- 1Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan, China
- 2State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 3College of Modern Agriculture Sciences, University of Chinese Academy of Sciences, Beijing, China
- 4Key Laboratory of Pesticide and Chemical Biology, Ministry of Education, Central China Normal University, Wuhan, China
- 5State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 6Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Immunoglobulin M (IgM) is the major antibody in teleost fish and plays an important role in humoral adaptive immunity. The N-linked carbohydrates presenting on IgM have been well documented in higher vertebrates, but little is known regarding site-specific N-glycan characteristics in teleost IgM. In order to characterize these site-specific N-glycans, we conducted the first study of the N-glycans of each glycosylation site of the grass carp serum IgM. Among the four glycosylation sites, the Asn-262, Asn-303, and Asn-426 residues were efficiently glycosylated, while Asn-565 at the C-terminal tailpiece was incompletely occupied. A striking decrease in the level of occupancy at the Asn-565 glycosite was observed in dimeric IgM compared to that in monomeric IgM, and no glycan occupancy of Asn-565 was observed in tetrameric IgM. Glycopeptide analysis with liquid chromatography-electrospray ionization tandem mass spectrometry revealed mainly complex-type glycans with substantial heterogeneity, with neutral; monosialyl-, disialyl- and trisialylated; and fucosyl-and non-fucosyl-oligosaccharides conjugated to grass carp serum IgM. Glycan variation at a single site was greatest at the Asn-262 glycosite. Unlike IgMs in other species, only traces of complex-type and no high-mannose glycans were found at the Asn-565 glycosite. Matrix-assisted laser desorption ionization analysis of released glycans confirmed the overwhelming majority of carbohydrates were of the complex-type. These results indicate that grass carp serum IgM exhibits unique N-glycan features and highly processed oligosaccharides attached to individual glycosites.
Introduction
Glycosylation represents a major post-translational modification of proteins involved in various biological processes, including transcription, differentiation, apoptosis, cell adhesion, receptor-ligand binding, as well as oncogenic transformation and immune response (1–4). Almost all proteins in the immune system are glycoproteins, the attached glycans are thought to be crucial to their structure and the immune effector mechanism (5, 6).
The two main types of glycan linkages to proteins are the N-linked and O-linked types. The N-linked oligosaccharide is covalently bonded with nitrogen of asparagine when it occurs in the sequence Asn-X-Ser/Thr or, more rarely, as part of an Asn-X-Cys motif (where X≠Pro) (7). N-linked oligosaccharides have been classified into three most common ones, being the high-mannose, hybrid, and complex types. All have a basic core structure of conserved pentasaccharide (GlcNAc2Man3) backbone but vary with respect to the structures attached to this core (8).
The N-linked oligosaccharides presenting on immunoglobulins have received particular attention because changes in the attached glycans can impact immunoglobulin solubility, structural stability, and biological function (9). In human IgG, differential of single monosaccharide at Asn-297 glycosite at the Fc fragment can drastically affect IgG binding to FcγR (10, 11) and affecting complement action (12). In human IgM, there are five putative glycosylation sites (Asn-171, Asn-332, Asn-395, Asn-402, and Asn-563) on the heavy chain. The glycans linked to each glycosites of IgM have been demonstrated to be involved in various biological functions, including B-cell maturation (Asn-171) (13), complement activation (Asn-402) (14, 15), and J-chain incorporation (Asn563) (16, 17).
In teleosts, IgM is the major antibody in serum, and it plays a key role in humoral adaptive immunity. Similar to mammalian IgM, teleost IgM consists of two identical heavy and two identical light chains (2H+2L). The heavy chain possesses four constant domains, CH1–CH4, containing the sites for the binding of effector cells (18), cytotoxic cells (19), or molecules such as complement system components (20). Bioinformatics analysis of teleost IgM has indicated the presence of 4–5 N-linked glycosylation sites in CH2, CH3, and CH4, while there is no glycosylation at CH1 (21, 22). The reported carbohydrate content was estimated to be approximately 12.5% for Atlantic salmon (Salmo salar) and 10% for Atlantic cod (Gadusmorhua) IgM (23, 24). Atlantic cod IgM contains oligosaccharides with N-acetylneuraminate and a core-fucose unit (24). A study of the IgM of the nurse shark (Ginglymostomacirratum), a cartilaginous fish, revealed that the most abundant glycan was high-mannose type. Moreover, the structures of both complex glycans and high-mannose on nurse shark IgM were very similar to the vertebrate species (25).
Compared to mammalian IgM, teleost IgM displays a significant degree of structural diversity, with different polymerization states, including monomers, dimers, trimers, and tetramers, being revealed (26–29). Structural diversity appears universal among teleost IgMs, the cause of the structural diversity has remained elusive for decades. The C-terminal tailpiece of secretory IgM is highly conserved, cysteine residue and a N-glycan Asn-X-Ser/Thr sequence have been found in all species sequenced thus far, including human (30), mouse (31), bovine (32), rabbit (33), chicken (34), hamster (35), frog (36), and fish (22). The C-terminal tailpiece contains crucial information for mammalian IgM assembly, as IgM polymers are influenced by the presence of glycans linked to Asn-563 in human IgM (37, 38). Furthermore, glycans linked to conserved glycosite at the C-terminal tailpiece has been demonstrated to influence the binding of joint chains and also mammalian IgM polymerization (16). A recent report revealed that replacement of Asn-563 with alanine (Ala) in human IgM prevents the attachment of N-glycans on IgM, resulting in the secretion of higher molecular weight hexameric, rather than pentameric, IgM complexes. This suggests that Asn-563 glycans at the C-terminal tailpiece play important roles in the process of IgM polymerization (39). Taken together, these findings indicate that glycosylation at conserved C-terminal tailpieces maybe an interesting field of study for analyzing the incorporation and formation of polymeric IgM differences in teleosts.
Thus far, however, no data have been reported on the site-specific N-glycan of teleost IgM. In order to characterize these site-specific N-glycans, we conducted the first comprehensive study of the carbohydrates conjugated at each N-glycosylation site, as well as the occupancy at the Asn-565 residue in the conserved C-terminal tailpiece, among grass carp IgM isoforms. These data expand the limited information currently available concerning teleost IgM glycosylation and provide a knowledge base for understanding the biological significance of glycosylation on teleost IgM.
Materials and Methods
Analytical Approach
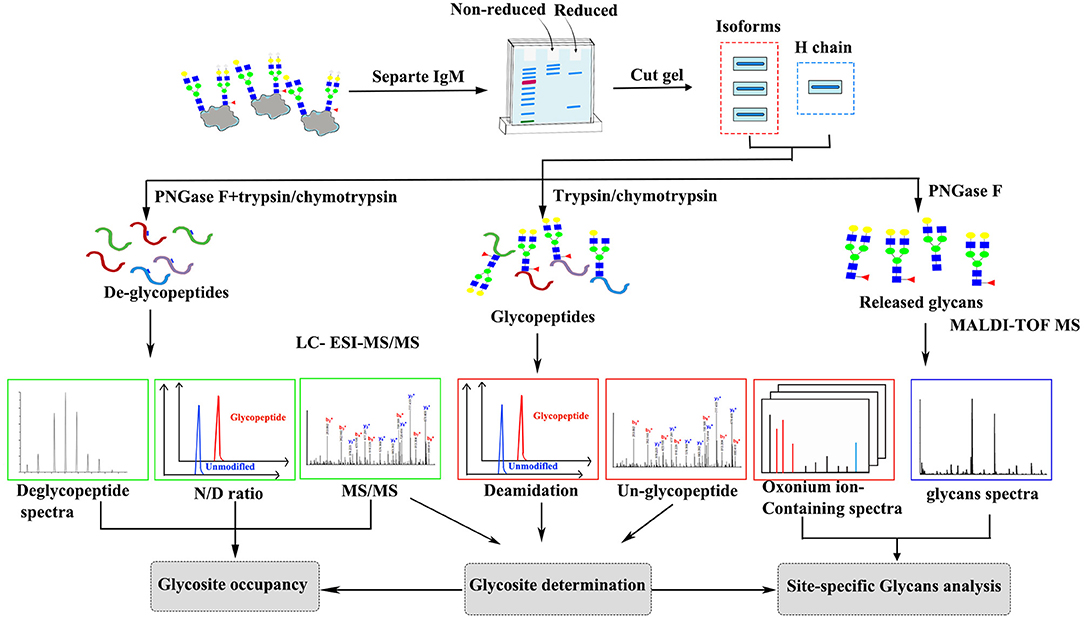
The general approach to characterizing IgM glycosylation structures is shown in Figure 1. First, liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) was used to map tryptic/chymotryptic deglyco-peptides and to identify the N-linked glycosylation sites. N-glycan occupancy at Asn-565 residues between monomeric, dimeric and tetrameric IgM were determined by comparing de-glycosylated and non-glycosylated N-565 peptides peak areas. LC-ESI–MS/MS analysis with in-source higher energy collision dissociation (HCD) was used to locate glycopeptides and for site specific glycoform characterization. Glycoform characterization was further confirmed by MALDI-TOF MS analysis of release glycans.
The Fish and Serum Collection
Grass carp (Ctenopharyngodonidella idella) 350–500 g, were bred and kept in 1,000-L tanks with recirculating and Uv-sterilized water. Fish were anesthetized with 10 ppm Benzocaine (Beijing, China National Pharmaceutical Group Corporation) for 3–5 min, blood was collected from the caudal vein from 10 naïve grass carp, allowed to clot for 1 h at room temperature and overnight at 4°C, serum was collected after centrifuging at 4,000 rpm for 10 min, pooled and stored at −80°C. All animal experiments were approved by the Institutional Animal Care and Use Committee of Central China Normal University.
Preparation of IgM
IgM was purified from pools of 10 individuals using a combination of polyethylene glycol (PEG) precipitation and ion-exchange chromatography as described previously (40). Briefly, Grass carp plasma was precipitated by added 16% (w/v) PEG (final concentration), stirring at 4°C for 30 min. The precipitated proteins were collected by centrifugation at 10,000 × g for 15 min. The collected protein were dialyzed against 10 mM sodium phosphate buffer (pH 7.5) containing 5 mM EDTA at 4°C overnight, the dialyzed buffer was changed for 3 times. The protein samples were passed through Source Q anion column (Amersham Biosciences) with 10 mM sodium phosphate buffer (PBS 0.01 M, pH 7.5) containing 5 mM EDTA. Bound proteins were eluted with a 0–1M linear NaCl gradient at a flow rate of 1 ml/min. Fraction containing grass carp IgM was collected and dialyzed against 10 mM sodium phosphate (pH 5.9), and further polished by pass through Source S cation exchange resin (Amersham Biosciences). Purity and size of IgM were checked by SDS-PAGE analysis and Coomassie Brilliant Blue G250 staining.
Gel Electrophoresis
To isolate the heavy and light chains of grass carp IgM, 4 ug of purified IgM was mixed with denatured loading buffer and loaded onto 4–10% gradient SDS-polyacrylamide gel, then subjected to electrophoresis at 25 mA till the dye reached the bottom of the gel. The gel was stained with Coomassie Brilliant blue (Sigma) and the heavy and light chains was determined according to molecular weight markers. Previously electrophoretic analysis revealed carp fish serum containing different populations as monomeric, dimeric, and tetrameric forms of IgM (41, 42). To verify of the oligomerization state of the grass carp serum IgM, 8 ug of purified IgM was mixed with loading buffer without β-mercaptoethanol and characterized by 6% SDS-polyacrylamide gel electrophoresis at 30 mA for about 2.5 h. The gel was stained with Coomassie Brilliant blue and the molecular band of monomer, dimer, and tetramer was estimated.
Glycosylation Site Determination
To determination the glycosylation site on grass carp IgM, coomassie-stained SDS-PAGE gel pieces containing IgM heavy chain were destained with a solution of 50 mM ammonium bicarbonate in 50% acetonitrile (1:1, vol/vol) with shaking, and followed by 100% acetonitrile. The sample pieces were reduced by incubating with 64 mM DTT at 56°C for 60 min, followed by alkylation with 130 mM iodoacetamide at R.T. for 45 min in the dark. The samples were washed with 50 mM ammonium bicarbonate and dehydrated in acetonitrile, dried in a SpeedVac centrifuge, rehydrated in 50 mM ammonium bicarbonate. N-glycans were released by incubating the sample with 100 Units/ml of PNGase F for 12 h at 37°C, the N-linked glycans were collected for further MALDI-TOF analysis. Gel piece was dehydrated in acetonitrile, rehydrated in 50 mM ammonium bicarbonate and treated with trypsin (Promega) at an enzyme/substrate ratio of 1:20 (w/w) or with chymotrypsin (Promega) at an enzyme/substrate ratio of 1:13 (w/w) overnight at 37°C. Peptides were extracted from the gel pieces with 50% acetonitrile in 5% formic acid. The resulting peptides were then extracted and desalted with a C18 tip (Millipore) for analysis by LC-ESI-MS/MS. Glycosylation site were determined by mass of deglycopeptide-peptide (Asn to Asp; Δm = +0.984 Da). Sequence and potential N-glycosylation sites for grass carp serum IgM were obtained from GenBank (accession No ABD76396.1).
N-Glycopeptide Enrichment by HILIC
The digested peptides were enriched with Unisol-Amide HILIC media as previously described with slight modifications (29, 43). Briefly, the gel band corresponding to the IgM heavy chain (approximately 4 μg) was excised, destained and digested with trypsin (Promega) or chymotrypsin (Promega) overnight. Unisol-Amide HILIC resin (Agela, China, 10 μm particle size, 200 Å pore size) were packed on top of a 200 μL C18 tip (Millipore) to an approximate column height of 5 mm. The column was washed with 1% formic acid followed by 1% formic acid in 80% acetonitrile. The trypsin or chymotrypsin digested peptide mixtures were dissolved with 20 μL 80% acetonitrile and loaded onto the column, the flow through fraction was reloaded onto the column twice to maximize the glycopeptide binding, the column was then washed thrice with 1% formic acid in 80% acetonitrile. The bound glycopeptides were eluted twice with 1% formic acid, and subsequent elution with 20 μL 1% formic acid in 80% acetonitrile was performed to elute any glycopeptides binding to the C18 tip during the elution. The enriched glycopeptides were dried and kept at −20°C till for analysis.
LC-ESI-MS/MS Analysis
Trypsin or chymotrypsin digested grass carp IgM de-glycopeptides and glycopeptides were analyzed using nanoflow HPLC instrument (EASY-nLC 1200 system, Thermo Fisher Scientific, USA) coupled on-line to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, USA) in positive polarity mode. Chromatography column was Acclaim PepMap, C18, 50 um × 150 mm, particle size 2 um, 100Å (Thermo Fisher Scientific, USA) with buffer A (95% water and 4.9% acetonitrile with 0.1% FA) and buffer B (90% acetonitrile and 9.9% water with 0.1% FA) at a flow rate of 300 nL/min. The gradient profile was set as follows: 3–5% B for 6 min, 5–25% B for 54 min, 25–40% B for 25 min and hold 5 min, 40–80% B for 5 min, and 80–100% B for 5 min. The electrospray voltage was 2.0 kV. Peptides were analyzed with high-energy collisional dissociation (HCD) fragmentation mode by data-dependent MS/MS acquisition. In MS1, the resolution was 70,000, the AGC target was 3e6, and the maximum injection time was 20 ms. In MS2, the resolution was 17,500, the AGC target was 5e4, and the maximum injection time was 50 ms, NCE stepped set 27. The scan range was set at a resolution of 350–2,000 m/z and the 20 most intense precursors were chosen analysis by data-dependent mode.
Analysis of De-Glycopeptide and Intact Glycopeptide MS Data
N-linked-glycosylation sites were determined in de-glycosylated peptides based on converting Asn to Asp by PNGase F treatment, the corresponding glycosylated peptides were confirmed by HCD tandem MS. The obtained de-glycopeptide MS data of were searched against grass carp IgM sequence (GenBank accession No ABD76396.1) using Proteome Discoverer V2.2 (Thermo Fisher Scientific). Carbamidomethylation of Cys was set as a fixed modification, oxidation of Met and converting of Asn to Asp were set as variable modifications. The mass tolerance was set at 10 ppm for precursor ions, and the MS/MS mass tolerance was set at 0.01Da. Trypsin or chymotrypsin was set with up to two missed cleavages. Annotation of the corresponding glycopeptide spectra was performed in using Byonic software v1.09 (Protein Metrics) (44). Searches were performed with the following modifications: precursor ion tolerance of 10 ppm, carbamidomethylation of Cys, protease treatment with up to two missed cleavages, and with the following variable modifications: oxidation of Met and glycosylation of Asn. The obtained grass carp IgM glycopeptide MS data were identified by Byonic software (44). Additionally, all assigned N-glycopeptide HCD spectra were then queried to ensure the presence of diagnostic oxonium ions at m/z 204.087 (HexNAc+), and at least one of the following oxonium ions as 186.076 (HexNAc-H2O+), 168.066 (HexNAc-2H2O+) and 366.140 (HexNAc-Hex+) within 50 ppm. The existence of peptide or peptide+HexNAc ions in MS/MS spectra was checked manually to determine the glycosite-containing peptide and glycan compositions of the glycopeptides. Spectra were also checked manually about b and y ion data from peptide portion of the corresponding glycopeptides.
MALDI-TOF MS Analysis Glycans
N-glycans released from grass carp serum IgM by PNGaseF treatment were analyzed by an Axima MALDI Resonance mass spectrometer (Shimadzu). N-glycans were dissolve in 10 μl 0.1% TFA followed by spotting 1 μl glycan sample with 1 μl 2,5-dihydroxybenzoic acid matrix (100 mg/ml DHB in 50% acetonitrile, 0.1 mM NaCl) onto a MALDI plate. Positive ion MALDI spectra were obtained with a Bruker ultraflextreme™ Waters-MicromassToF Spec 2E time-of-flight (TOF) mass spectrometer (Waters MS Technology Ltd, Manchester, UK) operated under flexControl 3.3 (Build 108; Bruker Daltonics). Spectra were obtained in positive linear ion mode in the range 1,000–3,200 m/z, using an accelerating voltage of 2.2 kV and a laser energy of 6.5 kV. The positive control in the MALDI-TOF MS analysis was authentic complex-type glycans from mAb rituximab, 2,5-dihydroxybenzoic acid matrix was used as negative control. The average MS spectra (1,000 profiles) were used for glycan assignment by GlycoMod software (http://web.expasy.org/glycomod/) (45). N-glycans identification was set by allowing a maximum mass deviation of 0.04 Da for mainly present [M+Na]+ ions. Searches were conducted with the following parameters: Hexose range 3-9, HexNAc range 2-9, deoxyhexose range 0-4, NeuAc range 0-4, NeuGc no, Pentose no, Sulfate/Phosphate no, KDN no, HexA no, UniCarbKB entries were listed separately.
Results
Purification of IgM and Analysis of N-Glycosylation Assignments
To determine the glycosylation status of grass carp IgM, IgM was purified by polyethylene glycol (PEG) precipitation and ion-exchange chromatography from serum. Purified IgM migrated two major protein bands with molecular weights of 75 kDa and 23 kDa under reducing conditions, representing the heavy and light chains, respectively (Figure 2A). To identify different isoforms of grass carp serum IgM, intact IgM molecules were resolved using non-reduced PAGE. Grass carp serum IgM migrated three protein bands larger than 170 kDa under non-reduced conditions (Figure 2B), which could represent variable polymerization into monomeric, dimeric, and tetrameric forms, as reported previously in common carp and Indian major carp (41, 42). A decrease in the molecular size after PNGase F digestion was apparent in the heavy chains of grass carp IgM, while the band corresponding to the light chain was not affected by PNGase F treatment (Figure 2C). This suggests that N-linked oligosaccharides are present on the grass carp IgM heavy chain but not on the light chain.
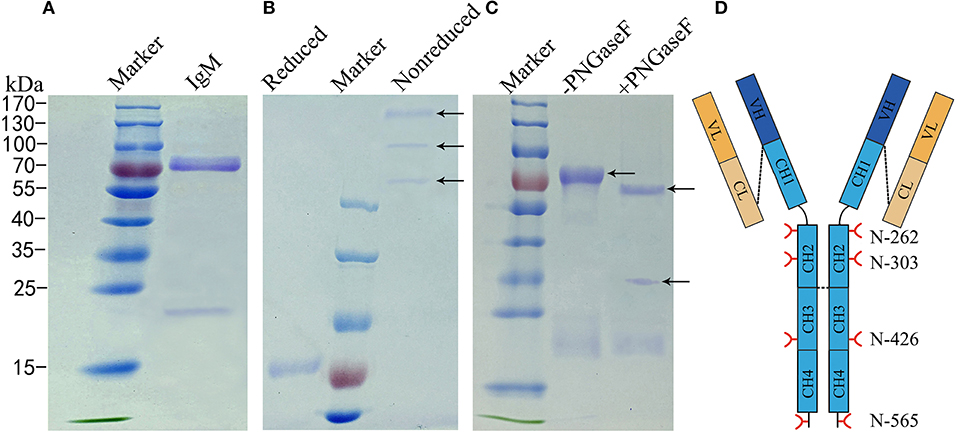
Figure 2. Analysis of N-glycans assignments on grass carp serum IgM. (A) A 4–10% uncontinued SDS-PAGE of reduced samples to show 75–80 kDa heavy chain band and 23 kDa light chain band of grass carp IgM. (B) A 6% SDS-PAGE stained Coomassie Blue with showing different grass carp IgM isoforms, in the non-reduced samples lane, from up to down, the tetramer, dimer, and monomer were marked by arrow. (C) IgM digested with (+) or without (–) PNGase F, arrow at the bottom indicate PNGase F. (D) Schematic of four N-glycosyationsites on grass carp IgM, red forks represent four putative N-glycosylation sites on heavy chain, black lines at the end of CH4 domain indicating 16–18 amino acid residue at the C-terminal tailpiece, possible inter-subunit disulfide bridges between heavy chains and covalently link the light chain are shown with black dash lines.
Bioinformatics analysis of the amino acid sequences of the light chain (GenBank No.AEH76780.1 and AEH76777.1) also indicated no N-linked carbohydrate attached to the grass carp IgM light chain (data not shown). Analysis indicated that each of the heavy chains contains four potential N-glycosylation sites restricted to the constant regions. Two of these are located at Asn-262 and Asn-303 in the CH2 domain, one at Asn-426 in the CH3 domain, and one at the Asn-565 residue within the conserved C-terminal tailpiece of the CH4 domain (Figure 2D).
Site Occupancy of Four Putative N-Glycosylation Sites
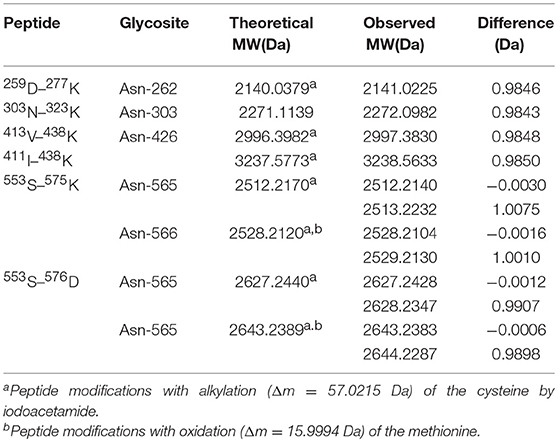
The occupancy at each glycosylation site was first checked via PNGase F digestion using LC-ESI-MS/MS. Treatment with PNGase F specifically releases N-glycans attached to the nitrogen of Asn, thereby converting Asn residue to Asp with mass increasing of 0.984 Da; thus, glycosylation sites can be deduced by amino acid sequence analysis after PNGase F treatment (30, 46, 47). Overall, tryptic peptides accounted for 72.74% of the total heavy chain amino acid sequence and 86.99% of the constant region, and the identified region spanning the four N-glycosylation sites on the heavy chain (Figure 3). The observed tryptic peptide masses derived from the Asn-262, Asn-303, and Asn-426 glycosylation sites exhibited increases of 0.9846–0.9850 Da compared with their theoretical mass weights, indicating that the Asn-262, Asn-303, and Asn-426 residues were glycosylated and converted to aspartic acid after PNGase F digestion (Figure 4A, Table 1). In contrast, both de-glycosylated and non-glycosylated Asn-565 sites were detected, with the tryptic peptide 553S–575K being observed in both forms in the dimer-isotopic mass (2512.2140 Da/2513.2232 Da) and with the additional oxidation at the methionine residue (2528.2104 Da/2529.2130 Da) (Table 1). Furthermore,553S–576D, with one trypsin digest missing, was also observed in the dimer-isotopic mass (2627.2428 Da/2628.2347 Da) and with methionine oxidation (2643.2383 Da/2644.2287 Da) (Figure 4B, Table 1), indicating that the Asn-565 residue is not completely utilized.
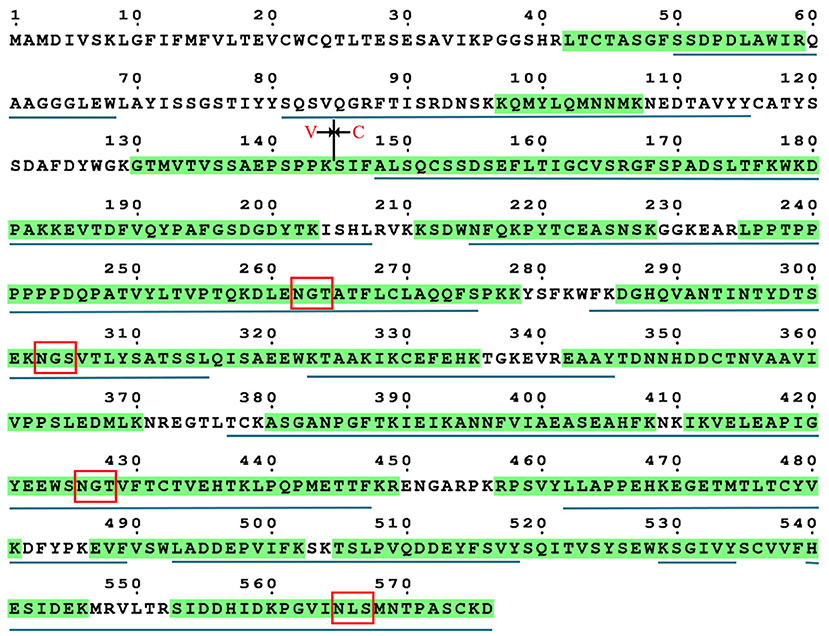
Figure 3. Sequence coverage of grass carp IgM heavy chain. The identified tryptic peptides are indicated by green shadow, four putative N-glycosylation sites are marked by red box. The identified chymotryptic peptides are indicated by black underline.
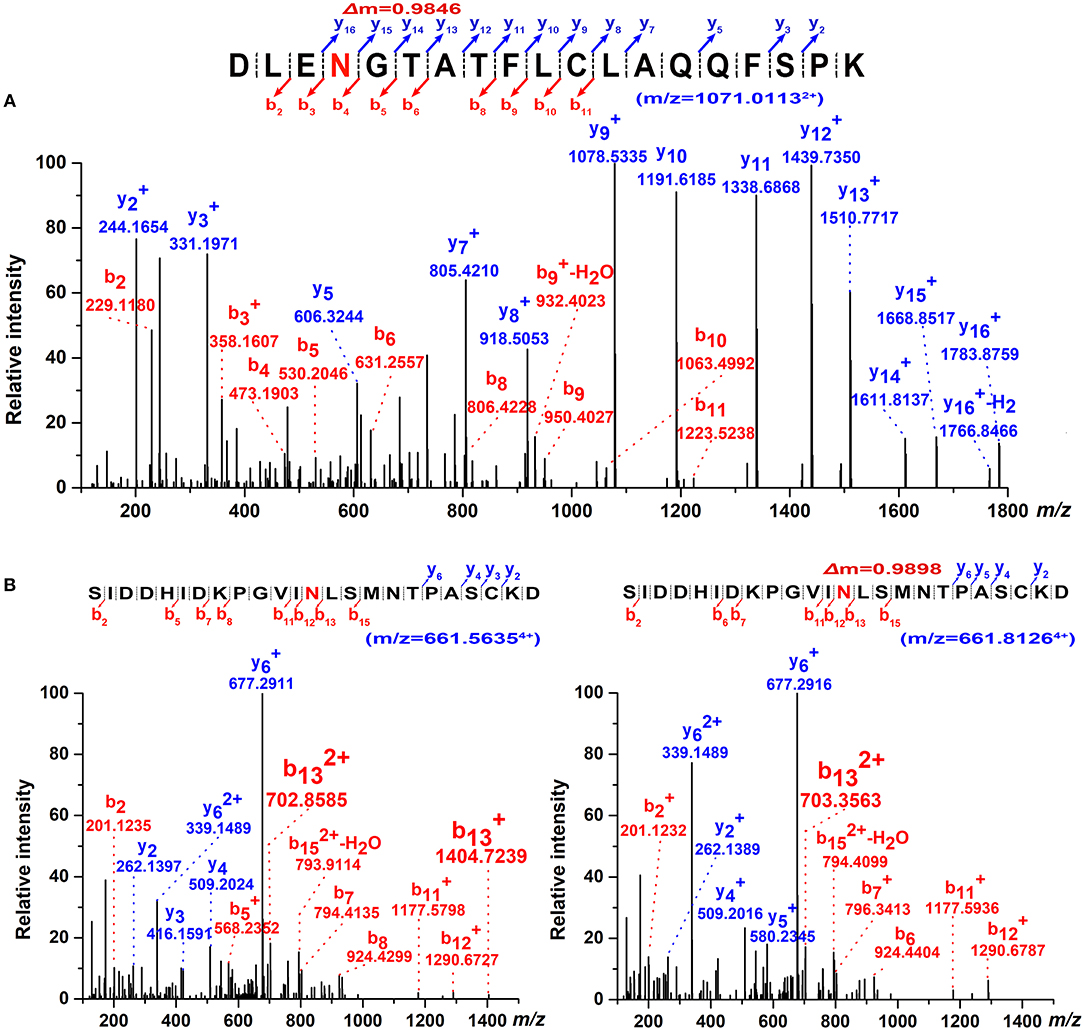
Figure 4. Determination of N-glycosylation sites by high-resolution ESI MS. (A) Representative MS/MS spectrum of the tryptic 259D–277Kpeptide (m/z = 1071.01132+) covering Asn-262 glycosite with PNGase F digestion. The mass increment of 0.9846 Da comparing to its theoretical mass weight is indicated. (B) Dimer-isotopic mass spectrum of 553S–576D peptide covering Asn-565 glycosite. Non-glycosylated 553S–576D peptide (m/z = 661.56354+) is shown in left panel, while de-glycosylated 553S–576D peptide (m/z = 661.81264+) with a mass increasing of 0.9898 Da due to the converting of Asn-565 residue to an Asp with PNGase F treatment is indicated in right panel.
Comparison of N-Glycan Occupancy at Asn-565 Between Monomeric, Dimeric, and Tetrameric IgM
To assess the glycosylation occupancy at the Asn-565 site among different IgM isoforms, the monomeric, dimeric, and tetrameric IgM bands (seen in Figure 1B) were excised and de-glycosylated by PNGase F. Then, the occupancy of this site was estimated by comparing the relative amounts of de-glycopeptide and non-glycopeptide. Although isotopic clusters of PNGaseF de-glycosylated peptides (Δm = 0.984 Da) could be clearly determined in comparison with non-glycosylated Asn-565 peptides via mass spectrometry (Figure 5A), some de-glycosylated peptide peaks overlapped with non-glycosylated peptides peak, making it difficult to estimate the peak areas between de-glycosylated and non-glycosylated Asn-565peptides. Occupation was then estimated by comparing the peptide spectral matches (PSMs) of de-glycosylated and non-glycosylated Asn-565 peptides. A striking decrease in de-glycosylated peptides was observed for dimeric IgM as compared with that for monomeric IgM, with the glycan occupancy at the Asn-565 site estimated to be approximately 36.34% for monomeric IgM and about 20.10% for dimeric IgM (Figures 5B,C). Moreover, no de-glycosylated Asn-565 peptide was detected on tetrameric IgM in three separate analyses, suggesting that Asn-565 is unoccupied by N-glycans on tetrameric IgM.
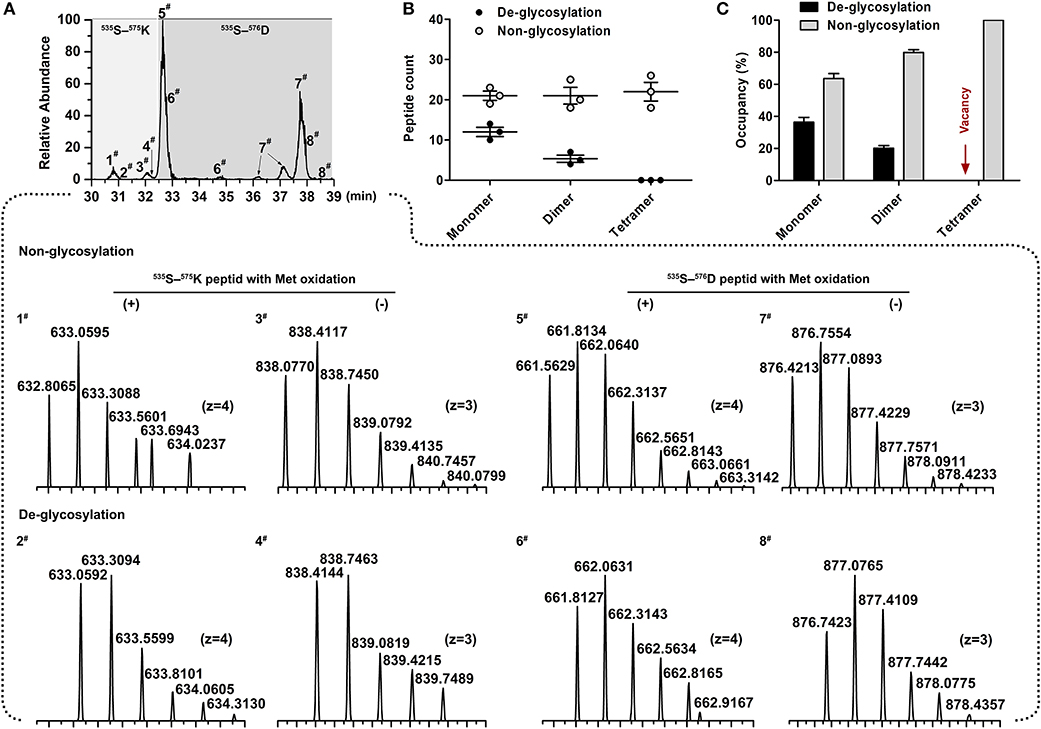
Figure 5. Determination of site occupancy for Asn-565 glycosite. (A) Overlaid chromatograms of non-glycosylated and de-glycosylated Asn-565 peptides. A total of 8 peptide variants were identified; 1# and 3#, 5# and 7#, non-glycosylated 553S–575K peptide and 553S–576D peptide without, and with methionineoxidation. 2# and 4#, 6# and 8#, De-glycosylated 553S–575K peptide and 553S–576D peptide without, and with methionineoxidation. (B) Asn-565 peptides spectral counts plot. Non-glycosylated or de-glycosylated Asn-565 peptides counts are shown by the sum of 553S–575K peptide and 553S–576D peptide. (C) Asn-565 glycosite occupancy. Occupancy was calculated by sum of the non-glycosylated or de-glycosylated peptide as a percentage of the total spectral counts of non-glycosylated and de-glycosylated peptide. Error bars in (B,C) indicate the standard deviation of the same serum pools with three repeats.
Identification of Glycopeptides
To determine site-specific glycan profiles at each of the four sites of the grass carp serum IgM, tryptic glycopeptides were investigated with LC-ESI-MS/MS by employing HCD fragmentation. Analysis of the grass carp IgM glycopeptides could provide site-specific glycans information. A typical LC-ESI-MS/MS base peak intensity chromatogram for individual tryptic glycopeptide family is shown in Figure 6. Compared with de-glycopeptide, individual glycopeptide family eluted over wider chromatographic intervals, which indicating that the retention time is influenced by the peptide and also modulated by the sugar moiety.
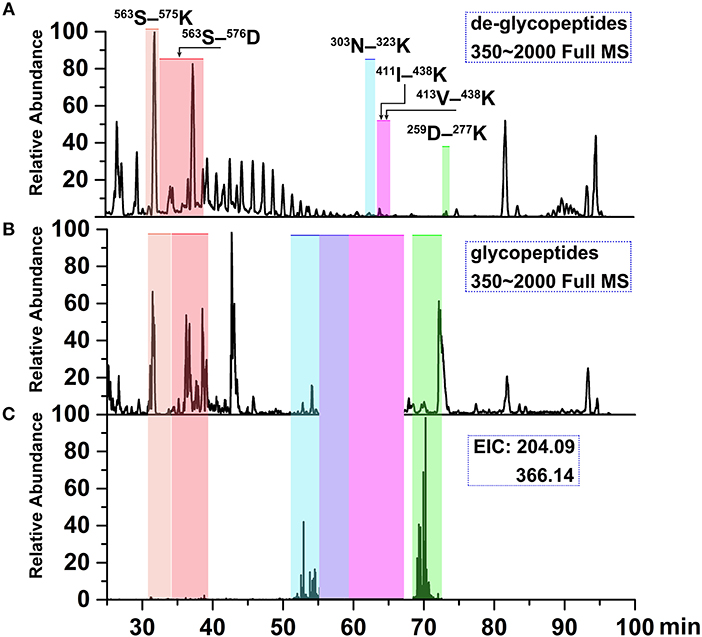
Figure 6. Ion chromatograms of LC-separated tryptic peptides. (A) Base peak chromatogram (BPC) of the tryptic de-glycopeptides with PNGase F digestion. (B) BPC of tryptic glycopeptides with HILIC enrichment. (C) Extracted ion chromatograms (EICs) of diagnostic oxonium fragment ions at m/z of 204.09 and 366.14. The shaded regions depict the identified deglycopeptides, glycopeptide clusters and Tables 2, 3 summarizes glycan heterogeneity at each site.
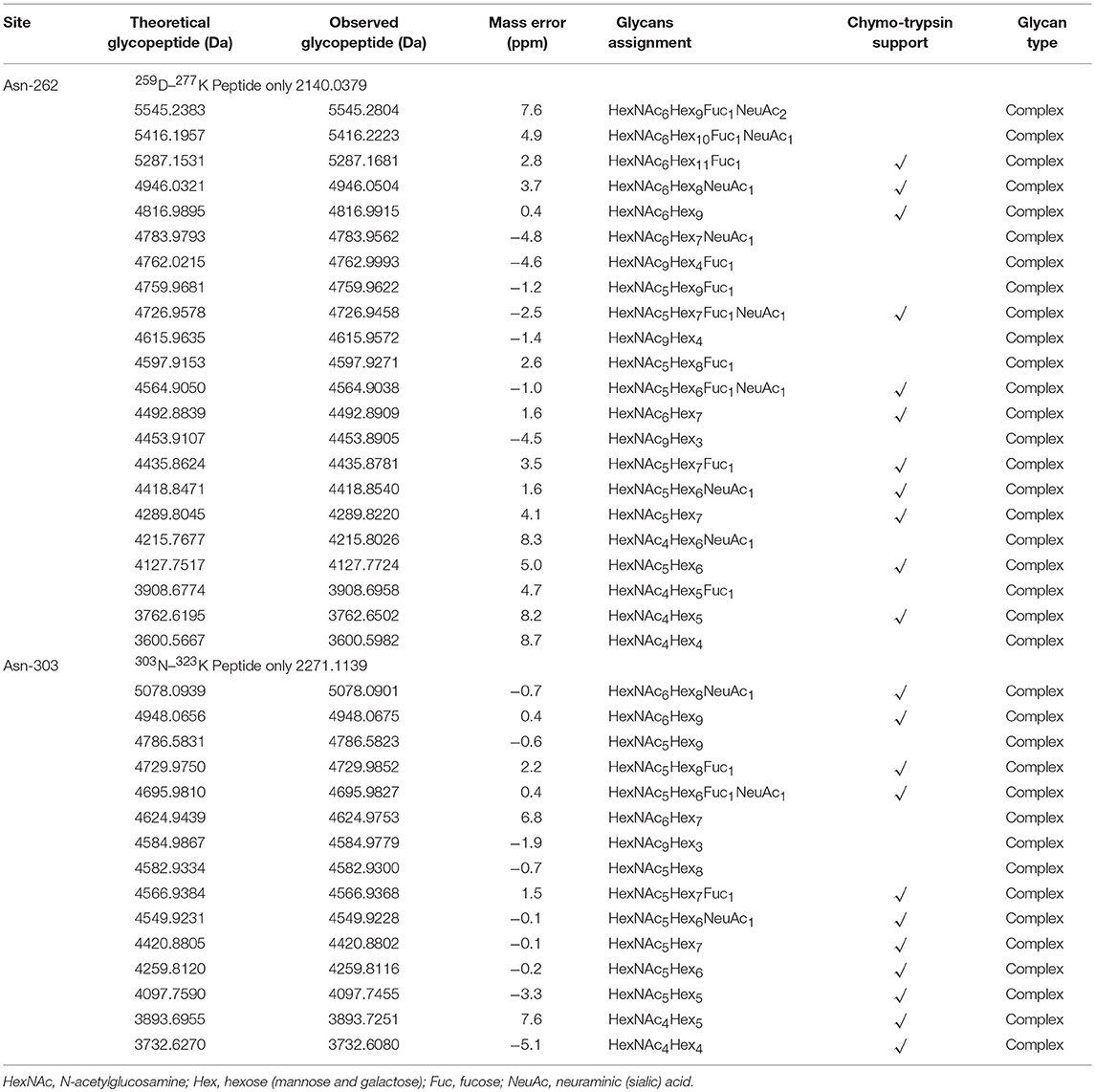
Three representative glycans detected on the 259D–277K (Asn-262) peptide are shown in Figure 7. The oxonium ionsat m/z 204.08 (HexNAc+), 186.07 (HexNAc-H2O+), 168.06 (HexNAc-2H2O+), and 366.14 (HexNAc-Hex+) are readily discernible within the fragmentation pattern. The composition of each glycan was identified by data-dependent MS/MS with an accuracy of 10 ppm or less. On the basis of previous information regarding fish IgM N-glycosylation (25, 48), the deoxyhexose residue was recognized as fucose in this study. Sialylated ions, such as those at m/z 292.09 (NeuAc+) and m/z 657.23 (NeuAc-Hex-HexNAc+), indicated the presence of sialylated glycans, with fragment ions corresponding to core-fucose on the peptide+HexNAc residue suggesting core-fucosylated glycans (Figures 7A,B). Glycan sequencing results for HexNAc-Hex-NeuAc, peptide+HexNAc, peptide+2HexNAc, peptide+HexNAc+Hex, as well as the intact peptide backbone and a set of b/y ions, confirmed the assignment of the glycan to the 259D–277K tryptic peptide fragment (Figures 7A–C). Notably, the fucose on the core-HexNAc residue, observed as a peptide+HexNAc+Fucion, was unique fucose residue observed by MS/MS, indicating the location of the fucose as a core-fucosylation on N-glycans. Glycopeptide identification also revealed that N-acetylneuraminic acid (NeuAc), rather than N-glycolylneuraminic acid (NeuGc), was the only form of sialic acid observed in grass carp serum IgM.
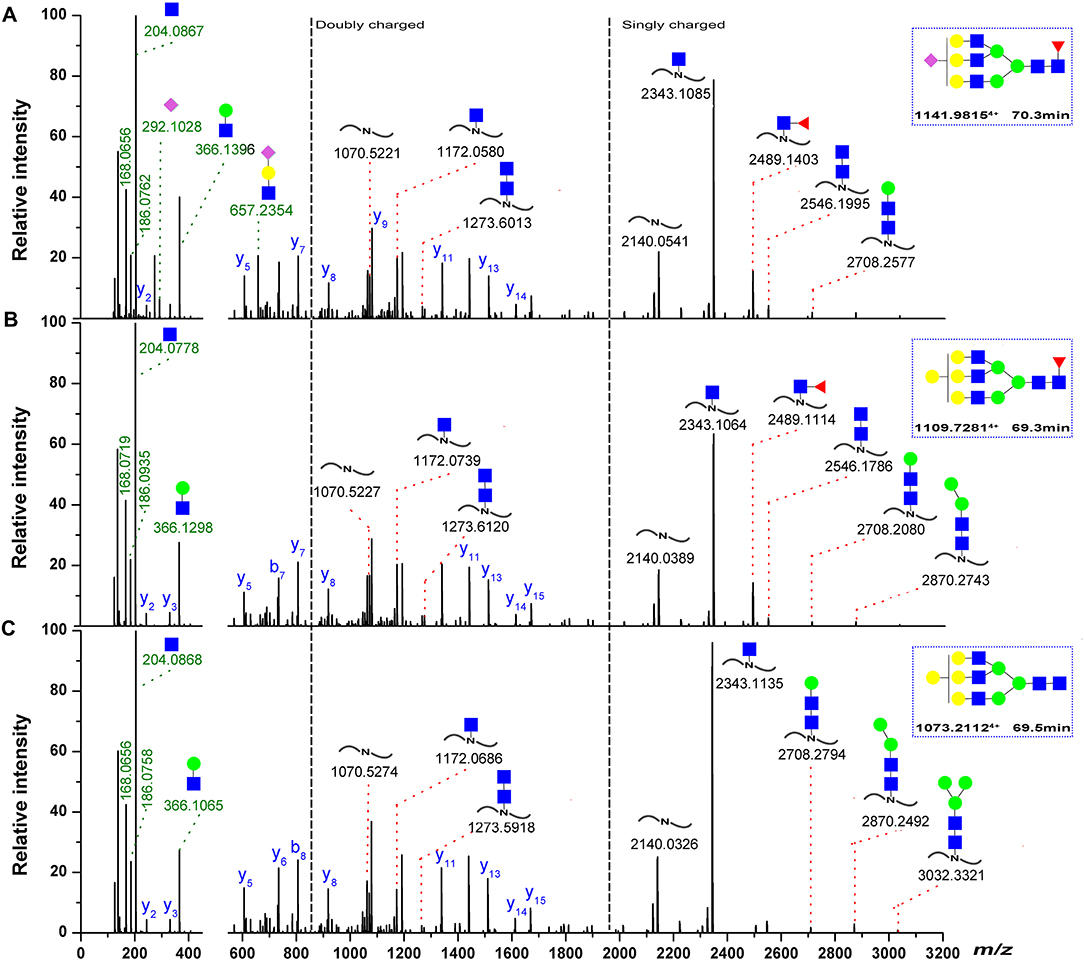
Figure 7. Representative spectra for Asn-262 glycopeptides with a sialylated glycan. (A) A neutral glycan with core-fucose (B) A neutral glycan without core-fucose glycan. (C) The oxonium ions were used to extract the intact glycopeptide spectrum, and the masses of the precursor and peptide/peptide + core GlcNAc ± Fuc ± Hex ions (red) can provide structural details, the b/y-ions (only y-ions were marked, blue) of the peptide portion were used to confirm peptide sequence. The proposed glycan structures were determined on the basis of monosaccharide composition, the monosaccharide linkages were not yet determined. Symbols as the following: blue box, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; red triangle, fucose; pink diamond, N-acetylneuraminic acid.
Site-Specific N-Glycan Analysis
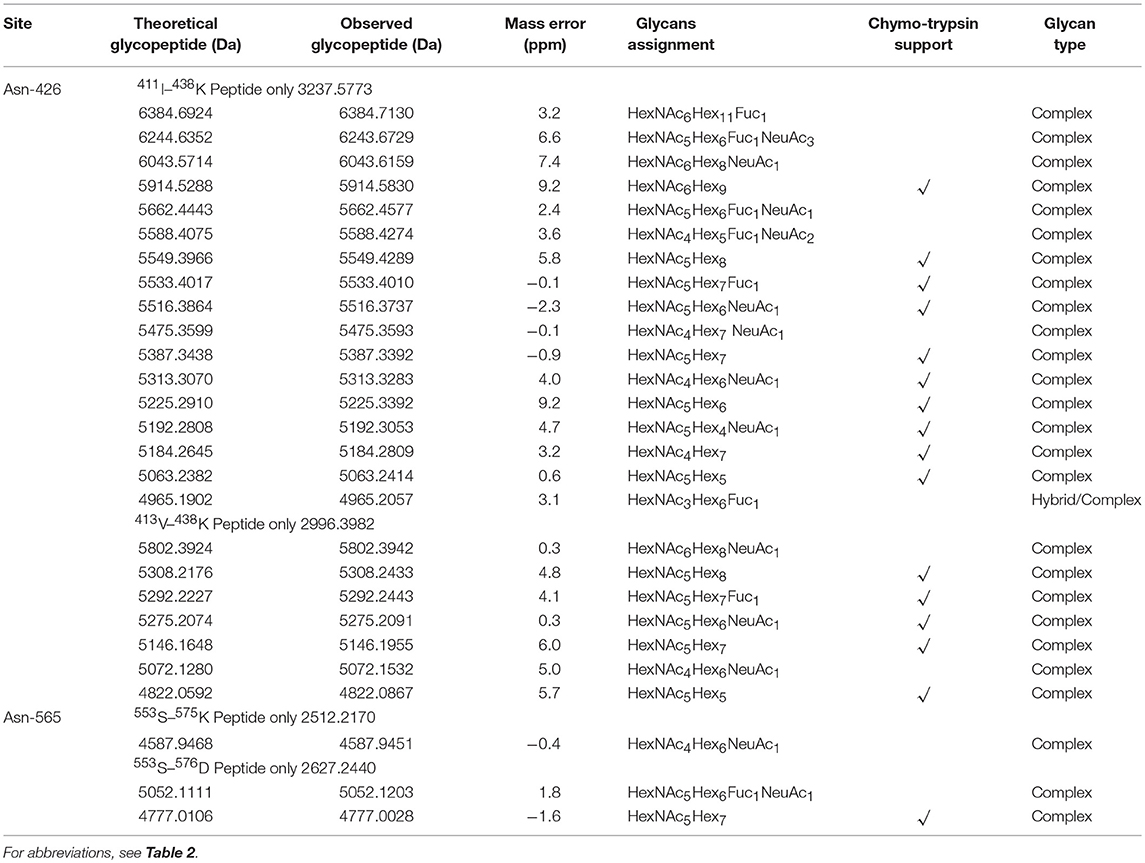
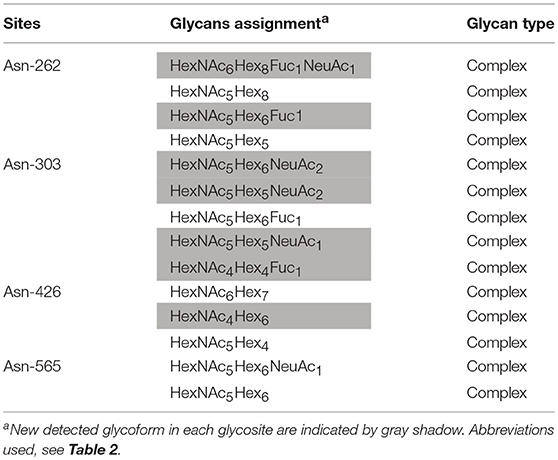
Assignments a set of glycans at each glycosite were achieved by selectively detecting tryptic glycopeptides, as shown in Tables 2, 3. Substantial heterogeneity was evident, with different glycan compositions detected at the four glycosites of grass carp serum IgM, including 22 forms at Asn-262, 15 at Asn-303, and 17 at Asn-426, indicating a considerable overlap in the glycoforms present at these three sites. In contrast, the Asn-565 glycosite was unique in having trace carbohydrate moieties and only three classes of glycans. In addition to trypsin, chymotrypsin digestion was also performed to increase sequence coverage and obtain more glycan information at each glycosite. The majority of glycans identified by chymotrypsin treatment were the same as those identified by trypsin digestion at the Asn-262, Asn-303, and Asn-426 glycosites, serving as a secondary verification of the site-specific glycosylation mapping (Table 4). Furthermore, chymotryptic glycopeptide analysis revealed 2–5 new glycans at each of the four glycosites, as shown in Table 4.
Of the 38 total glycan compositions observed on grass carp serum IgM by tryptic and chymotryptic glycopeptide analysis, 17 were sialylated and 21 were not. Notably, when the glycans were grouped into high-mannose, hybrid, complex types, the glycans identified on grass carp serum IgM were limited to complex-type carbohydrates, with the exception of a single carbohydrate with a HexNAc3Hex6Fuc1 composition on Asn-426 glycosite, for which the identification as a hybrid or complex type glycan was ambiguous. Heterogeneity was greatest at the Asn-262 glycosite, with both neutral and mono/disialylated, as well as small and large glycans, were observed. The Asn-303 and Asn-426 glycosites exhibited the next highest levels of variation. Three novel oligosaccharides, HexNAc9Hex3, HexNAc9Hex4, and HexNAc9Hex4Fuc1, with redundancy in the HexNAc monosaccharide (tentatively assigned as GalNAcβ1 → 4GlcNAc antennae, the relatively rare lacdiNAc motif) were identified on grass carp serum IgM with a mass accuracy within 5 ppm. No other glycan composition was possible with respect to the observed m/z values, suggesting the presence of glycans on grass carp serum IgM containing the diacetyllactosamine motif. Unlike IgMs in other higher vertebrates and some reported fish species, which possess major high-mannose glycans at the C-terminal tailpiece glycosite, the Asn-565 site of the grass carp serum IgM possesses exclusively complex-type N-glycans and is devoid of high-mannose structures. Together, these data indicate that the glycans on grass carp serum IgM are highly processed. It should also be pointed out that only a minority of Asn-565 peptides were conjugated with carbohydrates with both tryptic and chymotryptic glycopeptide analysis, further confirming the incomplete glycan occupancy at the Asn-565 glycosite.
MALDI-TOF MS Analysis of N-Glycans
Information from released glycans can be used to confirm the results of glycopeptide analysis and provide additional insight into the N-glycans present on grass carp serum IgM. The MALDI-TOF spectra of the released glycans revealed that the main carbohydrate moieties are HexNAc3−6Hex4−9Fuc1NeuAc1,3 complex-type glycans (Figure 8), which is in general agreement with the IgM glycopeptide analysis. Of these, the four most abundant peaks corresponded to HexNAc5Hex7, HexNAc6Hex9, HexNAc5Hex7Fuc1, and HexNAc5Hex6 glycan compositions. These four glycans were also prominent among those observed in the glycopeptide analysis, as HexNAc5Hx7 presented at each of the four glycosites; HexNAc6Hex9 and HexNAc5Hex7Fuc1 appeared at the Asn-262, Asn-303, and Asn-426 sites; and HexNAc5Hx6 appeared at the Asn-262 and Asn-426 sites. Some sialylated glycan structures, such as HexNAc4−5Hex4−6Fuc0−1NeuAc1,3, were detected by MALDI-TOF analysis. Notably, the HexNAc4Hex6, HexNAc6Hex8, and HexNAc6Hex9Fuc1 glycans, which were observed by MALDI-TOF but not glycopeptide analysis, may be, to some extent, artifactually generated forms of the HexNAc4Hex6NeuAc1, HexNAc6Hex8NeuAc1, and HexNAc6Hex9 Fuc1NeuAc1 glycans, respectively, derived from the loss of sialic acid residues during the MALDI analysis. Data from the released glycans also provided additional information not present in the glycopeptide analysis, as a HexNAc6Hex9Fuc1 glycan with an m/z >2800 was detected by MALDI-TOF MS. Overall, MALDI-TOF analysis of released glycans further confirmed that the main oligosaccharides present on grass carp serum IgM are complex-type glycans.

Figure 8. Glycans detected by MALDI-TOF. (A) Molecular masses are of unlabeled glycans, detected as [M + Na]+ in positive polarity mode. (B) The proposed glycan structures were determined on the basis of monosaccharide composition, as well as literature (24, 25, 48). For symbol definitions, seen Figure 6. N.D., not determined.
Discussion
In this study, a detailed site-specific N-glycan profile and glycan occupancy at the C-terminal tailpiece glycosite were described for the first time of the teleost IgM. The results revealed that grass carp serum IgM possesses primarily complex-type oligosaccharides that may be either neutral or sialylated. Glycan occupancy at the C-terminal tailpiece glycosite has significant different among grass carp IgM isoforms.
The N-linked carbohydrate moieties of carp IgM were found to be restricted to the heavy chain. The Asn-262 and Asn-303 glycosites in the CH2 domain and Asn-426 glycosite in CH3 were found to be fully occupied, while the Asn-565 site in the C-terminal tailpiece was incompletely occupied. It is a structural feature of teleost IgMs that a number of teleosts produce heterogeneous mixtures of IgM polymers. This has been attributed to the variation in the degree of disulfide polymerization, referred to as redox forms, and has implications for teleost IgM assembly processes (26, 49). Here, we found that the occupancy at the Asn-563 site is markedly different between monomeric and dimeric IgM forms, while the site is totally unoccupied in tetrameric IgM. This is the first evidence of variable site occupancy at the C-terminal tailpiece of teleost IgM isoforms.
In the site-specific analysis, the largest heterogeneity in oligosaccharides was observed at the Asn-262 glycosite, followed by Asn-303 in the CH1 domain and Asn-426 in the CH3 domain. The main oligosaccharides were found to be complex-type glycans with considerable heterogeneity, with neutral; monosialyl-, disialyl-, and trisialylated; and fucosyl-and non-fucosyl-oligosaccharides conjugated to these three sites. Interestingly, oligosaccharides with HexNAc9Hex3, HexNAc9Hex4Fuc1, and HexNAc9Hex4 compositions with redundancy in the HexNAc monosaccharide were identified at the Asn-262 and Asn-303 glycosites by glycopeptide analysis. This indicates that Asn-262 and Asn-303 are occupied by glycans with terminal HexNAc residues, possibly lacdiNAc motifs (GalNAcβ1 → 4GlcNAcantennae), as are common on invertebrate glycans as well as the N-glycans of zebrafish embryos (48, 50, 51). Compared to the C-terminal tailpieces of glycosites in higher vertebrates, which possess major high-mannose glycans (30, 43, 52), the C-terminal tailpiece glycosite of grass carp serum IgM possesses exclusively complex-type N-glycans and is devoid of high-mannose glycans. Although the minority of Asn-565 glycosites were conjugated with carbohydrates, grass carp serum IgM possesses sialylated oligosaccharides at the C-terminal tailpiece glycosite.
Previous studies of the oligosaccharides on the C-terminal tailpiece have documented their important roles in determining IgM polymerization in mammals (38). It has been found that human IgM polymers are influenced by the presence of glycans linked to Asn-563, as replacement of Asn-563 with Ala abolishes the attachment of glycan moieties and results in the secretion of hexameric and higher molecular weight species (39). It has been suggested that glycans linked to Asn-563, located in the tightly packed IgM core, may limit the number of subunits that can be incorporated into an oligomer or serve as a docking device for ERGIC53, a hexameric lectin shown to promote polymerization in non-lymphoid cells (53). Teleost IgM lacks J chain and tetrameric IgM was thought to polymerize by interchain disulfide linkages (26). Our observation of highly processed oligosaccharides and variable site occupancies at the C-terminal tailpiece glycosite of grass carp IgM isoforms may therefore have important biological implications. It may indicate that glycans linked to the C-terminal tailpiece have an impact on teleost IgM oligomer formation, and oligosaccharides presenting on the C-terminal tailpiece can interfere teleost IgM oligomer formation.
Glycosylation is highly susceptible to changes of the physiological conditions and environmental factors (54), changes related to age, gender, and pregnancy have also been described for mammalian Ig glycan (55, 56). Various B-cell stimuli have been shown to modulate Ig glycosylation, as CpG oligodeoxynucleotide and IL- 21 have been proved to increase Fc galactosylation and reduce bisecting GlcNAc levels. While, all-trans retinoic acid decreases galactosylation and sialylation levels (57). The transcription factor hepatocyte nuclear factor (HNF) 1a and its downstream target HNF4a have been identified as critical regulators of fucosyltransferase and fucose biosynthesis genes, thus influence N-glycan levels in plasma protein (58). It is known teleost produce both mucosal (mlg) and serum (slg) IgM. Previous report indicated glycosylation patterns may differences between mlgM and slg IgM, as mAb reacting with mlgM H chain carbohydrate determinant not or poorly with slg, which indicating differences in carbohydrate composition or structure (41). Knowledge on teleost mlgM glycosylation is scant so far, analysis of mIgM are expected to shed light on the mlg mucosa-associate Ig glycosylation pattern.
It has been found that differiental of sugar moiety influences mammalian antibody affinity to activating or inhibitory FcγR and thus influence antibody effector activity. IgG galactosylation has been found to be decreased in rhumatoid arthritis, and galactosylated IgG correlates with remission of arthritis during pregnancy rheumatoid arthritis patients (56, 59, 60). Changes in mouse IgG glycosylation are considered to affect the binding affinity of IgG to FcγR (61, 62). Absence of core fucose in human IgG was reported to alter binding affinity to FcγRIII, the effect of therapeutic anticancer mAb as trastuzumab with decreased fucosylation has been well recognized and is currently being harnessed in clinical trials (63, 64). Attachment of sialic acid on IgG appears to mediate the anti-inflammatory effects, in contrast, a lack of terminal sialic acid residue appears to mediate the inflammatory effect leading to osteoclast differentiation and bone loss by increases the affinity to activating FcγRIIand FcγRIII (65, 66). The N-glycans on IgM are of particular interest because IgM has a relatively high carbohydrate content compared with those of other immunoglobulin classes. Until now, very little has been known specifically about the role of the carbohydrate moiety in fish IgM. Thus, it would be of considerable interest to determine whether teleost IgM N-linked glycans have functional roles in interactions with natural biological partners, such as complement system components and/or FcR on immune cells. Work along these lines is now in progress in our laboratory.
In conclusion, our study highlights the importance of performing site-specific characterization of glycoproteins at multiple glycosites, as total glycosylation characterization of the entire protein will fail to reveal differences in the glycosylation distributions of individual sites. The site-specific characterization of the N-glycans of teleost IgM described here should facilitate targeted partial de-glycosylation approaches for subsequent functional analysis in order to obtain further insight into the functional effects of glycosylation on teleost IgM.
Data Availability Statement
The RAW MS data of grass carp serum IgM analysis, as well as the original search results of pGlyco 2.0 have been uploaded to the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with access codes: PXD010308.
Author Contributions
HG, Y-AZ, and BW conceived and designed the experiments. Y-LS, M-DH, Z-WC, and JW performed the experiments. Y-LS, QY, and HB analyzed the data. HG, Y-FC, LX, and C-HW wrote the manuscript. All authors reviewed the manuscript.
Funding
This work was supported by the Natural Science Foundation of China (21473065), Fundamental Research Funds for the Central University of Central China Normal University (CCNU16A02044), Hubei Province Natural Science Key Foundation Project (2014CFA079), and National Basic Research Program of China (2014CB138601).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, et al. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science (2004) 306:120–4. doi: 10.1126/science.1102109
2. Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell (2009) 139:1229–41. doi: 10.1016/j.cell.2009.12.008
3. Starossom SC, Mascanfroni ID, Imitola J, Cao L, Raddassi K, Hernandez SF, et al. Galectin-1 deactivates classically activated microglia and protects from inflammation-induced neurodegeneration. Immunity (2012) 37:249–63. doi: 10.1016/j.immuni.2012.05.023
4. Vasta GR, Ahmed H, Nita-Lazar M, Banerjee A, Pasek M, Shridhar S, et al. Galectins as self/non-self recognition receptors in innate and adaptive immunity: an unresolved paradox. Front Immunol. (2012) 3:199. doi: 10.3389/fimmu.2012.00199
5. Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science (2001) 291:2370–6. doi: 10.1126/science.291.5512.2370
6. Coombe DR, Parish CR. Editorial: carbohydrates: the yet to be tasted sweet spot of immunity. Front Immunol. (2015) 6:314. doi: 10.3389/fimmu.2015.00314
7. Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell (2010) 141:897–907. doi: 10.1016/j.cell.2010.04.012
8. Varki A, Cummings RD, Esko JD, Grewal PK. Essentials of Glycobiology (Chapter 9 N-Glycans). 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (2015).
9. Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. (2007) 25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702
10. Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. (2011) 108:12669–74. doi: 10.1073/pnas.1108455108
11. Ohmi Y, Ise W, Harazono A, Takakura D, Fukuyama H, Baba Y, et al. Sialylation converts arthritogenic IgG into inhibitors of collagen-induced arthritis. Nat Commun. (2016) 7:11205. doi: 10.1038/ncomms11205
12. Quast I, Keller CW, Maurer MA, Giddens JP, Tackenberg B, Wang LX, et al. Sialylation of IgG Fc domain impairs complement-dependent cytotoxicity. J Clin Invest. (2015) 125:4160–70. doi: 10.1172/JCI82695
13. Ubelhart R, Bach MP, Eschbach C, Wossning T, Reth M, Jumaa H. N-linked glycosylation selectively regulates autonomous precursor BCR function. Nat Immunol. (2010) 11:759–65. doi: 10.1038/ni.1903
14. Muraoka S, Shulman MJ. Structural requirements for IgM assembly and cytolytic activity. Effects of mutations in the oligosaccharide acceptor site at Asn402. J Immunol. (1989) 142:695–701. 142:695–701.
15. Wright JF, Shulman MJ, Isenman DE, Painter RH. C1 binding by mouse IgM. The effect of abnormal glycosylation at position 402 resulting from a serine to asparagine exchange at residue 406 of the mu-chain. J Biol Chem. (1990) 265:10506–13. 265:10506–13.
16. de Lalla C, Fagioli C, Cessi FS, Smilovich D, Sitia R. Biogenesis and function of IgM: the role of the conserved mu-chain tailpiece glycans. Mol Immunol. (1998) 35:837–45. doi: 10.1016/s0161-5890(98)00073-x
17. Wiersma EJ, Collins C, Fazel S, Shulman MJ. Structural and functional analysis of J chain-deficient IgM. J Immunol. (1998) 160:5979–89. 160:5979–89.
18. Nayak DK, Tang A, Wilson M, Miller NW, Bengten E. Channel catfish soluble FcmuR binds conserved linear epitopes present on Cmu3 and Cmu4. Mol Immunol. (2010) 47:1306–16. doi: 10.1016/j.molimm.2009.11.026
19. Shen L, Stuge TB, Evenhuis JP, Bengten E, Wilson M, Chinchar VG, et al. Channel catfish NK-like cells are armed with IgM via a putative FcmicroR. Dev Comp Immunol. (2003) 27:699–714. doi: 10.1016/S0145-305X(03)00042-9
20. Boshra H, Gelman AE, Sunyer JO. Structural and functional characterization of complement C4 and C1s-like molecules in teleost fish: insights into the evolution of classical and alternative pathways. J Immunol. (2004) 173:349–59. doi: 10.4049/jimmunol.173.1.349
21. Coscia MR, Morea V, Tramontano A, Oreste U. Analysis of a cDNA sequence encoding the immunoglobulin heavy chain of the Antarctic teleost Trematomus bernacchii. Fish Shellfish Immunol. (2000) 10:343–57. doi: 10.1006/fsim.1999.0244
22. Bilal S, Lie KK, Karlsen OA, Hordvik I. Characterization of IgM in Norwegian cleaner fish (lumpfish and wrasses). Fish Shellfish Immunol. (2016) 59:9–17. doi: 10.1016/j.fsi.2016.09.063
23. Magnadottir B, Gudmundsdottir BK, Gudmundsdottir S. The carbohydrate moiety of IgM from atlantic salmon (Salmo salar L.). Comp Biochem Physiol Part B (1997) 116:423–30. doi: 10.1016/S0305-0491(96)00264-7
24. Magnadottir B, Crispin M, Royle L, Colominas C, Harvey DJ, Dwek RA, et al. The carbohydrate moiety of serum IgM from Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. (2002) 12:209–27. doi: 10.1006/fsim.2001.0364
25. Harvey DJ, Crispin M, Moffatt BE, Smith SL, Sim RB, Rudd PM, et al. Identification of high-mannose and multiantennary complex-type N-linked glycans containing alpha-galactose epitopes from Nurse shark IgM heavy chain. Glycoconj J. (2009) 26:1055–64. doi: 10.1007/s10719-008-9226-5
26. Kaattari S, Evans D, Klemer J. Varied redox forms of teleost IgM: an alternative to isotypic diversity? Immunol Rev. (1998) 166:133–42. doi: 10.1111/j.1600-065X.1998.tb01258.x
27. Bromage ES, Ye J, Kaattari SL. Antibody structural variation in rainbow trout fluids. Comp Biochem Physiol B Biochem Mol Biol. (2006) 143:61–9. doi: 10.1016/j.cbpb.2005.10.003
28. Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. (2010) 11:827–35. doi: 10.1038/ni.1913
29. An Y, Cipollo JF. An unbiased approach for analysis of protein glycosylation and application to influenza vaccine hemagglutinin. Anal Biochem. (2011) 415:67–80. doi: 10.1016/j.ab.2011.04.018
30. Pabst M, Kuster SK, Wahl F, Krismer J, Dittrich PS, Zenobi R. A microarray-matrix-assisted laser desorption/ionization-mass spectrometry approach for site-specific protein N-glycosylation analysis, as demonstrated for human serum immunoglobulin M (IgM). Mol Cell Proteomics (2015) 14:1645–56. doi: 10.1074/mcp.O114.046748
31. Wang F, Nakouzi A, Angeletti RH, Casadevall A. Site-specific characterization of the N-linked oligosaccharides of a murine immunoglobulin M by high-performance liquid chromatography/electrospray mass spectrometry. Anal Biochem. (2003) 314:266–80. doi: 10.1016/s0003-2697(02)00693-0
32. Zhao Y, Kacskovics I, Rabbani H, Hammarstrom L. Physical mapping of the bovine immunoglobulin heavy chain constant region gene locus. J Biol Chem. (2003) 278:35024–32. doi: 10.1074/jbc.M301337200
33. Bernstein KE, Alexander CB, Reddy EP, Mage RG. Complete sequence of a cloned cDNA encoding rabbit secreted mu-chain of VHa2 allotype: comparisons with VHa1 and membrane mu sequences. J Immunol. (1984) 132:490–5. 132:490–5.
34. Dahan A, Reynaud CA, Weill JC. Nucleotide sequence of the constant region of a chicken mu heavy chain immunoglobulin mRNA. Nucleic Acids Res. (1983) 11:5381–9. doi: 10.1093/nar/11.16.5381
35. McGuire KL, Duncan WR, Tucker PW. Phylogenetic conservation of immunoglobulin heavy chains: direct comparison of hamster and mouse Cmu genes. Nucleic Acids Res. (1985) 13:5611–28. doi: 10.1093/nar/13.15.5611
36. Schwager J, Mikoryak CA, Steiner LA. Amino acid sequence of heavy chain from Xenopus laevis IgM deduced from cDNA sequence: implications for evolution of immunoglobulin domains. Proc Natl Acad Sci USA. (1988) 85:2245–9. doi: 10.1073/pnas.85.7.2245
37. Wormald MR, Wooten EW, Bazzo R, Edge CJ, Feinstein A, Rademacher TW, et al. The conformational effects of N-glycosylation on the tailpiece from serum IgM. Eur J Biochem. (1991) 198:131–9. doi: 10.1111/j.1432-1033.1991.tb15995.x
38. Muller R, Grawert MA, Kern T, Madl T, Peschek J, Sattler M, et al. High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc Natl Acad Sci USA. (2013) 110:10183–8. doi: 10.1073/pnas.1300547110
39. Pasalic D, Weber B, Giannone C, Anelli T, Muller R, Fagioli C, et al. A peptide extension dictates IgM assembly. Proc Natl Acad Sci USA. (2017) 114:E8575–84. doi: 10.1073/pnas.1701797114
40. Boshra H, Li J, Peters R, Hansen J, Matlapudi A, Sunyer JO. Cloning, expression, cellular distribution, and role in chemotaxis of a C5a receptor in rainbow trout: the first identification of a C5a receptor in a nonmammalian species. J Immunol. (2004) 172:4381–90. doi: 10.4049/jimmunol.172.7.4381
41. Rombout JH, Taverne N, van de Kamp M, Taverne-Thiele AJ. Differences in mucus and serum immunoglobulin of carp (Cyprinus carpio L.). Dev Comp Immunol. (1993) 17:309–17. doi: 10.1016/0145-305X(93)90003-9
42. Swain P, Dash S, Bal J, Routray P, Sahoo PK, Sahoo SK, et al. Passive transfer of maternal antibodies and their existence in eggs, larvae and fry of Indian major carp, Labeo rohita (Ham.). Fish Shellfish Immunol. (2006) 20:519–27. doi: 10.1016/j.fsi.2005.06.011
43. Moh ES, Lin CH, Thaysen-Andersen M, Packer NH. Site-specific N-glycosylation of recombinant pentameric and hexameric human IgM. J Am Soc Mass Spectrom. (2016) 27:1143–55. doi: 10.1007/s13361-016-1378-0
44. Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics (2012) Chapter 13:Unit 13.20. doi: 10.1002/0471250953.bi1320s40
45. Cooper CA, Gasteiger E, Packer NH. GlycoMod–a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics (2001) 1:340–9. doi: 10.1002/1615-9861(200102)1:2<340::aid-prot340>3.0.co;2-b
46. Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S, et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat Biotechnol. (2016) 34:84–8. doi: 10.1038/nbt.3403
47. Yang N, Goonatilleke E, Park D, Song T, Fan G, Lebrilla CB. Quantitation of site-specific glycosylation in manufactured recombinant monoclonal antibody drugs. Anal Chem. (2016) 88:7091–100. doi: 10.1021/acs.analchem.6b00963
48. Hanzawa K, Suzuki N, Natsuka S. Structures and developmental alterations of N-glycans of zebrafish embryos. Glycobiology (2017) 27:228–45. doi: 10.1093/glycob/cww124
49. Ye J, Kaattari IM, Ma C, Kaattari S. The teleost humoral immune response. Fish Shellfish Immunol. (2013) 35:1719–28. doi: 10.1016/j.fsi.2013.10.015
50. Kawar ZS, Van Die I, Cummings RD. Molecular cloning and enzymatic characterization of a UDP-GalNAc:GlcNAc(beta)-R beta1,4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J Biol Chem. (2002) 277:34924–32. doi: 10.1074/jbc.M206112200
51. Wuhrer M, Koeleman CA, Fitzpatrick JM, Hoffmann KF, Deelder AM, Hokke CH. Gender-specific expression of complex-type N-glycans in schistosomes. Glycobiology (2006) 16:991–1006. doi: 10.1093/glycob/cwl020
52. Arnold JN, Wormald MR, Suter DM, Radcliffe CM, Harvey DJ, Dwek RA, et al. Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. J Biol Chem. (2005) 280:29080–7. doi: 10.1074/jbc.M504528200
53. Anelli T, Ceppi S, Bergamelli L, Cortini M, Masciarelli S, Valetti C, et al. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. (2007) 26:4177–88. doi: 10.1038/sj.emboj.7601844
54. de Haan N, van Tol MJD, Driessen GJ, Wuhrer M, Lankester AC. Immunoglobulin G fragment crystallizable glycosylation after hematopoietic stem cell transplantation is dissimilar to donor profiles. Front Immunol. (2018) 9:1238. doi: 10.3389/fimmu.2018.01238
55. Pucic M, Knezevic A, Vidic J, Adamczyk B, Novokmet M, Polasek O, et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics (2011) 10:M111.010090. doi: 10.1074/mcp.M111.010090
56. Kapur R, Kustiawan I, Vestrheim A, Koeleman CA, Visser R, Einarsdottir HK, et al. A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy. Blood (2014) 123:471–80. doi: 10.1182/blood-2013-09-527978
57. Wang J, Balog CI, Stavenhagen K, Koeleman CA, Scherer HU, Selman MH, et al. Fc-glycosylation of IgG1 is modulated by B-cell stimuli. Mol Cell Proteomics (2011) 10:M110.004655. doi: 10.1074/mcp.M110.004655
58. Lauc G, Essafi A, Huffman JE, Hayward C, Knezevic A, Kattla JJ, et al. Genomics meets glycomics-the first GWAS study of human N-Glycome identifies HNF1α as a master regulator of plasma protein fucosylation. PLoS Genet. (2010) 6:e1001256. doi: 10.1371/journal.pgen.1001256
59. Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature (1985) 316:452–7. 316:452–7.
60. Förger F, Østensen M. Is IgG galactosylation the relevant factor for pregnancy-induced remission of rheumatoid arthritis? Arthritis Res Ther. (2010) 12:108. doi: 10.1186/ar2919
61. Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, et al. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim Biophys Acta (2006) 1760:693–700. doi: 10.1016/j.bbagen.2005.10.002
62. Anthony RM, Nimmerjahn F. The role of differential IgG glycosylation in the interaction of antibodies with FcγRs in vivo. Curr Opin Organ Transplant. (2011) 16:7–14. doi: 10.1097/MOT.0b013e328342538f
63. Shibata-Koyama M, Iida S, Okazaki A, Mori K, Kitajima-Miyama K, Saitou S, et al. The N-linked oligosaccharide at FcγRIIIa Asn-45: an inhibitory element for high FcγRIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology (2009) 19:126–34. doi: 10.1093/glycob/cwn110
64. Junttila TT, Parsons K, Olsson C, Lu Y, Xin Y, Theriault J, et al. Superior in vivo efficacy of afucosylated trastuzumab in the treatment of HER2-amplified breast cancer. Cancer Res. (2010) 70:4481–9. doi: 10.1158/0008-5472.can-09-3704
65. Anthony RM, Wermeling F, Karlsson MC, Ravetch JV. Identification of a receptor required for the anti-inflammatory activity of IVIG. Proc Natl Acad Sci USA. (2008) 105:19571–8. doi: 10.1073/pnas.0810163105
Keywords: teleost, grass carp, immunoglobulin M, N-glycan, liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS), glycosylation, matrix assisted laser desorption/ionization-time-of-flight-MS (MALDI-TOF-MS)
Citation: Su Y-L, Wang B, Hu M-D, Cui Z-W, Wan J, Bai H, Yang Q, Cui Y-F, Wan C-H, Xiong L, Zhang Y-A and Geng H (2018) Site-Specific N-Glycan Characterization of Grass Carp Serum IgM. Front. Immunol. 9:2645. doi: 10.3389/fimmu.2018.02645
Received: 04 May 2018; Accepted: 26 October 2018;
Published: 14 November 2018.
Edited by:
Geert Wiegertjes, Wageningen University & Research, NetherlandsReviewed by:
Irene Salinas, University of New Mexico, United StatesHai-peng Liu, Xiamen University, China
Copyright © 2018 Su, Wang, Hu, Cui, Wan, Bai, Yang, Cui, Wan, Xiong, Zhang and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Geng, genghui@mail.ccnu.edu.cn
Yong-An Zhang, yzhang@ihb.ac.cn
†These authors share first authorship
 Yi-Ling Su1†
Yi-Ling Su1† Zheng-Wei Cui
Zheng-Wei Cui Yong-An Zhang
Yong-An Zhang Hui Geng
Hui Geng