Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera
- 1Institut National de la Recherche Agronomique, Institute of Ecology and Environmental Sciences (iEES-Paris), Versailles, France
- 2Sorbonne Universités, Université Pierre et Marie Curie - Université Paris 6, iEES-Paris, Paris, France
- 3College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 4Genetics and Biology of Cancers Unit, Institut National de la Santé et de la Recherche Médicale U830, Institut Curie, PSL Research University, Paris, France
In moths, mate finding strongly rely on the detection of sex pheromones by pheromone receptors (PRs). Any modification in the functional properties of these receptors can have a drastic impact on reproduction. In the course of characterizing candidate PRs in the noctuid moth Spodoptera littoralis, we expressed them in Drosophila olfactory sensory neurons and stimulated them with a large panel of moth pheromone compounds. We found that two PRs detect (Z,E)-9,12-14:OAc, a minor component of the female pheromone blend. Whereas SlitOR6 is highly specific to this component, SlitOR13 is less sensitive and not strictly specific as it also detects (Z)9-14:OAc, another minor component of the sex pheromone. Interestingly, SlitOR13 expression is restricted to the distal part of male antennae, where we could identify a novel functional class of pheromone-sensitive neurons whose response spectrum matches that of SlitOR13. Based on a phylogenetic analysis of Lepidoptera PRs, we found that the ability to bind (Z,E)-9,12-14:OAc appeared independently within three paralogous lineages, and an analysis of selective pressures revealed sites under positive selection that could have played a role in the emergence of functional properties of OR6 and OR13 in Spodoptera species.
Introduction
In animals, various biological mechanisms prevent species from interbreeding with each other. One of the well-studied mechanisms of premating isolation is the sex pheromone communication system of moths (Cardé and Haynes, 2004). In those nocturnal insects, reproductive success largely depends on the long-distance detection of bouquets of air-borne chemicals usually emitted by the females. Most sex pheromones consist of a complex blend of a major component mixed with a few minor components, whose nature and precise relative ratios ensure a species-specific recognition (de Bruyne and Baker, 2008). In the male, antennae bear thousands of sensilla housing olfactory sensory neurons (OSNs) that detect the different components of the pheromone blend with various sensitivities and specificities (Kaissling, 1996). At the cellular level, this detection is mediated by transmembrane receptors belonging to the olfactory receptor (OR) family, and called pheromone receptors (PRs). Moth PR-encoding genes form a monophyletic group in the OR phylogeny (de Fouchier et al., 2014), suggesting that they all evolved from a single common ancestor. Numerous gene duplication events led to the variable number of putative PRs (usually from 4 to 9) found in the different species investigated (Krieger et al., 2004; Nakagawa et al., 2005; Wanner et al., 2010; Grosse-Wilde et al., 2011; Bengtsson et al., 2012; Zhang and Lofstedt, 2013; Steinwender et al., 2015).
The functional evolution of PRs is a key issue, because even slight modifications of their receptive range are expected to give rise to new reproductive barriers between sympatric populations, then to the emergence of new species. Since the discovery of the receptor for bombykol in the silk moth Bombyx mori (Sakurai et al., 2004), ligands of PRs have been identified in more than a dozen of species (for a review, see de Fouchier et al., 2014). These functional data highlight a rapid functional divergence among moth PRs resulting, at least in part, from increased evolutionary rates following gene duplications (Zhang and Lofstedt, 2013; Engsontia et al., 2014). At an extreme, some receptors belonging to the PR sub-family do not bind sex pheromone components but rather plant volatiles (Jordan et al., 2009; Bengtsson et al., 2014). Whereas functional properties are generally more conserved between orthologous genes from closely related species, only a few amino acid changes can modify these properties, sometimes drastically (Leary et al., 2012; Jiang et al., 2014; Steinwender et al., 2015).
Moths from the genus Spodoptera (Lepidoptera, Noctuidae) form a group of economically important crop pests. Several species are sympatric and synthesize partially overlapping pheromone blends, which make them an ideal model to study the links between the evolution of PRs and premating isolation. One of the most studied Spodoptera species is the cotton leafworm S. littoralis. It is a serious pest of more than 80 agricultural productions such as cotton, maize, rice, sorghum, alfalfa, soybean and vegetables, all over Africa, the Mediterranean Basin and the Middle East (Salama et al., 1971). Its sex pheromone blend, variable according to the strains and areas, is composed of up to eleven 14-carbon acetates, among which (Z,E)-9,11-14:OAc is always the major component (Muñoz et al., 2008; Saveer et al., 2014). Electrophysiology experiments on male antennae identified one OSN population specifically tuned to this component (Ljungberg et al., 1993; Quero et al., 1996) and housed in one class of long trichoid sensilla, further referred as LT1 sensilla. Another functional class of sensilla, here referred as LT2, house two OSNs, one of which tuned to the minor component (Z,E)-9,12-14:OAc (Ljungberg et al., 1993). As far as we know, no OSN detecting other S. littoralis pheromone components could be identified.
We previously identified four candidate PRs (named SlitOR6, 11, 13, and 16) in a S. littoralis male transcriptome, based on their clustering with other moth PRs in a phylogenetic analysis (Legeai et al., 2011). Among them, SlitOR6 has been characterized as a receptor for the minor component (Z,E)-9,12-14:OAc (Montagné et al., 2012). Through homology-cloning, the orthologs of these four PRs have been identified in S. litura (found throughout Asia and Oceania) and S. exigua (distributed worldwide) and functional data are available for some of them (Liu et al., 2013a; Zhang et al., 2015).
Here, we carried out a functional analysis of the four S. littoralis PR candidates, using a large panel of pheromone compounds, to investigate in depth the response specificity. We used in vivo heterologous expression in Drosophila OSNs housed in trichoid sensilla, an expression system that has been demonstrated to be suitable for studying moth PRs (Kurtovic et al., 2007; Syed et al., 2010; Montagné et al., 2012; Bengtsson et al., 2014). We identified two receptors to minor components with overlapping response spectra, both responding to (Z,E)-9,12-14:OAc, and localized the corresponding OSNs on male moth antenna. Evolutionary analyses revealed that the ability to bind (Z,E)-9,12-14:OAc emerged independently within three distinct paralogous lineages, two of which containing Spodoptera spp. sequences, and that positive selection acted on a few amino acid sites, which could have played a role in the evolution of PR response spectra in Spodoptera species.
Materials and Methods
Insect Rearing and Chemicals
Flies were reared on standard cornmeal-yeast-agar medium and kept in a climate- and light-controlled environment (25°C, 12 h light: 12 h dark cycle). S. littoralis were reared in the laboratory on a semi-artificial diet (Poitout and Buès, 1974) at 22°C, 60% relative humidity and under a 16 h light: 8 h dark cycle. Males and females were sexed as pupae and further reared separately. The pheromone compounds used in this study (see Supplementary Table S1) were either synthesized in the lab or purchased from Sigma-Aldrich (St Louis, MO, USA) and Pherobank (Wijk bij Duurstede, The Netherlands). Hexane was purchased from Carlo Erba Reagents (Val de Reuil, France).
Heterologous Expression of Slitors in Drosophila
The generation of transgenic flies expressing SlitOR6 has been described previously (Montagné et al., 2012). The same strategy was used for expression of SlitOR11, 13 and 16. Briefly, full-length open reading frames were first cloned into pCR®II-TOPO® (Invitrogen, Carlsbad, CA, USA) and subcloned into the pUAST vector. Plasmid constructs were purified from liquid cultures of One Shot® TOP10 E. coli using the EndoFree Plasmid Maxi kit (Qiagen, Venlo, Netherlands). Transformant UAS-SlitOr balanced fly lines were generated by BestGene Inc. (Chino Hills, California, USA), by injecting the pUAST-SlitOR plasmids into w1118 fly embryos. Only fly lines harboring a transgene insertion into the 2nd chromosome were used for further crossings.
UAS-SlitOr balanced lines were crossed to a line harboring a knock-in of the Gal4 ORF into the Or67d gene (Kurtovic et al., 2007), to obtain double homozygous flies (genotype w; UAS-SlitOr,w+; Or67dGal4) expressing a given SlitOR in at1 OSNs instead of the endogenous Drosophila receptor OR67d. The presence of the UAS-SlitOr transgenes was verified by PCR on genomic DNA extracted from two flies, and the correct expression of the SlitORs was verified by RT-PCR on total RNA extracted from ≥100 pairs of antennae.
Single-sensillum Recordings on Drosophila Antennae
Single-sensillum recordings were performed on at1 sensilla of transformed flies expressing a given SlitOR. For each experiment, a 2- to 6-day-old male fly was restrained in a pipette tip with only the head protruding. The tip was fixed on a microscope glass slide and one antenna was gently maintained using a glass capillary. The preparation was placed under a constant 1.5 L.min−1 flux of charcoal-filtered and humidified air delivered through a glass tube of a 7 mm diameter, and observed with a light microscope (BX51WI, Olympus, Tokyo, Japan) equipped with a 100 × magnification objective.
The SlitOR response spectra were established using a panel of 26 pheromone compounds (Supplementary Table S1). Stimulus cartridges were built by placing a 1 cm2 filter paper in the large opening end of a Pasteur pipette and dropping 10 μl of the pheromone solution onto the paper (1 μg/μl in hexane), or 10 μL of hexane as control. The cartridges were used at most one time on each fly and five times in total. Odorant stimulations were performed by inserting the tip of the pipette into a hole in the glass tube and generating a 500 ms air pulse (0.6 L.min−1), which reached the permanent air flux while going through the stimulation cartridge. Action potentials were recorded from at1 sensilla using electrolytically sharpened tungsten electrodes (TW5-6, Science Products, Hofheim, Germany). The reference electrode was inserted into the eye and the recording electrode was inserted at the base of the sensillum using a motor-controlled PatchStar micromanipulator (Scientifica, Uckfield, United Kingdom). The electrical signal was amplified using an EX-1 amplifier (Dagan Corporation, Minneapolis, MN, USA), high-pass (1 Hz) and low-pass (3 kHz) filtered and digitized (10 kHz) through a Digidata 1440A acquisition board (Molecular Devices, Sunnyvale, CA, USA) then recorded and analyzed using the pCLAMP™ 10 software (Molecular Devices). The responses of at1 OSNs were calculated by subtracting the spontaneous firing rate (in spikes.s−1) from the firing rate during the odorant stimulation. The time windows used to measure these two firing rates lasted for 500 ms and were respectively placed 500 ms before and 100 ms after the onset of stimulation (to take into account the time for the odorants to reach the antenna). The entire odorant panel was tested at least 5 times on each SlitOR. Odorants were considered as active if the response they elicited was statistically different from the response elicited by the solvent alone (P < 0.001; Mann–Whitney pairwise test). For all active odorants, dose-response experiments were conducted with quantities ranging from 10 μg down to the dose necessary to reach the response threshold. Each dilution of each pheromone was tested five times.
Single-sensillum Recordings on Spodoptera Antennae
Recordings of S. littoralis long trichoid sensilla were performed on 1- to 3-day-old male moths. Animals were restrained in a Styrofoam block and the antenna was visualized under a MZ16 stereomicroscope (Leica, Wetzlar, Germany). A humidified and charcoal-filtered airflow (1.2 L.min−1) was continuously directed to the preparation through a glass tube (7 mm of diameter). The stimulus panel consisted in the same 26 pheromone compounds as for recordings on Drosophila antennae (Supplementary Table S1). Stimulus cartridges were prepared as described above and were loaded with 1 μg of pheromone. During the stimulation, the tip of the Pasteur pipette was introduced through a lateral hole within the tube carrying the permanent humidified air flow onto the antennae at 15 cm from its outlet. The air pulse (0.2 L.min−1) lasted for 200 ms. Tungsten electrodes were prepared as described above and the recording electrode was inserted at the base of the sensillum of interest using a PatchStar micromanipulator (Scientifica). The biological signal was amplified (× 2000), high-pass (1 Hz) and low-pass (3 kHz) filtered using a CyberAmp 320 (Molecular Devices) and sampled at 10 kHz via a Digidata 1440A acquisition board (Molecular Devices). Recordings and analyses were performed with pCLAMP™ 10 (Molecular Devices). The responses were calculated as described above, using time windows of 200 ms. The stimulus panel was tested 3 times on each of the two functional classes of long trichoid sensilla investigated. For the cross adaptation experiment, sensilla were first stimulated during 5 s with either (Z)9-12:OAc, (Z)9-14:OAc or (Z,E)-9,12-14:OAc, then stimulated during 200 ms with the three compounds presented successively in a random order.
Quantitative Real-time PCR
Antennae from 2-day-old males were collected and cut into three pieces: a proximal part, a middle part and a distal part. Proximal parts from 60 individuals were pooled, as well as distal parts. These collections were repeated three times (biological replicates). Total RNA was extracted from each sample using RNeasy MicroKit (Qiagen), which included a DNase treatment. cDNA was synthesized using the Advantage® RT-for-PCR Kit (Clontech, Mountain View, CA, USA). Gene-specific primers for SlitOR6, SlitOR13 and the reference gene SlitORco have been previously described (Legeai et al., 2011). We used ORco, expected to be expressed equally in all OSNs (Larsson et al., 2004), as the reference gene because the number of sensilla (and thus the number of OSNs) clearly differs between the proximal (higher number of sensilla) and the distal parts (lower number of sensilla) of the antennae. Thus, normalization with a housekeeping gene would have biased relative expression calculation. qPCR mix was prepared in a total volume of 12 μL with 6 μL of iQ™ SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 3 μL of diluted cDNA (or water for the negative control or RNA for controlling the absence of genomic DNA) and 200 nM of each primer. qPCR assays were performed using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). The PCR program began with a cycle at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C, 30 s at 60°C. To assess the specificity of the PCR reactions, a dissociation curve of the amplified products was performed by gradual heating from 65°C to 95°C at 0.5°C.s−1. Standard curves were generated by a five-fold dilution series of a cDNA pool evaluating primer efficiency (E = 10(−1/slope)). For each case, the presence of only one amplified product was verified. All reactions were performed in duplicate for the three biological replicates. Expression levels between proximal and distal parts of the antennae were calculated relatively to the expression of the reference gene using the formula ratio = [(Etarget)ΔCptarget(control−sample)]/[(Eref)ΔCpref(control−sample)] (Pfaffl, 2001).
Phylogenetic Analysis
An amino acid sequence dataset was created, including every full-length candidate PR sequence identified in Lepidoptera (see Supplementary Table S2). Bombyx mori OR6 and Ctenopseustis obliquana OR22 were also included to serve as an external group. The 149 amino acid sequences were aligned using the online version of MAFFT v.7 (Katoh and Standley, 2013), with the G-INS-i algorithm (Katoh et al., 2005) and default parameters. Phylogenetic reconstruction was performed using the maximum likelihood method. The JTT+I+G+F substitution model (Jones et al., 1992), was determined as the best-fit model of protein evolution by ProtTest 2.4 (Abascal et al., 2005). Tree reconstruction was performed using PhyML 3.0 (Guindon et al., 2010), with both SPR (Subtree Pruning and Regrafting) and NNI (Nearest Neighbor Interchange) methods for topology improvement. Rate heterogeneity was set at four categories, and values calculated by ProtTest were used for the gamma distribution parameter and the proportion of invariable sites. Node support was estimated using a hierarchical likelihood-ratio test (Anisimova and Gascuel, 2006). The figure was created using the iTOL web server (Letunic and Bork, 2011).
Analysis of Selective Pressures
To study positive selection along the PR alignment, we first manually removed the sites which were poorly aligned or conserved. Next, we replaced the amino acids by their codons, and created a DNA alignment file that corresponded to the trimmed protein alignment. From that DNA alignment, we constructed a phylogenetic tree using PhyML 3.0 with the GTR substitution model, 4 substitution rate categories and an estimated gamma shape parameter. Both SPR and NNI-types tree modifications were allowed. Branch support values were generated using the Shimodaira-Hasegawa test (Shimodaira, 2002) as implemented in PhyML. We then used this tree and both protein and DNA alignments of OR sequences to perform branch-site tests of positive selection, which have proven to be more sensitive than branch-based tests, in regions where alignments are reliable (Yang and dos Reis, 2011). To do so we used the software fastcodeml (Valle et al., 2014), a recent program designed to reduce the calculation time of branch-site models developed in the codeml suite of programs (formerly defined in codeml using the parameters model = 2 and NSsites = 2). We split the full tree up into 6 independent clades having large support values (>0.95) and we performed the selection tests on each one of them. We made full use of fastcodeml parallelized algorithms by specifying computations with 8 threads on a computer cluster (parameter –nt 8). For each of the clades we reported the branches that have at least one site under positive selection and noted the associated probability that a diversifying selection occurred at this site along the branch under consideration.
Results
Response Spectra of Heterologously Expressed SlitORs
In order to systematically analyze the response spectra of SlitOR6, 11, 13 and 16 to a large panel of moth pheromone compounds, we expressed these receptors in Drosophila OSNs housed in at1 trichoid sensilla, in place of the endogenous PR DmelOR67d. RT-PCR experiments confirmed the correct expression of the SlitORs, except SlitOR16 (data not shown) that has not been studied further. Using the single-sensillum recording technique, we monitored the response of OSNs expressing SlitOR6, 11 and 13 to high doses of 26 pheromone compounds (Supplementary Table S1). This panel included all the compounds identified in the pheromone blend of S. littoralis and/or active on S. littoralis antennae (Ljungberg et al., 1993; Muñoz et al., 2008; Saveer et al., 2014), as well as closely related chemicals and ligands of previously characterized noctuid PRs (Wang et al., 2011). While SlitOR11-expressing OSNs did not display any response to the panel (data not shown), SlitOR6 and SlitOR13-expressing OSNs gave significant responses to two and three compounds, respectively (Figure 1A). As observed previously (Montagné et al., 2012), SlitOR6 responded strongly to (Z,E)-9,12-14:OAc (133 spikes.s−1, P < 0.001, Mann–Whitney). Here, we also observed a smaller response to (Z)9-12:OAc (35 spikes.s−1, P < 0.01). SlitOR13 displayed similar responses to (Z,E)-9,12-14:OAc and (Z)9-14:OAc (76 and 77 spikes.s−1, P < 0.001), as well as a smaller response to (Z)9-12:OAc (52 spikes.s−1, P < 0.001).
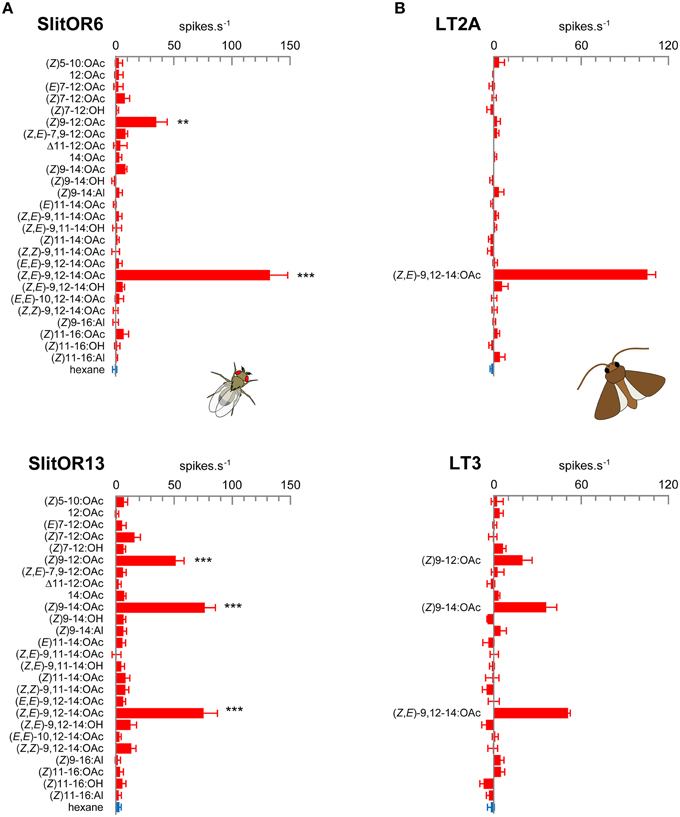
Figure 1. (A) Response spectra of SlitOR6 and SlitOR13 expressed in Drosophila at1 OSNs. (B) Response spectra of LT2A and LT3 OSNs from S. littoralis male antennae. Values correspond to the increase in the frequency of action potentials emitted by the OSN during the odorant stimulation. The stimulus panel consisted in 26 pheromone compounds (10 μg in the stimulus cartridge for single-sensillum recordings on Drosophila, 1 μg for recordings on Spodoptera). Error bars indicate SEM (n = 5–9 for SlitOR6, n = 7–12 for SlitOR13, and n = 3 for S. littoralis OSNs). ***P < 0.001, **P < 0.01 significantly different from the response to solvent (Mann-Whitney U-test).
To further analyze both the sensitivity and the selectivity of SlitOR6 and 13, we performed dose-response experiments with the ligands identified among the panel (Figure 2). The response of SlitOR6 to (Z)9-12:OAc was abolished at a dose of 1 μg loaded in the stimulus cartridge, while it still responded to (Z,E)-9,12-14:OAc at 0.1 μg (P < 0.05). SlitOR13 responded to (Z)9-14:OAc and (Z,E)-9,12-14:OAc down to 1 μg, with similar intensities. We thus identified two PRs detecting the same minor component of the S. littoralis pheromonal bouquet, albeit with different sensitivity and specificity.
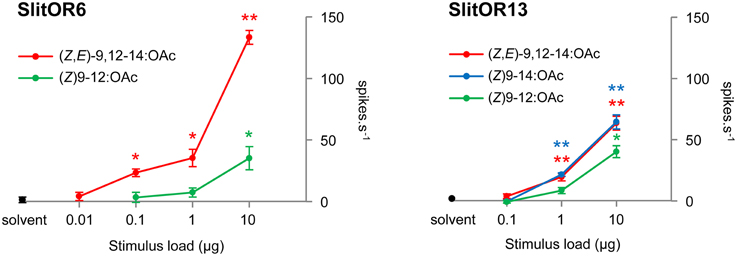
Figure 2. Dose-response experiments on SlitOR6 and SlitOR13 expressed in Drosophila at1 OSNs. Responses to previously identified ligands have been tested at doses from 10 μg down to 0.1 or 0.01 μg in the stimulus cartridge. Error bars indicate SEM (n = 5). **P < 0.001, *P < 0.01 significantly different from the response to solvent (Mann-Whitney U-test).
Identification of Two Different S. littoralis OSN Types Tuned to (Z,E)-9,12-14:OAc
Former studies on S. littoralis (Ljungberg et al., 1993; Quero et al., 1996) identified one pheromone-sensitive OSN type tuned to (Z,E)-9,12-14:OAc in type 2 long trichoid sensilla (here referred as the LT2A OSN type). The response spectrum of this OSN type matches that of SlitOR6, but no OSN detecting both (Z,E)-9,12-14:OAc and (Z)9-14:OAc, which would correspond to SlitOR13 response profile, was found. Considering that those electrophysiology studies focused on sensilla located on the proximal part of the antennae, where trichoid sensilla are abundant and easy to record, we investigated whether OSNs with a response profile matching that of SlitOR13 could be found on more distal parts of the antennae. We thus performed single-sensillum recordings all along the antennae, using the panel of 26 pheromone compounds previously used for the functional characterization of SlitORs. We found the previously described trichoid sensillum types LT1 and LT2 in both the proximal and distal part of the antennae, and LT2A OSNs displayed responses only to (Z,E)-9,12-14:OAc (106 spikes.s−1), thus confirming their specificity toward this compound (Figure 1B). Besides that, we identified a new functional type of long trichoid sensilla on the distal part of the antennae only, further referred as LT3. A cross-adaptation experiment determined that these sensilla housed only one OSN (data not shown). LT3 OSNs responded to (Z,E)-9,12-14:OAc (51 spikes.s−1), (Z)9-14:OAc (36 spikes.s−1) and to a lesser extent to (Z)9-12:OAc (20 spikes.s−1). The low number of recordings (LT3 were scarce and only three could be registered) did not allow statistical analysis. The LT2A and LT3 OSN detection spectra thus clearly matched those of Drosophila OSNs expressing SlitOR6 and SlitOR3, respectively (Figures 1A,B).
Expression Profiles of SlitOR6 and SlitOR13
We next verified whether expression profiles of SlitOR6 and 13 along the antennae would correlate with the localization of LT2 and LT3 sensilla. We used quantitative real-time PCR to compare the relative expression levels of the two PRs in the proximal and distal parts of male antennae (Figure 3). We found that SlitOR6 was expressed in both parts, with a two-fold enrichment in the proximal part. By contrast, SlitOR13 expression was found only in the distal part of the antennae, where LT3 were localized.
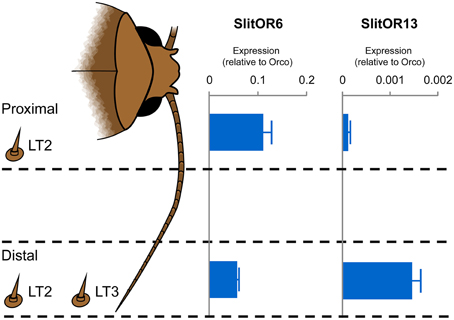
Figure 3. Relative expression levels of SlitOR6 and SlitOR13 in the proximal and the distal parts of the S. littoralis male antennae. Expression levels have been normalized to the expression of the obligate olfactory co-receptor ORco. The occurrence of the LT2 and LT3 trichoid sensilla in the proximal and distal parts is also indicated. Error bars indicate standard deviation.
Phylogenetic Analysis of Pheromone Receptors
In order to gather information about the evolutionary history of SlitOR6 and 13, we built a maximum-likelihood phylogeny of Lepidoptera candidate PRs, including 147 amino acid sequences from 37 species of moths and butterflies (Supplementary Table S2). These receptors grouped within five different paralogous lineages supported by the likelihood-ratio test, each lineage containing sequences from a various number of Lepidoptera super-families (Figure 4 and Supplementary Figure S1). In the Noctuoidea super-family, numerous gene duplications occurred in the lineage E and account for the large number of PRs observed in these species (Figure 4 and Supplementary Figure S1).
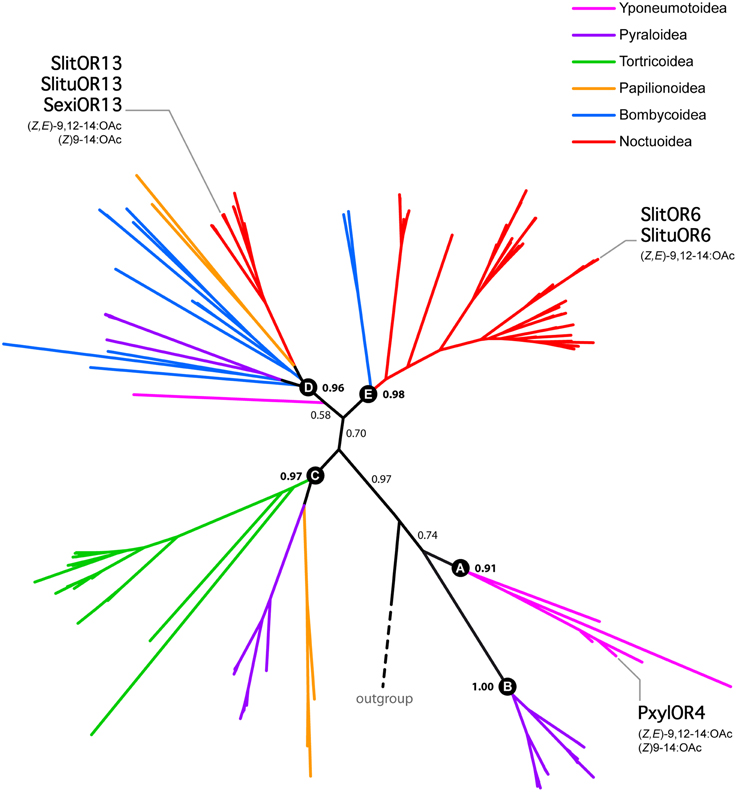
Figure 4. Unrooted maximum likelihood tree of Lepidoptera candidate PRs, highlighting the phylogenetic position of receptors tuned to (Z,E)-9,12-14:OAc. Branches are color-coded following the Lepidoptera super-families. Letters from A to E correspond to different paralogous lineages. Support values correspond to the result of the likelihood-ratio test.
The two S. littoralis receptors under scrutiny here, SlitOR6 and 13, belong to the paralogous lineages D and E, respectively. Apart from closely related orthologs in S. exigua and S. litura, no other PRs with similar functional properties have been found in these lineages. However, the receptor PxylOR4 from the diamondback moth Plutella xylostella (sub-family Yponeumotoidea) also binds (Z,E)-9,12-14:OAc, and belongs to the B lineage. This indicates that the ability to bind (Z,E)-9,12-14:OAc emerged three times independently, in three distinct paralogous lineages.
Amino Acid Sites under Positive Selection
Next, we searched for regions in Spodoptera spp. OR6 and OR13 that could be responsible for the functional divergence of these receptors. We carried out a large-scale analysis of selective pressures acting on Lepidoptera PR genes using fastcodeml (Valle et al., 2014), a software implementation of branch-site probabilistic models of evolution that were introduced in (Yang and Nielsen, 2002). We found evidence for positive selection in numerous branches of the PR phylogeny, notably in the one leading to the pair of orthologs SlitOR6/SlituOR6, which both detect (Z,E)-9,12-14:OAc (Figure 5A), and in the one leading to Spodoptera OR13 orthologs, detecting (Z,E)-9,12-14:OAc and (Z)9-14:OAc (Figure 5C). We then mapped the candidate sites under positive selection in those two branches, as inferred by the Bayes empirical Bayes approach, on the predicted topology of Spodoptera OR6 (Figure 5B) and OR13 (Figure 5D). For OR6, the four candidate sites were located within transmembrane domains (TM4 and TM5) as well as intra-cellular loops (IL2 and IL3). For OR13, six candidate sites were found, although with moderate support, in the intracellular N-terminus and in the IL1, EL1, TM4, and TM6 domains.
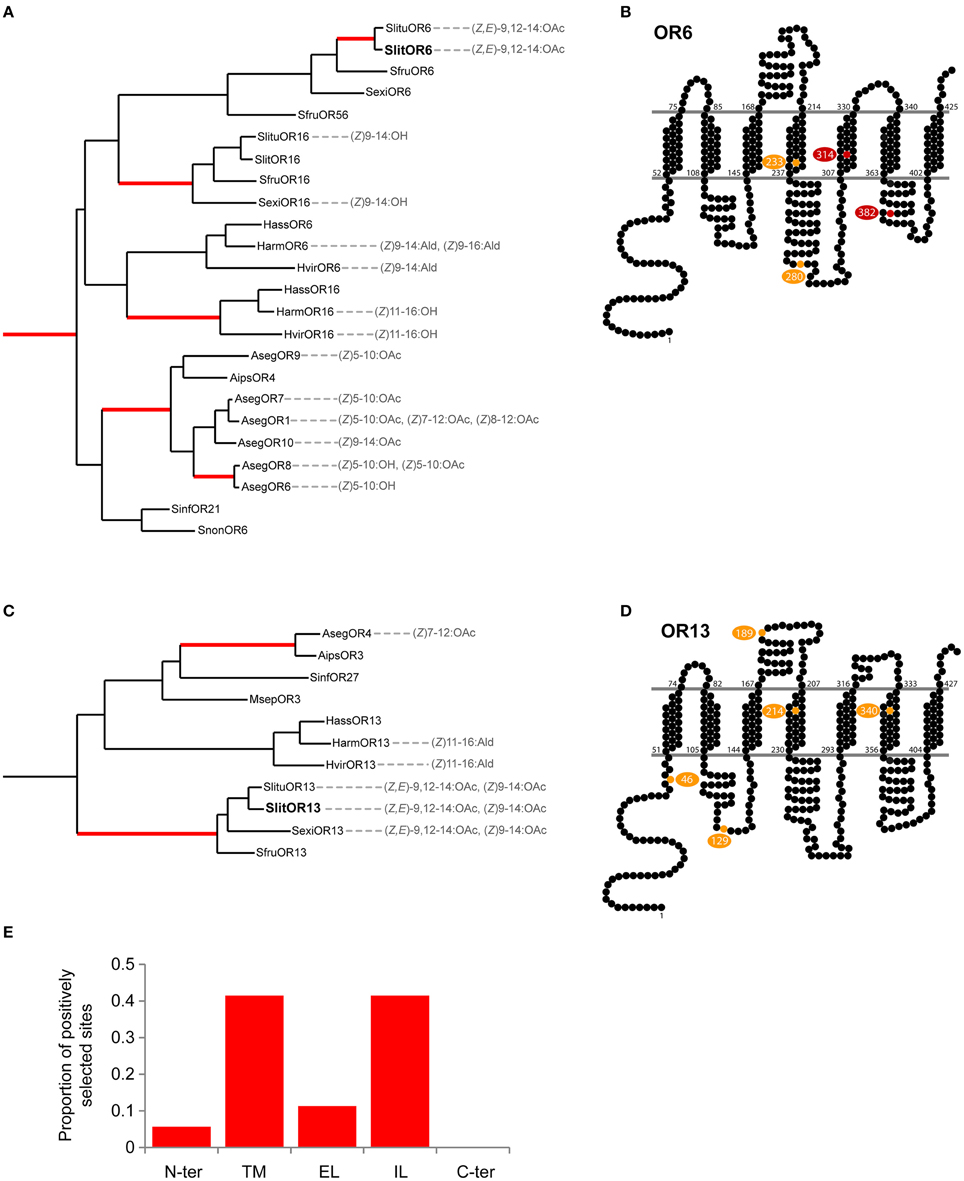
Figure 5. Analysis of positive selection acting on PR genes. (A,C) Detail of clades containing Spodoptera OR6 and OR13 orthologs, showing branches (in red) in which there is evidence of positive selection. Ligands indicated are the best ligands found during the functional characterization of PRs (Wang et al., 2011; Liu et al., 2013a,b; Zhang and Lofstedt, 2013; Zhang et al., 2015). (B,D) Putative positively selected sites in Spodoptera OR6 and OR13 orthologs mapped onto the predicted topologies (yellow, posterior probability > 0.5; red, PP > 0.9). (E) Distribution of the candidate positively selected sites found in Lepidoptera PRs by type of protein domains: N-ter, N-terminus; TM, Trans-membrane domains; EL, Extracellular loops; IL, Intracellular loops; and C-ter, C-terminus.
We then compared the localization of candidate positively selected sites in OR6 and OR13 with those found in other branches of the PR phylogeny. More than 80% of these sites were found in transmembrane domains, notably TM4, and intra-cellular loops, notably IL1 and IL2 (Figure 5E). This is fully consistent with what we have found for OR6, whereas our analysis for OR13 highlighted sites that fell outside of these common regions of positive selection.
Discussion
Moth PRs are often cited as striking examples of the sensitivity and specificity that can be ensured by insect ORs. However, most studies used a small panel of pheromone compounds for stimulation, usually restricted to those found in the species blend (Sakurai et al., 2004; Forstner et al., 2009) and at best in closely relatives (Mitsuno et al., 2008; Liu et al., 2013a,b; Zhang et al., 2015). Here, we have used a large panel of 26 compounds to functionally characterize four candidate PRs in S. littoralis. While SlitOR16 could not be expressed in Drosophila OSNs, thus precluding its functional characterization, its orthologs in S. litura and S. exigua bind preferentially (Z)9-14:OH (Liu et al., 2013a; Zhang et al., 2015). We can reasonably expect that SlitOR16 shares the same response spectrum. SlitOR11 could be expressed in Drosophila but remained silent, even after stimulation with such a large panel. Neither could OR11 orthologs be deorphanized in previous studies carried out in other noctuid species, including S. exigua and S. litura (Wang et al., 2011; Liu et al., 2013a,b; Jiang et al., 2014; Zhang et al., 2015). In D. melanogaster, it has been observed that OR67d-expressing neurons need the pheromone-binding protein (PBP) LUSH to detect its pheromone ligand (Ha and Smith, 2006). It could be hypothesized that SlitOR11 and its orthologs need a specific PBP to detect pheromone compounds. Another possible explanation is that OR11 is in fact not a PR, although it groups within the candidate PR clade in the Lepidoptera OR phylogeny (de Fouchier et al., 2014). In line with this hypothesis, it has been demonstrated that this PR clade actually includes receptors to plant odorants, such as the pear ester receptor of Cydia pomonella (Bengtsson et al., 2014).
The ligands we identified for SlitOR6 and SlitOR13 are the same as those previously identified for orthologous receptors in the closely related species, S. exigua and S. litura, although S. exigua OR6 could not be deorphanized (Liu et al., 2013a; Zhang et al., 2015). In addition, the larger panel used here (only 5 and 7 compounds were tested on S. exigua and S. litura ORs, respectively), confirms the narrow tuning of these receptors. Both SlitOR6 and SlitOR13 responded to the same minor component of the S. littoralis pheromone blend, (Z,E)-9,12-14:OAc, but with striking differences. Whereas SlitOR6 was highly specific and very sensitive, SlitOR13 detected (Z,E)-9,12-14:OAc, yet with less sensitivity than SlitOR6, and (Z)9-14:OAc, another minor component.
Using a combination of single-sensillum recordings along S. littoralis antennae and quantitative PCR, we could correlate the response spectra and the spatial distribution of SlitOR6 and OR13 with that of LT2A and LT3 OSNs, the latter type being described here for the first time. The difference observed between the responses of SlitOR6-expressing OSNs and LT2A OSNs to (Z)9-12:OAc could arise from the differences in the stimulus load (10 and 1 μg, respectively). Accordingly, SlitOR6 was not activated by lower amounts of (Z)9-12:OAc in dose-response experiments. We thus propose that SlitOR6 is expressed in the LT2A OSNs and that SlitOR13 is expressed in the newly described LT3 OSNs. The identification of a new type of pheromone-sensitive sensilla in S. littoralis, even after decades of electrophysiological studies (Muñoz et al., 2008; Binyameen et al., 2012), emphasizes the benefits of studying in detail both response spectra and expression patterns of ORs. Albeit difficult to perform, precise single-sensillum recordings at the distal part of the antennae deserve future attention and may lead to the identification of new functional types of sensilla. Whereas sucrose-detecting gustatory sensory neurons located in chaetic sensilla have been found to be especially abundant at the tip of Helicoverpa armigera antennae (Jørgensen et al., 2007), we report here for the first time such a specific localization of an OSN type. Whether such an atypical spatial repartition is important for moth orientation remains to be determined. In H. armigera, the relative abundance of sugar-detecting neurons in the distal region of the antenna has been linked with an ecological need to probe the nectar of the flowers (Jørgensen et al., 2007). In female S. littoralis, it has been hypothesized that the lowered response sensitivity of distal OSNs to several, but not all, odorants could participate in close range orientation to high concentration odor flux (Binyameen et al., 2012). Similarly, one can speculate that the distal localization of SlitOR13-expressing OSNs may play a role in a shorter range assessment of the pheromone blend composition, during the mating process.
The functional redundancy of PRs tuned to minor components could provide a mean for the precise coding of ratios within the pheromone blend. In S. littoralis, (Z,E)-9,12-14:OAc is present at a very low amount in the blend, and is even lacking in some strains (Muñoz et al., 2008). However, when added at a 1% ratio to the major pheromone component (Z,E)-9,11-14:OAc, it has a synergistic effect on the attraction of males, and increasing the ratio of (Z,E)-9,12-14:OAc in the blend drastically reduces male capture in pheromone traps (Kehat et al., 1976; Campion et al., 1980). Minute amounts of (Z,E)-9,12-14:OAc may enhance the attraction of males toward (Z,E)-9,11-14:OAc through the activation of the LT2A neurons (expressing the very sensitive SlitOR6). With increasing doses of (Z,E)-9,12-14:OAc, the LT3 OSN population (expressing the less sensitive SlitOR13) may also be recruited and would inhibit the attractive behavior triggered by (Z,E)-9,11-14:OAc. The antagonistic effect of high doses of (Z)9-14:OAc, the other ligand of SlitOR13, observed in field trapping studies (Campion et al., 1980) lends further support to this model. In nature, this precise detection of high doses of (Z,E)-9,12-14:OAc by S. littoralis males may constitute a mechanism to avoid interbreeding with the sympatric species S. exigua, whose sex pheromone contains high amounts of (Z,E)-9,12-14:OAc (Acin et al., 2010).
Focusing on OR13, it is worth noticing that this receptor exhibits different sensitivities toward its two ligands in the three Spodoptera species. Whereas SexiOR13 is more sensitive to (Z,E)-9,12-14:OAc than to (Z)9-14:OAc (Liu et al., 2013a), the opposite is observed for SlituOR13 (Zhang et al., 2015) and here SlitOR13 had the same sensitivity for both pheromone components. Such differences may be relevant for an efficient coding of ratios in the different Spodoptera species.
Even though the four candidate PRs have been studied in three different Spodoptera species (Liu et al., 2013a; Zhang et al., 2015; this study), no receptor has been identified for (Z,E)-9,11-14:OAc, the major pheromone component in S. littoralis and S. litura (Muñoz et al., 2008; Saveer et al., 2014). In a similar way, only two minor components of the S. littoralis pheromone blend, (Z)9-14:OAc and (Z,E)-9,12-14:OAc, could be assigned to a receptor, leaving all the other minor components orphans. This suggests that more PRs await further identification and/or that PBPs could modify Spodoptera PR response spectra as observed in P. xylostella (Sun et al., 2013).
As revealed by our phylogenetic analysis, the ability to bind (Z,E)-9,12-14:OAc appeared independently in three paralogous lineages. This prompted us to investigate whether similar evolutionary mechanisms occurred in each case. We detected episodes of positive selection in numerous branches of the PR phylogeny, and the putative positively selected sites were located mainly in TM and IL domains, which is consistent with studies on other insects. In ORs of the pea aphid, most positively selected sites have been found in TM domains (Smadja et al., 2009) and in Drosophila, such sites have been identified mostly in the IL domains (Guo and Kim, 2007). We found sites under positive selection in Spodoptera OR6 and OR13 orthologs, but no shared sites. This suggests that the capacity to bind (Z,E)-9,12-14:OAc emerged either due to positive selection acting on different amino acids in the two lineages or due to genetic drift rather than positive selection. As in the majority of other PRs, candidate positively selected sites in OR6 (and to a lesser extent in OR13) were located in TM and IL domains. What could be the functional significance of modifications occurring in these domains? Molecular modeling of the interactions between Drosophila ORs and their cognate ligands led to the hypothesis that the ligand-binding pocket is located on the extra-cellular sides of TM domains (Guo and Kim, 2010). There are experimental evidences that mutations in TM2, 3 and 4 domains affect ligand binding in Drosophila, mosquito or moth receptors (Nichols and Luetje, 2010; Pellegrino et al., 2011; Leary et al., 2012; Hughes et al., 2014) whereas mutations in TM5, 6 and 7 affect the ion channel function of a moth PR (Nakagawa et al., 2012). An in silico analysis of insect OR structural features also identified N-ter, EL2 and IL3 domains as the most evolutionary constrained (Hopf et al., 2015), which is consistent with their importance for the correct functioning of ORs (Benton et al., 2006; Jin et al., 2008; Xu and Leal, 2013). Further experimental validations are needed to confirm whether positive selection has indeed been a driving force for the diversification of moth PRs or not. This will shed light on the evolutionary mechanisms at the base of the evolution of pheromone communication and reproductive isolation in this major insect group.
Conflict of Interest Statement
The intellectual property rights of SlitOR6 have been licensed by INRA for the purposes of developing novel insect control agents. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr Barry Dickson for providing the Or67dGAL4 mutant knock-in fly line and Marie-Christine François for insect rearing and technical help. This work has been funded by INRA, Université Pierre et Marie Curie (UPMC) and an INRA doctoral fellowship to AdF. XS received a grant from the China Scholarship Council (CSC).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00095
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Acin, P., Rosell, G., Guerrero, A., and Quero, C. (2010). Sex pheromone of the spanish population of the beet armyworm Spodoptera exigua. J. Chem. Ecol. 36, 778–786. doi: 10.1007/s10886-010-9817-z
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagné, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bengtsson, J. M., Trona, F., Montagné, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 7:e31620. doi: 10.1371/journal.pone.0031620
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Binyameen, M., Anderson, P., Ignell, R., Seada, M. A., Hansson, B. S., and Schlyter, F. (2012). Spatial organization of antennal olfactory sensory neurons in the female Spodoptera littoralis moth: differences in sensitivity and temporal characteristics. Chem. Senses 37, 613–629. doi: 10.1093/chemse/bjs043
Campion, D. G., Hunter-Jones, P., Mcveigh, L. J., Hall, D. R., Lester, R., and Nesbitt, B. F. (1980). Modification of the attractiveness of the primary pheromone component of the Egyptian cotton leafworm, Spodoptera littoralis (Boisduval) (Lepidoptera: Noctuidae), by secondary pheromone components and related chemicals. Bull. Entomol. Res. 70, 417. doi: 10.1017/s0007485300007689
Cardé, R. T., and Haynes, K. F. (2004). “Structure of the pheromone communication channel in moths,” in Advances in Insect Chemical Ecology, eds R. T. Cardé and J. G. Millar (Cambridge: Cambridge University Press), 283–331.
de Bruyne, M., and Baker, T. C. (2008). Odor detection in insects: volatile codes. J. Chem. Ecol. 34, 882–897. doi: 10.1007/s10886-008-9485-4
de Fouchier, A., Montagné, N., Mirabeau, O., and Jacquin-Joly, E. (2014). “Current views on the function and evolution of olfactory receptors in Lepidoptera,” in Short Views on Insect Biochemistry and Molecular Biology, eds R. Chandrasekar, B. K. Tyagi, Z. Z. Gui, and G. R. Reeck (Manhattan, KS: International Book Mission), 385–408.
Engsontia, P., Sangket, U., Chotigeat, W., and Satasook, C. (2014). Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J. Mol. Evol. 79, 21–39. doi: 10.1007/s00239-014-9633-0
Forstner, M., Breer, H., and Krieger, J. (2009). A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 5, 745–757. doi: 10.7150/ijbs.5.745
Grosse-Wilde, E., Kuebler, L. S., Bucks, S., Vogel, H., Wicher, D., and Hansson, B. S. (2011). Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 108, 7449–7454. doi: 10.1073/pnas.1017963108
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, S., and Kim, J. (2007). Molecular evolution of Drosophila odorant receptor genes. Mol. Biol. Evol. 24, 1198–1207. doi: 10.1093/molbev/msm038
Guo, S., and Kim, J. (2010). Dissecting the molecular mechanism of drosophila odorant receptors through activity modeling and comparative analysis. Proteins 78, 381–399. doi: 10.1002/prot.22556
Ha, T. S., and Smith, D. P. (2006). A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 26, 8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006
Hopf, T. A., Morinaga, S., Ihara, S., Touhara, K., Marks, D. S., and Benton, R. (2015). Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat. Commun. 6, 6077. doi: 10.1038/ncomms7077
Hughes, D. T., Wang, G., Zwiebel, L. J., and Luetje, C. W. (2014). A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem. Senses 39, 761–769. doi: 10.1093/chemse/bju048
Jiang, X. J., Guo, H., Di, C., Yu, S., Zhu, L., Huang, L. Q., et al. (2014). Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect. Biochem. Mol. Biol. 48, 63–74. doi: 10.1016/j.ibmb.2014.02.010
Jin, X., Ha, T. S., and Smith, D. P. (2008). SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 10996–11001. doi: 10.1073/pnas.0803309105
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282.
Jordan, M. D., Anderson, A., Begum, D., Carraher, C., Authier, A., Marshall, S. D., et al. (2009). Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394. doi: 10.1093/chemse/bjp010
Jorgensen, K., Almaas, T. J., Marion-Poll, F., and Mustaparta, H. (2007). Electrophysiological characterization of responses from gustatory receptor neurons of sensilla chaetica in the moth Heliothis virescens. Chem. Senses 32, 863–879. doi: 10.1093/chemse/bjm057
Kaissling, K. E. (1996). Peripheral mechanisms of pheromone reception in moths. Chem. Senses 21, 257–268.
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kehat, M., Greenberg, S., and Tamaki, Y. (1976). Field evaluation of the synthetic sex pheromone, as an attractant for males of the cotton leafworm, Spodoptera littoralis (Boisd.), in Israel. Appl. Entomol. Zool. 11, 45–52. doi: 10.1303/aez.11.45
Krieger, J., Grosse-Wilde, E., Gohl, T., Dewer, Y. M., Raming, K., and Breer, H. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U.S.A. 101, 11845–11850. doi: 10.1073/pnas.0403052101
Kurtovic, A., Widmer, A., and Dickson, B. J. (2007). A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546. doi: 10.1038/nature05672
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E. Jr., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Legeai, F., Malpel, S., Montagné, N., Monsempes, C., Cousserans, F., Merlin, C., et al. (2011). An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12:86. doi: 10.1186/1471-2164-12-86
Letunic, I., and Bork, P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013a). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hubner). Insect. Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibmb.2013.05.009
Liu, Y., Liu, C., Lin, K., and Wang, G. (2013b). Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS ONE 8:e62094. doi: 10.1371/journal.pone.0062094
Ljungberg, H., Anderson, P., and Hansson, B. S. (1993). Physiology and morphology of pheromone-specific sensilla on the antennae of male and female Spodoptera littoralis (Lepidoptera, Noctuidae). J. Insest Physiol. 39, 253–260. doi: 10.1016/0022-1910(93)90096-A
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Montagné, N., Chertemps, T., Brigaud, I., Francois, A., Francois, M. C., de Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Muñoz, L., Rosell, G., Quero, C., and Guerrero, A. (2008). Biosynthetic pathways of the pheromone of the Egyptian armyworm Spodoptera littoralis. Physiol. Entomol. 33, 275–290. doi: 10.1111/j.1365-3032.2008.00645.x
Nakagawa, T., Pellegrino, M., Sato, K., Vosshall, L. B., and Touhara, K. (2012). Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS ONE 7:e32372. doi: 10.1371/journal.pone.0032372
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
Nichols, A. S., and Luetje, C. W. (2010). Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J. Biol. Chem. 285, 11854–11862. doi: 10.1074/jbc.M109.058321
Pellegrino, M., Steinbach, N., Stensmyr, M. C., Hansson, B. S., and Vosshall, L. B. (2011). A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478, 511–514. doi: 10.1038/nature10438
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Poitout, S., and Buès, R. (1974). Elevage de chenilles de vingt-huit espèces de Lépidoptères Noctuidae et de deux espèces d'Arctiidae sur milieu artificiel simple. Particularités de l'élevage selon les espèces. Ann. Zool. Ecol. Anim. 6, 431–441.
Quero, C., Lucas, P., Renou, M., and Guerrero, A. (1996). Behavioral responses of Spodoptera littoralis males to sex pheromone components and virgin females in wind tunnel. J. Chem. Ecol. 22, 1087–1102. doi: 10.1007/Bf02027947
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Salama, H. S., Dimetry, N. Z., and Salem, S. A. (1971). On the host preference and biology of the cotton leaf worm Spodoptera littoralis Bois. Z Angew. Entomol. 67, 261–266. doi: 10.1111/j.1439-0418.1971.tb02122.x
Saveer, A. M., Becher, P. G., Birgersson, G. R., Hansson, B. S., Witzgall, P., and Bengtsson, M. (2014). Mate recognition and reproductive isolation in the sibling species Spodoptera littoralis and Spodoptera litura. Front. Ecol. Evol. 2:18. doi: 10.3389/fevo.2014.00018
Shimodaira, H. (2002). An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508. doi: 10.1080/10635150290069913
Smadja, C., Shi, P., Butlin, R. K., and Robertson, H. M. (2009). Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 26, 2073–2086. doi: 10.1093/molbev/msp116
Steinwender, B., Thrimawithana, A. H., Crowhurst, R. N., and Newcomb, R. D. (2015). Pheromone receptor evolution in the cryptic leafroller species, Ctenopseustis obliquana and >C. herana. J. Mol. Evol. 80, 42–56. doi: 10.1007/s00239-014-9650-z
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS ONE 8:e62098. doi: 10.1371/journal.pone.0062098
Syed, Z., Kopp, A., Kimbrell, D. A., and Leal, W. S. (2010). Bombykol receptors in the silkworm moth and the fruit fly. Proc. Natl. Acad. Sci. U.S.A. 107, 9436–9439. doi: 10.1073/pnas.1003881107
Valle, M., Schabauer, H., Pacher, C., Stockinger, H., Stamatakis, A., Robinson-Rechavi, M., et al. (2014). Optimization strategies for fast detection of positive selection on phylogenetic trees. Bioinformatics 30, 1129–1137. doi: 10.1093/bioinformatics/btt760
Wang, G., Vásquez, G. M., Schal, C., Zwiebel, L. J., and Gould, F. (2011). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect. Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE 5:e8685. doi: 10.1371/journal.pone.0008685
Xu, P., and Leal, W. S. (2013). Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem. Biophys. Res. Commun. 435, 477–482. doi: 10.1016/j.bbrc.2013.05.015
Yang, Z., and dos Reis, M. (2011). Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 28, 1217–1228. doi: 10.1093/molbev/msq303
Yang, Z., and Nielsen, R. (2002). Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917. doi: 10.1093/oxfordjournals.molbev.a004148
Zhang, D. D., and Löfstedt, C. (2013). Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS ONE 8, e77345. doi: 10.1371/journal.pone.0077345
Keywords: insect, olfaction, olfactory receptor, sex pheromone, Spodoptera littoralis, heterologous expression, positive selection
Citation: de Fouchier A, Sun X, Monsempes C, Mirabeau O, Jacquin-Joly E and Montagné N (2015) Evolution of two receptors detecting the same pheromone compound in crop pest moths of the genus Spodoptera. Front. Ecol. Evol. 3:95. doi: 10.3389/fevo.2015.00095
Received: 18 May 2015; Accepted: 31 July 2015;
Published: 12 August 2015.
Edited by:
Astrid T. Groot, University of Amsterdam, NetherlandsReviewed by:
Ewald Grosse-Wilde, Max Planck Institute for Chemical Ecology, GermanyCesar Gemeno, University of Lleida, Spain
Copyright © 2015 de Fouchier, Sun, Monsempes, Mirabeau, Jacquin-Joly and Montagné. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas Montagné, Institute of Ecology and Environmental Sciences (iEES-Paris), Université Pierre et Marie Curie, Case courrier 1211, 7 quai Saint-Bernard, 75252 Paris, France, nicolas.montagne@upmc.fr
 Arthur de Fouchier
Arthur de Fouchier Xiao Sun1,2,3
Xiao Sun1,2,3  Christelle Monsempes
Christelle Monsempes Olivier Mirabeau
Olivier Mirabeau Emmanuelle Jacquin-Joly
Emmanuelle Jacquin-Joly Nicolas Montagné
Nicolas Montagné