- 1Department of Microbiology, Bose Institute, Kolkata, India
- 2Department of Microbiology, The University of Burdwan, Burdwan, India
- 3Division of Molecular Medicine, Bose Institute, Kolkata, India
- 4Department of Biotechnology, University of North Bengal, Siliguri, India
- 5Department of Chemistry, University of North Bengal, Siliguri, India
Exploration of the aquatic microbiota of several circum-neutral (6.0–8.5 pH) mid-temperature (55–85°C) springs revealed rich diversities of phylogenetic relatives of mesophilic bacteria, which surpassed the diversity of the truly-thermophilic taxa. To gain insight into the potentially-thermophilic adaptations of the phylogenetic relatives of Gram-negative mesophilic bacteria detected in culture-independent investigations we attempted pure-culture isolation by supplementing the enrichment media with 50 μg ml−1 vancomycin. Surprisingly, this Gram-positive-specific antibiotic eliminated the entire culturable-diversity of chemoorganotrophic and sulfur-chemolithotrophic bacteria present in the tested hot water inocula. Moreover, it also killed all the Gram-negative hot-spring isolates that were obtained in vancomycin-free media. Concurrent literature search for the description of Gram-negative thermophilic bacteria revealed that at least 16 of them were reportedly vancomycin-susceptible. While these data suggested that vancomycin-susceptibility could be a global trait of thermophilic bacteria (irrespective of their taxonomy, biogeography and Gram-character), MALDI Mass Spectroscopy of the peptidoglycans of a few Gram-negative thermophilic bacteria revealed that tandem alanines were present in the fourth and fifth positions of their muropeptide precursors (MPPs). Subsequent phylogenetic analyses revealed a close affinity between the D-alanine-D-alanine ligases (Ddl) of taxonomically-diverse Gram-negative thermophiles and the thermostable Ddl protein of Thermotoga maritima, which is well-known for its high specificity for alanine over other amino acids. The Ddl tree further illustrated a divergence between the homologs of Gram-negative thermophiles and mesophiles, which broadly coincided with vancomycin-susceptibility and vancomycin-resistance respectively. It was thus hypothesized that thermophilic Ddls have been evolutionarily selected to favor a D-ala-D-ala bonding. However, preference for D-ala-D-ala-terminated MPPs does not singlehandedly guarantee vancomycin susceptibility of thermophilic bacteria as the large and relatively-hydrophilic vancomycin molecule has to cross the outer membrane before it can inhibit peptidoglycan biosynthesis. Literature shows that many mesophilic Gram-negative bacteria also have D-ala-D-ala-terminated MPPs, but they still remain resistant to vancomycin due to the relative impermeability of their membranes. But the global vancomycin-susceptibility phenotype of thermophilic bacteria itself testifies that the drug crosses the membrane in all these cases. As a corollary, it seems quite likely that the outer membranes of thermophilic bacteria have some yet-unknown characteristic feature(s) that invariably ensures the entry of vancomycin.
Introduction
Relatives of phylogenetically diverse mesophilic bacteria are known to be present in hot spring waters alongside the typically thermophilic and hyperthermophilic prokaryotes (Jimenez et al., 2012; Wemheuer et al., 2013; Chan et al., 2015; Menzel et al., 2015). Our ongoing exploration of the hot water microbiota of geographically- and physicochemically-discrete hydrothermal areas of India also discovered rich diversities of Alpha-, Beta- and Gammaproteobacteria occurring in conjunction with diverse members of Bacteroidetes, Actinobacteria, Nitrospirae and Firmicutes. Interestingly, in all the explored hot water communities, diversity of such phylogenetic relatives of mesophilic bacteria surpassed the diversity of the truly thermophilic Aquificaea, Thermotogae and Thermodesulfobacteria (see Results section below). So far as truly thermophilic bacteria (growth optima above 80°C Kristjansson and Stetter, 1992) are concerned, attributes like protein architecture (Kumar et al., 2000), DNA topology (Dekker et al., 2003), membrane lipid composition (Koga, 2012) etc. have been implicated as crucial to their extremely high temperature adaptation. In contrast, little insight is available on the molecular basis of bacterial adaptation below the hyperthermophilic boundary. And whatever little information is available is confined to Gram-positive Firmicutes like Bacillus (Volker et al., 1992; Schumann, 2003; Endo et al., 2006; Sikorski et al., 2008), Geobacillus (Shih and Pan, 2011; Tripathy and Maiti, 2014; Wang et al., 2014) and Anoxybacillus (Burgess et al., 2009; Paul et al., 2012; Goh et al., 2014). This is despite the fact that taxonomically-diverse bacteria are known to grow facultatively at temperatures between 50 and 80°C (often in addition to their mesophilic growths below 50°C) (Moreira et al., 2000; Alves et al., 2003; Rainey et al., 2003). In this scenario, our laboratory intended to isolate moderately-thermophilic bacteria outside the phylum Firmicutes so that they can be used to elucidate the biology of mid-temperature adaptation in Gram-negative bacteria. So we supplemented all the isolation media with the Gram-positive-specific antibiotic vancomycin (Watanakunakorn, 1984; Lundstrom and Sobel, 2000; Nailor and Sobel, 2009) in order to potentially eliminate Firmicutes and allow Gram-negative taxa to appear in the isolation plates. Interestingly, addition of vancomycin to the culture media caused complete destruction of all the tested hot water inocula at 55 as well as 30°C. However, parallel inoculation of vancomycin-free media yielded flocculent growth of mixed consortia at either temperature. This phenomenon held true for all the hot water inocula that were tested from various geothermal districts of India. Concurrently, all the Gram-negative bacteria that we could isolate in vancomycin-free media (plus their sibling strains that had been isolated earlier from other parts of the world) were found to be susceptible to vancomycin. Again, when we scrutinized the descriptions of Gram-negative thermophilic bacteria in the literature we found that at least 16 of them had been reported as vancomycin-susceptible, while a large majority had never been tested for this phenotype. All these observations collectively suggested that vancomycin-susceptibility could be a global trait of thermophilic bacteria, irrespective of their taxonomy, biogeography and Gram-character. Subsequently, MALDI Mass Spectroscopic (MS) analysis of the peptidoglycans of a few Gram-negative thermophilic bacteria revealed the occurrence of tandem alanines in the fourth and fifth positions of their muro-pentapetide precursors (MPPs), which again was the reported target of vancomycin-binding in Gram-positive Firmicutes (Barna and Williams, 1984; Reynolds, 1989). This proved that vancomycin killed Gram-negative bacteria by the same mode of action as that which kills Gram-positive bacteria. In conclusion, maximum likelihood (ML) phylogeny of the D-alanine-D-alanine ligase protein was reconstructed to elucidate the ecological and evolutionary basis of the wide spread of vancomycin-susceptibility in Gram-negative thermophilic bacteria.
Materials and Methods
Sampling
For any given venting point, batches of 500 ml freshly-discharged hot water were passed through sterile 0.22 μm filters (4.7 cm radius). Filters were immediately put into cryovials containing 5 ml of either 50 mM:50 mM Tris:EDTA (TE, pH 7.8) or 15% Glycerol in 0.9% NaCl and transferred to the lab in dry ice. Upon reaching the lab the cryovials were stored at −20°C until further use, which was anyhow within 2 weeks from sampling. While the filters suspended in TE were used for the isolation of total environmental DNA, those put in glycerol stocks were used in various culture-based experiments (one filter per 80 ml of any medium).
Isolation of Environmental DNA
A given TE-suspended filter was cut into small pieces with sterile scissors and put back to the original cryovial. The vial was vortexed vigorously for 30 min, following which the filter shreds were discarded and the 5 ml TE distributed to five 1.5 ml microfuge tubes. All the five tubes were centrifuged at 10,800 g for 30 min, following which 900 μl TE was discarded from the top of each tube. The 100 μl TE remaining in each tube was vortexed vigorously for 15 min and the contents of all five tubes were pooled up into one microfuge tube. This pooled up 500 μl TE was again centrifuged at 10,800 g for 30 min, following which 400 μl was discarded from the top. The remaining 100 μl was presumably a suspension of all microbial cells recoverable from the 500 ml hot water sample in question. DNA was isolated from this 100 μl purported cell suspension by the QIAamp DNA Mini Kit (QIAGEN) following manufacturer's protocol.
Media, Culture Conditions, and Isolation of Cultured-Metagenomes
Chemoorganoheterotrophic growth experiments were performed in oligotrophic R2A medium while chemolithoautotrophic growth was checked in a modified basal mineral salt (MMS) medium [1 gm l−1 NH4Cl, 2 gm l−1 K2HPO4, 0.75 gm l−1 KH2PO4, 0.5 gm l−1 MgSO4 and 2 ml l−1 trace metals solution (Vishniac and Santer, 1957)] supplemented with 10 mM thiosulfate (MMST). Culturable diversity of chemoorganoheterotrophs or chemolithoautotrophs in a given hot water community was explored by incubating the glycerol stocked filters (at 55 as well as 30°C) in R2A or MMST broth respectively. From the resultant mixed consortia pure cultures were isolated by dilution plating 5 ml of cell suspensions in respective solid media. Vancomycin (50 μg ml−1) response of cultured consortia, or pure culture isolates, was tested in relevant media types at 55 as well as 30°C. For the isolation of metagenomes from the consortia cultured in R2A or MMST, 5 ml cell suspensions were harvested at appropriate time points and subjected to DNA preparation using the QIAamp DNA Mini Kit (QIAGEN) following manufacturer's protocol.
Amplification of 16S rRNA Gene Fragments and Sequencing by Ion PGM
V3 regions of all potential bacterial 16S rRNA genes present in an environmental or cultured metagenome were PCR-amplified by the “Fusion Primer Protocol” using Bacteria-specific universal oligonucleotides. In order to enable tandem sequencing of multiple PCR product pools on a single Ion chip a DNA template in question was subjected to PCR using a 16S forward primer prefixed with an Ion Torrent adapter and a unique sample-specific barcode or multiplex identifier in the following order in the 5′ to 3′ direction: (i) a 26-mer A-linker followed by a 4-mer A-linker key (bases represented in bold fonts), common for all sample primers, (ii) a 10-mer barcode unique to each sample primer followed by a common 3-mer barcode adaptor (all marked as stars), and finally (iii) the relevant domain-specific universal forward primer in its 5′ to 3′ direction (underlined bases). Reverse primers, in their turn, had (i) a common trP1 adapter (bases represented in italics), followed by (ii) the relevant domain-specific universal reverse primer in its 5′ to 3′ direction (underlined bases). Thus, V3 portions of all bacterial 16S rRNA genes present in a sample DNA were amplified using the forward primer 5′ – CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG *** *** *** *** *CC TAC GGG AGG CAG CAG – 3′ and the reverse primer 5'-CCT CTC TAT GGG CAG TCG GTG ATA TTA CCG CGG CTG CTG G - 3′ (where underlined portions represent the universal primers 341f and 515r respectively).
Each 50 μl PCR reaction contained 10 μl template (corresponding to ~100 ng metagenomic DNA), 5 μl 10X KOD DNA polymerase buffer, 5 μl dNTP (0.25 mM each), 2 μl MgCl2 (25 mM), 1.5 μl (3%) DMSO, 3 μl each of the forward and reverse primer (0.3 μM each), 19.5 μl dH2O and 1 μl KOD hot start polymerase enzyme (Novagen, USA). PCR products were amplified for 30 cycles as follows: 94°C for 15 s, 65°C for 30 s and 68°C for 60 s. After amplification, all PCR products were electrophoresed on 2.5% w/v agarose gel, purified by size selection, and adjusted to final concentrations of 10 ng μl−1 using molecular grade water. PCR products from multiple samples were pooled up at equal concentrations for Ion PGM sequencing.
Before Ion PGM sequencing, size distribution and DNA concentration in the pooled-up amplicon mixture was examined using a Bioanalyzer 2100 (Agilent Technologies, USA). The mixed sample was adjusted to a final concentration of 26 pM and attached to the surface of Ion Sphere Particles (ISPs) using an Ion Onetouch 200 Template kit (Life Technologies, USA) according to the manufacturer's instructions. Manual enrichment of the resulting ISPs resulted in >95% templated-ISPs, which were then sequenced on Ion 316 Chips using the Ion PGM (Ion Express Template 200 chemistry) for 500 flows that gives an expected average read length of >220 bp. Sequencing was done upto such depths which ensured plateaus in rarefaction curves. Post sequencing, individual sequence reads were filtered by the PGM software to remove low quality and polyclonal sequences. Sequences matching the PGM 3′ adaptor were also automatically trimmed. All the data quality-filtered on the PGM were exported as fastq files for downstream applications.
The sequence files generated from PCR upon environmental DNA samples were deposited to the NCBI Sequence Read Archive (SRA) with the Run and BioProject accession numbers cited in Table 1. Similarly, sequence files obtained by PCR upon cultured metagenomes were deposited with the Run accession numbers cited in Table 2 and Supplementary Table S15 under the same BioProject as that used for sequences generated from environmental DNAs.
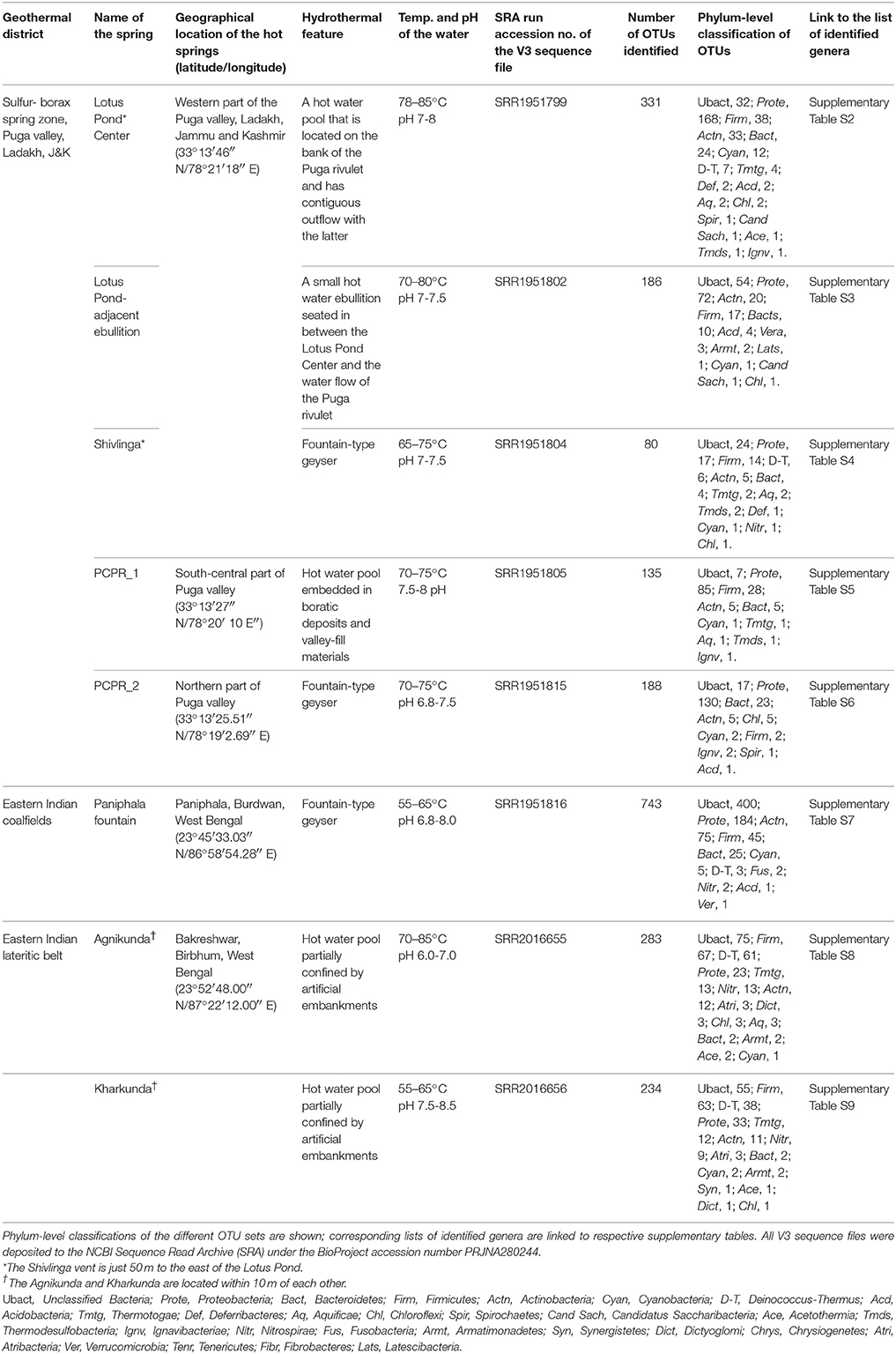
Table 1. Bacterial taxonomic diversity detected in the hot waters of various geothermal vents surveyed in this study.

Table 2. Bacterial taxonomic diversity detected after incubating vent water inocula in R2A or MMST medium.
Taxonomic Diversity Estimation
Raw V3 region-specific reads were first filtered for high quality value (QV 20) and length threshold of 100 bp. Selected reads were then converted to fasta from fastq using Fastx_toolkit (v0.0.13.2). Operational taxonomic units (OTUs) were created at 97% identity level using the various modules of the UPARSE (Edgar, 2013) OTU clustering methods. Singletons were discarded from the data sets. A Perl script was used to get the ACE, Chao, Shannon and Simpson's diversity and abundance indices. Rarefaction analysis was done using a module of R-package and graph was created for reads taking part in OTU formation vs. number of OTUs formed. All consensus sequences generated for a given dataset were taxonomically classified with the help of the “RDP Classifier” located at http://rdp.cme.msu.edu/classifier/classifier.jsp.
Peptidoglycan Isolation
Peptidoglycans of various bacterial strains were prepared by methods described earlier (Komagata and Suzuki, 1988; Schumann, 2011). For vancomycin-untreated cells, peptidoglycan was prepared directly from mid-log phase cultures (OD600 = 0.6), whereas for vancomycin-treated counterparts the antibiotic (300 μg ml−1) was added to actively growing cultures when their OD600 was 0.3–0.4. The latter sets were then incubated for four more hours before cells were harvested.
Cells were collected by centrifugation and the cell pellet (2 gm wet weight) was washed twice with 5 ml phosphate buffer (0.05 M, pH 7.2). Then the cell pellet was resuspended in 6 ml 0.05 M phosphate buffer (pH 7.2) and cells were disrupted by sonication on ice using 20 s pulse for 4 times. The cell lysate was centrifuged at 1800 g for 10 min and the supernatant was transferred to a fresh centrifuge tube. It was then centrifuged at 12,000 g for 1 h. Supernatant was discarded and pellet was resuspended in 5 ml phosphate buffer. 1 ml of 5% sodium dodecyl sulfate was added and incubated at 100°C for 40 min, followed by centrifugation at 12,000 g for 30 min at 30°C. Then the pellet was washed four to five times with 5 ml 60°C distilled water. It was then washed with 5 ml 0.05 M phosphate buffer (pH 7.6). The pellet was then resuspended in 2 ml phosphate buffer (0.05 M, pH 7.6) and 100 μl pronase E (1 mg ml−1) was added. The soup was then incubated at 37°C for 2 h. Pellet was collected by centrifugation at 12,000 g for 30 min. It was further washed twice 2 ml phosphate buffer (0.05 M, pH 7.6). After that, pellet was resuspended in 2 ml of 5% TCA (Trichloroacetic Acid) and boiled at 100°C for 20 min. The suspension was cooled at room temperature and transferred to glass centrifuge tubes. It was then centrifuged at 12,000 g for 30 min. Pellet was further washed thrice with 2 ml phosphate buffer (0.05 M, pH 7.6), once each with 2 ml ethanol (95%) and 2 ml diethyl ether (99%), and finally air dried at 60°C for 3 h before further analysis.
MALDI-MS
Extracted peptidoglycans were digested with lysozyme (40 mg ml−1) for 2 h at 37°C, following which deactivation was done for 20 min at 70°C. Digested products were lyophilized, resuspended in 100 μl 99% methanol, and directly used for MALDI-MS without any more purification. DHA was used as the MALDI matrix. MALDI-MS was carried out using an AutoFlex II tandem time of flight (TOF/TOF) MALDI-mass spectrometer (Bruker Daltonics) equipped with a pulsed N2 laser (λ-337 nm, 50 Hz). The machine was calibrated for reflector mode mass spectra using a mixture of standard peptides (having mass 750 to 3150) in the positive ion mode. MS spectra were analyzed using the Flex Analysis software V2.4.
Phylogeny
ML trees were constructed using MEGA6 (Tamura et al., 2013) plus PhyML (Guindon et al., 2010). The best substitution model used for likelihood analysis (general time reversible and gamma) was selected by Bayesian as well as corrected Akaike information criteria. After the starting NJ tree was obtained heuristic searches for likelihood were performed using the Nearest–Neighbor–Interchange as well as Close–Neighbor–Interchange branch swapping algorithms.
Results and Discussion
Rich Diversity of Phylogenetic Relatives of Mesophilic Bacteria in Circumneutral Hot Springs
Over the past few years we have investigated the taxonomic diversity (species richness) of the aquatic bacterial community of several circumneutral hot springs of Northern and Eastern India by analyzing amplified 16S rRNA gene fragments. V3 regions of all potential bacterial 16S rRNA genes present in the total environmental DNA isolated from thermal water samples were PCR-amplified using Bacteria–specific primers and sequenced by Ion PGM. Since many of the explored springs had similar taxonomic compositions only eight representative communities (five from Northern and three from Eastern India) are described in this paper. Statistics of clustering operational taxonomic units (OTUs) from the Ion PGM readsets is given in Supplementary Table S1. Table 1 shows the spring identities and the phylum-level classifications of their respective OTU sets. List of genera identified in these eight OTU sets are separately given in Supplementary Table S2 through Supplementary Table S9.
Notwithstanding the discrete locations and physicochemical characters of the studied springs, taxonomic structure of their hot water communities was unified by the occurrence of several such bacterial taxa that are otherwise unexpected in high-temperature habitats. While most of the communities encompassed maximum OTUs from the Alpha, Beta, and Gamma subclasses of Proteobacteria, the typical thermophilic phyla Aquificae, Thermotogae and Thermodesulfobacteria had very few OTUs affiliated to them (Table 1). The only exceptions to this trend were the Agnikunda and Kharkunda vents where taxonomic diversity was dominated by Firmicutes. Concurrent to these observations, a close inspection of the lists of genera identified in the described hot water communities revealed several such bacteria that have no report of laboratory growth above 45°C. While the hot water community of Agnikunda encompassed the lowest proportion (~52%) of such apparently-mesophilic genera, that of the Paniphala vent had the highest percentage (89%). So far as the actual numbers were concerned, the Lotus Pond Center (84) had the maximum count of supposedly-mesophilic genera, followed by Paniphala (81). In contrast, PCPR 1 and Shivlinga had the lowest number (11 and 15 respectively) of such genera, presumably because these vents as such had the lowest overall count of OTUs and genera.
In this scenario we sought to know how this large variety of purportedly-mesophilic genera survived in these high temperature habitats. First it was imperative to check whether they could at all grow at high temperatures or were only thermo-enduring entities. Alternatively, it was also plausible that many of them were stochastically introduced into these habitats in recent times and were not at all equipped to cope with thermal stress. Since only pure culture isolates could answer these queries we attempted to get the same in chemoorganoheterotrophic (R2A) as well as chemolithoautotrophic [modified minimal salts supplemented with thiosulfate, MMST (Ghosh and Roy, 2006)] media at various incubation temperatures between 30 and 70°C.
Preponderance of Firmicutes in R2A Plates
Strains related to Bacillus, Geobacillus, Anoxybacillus and Brevibacillus crowded all the R2A isolation plates incubated at temperatures ≥50°C. Consequently, very few non-Firmicutes (e.g., strains of known moderate-thermophiles like Thermomonas, Porphyrobacter, Meiothermus etc.) could be obtained even after several rounds of isolation from the various inoculum samples. May be, during high-temperature growth in R2A the Firmicutes out-competed other moderate-thermophiles by virtue of their metabolic versatility, faster growth rate and better thermal adaptations. So far as isolation in MMST was concerned, all the tested inocula acidified the enrichment broths and produced sulfate with concomitant disappearance of thiosulfate. However, only a few moderately-thermophilic Proteobacteria (e.g., Thermithiobacillus tepidarius) kept recurring when the enriched broths were plated in MMST-agar to obtain single colonies. In addition, a host of such apparently thermo-enduring Paracoccus strains got isolated (in MMST at 37°C) that despite failing to grow above 45°C did not lose substantial viability (measured by drop in CFU count of experimental cultures) even at 60°C over an exposure period of ~4–6 h. Now, our prime objective was to isolate thermophilic siblings of the reportedly-mesophilic Gram-negative bacteria listed in Supplementary Table S2 through Supplementary Table S9. Although success of such undertakings depended on the actual cultivability of the unexpected bacteria in question, failures could never ascertain whether the negative results were due to short-comings or intrinsic-limitations of the isolation procedures or whether the Gram-negative mesophiles surfacing in diversity analyses were not thermally-adapted at all. So we initiated another round of isolation by supplementing all media types with vancomycin so that Firmicutes were eliminated and Gram-negative taxa got a better chance to appear in the isolation plates.
Eradication of Culturable Diversity by Vancomycin
Supplementing R2A as well as MMST with 50 μg ml−1 vancomycin caused complete destruction of the corresponding culturable-diversities of all the explored hot water communities. However, to keep it brief, the data from three North Indian and two East Indian vents will be presented in detail. As shown in Table 2, zero or near-zero OD600 values were registered for all spent vancomycin-containing media after prolonged incubation at 55 as well as 30°C. No production of sulfuric acid in spent vancomycin-containing MMST media was taken as the main evidence of destruction of the culturable chemolithotrophic diversity. Concurrently-recorded CFU counts were always < 102 ml−1 of the spent media. This appeared to tally with spontaneous mutation rates (John et al., 1998) as parallel vancomycin-free cultures yielded rich growth of mixed consortia at 55 as well as 30°C. For all the tested inocula, OD600 of vancomycin-free R2A reached 0.8 (CFU counts ~108 ml−1 of spent medium) within 12–16 h of incubation, while that of vancomycin-free MMST reached 0.3–0.4 (CFU counts 104–105 ml−1 of spent medium) with concomitant acidification of spent media within 16–72 h of incubation. With regard to growth in MMST it must be appreciated that the observed OD600 values were not exclusively due to sulfur-oxidizing chemolithotrophs. Even though their active presence was evidenced by sulfuric acid production, the final cell masses recovered from spent MMST media were quite likely to contain organoheterotrophic secondary consumers.
The above data clearly implied that all the bacteria growing in the two vancomycin-free media types, irrespective of their taxonomic identity and Gram-property, were susceptible to this so-called Gram-positive-specific antibiotic. To know the taxonomic identity of these cultured consortia we isolated their total genomes, amplified the V3 regions of all 16S rRNA genes present therein, and sequenced the amplicon pools by Ion PGM. The obtained V3 readsets were analyzed by OTU-clustering, statistics of which are given in Supplementary Table S1. Table 2 shows the phylum-level classification of the respective OTU sets.
Corroborating the outcome of the isolation experiments, almost all the vancomycin-free cultured consortia (irrespective of the media type) encompassed maximum OTUs from Firmicutes (albeit after the unclassifiable ones). Nevertheless, Actinobacteria, Deinococcus-Thermus and Proteobacteria had maximum OTUs in the consortia obtained by incubating the Shivlinga Vent water in R2A at 30 and 55°C, and MMST at 30°C respectively. The most interesting attribute of these datasets was the affiliation of a large number of OTUs to Gram-negative phyla such as Proteobacteria, Bacteroidetes, Deinococcus-Thermus, Thermodesulfobacteria, Acidobacteria, Cyanobacteria, and also Actinobacteria, which has Gram-negative as well as positive members. The lists of genera identified in these OTU sets (Supplementary Table S10 through Supplementary Table S14) also showed that the vancomycin-free cultured consortia encompassed at least one (for Lotus Pond Center inoculum incubated in MMST at 30°C) to at most 46 (for Shivlinga inoculum incubated in MMST at 30°C) Gram-negative genera. Only when the Lotus Pond Center inoculum was incubated in R2A at 30°C, or the Agnikunda inoculum was incubated in R2A at 30 or 55°C, the resultant consortium encompassed no Gram-negative genus.
The above data summarily indicated that the Gram-negative components of hot water communities were as susceptible to vancomycin as their Gram-positive counterparts. The wide taxonomic as well as geographic spread of these community analyses further hinted that the association between vancomycin-susceptibility and thermal adaptation could well be a global phenomenon. Significantly again, this comprehensive vancomycin-susceptibility of all taxonomic- and Gram-types was not detected when the above experiments were repeated with mesophilic (in situ temperature 30°C) lake water inocula (see Supplementary Tables S15, S15A through Supplementary Table S15D). This buttressed our assumption that the phenomenon was indeed an exclusive hallmark of hydrothermal communities. Subsequently, to cross-examine this supposition, vancomycin challenge was extended to all the hot spring isolates that were there at our disposal. On top of that we also scrutinized the species descriptions of several Gram-negative thermophilic bacteria in the literature to see whether any information existed about their vancomycin response.
Vancomycin-Susceptibility is Widespread among Gram-Negative Thermophiles
All the current hot-spring isolates died (showing near-zero CFU counts) in the presence of 50 μg ml−1 vancomycin at both higher and lower incubation temperatures. Susceptible phenotype was also exhibited by the tested siblings strains of the present isolates reported from other parts of the world. Although the strains in question took negative Gram stain, their response to vancomycin challenge was exactly same as that of the Bacillus controls. On the other hand, mesophilic Gram-negative Alpha-, Beta- and Gammaproteobacteria like Pseudaminobacter salicylatoxidans KCT001, Advenella kashmirensis WT001 and E. coli BL21 did not show any growth perturbation in the presence of vancomycin. Relevant growth experiment data for some of these organisms are shown in Table 3.
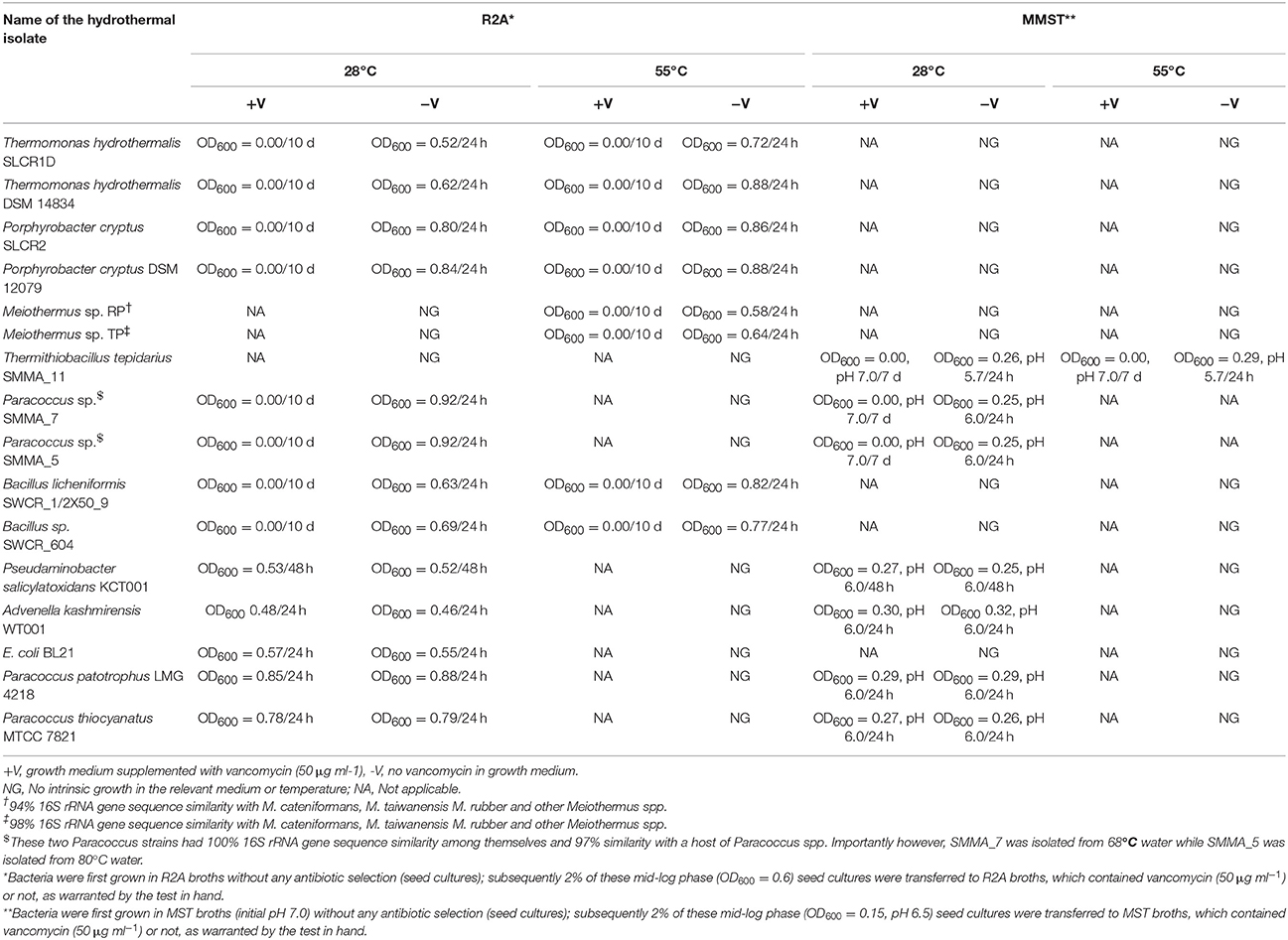
Table 3. Pure culture isolates of Gram negative bacteria tested in the present study for their vancomycin response.
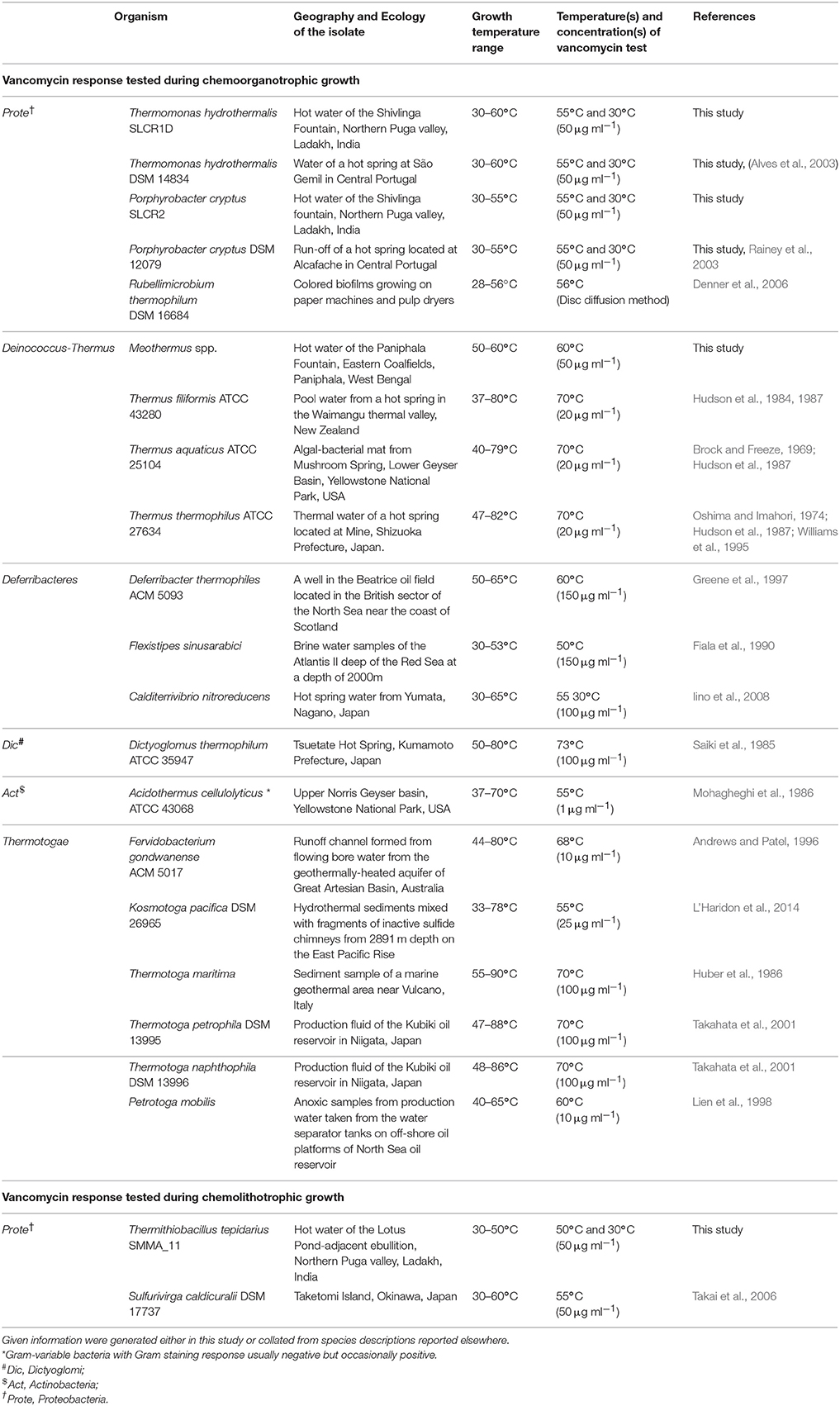
The most remarkable of all these observations was the susceptibility of the thermo-enduring Paracoccus isolates. They were all killed by 50 μg ml−1 vancomycin at the same time as standard strains of Paracoccus pantotrophus (LMG 4218) and Paracoccus thiocyanatus (MTCC 7821) isolated from various mesophilic habitats remained unaffected (Table 3). This evoked the conjecture that vancomycin-susceptibility of Gram-negative bacteria could be a function of high temperature exposure rather than actual growth efficiency at elevated temperatures. In tandem with these experiments we also scrutinized the descriptions of Gram-negative thermophilic bacteria in the literature and found at least 16 such species that were reported as susceptible to vancomycin (Table 4). For obvious reasons, Gram-negative bacteria are seldom tested for vancomycin response; hence scarcity of such data was understandable. In such a scenario, the taxonomy and biogeography of the bacteria listed in Table 4 could be considered diverse enough to implicate vancomycin susceptibility as a global trait of Gram-negative thermophiles.
Vancomycin Susceptibility of Gram-Negative Thermophiles Stems from Di-Alanine-Terminated MPPs
In view of the above data it was deemed imperative to know whether the typical vancomycin mode of action that works against Gram-positive bacteria (Healy et al., 2000) was also instrumental in the killing of Gram-negative thermophiles. It may be recalled that transpeptidase enzymes cross-link MPPs to provide structural integrity and strength to the peptidoglycan layer of the cell wall (Waxman and Strominger, 1983). Gram-positive bacteria generally have tandem D-ala-D-ala residues at the N-terminal end of their MPPs. These two amino acid residues afford the high-affinity target of vancomycin (Perkins and Nieto, 1974; Healy et al., 2000), which out-competes the transpeptidases in the race for binding with the substrate. In the process peptidoglycan cross-linking fails, causing the cell wall to eventually disintegrate and collapse under extraneous pressure. In contrast, a majority of Gram-negative bacteria as well as vancomycin-resistant Gram-positive bacteria have the last D-alanine replaced by D/L-lactate/lysine/glycine etc, and thus remain unscathed by vancomycin (Reynolds, 1989; Arthur et al., 1996; Courvalin, 2006). However, many others do have tandem D-ala-D-ala residues in their MPPs but impermeability of their outer membrane toward vancomycin confers them resistance to this drug. As such, the normal mechanism of vancomycin resistance starts with a permeability barrier (Shlaes et al., 1989) even as its mechanism of action is invariably rendered through binding of the terminal D-ala-D-ala residues resulting in steric hindrance of further addition to the growing peptidoglycan chain (Perkins and Nieto, 1974).
Peptidoglycan was extracted from vancomycin-treated as well as -untreated cells of the current Gram-negative thermophilic isolates and analyzed by MALDI MS. The resultant data suggested that the organisms in question were all susceptible to this antibiotic in the same mechanistic way as the Gram-positive Firmicutes (that is on account of having tandem alanines in the N-termini of their MPPs). As such, details of the experiment with Thermomonas hydrothermalis SLCR_1D is described below.
Peptidoglycan was extracted from T. hydrothermalis SLCR_1D, and analyzed in comparison with the isolates Bacillus licheniformis SWCR_1/2X50_9 and A. kashmirensis WT 001. Extraction was done in such a way that MPPs were represented abundantly. When the OD600 of a given culture reached 0.3–0.4, excess vancomycin (300 μg ml−1) was added to the growth media, following which they were incubated for another 4 h before cells were harvested for peptidoglycan extraction. Vancomycin-untreated cultures were passed through the same steps as above, except antibiotic addition. It was reasoned that if a test organism actually afforded the typical vancomycin target in its MPPs, this treatment would stop the cross-linking of its peptidoglycan backbone and enrich the precursors in the extract. As a final point, the extracts were digested with the muramidase, lysozyme to obtain peptidoglycan mono- or oligo-mers suitable for MALDI MS detection.
MALDI mass spectra of the SLCR_1D peptidoglycan extracted after incubation in the presence as well as absence of vancomycin encompassed a common peak (m/z ~1008) [M-3H]+ attributable to the monomeric precursor GlcNAc-MurNAc-ala-glu-A2pm-ala-ala (having a calculated mass average of 1011.00; Figures 1A,B). On the other hand, a unique peak of m/z 1864.782 characterized the spectrum of SLCR_1D peptidoglycan extracted after incubation without vancomycin (Figure 1B). This peak matched the calculated mass average (1861.83) of a potentially protonated [M+3H]+ form of the “tetrapeptide-tetrapeptide cross-linked peptidoglycan dimer.” Detectable fragmented ion peaks, or for that matter peaks arising from various modifications of the above two molecular ions of basic peptidoglycan moieties are accounted for in Table 5.
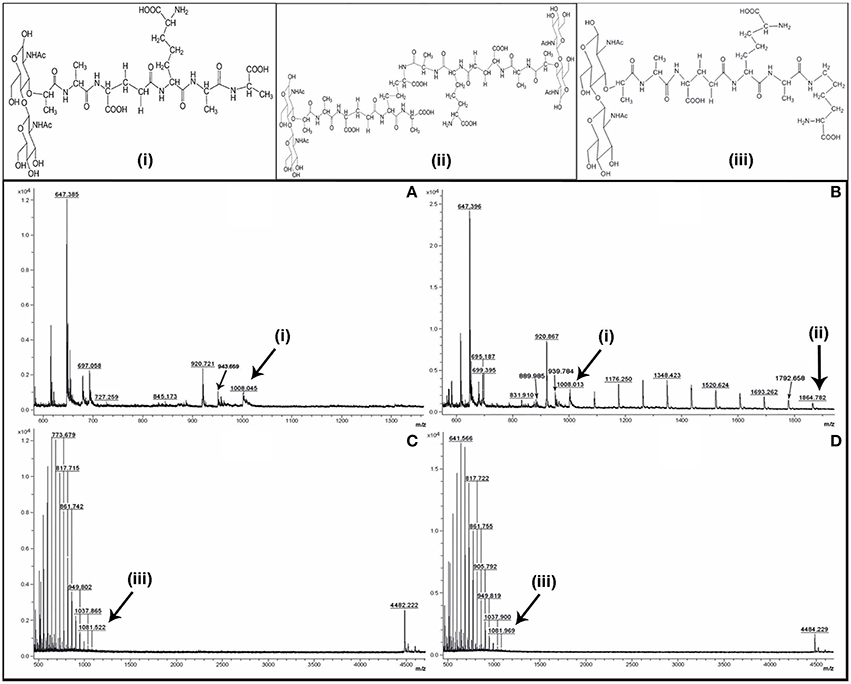
Figure 1. Positive reflector ion MALDI mass spectrum of the digested peptidoglycan fragments of vancomycin-treated (A,C) and -untreated (B,D) cells of T. hydrothermalis SLCR_1D (Upper panels A,B) and A. kashmirensis WT 001 (Lower panels C,D). Structure (i) corresponds to muro-pentapeptide precursor having terminal alanine-alanine dipeptide, (ii) corresponds to tetrapeptide-tetrapeptide cross linking, whereas structure (iii) represents muro-pentapeptide precursor having terminal alanine-lysine dipeptide.
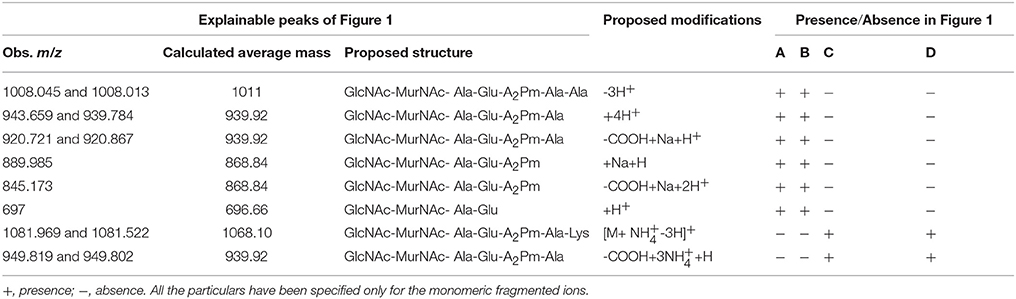
Table 5. Observed and calculated average m/z's along with their proposed structures and proposed modifications as present in the Figure 1.
The most significant information revealed by the Figures 1A,B spectra was the presence of two tandem alanine residues at the N-terminal end of the MPP of SLCR_1D. This unambiguously accounted for the basis of vancomycin susceptibility of this Gram-negative organism. On the flip side, the absence of the “tetrapeptide-tetrapeptide cross-linked peptidoglycan dimer” in the Figure 1A confirmed that vancomycin indeed rendered total or near-total inhibition of peptidoglycan cross-linking in SLCR_1D, leading to the complete killing of this bacterium. Concurrent to these suppositions, mass spectra identical to those shown in Figures 1A,B were also obtained for B. licheniformis peptidoglycans extracted after incubation in the presence and absence of vancomycin respectively (data not shown).
Unlike T. hydrothermalis, A. kashmirensis yielded identical mass spectra for peptidoglycans extracted after incubation with or without vancomycin (compare Figures 1C,D). Out of the series of common peaks present in the two spectra, the one having m/z 1081.776 could be attributable [M+ NH-3H]+ to the monomeric MPP, GlcNAc-MurNAc-ala-glu-A2pm-ala-lys, which has a calculated mass average of 1068.10. Notably, two mass peaks of m/z 4482.22 and 4484.23 present in Figures 1C,D respectively could not be explained satisfactorily. Presumably, these peaks represented some higher polymer of peptidoglycan. Fragmented ion peaks as well as peaks arising from other modifications of the aforesaid molecular ions are accounted for in Table 5.
Concluding Remarks
All the present observations unanimously indicated that vancomycin-susceptibility was widespread in thermophilic (and perhaps also thermo-enduring) bacteria, irrespective of their taxonomic affiliation and Gram phenotype. It was also confirmed that this susceptibility stemmed from the predominance of alanine-terminated MPPs in the concerned organisms. In this regard it would be pertinent to recall that the D-ala-D-ala ligase (Ddl) of Thermotoga maritima (TmDdl), which can grow in the laboratory between 55 and 90°C, has been previously shown to retain its activity over a wide range of temperature (10–90°C; Sato et al., 2006). Specific-activity of this thermostable enzyme is reportedly much higher for alanine than for other amino acids (Sato et al., 2006). Interestingly, in blastp, the TmDdl (NP_228072) showed closest homology with Ddl homologs from Gram-negative thermophilic bacteria as diverse as species of Thermotoga, Kosmotoga, Petrotoga, Mesotoga, Fervidobacterium (phylum Thermotogae); Flexistipes, Deferribacter, Calditerrivibrio (phylum Deferribacteres); Aquifex, Hydrogenivirga, Persephonella, Sulfurihydrogenibium (phylum Aquificae); Methylacidiphilum (Verrucomicrobia); Thermodesulfovibrio thiophilus (Nitrospirae); Dictyoglomus (Dictyoglomi) etc. ML phylogeny not only reiterated the close affinity of these homologs but also pointed out their clear divergence from the Ddl sequences of mesophilic Gram-negative bacteria (Figure 2). Notably, among the Gram-negative thermophiles mentioned above, all the tested ones except Mesotoga prima have been found susceptible to vancomycin in previous studies. Since a close scrutiny of the genomes of these thermophiles revealed no second Ddl homolog it was obvious that their Ddl proteins, like the T. maritima prototype, were also adapted to favor a D-ala-D-ala bonding in MPPs, which in turn caused the handicap of vancomycin-susceptibility. In this context it was further noteworthy that the Ddl homologs of Thermus and Meiothermus species not only had lower (≤35%) identities with the TmDdl but were also widely diverged from the latter in ML phylogeny. As such, it was not surprising that previous mass spectroscopic studies had reported considerable variability in the proportion of D-ala-D-ala-terminated MPPs in Thermus species (Quintela et al., 1999). However, despite potential infidelity in the choice of the fifth amino acid of their MPPs, it is noteworthy that all tested Thermales members have till date been found susceptible to vancomycin (see Tables 3, 4). This implied that alanine was still selected as the predominant amino acid species in the fifth position of the MPPs of the Thermales members.
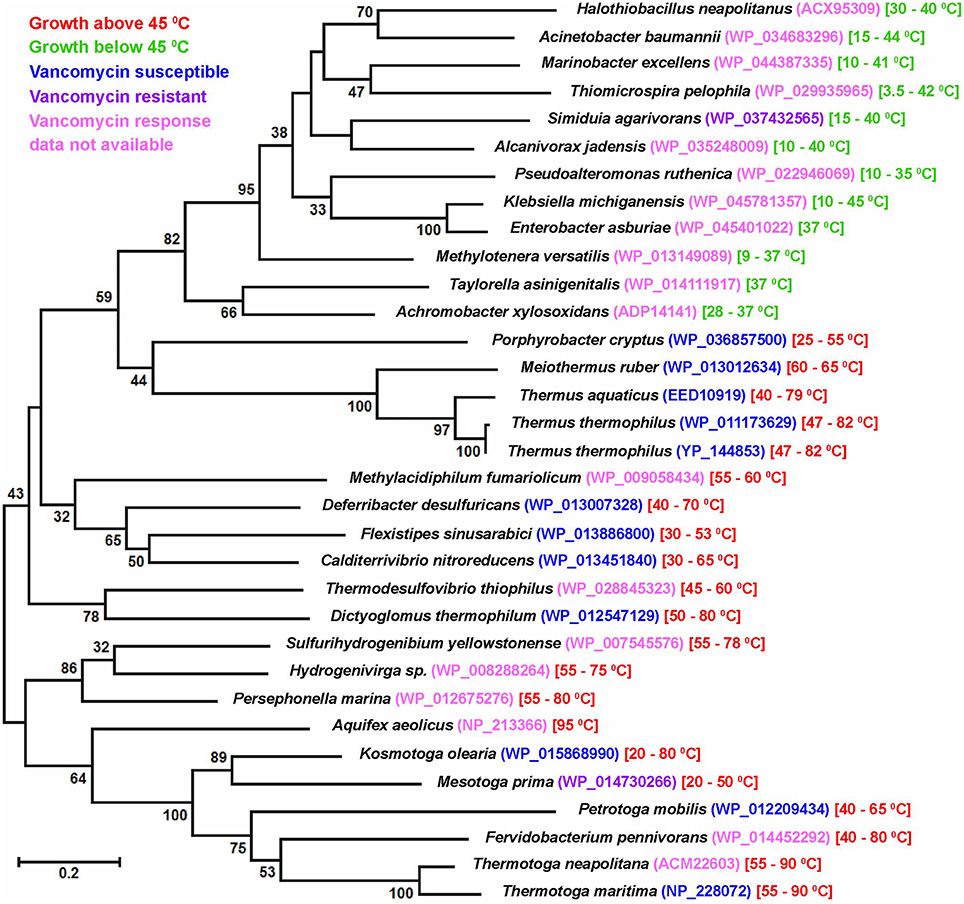
Figure 2. Maximum likelihood tree constructed with D-alanine-D-alanine ligase (Ddl) sequences of Gram-negative thermophilic as well as mesophilic bacteria. Growth temperature range of a given species is mentioned following the protein IDs of the Ddl homologs. Bootstrap support >30 (among 100 replicates) are given at the nodes. The scale bar represents two substitutions per 100 sites for a unit branch length.
When our current findings were juxtaposed with a decades-old report showing accumulation of excess DL-alanine by thermophilic bacterial cells (Matsumoto et al., 1967) it provoked the conjecture that thermophilic processes involving freedom of amino acid choice could, in general, be skewed in favor of alanine. However, it must be acknowledged that the potential thermo-adaptive advantage (if any) conferred by D-ala-D-ala-terminated MPPs is still not clear. But, pending biophysical verification, it seems obvious that such dipeptides were preferred by bacteria in high temperature environments and vancomycin susceptibility was just a collateral consequence of this thermodynamic compulsion.
In this context it must also be appreciated that an apparently-universal preference for D-ala-D-ala-terminated MPPs still does not guarantee vancomycin susceptibility of thermophilic bacteria as a relatively-hydrophilic molecule as large as vancomycin still has to cross the outer membrane before it can inhibit peptidoglycan biosynthesis. Notably, many mesophilic Gram-negative bacteria (unlike the case of A. kashmirensis stated above) also have D-ala-D-ala-terminated MPPs, but they still remain resistant to vancomycin due to the relative impermeability of their outer membrane toward this large glycopeptide molecule (Vollmer et al., 2008; Gordon et al., 2010). As such, the above-observed vancomycin resistance of the other two Gram-negative mesophiles Pseudaminobacter salicylatoxidans KCT001 and E. coli BL21 could also be due to impermeability toward vancomycin. But keeping in mind the global vancomycin-susceptibility phenotype of thermophilic bacteria it is obvious that the drug manages to cross the membrane in all these cases. As a corollary of this, it also seems quite likely that the composition of the outer membranes of thermophilic bacteria have some yet-unknown characteristic feature(s) that invariably ensures the entry of such relatively-hydrophilic large molecules as vancomycin.
With regard to the apparent preference of thermophilic bacteria for D-ala-D-ala-terminated MPPs two intriguing facts remain to be clarified at length. One is the vancomycin susceptibility of the mesophilic but apparently thermo-enduring Paracoccus strains isolated in this study and the other is the reported vancomycin resistance of at least two Gram-negative thermophilic species in the literature (viz. Mesotoga prima and Geoalkalibacter subterraneus) (Greene et al., 2009; Nesbø et al., 2012). It is noteworthy that despite its inability to grow above 45°C, the Paracoccus strains were native to an ambient temperature of 70–85°C. Since such strains got frequently isolated over multiple rounds of sampling at the Lotus Pond spring system they cannot be discounted as mere stochastic introductions at the time of sampling. As such, they must be migrating intermittently from the Puga rivulet to the adjoining Lotus Pond waters and in the process getting acclimatized to elevated temperatures. In contrast, Mesotoga prima was isolated from an apparently mesophilic sample of unknown temperature (viz., Baltimore harbor sediment Nesbø et al., 2012), while Geoalkalibacter subterraneus was retrieved from formation water of an oil-well (in Redwash oilfield of Utah) that had a temperature of only 52°C (Greene et al., 2009). Additionally, Mesotoga prima was enriched via several months of incubation and serial sub-culturing at 22 and 30°C, whereas the current Paracoccus strains were isolated immediately upon retrieval of hydrothermal samples to the laboratory. In the light of the above facts vancomycin-susceptibility, and as a corollary the D-ala-D-ala-specificity of Ddl homologs, seems to be a function of thermal-conditioning of a bacterium rather than the actual upper limit of its growth temperature. As such, it would not be surprising if our Paracoccus isolates become vancomycin-resistant after prolonged maintenance at 28 or 37°C, or for that matter the Mesotoga and Geoalkalibacter strains become vancomycin-susceptible after several sub-cultures at ≥50°C. This kind of subtle functional switching over short evolutionary times does not seem improbable when we keep in mind the high levels of sequence divergence existing between Ddl homologs of even closely-related genera. For example, Ddl sequences of Thermotogaceae members Thermotoga neapolitana (ACM22603) and Kosmotoga olearia (WP_015868990) have only 43% identity, which is quite low by any “house-keeping gene” standard.
Author Contributions
CR anchored the whole work and participated in all experiments and manuscript writing. WG conceived the program, interpreted the results and wrote the manuscript. On site samplings were done by CR, MA, PH, TM, SKM, and WG. CR, MA, SM, PH, SB, TM, RR, MR, RC, and SKM did metagenomics and bioinformatics analyses. Pure culture microbiology was done by CR, MA, SM, SB, and TM. Organic chemistry experiments and data analyses were done by CR, MA, AM, RC, AN, and WG. AM, RC, AN, and SKM also contributed in improving the intellectual content of the work as well as the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The extensive field work and DNA sequencing involved in this paper would have never been possible without the pro-active espousal of the Director of Bose Institute, Professor Sibaji Raha. Financially, the work was supported by the Bose Institute as well as the Science and Engineering Research Board, Department of Science and Technology (DST), Government of India (GOI), with the latter grant having the number SR/FT/LS-204/2009. CR and MR received fellowships from the UGC, GOI. SM received a fellowship from the DST, GOI. SB was awarded a fellowship by Bose Institute, DST.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00412
References
Alves, M. P., Rainey, F. A., Nobre, M. F., and da Costa, M. S. (2003). Thermomonas hydrothermalis sp. nov., a new slightly thermophilic gammaproteobacterium isolated from a hot spring in central portugal. Syst. Appl. Microbiol. 26, 70–75. doi: 10.1078/072320203322337335
Andrews, K. T., and Patel, B. K. (1996). Fervidobacterium gondwanense sp. nov., a new thermophilic anaerobic bacterium isolated from nonvolcanically heated geothermal waters of the Great Artesian Basin of Australia. Int. J. Syst. Bacteriol. 46, 265–269. doi: 10.1099/00207713-46-1-265
Arthur, M., Reynolds, P., and Courvalin, P. (1996). Glycopeptide resistance in enterococci. Trends Microbiol. 4, 401–407. doi: 10.1016/0966-842X(96)10063-9
Barna, J. C., and Williams, D. H. (1984). The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38, 339–357. doi: 10.1146/annurev.mi.38.100184.002011
Brock, T. D., and Freeze, H. (1969). Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 98, 289–297.
Burgess, S. A., Brooks, J. D., Rakonjac, J., Walker, K. M., and Flint, S. H. (2009). The formation of spores in biofilms of Anoxybacillus flavithermus. J. Appl. Microbiol. 107, 1012–1018. doi: 10.1111/j.1365-2672.2009.04282.x
Chan, C. S., Chan, K. G., Tay, Y. L., Chua, Y. H., and Goh, K. M. (2015). Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front. Microbiol. 6:177. doi: 10.3389/fmicb.2015.00177
Courvalin, P. (2006). Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1), S25–S34. doi: 10.1086/491711
Dekker, N. H., Viard, T., de La Tour, C. B., Duguet, M., Bensimon, D., and Croquette, V. (2003). Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 329, 271–282. doi: 10.1016/S0022-2836(03)00320-6
Denner, E. B., Kolari, M., Hoornstra, D., Tsitko, I., Kampfer, P., Busse, H. J., et al. (2006). Rubellimicrobium thermophilum gen. nov., sp. nov., a red-pigmented, moderately thermophilic bacterium isolated from coloured slime deposits in paper machines. Int. J. Syst. Evol. Microbiol. 56(Pt 6), 1355–1362. doi: 10.1099/ijs.0.63751-0
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Endo, A., Sasaki, M., Maruyama, A., and Kurusu, Y. (2006). Temperature adaptation of Bacillus subtilis by chromosomal groEL replacement. Biosci. Biotechnol. Biochem. 70, 2357–2362. doi: 10.1271/bbb.50689
Fiala, G., Woese, C., Langworthy, T., and Stetter, K. (1990). Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea. Arch. Microbiol. 154, 120–126. doi: 10.1007/BF00423320
Ghosh, W., and Roy, P. (2006). Mesorhizobium thiogangeticum sp. nov., a novel sulfur-oxidizing chemolithoautotroph from rhizosphere soil of an Indian tropical leguminous plant. Int. J. Syst. Evol. Microbiol. 56(Pt 1), 91–97. doi: 10.1099/ijs.0.63967-0
Goh, K. M., Gan, H. M., Chan, K. G., Chan, G. F., Shahar, S., Chong, C. S., et al. (2014). Analysis of anoxybacillus genomes from the aspects of lifestyle adaptations, prophage diversity, and carbohydrate metabolism. PLoS ONE 9:e90549. doi: 10.1371/journal.pone.0090549
Gordon, N. C., Png, K., and Wareham, D. W. (2010). Potent synergy and sustained bactericidal activity of a vancomycin-colistin combination versus multidrug-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 5316–5322. doi: 10.1128/AAC.00922-10
Greene, A. C., Patel, B. K. C., and Yacob, S. (2009). Geoalkalibacter subterraneus sp. nov., an anaerobic Fe(III)- and Mn(IV)-reducing bacterium from a petroleum reservoir, and emended descriptions of the family Desulfuromonadaceae and the genus Geoalkalibacter. Int. J. Syst. Evol. Microbiol. 59, 781–785. doi: 10.1099/ijs.0.001537-0
Greene, A. C., Patel, B. K., and Sheehy, A. J. (1997). Deferribacter thermophilus gen. nov., sp. nov., a novel thermophilic manganese- and iron-reducing bacterium isolated from a petroleum reservoir. Int. J. Syst. Bacteriol. 47, 505–509. doi: 10.1099/00207713-47-2-505
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Healy, V. L., Lessard, I. A., Roper, D. I., Knox, J. R., and Walsh, C. T. (2000). Vancomycin resistance in enterococci: reprogramming of the D-ala-D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 7, R109–R119. doi: 10.1016/s1074-5521(00)00116-2
Huber, R., Langworthy, T., Kanig, H., Thomm, M., Woese, C., Sleytr, U., et al. (1986). Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144, 324–333. doi: 10.1007/BF00409880
Hudson, J. A., Morgan, H. W., and Daniel, R. M. (1984). Isolation and characterisation of a new caldoactive filamentous bacterium. FEMS Microbiol. Lett. 22, 149–153. doi: 10.1111/j.1574-6968.1984.tb00714.x
Hudson, J. A., Morgan, H. W., and Daniel, R. M. (1987). Thermus filiformis sp. nov. a Filamentous Caldoactive Bacterium. Int. J. Syst. Bacteriol. 37, 431–436. doi: 10.1099/00207713-37-4-431
Iino, T., Nakagawa, T., Mori, K., Harayama, S., and Suzuki, K. (2008). Calditerrivibrio nitroreducens gen. nov., sp. nov., a thermophilic, nitrate-reducing bacterium isolated from a terrestrial hot spring in Japan. Int. J. Syst. Evol. Microbiol. 58(Pt 7), 1675–1679. doi: 10.1099/ijs.0.65714-0
Jimenez, D. J., Andreote, F. D., Chaves, D., Montana, J. S., Osorio-Forero, C., Junca, H., et al. (2012). Structural and functional insights from the metagenome of an acidic hot spring microbial planktonic community in the Colombian Andes. PLoS ONE 7:e52069. doi: 10.1371/journal.pone.0052069
John, W. D., Brian, C., Deborah, C., and James, F. C. (1998). Rates of spontaneous mutation. Genetics 148, 1667–1686.
Koga, Y. (2012). Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012, 789652. doi: 10.1155/2012/789652
Komagata, K., and Suzuki, K.-I. (1988). “Lipid and cell-wall analysis in bacterial systematics,” in Methods in Microbiology, eds R. R. Colwell and R. Grigorova (Orlando, FL: Academic Press), 161–207.
Kristjansson, J. K., and Stetter, K. O. (1992). “Introduction,” in Thermophilic Bacteria, ed J. K. Kristjansson (London: CRC Press Inc.), 2–18.
Kumar, S., Tsai, C. J., and Nussinov, R. (2000). Factors enhancing protein thermostability. Protein Eng. 13, 179–191. doi: 10.1093/protein/13.3.179
L'Haridon, S., Jiang, L., Alain, K., Chalopin, M., Rouxel, O., Beauverger, M., et al. (2014). Kosmotoga pacifica sp. nov., a thermophilic chemoorganoheterotrophic bacterium isolated from an East Pacific hydrothermal sediment. Extremophiles 18, 81–88. doi: 10.1007/s00792-013-0596-7
Lien, T., Madsen, M., Rainey, F. A., and Birkeland, N. K. (1998). Petrotoga mobilis sp. nov., from a North Sea oil-production well. Int. J. Syst. Bacteriol. 48(Pt 3), 1007–1013. doi: 10.1099/00207713-48-3-1007
Lundstrom, T. S., and Sobel, J. D. (2000). Antibiotics for gram-positive bacterial infections. Vancomycin, teicoplanin, quinupristin/dalfopristin, and linezolid. Infect. Dis. Clin. North Am. 14, 463–474. doi: 10.1016/S0891-5520(05)70258-0
Matsumoto, T., Yano, T., and Yamada, K. (1967). Studies on dl-alanine formation by thermophilic bacteria. Agric. Biol. Chem. 31, 1381–1388. doi: 10.1271/bbb1961.31.1381
Menzel, P., Gudbergsdóttir, S. R., Rike, A. G., Lin, L., Zhang, Q., Contursi, P., et al. (2015). Comparative metagenomics of eight geographically remote terrestrial hot springs. Microb. Ecol. 70, 411–424. doi: 10.1007/s00248-015-0576-9
Mohagheghi, A., Grohmann, K., Himmel, M., Leighton, L., and Updegraff, D. M. (1986). Isolation and characterization of acidothermus cellulolyticus gen. nov., sp. nov., a New Genus of Thermophilic, Acidophilic, Cellulolytic Bacteria. Int. J. Syst. Bacteriol. 36, 435–443. doi: 10.1099/00207713-36-3-435
Moreira, C., Rainey, F. A., Nobre, M. F., da Silva, M. T., and da Costa, M. S. (2000). Tepidimonas ignava gen. nov., sp. nov., a new chemolithoheterotrophic and slightly thermophilic member of the beta-Proteobacteria. Int. J. Syst. Evol. Microbiol. 50(Pt 2), 735–742. doi: 10.1099/00207713-50-2-735
Nailor, M. D., and Sobel, J. D. (2009). Antibiotics for gram-positive bacterial infections: vancomycin, teicoplanin, quinupristin/dalfopristin, oxazolidinones, daptomycin, dalbavancin, and telavancin. Infect. Dis. Clin. North Am. 23, 965–982. doi: 10.1016/j.idc.2009.06.010
Nesbø, C. L., Bradnan, D. M., Adebusuyi, A., Dlutek, M., Petrus, A. K., Foght, J., et al. (2012). Mesotoga prima gen. nov., sp. nov., the first described mesophilic species of the Thermotogales. Extremophiles 16, 387–393. doi: 10.1007/s00792-012-0437-0
Oshima, T., and Imahori, K. (1974). Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a Nonsporulating Thermophilic Bacterium from a Japanese Thermal Spa. Int. J. Syst. Bacteriol. 24, 102–112. doi: 10.1099/00207713-24-1-102
Paul, S. C., Jain, P., Mitra, J., Dutta, S., Bhattacharya, P., Bal, B., et al. (2012). Induction of Cr(VI) reduction activity in an Anoxybacillus strain under heat stress: a biochemical and proteomic study. FEMS Microbiol. Lett. 331, 70–80. doi: 10.1111/j.1574-6968.2012.02555.x
Perkins, H. R., and Nieto, M. (1974). The chemical basis for the action of the vancomycin group of antibiotics. Ann. N. Y. Acad. Sci. 235, 348–363. doi: 10.1111/j.1749-6632.1974.tb43276.x
Quintela, J. C., Zollner, P., Garcia-del Portillo, F., Allmaier, G., and de Pedro, M. A. (1999). Cell wall structural divergence among Thermus spp. FEMS Microbiol. Lett. 172, 223–229. doi: 10.1111/j.1574-6968.1999.tb13472.x
Rainey, F. A., Silva, J., Nobre, M. F., Silva, M. T., and da Costa, M. S. (2003). Porphyrobacter cryptus sp. nov., a novel slightly thermophilic, aerobic, bacteriochlorophyll a-containing species. Int. J. Syst. Evol. Microbiol. 53, 35–41. doi: 10.1099/ijs.0.02308-0
Reynolds, P. E. (1989). Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8, 943–950. doi: 10.1007/BF01967563
Saiki, T., Kobayashi, Y., Kawagoe, K., and Beppu, T. (1985). Dictyoglomus thermophilum gen. nov., sp. nov., a Chemoorganotrophic, Anaerobic, Thermophilic Bacterium. Int. J. Syst. Bacteriol. 35, 253–259. doi: 10.1099/00207713-35-3-253
Sato, M., Kirimura, K., and Kino, K. (2006). Substrate specificity of thermostable D-alanine-D-alanine ligase from Thermotoga maritima ATCC 43589. Biosci. Biotechnol. Biochem. 70, 2790–2792. doi: 10.1271/bbb.60307
Schumann, P. (2011). “Peptidoglycan structure,” in Methods in Microbiology, ed F. Rainey and A. Oren (London: Academic Press), 101–129.
Shih, T. W., and Pan, T. M. (2011). Stress responses of thermophilic Geobacillus sp. NTU 03 caused by heat and heat-induced stress. Microbiol. Res. 166, 346–359. doi: 10.1016/j.micres.2010.08.001
Shlaes, D. M., Shlaes, J. H., Davies, J., and Williamson, R. (1989). Escherichia coli susceptible to glycopeptide antibiotics. Antimicrob. Agents Chemother. 33, 192–197. doi: 10.1128/AAC.33.2.192
Sikorski, J., Brambilla, E., Kroppenstedt, R. M., and Tindall, B. J. (2008). The temperature-adaptive fatty acid content in Bacillus simplex strains from ‘Evolution Canyon’, Israel. Microbiology 154(Pt 8), 2416–2426. doi: 10.1099/mic.0.2007/016105-0
Takahata, Y., Nishijima, M., Hoaki, T., and Maruyama, T. (2001). Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. Int. J. Syst. Evol. Microbiol. 51(Pt 5), 1901–1909. doi: 10.1099/00207713-51-5-1901
Takai, K., Miyazaki, M., Nunoura, T., Hirayama, H., Oida, H., Furushima, Y., et al. (2006). Sulfurivirga caldicuralii gen. nov., sp. nov., a novel microaerobic, thermophilic, thiosulfate-oxidizing chemolithoautotroph, isolated from a shallow marine hydrothermal system occurring in a coral reef, Japan. Int. J. Syst. Evol. Microbiol. 56(Pt 8), 1921–1929. doi: 10.1099/ijs.0.64297-0
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tripathy, S., and Maiti, N. K. (2014). Construction of Geobacillus thermoglucosidasius cDNA library and analysis of genes expressed in response to heat stress. Mol. Biol. Rep. 41, 1639–1644. doi: 10.1007/s11033-013-3011-7
Volker, U., Mach, H., Schmid, R., and Hecker, M. (1992). Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J. Gen. Microbiol. 138, 2125–2135. doi: 10.1099/00221287-138-10-2125
Vollmer, W., Blanot, D., and de Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Wang, W., Ma, T., Zhang, B., Yao, N., Li, M., Cui, L., et al. (2014). A novel mechanism of protein thermostability: a unique N-terminal domain confers heat resistance to Fe/Mn-SODs. Sci. Rep. 4:7284. doi: 10.1038/srep07284
Watanakunakorn, C. (1984). Mode of action and in-vitro activity of vancomycin. J. Antimicrob. Chemother. 14(Suppl. D), 7–18. doi: 10.1093/jac/14.suppl_D.7
Waxman, D. J., and Strominger, J. L. (1983). Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu. Rev. Biochem. 52, 825–869. doi: 10.1146/annurev.bi.52.070183.004141
Wemheuer, B., Taube, R., Akyol, P., Wemheuer, F., and Daniel, R. (2013). Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka Peninsula. Archaea 2013:136714. doi: 10.1155/2013/136714
Williams, R. A. D., Smith, K. E., Welch, S. G., Micallef, J., and Sharp, R. J. (1995). DNA relatedness of thermus strains, description of thermus brockianus sp. nov., and Proposal To Reestablish Thermus thermophilus (Oshima and Imahori). Int. J. Syst. Bacteriol. 45, 495–499. doi: 10.1099/00207713-45-3-495
Keywords: vancomycin susceptibility, thermophilic bacteria, MALDI-TOF mass spectrometry, NGS, peptidoglycan, D-alanine-d-alanine ligase
Citation: Roy C, Alam M, Mandal S, Haldar PK, Bhattacharya S, Mukherjee T, Roy R, Rameez MJ, Misra AK, Chakraborty R, Nanda AK, Mukhopadhyay SK and Ghosh W (2016) Global Association between Thermophilicity and Vancomycin Susceptibility in Bacteria. Front. Microbiol. 7:412. doi: 10.3389/fmicb.2016.00412
Received: 29 December 2015; Accepted: 14 March 2016;
Published: 31 March 2016.
Edited by:
Octavio Luiz Franco, Universidade Catolica de Brasilia, BrazilReviewed by:
Dmitri Debabov, NovaBay Pharmaceuticals, USASanti M. Mandal, Vidyasagar University, India
Celio De Faria Junior, Central Public Health Laboratory (LACEN) of the Brazilian Federal District, Brazil
Copyright © 2016 Roy, Alam, Mandal, Haldar, Bhattacharya, Mukherjee, Roy, Rameez, Misra, Chakraborty, Nanda, Mukhopadhyay and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wriddhiman Ghosh, wriman@jcbose.ac.in; wriman@rediffmail.com
 Chayan Roy
Chayan Roy Masrure Alam
Masrure Alam Subhrangshu Mandal1
Subhrangshu Mandal1 Sabyasachi Bhattacharya
Sabyasachi Bhattacharya Moidu J. Rameez
Moidu J. Rameez Ranadhir Chakraborty
Ranadhir Chakraborty Wriddhiman Ghosh
Wriddhiman Ghosh