify the natal origin of Granddad. We used a combination of genome-wide association study (GWAS) and machine learning as previously described to identify the minimum SNPs for population differentiation (Seo et al., 2021).
PLINK v1.9 was used to perform association tests between populations (Purcell et al., 2007). SNPs with Bonferroni correction p-value < 0.05 were considered significant. The significant SNPs were used as part of a Random Forest model to identify the minimum number of SNPs for population differentiation (Kuhn, 2008a). The caret R package was used to carry out the Random Forest modelling with a 10-fold cross validation (Kuhn, 2008b). 70% of the samples were randomly assigned to a training data set, with the remaining assigned to an independent testing data set. The performance of the model was assessed with an area under curve (AUC). A principal component analysis (PCA) was also used to assess the differentiation of the populations by the SNPs returned from the Random Forest modelling. All analyses were carried out using R version 4.0.4 (R Core Team, 2014).
Targeted SNP Genotyping
The SNPs returned from the Random Forest modelling were collectively used to design an assay for the MassARRAY System (Ellis and Ong, 2017). The Agena Bioscience MassARRAY System enables genotyping of multiple targeted SNPs on small scale experiments. The MassARRAY genotyping was carried out at the Australian Genome Research Facility (AGRF). Multiplex PCR conditions were carried out as previously described (Gabriel et al., 2009).
Results
Granddad Age at Death
In total 32,328 reads were aligned to a partitioned genome with an alignment success rate of 97.5%. The level of methylation at each CpG site is given in Supplementary Table S5. Using the model previously developed for Neoceratodus forsteri, the age at death of Granddad was found to be 109 years (103–115 years, within a 95% confidence interval).
The revised maximum age estimate for N. forsteri places the species as the 21st oldest living animal, out of 3,761 longevity entries in the AnAge database (de Magalhães and Costa, 2009). Of the 952 fish (aquatic vertebrates with gills) records in the database, N. forsteri ranks as the 12th oldest living fish species in the world (Figure 2). This “centenarian guild” includes well-known marine species such as the Greenland shark (Somniosus microcephalus) which is the current record holder at 392 years; several Rock fish species (Sebastes sp) ranging in maximum age between 116 and 205 years; and the Orange Roughy (Hoplostethus atlanticus) at 149 years. Out of the 11 centenarian fish older than N. forsteri, all except one, the Lake Sturgeon (Acipenser fulvescens), inhabit marine waters, or utilise marine habitats for part of their life cycle. At 152 years maximum age, A. fulvescens is the only other obligate freshwater fish within the centenarian guild, however, N. forsteri now holds the record for the longest living freshwater sub-tropical fish species in the world.
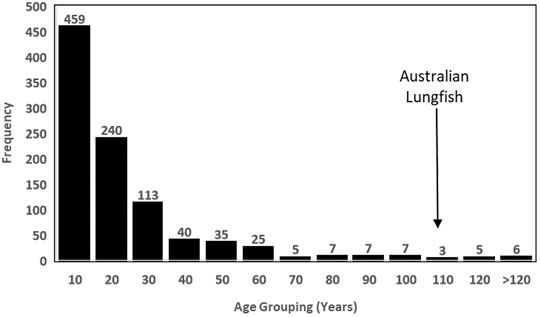
FIGURE 2. Longevity frequency distribution of all 952 fish species with longevity estimates provided in the Animal Ageing and Longevity database [AnAge (Tacutu et al., 2018)] demonstrating the position of N. forsteri as a rare centenarian, and within the top 12 longest living fish, and the second oldest freshwater fish, in the world.
Granddad River of Origin
On average per sample, 1,994,292 reads aligned to the Australian lungfish reference genome with an alignment rate of 96.1%. The GATK returned a total of 48,386 SNPs which were quality checked and passed the default parameters in PLINK. In total, 2,610 SNPs were found to be associated with either one of the three extant river populations (Supplementary Figure S1). These associated SNPs were added to the random forest model to rank SNPs in terms of importance for river identification. The random forest model resulted in 35 SNPs that could be used to differentiate populations (AUC = 1, Supplementary Figure S2, and Supplementary Table S6). The performance in both the training and testing data set of the model was 100%. The random forest model predicted Granddad as originating from the Burnett River (Figure 3). This is also demonstrated with an unsupervised PCA with the 35 SNPs (Figure 3). This analysis highlights three distinctive extant river populations of the Australian lungfish with 35 SNPs.
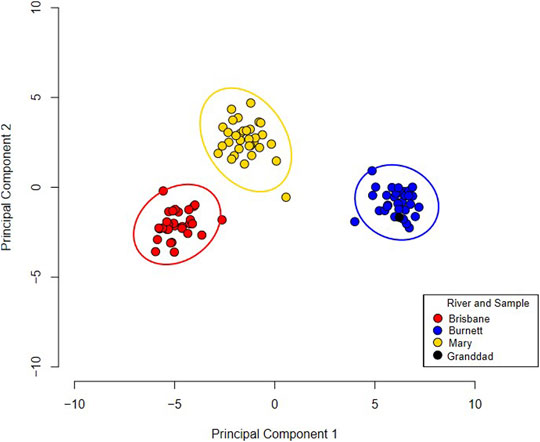
FIGURE 3. A principal component analysis of three river populations of Australian lungfish using a 35 single nucleotide polymorphism profile. Granddad is coloured black in the plot and is clustering with the Burnett River samples. Ellipses around each river population represent a 95% confidence interval.
Discussion
Australian lungfish (Neoceratodus forsteri) are considered the oldest surviving species from an ancient lineage of air-breathing, lobe-finned fish but face many modern anthropogenic threats which have led to contemporary population declines and increased extinction risk. Addressing key biological knowledge gaps to inform quantitative ecological risk assessment is critical to conservation efforts. Here we apply a novel epigenetic clock to determine the age of Granddad as 109 (±6) years and identify its natal origin as the Burnett River, Australia. At time of death, Granddad was likely the longest-lived N. forsteri known in a zoological park and confirms the species as a true centenarian. These findings provide a revised maximum age to aid in the conservation management of this endangered species. We also show that genotyping can effectively determine the natal origin of lungfish specimens, demonstrating a useful tool for forensic studies in the potential illegal trade of a CITES (Convention on International Trade in Endangered Species) species.
Recent population viability analyses (PVA) developed by the authors (unpublished data) to inform management of wild N. forsteri populations, used a previous maximum age estimate of 77 years (Fallon et al., 2019). Our findings extend the maximum age estimate for N. forsteri by 30% over previous estimates, to 109 (±6) years of age. This increase can affect many vital rates within quantitative population modelling tools, such as mortality probabilities, net reproduction, and fecundity potential, by extending the population age at first maturity through to reproductive senescence. N. forsteri can take at least 15 years to mature (Brooks and Kind, 2002), therefore extending the maximum age estimate from 77 to 109 years effectively adds two additional generations into a population model. Maximum age, however, can be artificially augmented in captive settings through improved husbandry, medical treatment and attenuation of key biological processes such as predation and competition (Mason, 2010). In addition, species maximum age estimates derived from single records also come with caution (Baylis et al., 2014; de Magalhães and Costa, 2009). Although captive animals such as Granddad can provide invaluable insight into species biology and behaviour, maximum age estimates for Australian lungfish, as with any long lived species, should utilise a broad sample of both captive and wild individuals to inform population modelling and other wildlife assessments (de Magalhães et al., 2007; Koons et al., 2006).
Epigenetic age clocks are also limited by the range of ages used in calibration, with decreasing confidence in estimates for animals outside the calibrated age range (Mayne et al., 2021a). The Lungfish clock used here included “known-age” and “radiocarbon-aged” N. forsteri up to 77 years (Mayne et al., 2021b). However, epigenetic clocks still offer a valuable tool for predicting ages beyond the calibrated range, albeit with lower certainty. Epigenetic clocks can also be improved as more samples within target age ranges become available and are included into the calibration data. Improving epigenetic clocks for older age classes is an important scientific activity that zoos and aquariums can support, by establishing an age for living specimens in exhibits that are within the calibrated age range and providing ongoing known-age samples beyond the calibrated range of existing clocks. This is particularly important for N. forsteri that exhibit highly variable growth rates in the wild and have populations heavily skewed towards adult fish (Fallon et al., 2019). In addition, the Lungfish clock provides a cost-effective ageing technique for effective management of wild populations through monitoring of sporadic recruitment events (Espinoza et al., 2013).
Our findings have broader implications beyond new scientific knowledge, by enhancing social context for promoting conservation and awareness of N. forsteri. The benefits of human connection for compelling action to conserve a species cannot be underestimated. Studies show that zoo and aquarium exhibits can improve conservation ethics of visitors which can be further enhanced with specific information about animals that appeal to people such as animal age, longevity, place of origin or extinction status (Moss and Esson, 2010; Fraser et al., 2020). Non-human charisma is generally afforded to more conspicuous megafauna such as mammals (Lorimer, 2007; Small, 2012; Root-Bernstein et al., 2013), however, certain flagship species, like N. forsteri (Ebner et al., 2016), are also powerful attractions for capturing the public interest. Granddad, was arguably the world’s first iconic freshwater flagship species connecting to over 100 million visitors at Chicago’s John G. Shedd Aquarium and rightfully held the mantle for the oldest living fish in a zoological park (Johnson, 2017). This study identifies the important contribution that public aquariums and zoos can make to both scientific research and community conservation outcomes, while providing an improved educational experience for visitors.
With freshwater biodiversity in decline and recognition that threats are inadequately addressed by policy and management actions (Dudgeon et al., 2006), global biodiversity advocates have called for specific actions to support freshwater biodiversity conservation, including the need to identify and promote freshwater flagship species, such as N. forsteri, in public education programs (Ebner et al., 2016; Tickner et al., 2020; Rees et al., 2021). Granddad the lungfish was a pivotal ambassador for N. forsteri, a species under threat of extinction. Given the centenarian status of N. forsteri, living specimens on exhibit in zoological parks could continue roles as ambassadors for global freshwater biodiversity conservation. Other living specimens of N. forsteri, such as “Methuselah” at the California Academy of Sciences’ Steinhart Aquarium in San Francisco, could also exceed the current record set by Granddad and contribute to science and education.
In life, the N. forsteri known as Granddad was seen by over 100 million visitors at the John G. Shedd Aquarium in Chicago (Johnson, 2017). Even after death, Granddad’s DNA has contributed to conservation efforts and raising public awareness by revealing new biological insights into the species. When alive, Granddad secured his place as the oldest freshwater fish in a zoological park and possibly the world, and propelled N. forsteri into the rare class of true centenarians within the animal kingdom. In reference to a famous quote by the late biologist Dr Stephen Wainwright, “Nature reveals great secrets in her extreme forms”, Granddad represents one of nature’s most extreme living forms, and our application of new non-lethal molecular-based techniques has revealed some of N. forsteri’s greatest secrets including maximum age and geographic origin of Granddad. These findings will not only facilitate research, management actions and future policy to underpin conservation of wild and captive-raised lungfish, but also inform global conservation of freshwater biodiversity efforts.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA477902.
Author Contributions
BM performed the epigenetic age, river origin prediction, and reviewed the manuscript. TE and DR prepared the manuscript and sourced biological samples and funding for the genotyping of Granddad.
Funding
The authors would also like to thank the Burnett Mary Regional Group (BMRG) for funding the genotyping of Granddad. BM is supported by the CSIRO Environomics Future Science Platform and the North West Shelf Flatback Turtle Conservation Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank all authors and contributors to the Australian lungfish RAD-seq data (BioProject Accession# PRJNA477902) which was used in this study. The authors acknowledge the contribution of staff at the John G. Shedd Aquarium for kindly donating the DNA material to enable this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.931467/full#supplementary-material
References
Arthington, A. H. (2009). Australian Lungfish, Neoceratodus forsteri, Threatened by a New Dam. Environ. Biol. Fish. 84 (2), 211–221. doi:10.1007/s10641-008-9414-y
Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy‐Moonshine, A., et al. (2013). From FastQ Data to High‐Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinforma. 43 (1110). doi:10.1002/0471250953.BI1110S43
Baylis, S. M., de Lisle, M., and Hauber, M. E. (2014). Inferring Maximum Lifespan from Maximum Recorded Longevity in the Wild Carries Substantial Risk of Estimation Bias. Ecography 37 (8), 770–780. doi:10.1111/ECOG.00507
Bell, C. G., Lowe, R., Adams, P. D., Baccarelli, A. A., Beck, S., Bell, J. T., et al. (2019). DNA Methylation Aging Clocks: Challenges and Recommendations. Genome Biol. 20 (1). doi:10.1186/S13059-019-1824-Y
Bird, A. P. (1993). Functions for DNA Methylation in Vertebrates. Cold Spring Harb. Symposia Quantitative Biol. 58, 281–285. doi:10.1101/SQB.1993.058.01.033
Bishop, C. R., Hughes, J. M., and Schmidt, D. J. (2017). Mitogenomic Analysis of the Australian Lungfish (Neoceratodus forsteri) Reveals Structuring of Indigenous Riverine Populations and Late Pleistocene Movement between Drainage Basins. Conserv. Genet. 19, 587–597. doi:10.1007/s10592-017-1034-7
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a Flexible Trimmer for Illumina Sequence Data. Bioinforma. Oxf. Engl. 30 (15), 2114–2120. doi:10.1093/BIOINFORMATICS/BTU170
Brooks, S., Espinoza, T., Mark, J. K., Arthington, A. H., and Roberts, D. T. (2019). Neoceratodus forsteri. The IUCN Red List Of Threatened Species 2019. E.122899816A123382021.
Brooks, S. G., and Kind, P. K. (2002). Ecology and Demography of the Queensland Lungfish (Neoceratodus Fosteri) in the Burnett River, Queensland with Reference to the Impacts of Walla Weir and Future Water Infrastructure Development : Final Report. Brisbane: Dept. of Primary Industries, Queensland Agency for Food and Fibre Sciences.
Cavin, L., and Guinot, G. (2014). Coelacanths as €œalmost Living Fossilsâ€. Front. Ecol. Evol. 2 (AUG), 49. doi:10.3389/FEVO.2014.00049/ABSTRACT
Clark, S. J., Statham, A., Stirzaker, C., Molloy, P. L., and Frommer, M. (2006). DNA Methylation: Bisulphite Modification and Analysis. Nat. Protoc. 1 (5), 2353–2364. doi:10.1038/NPROT.2006.324
de Magalhães, J. P., and Costa, J. (2009). A Database of Vertebrate Longevity Records and Their Relation to Other Life‐history Traits. J. Evol. Biol. 22 (8), 1770–1774. doi:10.1111/J.1420-9101.2009.01783.X
de Magalhães, J. P. d., Costa, J., and Church, G. M. (2007). An Analysis of the Relationship between Metabolism, Developmental Schedules, and Longevity Using Phylogenetic Independent Contrasts. Journals Gerontology Ser. A 62 (2), 149–160. doi:10.1093/GERONA/62.2.149
De Paoli-Iseppi, R., Deagle, B. E., McMahon, C. R., Hindell, M. A., Dickinson, J. L., and Jarman, S. N. (2017). Measuring Animal Age with DNA Methylation: From Humans to Wild Animals. Front. Genet. 8 (AUG), 106. doi:10.3389/FGENE.2017.00106
Dudgeon, D., Arthington, A. H., Gessner, M. O., Kawabata, Z., Knowler, D. J., Lévêque, C., et al. (2006). Freshwater Biodiversity: Importance, Threats, Status and Conservation Challenges. Biol. Rev. Camb Philos. Soc. 81 (2), 163–182. doi:10.1017/S1464793105006950
Ebner, B. C., Morgan, D. L., Kerezsy, A., Hardie, S., Beatty, S. J., Seymour, J. E., et al. (2016). Enhancing Conservation of Australian Freshwater Ecosystems: Identification of Freshwater Flagship Fishes and Relevant Target Audiences. Fish. Fish. 17 (4), 1134–1151. doi:10.1111/faf.12161
Ellis, J. A., and Ong, B. (2017). The MassARRAY System for Targeted SNP Genotyping. Methods Mol. Biol. Clift. N.J.), 77–94. doi:10.1007/978-1-4939-6442-0_5
Espinoza, T., Marshall, S. M., and McDougall, A. J. (2013). Spawning of the Endangered Australian Lungfish (Neoceratodus Forsteri ) in A Heavily Regulated River: A Pulse for Life. River Res. applic. 29, 1215–1225. doi:10.1002/rra.2607
Fallon, S. J., McDougall, A. J., Espinoza, T., Roberts, D. T., Brooks, S., Kind, P. K., et al. (2019). Age Structure of the Australian Lungfish (Neoceratodus forsteri). PLoS ONE 14 (1), e0210168. doi:10.1371/journal.pone.0210168
Field, A. E., Robertson, N. A., Wang, T., Havas, A., Ideker, T., and Adams, P. D. (2018). DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Mol. Cell. 71 (6), 882–895. doi:10.1016/J.MOLCEL.2018.08.008
Fraser, J., Bicknell, J., Sickler, J., and Taylor, A. (2020). What Information Do Zoo & Aquarium Visitors Want on Animal Identification Labels? J. Interpretation Res. 14 (2), 7–18. doi:10.1177/109258720901400202
Gabriel, S., Ziaugra, L., and Tabbaa, D. (2009). SNP Genotyping Using the Sequenom MassARRAY iPLEX Platform. Curr. Protoc. Hum. Genet. 60 (Suppl. L), 60. doi:10.1002/0471142905.HG0212S60
Hughes, J. M., Schmidt, D. J., Huey, J. A., Real, K. M., Espinoza, T., McDougall, A., et al. (2015). Extremely Low Microsatellite Diversity but Distinct Population Structure in a Long-Lived Threatened Species, the Australian Lungfish Neoceratodus forsteri (Dipnoi). PLoS ONE 10 (4), e0121858–14. doi:10.1371/journal.pone.0121858
Johnson, S. (2017). Australian Lungfish “Granddad,” the Oldest Zoo Animal in Chicago, Dies - Chicago Tribune. Chicago Tribune. Available at: https://www.chicagotribune.com/entertainment/museums/ct-granddad-fish-shedd-acquarium-dead-20170206-column.html.
Kemp, A. (1997). A Revision of Australian Mesozoic and Cenozoic Lungfish of the Family Neoceratodontidae (Osteichthyes:Dipnoi), with a Description of Four New Species. J. Paleontol. 71 (4), 713–733. doi:10.1017/s0022336000040166
Kind, P. (2010). “The Natural History of the Australian Lungfish neoceratodus Forsteri (Krefft, 1870),” in The Biology of Lungfishes, 61–95. doi:10.1201/b10357-4
Kind, P. (2011). The Natural History of the Australian Lungfish Neoceratodus Forsteri (Krefft, 1870). Biol. Lungfishes, 61–95. doi:10.1201/b10357-4
Koons, D. N., Rockwell, R. F., and Grand, J. B. (2006). Population Momentum: Implications for Wildlife Management. J. Wildl. Manag. 70 (1), 19–26. doi:10.2193/0022-541X10.2193/0022-541x(2006)70[19:pmifwm]2.0.co;2
Krefft, G. (1870). Ceratodus Forsteri - to the Editor of the Herald. Sydney: The Sydney Morning Herald. doi:10.1038/003107d0
Krueger, F., and Andrews, S. R. (2011). Bismark: a Flexible Aligner and Methylation Caller for Bisulfite-Seq Applications. Bioinforma. Oxf. Engl. 27 (11), 1571–1572. doi:10.1093/BIOINFORMATICS/BTR167
Kuhn, M. (2008b). Building Predictive Models inRUsing thecaretPackage. J. Stat. Soft. 28 (5), 1–26. doi:10.18637/JSS.V028.I05
Kuhn, M. (2008a). Journal of Statistical Software Building Predictive Models in R Using the Caret Package. Available at: http://www.jstatsoft.org/.
Langmead, B., and Salzberg, S. L. (2012). Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/NMETH.1923
Lee, J., Alrubaian, J., and Dores, R. M. (2006). Are Lungfish Living Fossils? Observation on the Evolution of the Opioid/orphanin Gene Family. General Comp. Endocrinol. 148 (3), 306–314. doi:10.1016/J.YGCEN.2006.07.010
Marshall, S. M., Espinoza, T., and Mcdougall, A. J. (2014). Effects of Water Level Fluctuations on Spawning Habitat of an Endangered Species, the Australian Lungfish (neoceratodus Forsteri). River Res. Appl. Available at: http://www.scopus.com/inward/record.url?eid=2-s2.0-84899043079&partnerID=40&md5=22a22e6621fa92ad29e871a02710b242.
Mason, G. J. (2010). Species Differences in Responses to Captivity: Stress, Welfare and the Comparative Method. Trends Ecol. Evol. 25 (12), 713–721. doi:10.1016/J.TREE.2010.08.011
Mayne, B., Berry, O., and Jarman, S. (2021). Optimal Sample Size for Calibrating DNA Methylation Age Estimators. Mol. Ecol. Resour. 21 (7), 2316–2323. doi:10.1111/1755-0998.13437
Mayne, B., Berry, O., and Jarman, S. (2020). Redefining Life Expectancy and Maximum Lifespan for Wildlife Management. Austral Ecol. 45 (7), 855–857. doi:10.1111/AEC.12931
Mayne, B., Espinoza, T., Roberts, D., Butler, G. L., Brooks, S., Korbie, D., et al. (2021). Nonlethal Age Estimation of Three Threatened Fish Species Using DNA Methylation: Australian Lungfish, Murray Cod and Mary River Cod. Mol. Ecol. Resour. 21, 2324–2332. doi:10.1111/1755-0998.13440
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 20 (9), 1297–1303. doi:10.1101/GR.107524.110
Meyer, A., Schloissnig, S., Franchini, P., Du, K., Woltering, J. M., Irisarri, I., et al. (2021). Giant Lungfish Genome Elucidates the Conquest of Land by Vertebrates. Nature 590, 284–289. doi:10.1038/s41586-021-03198-8
Moss, A., and Esson, M. (2010). Visitor Interest in Zoo Animals and the Implications for Collection Planning and Zoo Education Programmes. Zoo. Biol. 29 (6), 715–731. doi:10.1002/ZOO.20316
O’Connor, D. (1896). Report on Preservation of Ceratodus. Brisbane: Proceedings of the Royal Society of Queensland, 101–102.
Olden, J. D., Fallon, S. J., Roberts, D. T., Espinoza, T., and Kennard, M. J. (2019). Looking to the Past to Ensure the Future of the World's Oldest Living Vertebrate: Isotopic Evidence for Multi‐decadal Shifts in Trophic Ecology of the Australian Lungfish. River Res. Applic 35 (10), 1629–1639. doi:10.1002/rra.3369
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 81 (3), 559–575. doi:10.1086/519795
Pusey, B. J., Kennard, M., and Arthington, A. (2004). Freshwater Fishes of North-eastern Australia. Collingwood: CSIRO Publishing.
Roberts, D. T., Mallett, S., Krück, N. C., Loh, W., and Tibbetts, I. (2014). Spawning Activity of the Australian lungfishNeoceratodus Forsteriin an Impoundment. J. Fish. Biol. 84, 163–177. doi:10.1111/jfb.12264
Root-Bernstein, M., Douglas, L., Smith, A., and Veríssimo, D. (2013). Anthropomorphized Species as Tools for Conservation: Utility beyond Prosocial, Intelligent and Suffering Species. Biodivers. Conserv. 22, 1577–1589. doi:10.1007/s10531-013-0494-4
Schmidt, D. J., Fallon, S., Roberts, D. T., Espinoza, T., McDougall, A., Brooks, S. G., et al. (2018). Monitoring Age‐related Trends in Genomic Diversity of Australian Lungfish. Mol. Ecol. 27 (16), 3231–3241. doi:10.1111/mec.14791
Seo, D., Cho, S., Manjula, P., Choi, N., Kim, Y.-K., Koh, Y. J., et al. (2021). Identification of Target Chicken Populations by Machine Learning Models Using the Minimum Number of SNPs. Animals 11 (1), 241–259. doi:10.3390/ANI11010241
Shen, W., Le, S., Li, Y., and Hu, F. (2016). SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PloS One 11 (10), e0163962. doi:10.1371/JOURNAL.PONE.0163962
Small, E. (2012). The New Noah's Ark: Beautiful and Useful Species Only. Part 1. Biodiversity Conservation Issues and Priorities. Biodiversity 12 (4), 232–247. doi:10.1080/14888386.2011.642663
Tacutu, R., Thornton, D., Johnson, E., Budovsky, A., Barardo, D., Craig, T., et al. (2018). Human Ageing Genomic Resources: New and Updated Databases. Nucleic Acids Res. 46 (D1), D1083–D1090. doi:10.1093/NAR/GKX1042
Tickner, D., Opperman, J. J., Abell, R., Acreman, M., Arthington, A. H., Bunn, S. E., et al. (2020). Bending the Curve of Global Freshwater Biodiversity Loss: An Emergency Recovery Plan. BioScience 70 (4), 330–342. doi:10.1093/biosci/biaa002
Todd, C. R., Koehn, J. D., Pearce, L., Dodd, L., Humphries, P., and Morrongiello, J. R. (2017). Forgotten Fishes: What Is the Future for Small Threatened Freshwater Fish? Population Risk Assessment for Southern Pygmy Perch, Nannoperca Australis. Aquat. Conserv. Mar. Freshw. Ecosyst. 27 (6), 1290–1300. doi:10.1002/AQC.2808
Todd, H. J. (1978). A Look at the Basic Correlates of Longevity: Is Longevity a Superficial Trait? Mech. Ageing Dev. 7 (C), 33–52. doi:10.1016/0047-6374(78)90051-9
Tokita, M., Okamoto, T., and Hikida, T. (2005). Evolutionary History of African Lungfish: a Hypothesis from Molecular Phylogeny. Mol. Phylogenetics Evol. 35 (1), 281–286. doi:10.1016/J.YMPEV.2004.11.025
van Rees, C. B., Waylen, K. A., Schmidt‐Kloiber, A., Thackeray, S. J., Kalinkat, G., Martens, K., et al. (2021). Safeguarding Freshwater Life beyond 2020: Recommendations for the New Global Biodiversity Framework from the European Experience. Conserv. Lett. 14 (1), e12771. doi:10.1111/CONL.12771
Watson, F. J. (1933). “Vocabularies of Four Representative Tribes of South Eastern Queensland with Grammatical Notes Thereof and Some Notes on Manners and Customs,” in Also, a List of Aboriginal Names and Their Derivations (Queensland, Brisbane: Royal Geographical Society of Australasia).
Wright, B., Farquharson, K. A., McLennan, E. A., Belov, K., Hogg, C. J., and Grueber, C. E. (20192019). From Reference Genomes to Population Genomics: Comparing Three Reference-Aligned Reduced-Representation Sequencing Pipelines in Two Wildlife Species. BMC Genomics 20 (11), 1–10. doi:10.1186/S12864-019-5806-Y
Keywords: maximum age, long-lived, epigenetics, natal origin, lifespan, longevity, centenarian
Citation: Mayne B, Espinoza T and Roberts D (2022) Tell Us a Story Granddad: Age and Origin of an Iconic Australian Lungfish. Front. Environ. Sci. 10:931467. doi: 10.3389/fenvs.2022.931467
Received: 29 April 2022; Accepted: 25 May 2022;
Published: 21 June 2022.
Edited by:
Benjamin Cook, FRC Environmental, AustraliaReviewed by:
Peter John Unmack, University of Canberra, AustraliaLucian Pârvulescu, West University of Timișoara, Romania
Copyright © 2022 Mayne, Espinoza and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Roberts, david.t.roberts@seqwater.com.au
 Benjamin Mayne
Benjamin Mayne Tom Espinoza
Tom Espinoza David Roberts
David Roberts