- 1Department of Otolaryngology, Miller School of Medicine, University of Miami, Miami, FL, USA
- 2Department of Human and Molecular Genetics, Herbert Wertheim College of Medicine, Florida International University, Miami, FL, USA
- 3Chemistry Department, Center for Advanced Microscopy, University of Miami, Coral Gables, FL, USA
- 4Rosenstiel School of Marine and Atmospheric Science, University of Miami, Key Biscayne, FL, USA
- 5Department of Biochemistry and Molecular Biology, Miller School of Medicine, University of Miami, Miami, FL, USA
- 6Global Health Consortium and Biomolecular Science Institute, Florida International University, Miami, FL, USA
- 7Health Sciences Department, University of Sagrado Coração, Bauru, Brazil
Otitis media (OM) is a broad term describing a group of infectious and inflammatory disorders of the middle ear. Despite antibiotic therapy, acute OM can progress to chronic suppurative otitis media (CSOM) characterized by ear drum perforation and purulent discharge. Pseudomonas aeruginosa is the most common pathogen associated with CSOM. Although, macrophages play an important role in innate immune responses but their role in the pathogenesis of P. aeruginosa-induced CSOM is not known. The objective of this study is to examine the interaction of P. aeruginosa with primary macrophages. We observed that P. aeruginosa enters and multiplies inside human and mouse primary macrophages. This bacterial entry in macrophages requires both microtubule and actin dependent processes. Transmission electron microscopy demonstrated that P. aeruginosa was present in membrane bound vesicles inside macrophages. Interestingly, deletion of oprF expression in P. aeruginosa abrogates its ability to survive inside macrophages. Our results suggest that otopathogenic P. aeruginosa entry and survival inside macrophages is OprF-dependent. The survival of bacteria inside macrophages will lead to evasion of killing and this lack of pathogen clearance by phagocytes contributes to the persistence of infection in CSOM. Understanding host–pathogen interaction will provide novel avenues to design effective treatment modalities against OM.
Introduction
Innate immune system serves as the first-line of defense against invading pathogens during infection (Alberts et al., 2002; Clarke, 2014). One of the principal components of the innate immune system are macrophages (Tam and Aderem, 2014; Divangahi et al., 2015; Hume, 2015; Schultze et al., 2015). They are considered as “professional phagocytes” that play a crucial role in eradication of pathogens through phagocytosis leading to clearance of infection (Aderem, 2003). Stimuli like infections lead to activation of macrophages. In addition, they can rapidly attract neutrophils, monocytes and other immune cells from the blood and hematopoietic tissues to the site of infection by the release of cytokines and chemotactic substances (Zhang and Wang, 2014). This leads to an efficient orchestration of both innate and adaptive host immune responses. Given the prominent role of macrophages as an effector cell type in the host immune responses, it is not surprising that certain pathogens have evolved strategies to evade killing and use macrophages as a shield against cell-mediated and humoral immune responses to cause infection in humans. Some pathogens inhibit phagocytosis and subsequently activation of macrophages (Fällman et al., 2002). However, no information is available regarding host–pathogen interplay in the pathogenesis of otitis media (OM).
Otitis media refers to any inflammatory or infectious process involving the middle ear (Cunningham et al., 2012; Minovi and Dazert, 2014; Atkinson et al., 2015; Wallis et al., 2015). Chronic suppurative OM (CSOM) is a chronic inflammation of the middle ear characterized by persistent middle ear drainage through the perforated tympanic membrane for more than 6 weeks (Bluestone, 1998; Qureishi et al., 2014; Mittal et al., 2015). The most common cause of CSOM is the bacterial infection (Meyerhoff, 1988; Mittal et al., 2015). Pseudomonas aeruginosa is one of the leading causes of CSOM (Yeo et al., 2007; Dayasena et al., 2011; Madana et al., 2011; Afolabi et al., 2012). CSOM is one of the largest public health burdens worldwide leading to hearing loss and life-threatening central nervous system complications, including brain abscess and meningitis (Chew et al., 2012; Yorgancılar et al., 2013; Sun and Sun, 2014). There is an urgent need to develop new therapies to combat this disease and help prevent complications associated with it. Understanding the role of host immunity in the pathogenesis of CSOM will open up novel avenues of treatment against the disease other than antibiotics.
Macrophages are an integral component of innate immunity and provide efficient protection against pathogens; however, the role of macrophages in CSOM has not been elucidated. In this study, we characterized the interaction of otopathogenic P. aeruginosa with primary human monocyte-derived macrophages (MDMs) and mouse bone marrow-derived macrophages (BMMϕ), in vitro. Our data suggests that otopathogenic P. aeruginosa enters and survives inside human MDMs and mouse BMMϕ. Since bacterial outer membrane proteins (OMPs) play a crucial role in interaction of pathogens with immune cells (Confer and Ayalew, 2013), we determined the role of P. aeruginosa OprF in bacterial survival inside macrophages. OprF is the major OMP of P. aeruginosa and has been demonstrated to play an important role in interaction of this pathogen with host cells (Nestorovich et al., 2006; Bouffartigues et al., 2012; Reusch, 2012; Mishra et al., 2015). We observed that survival of otopathogenic P. aeruginosa inside macrophages requires bacterial oprF expression. The ability of P. aeruginosa to survive inside macrophages enables it to escape killing by potent host immune responses and this lack of clearance by phagocytes contributes to the persistence of infection in CSOM.
Materials and Methods
Cell Culture
Mouse BMMϕ were generated by harvesting bone marrow cells from murine tibias and femurs as described earlier (Godek et al., 2006; Weischenfeldt and Porse, 2008). Briefly, bone marrow cells were flushed from mouse bones (C57 BL6), and then differentiated into a macrophage phenotype by incubating in complete DMEM (cDMEM, 10% heat inactivated fetal bovine serum (FBS, Hyclone®, Logan, UT, USA), 30% L-929 fibroblast conditioned medium, 1% penicillin–streptomycin (Gibco, Carlsbad, CA, USA), 0.01 M Hepes buffer, 1 mM sodium pyruvate, and 1% of a × 100 MEM non-essential amino acids solution (all from Sigma, St. Louis, MO, USA) in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech, Herndon, VA, USA). Cells were differentiated for 7 days with media changes every 2 days. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Miami.
Human MDMs were generated by separating monocytes from peripheral blood obtained from healthy blood donors, as previously described (Welin et al., 2008; Eklund et al., 2010; Meng et al., 2015; Shinzaki et al., 2016). Donors were between age groups of 25–50 years, of either sex and were free from blood-borne diseases. In brief, monocytes were obtained by layering blood onto a density gradient, followed by centrifugation and isolation of the monocyte/lymphocyte fraction. Cells were seeded in culture flasks and allowed to adhere for 2 h before non-adherent lymphocytes were washed away. The monocytes were allowed to differentiate into human MDMs for 5–8 days in DMEM (Gibco, Carlsbad, CA, USA) containing 25 mM Hepes, 100 U/ml penicillin, 100 μg/ml streptomycin and 10% active human serum (all Sigma, St. Louis, MO, USA). All cell cultures were incubated under 37°C, 5% CO2, 98% humidity. Human whole blood was obtained from commercial vendors including Interstate blood bank and followed NIH guidelines for the protection of human subjects including informed signed consent from all subjects by these vendors. All the information regarding human subjects was deidentified and investigators were having no access to any human subject information. The institutional review board (IRB) of the University of Miami approved the study protocol.
Bacterial Strains
The clinical strains of P. aeruginosa (10 strains) isolated from CSOM patients attending University of Miami Hospital or Hospital for Rehabilitation of Craniofacial Anomalies of the University of São Paulo, Bauru, Brazil were used in this study. The patients exhibited persistent chronic purulent otorrhoea through perforated tympanic membrane for more than 6 weeks confirmed by otolaryngologic diagnosis. The isolation and identification of P. aeruginosa was performed using standard methods (MacFaddin, 1976; Forbes et al., 1998). The oprF mutant and complemented strain of P. aeruginosa was generated as described in previous studies (Woodruff and Hancock, 1989; Horton et al., 1990; Schweizer and Hoang, 1995; Rietsch et al., 2005; Balasubramanian et al., 2012; Yakhnina et al., 2015) (Supplementary Materials). Bacteria were grown overnight at 37°C in Luria broth (LB) (Teknova, Hollister, CA, USA) in a rotary shaker.
Invasion Assays
Gentamicin protection assays were used to quantify the extent of human MDM and mouse BMMϕ invasion and survival by otopathogenic P. aeruginosa. Briefly, cells were infected with bacteria at 1:1, 5:1, 10:1, 25:1, 50:1, 100:1 multiplicity of infection (MOI) (bacteria:cell) for 1, 2, 4, and 6 h. After incubation, the cells were washed five times with warm RPMI medium (Corning, NY, USA) followed by addition of medium containing gentamicin (200 μg/ml) (Life Technologies, Carlsbad, CA, USA) and further incubated for 1 h at 37°C. All P. aeruginosa isolates we used in this study were sensitive to gentamicin at this concentration. The cells were washed three times with RPMI and then lysed with 1% saponin (Sigma, St. Louis, MO, USA) to release intracellular bacteria. Serial dilutions were then plated on blood agar plates and bacterial colonies were counted the next day. In some experiments, bacteria were pretreated with 20% human or mouse pooled serum or heat inactivated pooled serum and then used to infect human MDMs and mouse BMMϕ, respectively. In some experiments, bacteria were pretreated with monoclonal anti-OprF antibody (kindly provided by Dr. Hancock) and then used in the invasion assay. The monoclonal antibody (mAb) was specific to surface epitopes of OprF and was generated as described previously (Finnen et al., 1992; Rawling et al., 1995). To determine the effect of cytoskeletal inhibitors, macrophages were pretreated with different concentrations of cytochalasin D, vinblastine, nocodazole, or colchicine (all from Sigma, St. Louis, MO, USA) for 30 min before infecting with bacteria, and maintained in the medium for the entire infection period.
Scanning Electron Microscopy (SEM)
Human MDMs and mouse BMMϕ were cultured on glass cover slips and were infected with bacteria for 30 min to 8 h. After incubation, the cells were washed five times with warm phosphate buffer saline (PBS) buffer (Sigma, St. Louis, MO, USA) to remove unbound bacteria and were then processed for scanning electron microscopy (SEM). Samples were fixed in 2% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) in PBS buffer followed by three changes of PBS buffer for 10 min each. The samples were then post-fixed in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA, USA) in PBS buffer for 45 min and rinsed in three changes of PBS buffer for 10 min each. The samples were dehydrated in a graded series of ethanol, dried in hexamethyldisilazane (HMDS) (Electron Microscopy Sciences, Hatfield, PA, USA) and mounted on carbon adhesive tabs fixed to metal stubs. The samples were coated with palladium in a plasma sputter coater and viewed in a SEM (FEI, ESEM-FEG XL-30).
Transmission Electron Microscopy (TEM)
Human MDMs and mouse BMMϕ were infected with bacteria for 30 min to 8 h. After incubation, the cells were washed with PBS and fixed using 2% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA). The samples were rinsed in three washes of PBS buffer then post-fixed in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA, USA) in 0.1 M phosphate buffer for 1 h. After buffer rinses, specimens were dehydrated through a series of graded ethanol, placed in two rinses of propylene oxide (Electron Microscopy Sciences, Hatfield, PA, USA) for 5 min each and then put in a 1:1 mixture of propylene oxide: EMbed/Araldite resin (Electron Microscopy Sciences, Hatfield, PA, USA) for overnight incubation at room temperature. Next day, the pellets were placed in fresh EMbed/Araldite and put in a vacuum desiccator for 2–4 h. The samples were changed to fresh EMbed/Araldite and polymerized overnight. Silver/gold sections were then cut on a Leica Ultracut E (Leica, Buffalo Grove, IL, USA), stained in uranyl acetate (Electron Microscopy Sciences, Hatfield, PA, USA) and lead citrate (Electron Microscopy Sciences, Hatfield, PA, USA), and viewed in a JEOL 1400 electron microscope (JEOL, Peabody, MA, USA) with Gatan Orius SC1000 camera (Gatan, Pleasanton, CA, USA).
Immunofluorescence
For staining of bacteria and actin, human MDMs and mouse BMMϕ were cultured in 8-well chamber slides and infected with P. aeruginosa for 30 min to 8 h. After incubation, cells were washed three times with PBS buffer and then fixed and permeabilized with BD cytofix and cytoperm reagent (BD Biosciences, San Jose, CA, USA) for 30 min. After washing, the cells were blocked with 3% normal goat serum (NGS) (Life Technologies, Carlsbad, CA, USA) for 20 min and then incubated with anti-P. aeruginosa antibody (1:200) (Abcam, Cambridge, MA, USA) for 45 min followed by Alexa Fluor 488 antibody (1:1000) (Life Technologies, Carlsbad, CA, USA). After washing, cells were counterstained for actin with rhodamine phalloidin (Life Technologies, Carlsbad, CA, USA) for 45 min, washed and mounted in an antifade Vectashield solution containing 4, 6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA). The cells were viewed with a Zeiss LSM 710 microscope (Carl Zeiss, Germany) and images were assembled using Adobe photoshop 7.0.
Cell Viability
To evaluate the cytotoxic effects of P. aeruginosa on human MDMs and mouse BMMϕ during the course of infection, cells were seeded in 96 well plates and infected with bacteria for 2–24 h at an MOI of 10. Lactate dehydrogenase (LDH) levels were then determined in cell culture supernatants using commercially available kit, as per the manufacturer’s instructions (Cayman Chemical, Ann Arbor, MI, USA). Uninfected cells were included as negative control. Maximum LDH release induced by treatment of macrophages with 1% Triton X-100 (Sigma, St. Louis, MO, USA) was used as a positive control. Results were expressed as percentage LDH release compared to the positive control.
The viability of P. aeruginosa infected human MDMs and mouse BMMϕ was also determined by ethidium homodimer-1 staining (Eidet et al., 2015; McCanna et al., 2015; Yoeruek et al., 2016). Ethidium homodimer-1 enters inside dead cells through the disrupted plasma membrane and binds to nucleic acids that increase its fluorescence intensity leading to the production of red fluorescence. Live cells exclude ethidium homodimer-I staining due to the intact plasma membrane. Human MDMs and mouse BMMϕ were infected with bacteria for 2–24 h time periods at an MOI of 10 and then stained with ethidium homodimer-1. Fluorescence was then determined using a microplate reader with appropriate filters (excitation at 535 nm and emission at 620 nm). The percentage dead cells were then calculated using the following formula:
where F(620 nm) is fluorescence intensity at a wavelength of 620 nm, F(620 nm)sample is fluorescence intensity of human MDMs and mouse BMMϕ infected with P. aeruginosa, F(620 nm)minimum is fluorescence intensity of uninfected human MDMs and mouse BMMϕ, and F(620 nm)maximum is fluorescence intensity of human MDMs and mouse BMMϕ treated with 1% Triton X-100 (Sigma, St. Louis, MO, USA).
Statistical Analysis
Statistical significance was determined by a paired, two-tailed Student’s t-test and ANOVA using SPSS software. Values of P < 0.05 were considered to be statistically significant.
Results
Otopathogenic P. aeruginosa Enters and Survives Inside Primary Human MDMs and Mouse BMMϕ
Since P. aeruginosa is the major etiological agent of CSOM, we hypothesize that this pathogen hijacks host innate immunity that forms the first line of defense. To test this hypothesis, we examined the ability of a clinical isolate of P. aeruginosa from CSOM patient to enter and survive inside primary human and mouse macrophages by gentamicin protection assay at different MOIs for 2 h. We observed that P. aeruginosa is able to enter and survive inside macrophages. At an MOI of 1, log 1.45 colony forming unit (cfu) bacteria were recoverable from human MDMs whereas log 1.12 cfu bacteria were demonstrable inside mouse BMMϕ (Figure 1a). The number of bacteria inside human MDMs increased from 2.23 to 4.11 log cfu with a corresponding increase in MOI from 5 to 10 (Figure 1a). Similar increase in number of bacteria was observable inside mouse BMMϕ. The number of bacteria increased from log 2.11 log cfu at an MOI of 5 to log 3.48 cfu at an MOI of 10 (Supplementary Figure S1a). Further increase in MOI to 25, 50, and 100 caused only marginal increase in number of bacteria inside human MDMs and mouse BMMϕ (P > 0.05). Therefore, we selected a MOI of 10 and post-infection time period of 2 h for further experiments.
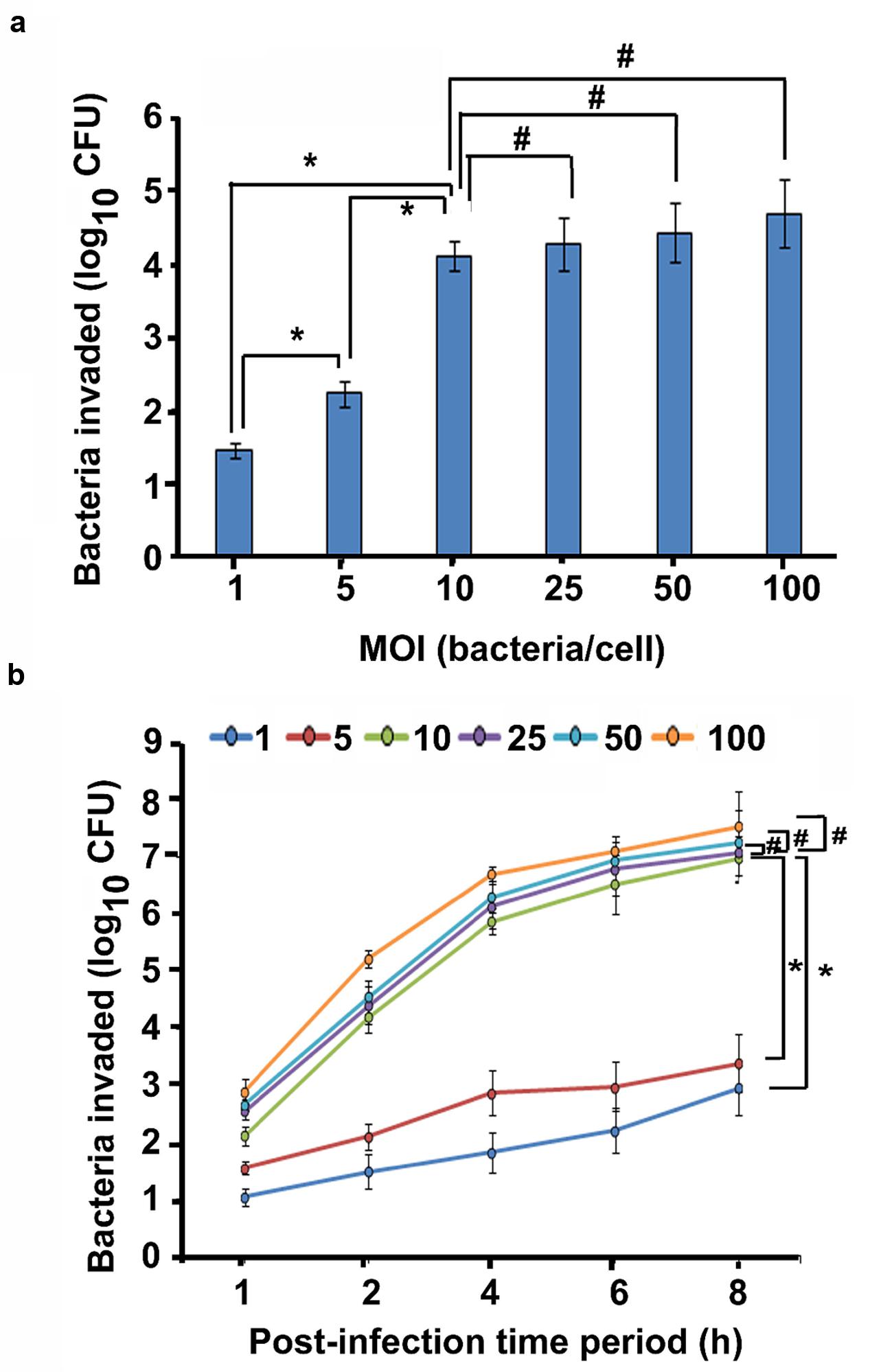
FIGURE 1. Otopathogenic Pseudomonas aeruginosa enters and survives inside macrophages. Human MDMs were infected with a clinical isolate of P. aeruginosa for 2 h and intracellular survival was determined by gentamicin protection assay (a). In separate experiments, human MDMs were infected with P. aeruginosa at different MOIs for 1–8 h and phagocytosis was determined (b). Data represents mean ± SD and is representative of five individual experiments carried out in triplicate. ∗P < 0.01 or #P > 0.05 by Student’s t-test and ANOVA.
In order to determine the effect of increasing infection time on the cell invasion, macrophages and P. aeruginosa were incubated for several time-points. By 1 h post-infection time period, 1.04 log cfu bacteria were demonstrable inside human MDMs that increased to 2.96 log cfu bacteria by 8 h post-infection at an MOI of 1 (Figure 1b). At an MOI of 5 and 10, there were 1.56 and 2.12 log cfu bacteria at 1 h post-infection that increased to 3.39 and 6.98 at 8 h post-infection time period respectively. At a high MOI of 100, 7.55 log cfu bacteria were recoverable from human MDMs at 8 h post-infection time period. Mouse BMMϕ also demonstrated increase in bacterial load with the corresponding increase in post-infection time period from 1 to 8 h at all MOIs (Supplementary Figure S1b). Similar results of human MDM and mouse BMMϕ invasion was observed with nine additional P. aeruginosa CSOM clinical isolates at an MOI of 10 (Supplementary Figures S2 and S3). There was increase in bacterial load in both human MDMs and mouse BMMϕ with increase in post-infection time period from 2 to 6 h. Taken together, these results suggest that otopathogenic P. aeruginosa invades human MDMs and mouse BMMϕ in a time and dose dependent manner.
Opsonization Has No Effect on the Invasion of Macrophages by P. aeruginosa
Complement proteins are constitutively present in the serum and can opsonize bacteria non- specifically promoting pathogen killing and clearance of infection (Daha, 2010; Merle et al., 2015; Varela and Tomlinson, 2015). Therefore, we evaluated whether complement affects the phagocytosis of P. aeruginosa by macrophages. To determine the effect of opsonins on the survival of P. aeruginosa, two CSOM bacterial isolates were opsonized with fresh normal or heat-inactivated human or mouse pooled serum or were treated with medium alone prior to infection of human MDMs and mouse BMMϕ. Interestingly, there was no statistical difference in the number of non-opsonized and opsonized bacteria in human MDMs and mouse BMMϕ (P > 0.05; Figures 2a,b). The heat-inactivated serum also had no significant effect on the ability of otopathogenic P. aeruginosa to invade macrophages (P > 0.05). These results suggest that complement did not play a significant role in invasion of human MDMs and mouse BMMϕ by P. aeruginosa.
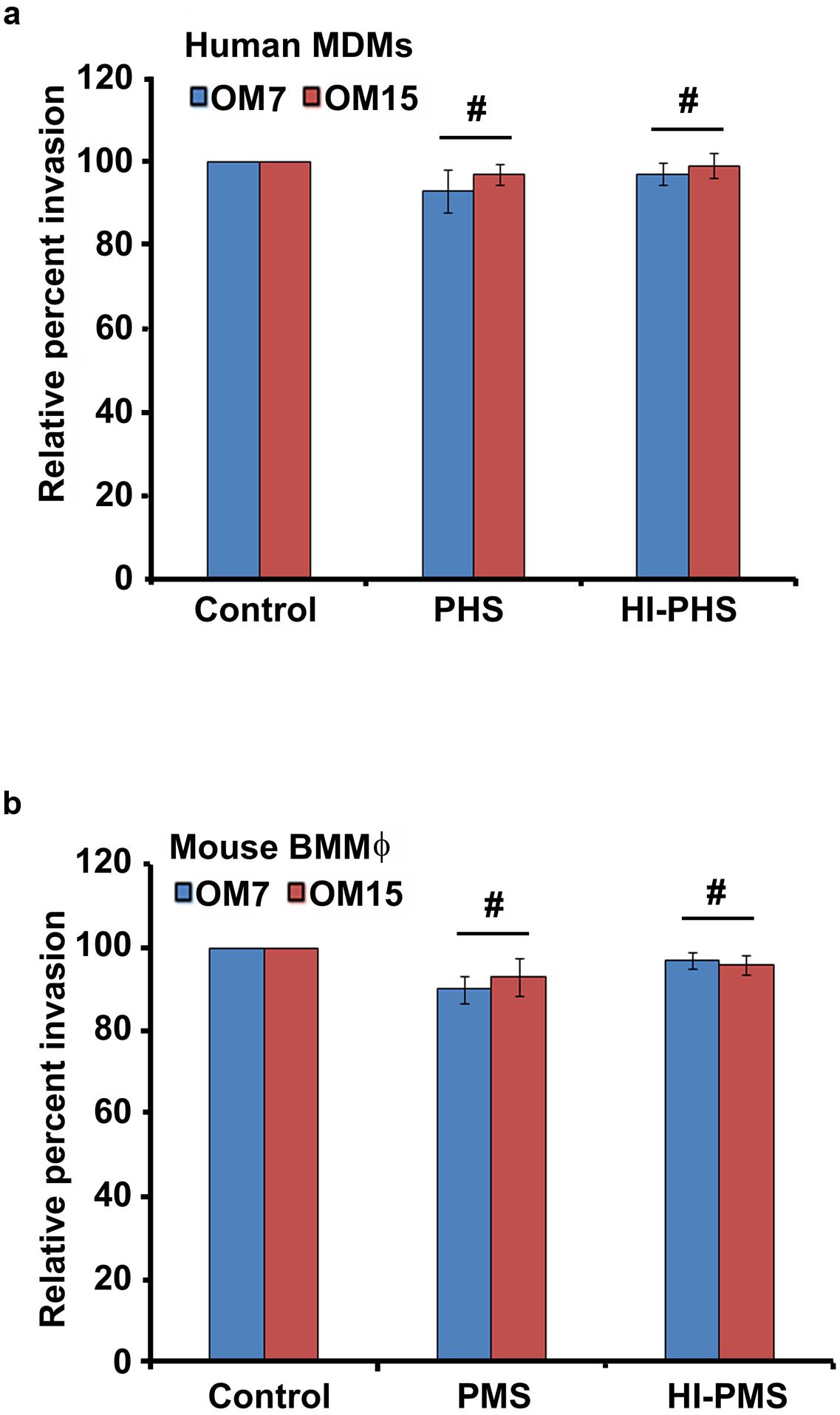
FIGURE 2. Bacterial opsonization has no significant effect on internalization of P. aeruginosa by macrophages. Otopathogenic P. aeruginosa isolates (OM7 and OM15) were pretreated with 20% pooled human serum (PHS) or pooled mouse serum (PMS) or heat-inactivated serum (HI-PHS or HI-PMS) or left untreated (control) and then used to infect human MDMs (a) and mouse BMMϕ (b). The invasion of macrophages by bacteria was then determined by gentamicin protection assay. Results were expressed as percentage invasion relative to control group. Data represents mean ± SD and is representative of four individual experiments carried out in triplicate. #P > 0.05 compared to control by Student’s t-test and ANOVA.
Ultrastructural Examination of P. aeruginosa Infected Human MDMs and Mouse BMMϕ
To examine the interaction of P. aeruginosa with macrophages in detail, human MDMs and mouse BMMϕ were subjected to SEM. During the first 15 min of post-infection, P. aeruginosa was observed to adhere to human MDMs through pseudopod like structures (Figure 3a). There was an increase in the formation of these pseudopod like structures at 30 min post-infection (Figure 3b). Bacteria were seen to be adhered to the human MDMs mostly in clusters. However, individually attached bacteria were also observed. By 1 h post-infection, few bacteria were demonstrable on the surface of human MDMs accompanied with membrane ruffling (Figure 3c). The number of bacteria on the surface of human MDMs increased with increase in post-infection time period (Figures 3d,e). A large number of bacteria were observed on the surface of macrophages by 8 h post-infection (Figure 3f). Similarly few bacteria were demonstrable on the surface of mouse BMMϕ at 60 min post-infection that increased in number with increase in post-infection time period to 8 h. As observed with human MDMs, we also observed pseudopod formation and membrane ruffling in P. aeruginosa infected mouse BMMϕ (Supplementary Figure S4).
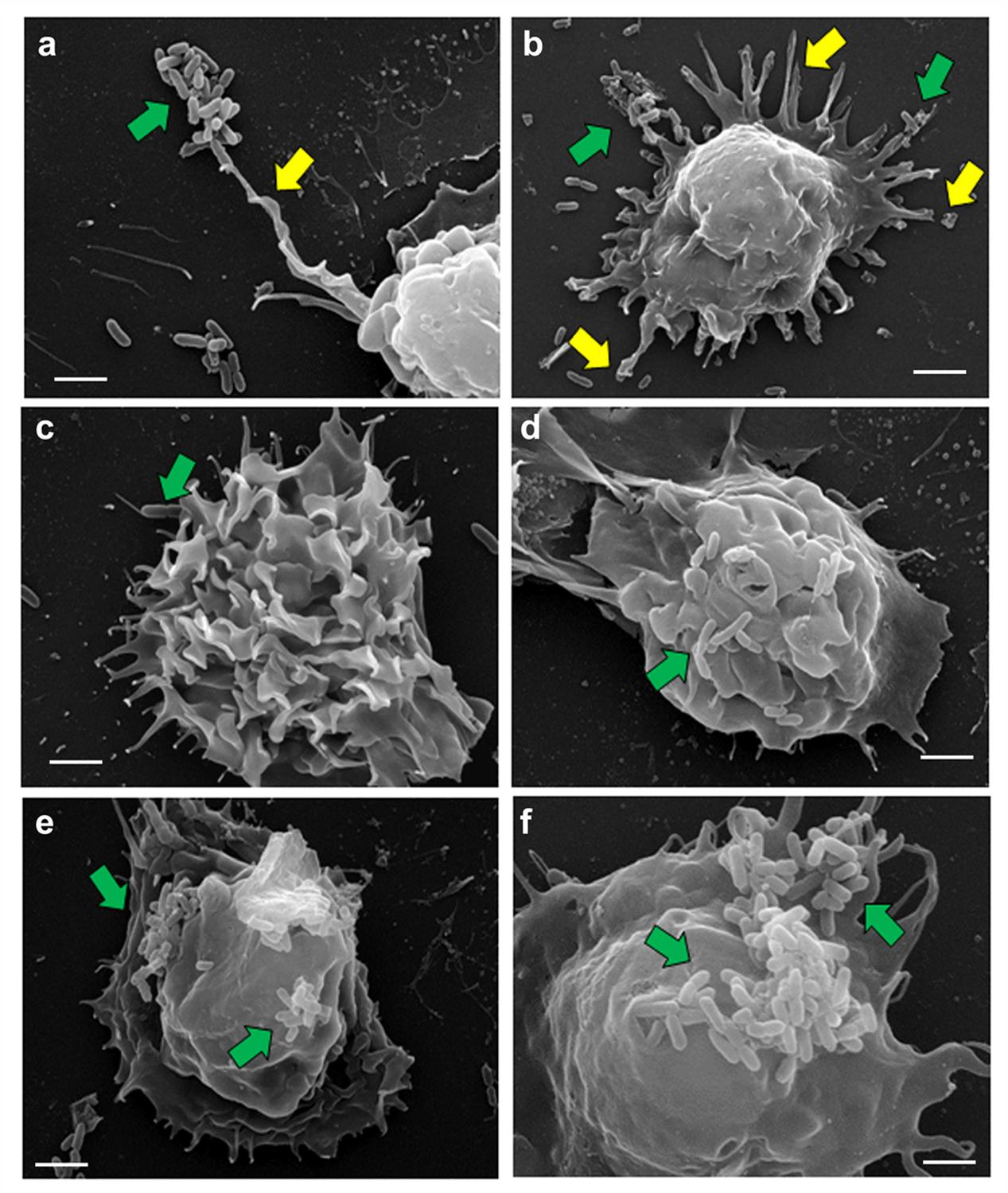
FIGURE 3. Scanning electron micrographs of human MDMs infected with P. aeruginosa. Human MDMs were infected with P. aeruginosa for 15 min (a), 30 min (b), 1 h (c), 2 h (d), 4 h (e), and 8 h (f) and subjected to SEM. We observed that P. aeruginosa (green arrows) attach to pseudopod like structures (yellow arrows) on human macrophages. Results are representative of three individual experiments. Scale bars 2 μm.
Transmission electron microscopy (TEM) demonstrated that human MDMs form plasma membrane protrusions in response to P. aeruginosa infection that can act as bacterial adhesion sites (Figure 4a). The bacteria were found in the enclosed membrane protrusions by 30 min post-infection in human MDMs (Figure 4b). At 60 min post-infection, P. aeruginosa was internalized into human MDMs inside membrane bound vacuoles (Figure 4c). In some vacuoles, the membrane was closely apposed to the bacteria whereas in others there was a space between bacteria and the membrane. In addition, actively dividing bacteria inside human MDMs were also observed (Figure 4d). There was increase in bacterial number inside human MDMs at 2 h post-infection (Figure 4e). At 4 h post-infection, the vacuoles containing bacteria increased in size (Figure 4f). At 8 h post-infection, there was clustering of bacteria inside human MDMs. In some vacuoles, multiple bacteria were demonstrable inside them (Figure 4g). At this post-infection time-period, P. aeruginosa disrupted the membrane and many free bacteria were observed in the cytoplasm of human MDMs (Figure 4h). Mouse BMMϕ displayed similar phenotypic changes in response to otopathogenic P. aeruginosa infection (Supplementary Figure S5).
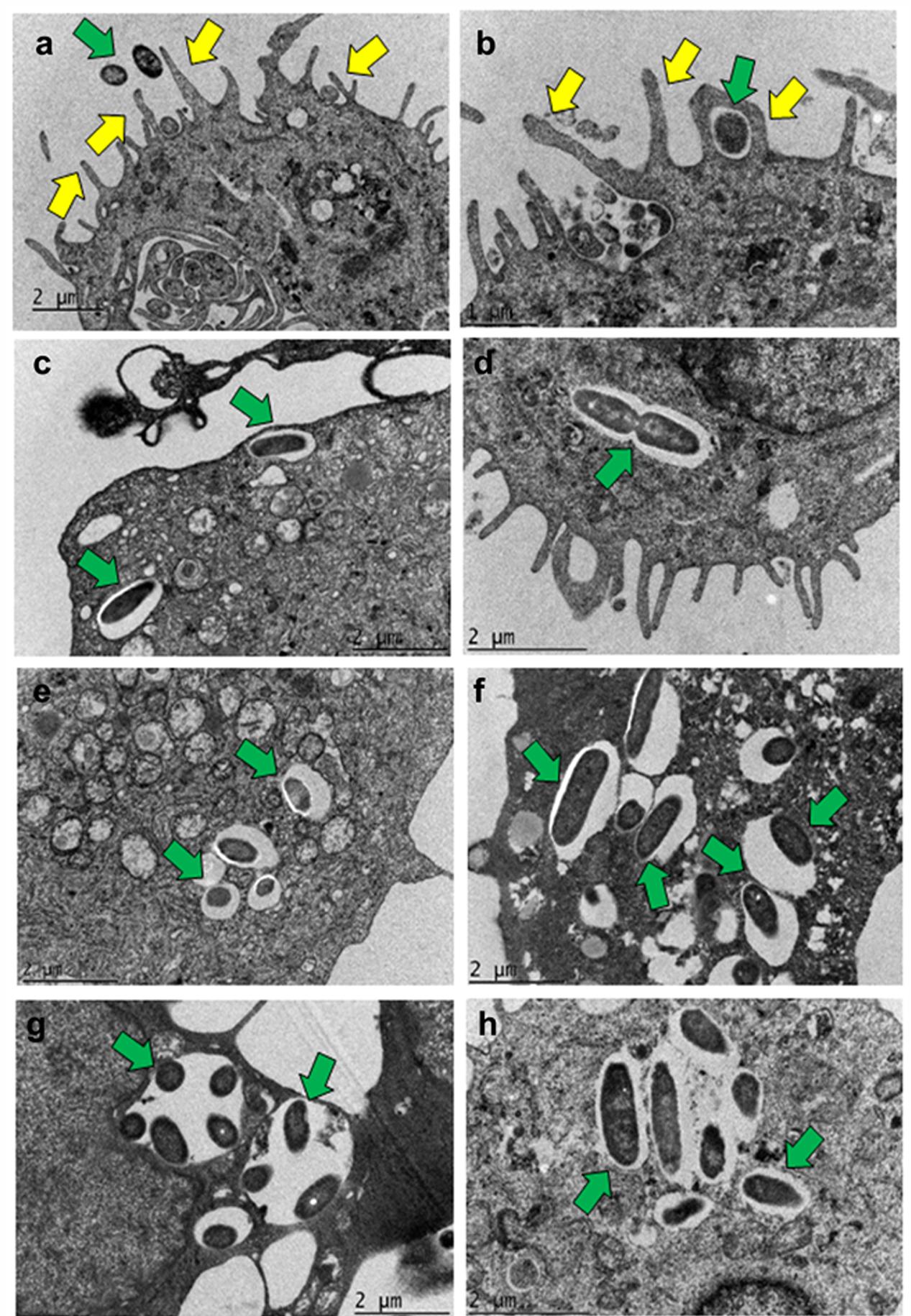
FIGURE 4. Transmission electron micrographs demonstrating phagocytosis of P. aeruginosa by macrophages. Human MDMs were infected with P. aeruginosa for 15 min (a), 30 min (b), 1 h (c,d), 2 h (e), 4 h (f), and 8 h (g,h) and subjected to TEM. Bacteria were demonstrable inside membrane bound vacuoles in human MDMs which eventually were disrupted by 8 h post-infection. Yellow arrows indicate pseudopod like structures and green arrows indicate bacteria. Results are representative of three individual experiments. Scale bars (a,c–h) 2 μm; (b) 1 μm.
Entry of Otopathogenic P. aeruginosa Is Dependent on Both Microfilament and Microtubule Associated Pathway
Host cytoskeleton can play a crucial role in bacterial cell invasion. Therefore, we determined whether invasion of macrophages by P. aeruginosa relies on microfilament and microtubule dependent pathways. Human MDMs and mouse BMMϕ were infected with P. aeruginosa in the presence of increasing concentrations of cytochalasin D, an inhibitor of actin polymerization. There was a significant dose-dependent decrease in the invasion of human MDMs and mouse BMMϕ in the presence of cytochalsin D compared to DMSO treated or untreated cells (P < 0.01) (Figure 5a). A 30% decrease in invasion in the presence of 2 μM cytochalasin D was observed whereas 90% decrease in invasion was observable at a concentration of 20 μM in human MDMs (Figure 5a). Microtubule disrupting compounds, vinblastine, colchicine, and nocodazole also caused a dose-dependent decrease in the invasion of human MDMs by P. aeruginosa (P < 0.01). Human MDMs pretreated with 10 μM vinblastine showed 40% decrease in invasion whereas cells pretreated with 50 μM demonstrated more than 90% decrease in invasion compared to DMSO treated or untreated macrophages (Figure 5b). Colchicine caused an 80% decreased in invasion at a concentration of 25 μM, whereas a 95% decrease in invasion was observed in the presence of 30 μM nocodazole (Figures 5c,d). Similar significant decrease in invasion of mouse BMMϕ was observed in the presence of cytochalsin D, vinblastine, colchicine, and nocodazole (P < 0.01) (Figures 5a–d). We observed that there were no toxic effects of these reagents on bacteria or on cells at the tested concentrations (data not shown). These results with inhibitory compounds suggest that P. aeruginosa invades human MDMs and mouse BMMϕ through both microfilament and microtubule dependent uptake mechanisms.
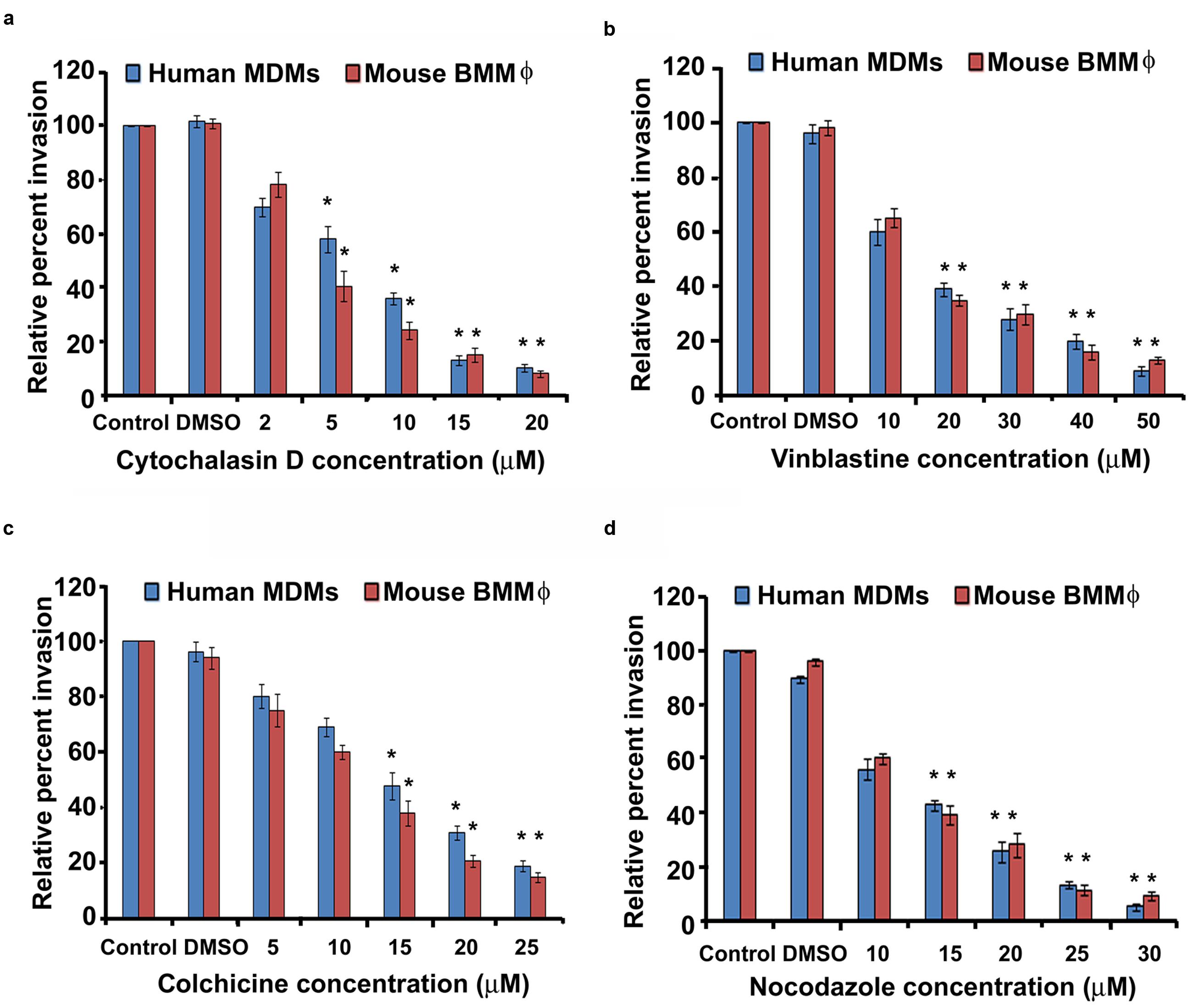
FIGURE 5. Macrophages phagocytose P. aeruginosa through microfilament and microtubule dependent uptake mechanisms. Human MDMs or mouse BMMϕ were pretreated with cytochalasin D (a), vinblastine (b), colchicine (c), and nocodazole (d) and then infected with P. aeruginosa. Phagocytosis of P. aeruginosa by macrophages was determined by gentamicin protection assay. Data represents mean ± SD and is representative of four individual experiments carried out in triplicate. ∗P < 0.01 compared to control by Student’s t-test and ANOVA.
P. aeruginosa Induces Actin Cytoskeleton Rearrangements during Invasion of Macrophages
Actin cytoskeleton rearrangement is a common strategy employed by pathogens to invade host cells (Yang et al., 2012; Navarro-Garcia et al., 2013; de Souza Santos and Orth, 2015; Zheng et al., 2015). To determine whether P. aeruginosa induces alterations to the cytoskeleton during invasion of human MDMs and mouse BMMϕ, the distribution of F-actin in macrophages was examined. Human MDMs and mouse BMMϕ were infected with P. aeruginosa and stained with rhodamine phalloidin to detect F-actin. Uninfected human MDMs showed spatial distribution of F-actin throughout the cell (Figures 6a–d). However, infected human MDMs showed actin condensation in response to otopathogenic P. aeruginosa infection (Figure 6). At 30-min post-infection, a lot of actin accumulation underneath the bacterial binding sites was observed (Figures 6e–h). There was a lot of interaction of bacteria with the actin filaments. At 60-min, there was further increase in actin accumulation that colocalized with the bacteria as indicated by the yellow color (Figures 6i–l). Similar pattern of actin accumulation was observed in mouse BMMϕ infected with otopathogenic P. aeruginosa (data not shown). Taken together, these findings suggest that P. aeruginosa induces actin cytoskeleton rearrangements that facilitate its entry inside macrophages.
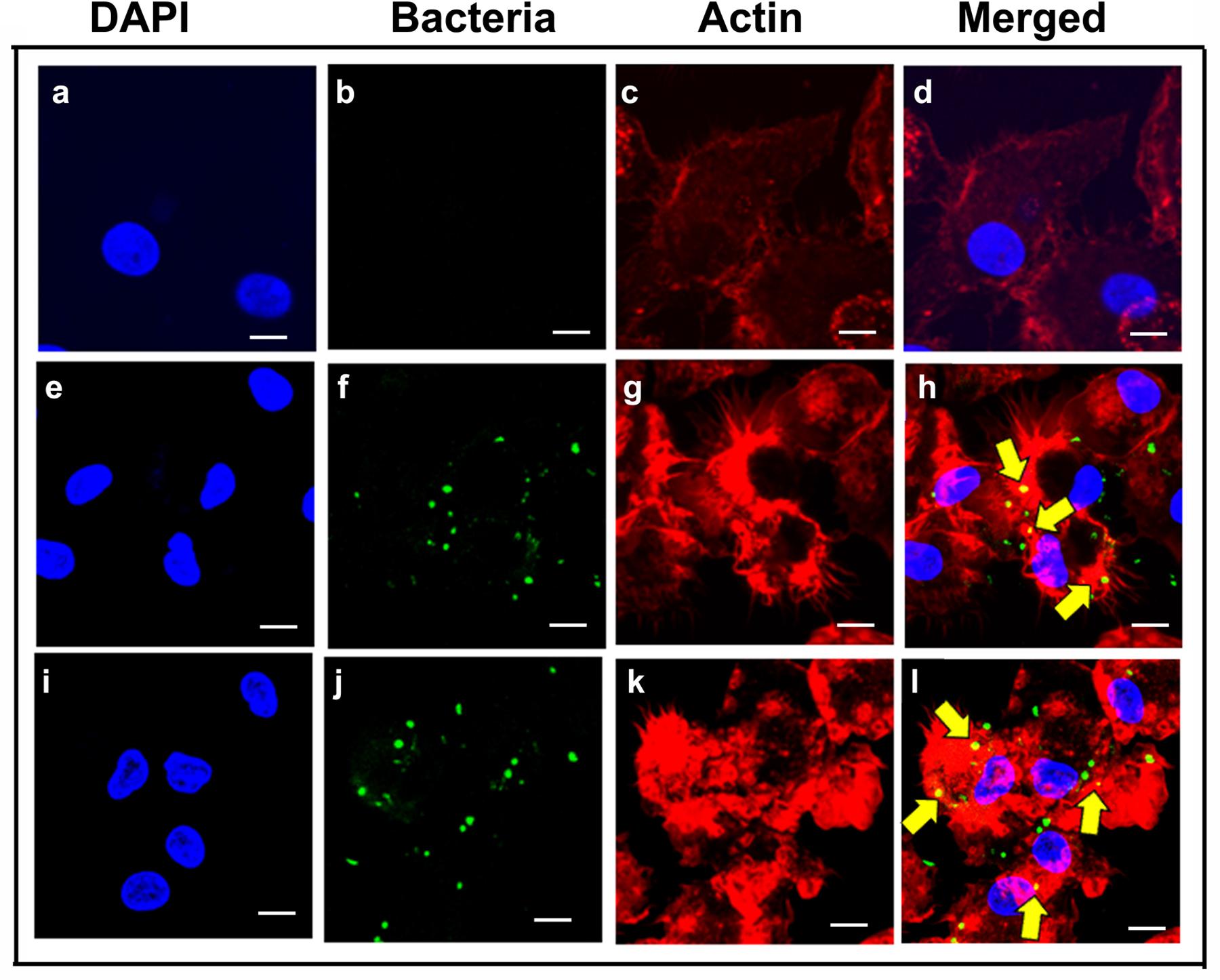
FIGURE 6. Pseudomonas aeruginosa induces actin cytoskeletal rearrangements during invasion of macrophages. Human MDMs were infected with P. aeruginosa for 30 min (e–h) or 60 min (i–l) or left uninfected (a–d) and then stained with anti-P. aeruginosa antibody followed by Alexa Fluor 488 (green). Cells were counterstained with rhodamine phalloidin to visualize actin (red) and DAPI for cell nuclei (blue). Results are representative of three individual experiments. Scale bars 10 μm.
Bacterial OprF Expression Plays a Crucial Role in Invasion of Macrophages by P. aeruginosa
Bacterial OMPs play a crucial role in interaction of pathogens with host cells (Toma et al., 2014; Alzahrani et al., 2015; Bulir et al., 2015). OprF is the major OMP/porin of P. aeruginosa that has been demonstrated to facilitate biofilm formation under anaerobic conditions and adhesion to host cells (Azghani et al., 2002; Yoon et al., 2002). Therefore, we examined whether OprF play a role in the invasion of macrophages by P. aeruginosa. Both human and mouse macrophages were infected with wild-type (WT), ΔoprF and trans-complemented (pOprF) strains of P. aeruginosa. At 2 h post-infection, there was no significant difference (P > 0.05) in the phagocytosis of ΔoprF mutant strain of P. aeruginosa compared to WT or pOprF strains (Figures 7a,b). However, the number of ΔoprF bacterial mutant decreased significantly inside both human MDMs and mouse BMMϕ at 4 h post-infection (P < 0.01). At 6 h post-infection, no viable ΔoprF bacteria were recoverable from human MDMs and very low number of bacteria from mouse BMMϕ. In contrast, high numbers of bacteria were recoverable from WT or pOprF infected human MDMs and mouse BMMϕ.
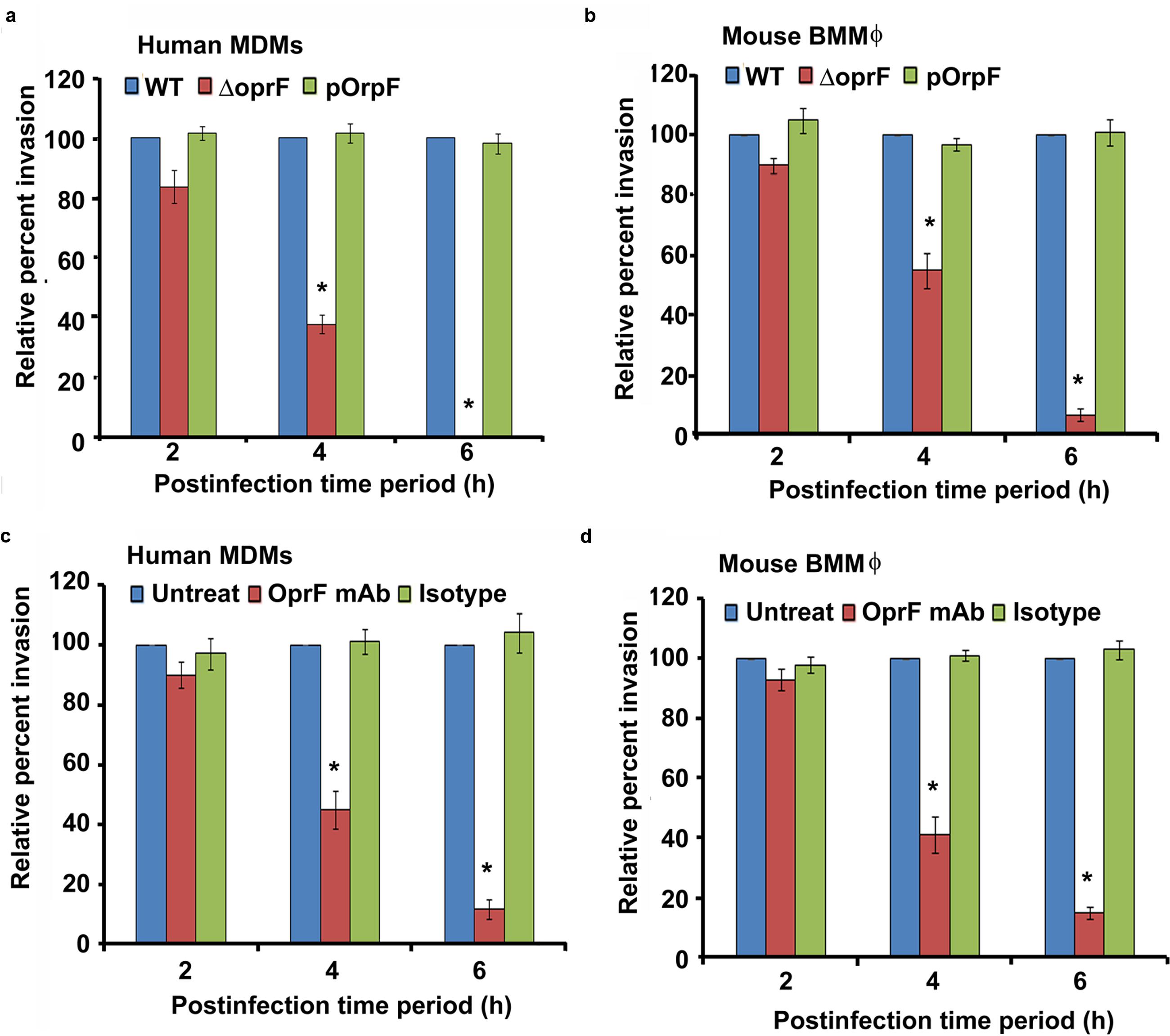
FIGURE 7. Intracellular survival of P. aeruginosa inside macrophages requires bacterial oprF expression. Human MDMs and mouse BMMϕ were infected with wild-type (WT), ΔoprF mutant or plasmid complemented (pOprF) strains of P. aeruginosa. Phagocytosis of bacteria by human MDMs (a) and mouse BMMϕ (b) was determined by gentamicin protection assay. In separate experiments, bacteria were pretreated with anti-oprF monoclonal antibody or left untreated and then used to infect human MDMs (c) and mouse BMMϕ (d). Results were expressed as percentage compared to the phagocytosis of the WT strain. Data represents mean ± SD and is representative of four individual experiments carried out in triplicate. ∗P < 0.01 compared to control by Student’s t-test and ANOVA.
To further confirm the role of OprF in invasion of macrophages by P. aeruginosa, WT bacteria were pretreated with anti-OprF monoclonal antibody or isotype control or left untreated and then used to infect human MDMs and mouse BMMϕ. Pretreatment of bacteria with anti-OprF monoclonal antibody significantly decreased the number of bacteria recoverable from human MDMs at 4 and 6 h post-infection compared to isotype control treated or untreated bacteria (P < 0.01) (Figure 7c). Similar decrease in number of intracellular P. aeruginosa was observed at 4 and 6 h post-infection when pretreated bacteria were used to infect mouse BMMϕ (Figure 7d). These results suggest that OprF plays a critical role in the survival of P. aeruginosa inside macrophages.
Otopathogenic P. aeruginosa Exerts Cytopathic Effect on Macrophages
To determine whether infection of macrophages with P. aeruginosa causes cell death. Human MDMs and mouse BMMϕ were infected with P. aeruginosa at an MOI of 10 and the levels of LDH released were determined in the cell culture supernatants. LDH release is the most acceptable and reliable marker to determine cell viability34-36. There was not much cell damage up to 8 h post-infection as demonstrated by minimal LDH release in cell culture supernatants of both human MDMs and mouse BMMϕ infected with WT otopathogenic P. aeruginosa (Figures 8a,b). However, a further increase in time resulted in significant cell death (P < 0.01). LDH levels increased from 12.5% at 10 h post-infection to 46.7% by 16 h post-infection in culture supernatants of infected human MDMs. Similarly, LDH levels increased from 15.7% at 8 h post-infection to 39.8% at 16 h post-infection in mouse BMMϕ. At 24 h post-infection, there was even a higher level of cell death, as indicated by high LDH levels in human MDMs and mouse BMMϕ. Interestingly, deletion of oprF abrogated the ability of otopathogenic to induce significant cell death in macrophages (P < 0.01). However, complementation with the pOprF plasmid restored the ability of mutant strain to exert cytopathic effects on human MDMs and mouse BMMϕ.
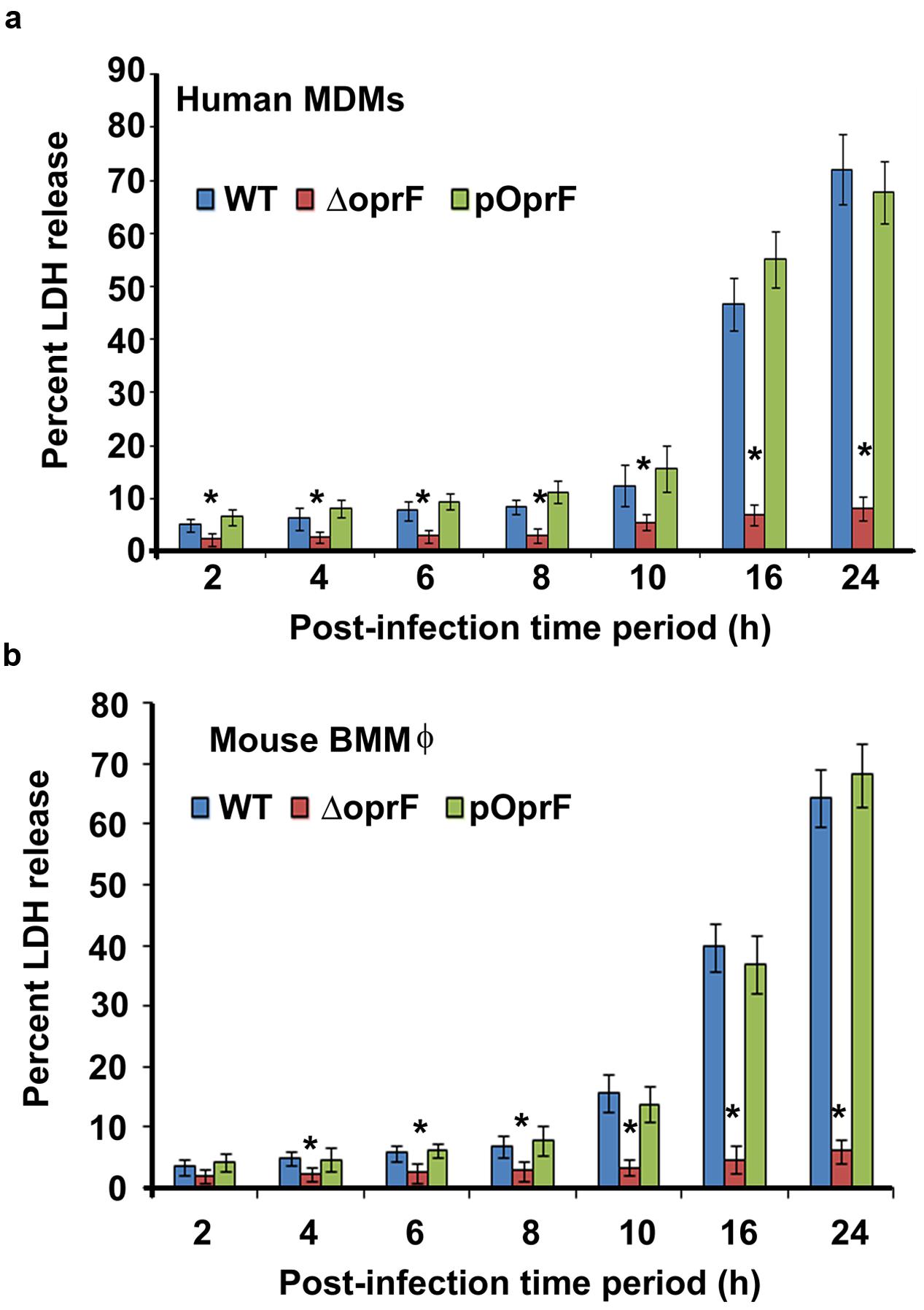
FIGURE 8. Otopathogenic P. aeruginosa affect viability of macrophages. Human MDMs (a) or mouse BMMϕ (b) were infected with P. aeruginosa at an MOI of 10 and viability of the macrophages was determined by measuring LDH levels in cell culture supernatants. Results were expressed as the percentage compared with maximum LDH release by lysed cells. Data represents mean ± SD and is representative of four individual experiments carried out in triplicate. ∗P < 0.01 compared to WT or pOprF by Student’s t-test and ANOVA.
To further confirm that macrophages undergo apoptosis, infected human MDMs and mouse BMMϕ were stained with ethidium homodimer-1, a high affinity, membrane-impermeant dye that exclusively stains the DNA of dead cells (Eidet et al., 2015; McCanna et al., 2015; Yoeruek et al., 2016). In agreement with the LDH assay results, at 8 h post-infection very little ethidium homodimer-1 staining of infected human MDMs was observed (P > 0.05) (Supplementary Figure S6). However, 25% of human MDMs were stained with membrane-impermeant dye at 10 h post-infection. At 16 h post-infection, 50% of infected MDMs failed to exclude the dye. By 24 h post-infection, almost 75% of human MDMs had been stained with ethidium homodimer-1 suggesting considerable cell death. Similar results were obtained with mouse BMMϕ demonstrating extensive cell death at 24 h post-infection (Supplementary Figure S6). On par with our LDH data, human MDMs and mouse BMMϕ infected with ΔoprF mutant strain did not demonstrate significant ethidium homodimer-1 staining even at 24 h post-infection compared to WT infected cells (P < 0.01). This phenotype was restored upon complementation with pOprF. These results suggest that otopathogenic P. aeruginosa exerts cytopathic effects on macrophages for which bacterial OprF expression is necessary.
Discussion
Despite advances in medical therapy, CSOM is still a clinically challenging disease. Antibiotics are the only available treatment modalities against CSOM at present but these have moderate efficacy against the disease, and at times are not effective at all. The antibiotics used to treat CSOM include neomycin, ciprofloxacin, cefepime, carbapenem, levofloxacin, and ceftazidime (Saunders J. et al., 2011; Mittal et al., 2015). However, P. aeruginosa has been demonstrated to be resistant to a wide variety of antibiotics including β lactams (penicillins, cephalosporins, and carbapenems), fluoroquinolones (ciprofloxacin), polymyxins and macrolides (erythromycin and azithromycin) (Nordmann and Guibert, 1998; Livermore, 2002; Walsh et al., 2003; Jang and Park, 2004; Poole, 2004, 2011; Saunders J. et al., 2011; Morita et al., 2014; Mittal et al., 2015). P. aeruginosa exhibits some degree of sensitivity to aminoglycosides but this class of antibiotics has significant ototoxicity and is not recommended for the treatment of CSOM (Black et al., 2004; Jing et al., 2015; Koo et al., 2015; Leis et al., 2015). A better knowledge of the interaction of pathogens with immune cells will provide new opportunities to design effective novel therapeutic strategies against CSOM. Although immune cells play an important role in clearance of infection, the interaction of otopathogenic P. aeruginosa with macrophages has never been investigated. The results of the present study contribute to our understanding of the interaction between otopathogenic P. aeruginosa and primary macrophages.
Macrophages form an important line of host defense in innate immune system against infections (Aderem, 2003; Tam and Aderem, 2014; Zhang and Wang, 2014; Divangahi et al., 2015; Hume, 2015; Schultze et al., 2015). Some studies employ macrophage cell lines to understand the interaction of pathogens with immune cells that may not mimic the true characteristics of primary cells. These cell lines are fundamentally different from the primary cells in that they grow continuously in culture due to permanent alterations in their genes. Such changes could have an effect on the signaling cascades that are activated following interaction of immune cells with pathogens. The results of studies utilizing primary cells have been instrumental in developing our understanding regarding immune cell response to infection. Therefore, in the present study primary human MDMs and mouse BMMϕ have been used to investigate the interaction between otopathogenic P. aeruginosa and macrophages. This study for the first time demonstrated that otopathogenic P. aeruginosa enters and survives inside primary human MDMs and mouse BMMϕ in a dose and time dependent manner in context of ear infections. We also observed that serum opsonization has no significant effect on invasion of macrophages by P. aeruginosa. Since opsonization of pathogens by serum components facilitate phagocytosis through Fc gamma receptors on macrophages, these results suggest that otopathogenic P. aeruginosa employs unique mechanisms to enter inside macrophages. SEM demonstrated that otopathogenic P. aeruginosa adheres to the primary macrophages through the formation of pseudopod like structures. TEM confirmed the internalization of bacteria inside human MDMs and mouse BMMϕ. We also observed that survival of otopathogenic P. aeruginosa inside depends on the expression of OprF, the most abundant bacterial OMP/porin. On par with these findings, pretreatment of bacteria with anti-OprF monoclonal antibody significantly decreased the invasion of macrophages by P. aeruginosa, highlighting the crucial role of OprF in cell invasion. The ability of otopathogenic P. aeruginosa to survive inside macrophages provides protection against complement, lysozyme activity and other host immune defenses. This enables P. aeruginosa to evade killing and hence may constitute an important virulence trait.
Macrophages are a preferred niche for certain pathogens as they are highly phagocytic and long lived, providing protection for a prolonged period. Pathogens including Candida glabrata, Enterococcus faecalis, Brucella sp., Mycobacterium tuberculosis, Escherichia coli and Group B streptococci, have also been demonstrated to enter and survive inside macrophages, which has been correlated with their ability to cause infection (Valentin-Weigand et al., 1996; Cornacchione et al., 1998; Pieters and Gatfield, 2002; Celli, 2006; Miramón et al., 2013; Miskinyte and Gordo, 2013; Elliott et al., 2015; Kasper et al., 2015; Sabatino et al., 2015). Other pathogens such as Yersinia spp. avoid uptake and phagocytosis preventing activation of macrophages thus avoiding potent host immune responses (Fällman et al., 2002). It has been demonstrated that tyrosine phosphorylation plays a crucial role in phagocytosis and subsequent activation of professional phagocytes (Park et al., 2011). However, pathogens produce effector proteins to neutralize tyrosine phosphorylation that promotes inhibition of phagocytosis and activation of macrophages. The effector protein, YopH, of Yersinia spp. possess tyrosine phosphatase activity thus counteracting the activating tyrosine phosphorylation signals of the host cell (Yuan et al., 2005). This protein is delivered into host cells through bacterial type III secretion system (T3SS) and prevents Yersinia internalization by macrophages by dephosphorylating the adaptor protein p130Cas, among other targets (Andersson et al., 1996). Salmonella typhimurium secretes the tyrosine phosphatase SptP, which is also injected into host cells through a T3SS (Kaniga et al., 1996). SptP deactivates Rho-family GTPases and subsequently abrogates activation of macrophages. In addition, in case of adherent S. aureus, it has been demonstrated that surface of biomaterial plays a crucial role in bacterial phagocytosis by macrophages (Domingues et al., 2015). S. aureus adhered to hydrophilic surfaces had a lowest rate of phagocytosis while bacteria adhered to common biomaterials such as silicone rubber, tissue culture polystyrene and stainless steel has intermediate rate of phagocytosis by J774A macrophage cell line. It was concluded that hydrophobicity is a necessary surface condition for effective phagocytosis of S. aureus by J774A macrophage cell line. Our previous studies have demonstrated that P. aeruginosa can invade human middle ear epithelial cells (HMEECs) (Mittal et al., 2014). However, the interaction of otopathogenic P. aeruginosa with primary macrophages has never been explored in previous studies.
Pathogens including Shigella dysenteriae utilize cytoskeletal rearrangement in order to gain entry inside host cells (Rottner et al., 2005). However, each pathogen utilizes unique mechanism to invade immune cells. We observed that internalization of P. aeruginosa inside human MDM and mouse BMMϕ relies on both microfilament and microtubule dependent uptake mechanisms. Pretreatment of human MDMs and mouse BMMϕ with actin polymerization inhibitors or microtubule-destabilizing agents led to a significant decrease in invasion of macrophages by P. aeruginosa in a dose-dependent manner. Some pathogens utilize only microfilaments whereas others employ microfilaments to gain entry inside host cells that are also cell type specific (Oelschlaeger et al., 1993; Kopecko et al., 2001; Mazon Moya et al., 2014; Valencia-Gallardo et al., 2015). However, the entry of other pathogens requires both microfilament and microtubule dependent uptake mechanisms (Ferrero et al., 2009; Taylor et al., 2010) as observed for otopathogenic P. aeruginosa in the present study.
Pathogens employ various strategies in order to survive inside the host and cause infection. Some pathogens trigger anti-apoptotic mechanisms to prevent host cell death. Pathogens including Enterococcus faecalis, Toxoplasma gondii, Brucella sp., Neisseria meningitidis, and Neisseria gonorrhoeae inhibit apoptosis of host cells, providing a niche where they can survive and replicate (Beck and Meyer, 2000; Tunbridge et al., 2006; Cai et al., 2014; Cui et al., 2014; Zou and Shankar, 2014). On the contrary, other microbes including Mycobacterium tuberculosis, Legionella pneumophila, Bordetella pertussis, Listeria monocytogenes, Corynebacterium diphtheriae, Shigella flexneri, and Salmonella typhimurium, induces cell death that allow the pathogens to efficiently exit the host cell, spread to neighboring cells, evade immune cells, and/or to gain nutrients (Haimovich and Venkatesan, 2006; Hewlett et al., 2006; Cervantes et al., 2008; dos Santos et al., 2010; Morinaga et al., 2010; Welin et al., 2011; Ashida et al., 2014; Behnsen et al., 2015). Thus, knowledge of the mechanisms employed by pathogens in the progression of disease is critical to have a better understanding of virulence and host defense. In this study, we observed that otopathogenic P. aeruginosa induces death of human MDMs and mouse BMMϕ as determined by LDH release in the cell culture supernatants and ethidium homodimer-I staining. LDH is the most acceptable and reliable marker for determining host cell death (Kwon et al., 2015; Park et al., 2015; Lv et al., 2016). Ethidium homodimer-I is a fluorescent nuclear stain that penetrates dead cells due to disrupted plasma membrane and increases intensity after binding to DNA (Eidet et al., 2015; McCanna et al., 2015; Yoeruek et al., 2016). The ability of otopathogenic P. aeruginosa to induce cell death in macrophages enables this pathogen to escape potent host defenses and will enable it to cause infection.
In summary, the results presented here demonstrate for the first time that otopathogenic P. aeruginosa are able to enter and survive inside macrophages in context of ear infections. Studies are in progress in our laboratory to elucidate the molecular mechanisms through which otopathogenic P. aeruginosa escape from macrophage killing. The uptake of otopathogenic P. aeruginosa by human MDMs and mouse BMMϕ relies on actin polymerization and microtubule dependent process. Bacterial OprF expression plays a crucial role in the intracellular survival of P. aeruginosa inside human MDMs and mouse BMMϕ. Future studies comparing the level of oprF gene expression between strains isolated from CSOM patients and the other diseases such as pneumonia or urinary infection will help in confirming the role of bacterial oprF for the establishment of CSOM. In addition, it has been demonstrated that pathogens form biofilms on middle ear during CSOM (Saunders J.E. et al., 2011; Kaya et al., 2013; Gu et al., 2014). Therefore, future investigations are warranted to determine the interaction of biofilm cells of otopathogenic P. aeruginosa with primary macrophages. Our findings suggest that otopathogenic P. aeruginosa is recognized by phagocytic cells but remain impermeable to attacks by their antimicrobial components and is able to exert cytotoxic effects on macrophages. Further studies employing mouse model of CSOM are warranted to delineate the role of macrophages in the disease process. Understanding host–pathogen interactions will provide novel avenues to design effective treatment modalities against CSOM, and hence, prevent consequent hearing loss as well as life-threatening CNS complications.
Author Contributions
RM, CL, HK, MG, CJ, and PB performed the experiments. RM, KM, MG, and PB wrote the manuscript. RM, KM, PW, CJ, DY, and XL designed and supervised the study. All authors approved the final version of the manuscript.
Funding
The research work in Dr. XL’s laboratory is supported by grants R01 DC05575, R01 DC01246, and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. R. E. W. Hancock for kindly providing anti-OprF monoclonal antibody. We are grateful to April Mann for critical reading of the manuscript. We are thankful to Electron Microscopy Core Facility at Miller School of Medicine, University of Miami for assisting in electron microscopy experiments. We are also thankful to Dr. Marcia Boulina, University of Miami Analytical Imaging Core Facility, for help in confocal microscopy experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01828/full#supplementary-material
References
Aderem, A. (2003). Phagocytosis and the inflammatory response. J. Infect. Dis. 187(Suppl. 2), S340–S345. doi: 10.1086/374747
Afolabi, O. A., Salaudeen, A. G., Ologe, F. E., Nwabuisi, C., and Nwawolo, C. C. (2012). Pattern of bacterial isolates in the middle ear discharge of patients with chronic suppurative otitis media in a tertiary hospital in North central Nigeria. Afr. Health Sci. 12, 362–367.
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular Biology of the Cell, 4th Edn. New York, NY: Garland Science.
Alzahrani, H., Winter, J., Boocock, D., De Girolamo, L., and Forsythe, S. J. (2015). Characterization of outer membrane vesicles from a neonatal meningitic strain of Cronobacter sakazakii. FEMS Microbiol. Lett. 362:fnv085. doi: 10.1093/femsle/fnv085
Andersson, K., Carballeira, N., Magnusson, K. E., Persson, C., Stendahl, O., Wolf-Watz, H., et al. (1996). YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol. Microbiol. 20, 1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x
Ashida, H., Kim, M., and Sasakawa, C. (2014). Manipulation of the host cell death pathway by Shigella. Cell. Microbiol. 16, 1757–1766. doi: 10.1111/cmi.12367
Atkinson, H., Wallis, S., and Coatesworth, A. P. (2015). Acute otitis media. Postgrad. Med. 127, 386–390. doi: 10.1080/00325481.2015.1028872
Azghani, A. O., Idell, S., Bains, M., and Hancock, R. E. (2002). Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33, 109–114. doi: 10.1006/mpat.2002.0514
Balasubramanian, D., Schneper, L., Merighi, M., Smith, R., Narasimhan, G., Lory, S., et al. (2012). The regulatory repertoire of Pseudomonas aeruginosa AmpC ß-lactamase regulator AmpR includes virulence genes. PLoS ONE 7:e34067. doi: 10.1371/journal.pone.0034067
Beck, S. C., and Meyer, T. F. (2000). IgA1 protease from Neisseria gonorrhoeae inhibits TNFalpha-mediated apoptosis of human monocytic cells. FEBS Lett. 472, 287–292. doi: 10.1016/S0014-5793(00)01478-2
Behnsen, J., Perez-Lopez, A., Nuccio, S. P., and Raffatellu, M. (2015). Exploiting host immunity: the Salmonella paradigm. Trends Immunol. 36, 112–120. doi: 10.1016/j.it.2014.12.003
Black, F. O., Pesznecker, S., and Stallings, V. (2004). Permanent gentamicin vestibulotoxicity. Otol. Neurotol. 25, 559–569. doi: 10.1097/00129492-200407000-00025
Bluestone, C. D. (1998). Epidemiology and pathogenesis of chronic suppurative otitis media: implications for prevention and treatment. Int. J. Pediatr. Otorhinolaryngol. 42, 207–223. doi: 10.1016/S0165-5876(97)00147-X
Bouffartigues, E., Gicquel, G., Bazire, A., Bains, M., Maillot, O., Vieillard, J., et al. (2012). Transcription of the oprF gene of Pseudomonas aeruginosa is dependent mainly on the SigX sigma factor and is sucrose induced. J. Bacteriol. 194, 4301–4311. doi: 10.1128/JB.00509-12
Bulir, D. C., Waltho, D. A., Stone, C. B., Liang, S., Chiang, C. K., Mwawasi, K. A., et al. (2015). Chlamydia Outer Protein (Cop) B from Chlamydia pneumoniae possesses characteristic features of a type III secretion (T3S) translocator protein. BMC Microbiol. 15:163. doi: 10.1186/s12866-015-0498-1
Cai, Y., Chen, H., Mo, X., Tang, Y., Xu, X., Zhang, A., et al. (2014). Toxoplasma gondii inhibits apoptosis via a novel STAT3-miR-17-92-Bim pathway in macrophages. Cell. Signal. 26, 1204–1212. doi: 10.1016/j.cellsig.2014.02.013
Celli, J. (2006). Surviving inside a macrophage: the many ways of Brucella. Res. Microbiol. 157, 93–98. doi: 10.1016/j.resmic.2005.10.002
Cervantes, J., Nagata, T., Uchijima, M., Shibata, K., and Koide, Y. (2008). Intracytosolic Listeria monocytogenes induces cell death through caspase-1 activation in murine macrophages. Cell. Microbiol. 10, 41–52.
Chew, Y. K., Cheong, J. P., Khir, A., Brito-Mutunayagam, S., and Prepageran, N. (2012). Complications of chronic suppurative otitis media: a left otogenic brain abscess and a right mastoid fistula. Ear Nose Throat J. 91, 428–430.
Clarke, T. B. (2014). Microbial programming of systemic innate immunity and resistance to infection. PLoS Pathog. 10:e1004506. doi: 10.1371/journal.ppat.1004506
Confer, A. W., and Ayalew, S. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet. Microbiol. 163, 207–222. doi: 10.1016/j.vetmic.2012.08.019
Cornacchione, P., Scaringi, L., Fettucciari, K., Rosati, E., Sabatini, R., Orefici, G., et al. (1998). Group B streptococci persist inside macrophages. Immunology 93, 86–95. doi: 10.1046/j.1365-2567.1998.00402.x
Cui, G., Wei, P., Zhao, Y., Guan, Z., Yang, L., Sun, W., et al. (2014). Brucella infection inhibits macrophages apoptosis via Nedd4-dependent degradation of calpain2. Vet. Microbiol. 174, 195–205. doi: 10.1016/j.vetmic.2014.08.033
Cunningham, M., Guardiani, E., Kim, H. J., and Brook, I. (2012). Otitis media. Future Microbiol. 7, 733–753. doi: 10.2217/fmb.12.38
Daha, M. R. (2010). Role of complement in innate immunity and infections. Crit. Rev. Immunol. 30, 47–52. doi: 10.1615/CritRevImmunol.v30.i1.30
Dayasena, R., Dayasiri, M., Jayasuriya, C., and Perera, D. (2011). Aetiological agents in chronic suppurative otitis media in Sri Lanka. Australas. Med. J. 4, 101–104. doi: 10.4066/AMJ.2011.549
de Souza Santos, M., and Orth, K. (2015). Subversion of the cytoskeleton by intracellular bacteria: lessons from Listeria, Salmonella and Vibrio. Cell. Microbiol. 17, 164–173. doi: 10.1111/cmi.12399
Divangahi, M., King, I. L., and Pernet, E. (2015). Alveolar macrophages and type I IFN in airway homeostasis and immunity. Trends Immunol. 36, 307–314. doi: 10.1016/j.it.2015.03.005
Domingues, J. F., Roest, S., Wang, Y., van der Mei, H. C., Libera, M., van Kooten, T. G., et al. (2015). Macrophage phagocytic activity toward adhering staphylococci on cationic and patterned hydrogel coatings versus common biomaterials. Acta Biomater. 18, 1–8. doi: 10.1016/j.actbio.2015.02.028
dos Santos, C. S., dos Santos, L. S., de Souza, M. C., dos Santos, Dourado, F., de Oliveira Dias, A. A., et al. (2010). Non-opsonic phagocytosis of homologous non-toxigenic and toxigenic Corynebacterium diphtheriae strains by human U-937 macrophages. Microbiol. Immunol. 54, 1–10. doi: 10.1111/j.1348-0421.2009.00179.x
Eidet, J. R., Utheim,Ø. A., Islam, R., Lyberg, T., Messelt, E. B., Dartt, D. A., et al. (2015). The impact of storage temperature on the morphology, viability, cell number and metabolism of cultured human conjunctival epithelium. Curr. Eye Res. 40, 30–39. doi: 10.3109/02713683.2014.909497
Eklund, D., Welin, A., Schön, T., Stendahl, O., Huygen, K., and Lerm, M. (2010). Validation of a medium-throughput method for evaluation of intracellular growth of Mycobacterium tuberculosis. Clin. Vaccine Immunol. 17, 513–517. doi: 10.1128/CVI.00446-09
Elliott, T. R., Elliott, T. R., Hudspith, B. N., Rayment, N. B., Prescott, N. J., Petrovska, L., et al. (2015). Defective macrophage handling of Escherichia coli in Crohn’s disease. J. Gastroenterol. Hepatol. 30, 1265–1274. doi: 10.1111/jgh.12955
Fällman, M., Deleuil, F., and McGee, K. (2002). Resistance to phagocytosis by Yersinia. Int. J. Med. Microbiol. 291, 501–509. doi: 10.1078/1438-4221-00159
Ferrero, M. C., Fossati, C. A., and Baldi, P. C. (2009). Smooth Brucella strains invade and replicate in human lung epithelial cells without inducing cell death. Microbes Infect. 11, 476–483. doi: 10.1016/j.micinf.2009.01.010
Finnen, R. L., Martin, N. L., Siehnel, R. J., Woodruff, W. A., Rosok, M., and Hancock, R. E. (1992). Analysis of the Pseudomonas aeruginosa major outer membrane protein OprF by use of truncated OprF derivatives and monoclonal antibodies. J. Bacteriol. 174, 4977–4985.
Forbes, B. A., Sahm, D. F., and Weissfeld, A. S. (1998). Bailey and Scott’s Diagnostic Microbiology, 10th Edn. St. Louis, MO: Mosby Inc.
Godek, M. L., Sampson, J. A., Duchsherer, N. L., McElwee, Q., and Grainger, D. W. (2006). Rho GTPase protein expression and activation in murine monocytes/macrophages is not modulated by model biomaterial surfaces in serum-containing in vitro cultures. J. Biomater. Sci. Polym. Ed. 17, 1141–1158. doi: 10.1163/156856206778530731
Gu, X., Keyoumu, Y., Long, L., and Zhang, H. (2014). Detection of bacterial biofilms in different types of chronic otitis media. Eur. Arch. Otorhinolaryngol. 271, 2877–2883. doi: 10.1007/s00405-013-2766-8
Haimovich, B., and Venkatesan, M. M. (2006). Shigella and Salmonella: death as a means of survival. Microbes Infect. 8, 568–577. doi: 10.1016/j.micinf.2005.08.002
Hewlett, E. L., Donato, G. M., and Gray, M. C. (2006). Macrophage cytotoxicity produced by adenylate cyclase toxin from Bordetella pertussis: more than just making cyclic AMP! Mol. Microbiol. 59, 447–459.
Horton, R. M., Cai, Z. L., Ho, S. N., and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528–535.
Hume, D. A. (2015). The many alternative faces of macrophage activation. Front. Immunol. 6:370. doi: 10.3389/fimmu.2015.00370
Jang, C. H., and Park, S. Y. (2004). Emergence of ciprofloxacin-resistant Pseudomonas in chronic suppurative otitis media. Clin. Otolaryngol. Allied Sci. 29, 321–323. doi: 10.1111/j.1365-2273.2004.00835.x
Jing, W., Zongjie, H., Denggang, F., Na, H., Bin, Z., Aifen, Z., et al. (2015). Mitochondrial mutations associated with aminoglycoside ototoxicity and hearing loss susceptibility identified by meta-analysis. J. Med. Genet. 52, 95–103. doi: 10.1136/jmedgenet-2014-102753
Kaniga, K., Uralil, J., Bliska, J. B., and Galán, J. E. (1996). A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol. Microbiol. 21, 633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x
Kasper, L., Seider, K., and Hube, B. (2015). Intracellular survival of Candida glabrata in macrophages: immune evasion and persistence. FEMS Yeast Res. 15:fov042. doi: 10.1093/femsyr/fov042
Kaya, E., Dag, I., Incesulu, A., Gurbuz, M. K., Acar, M., and Birdane, L. (2013). Investigation of the presence of biofilms in chronic suppurative otitis media, nonsuppurative otitis media, and chronic otitis media with cholesteatoma by scanning electron microscopy. Sci. World J. 2013:638715. doi: 10.1155/2013/638715
Koo, J. W., Quintanilla-Dieck, L., Jiang, M., Liu, J., Urdang, Z. D., Allensworth, J. J., et al. (2015). Endotoxemia-mediated inflammation potentiates aminoglycoside-induced ototoxicity. Sci. Transl. Med. 7:298ra118. doi: 10.1126/scitranslmed.aac5546
Kopecko, D. J., Hu, L., and Zaal, K. J. (2001). Campylobacter jejuni–microtubule-dependent invasion. Trends Microbiol. 9, 389–396. doi: 10.1016/S0966-842X(01)02107-2
Kwon, S. H., Ma, S. X., Hwang, J. Y., Lee, S. Y., and Jang, C. G. (2015). Involvement of the Nrf2/HO-1 signaling pathway in sulfuretin-induced protection against amyloid beta 25-35 neurotoxicity. Neuroscience 304, 14–28. doi: 10.1016/j.neuroscience.2015.07.030
Leis, J. A., Rutka, J. A., and Gold, W. L. (2015). Aminoglycoside-induced ototoxicity. CMAJ 187, E52. doi: 10.1503/cmaj.140339
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34, 634–640. doi: 10.1086/338782
Lv, J. M., Guo, X. M., Chen, B., Lei, Q., Pan, Y. J., and Yang, Q. (2016). The noncompetitive AMPAR antagonist perampanel abrogates brain endothelial cell permeability in response to ischemia: involvement of claudin-5. Cell. Mol. Neurobiol. 36, 745–753. doi: 10.1007/s10571-015-0257-8
MacFaddin, J. (1976). Biochemical Tests for Identification of Medical Bacteria, 3rd Edn. Philadelphia, PA: Lippincott Williams and Wilkins.
Madana, J., Yolmo, D., Kalaiarasi, R., Gopalakrishnan, S., and Sujatha, S. (2011). Microbiological profile with antibiotic sensitivity pattern of cholesteatomatous chronic suppurative otitis media among children. Int. J. Pediatr. Otorhinolaryngol. 75, 1104–1108. doi: 10.1016/j.ijporl.2011.05.025
Mazon Moya, M. J., Colucci-Guyon, E., and Mostowy, S. (2014). Use of Shigella flexneri to study autophagy-cytoskeleton interactions. J. Vis. Exp. 91:e51601. doi: 10.3791/51601
McCanna, D. J., Barthod-Malat, A. V., and Gorbet, M. B. (2015). In vitro methods of assessing ocular biocompatibility using THP-1-derived macrophages. Cutan. Ocul. Toxicol. 34, 89–100. doi: 10.3109/15569527.2014.908205
Meng, L., Jin, W., and Wang, X. (2015). RIP3-mediated necrotic cell death accelerates systematic inflammation and mortality. Proc. Natl. Acad. Sci. U.S.A. 112, 11007–11012. doi: 10.1073/pnas.1514730112
Merle, N. S., Noe, R., Halbwachs-Mecarelli, L., Fremeaux-Bacchi, V., and Roumenina, L. T. (2015). Complement system part II: role in immunity. Front. Immunol. 6:257. doi: 10.3389/fimmu.2015.00257
Meyerhoff, W. L. (1988). Pathology of chronic suppurative otitis media. Ann. Otol. Rhinol. Laryngol. 97, 21–24.
Minovi, A., and Dazert, S. (2014). Diseases of the middle ear in childhood. GMS Curr. Top. Otorhinolaryngol. Head Neck Surg. 13:Doc11. doi: 10.3205/cto000114
Miramón, P., Kasper, L., and Hube, B. (2013). Thriving within the host: Candida spp. interactions with phagocytic cells. Med. Microbiol. Immunol. 202, 183–195. doi: 10.1007/s00430-013-0288-z
Mishra, M., Ressler, A., Schlesinger, L. S., and Wozniak, D. J. (2015). Identification of OprF as a complement component C3 binding acceptor molecule on the surface of Pseudomonas aeruginosa. Infect. Immun. 83, 3006–3014. doi: 10.1128/IAI.00081-15
Miskinyte, M., and Gordo, I. (2013). Increased survival of antibiotic-resistant Escherichia coli inside macrophages. Antimicrob. Agents Chemother. 57, 189–195. doi: 10.1128/AAC.01632-12
Mittal, R., Grati, M., Gerring, R., Blackwelder, P., Yan, D., Li, J. D., et al. (2014). In vitro interaction of Pseudomonas aeruginosa with human middle ear epithelial cells. PLoS ONE 9:e91885. doi: 10.1371/journal.pone.0091885
Mittal, R., Lisi, C. V., Gerring, R., Mittal, J., Mathee, K., Narasimhan, G., et al. (2015). Current concepts in the pathogenesis and treatment of chronic suppurative otitis media. J. Med. Microbiol. 64, 1103–1116. doi: 10.1099/jmm.0.000155
Morinaga, Y., Yanagihara, K., Nakamura, S., Hasegawa, H., Seki, M., Izumikawa, K., et al. (2010). Legionella pneumophila induces cathepsin B-dependent necrotic cell death with releasing high mobility group box1 in macrophages. Respir. Res. 11:158. doi: 10.1186/1465-9921-11-158
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. doi: 10.3389/fmicb.2013.00422
Navarro-Garcia, F., Serapio-Palacios, A., Ugalde-Silva, P., Tapia-Pastrana, G., and Chavez-Dueñas, L. (2013). Actin cytoskeleton manipulation by effector proteins secreted by diarrheagenic Escherichia coli pathotypes. Biomed. Res. Int. 2013:374395. doi: 10.1155/2013/374395
Nestorovich, E. M., Sugawara, E., Nikaido, H., and Bezrukov, S. M. (2006). Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281, 16230–16237. doi: 10.1074/jbc.M600650200
Nordmann, P., and Guibert, M. (1998). Extended-spectrum β-lactamase in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 42, 128–132. doi: 10.1093/jac/42.2.128
Oelschlaeger, T. A., Guerry, P., and Kopecko, D. J. (1993). Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. U.S.A. 90, 6884–6888. doi: 10.1073/pnas.90.14.6884
Park, H., Ishihara, D., and Cox, D. (2011). Regulation of tyrosine phosphorylation in macrophage phagocytosis and chemotaxis. Arch. Biochem. Biophys. 510, 101–111. doi: 10.1016/j.abb.2011.02.019
Park, S. Y., Choi, Y. H., Park, G., and Choi, Y. W. (2015). Neuroprotective effects of α-iso-cubebenol on glutamate-induced neurotoxicity. Environ. Toxicol. Pharmacol. 40, 549–556. doi: 10.1016/j.etap.2015.08.008
Pieters, J., and Gatfield, J. (2002). Hijacking the host: survival of pathogenic mycobacteria inside macrophages. Trends Microbiol. 10, 142–146. doi: 10.1016/S0966-842X(02)02305-3
Poole, K. (2004). Resistance to beta-lactam antibiotics. Cell. Mol. Life Sci. 61, 2200–2223. doi: 10.1007/s00018-004-4060-9
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Qureishi, A., Lee, Y., Belfield, K., Birchall, J. P., and Daniel, M. (2014). Update on otitis media - prevention and treatment. Infect. Drug Resist. 7, 15–24. doi: 10.2147/IDR.S39637
Rawling, E. G., Martin, N. L., and Hancock, R. E. (1995). Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect. Immun. 63, 38–42.
Reusch, R. N. (2012). Biogenesis and functions of model integral outer membrane proteins: Escherichia coli OmpA and Pseudomonas aeruginosa OprF. FEBS J. 279, 893. doi: 10.1111/j.1742-4658.2012.08486.x
Rietsch, A., Vallet-Gely, I., Dove, S. L., and Mekalanos, J. J. (2005). ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011. doi: 10.1073/pnas.0503005102
Rottner, K., Stradal, T. E., and Wehland, J. (2005). Bacteria-host-cell interactions at the plasma membrane: stories on actin cytoskeleton subversion. Dev. Cell 9, 3–17. doi: 10.1016/j.devcel.2005.06.002
Sabatino, R., Di Cesare, A., Pasquaroli, S., Vignaroli, C., Citterio, B., Amiri, M., et al. (2015). Adherence and intracellular survival within human macrophages of Enterococcus faecalis isolates from coastal marine sediment. Microbes Infect. 17, 660–664. doi: 10.1016/j.micinf.2015.06.001
Saunders, J., Murray, M., and Alleman, A. (2011). Biofilms in chronic suppurative otitis media and cholesteatoma: scanning electron microscopy findings. Am. J. Otolaryngol. 32, 32–37. doi: 10.1016/j.amjoto.2009.09.010
Saunders, J. E., Raju, R. P., Boone, J. L., Hales, N. W., and Berryhill, W. E. (2011). Antibiotic resistance and otomycosis in the draining ear: culture results by diagnosis. Am. J. Otolaryngol. 32, 470–476. doi: 10.1016/j.amjoto.2010.09.009
Schultze, J. L., Schmieder, A., and Goerdt, S. (2015). Macrophage activation in human diseases. Semin. Immunol. 27, 249–256. doi: 10.1016/j.smim.2015.07.003
Schweizer, H. P., and Hoang, T. T. (1995). An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158, 15–22. doi: 10.1016/0378-1119(95)00055-B
Shinzaki, S., Ishii, M., Fujii, H., Iijima, H., Wakamatsu, K., Kawai, S., et al. (2016). N-Acetylglucosaminyltransferase V exacerbates murine colitis with macrophage dysfunction and enhances colitic tumorigenesis. J. Gastroenterol. 51, 357–369. doi: 10.1007/s00535-015-1119-3
Sun, J., and Sun, J. (2014). Intracranial complications of chronic otitis media. Eur. Arch. Otorhinolaryngol. 271, 2923–2926. doi: 10.1007/s00405-013-2778-4
Tam, V. C., and Aderem, A. (2014). Macrophage activation as an effector mechanism for cell-mediated immunity. J. Immunol. 193, 3183–3184. doi: 10.4049/jimmunol.1402046
Taylor, J. B., Hogue, L. A., LiPuma, J. J., Walter, M. J., Brody, S. L., and Cannon, C. L. (2010). Entry of Burkholderia organisms into respiratory epithelium: CFTR, microfilament and microtubule dependence. J. Cyst. Fibros. 9, 36–43. doi: 10.1016/j.jcf.2009.10.002
Toma, C., Murray, G. L., Nohara, T., Mizuyama, M., Koizumi, N., Adler, B., et al. (2014). Leptospiral outer membrane protein LMB216 is involved in enhancement of phagocytic uptake by macrophages. Cell. Microbiol. 16, 1366–1377. doi: 10.1111/cmi.12296
Tunbridge, A. J., Stevanin, T. M., Lee, M., Marriott, H. M., Moir, J. W., Read, R. C., et al. (2006). Inhibition of macrophage apoptosis by Neisseria meningitidis requires nitric oxide detoxification mechanisms. Infect. Immun. 74, 729–733. doi: 10.1128/IAI.74.1.729-733.2006
Valencia-Gallardo, C. M., Carayol, N., and Nhieu, G. (2015). Cytoskeletal mechanics during Shigella invasion and dissemination in epithelial cells. Cell. Microbiol. 17, 174–182. doi: 10.1111/cmi.12400
Valentin-Weigand, P., Benkel, P., Rohde, M., and Chhatwal, G. S. (1996). Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64, 2467–2473.
Varela, J. C., and Tomlinson, S. (2015). Complement: an overview for the clinician. Hematol. Oncol. Clin. North Am. 29, 409–427. doi: 10.1016/j.hoc.2015.02.001
Wallis, S., Atkinson, H., and Coatesworth, A. P. (2015). Chronic otitis media. Postgrad. Med. 127, 391–395. doi: 10.1080/00325481.2015.1027133
Walsh, T. R., Toleman, M. A., Hryniewicz, W., Bennett, P. M., and Jones, R. N. (2003). Evolution of an integron carrying blavim2 in Eastern Europe: report from the SENTRY antimicrobial surveillance program. J. Antimicrob. Chemother. 52, 116–119. doi: 10.1093/jac/dkg299
Weischenfeldt, J., and Porse, B. (2008). Bone Marrow-Derived Macrophages (BMM): isolation and applications. CSH Protoc. 2008:pdb.rot5080. doi: 10.1101/pdb.prot5080
Welin, A., Eklund, D., Stendahl, O., and Lerm, M. (2011). Human macrophages infected with a high burden of ESAT-6-expressing M. tuberculosis undergo caspase-1- and cathepsin B-independent necrosis. PLoS ONE 6:e20302. doi: 10.1371/journal.pone.0020302
Welin, A., Winberg, M. E., Abdalla, H., Särndahl, E., Rasmusson, B., Stendahl, O., et al. (2008). Incorporation of Mycobacterium tuberculosis lipoarabinomannan into macrophage membrane rafts is a prerequisite for the phagosomal maturation block. Infect. Immun. 76, 2882–2887. doi: 10.1128/IAI.01549-07
Woodruff, W. A., and Hancock, R. E. (1989). Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J. Bacteriol. 171, 3304–3309.
Yakhnina, A. A., McManus, H. R., and Bernhardt, T. G. (2015). The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability. Mol. Microbiol. 97, 957–973. doi: 10.1111/mmi.13077
Yang, G., Xiao, X., Yin, D., and Zhang, X. (2012). The interaction between viral protein and host actin facilitates the virus infection to host. Gene 507, 139–145. doi: 10.1016/j.gene.2012.06.026
Yeo, S. G., Park, D. C., Hong, S. M., Cha, C. I., and Kim, M. G. (2007). Bacteriology of chronic suppurative otitis media-a multicentre study. Acta Otolaryngol. 127, 1062–1067. doi: 10.1080/00016480601126978
Yoeruek, E., Bartz-Schmidt, K. U., and Hofmann, J. (2016). Impact of the radius of the injector system on the cell viability in descemet membrane endothelial keratoplasty: an ex vivo experimental study. Acta Ophthalmol. 94, e1–e5. doi: 10.1111/aos.12787
Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., et al. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603. doi: 10.1016/S1534-5807(02)00295-2
Yorgancılar, E., Yildirim, M., Gun, R., Bakir, S., Tekin, R., Gocmez, C., et al. (2013). Complications of chronic suppurative otitis media: a retrospective review. Eur. Arch. Otorhinolaryngol. 270, 69–76. doi: 10.1007/s00405-012-1924-8
Yuan, M., Deleuil, F., and Fällman, M. (2005). Interaction between the Yersinia tyrosine phosphatase YopH and its macrophage substrate, Fyn-binding protein, Fyb. J. Mol. Microbiol. Biotechnol. 9, 214–223. doi: 10.1159/000089649
Zhang, L., and Wang, C. C. (2014). Inflammatory response of macrophages in infection. Hepatobiliary Pancreat. Dis. Int. 13, 138–152. doi: 10.1016/S1499-3872(14)60024-2
Zheng, K., Kitazato, K., Wang, Y., and He, Z. (2015). Pathogenic microbes manipulate cofilin activity to subvert actin cytoskeleton. Crit. Rev. Microbiol. 8, 1–19. doi: 10.3109/1040841X.2015.1010139
Keywords: otopathogenic P. aeruginosa, monocyte-derived macrophages, mouse bone marrow-derived macrophages, cell viability
Citation: Mittal R, Lisi CV, Kumari H, Grati M, Blackwelder P, Yan D, Jain C, Mathee K, Weckwerth PH and Liu XZ (2016) Otopathogenic Pseudomonas aeruginosa Enters and Survives Inside Macrophages. Front. Microbiol. 7:1828. doi: 10.3389/fmicb.2016.01828
Received: 05 June 2016; Accepted: 31 October 2016;
Published: 18 November 2016.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Gabriele Margos, Bavarian Health and Food Safety Authority, GermanyShreekumar R. Pillai, Alabama State University, USA
Ruth Thornton, University of Western Australia, Australia
Yi Wang, U.S. Food and Drug Administration, USA
Copyright © 2016 Mittal, Lisi, Kumari, Grati, Blackwelder, Yan, Jain, Mathee, Weckwerth and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Z. Liu, xliu@med.miami.edu Rahul Mittal, r.mittal11@med.miami.edu
 Rahul Mittal
Rahul Mittal Christopher V. Lisi
Christopher V. Lisi Hansi Kumari
Hansi Kumari M’hamed Grati1
M’hamed Grati1 Denise Yan
Denise Yan Chaitanya Jain
Chaitanya Jain Kalai Mathee
Kalai Mathee Paulo H. Weckwerth
Paulo H. Weckwerth Xue Z. Liu
Xue Z. Liu