- College of Bioscience and Biotechnology, Yangzhou University, Yangzhou, China
The proteins encoded by three Agrobacterial genes, atu5117, atu4860, and atu4856, are highly homologous to each other in amino acid sequence. All three proteins can bind to VirD2 and are named VBP1, VBP2, and VBP3 (VirD2-binding protein), respectively. VBP is involved in T-DNA transfer by recruiting the T-complex from the cytosol to the polar transport apparatus T4SS (type IV secretion system) and is defined as the “T-complex recruiting protein.” However, it remains unknown how these three homologous genes co-exist in a relatively small prokaryotic genome. To understand whether these three homologous genes are expressed differentially under virulence induction conditions, we examined the effects of virulence induction conditions, including various pH values, temperatures and acetosyringone (AS, an effective virulence inducer to Agrobacterium tumefaciens) concentrations, on the expression of the three VBP-encoding genes. Our data showed that vbp1 (atu5117) and vbp3 (atu4856) maintained constant expression under the tested induction conditions, whereas the expression of vbp2 (atu4860) was affected by the conditions. Culture conditions favorable to the expression of vbp2 differed from the reported induction conditions for other virulence proteins. In particular, the pH value was a crucial factor for the expression of vbp2. In addition, the deletion of vbp1 affected the expression of vbp2. Taken together, these results suggest that the mechanisms regulating the expression of these three homologous genes are different from the virulence induction mechanism and that VBP homologs are presumably involved in other biological processes in addition to T-complex recruitment.
Introduction
Agrobacterium tumefaciens is a well-known phytopathogen that causes crown gall tumor disease in various dicotyledonous plants. Pathogenicity is achieved by the transfer of a T-DNA fragment from the bacterial tumor-inducing (Ti) plasmid into host cells, genetically transforming the host. Agrobacterium uses the VirB/D4 T4SS to transfer the T-DNA in the form of a VirD2-T-DNA nucleoprotein complex (called T-complex; Guo et al., 2011; Pacurar et al., 2011; Chandran, 2013; Kado, 2014). The T-DNA transfer process has been largely described, and accumulating data have revealed that many Vir proteins are involved in T-DNA transfer. VirD2, one of the Vir proteins, is a relaxase that can cleave the bottom strand of the T-DNA and covalently attach to the 5′ end of the single-stranded T-DNA, thereby forming the VirD2-T-DNA nucleoprotein complex. In 2007, we used VirD2 as a pull-down bait to identify a VBP; (Guo et al., 2007a). An Agrobacterial genome-wide search demonstrated that in addition to the identified VBP-encoding gene atu5117, two other A. tumefaciens C58 genes, atu4860 and atu4856, can encode two VBP homologs. All three VBP homologs were confirmed to be able to bind VirD2 and thus designated VBP1 (encoded by atu5117), VBP2 (encoded by atu4860), and VBP3 (encoded by atu4856; Guo et al., 2007a,b). Our further investigation showed that VBPs are able to recruit the T-complex from the cytosol to the polar VirB/D4 transport apparatus T4SS (Guo et al., 2007a). Thus, VBPs were also defined as being T-complex recruiting proteins.
Bioinformatics investigations have demonstrated that three VBPs are highly conversed with regard to their functional domains, and all of the VBPs contain a putative nucleotidyltransferase domain near the N-terminus and a putative higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain near the C-terminus (Guo et al., 2007b; Gao et al., 2013). Structural studies have shown that VBP is a dimer and that the C-terminal HEPN domain is the dimerization domain of VBP. Associated functional studies of the HEPN domain of VBP have demonstrated that the dimerization of VBP is essential for the induction of tumors in plants (Padavannil et al., 2014). Our recent experimental data confirmed that VBP1 (encoded by atu5117) is an NTPase that might energize the recruitment of T-complex to the transport site (Gao et al., 2013).
As a ubiquitous soil-born bacterium, Agrobacterium has two lifestyles: independent free-living or acting as a phytopathogen. Its pathogenicity is not indispensable for its life cycle (Baek and Shapleigh, 2005; Gao and Lynn, 2005). However, it is currently not understood how a relatively small prokaryotic genome maintains three homologs for a non-essential biological function despite the ability of Agrobacterium tumorigenesis to be attenuated only by inactivating all three vbp genes. In addition, all three VBPs can complement each other in recruiting the T-complex (Guo et al., 2007a). True genetic redundancy is evolutionarily unstable (Brookfield, 1992; Nowak et al., 1997; Kafri et al., 2008), and bacterial genes that are redundant and not under efficient selection could be rapidly lost (Mira et al., 2001). Intuitively, VBPs may potentially be involved in other biological processes as well as in T-complex recruitment. To explore this possibility, many questions remain unanswered, but one important question is how the expression of three vbp genes respond to the virulence induction conditions.
Because VBPs are involved in Agrobacterium tumorigenesis, we investigated whether the growth conditions that induce tumorigenesis could affect the expression of the three vbp genes. Phenolic compounds and sugar compounds released by wounded plant tissue can be sensed and recognized by A. tumefaciens, and induce Agrobacterium to express vir genes. The induced Agrobacterium cells can then transfer the T-DNA to host plant cells, resulting in the formation of a tumor (Pitzschke and Hirt, 2010). Of these compounds, AS is the most effective inducing agent to Agrobacterium tumorigenesis. Although the effects of AS concentration, pH and temperature during the inducing process on the transformation efficiency vary in different reports (Baron et al., 2001; Gelvin, 2006; McCullen and Binns, 2006; Sudarshana et al., 2006), it has been proposed that acidic pH (4.8–5.5), moderate temperature (25°C), and a relatively high AS concentration (200 μg/ml) could induce most tumor tissues (Holford et al., 1992; McCullen and Binns, 2006). Thus, we investigated the effects of tumor-inducing conditions, including AS concentration, pH and temperature, on the expression of the three vbp genes using western blot analyses. Our data showed that the expression levels of vbp1 and vbp3 were nearly unchangeable, independent of the variable induction conditions. However, the expression of vbp2 was controlled by the culture conditions. Both the temperature and pH optimal for vbp2 expression were higher than those for virulence induction. The most effective virulence inducer, AS, appears to inhibit the expression of vbp2. Among these three tested factors, pH plays an important role in regulating the expression of vbp2. The expression of vbp2 was also affected by vbp1-deletion. These results indicate that the expression of three vbp genes may be regulated by a novel unknown pathway, which is contradictory from the reported virulence induction pathway. This unknown vbp-regulating pathway may be involved in the regulation of Agrobacterium tumorigenesis. Taken together, these results also provide additional details for further elucidating the potential versatile functions of VBP homologs.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains and plasmids used in the present study are listed in Supplementary Table S1. Escherichia coli strains were cultured in Luria broth at 37°C. A. tumefaciens strains were cultured in YEP medium, AB-sucrose medium or IB medium at different culture steps, as previously described (please refer to the text; Gelvin, 2006; Guo et al., 2007a). Corresponding antibiotics were added to the culture medium according to Supplementary Table S1.
DNA Manipulations
The vbp1 gene fragment was amplified using primers, vbp1-F and vbp1-R (Supplementary Table S1), and cloned into the sites of Xho I and Hind III endonucleases of the expression vector pRSET-A (The corresponding restriction sites of the vbp1 gene fragment are underlined in the primers). The resulting plasmid pR-vbp1 was transformed into E. coli BL21 (DE3) using the heat shock method (Sambrook et al., 1989). Plasmids expressing vbp2 and vbp3 genes were previously constructed (Guo et al., 2007b).
Heterogeneous Production of Three VBPs
Escherichia coli BL21 harboring the corresponding plasmid with the His-tag fused vbp gene was grown to an OD600 of approximately 0.5 and was subsequently induced by IPTG with a final concentration of 0.3 mM for 1-h to express the vbp gene. The induced E. coli BL21 cells were harvested by centrifugation (13,000 × g, 4°C, 10 min) and washed twice with phosphate-buffered solution (PBS, 10 mM, pH 7.2). Cell pellets from 10 ml of culture were lysed with 2 ml of sodium dodecyl sulfate (SDS)-loading buffer (0.1 M Tris-Cl, pH 6.8, 4% SDS, 0.1% bromophenol blue, 20% glycerol, 200 mM dithiothreitol) and stored at -20°C until further analysis.
Expression of Three vbp Genes in A. tumefaciens
To test the responses of vbp genes to the virulence induction condition, the preparation of medium for culturing A. tumefaciens and the induction culture for different A. tumefaciens strains was performed according to ref. 16. For normal virulence induction, cells of different A. tumefaciens strains were first inoculated into YEP liquid medium containing appropriate antibiotics and grown overnight at 28°C (while shaking at 250 rpm) and diluted (in the ratio of 1/100, v/v) into AB-sucrose medium containing appropriate antibiotics. After A. tumefaciens in AB-sucrose medium grew to OD600 approaching 0.8, the cells were harvested by centrifugation (∼3,800 × g, 10 min) and washed two times with IB medium. The cell pellets were resuspended in IB medium (pH 5.5) containing 100 μg/ml AS (The cell concentration in IB medium was adjusted to OD600 = 0.4) and induced at 25°C for 15–16 h, while shaking at 50 rpm. The induced Agrobacterium cells were harvested by centrifugation as described above. Agrobacterium cells from 10 ml of IB medium culture were lysed by 1 ml of SDS-loading buffer and stored at -20°C until further protein analysis. Meanwhile, another sample of Agrobacterium cells from equal volume of IB medium culture was prepared for the determination of protein concentration in the Agrobacterium sample, so that each Agrobacterium sample can be adjusted to the same protein concentration for running SDS-polyacrylamide gel electrophoresis (PAGE).
To test the effect of pH on the expression of three vbp genes, the pH in IB medium was adjusted to 5.0, 5.5, 6.0, 6.5, 7.0, and 7.5, respectively, and the AS concentration in IB medium was 100 μg/ml. Agrobacterium cells were induced in IB medium with different pH and 100 μg/ml AS at 25°C for 15–16 h. To test the effect of temperature on the expression of three vbp genes, the pH of the IB medium was adjusted to 5.5; the AS concentration of the IB medium was 100 μg/ml. Agrobacterium cells were induced in IB medium at the following five different temperatures: 19, 25, 28, 32, and 37°C. To test the effect of AS concentration on the expression of three vbp genes, four different AS concentrations, 0, 50, 100, and 200 μg/ml, were chosen to induce the Agrobacterium cells; the pH of the IB medium was adjusted to 5.5. Agrobacterium cells were induced in IB medium at different AS concentrations at 25°C for 15–16 h.
Generation of Antibodies Against Three VBPs
Polyclonal antibodies against three VBPs were supplied by Jinsirui Bio (GenScript-China, Nanjing, China). To obtain the polyclonal antibodies that can discriminate three highly homologous VBPs, peptide fragments from the variable sequences of three VBPs (Supplementary Figure S1) were artificially synthesized and used as antigens to generate polyclonal antibodies against VBPs in rabbits.
Protein Analysis
Proteins were analyzed using SDS-PAGE and Western blotting analyses. Each protein sample was adjusted to the same concentration. An equal volume of each protein sample was mixed with an equal volume of 2x loading buffer (0.1 M Tris-Cl [pH 6.8], 4% SDS, 0.1% bromophenol blue, 20% glycerol, 200 mM dithiothreitol) and incubated at 100°C for 5 min before loading. After electrophoresis, the gels were stained with Coomassie blue R-250 solution to visualize the protein bands, so that we could further confirm that every sample was loaded equal amount of protein and the protein band pattern of each sample was comparable. For Western blotting, the proteins in gels, which were run in parallel with the Coomassie blue-stained gels, were electrophoretically transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA) and detected using the BCIP/NBT alkaline phosphatase color development kit (Beyotime Corp., China) according to the procedure recommended by the manufacturer. Polyclonal antibodies against the variable sequence regions of three VBPs were used as the primary antibodies for detection.
Results
Effective Discrimination of Three VBPs
Western blotting is a routine method to assess protein expression. However, the high homology of amino acid sequences of three VBPs (Supplementary Figure S1) suggests that the polyclonal antibody generated by any full-length VBP would most likely cause cross-reaction with each of the three VBPs, resulting in non-specific results. Consequently, the peptide fragments from the variable sequence regions of the three VBPs (283–313 amino acid residues of VBP1, 163–191 amino acid residues of VBP2, and 161–189 amino acid residues of VBP3; Supplementary Figure S1) were artificially synthesized and used as antigens to generate antibodies that could differentiate between the three VBPs. As shown in Figure 1, three VBPs heterogeneously produced by E. coli could be specifically identified by their corresponding antibodies.
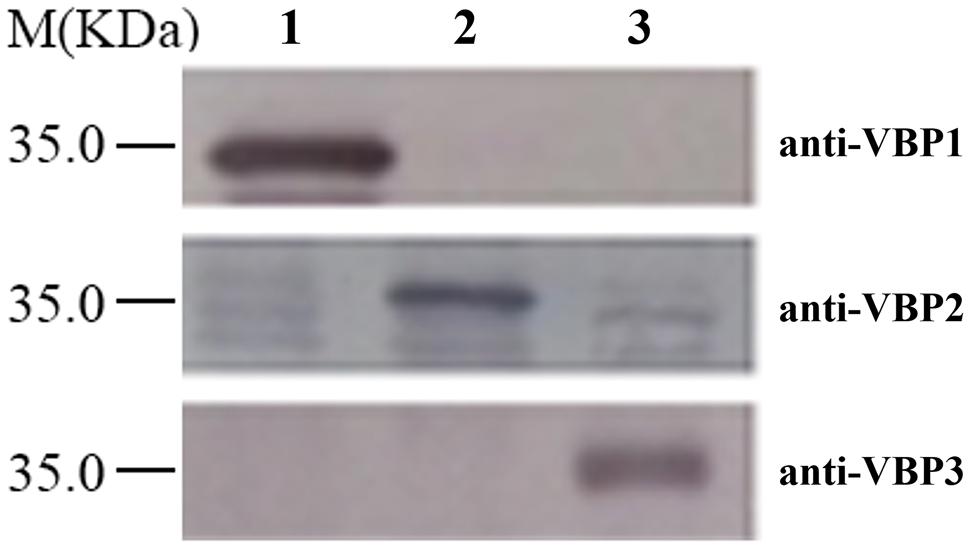
FIGURE 1. Discrimination of three virD2-binding proteins (VBPs) by Western blot assay. Crude extracts from Escherichia coli cells expressing different vbp genes were separated using SDS-PAGE in the indicated lanes and then analyzed by Western blotting using antibodies against the variable sequence regions of the three VBPs. M: Molecular weight marker. The antibodies used for each assay are listed on the right. Lane 1: crude extracts from E. coli cells expressing vbp1. Lane 2: crude extracts from E. coli cells expressing vbp2. Lane 3: crude extracts from E. coli cells expressing vbp3.
Expression of Three vbp Genes in Wild-type Strain A. tumefaciens C58 Differ in Response to vir Gene Induction Conditions
Agrobacterium tumefaciens C58 is a wild-type strain isolated from a cherry tree tumor; it was completely sequenced in 2001 (Goodner et al., 2001; Wood et al., 2001). The A. tumefaciens C58 genome contains a circular chromosome, a linear chromosome, a Ti plasmid pTiC58 and a cryptic megaplasmid pAtC58. In the genome of A. tumefaciens C58, vbp1 is located on the plasmid pAtC58, whereas vbp2 and vbp3 are located on the linear chromosome (Guo et al., 2007b). The expression of homologous genes located at different loci may be regulated by different mechanisms. This encourages the exploration of the molecular mechanisms that control the expression of three vbp genes.
To test the responses of the three vbp genes to vir gene induction conditions, A. tumefaciens C58 strain was induced by different pHs (5.0, 5.5, 6.0, 6.5, 7.0, and 7.5), AS concentrations (0, 50, 100, and 200 μg/ml) or temperatures (19, 25, 28, 32, and 37°C), and vbp gene expression in the differentially induced Agrobacterium cells were examined using Western blotting (Figure 2). The results showed that the responses to vir gene induction conditions differed among the three vbp genes. Both vbp1 and vbp3 were expressed at any of the tested pH, temperature or AS concentration conditions, whereas the vbp2 gene was expressed only under some specific induction conditions. Strikingly, Agrobacterium cells that were not induced by IB medium did not express vbp2, and the optimal pH for vbp2 expression was within the range of 6.0–7.5, which was significantly higher than that for vir gene expression. Furthermore, vir gene induction is maximal in the pH range of 5.2–6.0, whereas this pH range was not optimal for the expression of vbp2 (Figure 2A). The results in Figure 2B show that vbp2 was expressed only in the temperature range of 28–32°C, which was higher than the optimal temperature for virulence gene expression. The most effective virulence inducer AS also affected the expression of vbp2, but the effect of AS on vbp2 expression inhibited the expression of vbp2. When the concentration of AS was higher than 100 μg/ml, the expression of vbp2 gene was fully inhibited. Combining all of the results in Figure 2, we conclude that the culture conditions optimal for virulence gene induction were not very favorable to the expression of vbp2. When comparing the results in Figure 2, we found little inconsistency in the expression of vbp2. The results in Figure 2A show that vbp2 was weakly expressed under the culture condition of pH 5.5, temperature 25°C and AS concentration 100 μg/ml, but the results shown in both Figures 2B,C show that vbp2 was not expressed under the same culture condition. This slight difference for vbp2 expression suggested that one of the three factors (pH, temperature, and AS concentration) is critical to the expression of vbp2 and that any slight fluctuation of the key factor under this culture condition could result in the differential expression of vbp2. Data (Figure 2B) also showed that the expression of the vbp2 gene was inhibited at a temperature of 37°C, which was higher than the optimal temperature (28–32°C) for the vegetative growth of Agrobacterium. In conclusion, these data indicated that the expression of three vbp genes were different from each other under the above-tested induction conditions. Both vbp1 and vbp3 genes were expressed constantly, whereas the expression of vbp2 gene was selective and only occurred near a neutral pH, temperature of 28–32°C and AS concentration lower than 100 μg/ml.
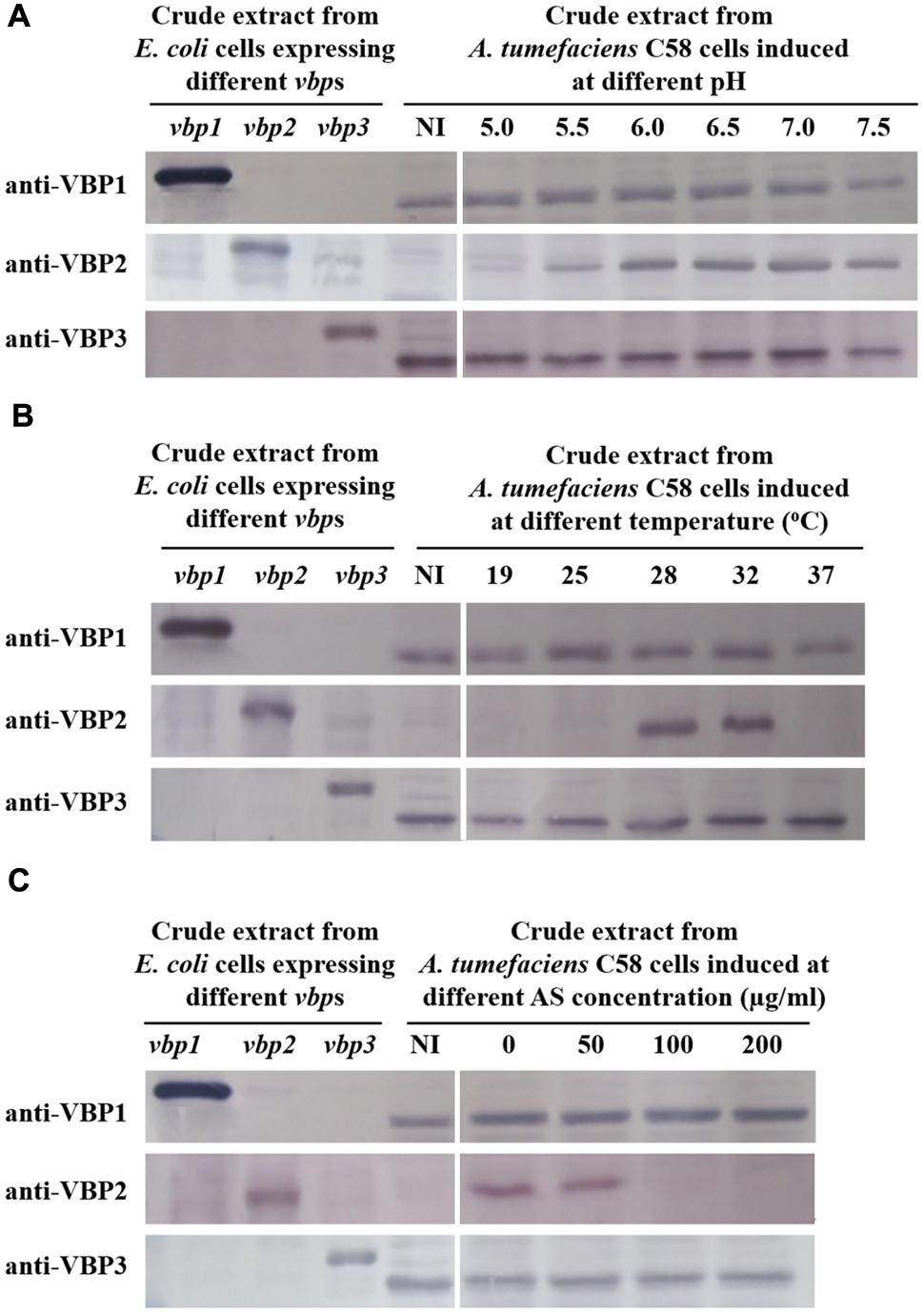
FIGURE 2. Expression of three vbp genes in Agrobacterium tumefaciens strain C58 at different pHs (A), temperatures (B) and AS concentrations (C). A. tumefaciens strain C58 cells were induced under the indicated pH, temperature or AS concentration. Crude extracts from the differentially induced C58 cells were separated using SDS-PAGE and then analyzed by Western blotting. Crude extracts from E. coli cells expressing different vbp genes with His-tag were used as positive controls. The antibodies used in this study are listed on the left. NI: crude extracts from C58 cells not induced in IB medium. (A) Effect of pH on the expression of vbps in A. tumefaciens C58. The temperature is 25°C and AS concentration is 100 μg/ml. (B) Effect of temperature on the expression of vbps in A. tumefaciens C58. The pH is 5.5 and AS concentration is 100 μg/ml. (C) Effect of AS concentration on the expression of vbps in A. tumefaciens C58. The pH is 5.5 and temperature is 25°C. Some differences in protein size of E. coli recombinant VBP2 protein (the positive control band in panel C immigrated faster than that in other panels) are likely attributable to prolonged storage.
Deletion of the vbp1 Gene Affects the Regulation of vbp2 Expression by vir Gene Induction Conditions
As the expression of the three vbp genes differed under vir gene induction conditions, we investigated whether some regulatory relationships exist among these three homologous genes. To examine the effect of vbp1 deletion on the expression of vbp2 and vbp3, Agrobacterium strain GMI9017, in which the vbp1 gene was deleted, was grown under the same induction conditions as the A. tumefaciens C58 strain.
The data showed that deletion of vbp1 did not affect the expression of vbp3. As shown in Figure 3, vbp3 in GMI9017 was expressed under all of the tested induction conditions, similar to C58. However, the expression of vbp2 displayed a greater difference in A. tumefaciens GMI9017 compared with A. tumefaciens C58. The vbp1-deleted GMI9017 strain was able to express the vbp2 gene in the pH range of 6.0–7.5, which was similar to the C58 strain, but it did not express vbp2 at the acidic pH 5.5, regardless of the temperature and AS concentration (Figures 3B,C). Unlike the GMI9017 strain, wild-type strain C58 was able to express vbp2 at pH 5.5 when grown in a temperature range of 28–32°C or with an AS concentration of less than 50 μg/ml (Figures 2B,C). The effects of pH, temperature or AS concentration on the expression of vbp2 in GMI9017 were different from those in the wild-type strain C58, indicating that deletion of vbp1 affects the responses of vbp2 expression to pH, temperature and AS concentration. In other words, vbp1 is involved in the regulation of vbp2 expression by pH, temperature and AS concentration, though the mechanism underlying this phenomenon is unclear. When grown at pH 5.5, which is not the pH optimum for vbp2 expression, GMI9017 did not express vbp2 at any tested temperature and AS concentration, suggesting that pH may be a crucial external factor in regulating the expression of vbp2. Combined with the analysis on the inconsistency of vbp2 expression at pH 5.5 in C58 (Figure 2), we speculated that pH 5.5 might be the pH critical point of vbp2 expression.
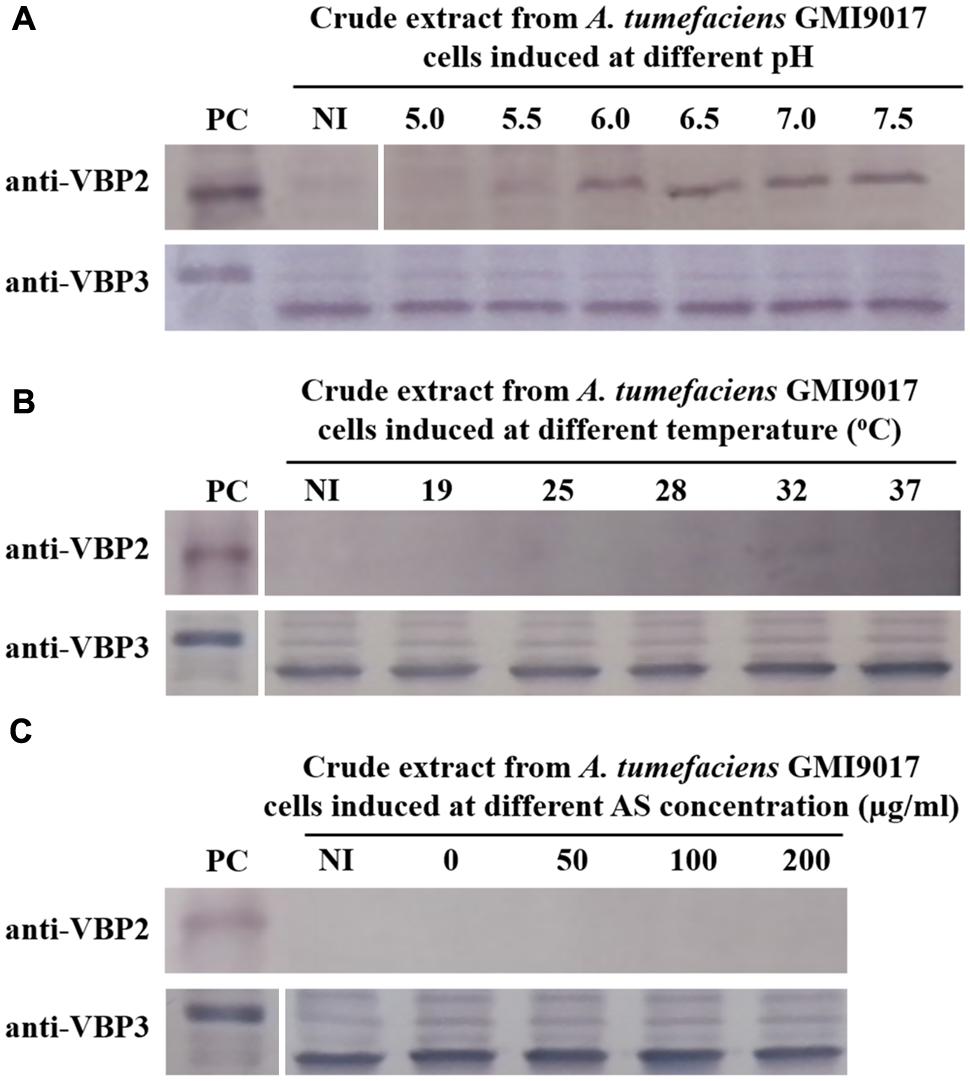
FIGURE 3. Expression of vbp2 and vbp3 in A. tumefaciens strain GMI9017 at different pHs (A), temperatures (B), and AS concentrations (C). A. tumefaciens strain GMI9017 cells were induced under the indicated pH, temperature or AS concentration. Crude extracts from the differentially induced GMI9017 cells were separated using SDS-PAGE and then analyzed by Western blotting. PC, positive control, crude extracts from E. coli cells expressing vbp2 (up) or vbp3 (down) with His-tag. The antibodies used in this study are listed on the left. NI: crude extracts from GMI9017 cells not induced in IB medium. (A) Effect of pH on the expression of vbp2 and vbp3 in A. tumefaciens GMI9017. The temperature is 25°C and AS concentration is 100 μg/ml. (B) Effect of temperature on the expression of vbp2 and vbp3 in A. tumefaciens GMI9017. The pH is 5.5 and AS concentration is 100 μg/ml. (C) Effect of AS concentration on the expression of vbp2 and vbp3 in A. tumefaciens GMI9017. The pH is 5.5 and temperature is 25°C.
To eliminate any uncontrollable error that is potentially caused by bacterial culture and induction, the wild-type strain C58 and vbp1-deleted strain GMI9017 were induced under different pHs (5.5, 6.5, and 7.5), temperatures (25 and 28°C) or AS concentrations (0 and 100 μg/ml) in parallel. As expected, the results shown in Figure 4 were consistent with the results in Figures 2 and 3. This further confirmed that vbp1 deletion affected the responses of vbp2 expression to vir gene induction conditions (pH, temperature and AS concentration).
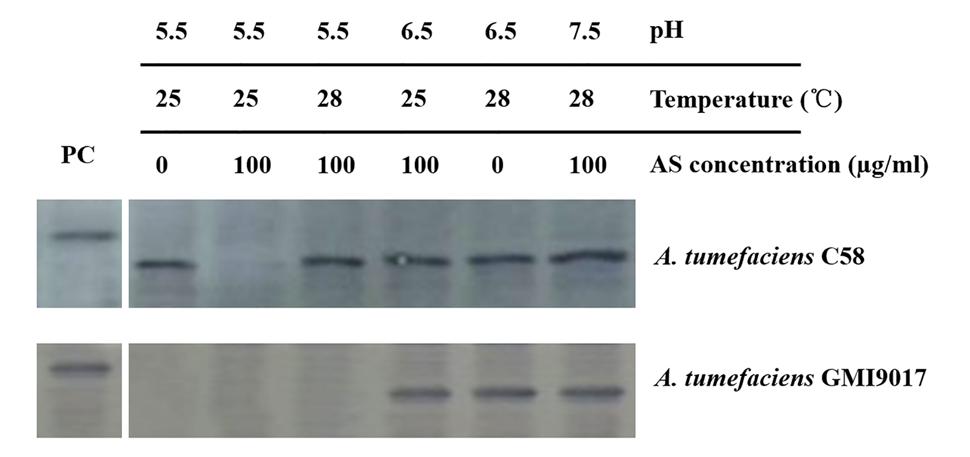
FIGURE 4. Comparison of the expression of vbp2 in A. tumefaciens C58 and A. tumefaciens GMI9017 at different pHs, temperatures and AS concentrations. Both the A. tumefaciens strain C58 cells and GMI9017 cells were induced in parallel under the indicated pH, temperature or AS concentration. Crude extracts from the differentially induced Agrobacterium cells were separated using SDS-PAGE and then analyzed by Western blotting using antibody against VBP2. PC, positive control, crude extract from E. coli cells expressing vbp2 with His-tag.
The Expression of vbp3 Gene is Not Affected by the Double Deletion of vbp1 and vbp2
Although the expression of vbp3 is not affected by the vir gene induction conditions and deletion of vbp1, we determined whether a double deletion of vbp1 and vbp2 affects the expression of vbp3. Thus, we examined the expression of vbp3 in a double-mutant strain of vbp1 and vbp2, GMV12 under different pHs, temperatures or AS concentrations. No significant change was observed in the expression level of the vbp3 gene of the GMV12 strain under all of the tested vir gene-inducing conditions (Figure 5), indicating that in addition to the virulence induction conditions, the homologous genes vbp1 and vbp2 do not regulate the expression of vbp3.
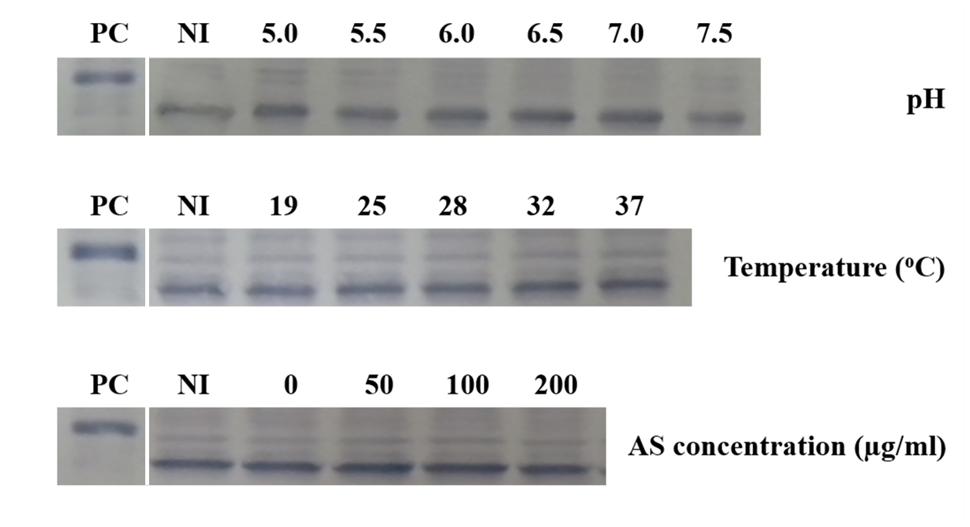
FIGURE 5. Expression of vbp3 in the double-mutant (vbp1 and vbp2) strain GMV12 at different pHs, temperatures and AS concentrations. A. tumefaciens strain GMV12 cells were induced under the indicated pH, temperature or AS concentration. Crude extracts from the differentially induced GMV12 cells were separated using SDS-PAGE and then analyzed by Western blotting with antibody against VBP3. PC, positive control, crude extract from E. coli cells expressing vbp3 with His-tag. NI: crude extracts from GMV12 cells not induced in IB medium.
Discussion
The responses of three VBP-encoding genes to pH, temperature, and AS concentration are very different from the responses of other reported virulence genes to these three virulence induction factors. According to our data, both vbp1 and vbp3 could be expressed ubiquitously, despite varying induction conditions, indicating that neither of these genes are affected by well-known virulence induction conditions. However, vbp2 showed selective expression under diverse induction conditions. Importantly, pH is a crucial factor for the expression of vbp2 and is favorable near 7.0. The vbp2 promoter region was subcloned in front of a promoterless egfp gene, and the expression of the reporting gene egfp further confirmed the response of the vbp2 promoter to pH (unpublished data). However, the pH optimal for the expression of virulence genes is approximately 5.5. In A. tumefaciens, the reported virulence genes that were induced by acidic conditions (pH 5.5) include all virulence genes located on the Ti plasmid, chromosome virulence genes (chvG and chvI), and some other genes involved in tumorigenesis, such as katA and aopB (Xu and Pan, 2000; Jia et al., 2002; Li et al., 2002; Yuan et al., 2008). The induction of vir genes by acidic conditions is regulated by a two-component system VirA-VirG. VirA perceives acidic, phenolic or monosaccharide signals, processes all these signals and finally phosphorylates VirG. Subsequently, phosphorylated VirG binds to a conserved 12-bp AT-rich sequence (vir box) in the promoter regions of all vir genes and initiates their transcription (Pazour and Das, 1990; Yuan et al., 2008). DNA sequence analysis revealed no “vir box” in the promoter regions of vbp genes, indicating that the vbp promoters cannot bind phosphorylated VirG. These findings suggest that vbp transcription is not regulated by VirA-VirG-controlling acidic induction. This silica prediction is consistent with our experimental data. In addition, a recent study on the transcriptome of A. tumefaciens in response to acidic conditions did not demonstrate that vbp transcription was significantly affected by acidic conditions and a deep sequence analysis on the promoters of all four replicons of A. tumefaciens did not identify any small RNA targeting sequence in vbp gene promoters (Yuan et al., 2008; Wilms et al., 2012). Thus, we conclude that the expression of vbp1 and vbp3 is not regulated by vir-inducing conditions and that the expression of vbp2 is regulated by a mechanism different from the reported pH-regulating mechanism. The molecular mechanism regulating the expression of vbp2 is requires further study.
VBP proteins are able to recruit the VirD2-T-DNA complex to the T4SS apparatus and are thus important for the pathogenicity of A. tumefaciens (Guo et al., 2007a). The similarity of three VBPs in the amino acid sequence is over 70% and three VBPs are redundant for the function of T-complex recruitment. However, from an evolutionary perspective, functional overlaps of homologous proteins are inherently unstable. If a protein’s function can be fully compensated for by a redundant homolog, then the mutations in the protein-encoding gene would have no effect on the phenotype. Consequently, such mutations would not remain, and the redundancy would be gradually eliminated (Lynch and Conery, 2000). Homologous genes can be obtained from duplications and lateral acquisitions. A recent study declared that the secondary chromosome (linear chromosome) of A. tumefaciens C58 originated from an intragenomic transfer from the primary chromosome (circular chromosome) to an ancestral plasmid (Slater et al., 2009). Both vbp2 and vbp3 genes are located on the linear chromosome, indicating that these two homologous genes were likely to originate from duplication, whereas, the vbp1 gene is located on the cryptic megaplasmid pAtC58, indicating that vbp1 was potentially obtained from lateral acquisition. Several homologs inhabit the same genome stably, indicating that either their functions or their cis-regulatory motifs have diversified (Gu et al., 2004). The difference between the expression of vbp2 and vbp3 genes demonstrated that the cis-regulatory motifs of these two homologs have diversified. However, the expression of vbp1 is very similar to the expression of vbp3, implying that both promoter regions of vbp1 and vbp3 may have similar cis-regulatory motifs. We used several databases1 to analyze the promoter regions of vbp1 and vbp3. The analysis showed that both promoter regions of vbp1 and vbp3 have the same transcription factor-binding motifs (data not show). The co-inhabitancy of vbp1 with its homologs in the same genome indicates that the function of vbp1 has diversified from its homologs. Therefore, these three vbp homologs may provide a good example for the research of homologous gene evolution. Recent bioinformatics studies have shown that redundant protein partners are significantly more frequently associated with the essential core proteins of protein-interaction networks (Kafri et al., 2008; Plata and Vitkup, 2014). T-DNA transfer is not necessary for the life cycle of A. tumefaciens. Nevertheless, the A. tumefaciens mutant of the vbp triple-deletion was difficult to construct (Guo et al., 2007b), demonstrating that VBP may be involved in some essential biological process as well as T-complex recruitment. Evidence that the expression of three vbp genes is not involved in the expression of other virulence genes suggests that the major function of VBP homologs is not T-complex recruitment. Taken together, these data suggest that redundant VBPs are versatile proteins and are involved in biological processes other than T-complex recruitment. It is possible that VBP1 has diversified to specify T-complex recruitment. However, the biological processes involving VBP require further investigation.
Author Contributions
MG planned the experiments. JY, MW, XZ, and ZH prepared and performed the experiments. JY, MG, and MW analyzed the data. JY and MG wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was funded by the National Natural Science Foundation of China (Grant No. 30870054, 31170073), the Natural Science Foundation of Jiangsu Province (Grant No. BK20150445), and the Natural Science Fund for Universities of Jiangsu Province (Grant No. 15KJB180020).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01379
Footnotes
Abbreviations
AS, acetosyringone; HEPN domain, higher eukaryotes and prokaryotes nucleotide-binding domain; IB medium, inducing broth medium; T4SS, type IV secretion system; Ti plasmid, tumor-inducing plasmid; VBP, VirD2-binding protein; Vir proteins, virulence proteins.
References
Baek, S. H., and Shapleigh, J. P. (2005). Expression of nitrite and nitric oxide reductases in free-living and plant-associated Agrobacterium tumefaciens C58 cells. Appl. Environ. Microbiol. 71, 4427–4436. doi: 10.1128/AEM.71.8.4427-4436.2005
Baron, C., Domke, N., Beinhofer, M., and Hapfelmeier, S. (2001). Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 183, 6852–6861. doi: 10.1128/JB.183.23.6852-6861.2001
Brookfield, J. (1992). Can genes be truly redundant? Curr. Biol. 2, 553–554. doi: 10.1016/0960-9822(92)90036-A
Chandran, V. (2013). Type IV secretion machinery: molecular architecture and function. Biochem. Soc. Trans. 41, 17–28. doi: 10.1042/BST20120332
Gao, D., Bian, X., Guo, M., Wang, J., and Zhang, X. (2013). Identification and characterization of the biochemical function of Agrobacterium T-complex-recruiting protein Atu5117. FEBS J. 280, 4865–4875. doi: 10.1111/febs.12460
Gao, R., and Lynn, D. G. (2005). Environmental pH sensing: resolving the VirA/VirG two-component system inputs for Agrobacterium pathogenesis. J. Bacteriol. 187, 2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005
Goodner, B., Hinkle, G., Gattung, S., Miller, N., Blanchard, M., Qurollo, B., et al. (2001). Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294, 2323–2328. doi: 10.1126/science.1066803
Gu, Z., Rifkin, S. A., White, K. P., and Li, W.-H. (2004). Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 36, 577–579. doi: 10.1038/ng1355
Guo, M., Bian, X., Wu, X., and Wu, M. (2011). “Agrobacterium-mediated genetic transformation: history and progress,” in Genetic Transformation, ed. M. Alvarez (Rijeka: InTech), 3–28.
Guo, M., Hou, Q., Hew, C. L., and Pan, S. Q. (2007a). Agrobacterium VirD2-binding protein is involved in tumorigenesis and redundantly encoded in conjugative transfer gene clusters. Mol. Plant Microbe Interact. 20, 1201–1212. doi: 10.1094/MPMI-20-10-1201
Guo, M., Jin, S., Sun, D., Hew, C. L., and Pan, S. Q. (2007b). Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. U.S.A. 104, 20019–20024. doi: 10.1073/pnas.0701738104
Holford, P., Hernandez, N., and Newbury, H. J. (1992). Factors influencing the efficiency of T-DNA transfer during co-cultivation of Antirrhinum majus with Agrobacterium tumefaciens. Plant Cell Rep. 11, 196–199. doi: 10.1007/BF00232532
Jia, Y. H., Li, L. P., Hou, Q. M., and Pan, S. Q. (2002). An Agrobacterium gene involved in tumorigenesis encodes an outer membrane protein exposed on the bacterial cell surface. Gene 284, 113–124. doi: 10.1016/S0378-1119(02)00385-2
Kado, C. I. (2014). Historical account on gaining insights on the mechanism of crown gall tumorigenesis induced by Agrobacterium tumefaciens. Front. Microbiol. 5:340. doi: 10.3389/fmicb.2014.00340
Kafri, R., Dahan, O., Levy, J., and Pilpel, Y. (2008). Preferential protection of protein interaction network hubs in yeast: evolved functionality of genetic redundancy. Proc. Natl. Acad. Sci. U.S.A. 105, 1243–1248. doi: 10.1073/pnas.0711043105
Li, L. P., Jia, Y. H., Hou, Q. M., Charles, T. C., Nester, E. W., and Pan, S. Q. (2002). A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. U.S.A. 99, 12369–12374. doi: 10.1073/pnas.192439499
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate Genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
McCullen, C. A., and Binns, A. N. (2006). Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 22, 101–127. doi: 10.1146/annurev.cellbio.22.011105.102022
Mira, A., Ochman, H., and Moran, N. A. (2001). Deletional bias and the evolution of bacterial genomes. Trends Genet. 17, 589–596. doi: 10.1016/S0168-9525(01)02447-7
Nowak, M. A., Boerlijst, M. C., Cooke, J., and Smith, J. M. (1997). Evolution of genetic redundancy. Nature 388, 167–171. doi: 10.1038/40618
Pacurar, D. I., Thordal-Christensen, H., Pacurar, M. L., Pamfil, D., Botez, C., and Bellini, C. (2011). Agrobacterium tumefaciens: from crown gall tumors to genetic transformation. Physiol. Mol. Plant Pathol. 76, 76–81. doi: 10.1016/j.pmpp.2011.06.004
Padavannil, A., Jobichen, C., Yang, Q. H., Seetharaman, J., Velazquez-Campoy, A., Yang, L., et al. (2014). Dimerization of VirD2 binding protein is essential for Agrobacterium induced tumor formation in plants. PLoS Pathog. 10:e1003948. doi: 10.1371/journal.ppat.1003948
Pazour, G. J., and Das, A. (1990). Characterization of the VirG binding site of Agrobacterium tumefaciens. Nucleic Acids Res. 18, 6909–6913. doi: 10.1093/nar/18.23.6909
Pitzschke, A., and Hirt, H. (2010). New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 29, 1021–1032. doi: 10.1038/emboj.2010.8
Plata, G., and Vitkup, D. (2014). Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Res. 42, 2405–2414. doi: 10.1093/nar/gkt1200
Sambrook, J., Maniatis, T., and Fritsch, E. F. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Slater, S. C., Goldman, B. S., Goodner, B., Setubal, J. C., Farrand, S. K., Nester, E. W., et al. (2009). Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191, 2501–2511. doi: 10.1128/JB.01779-08
Sudarshana, P., Mcclean, A. E., and Kluepfel, D. A. (2006). Effect of temperature and detergents on Agrobacterium tumefaciens, the causal pathogen of crown gall disease of walnut. Phytopathology 96, S111.
Wilms, I., Overloper, A., Nowrousian, M., Sharma, C. M., and Narberhaus, F. (2012). Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 9, 446–457. doi: 10.4161/rna.17212
Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323. doi: 10.1126/science.1066804
Xu, X. Q., and Pan, S. Q. (2000). An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35, 407–414. doi: 10.1046/j.1365-2958.2000.01709.x
Yuan, Z. C., Liu, P., Saenkham, P., Kerr, K., and Nester, E. W. (2008). Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J. Bacteriol. 190, 494–507. doi: 10.1128/JB.01387-07
Keywords: Agrobacterium, virD2-binding protein (VBP), virulence induction, gene expression, T-complex recruiting protein
Citation: Yang J, Wu M, Zhang X, Guo M and Huang Z (2015) Expression of Agrobacterium Homolog Genes Encoding T-complex Recruiting Protein under Virulence Induction Conditions. Front. Microbiol. 6:1379. doi: 10.3389/fmicb.2015.01379
Received: 27 January 2015; Accepted: 20 November 2015;
Published: 02 December 2015.
Edited by:
Aurelio Ciancio, Istituto per la Protezione Sostenibile delle Piante del CNR, ItalyReviewed by:
Seiichi Toki, National Institute of Agrobiological Sciences, JapanAndrea Pitzschke, University of Salzburg, Austria
Luigi Ruggiero Ceci, Institute of Biomembranes and Bioenergetics - CNR, Italy
Copyright © 2015 Yang, Wu, Zhang, Guo and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minliang Guo, guoml@yzu.edu.cn; GUO_M_L@hotmail.com
 Jing Yang
Jing Yang Meixia Wu
Meixia Wu Minliang Guo
Minliang Guo