- 1Laboratory of Microbiology, Faculty of Fisheries Sciences, Hokkaido University, Hakodate, Japan
- 2Institute of Biology, SAGE-COPPE, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
- 3Department of Nutrition, Hakodate Junior College, Hakodate, Japan
Coral reefs perform a major role in regulating marine biodiversity and serve as hotspot for highly dynamic and diverse microbiomes as holobionts. Corals around Ishigaki, however, are at risk due to tremendous stressors including elevation of seawater temperature, eutrophication and so on. However, no information is currently available on how Vibrio diversity fluctuates spatially and temporally due to environmental determinants in Ishigaki coral reef ecosystems. The aim of this study is to elucidate spatiotemporal Vibrio diversity dynamic at both community and population levels and to assess the environmental drivers correlated to Vibrio abundance and diversity. The Vibrio community identified based on pyrH gene phylogeny of 685 isolates from seawater directly connecting to Ishigaki coral holobionts consisted of 22 known and 12 potential novel Vibrionaceae species. The most prominent species were V. hyugaensis, V. owensii and V. harveyi followed by V. maritimus/V. variabillis, V. campbellii, V. coralliilyticus, and Photobacterium rosenbergii. The Vibrio community fluctuations, assessed by PCoA with UniFrac distance and clustering with Euclidiean distance were varied less not only by year but also by site. Interestingly, significant positive correlation was observed between rising seawater temperature and the abundance of V. campbellii (r = 0.62; P < 0.05) whereas the opposite was observed for V. owensii (r = -0.58; P < 0.05) and the C6 group of V. hyugaensis (r = -0.62; P < 0.05). AdaptML-based microhabitat differentiation revealed that V. harveyi, V. campbellii, P. rosenbergii, and V. coralliilyticus populations were less-ecologically distinctive whereas V. astriarenae and V. ishigakensis were ecologically diverse. This knowledge could be important clue for the future actions of coral conservation.
Introduction
Coral reef ecosystem consists of a flexible consortium of eukaryotic and prokaryotic organisms as a holobiont and are rich in compounds for interacting each other as cross talks, protection of their territory and as triggers for these symbiotic association dynamics (Kooperman et al., 2007; Shnit-Orland and Kushmaro, 2009; Kvennefors et al., 2012). Coral hosts large, diverse and specific microbial populations, which have both important beneficial and harmful roles for the host (Ritchie and Smith, 2004; Rosenberg et al., 2007). More specifically, corals provide three habitats for bacteria (the surface mucus layer, coral tissue, and calcium carbonate skeleton) each of which harbors a distinct bacterial population (Koren and Rosenberg, 2006). Among the bacterial populations, vibrios have been recognized as important members of coral holobionts (Rohwer and Kelly, 2004; Vezzulli et al., 2010, 2012; Baker-Austin et al., 2013; Moreira et al., 2014; Rubio-Portillo et al., 2014; Munn, 2015). Some vibrios may establish mutualistic partnership with corals by releasing nutrients and secondary metabolites (Ritchie, 2006; Chimetto et al., 2008, 2009), while others play a major role in the disruption of coral health (Munn, 2015).
Coral and coral reefs are in steep decline worldwide due to the combined effects of various stressors such as global warming, pollution, overfishing and infectious diseases (Hughes et al., 2003; Weil et al., 2006; Dinsdale et al., 2008; Vezzulli et al., 2012; Baker-Austin et al., 2013). Rising seawater temperature related to global climate change is a big threat to coral health and is ultimately linked to increasing coral disease such as bleaching events (Tout et al., 2015). Impressive progress has been reported regarding characterization of the coral microbiota over the last decade with its altered composition often correlated to the appearance of signs and diseases and/or bleaching that ultimately suggest a link between microbes, global coral health and stability of reef ecosystems (Krediet et al., 2013). Among them, coral vibriosis is a well-known disease, of which occurrences are greatly influenced by increasing seawater temperature (Vezzulli et al., 2012; Baker-Austin et al., 2013; Munn, 2015). In particular, at warm temperatures, V. shiloi expresses a cell-surface adhesion protein that is required for bacterial attachment to the coral surface and simultaneously expresses Toxin-P that ultimately inhibits photosynthesis of the coral-endosymbiotic algae and super-oxide dismutase required for survival of this pathogen inside the coral (Rosenberg et al., 2007; Munn, 2015).
The coral reef ecosystem in Ishigaki Island in southwestern Japan is surrounded by well-developed fringing corals with a variety of mangroves and sandy or rocky shores (Sano, 2000; Shimomura and Naruse, 2015). Recently, these reefs have been severely stressed due to typhoons, coral bleaching and soil pollution. Red soil derived turbid seawater in Okinawa, Ishigaki, and Iriomote coral reef seawater adversely affects the photosynthesis of coral symbiont algae (Zooxanthelae) and also interfered with the settlement of coral larvae on the ocean bed which ultimately influenced coral damage (Kakuma and Kamimura, 2011). Due to this, eutrophication including inflow of red soil is an important threat to Ishigaki corals. More specifically, accumulation of red soil and high nutrient inputs due to land development since 1972, have severely affected the Ishigaki coral reef (Hasegawa, 2011). Rivers from the northern part of Ishigaki coral reef, originating from agricultural watershed may have detrimental effects on corals because exposure to silt and nutrient rich sediments may badly stress corals (Weber et al., 2006). In addition, run-off from soils after typhoons can easily damage coral as well (Harii et al., 2014). Presently, several reports are available regarding coral-microbiota from different parts of the world (Rohwer et al., 2002; Wild et al., 2004; Rosenberg et al., 2007; Chimetto et al., 2009; Alves et al., 2010; Krediet et al., 2013; Chimetto Tonon et al., 2015) but little is known about the Ishigaki coral reef microbial communities.
Therefore, the aims of this study are to characterize the Vibrio diversity spatially and temporally, to elucidate which environmental determinants [temperature or dissolved organic carbon (DOC)] correlate the Vibrio diversity dynamics and to characterize antimicrobial resistance profiles of potential coral pathogens.
Materials and Methods
Water Sampling and Isolation of Vibrios
Seawater samples were collected in 2012, 2013, and 2014 (June–July) from five sites (Figure 1) off the Ishigaki coral reef, Okinawa, Japan. The sampling sites were Miyara (24°20.5489′ N, 124°13.0408′ E), Osaki (24°25.4171′ N, 124°04.4956′ E), Taketomi (24°20.5260′ N, 124°05.6443′ E), Sekisei outer (24°21.7557′ N, 124°02.7190′ E), and Iriomote (24°23.208′ N, 123°55.681′ E) at depths of 3–4 m. More specifically, these seawater samples were within 2 m of coral holobionts and were collected by SCUBA diving and stored in clean containers and/or sterilized tubes. No specific permissions were required for the water sampling activities in these locations. The seawater samples were then brought back to the laboratory for bacterial isolation. A total of 0.2 mL of the seawater sample was directly spread on to thiosulfate-citrate-bile salt-sucrose (TCBS) agar plate (Nissui Pharmacy, Tokyo, Japan) and incubated at 25°C for 48 h. After incubation, numbers of individual colonies were counted manually to estimate ‘viable Vibrio counts.’ All colonies except those showing lack of growth were purified twice on the TCBS agar plates. The purity was checked using the ZoBell 2216E agar plate at 25°C. These strains were preserved in cryo-vials using the ZoBell broth supplemented with 20% (v/v) glycerol at -80°C.
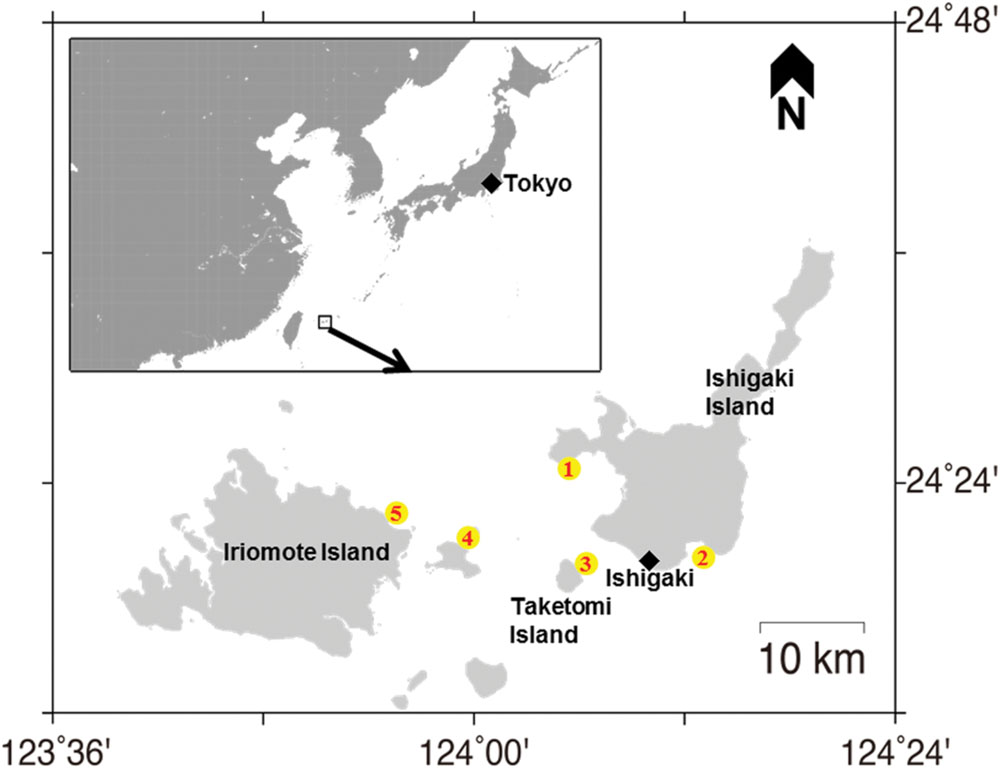
FIGURE 1. Map showing the sampling locations. Sites are marked with a yellow circle. (1) Osaki; (2) Miyara; (3) Taketomi; (4) Sekisei outer; (5) Iriomote in Ishigaki coral reef ecosystem.
Molecular Phylogenetic Affiliation on the Basis of pyrH Gene Sequences to Vibrionaceae Species Groups
Molecular phylogenetic affiliation of all Vibrio isolates (n = 685) was performed on the basis of pyrH (uridylate kinase) gene sequences. A 550 bp of pyrH gene was amplified using pyrH80F and pyrH530R primers (Sawabe et al., 2007) as a colony suspension or a purified DNA as a PCR template. Genomic DNA was purified using a Promega Wizard Genomic DNA extraction system according to the protocol provided by the manufacturer. The PCR cycle consisted of an initial denaturation step at 95°C for 180 s and 30 cycles of an denaturation (95°C for 60 s), an annealing step (50°C for 60 s) and an extension step (72°C for 60 s). The PCR products were analyzed on 1.5% agarose gel and the PCR products producing a single band on agarose gels were purified using Promega Gel and PCR purification system (Promega, Madison, WI, USA). Approximately, 50 ng of template was directly sequenced using the pyrH80F or pyrH530R primer and a BigDye terminator sequencing kit version 3.1 (Life Technologies, Carlsbad, CA, USA) according to the protocol recommended by the manufacturer. DNA sequencing was performed with an Applied Biosystems model 373A automated sequencer. The same primers (pyrH80F or pyrH530R) were used for the sequencing.
Using our well-curated pyrH gene sequences, which could cover more than 145 known described species of Vibrionaceae, molecular phylogenetic affiliations of Ishigaki Vibrio isolates were performed. To retain the pyrH tree topology within Vibrionaceae, pyrH gene sequence of Escherichia coli W3110 (NC_010473) was included in the dataset as an out group. The sequences were aligned using Clustal X version 2.1 (Larkin et al., 2007) and the alignment was checked by eye and corrected manually. The position used for the affiliation corresponded to positions 171 to 543 in the V. cholera O1 Eltor N16961 (AE003852). pyrH gene phylogeny was reconstructed by neighbor-joining method (Saitou and Nei, 1987) with bootstrap values of 500 replicates implemented to MEGA programs version 6.06 (Tamura et al., 2013). Evolutionary distances were corrected using the Jukes-Cantor method. Clustering to a described species with more than 98.4% pyrH gene similarity was used for the final affiliation for species identification of Ishigaki coral reef isolates. For the affiliation of closed species in clusters harboring V. harveyi, V. owensii, V. hyugaensis, V. communis, V. campbellii, and P. damselae subsp. piscicida, a >99.7% cutoff was adopted to affiliate species. All pyrH and 16S rRNA genes sequences used in this study have been deposited in BaMBa under data package identification number pmeirelles.20.1. To further affiliate unassigned pyrH clusters to new species candidates, 16S rRNA gene sequences of 38 representative strains from pyrH unassigned clusters were obtained according to Al-saari et al. (2015). In brief, the amplification primers (24F and 1509R) used for PCR amplification gave a 1.5 kb long PCR product and corresponded to positions 25–1521 in the E. coli sequence. A 99.7% cut-off was used for the final affiliation of Ishigaki coral reef isolates to the new Vibrionaceae species candidate.
Inferred Habitat Associations of Vibrio Community
To infer habitat associations of the Vibrio community in Ishigaki coral reef during a 3-years (2012–2014) assessment, PCoA based on unweighted UniFrac distance, hierarchical cluster analysis of Vibrio community composition and AdaptML was performed.
UniFrac distance was calculated using 658 pyrH gene sequences of Vibrio isolates and Quantitative Insights Into Microbial Ecology (QIIME) 1.9 software package (Caporaso et al., 2010) according to Yamazaki et al. (2016) except the use of pyrH gene phylogenetic tree. Relationships among Vibrio communities in accordance with sampling sites and years were visualized using PCoA. Hierarchical clustering analyses were performed with R Development Core Team (2011), except where indicated. Abundance and multivariate figures were plotted with packages ggplot2 and reshape (Wickham, 2007, 2009). The hierarchical cluster shown in Figure 3B was constructed using Ward’s minimum variance method (performed using “hclust” R function), which aims at finding compact, spherical clusters, on a Euclidian dissimilarity matrix (calculated using “dist” R function). Vibrio diversity indices (Shannon entropy and Shannon evenness [i.e., Hill’s Ratio]) and richness were calculated with the vegan R package (Oksanen et al., 2005).
The mathematical model (AdaptML), which uses a Hidden Markov Model, was used to predict the phylogenetic bounds of ecologically distinct Vibrio populations and their habitat composition (distribution among environmental categories). ‘Clusters of vibrios’ sequences were obtained using the software AdaptML as described previously (Hunt et al., 2008). In brief, the software combines genetic information embedded in sequence-based phylogenies and information about the ecology, herein the source and place of isolation, in order to identify genetically and ecologically distinct bacterial populations. AdaptML algorithms can account for environmental parameter discretization schemes and are based on the model concept of habitat (a place and related features that determines microbial distribution). Habitats are characterized by discrete probability distributions describing the likelihood that a strain adapted to a habitat will be sampled from a given ecological state, i.e., at a particular location in the water column). A maximum likelihood model is used for the evolution of habitat association on the tree (Hunt et al., 2008). The habitat-learning and clustering steps of AdaptML were performed using the default settings. Confident assignments are shown to fit ecological populations predicted by the model. The model threshold value was set at 0.05 and E. coli W3110 was used as an out-group. The bootstrap percentages analysis were rerun 100 times with the same phylogenetic tree to verify the stability of the predictions. The circular tree figure was drawn using online iTOL software (Letunic and Bork, 2007). To prevent numerical instabilities in AdaptML’s maximum likelihood computations, branches with zero length were assigned to the minimal observed non-zero branch length: 0.001. Clades supported in 80% of bootstraps are shown.
Correlation between Vibrionaceae Species Groups and Environmental Determinants
Seawater environmental determinants including temperature and DOC were assessed. Water temperature was measured directly using a thermometer. DOC was determined using the method described by Grasshoff et al. (2009). Pearson correlation between each Vibrio abundance and environmental determinant was calculated using Microsoft Excel software.
Results
Diversity of Vibrios in Ishigaki Coral Reef Seawater
Our investigation of Vibrio diversity in Ishigaki coral reef seawater using pyrH gene phylogeny discovered a total of 44 clusters (Supplementary Table S1; Figure 2); 32 clusters were affiliated to 22 known Vibrionaceae species including 10 V. hyugaensis and 2 P. damselae subsp. piscicida subclusters, and the other 12 were affiliated to novel species candidates in Vibrionaceae. A total of 658 strains were identified as known Vibrionaceae species to V. alfacsensis, V. alginolyticus, V. astriarenae, V. azureus, V. campbellii, V. communis, V. coralliilyticus/V. neptunis, V. harveyi, V. hyugaensis, V. ishigakensis, V. mediterranei, V. nigripulchritudo, V. orientalis, V. owensii, V. ponticus, V. pelagius, V. rotiferianus, V. tubiashii, V. variabillis/maritimus, Photobacterium aphoticum, P. damselae subsp. piscicida, and Photobacterium rosenbergii.
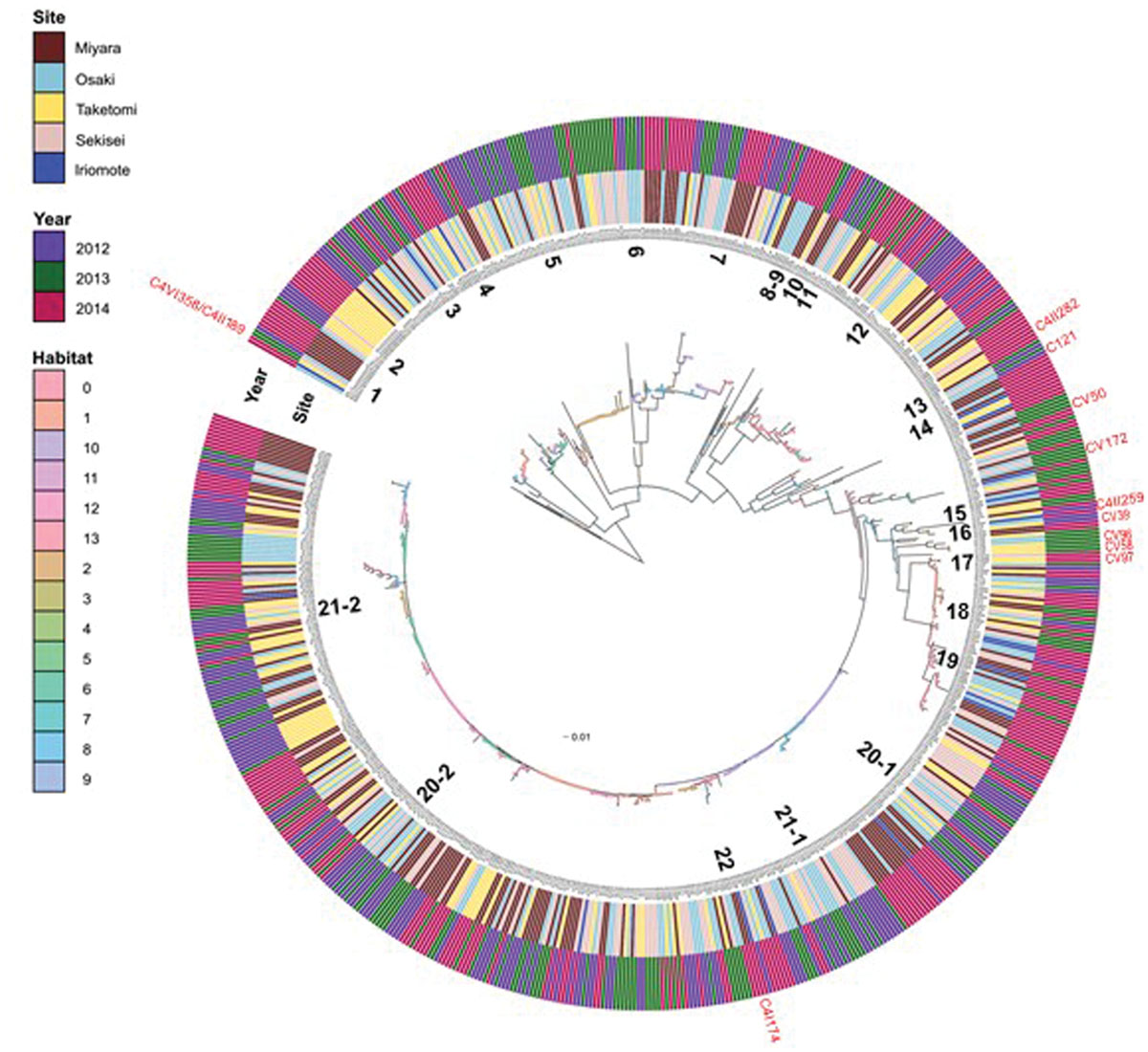
FIGURE 2. Inferred habitats predicted by AdaptML model using pyrH gene sequences of Vibrio isolates from Ishigaki coral reef seawater. Inner and outer ring indicate sampling sites and sampling years respectively. Predicted habitats are shown as nodes on the tree (habitat legend matches the colored circles). The closest named Vibrio species to numbered Vibrio populations’ are as follows- (1) P. aphoticum; (2) V. astriarenae; (3) V. ishigakensis; (4) V. pelagius; (5) P. rosenbergii; (6) V. mediterranei; (7) V. variabillis/maritimus; (8) V. nigripulchritudo; (9) V. orientalis; (10) V. tubiashii; (11) V. alginolyticus; (12) V. coralliilyticus/neptunis; (13) V. ponticus; (14) V. azurius; (15) V. alfacsensis; (16) V. orientalis; (17) P. damsella subsp. piscicida; (18) V. harveyi; (19) V. campbellii; (20-1) V. owensii-1; (20-2) V. owensii-2; (21-1) V. hyugaensis; (21-2) V. hyugaensis; (22) V. communis. Red marked strains, outside of the outer ring indicate the positions of putative new species candidates.
However, 26 strains belonged to C121 (n = 5), CV39 (n = 2), CV50 (n = 4), CV58 (n = 1), CV96 (n = 1), CV97 (n = 1), CV172 (n = 1), C4I174 (n = 4), C4II189, C4II259 (n = 1), C4III282 (n = 3) and C4V358 clusters which were not affiliated to any known vibrios species (Figure 2; Supplementary Table S1). V. hyugaensis was the largest group of the Vibrio community in Ishigaki coral reef seawater (n = 200, 29.2%) and was mostly diversified into 10 different pyrH clusters (Supplementary Table S1). The occupation rates of each V. hyugaensis cluster were C71 (2.0%), C68 (4.5%), C6 (7.6%), C16 (1.9%), C49 (1.9%), C58 (3.7%), C46 (6.0%), C156 (0.1%), C164 (1.3%), and C4I100 (0.1%). V. owensii, a potent coral pathogen, was present in 21.6% of the total isolated vibrios. The third most abundant Vibrio group was V. variabilis/maritimus which was recorded in 8.8% of the isolated vibrios. Other prominent vibrios were P. rosenbergii (6.0%), V. campbellii (5.3%), and V. harveyi (4.5%). Some other recognized potential coral pathogens, V. coralliilyticus (n = 49, 7.2%) and V. alginolyticus (n = 5) were also found. Less significant amounts of identified vibrios (Supplementary Table S1) were V. ponticus (n = 3, 0.4%), V. pelagius (n = 9, 1.3%), V. tubiashii (n = 5, 0.7%), V. rotiferianus (n = 3, 0.4%), P. aphoticum (n = 3, 0.4%) and two recent newly described species, V. astriarenae (3.4%) and V. ishigakensis (4.2%). V. nigripulchritudo (n = 1), V. alfacsensis (n = 1), V. orientalis (n = 1), V. azureus (n = 1), and V. mediterranei (n = 1) were less highlighted vibrios.
Vibrio Community Dynamics
Unweighted FastUniFrac analysis revealed no-apparent grouping of Vibrio communities not only site by site but also year by year (Figure 3A). However, a simple cladogram constructed on the basis of relative abundances in the Vibrio community illustrated two apparent clusters in the Vibrio communities of Iriomote 2014, Osaki 2014, Sekisei 2014, Taketomi 2014 and Miyara 2013 and those of Taketomi 2012, Miyara 2014, Sekisei 2012, and Osaki 2013 (Figure 3B). Signatures of the former and the latter communities showed significant abundances (P < 0.05) of V. campbellii, V. hyugaensis (C6 group), and V. owensii respectively. Vibrios from Sekisei 2013 and Osaka 2012 were sub-clustered using a relative higher abundance of P. rosenbergii.
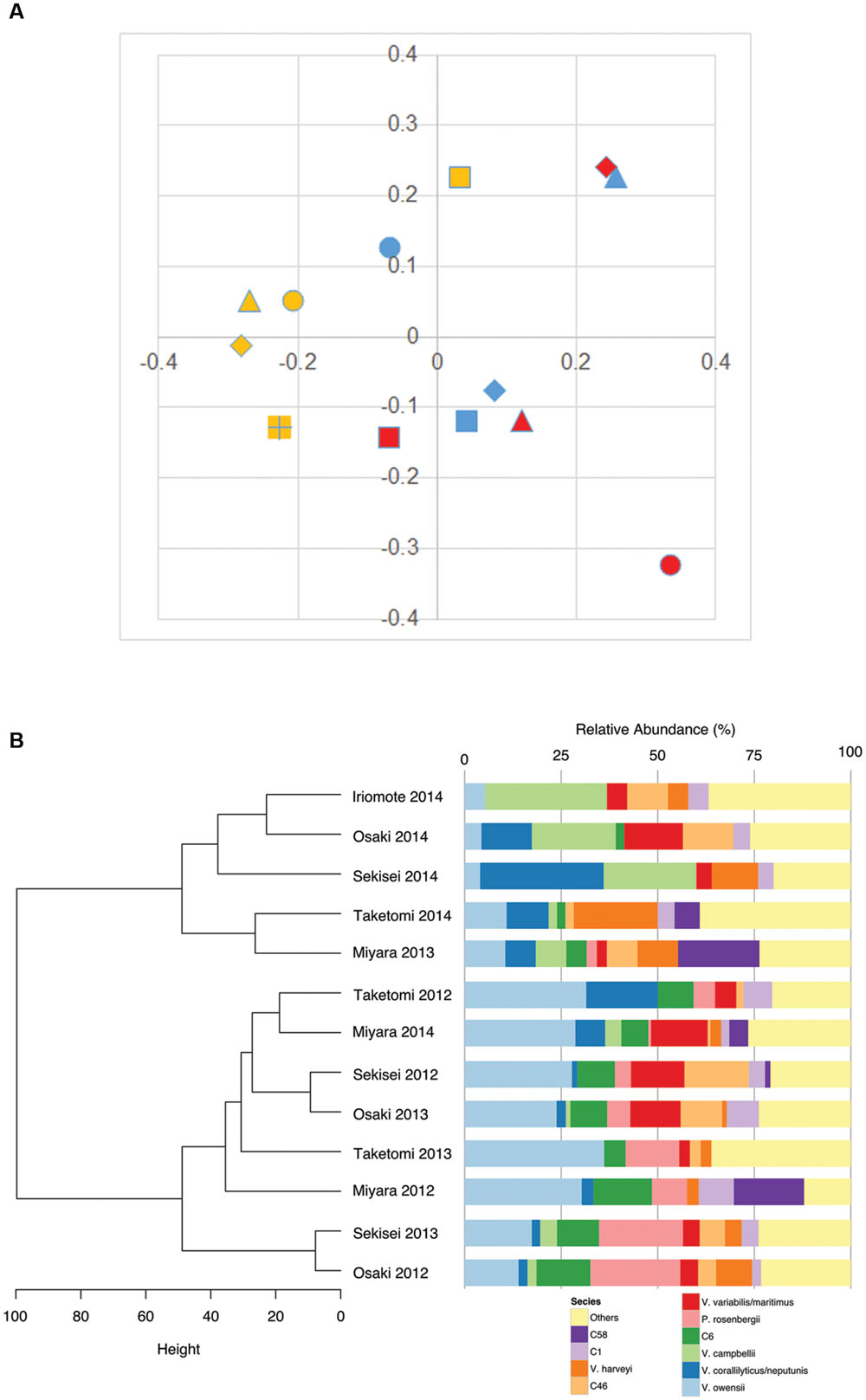
FIGURE 3. Vibrio community diversity dynamics in Ishigaki coral reef seawater. (A) Multidimensional scaling plot (FastUnifrac:PCoA), isolates indicating colors and shapes: blue, 2012; red, 2013; yellow, 2014; dot, Taketomi; square, Miyara; diamond, Osaki; triangle, Sekisei; square (+), Iriomote; (B) dendrogram clustering of vibrios according to their relative abundances.
Correlation between Environmental Variables and Vibrio Abundances
Vibrio campbellii abundance significantly increased (r = 0.62; P < 0.05) along with rising seawater temperature whereas V. owensii (r = -0.58; P < 0.05) and the C6 group of V. hyugaensis (r = -0.62; P < 0.05) abundances significantly decreased with an increase in seawater temperature (Table 1). Moreover, the correlation assessment between Vibrio abundances and DOC availability illustrated (Table 1) that P. rosenbergii and the V. hyugaensis (C6 group) increased along with increased DOC whereas V. owensii, V. campbellii, and V. coralliilyticus showed the opposite results.
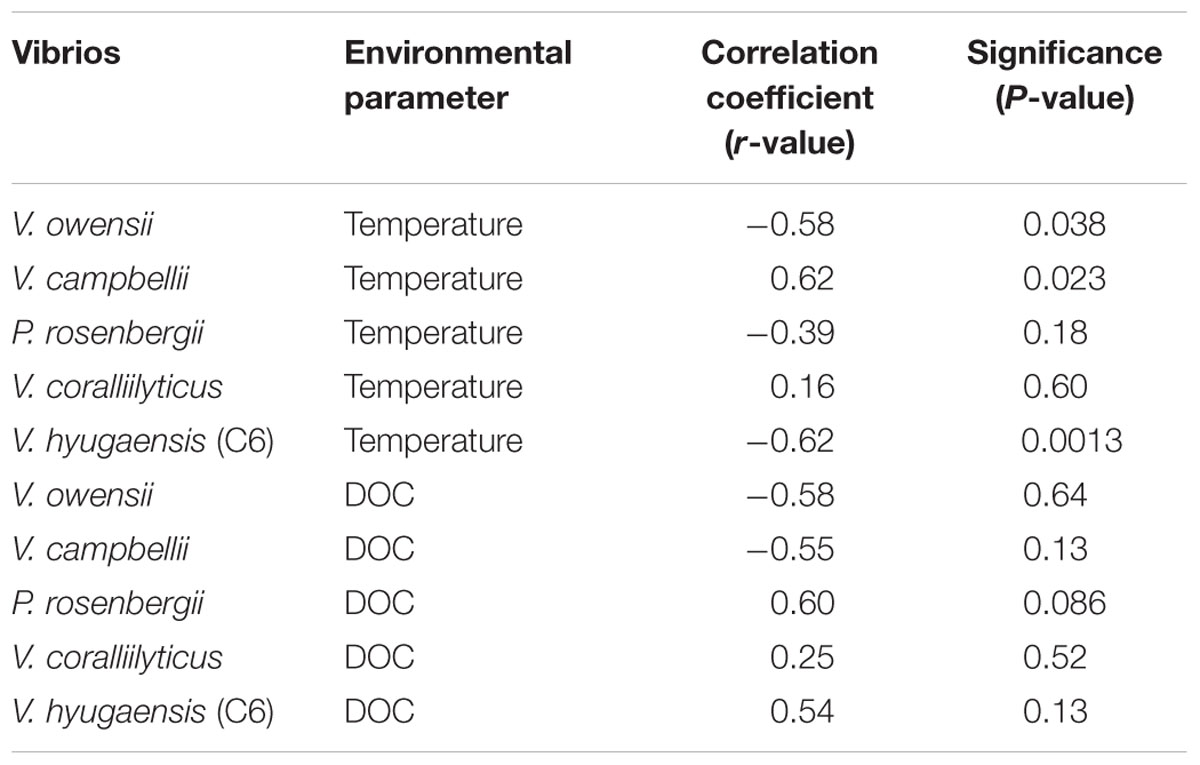
TABLE 1. Correlation coefficient (r) and significant levels (P-values) obtained from the correlation analysis between major Vibrio abundances and environmental variables.
Ecologically Distinct Populations among Vibrio Isolates
The AdaptML analysis showed that the Vibrio isolates are distributed in eight habitat spectra (H-0, H-1, H-2, H-6, H-8, H-10, H-12, and H-13; Figures 2 and 4). Habitats are the part of an ecosystem (a spectrum of environment types) from where microbial populations are isolated. V. owensii, V. harveyi, V. hyugaensis (C71, C46, C6, C16, C49, C58, and C68 groups), V. campbellii, V. ishigakensis, V. variabillis/V. maritimus, V. coralliilyticus/V. neptunis, P. rosenbergii, and V. pelagius were observed from all the sites although V. alfacsensis, and V. nigripulchritudo were found only from Taketomi and Miyara respectively (Figure 4). Another two vibrios, V. azureus and V. mediterranei were specific for Osaki and one more single Vibrio, V. orientalis was recorded solely from Sekisei reef site. The spatiotemporal distribution of V. campbelli was likely to support the temperature dependent abundance from 2014 in these study areas (Figure 4). From all the predicted habitat spectra, H-10 and H-12 showed the strains which were isolated from all the sampling sites and sampling years with major isolates from Sekisei-2012 (41.7%) and Miyara-2014 (41.2%), respectively. Other habitats (H-0, H-1, H-2, H-6, H-8, H-13) showed the isolates (Supplementary Figure S1) from several sampling sites and years but not from all. The major isolates of H-0, H-1, H-2, H-6, H-8, and H-13 were obtained from Osaki-2014 (34.9%), Taketomi-2014 (34.9%), Osaki-2012 (20.2%), Osaki-2013 (33.7%), Miyara-2014 (78.6%), and Taketomi-2012 (55%) respectively.
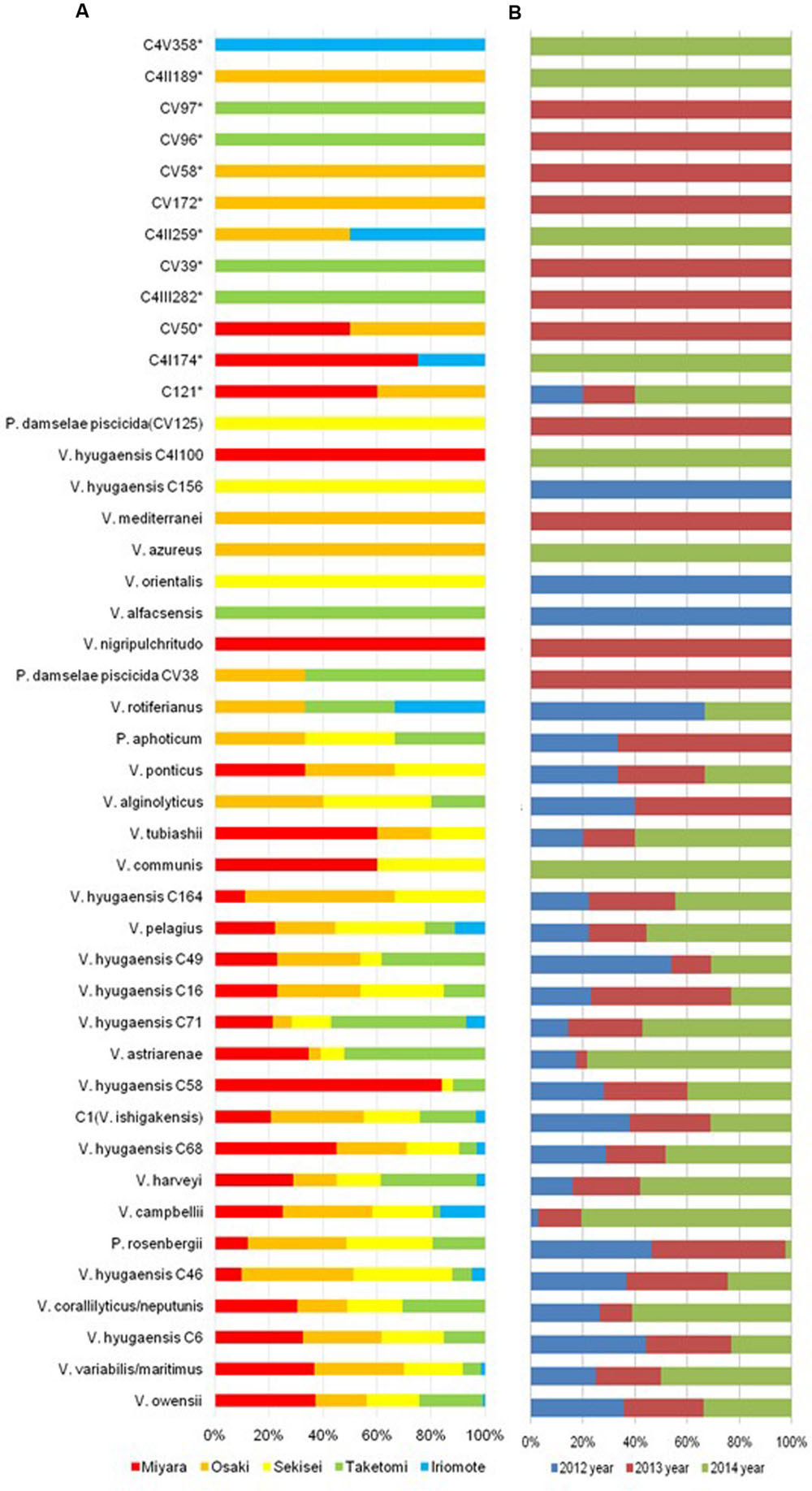
FIGURE 4. Spatiotemporal Vibrio population dynamics in Ishigaki coral reef seawater. (A) Spatial Vibrio population dynamics (red, brown, yellow, green, sky blue colors indicate sampling sites of Miyara, Osaki, Sekisei Outer, Taketomi, and Iriomote respectively); (B) Temporal Vibrio population dynamics (blue, violate, and green colors indicate sampling year of 2012, 2013, and 2014 accordingly).
The adapted predominant Vibrio species (Supplementary Table S2) in H-0 were V. harveyi (9.7%), V. campbellii (94.4%), V. ishigakensis (6.9%), V. variabillis/V. maritimus (11.7%), V. coralliilyticus/V. neptunis (32.7%), and V. pelagius (44.4%). The highlighted vibrios in H-1 were V. harveyi (80.6%) and V. hyugaensis C6 group (42.3%), V. hyugaensis C58 group (48.0%), V. hyugaensis C46 group (26.8%) and V. astriarenae (43.5%). H-2 comprised mostly of V. owensii (8.8%), V. hyugaensis C16 group (76.9%) and P. rosenbergii (97.6%). Two prevalent vibrios in H-6 were V. owensii (24.3%) and V. ishigakensis (44.8%). The most abundant vibrios in H-8 were V. owensii (16.2%) and V. variabillis/V. maritimus (28.3%). The dominant vibrios in H-10 were V. owensii (17.6%), V. hyugaensis C68 group (32.3%), V. hyugaensis C46 group (56.1%), V. ishigakensis (31.0%) and V. variabillis/V. maritimus (41.7%). In H-12 the preeminent vibrios were V. owensii (29.1%) and V. hyugaensis-C6 (30.8%). H-13 was found to contain V. coralliilyticus–V. neptunis (40.8%) and V. astriarenae (30.4%). Moreover, several potential novel vibrios (CV39, CV58, CV96, CV97) and one opportunistic coral pathogen V. alginolyicus (100%) were retrieved specifically from H-2.
AdaptML could also estimate the evolutionary history of ecological differentiation (Hunt et al., 2008). Interestingly, the recent ongoing adaptive radiations (with a shallow branch: meaning frequently changing adaptations) were observed as more likely to be in V. astriarenae, V. ishigakensis, V. mediterranei, V. variabilis/V. maritimus, V. coralliilyticus/V. neputunis, and V. communis/V. owensii/V. hyugaensis, however, no such radiations were observed in V. campbellii, V. harveyi, and P. rosenbergii (Figure 2).
Discussion
Ishigaki coral reef ecosystems are known to be the largest and the most diverse reef ecosystems in Japan, but are in now extremely vulnerable. In this era of global warming, the surrounding seawater of coral reefs are one of the most important ecological niches as well as heat carriers. It is worth investigating the evolution of spatiotemporal Vibrio dynamics in oceanic fields not only for the conservation of fragile coral reefs but also for public health and aquaculture issues (Rubio-Portillo et al., 2014; Tout et al., 2015). We performed a 3 years survey to elucidate Vibrio diversity using well-curated pyrH gene sequence set to achieve fine scale discrimination (Thompson J.R. et al., 2005; Thompson et al., 2005a, 2007; Tall et al., 2013; Chimetto Tonon et al., 2015). The pyrH gene set allows us not only to affiliate Vibrio isolates to known described Vibrionaceae species or currently unknown new species candidates but also to elucidate the structure of regional populations of specific species and how they have diverged. Our study demonstrated that the Vibrio community of the coral surrounding seawater consisted of at least 22 known described species and 12 undescribed species candidates (26 isolates) which is likely to be a more diversified Vibrio community compared to those reported from tropical and temperate coral reef environments (Chimetto et al., 2008; Raina et al., 2009; Alves et al., 2010; Chimetto Tonon et al., 2015). Unfortunately, we did not assess the Ishigaki coral reef microbiota due to lack of coral samples, the seawater around Ishigaki coral reefs is likely to share major Vibrionaceae communities of reported microbiomes V. communis/V. owensii, V. mediterranei (V. shiloi), V. harveyi, V. alginolyticus, V. campbellii, V. maritimus/V. variabilis, V. tubiashii, V. coralliilyticus, V. pelagius consisting of coral holobionts (Alves et al., 2010; Chimetto Tonon et al., 2015). High abundance of Harveyi clade (V. hyugaensis, V. communis, V. owensii, V. harveyi, V. campbellii) and lower abundance of Splendidus clade species (V. pelagius) are the specific clues to help define subtropical Ishigaki coral reef microbiota (Hunt et al., 2008; Tall et al., 2013). Coralliilyticus clade species (V. coralliilyticus, V. neptunis), Mediterranei clade species (V. mediterranei, V. maritimus, V. variabillis) and P. rosenbergii are secondary significant members (>5% constitution) of the Ishigaki coral reef seawaters.
The availability of at least 12 novel species candidates (CV172, CV50, C4I174, CV96, CV58, CV39, CV97, C4II282, C121, C4II259, C4II189, and C4V358) in the Ishigaki coral reef ecosystem suggests important evidence of Vibrionaceae evolution. Relevant subsequent studies (Thompson et al., 2005b; Rosenberg et al., 2007; Reis et al., 2009; Moreira et al., 2014; Chimetto Tonon et al., 2015) reported that coral and surrounding seawater are prime sources in isolating novel vibrios and is an important habitat for studying the microevolution of vibrios. Comparing previously recorded vibrios from coral and reef ecosystems (Chimetto et al., 2009; Alves et al., 2010; Chimetto Tonon et al., 2015) we found mostly diversified populations including numerous vibrios unique to our studied reef locations. The retrieved unique Vibrio populations were V. hyugaensis, V. ishigakensis, V. astriarenae, P. rosenbergii, P. damsella subsp. piscicida, P. aphoticum, V. nigripulchritudo, V. alfacsensis, and V. azureus suggesting that Ishigaki coral reef locations are hotspots for versatile and dynamic Vibrio populations and may serve as diversified ecological niches.
This spatiotemporal survey also revealed that the community and population level dynamics in the surrounding seawater of coral holobionts in Ishigaki reef ecosystems can determine generalists and specialists (Figures 2 and 4). V. owensii and V. hyugaensis populations were considered to be the dominant generalist populations in the coral seawater ecosystems. The other prevalent generalists (Supplementary Table S1; Figure 2) in Ishigaki reef seawater were V. variabillis, V. harveyi, V. campbellii, P. rosenbergii, V. coralliilyticus, V. astriarenae, and V. ishigakensis.
Numerous previous reports suggested that vibrios from Harveyi clade appear to have ecologically diversified passively by invading new niches (Chimetto et al., 2008; Preheim et al., 2011; Chimetto Tonon et al., 2015). The observed diversities of V. owensii and V. hyugaensis populations were likely to be higher than those of V. harveyi and V. campbellii (Figure 2). V. owensii was isolated from cultured crustaceans in Australia as a member of Harveyi clade (Cano-Gomez et al., 2010). This species was also isolated from Acropora white syndrome lesions in American Samoa and is considered to be a putative coral pathogen (Ushijima et al., 2012; Wilson et al., 2012) and was also isolated from ‘Montipora capitata’ diseased-coral (Tissue loss disease: Montipora White Syndrome) in Hawaiian coral reefs. V. hyugaensis was isolated from a seawater sample collected in Miyazaki prefecture in Japan (Urbanczyk et al., 2015). It is a luminous Vibrio and several light-producing bacteria were reported from seawater samples taken from different sites in Miyazaki prefecture between 2010 and 2012 (Urbanczyk et al., 2014).
Specialist Vibrio populations in Ishigaki reef seawater showed site-specific distributions (Supplementary Table S1). V. azureus and V. mediterranei were isolated specifically from Osaki whereas, V. nigripulchritudo, V. alfacsensis, and V. orientalis were noted as region-specific in Miyara, Taketomi and Sekisei, respectively. Bacteria as coral holobionts may be host-specific or may display spatial variability which might show protective activities against pathogens or other detrimental agents by occupying entry niches and space (as competitors) or by production of antibiotic compounds (Rohwer et al., 2002; Mullen et al., 2004; Kelman et al., 2006; Ritchie, 2006). Moreover, the mathematical model AdaptML generated the environmental grouping (Supplementary Table S2) of the Ishigaki reef seawater Vibrio populations and detected both ecologically restricted and ecologically distinctive (Hunt et al., 2008) populations. Two proposed novel vibrios isolated from this location V. astriarenae, Agarivorans clade (Al-saari et al., 2015) and V. ishigakensis, Halioticoli clade (Gao et al., 2016) were found in more ecologically diverse populations (Figure 2). The former is agarolytic whereas the latter is non-motile and alginolytic. Benthic flora might determine the specific niches for these species.
One of the more intriguing findings disclosed by this study was the availability of versatile and dominant putative coral pathogenic vibrios, V. owensii and V. coralliilyticus (Supplementary Table S1; Figure 2) occupied 21.6 and 7.2%, respectively in our total analyzed vibrios. This may be due to higher global oceanic temperatures. Temperature influenced pathogenic potential of coral pathogenic vibrios may lead to an increase in incidences of diseases in marine environments. Swimming motility, swarming motility and protease activity have been shown to be related to the virulence of V. owensii (Ushijima et al., 2012). According to Kimes et al. (2012) for V. coralliilyticus, an increase in seawater temperature to more than 27°C plays a direct role in triggering virulence genes and other factors related to host degradation, secretion, antimicrobial resistance, and motility. The mechanism of virulence includes motility and chemotaxis that involve post-colonization with subsequent production of coral tissue damaging ‘zink-metalloproteases’ (Ben-Haim et al., 2003). Recently, Tout et al. (2015) demonstrated that raised-seawater temperature increase natural Vibrio abundances, particularly the notorious coral pathogen V. coralliilyticus, in coral ecosystems. However, our findings on V. coralliilyticus abundance were not significantly correlated to seawater temperature rises. Beyond the previously mentioned potential coral pathogens, we also identified V. harveyi (4.5%) and V. alginolyticus (0.7%) from the seawater. V. alginolyticus was found only in the single habitat H-2 suggesting that this bacterium may have a new ecological niche-partitioning in Ishigaki reef seawater. Other results associated with coral pathogenic vibrios (Luna et al., 2010; Zhenyu et al., 2013; Munn, 2015) show that most of them are opportunistic in nature and show their virulence under certain environmental conditions by overwhelming the host-defense, overgrowth and tissue destruction.
Environmental variables, particularly temperature are considered to be the prominent triggering factor in Vibrio ecology, population dynamics, physiological stress response and evolution (Sato et al., 2009). Elevated temperatures and high loads of nutrients particularly and DOC have direct impacts on coral holobiont microbial communities and ultimately make them diverse, abundant and even niche or host specific (Kline et al., 2006; Paz et al., 2007). Other findings similarly demonstrate that elevated nutrients (i.e., phosphate, nitrate, ammonia) and DOC in coastal waters have been a cause of reef decline (Bruno et al., 2003; Kline et al., 2006). In our assessment, only specific Vibrionaceae populations correlated with environmental parameters (e.g., temperature, DOC). V. campbellii abundance was significantly correlated to rises in seawater temperatures, V. owensii and C6 group of V. hyugaensis showed significant negative correlations with increasing seawater temperature. The optimum growth temperature of V. hyugaensis was reported to be 26°C and it could not grow at 37°C, which might reflect its low tolerance to high temperatures. These findings corroborated previous reports that thermal anomalies and nutrient-rich environment including DOC are well-known ecological triggers, involved in proliferation and increased abundance of holobiont microbial communities in coastal environments globally (Ducklow et al., 1986; Wild et al., 2004; Barott and Rohwer, 2012) along with reef ecosystems in southern Japan (Goto et al., 2010). Roder et al. (2014) also noted that coral associated bacterial communities including potential pathogens emerge and dominate during environmental stress and the shift of bacterial community may be a direct effect of temperature on growth of specific members of the microbial community (Garren et al., 2014).
Conclusion
Our curated data set of Vibrionaceae pyrH gene sequences allows us to perform the first characterization of Vibrio diversity and assessment of spatiotemporal diversity dynamics of Vibrio populations both at community and population levels along with the relationship to environmental determinants from Ishigaki coral reef seawater. The determined Vibrio community of seawater directly connecting to Ishigaki coral holobionts included at least 22 known described species and 12 as yet undescribed species, which are more diversified compared to reported tropical and temperate reef vibrios. Several vibrios such as, V. campbellii, V. owensii and the C6 group of V. hyugaensis abundances seem to correlate with seawater temperature. V. owensii and C6 group of V. hyugaensis show significant negative correlations with increasing seawater temperature but V. campbellii is positively significant in this regard. This study also demonstrates the presence of most of the globally recognized opportunistic potential coral-pathogenic vibrios (V. owensii, V. harveyi, V. alginolyticus). This pioneering study on Ishigaki coral reef seawater vibrios both at community and population levels will be the important gateway for further deep research on vibrios in coral and reef systems.
Author Contributions
Conceived and designed the experiments: AA, FT, and TomooS. Performed the experiments: AA, GF, NA, PM, YY, and SM. Analyzed the data: AA, GF, NA, PM, YY, and TomooS. Contributed reagents/materials/analysis tools: AA, GF, NA, PM, YY, SM, FT, TokoS, and TomooS. Wrote the paper: AA and TomooS. Critical review, ideas and suggestion given during manuscript preparation: GF, NA, PM, YY, SM, FT, and TokoS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Strategic Japanese-Brazilian Cooperative Program, Biomass and Bioenergy (TomooS and FT), JSPS-CAPES bilateral cooperative program (TomooS and FT), and Kaken (26660168; TomooS). We gratefully thank Mr. Tomioka for collecting seawater samples. FT thanks CAPES, CNPq, and FAPERJ for funding. PM thanks CAPES for the Ph.D. scholarship (4848-14-9 CAPES/JSPS) and MARA for the Ph.D. education loan to NA.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01185
References
Al-saari, N., Gao, F., Amin, A. K. M. R., Sato, K., Sato, K., Mino, S., et al. (2015). Advanced microbial taxonomy combined with genome based approaches reveals that Vibrio astriarenae sp. nov., an agarolytic marine bacterium, forms a new clade in Vibrionaceae. PLoS ONE. 10:e0136279. doi: 10.1371/journal.pone.0136279
Alves, N. Jr., Neto, O. S. M., Silva, B. S. O., de Moura, R. L., Francini-Filho, R. B., Castro, C. B. E., et al. (2010). Diversity and pathogenic potential of vibrios isolated from Abrolhos Bank corals. Environ. Microbiol. Rep. 2, 90–95. doi: 10.1111/j.1758-2229.2009.00101.x
Baker-Austin, C., Trinanes, J. A., Taylor, N. G. H., Hartnell, R., Siitonen, A., and Martinez-Urtaza, J. (2013). Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 3, 73–77. doi: 10.1038/nclimate1628
Barott, K. L., and Rohwer, F. L. (2012). Unseen players shape benthic competition on coral reefs. Tren. Microbiol. 20, 621–628. doi: 10.1016/j.tim.2012.08.004
Ben-Haim, Y., Zicherman-keren, M., and Rosenberg, E. (2003). Temperature regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen V. coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003
Bruno, J. F., Petes, L. E., Harvell, C. D., and Hettinger, A. (2003). Nutrient enrichment can increase the severity of coral diseases. Ecol. Lett. 6, 1056–1061. doi: 10.1046/j.1461-0248.2003.00544.x
Cano-Gomez, A., Goulden, E. F., Owens, L., and Hoj, L. (2010). Vibrio owensii sp. nov., isolated from cultured crustaceans in Australia. FEMS Microbiol. Lett. 302, 175–181. doi: 10.1111/j.1574-6968.2009.01850.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1093/jac/dkr082
Chimetto, L. A., Brocchi, M., Gondo, M., Thompson, C. C., Gomez-Gil, B., and Thompson, F. L. (2009). Genomic diversity of vibrios associated with the Brazilian coral Mussismilia hispida and its sympatric zoanthids (Palythoa caribaeorum, Palythoa variabilis and Zoanthus solanderi). J. Appl. Microbiol. 106, 1818–1826. doi: 10.1111/j.1365-2672.2009.04149.x
Chimetto, L. A., Brocchi, M., Thompson, C. C., Martins, R. C. R., Ramos, H. R., and Thompson, F. L. (2008). Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst. Appl. Microbiol. 31, 312–319. doi: 10.1016/j.syapm.2008.06.001
Chimetto Tonon, L. A., Silva, B. S. O., Moreira, A. P. B., Valle, C., Alves, N. Jr., Cavalcanti, G., et al. (2015). Diversity and ecological structure of vibrios in benthic and pelagic habitats along a latitudinal gradient in the southwest Atlantic ocean. Peer J. 3, e741. doi: 10.7717/peerj.741
Dinsdale, E. A., Pantos, O., Smriga, S., Edwards, R. A., Haynes, M., Krause, L., et al. (2008). Microbial ecology of four coral atolls in the northern line Islands. PLoS ONE 3:e1584. doi: 10.1371/journal.pone.0001584
Ducklow, H. W., Purdie, D. A., Williams, P. J. L., and Davies, J. M. (1986). Bacterioplankton: a sink for carbon in a coastal marine plankton community. Science 232, 865–867. doi: 10.1126/science.232.4752.865
Gao, F., Al-saari, N., Rohul Amin, A. K., Sato, K., Mino, S., Suda, W., et al. (2016). Vibrio ishigakensis sp. nov., the first free-living species in Halioticoli clade isolated from seawater in Okinawa coral reef area, Japan. Syst. Appl. Microbiol. 39, 330–335. doi: 10.1016/j.syamp.2016.04.002
Garren, M., Son, K., Raina, J. B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Goto, K., Miyagi, K., Kawamata, H., and Imamura, F. (2010). Discrimination of boulders deposited by tsunamis and long waves at Ishigaki Island. Japan. Mar. Geol. 269, 34–45. doi: 10.1016/j.margeo.2009.12.004
Grasshoff, K., Kremling, K., and Ehrhard, M. (2009). Methods of Seawater Analysis. Hoboken, NJ: John Wiley & Sons.
Harii, S., Hongo, C., Ishihara, M., Ide, Y., and Kayanne, H. (2014). Impacts of multiple disturbances on coral communities at Ishigaki Island, Okinawa, Japan, during a 15 year survey. Mar. Ecol. Prog. Ser. 509, 171–180. doi: 10.3354/meps10890
Hasegawa, H. (2011). The decline of coral reef conditions caused by the extensive land modification: a case study of the Shiraho area on Ishigaki Island, Okinawa, Japan. J. Remote Sens. Soc. Japan 31, 73–86.
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Sci. Rev. 301, 929–933. doi: 10.1126/science.1085046
Hunt, D. E., David, L. A., Gevers, D., Preheim, S. P., Alm, E. J., and Polz, M. F. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320, 1081–1085. doi: 10.1126/science.1157890
Kakuma, S., and Kamimura, M. (2011). “Okinawa: Effective conservation practices from satoumi in a coral reef ecosystem” in biological and cultural diversity in coastal communities: exploring the potential of satoumi for implementing the ecosystem approach in the Japanese Archipelago. Secret. Conven. Biol. Diver. Tech. Ser. 61, 86–93.
Kelman, D., Kashman, Y., Rosenberg, E., Kushmaro, A., and Loya, Y. (2006). Antimicrobial activity of red-sea corals. Mar. Biol. 149, 357–363. doi: 10.1007/s00227-005-0218-8
Kimes, N. E., Grim, C. J., Johnson, W. R., Hasan, N. A., Tall, B. D., and Kothary, M. H. (2012). Temperature regulation of virulence factors in the pathogenic Vibrio coralliilyticus. ISME J. 6, 835–846. doi: 10.1038/ismej.2011.154
Kline, D. I., Kuntz, N. M., Breitbart, M., Knowlton, N., and Rohwer, F. (2006). Role of elevated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125. doi: 10.3354/meps314119
Kooperman, N., Ben-Dov, E., Kramarsky-Winter, E., Barak, Z., and Kushmaro, A. (2007). Coral mucus associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276, 106–113. doi: 10.1111/j.1574-6968.2007.00921.x
Koren, O., and Rosenberg, E. (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72, 5254–5259. doi: 10.1128/AEM.00554-06
Krediet, C. J., Ritchie, K. B., Paul, V. J., and Teplitski, M. (2013). Coral–associated microorganisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. B 280, 20122328. doi: 10.1098/rspb.2012.2328
Kvennefors, E. C. E., Sampayo, E., Kerr, C., Vieira, G., Roff, G., and Barnes, A. C. (2012). Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 63, 605–618. doi: 10.1007/s00248-011-9946-0
Larkin, M. A., Blachshields, G., Brown, N. P., Chenna, R., Mc Gettigan, P. A., Mc William, H., et al. (2007). Clustal W and Clustal X version 2.o. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., and Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Luna, G. M., Bongiorni, L., Gili, C., Biavasco, F., and Danovaro, R. (2010). Vibrio harveyi as a causative agent of the white syndrome in tropical stony corals. Environ. Microbiol. Rep. 2, 120–127. doi: 10.1111/j.1758-2229.2009.00114.x
Moreira, A. P. B., Duytschaever, G., Tonon, L. A. C., Froes, A. M., de Oliveira, L. S., Amado-Filho, G. M., et al. (2014). Photobacterium sanctipauli sp. nov. isolated from bleached Madracis decactis (Scleractinia) in the St Peter & St Paul Archipelago, mid-Atlantic ridge, Brazil. peerJ 2, e427. doi: 10.7717/peerj.427
Mullen, K., Peters, E. C., and Harvell, C. D. (2004). “Coral resistance to disease,” in Coral Health and Diseases, eds E. Rosenberg and Y. Loya (Berlin, NY: Springer), 377–399. doi: 10.1007/978-3-662-06414-6
Munn, C. B. (2015). The role of vibrios in diseases of corals. Microbiol. Spectr. 3, 4. doi: 10.1128/microbiolspec.VE-0006-2014
Oksanen, J., Kindt, R., and O’Hara, B. (2005). Vegan: R Functions for Vegetation Ecologists. Available at: http://cc.oulu.fi/~jarioksa/softhelp/vegan.html
Paz, S., Bisharat, N., Paz, E., Kidar, O., and Cohen, D. (2007). Climate change and the emergence of Vibrio vulnificans diseases in Israel. Environ. Res. 103, 390–396. doi: 10.1016/j.envres.2006.07.002
Preheim, S. P., Boucher, Y., Wildschutte, H., David, L. A., Veveziano, D., Alm, E. J., et al. (2011). Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13, 265–275. doi: 10.1111/j.1462-2920.2010.02328.x
R Development Core Team (2011). R: A Language and Environment for Statistical Computing. http://www.r-project.org/
Raina, J. B., Tapiolas, D., Wills, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Reis, A. M. M., Araujo, S. D. Jr., Moura, R. L., Francini-Filho, R. B., Pappas, G. Jr., Coelho, A. M. A., et al. (2009). Bacteria diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis. Appl. Microbiol. 106, 1378–1387. doi: 10.1111/j.1365-2672.2008.04106.x
Ritchie, K. B. (2006). Regulation of marine microbioes by coral mucus and mucus associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Ritchie, K. B., and Smith, G. W. (2004). “Microbial communities of coral surface mucopolysaccharide layers,” in Coral Health and Diseases,eds E. Rosenberg and Y. Loya (Berlin, NY: Springer Press), 259–264.
Roder, C., Arif, C., Bayer, T., Aranda, M., Daniels, C., Shibl, A., et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8, 31–39. doi: 10.1038/ismej.2013.127
Rohwer, F., and Kelly, S. (2004). “Culture-independent analyses of coral-associated microbes,” in Coral Health and Diseases, eds E. Rosenberg and Y. Loya (Berlin, NY: Springer Press), 265–277.
Rohwer, F., Seguritan, V., Azam, F., and Knolton, N. (2002). Diversity and distribution of coral associated bacteria. Mar. Ecol. Prog Ser. 243, 1–10. doi: 10.3354/meps243001
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rubio-Portillo, E., Yarza, P., Penalver, C., Ramos-Eapla, A. A., and Anton, J. (2014). New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J. 8, 1794–1807. doi: 10.1038/ismej.2014.33
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sano, M. (2000). Stability of reef fish assemblages: responses to coral recovery after catastrophic predation by Acanthaster planci. Mar. Ecol. Prog. Ser. 198, 121–130. doi: 10.3354/meps198121
Sato, Y., Bourne, D. G., and Willis, B. L. (2009). Dynamics of seasonal outbreaks of black band diseases in an assemblage of Montipora species at Pelorus Island (Great Barrier Reef. Australia). Proc. R. Soc. B 276, 2795–2803. doi: 10.1098/rspb.2009.0481
Sawabe, T., Kita-Tsukamoto, K., and Thompson, F. L. (2007). Inferring the evolutionary history of Vibrios by means of multilocus sequence analysis. J. Bacteriol. 189, 7932–7936. doi: 10.1128/JB.00693-07
Shimomura, M., and Naruse, T. (2015). Two new species of Asellota (Crustacea, Isopoda) from coral reefs on Iriomote Island, Okinawa, Japan. ZooKeys 520, 27–40. doi: 10.3897/zookeys.520.5943
Shnit-Orland, M., and Kushmaro, A. (2009). Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol. Ecol. 67, 371–380. doi: 10.1111/j.1574-6941.2008.00644.x
Tall, A., Hervio-Heath, D., Teillon, A., Boisset-Helbert, C., Delesmont, R., Bodilis, J., et al. (2013). Diversity of Vibrio spp. isolated at ambient environmental temperature in the eastern english channel as determined by pyrH sequencing. J. Appl. Microbiol. 114, 1713–1724. doi: 10.1111/jam.12181
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, F. L., Gevers, D., Thopson, C. C., Dawyndt, P., Naser, S., Hoste, B., et al. (2005a). Phylogeny and molecular identification of Vibrios on the basis of multilocus sequence analysis. Appl. Environ. Micribiol. 71, 5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005
Thompson, F. L., Gomez-Gil, B., Vasconselos, A. T. R., and Sawabe, T. (2007). Multilocus sequence analysis reveals that Vibrio harveyi and V. campbellii are distinct species. Appl. Environ. Micribiol. 73, 4279–4285. doi: 10.1128/AEM.00020-07
Thompson, F. L., Thompson, C. C., Naser, S., Hoste, B., Vandemeulebroecke, K., Munn, C., et al. (2005b). Photobacterium rosenbergii sp. nov. and Enterovibrio coralii sp. nov., Vibrio associated with coral bleaching. Int. J. Evol. Microbiol. 55, 913–917. doi: 10.1099/ijs.0.63370-0
Thompson, J. R., Pacocha, S., Pharino, C., Klepac-Ceraj, V., Hunt, D. E., Benoit, J., et al. (2005). Genotypic diversity within a natural coastal bacterioplankton population. Science 307, 1311–1313. doi: 10.1126/science.1106028
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Urbanczyk, H., Ogura, Y., and Hayashi, T. (2014). Contrasting inter- and intraspecies recombination patterns in the “Harveyi clade” vibrio collected over large spatial and temporal scales. Genome. Biol. Evol. 7, 71–80. doi: 10.1093/gbe/evu269
Urbanczyk, Y., Ogura, Y., Hayashi, T., and Urbanczyk, H. (2015). Description of novel marine bacterium, Vibrio hyugaensis sp. nov., based on genomic and phenotypic characterization. Syst. Appl. Microbiol. 38, 300–304. doi: 10.1016/j.syapm.2015.04.001
Ushijima, B., Smith, A., Aeby, G. S., and Callahan, S. M. (2012). Vibrio owensii induces the tissue loss disease Montipora white syndrome in the Hawaiian reef coral Montipora capitata. PLoS ONE 7:e46717. doi: 10.137/journal.pone.0046717
Vezzulli, L., Brettar, I., Pezzati, E., Reid, P. C., Colwell, R. R., Hofle, M. G., et al. (2012). Long-term effects of ocean warming on the prokaryotic community: evidence from the Vibrios. ISME J. 6, 21–30. doi: 10.1038/ismej.2011.89
Vezzulli, L., Previati, M., Pruzzo, C., Marchese, A., Bourne, D. G., and Cerrano, C. (2010). Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ. Microbiol. 12, 2007–2019. doi: 10.1111/j.1462-2920.2010.02209.x
Weber, M., Lott, C., and Fabricius, K. E. (2006). Sedimentation stress in a scleractinian coral exposed to terrestrial and marine sediments with constracting physical, organic and geo-chemical properties. J. Exp. Mar. Biol. Ecol. 336, 18–32. doi: 10.1016/j.jembe.2006.04.007
Weil, E., Smith, G., and Gil-Agudelo, D. L. (2006). Status and progress in coral reef disease research. Dis. Aquat. Organ. 69, 1–7. doi: 10.3354/dao069001
Wickham, H. (2007). Reshaping data with the reshape package. J. Stat. Soft. 21, 1–20. doi: 10.3978/j.issn.2305-5839.2016.01.33
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y., and Jørgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Wilson, B., Aeby, G. S., Work, T. M., and Bourne, D. G. (2012). Bacterial communities associated with healthy and Acropora white syndrome-affected corals from American Samoa. FEMS. Microbiol. Ecol. 80, 509–520. doi: 10.1111/j.1574-6941.2012.01319.x
Yamazaki, Y., Meirelles, P. M., Mino, S., Suda, W., Oshima, K., Hattori, M., et al. (2016). Individual Apostichopus japonicas fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci. Rep. 6, 21631. doi: 10.1038/asrep21631
Keywords: vibrios, diversity dynamics, coral reef, seawater, environmental determinants
Citation: Amin AKMR, Feng G, Al-saari N, Meirelles PM, Yamazaki Y, Mino S, Thompson FL, Sawabe T and Sawabe T (2016) The First Temporal and Spatial Assessment of Vibrio Diversity of the Surrounding Seawater of Coral Reefs in Ishigaki, Japan. Front. Microbiol. 7:1185. doi: 10.3389/fmicb.2016.01185
Received: 26 April 2016; Accepted: 18 July 2016;
Published: 08 August 2016.
Edited by:
Learn-Han Lee, Monash University Malaysia Campus, MalaysiaReviewed by:
Vengadesh Letchumanan, University of Malaya, MalaysiaJennifer Ronholm, Health Canada, Canada
Copyright © 2016 Amin, Feng, Al-saari, Meirelles, Yamazaki, Mino, Thompson, Sawabe and Sawabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoo Sawabe, sawabe@fish.hokudai.ac.jp
 A.K. M. R. Amin1
A.K. M. R. Amin1 Pedro M. Meirelles
Pedro M. Meirelles Fabiano L. Thompson
Fabiano L. Thompson Tomoo Sawabe
Tomoo Sawabe